Abstract
Foodborne infection is one of the leading sources of infections spreading across the world. Foodborne pathogens are recognized as multidrug-resistant (MDR) pathogens posing a significant problem in the food industry and healthy consumers resulting in enhanced economic burden, and nosocomial infections. The continued search for enhanced microbial detection tools has piqued the interest of the CRISPR-Cas system and Nanoparticles. CRISPR-Cas system is present in the bacterial genome of some prokaryotes and is repurposed as a theragnostic tool against MDR pathogens. Nanoparticles and composites have also emerged as an efficient tool in theragnostic applications against MDR pathogens. The diagnostic limitations of the CRISPR-Cas system are believed to be overcome by a synergistic combination of the nanoparticles system and CRISPR-Cas using nanoparticles as vehicles. In this review, we have discussed the diagnostic application of CRISPR-Cas technologies along with their potential usage in applications like phage resistance, phage vaccination, strain typing, genome editing, and antimicrobial. we have also elucidated the antimicrobial and detection role of nanoparticles against foodborne MDR pathogens. Moreover, the novel combinatorial approach of CRISPR-Cas and nanoparticles for their synergistic effects in pathogen clearance and drug delivery vehicles has also been discussed.
Keywords: Antimicrobial activity, Diagnosis, CRISPR-Cas system, Nanoparticle and MDR food-Borne- pathogens
Graphical abstract
Highlights
-
•
Bacterial CRISPR Cas system are repurposed as a thergodiganostic tool against MDR pathogen.
-
•
Combinatorial approach of CRISPR-Cas and Nanoparticle is used as delivery vehicle and clearing pathogens.
-
•
CRISPR-Cas and Nanoparticles is a tool for the food safety profiling of MDR food-borne pathogen.
1. Introduction
Foodborne MDR pathogens are primarily recognized as one of the main causes of infectious diseases worldwide considering their presence in varieties of food as an important carrier for transmission [1,2]. According to World Health Organization (WHO), foodborne diseases are caused by MDR pathogens which include Salmonella spp., Staphylococcus aureus, Listeria monocytogenes, Shigella spp., Clostridium botulinum, Escherichia coli O157:H7 [3]. The WHO estimates around 600 million cases due to foodborne diseases causing approximately 42,00,00 deaths thereby resulting in the loss of 33 million healthy life years annually [3]. The rapid increase in foodborne diseases, coupled with the emergence and re-emergence of foodborne pathogens, have put food safety at high risk for public health [4]. These foodborne pathogen accounts for almost two-thirds of the human foodborne diseases worldwide with a considerable amount of risk in the developed countries; which thereby necessitates the development of novel therapies for combating this MDR pathogen [[4], [5], [6]]. The WHO has reported the rapid rise in infections was mostly due to methicillin-resistant Staphylococcus aureus, cephalosporin, and fluoroquinolone-resistant Escherichia coli, fluoroquinolone-resistant Campylobacter species, and streptomycin, gentamicin, tetracycline-resistant Salmonella species [4,7]. Food-borne pathogens, being diverse, accounts for most of the food-borne diseases creating major health problems in human. It is more at risk for infants, elderly people and immunocompromised individuals through ingestion of contaminated food, causing diarrhoea, stomach ache and vomiting as symptoms post-manifestation [8,9]. Food contamination of meat, eggs and milk products is major an issue which occurs at any stage of the food chain and requires to be dealt to prevent outbreaks like 2017(reported by EFSA and CDC) [10] (see Table 1, Table 2).
Table 1.
Application of NPs as an antimicrobial and detection tool.
| NPs | Characterization | Type of preclinical study | Pathogen | Result | References |
|---|---|---|---|---|---|
| Ag-NPs | Size-75nm(spherical) and 8–20 nm (triangular) | RCT |
E.Coli (Gm-ve), Staphylococcus aureus(Gm + ve), Salmonella typhi (Gm-ve) |
The highest antimicrobial activity of Ag-NPs was shown against E.coli while S.typhi showed moderate activity and it was low for S.aureus. | [151] |
| Ag-NPs | Size-8-15nm | RCT |
E.coli O157:H7 (Gm-ve), Listeria monocytogenes (Gm + ve) |
The material containing Ag-NPs exerted greater antimicrobial activity against L.monocytogenes than E.coli because of greater cell wall thickness of Gram-positive bacteria. | [27] |
| ZnO-NPs | Size-8nm | In vitro | E.coli(Gm-ve) and S.aureus(Gm + ve) | The material containing ZnO-NPs inhibited the growth of both E. coli and S.aureus | [152] |
| ZnO-NPs | Surface-volume ratio | In vitro | E.coli(Gm-ve) and S.aureus(Gm + ve) | Antimicrobial activity against both E.coli and S.aureus increased with increase in surface to volume ratio with decrease in particles size of ZnO-NPs. | [152] |
| Nano-Colloidal Silver | MIC/MBC conc. | In vitro |
E.coli(Gm-ve), S.aureus(Gm + ve), Listeria monocytogenes (Gm-ve), Salmonella enterica (Gm-ve) |
The MIC values showed average range of 7–25 ppm whereas the MBC values for all the bacteria were above 100 ppm which showed good inhibitory effect but poor bactericidal effect of Nano-Colloidal Silver | [108] |
| Graphene nanosheets | Molecular dynamics stimulation | In vitro | E.coli (Gm-ve) | Graphene nanosheets induce degradation of the cell membrane with the extraction of phospholipid molecules from the membrane leading to bacterial inactivation. | [153] |
| Pd-Nps | Size - 2 nm | In vitro | E.coli (Gm-ve), S.aureus (Gm + ve) | Greater antimicrobial activity of Pd-NPs was observed ast concentrations as low as 2.5 nm in case of Gm + ve S.aureus as compared to Gm-ve E.coli | [154] |
| Prismatic shaped Y2O3-NPs | Shape | In vitro | S.aureus (Gm + ve) | Greater antimicrobial activity was shown against S.aureus which is mainly influenced by the shape of the Y2O3-NPs occurring due to direct interaction between prismatic shaped Y2O3-NPs and bacterial cell membrane leading to breakage of the cell membrane. | [155] |
| Silver NPs and streptavidin-coated magnetic beads | Size – 20 nm | In vitro | S.aureus (Gm + ve) | Detection sensitivity of 10–106 CFU/mL, detected by an electrochemical immunosensor through sandwich immunoassay on biotinylated primary anti S.aureus aptamer and secondary anti S.aureus aptamer | [156] |
| Mesosporous TiO2 coated magnetic NP | Size - 200 nm | In vitro | E.coli (Gm-ve) and S.aureus(Gm + ve) | Detection sensitivity of 10–2000 CFU/mL, detected through fluorescent imaging with aptamer based recognition. | [157] |
| Gold NP | Size – 20 nm | In vitro | S.typhimurium (Gm-ve) | High level of sensitivity and specificity for detection of S.typhimurium in pork meat detected by LSPR signal sensing with aptamer based recognition | [158] |
Table 2.
Thero-diganostic Application of CRISPR-Cas system.
| CRISPR Cas | Type of preclinical study | Pathogen | Result | References |
|---|---|---|---|---|
| Cas9 | In vivo | Staphylococcus aureus | Phagemid targetting methicilin resistance gene mecA to selectively eradicate virulent and avirulent S.aureus | [77] |
| casABCDE and cas3 | In vitro | Escherichia coli, Salmonella enterica | Examined selective removal of individual strains in pure and mixed cultures | [73] |
| Cas9 | In vivo and In vitro | Enterohemorrhagic E. coli (EHEC), E. coli |
Phagemid delivery resulted in 20 fold reduction in EHEC in case of in vitro and in case of in vivo it showed increase in survival rates in Galleria mellonella | [75] |
| Cas13a | In vitro | Staphylococcus aureus | A novel CRISPR-Cas13a based bacterial detection method, CCB detection tool successfully detects the target gDNA with a dynamic selction range revealing enhanced selectivity of S.aureus by rapid detection assay. | [17] |
| dCas9 | In vitro | Escherichia coli | Genome wide screening of CRISPR-dCas9 by utilizing sgRNAs targeting random E.coli chromosome locations, utilizing CRISPRi during gene essentiality prediction in the E.coli genome thereby encompassing the full phage cycle. | [50] |
| Cas13a | In vitro | Salmonella enteritidis | A detection technique called APC-Cas utilizes a combination of allosteric probes based on nucleic acids and CRISPR-Cas13a to detect low single cells of S.enteritidis with low cross reactivity with other bacteria. | [59] |
The MDR pathogens which causes the diseases have different molecular strategies to overcome the effect of the antimicrobial drugs induced for their killing. Some of these mechanism includes the resistant to the inhibition of the nucleotide synthesis, disruption of cell membrane, and reduced protein synthesis caused by the induced antimicrobial drugs. Another antimicrobial resistant (AMR) mechanism exhibited by these MDR pathogens includes fluoroquinolone resistance due to mutations in DNA gyrase and topoisomerase IV genes. Similarly, tetracycline resistance mechanisms include transport-based mechanisms by protecting ribosomal binding site of tetracycline via RNA-binding proteins (RBP) in MDR pathogens [11]. Even though traditional microbiological approaches for detection and mitigation of MDR related problems are considered to be gold standard methods in food industry, yet they poses rate limitation [12]. The rapid detection and identification of pathogens is crucial in the theragnostic applications. Which poses a varied widespread opportunity for utilizing CRISPR-Cas complex techniques for regulating food safety and environmental monitoring. The CRISPR-Cas complex serves as an excellent defense system for adaptive immune responsive tool in prokaryotes, regardless of archaea (∼90%)) or bacteria (∼40%). It is useful to identify reoccurring infections by integrating short sequences of invading genomes, known as spacer sequence [13]. Most bacterial species possess an inherent noble CRISPR-Cas9 system, which makes it a promising detection tool over other traditional approaches for uncovering of food pathogens. Till date, CRISPR-Cas systems can be classified into two main category based on the occurrence of effector Cas protein, which further can be segregated into six different types and subtypes [[14], [15], [16]]. Despite of having high level of diversification, CRISPR-Cas9 works in a similar fashion based on sequence specificity by recognizing and cleaving foreign DNA/RNA. The mechanism can be staged in to three major events; i) integration into CRISPR array or spacer acquisition, ii) crRNA maturation and iii) target interference as illustrated in Fig. 1 [13]. Besides, the application of CRISPR-Cas9 system (pCasSA) extends beyond genome editing; that includes high resolution typing of pathogens, manipulation of MDR genes, designing vaccine of starter culture against phages. This further pushes the boundary of deciphering new mechanisms of different CRISPR-Cas systems with new cas genes and enzymatic activities. The major focus is to use the CRISPR system as an antimicrobial and detection tool for pathogen clearance. CRISPR-Cas9 editing system has also been used to successfully demonstrate the sequence specific removal of AMR gene bearing plasmid thus making the bacteria susceptible to antibiotics. Moreover, CRISPR-Cas system presents in the genome of the pathogens aids as a detection marker for food borne pathogen. The major issue with CRISPR-Cas system is its delivery. The use of novel non-viral delivery strategies using nanoparticle aids to transport the CRISPR-Cas cargoes to the desired bacterial strain enhancing the CRISPR-Cas efficiency [17].
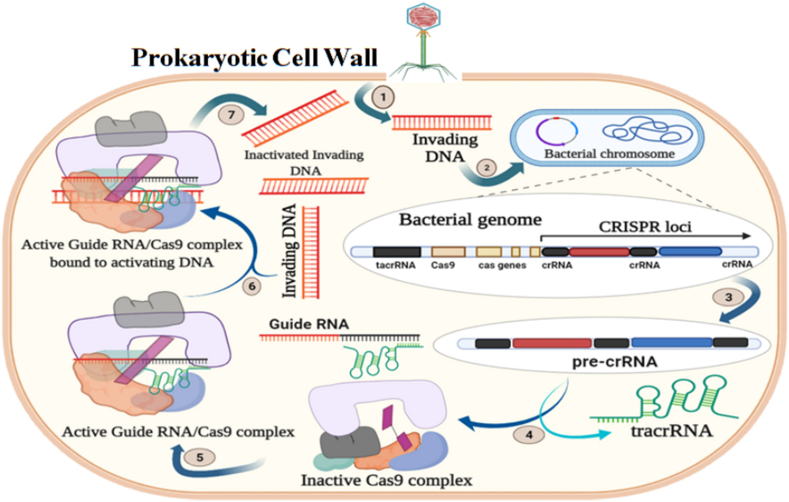
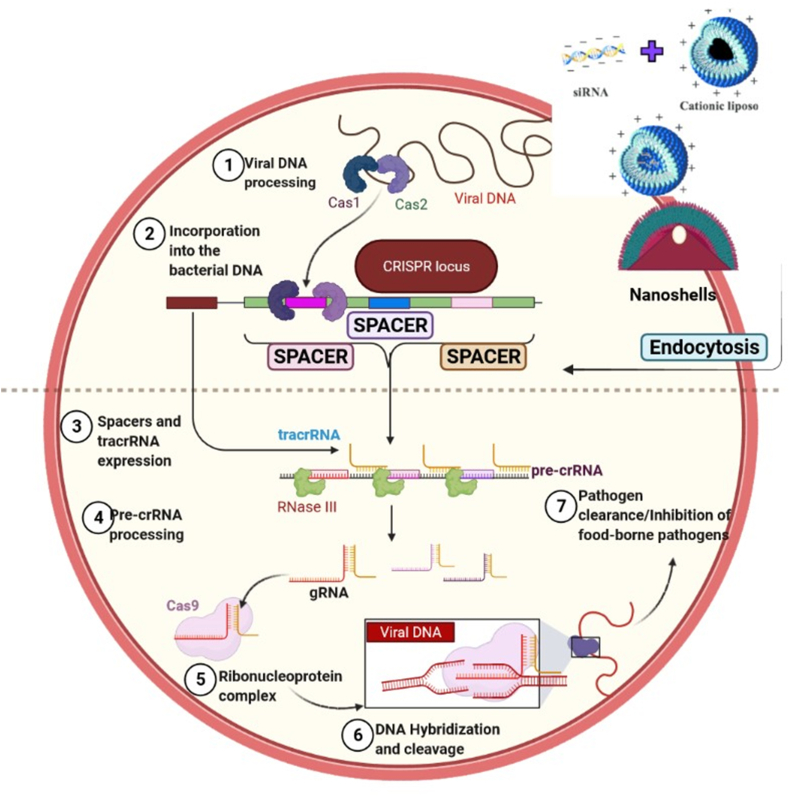
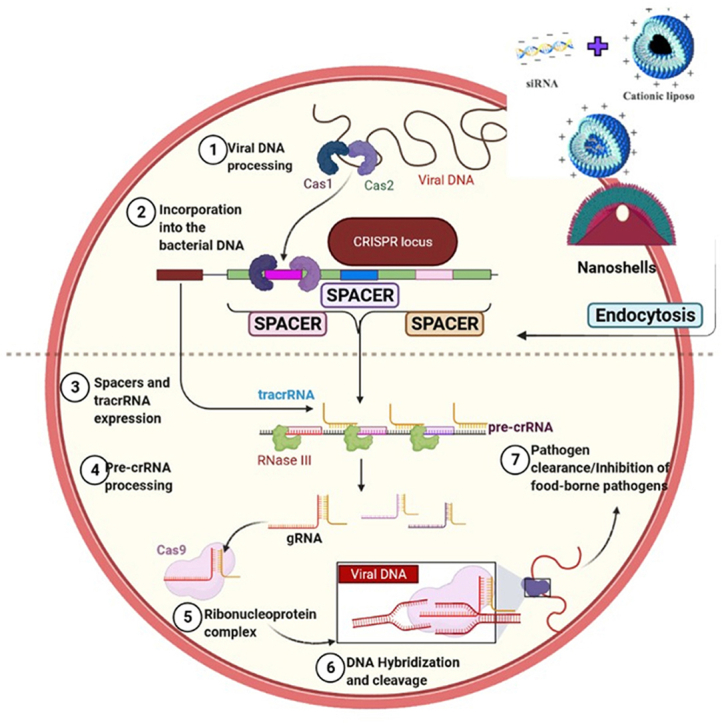
Fig. 1.
Illustrative representation of CRISPR-Cas general mechanism. 1. Invasion of DNA where a foreign DNA from a plasmid invades the cell, 2. Incorporation of the DNA fragments from the invading DNA into the CRISPR locus as spacers, 3. pre-cRNA transcription occurs where the cell constitutively transcribes a spacer group, 4. Formation of guide RNA takes place where transactivating (tracrRNA) base pairs with the CRISPR repeat sequences on the pre-crRNA, 5. Activation of Cas9 protein takes place where inactivated Cas9 protein complex binds to the guide RNA and thus gets activated, 6. The activated guide RNA/Cas9 protein complex binds with the target DNA, 7. Inactivation of the target DNA which is cleaved by the Cas9 protein.
In recent time, nanotechnology has been revolutionizing the food industry with its various showcased applications in different dimensions such as use of low-cost nano-sensors for detection, quantification of pathogens, alteration of food composition, food deterioration control, and use of target delivery of nutrients or bioactive molecules through nano-encapsulation [18,19]. Nanoparticles has been a promising candidate in food industry owing to its widespread application against MDR pathogens due to its peculiarity in variation in attainable size and shape that can penetrates directly to the bacterial cell [20,21]. The underlying mechanism of antimicrobial effects of NPs has been described to be mediated through different ways such as: i) generation of ROS [22], ii) disruption of bacterial cell membrane [23], iii) penetration of bacterial cell membrane, iv) influencing the physiological process through interactions with DNA and proteins [24,25]. Nanoparticles has been portrayed as vector for the CRISPR-Cas delivery and a potential detection tool for pathogens [26]. Concerning the potential usage and increasing popularity of nanotechnology [27] and CRISPR-Cas technique, it is the need of hour to review in detail about the two strategies and the potential combinatorial approach for a higher output in food industry in respect of efficiency and speed.
The reviews discuss the application of nanoparticle and CRIPSR-Cas to combat food borne pathogens in the management of their manifested infections. Herein, we have not only discussed the theragnostic application of CRISPR-Cas system and NPs as detection tools and antimicrobial against foodborne pathogens but also highlighted a novel combinatorial approach to their synergy against foodborne pathogens. Moreover, we have elucidated the application of the CRISPR-Cas system as phage vaccination, plasmid resistance, and genetic editing tool. This review will pave the way for researchers to revolutionize personalized medicine through the use of nanomedicine and CRISPR-Cas system both alone as well as in synergy. These will advocate the early onset diagnosis and selective killing of food borne pathogens.
2. CRISPR cas complex as a detection tool against food pathogens
The seriousness of food-borne infections caused by MDR pathogens or their toxins necessitates the rapid development of a precise and sensitive technology for combating these pathogens [17]. Conventional microbiological methods are considered as the gold standard approach because of their reliability and accuracy. However, their limitations of prolonged, laborious, and destructive mode of application impose issues for their accuracy and long term usage in the food industry [12]. Similarly, the other famous approaches which are based on immunological methods and their combination also have limited utility for food pathogen identification [28]. Additionally, the other traditional pathogen detection techniques which are biosensor-based and nucleic acid-based are used as an effective tools, despite their limited application in the food sector [28]. Concerning to these limitations, researchers are always being in search of new, advanced and powerful techniques for pathogen diagnostics. One of the most recent technique is the use of CRISPRs-Cas systems and nanotechnological inventions to design new diagnostics methods.
Most of the sequenced bacterial genomes (45%) CRISPRs-Cas systems act as an adaptive immunity for them [29]. These CRISPR/Cas9 system (pCasSA) has been designed as a powerful tool to inhibit transcription in gene knockdown strategies, genome-wide screening, and genomic modifications through mutations. The techniques using inhibition of transcription through CRISPR/Cas9 system for gene-editing techniques has progressed significantly in recent years [30]. The utilization of Cas proteins having variable functionality has resulted in more sensitive and reliable nucleic acid detection systems. The extremely variable spacer sequences residing within the CRISPR array are separated from one another by invariant direct repeat sequences of CRISPR-Cas-based detection techniques [31]. Recent studies have showed the potential of using these new CRISPR-Cas technologies for cost-effective pathogen detection tools as illustrated in Fig. 2 [32]. Some of the listed one includes specific to the some MDR pathogens like Staphylococcus aureus, Salmonella typhi etc. as described; Staphylococcus aureus is a MDR food borne pathogen belonging to the Micrococcaceae family that causes several deadly infectious disorders with a high fatality rate [33]. It is present in humans as a natural resident exhibiting colonization effects in 30–50% of healthy persons while 10–20% of the human population are persistently colonized by MDR strains like methicillin-sensitive and methicillin-resistant strains(MRSA) [[34], [35], [36]]. The colonization establish with the phenomenon of biofilm formation which makes them resistant against the immunological action in body, thus making the infection more challenging. Moreover, further dispersal of bacterial cells from the biofilm during infection leads to their transmission to other body secondary sites [37]. Though vancomycin (a glycopeptide), daptomycin (a lipopeptide), and linezolid (an oxazolidinone) are deemed effective against MRSA infection, its long-term usage renders the strains resistant to deadly conditions that needs to be detected and diagnosed for treatment [38]. Although genetic alternation of S. aureus is tedious and laborious, it is an effective tool for investigation of pathogens physiology. A bacterial sensing technique called CCB-Detection has been developed, by utilizing advantage of the crRNA-based programmability of the CRISPR-Cas13a system and its RNase activity to target RNA recognition against Staphylococcus aureus [17]. CCB-Detection is capable enough to target gDNA successfully with a dynamic detection range (100–107 CFU/mL) for food samples containing known and unknown concentrations of bacteria (spiked and non-spiked). The technique further reveals enhanced selectivity for S. aureus by rapid detection assay [17].
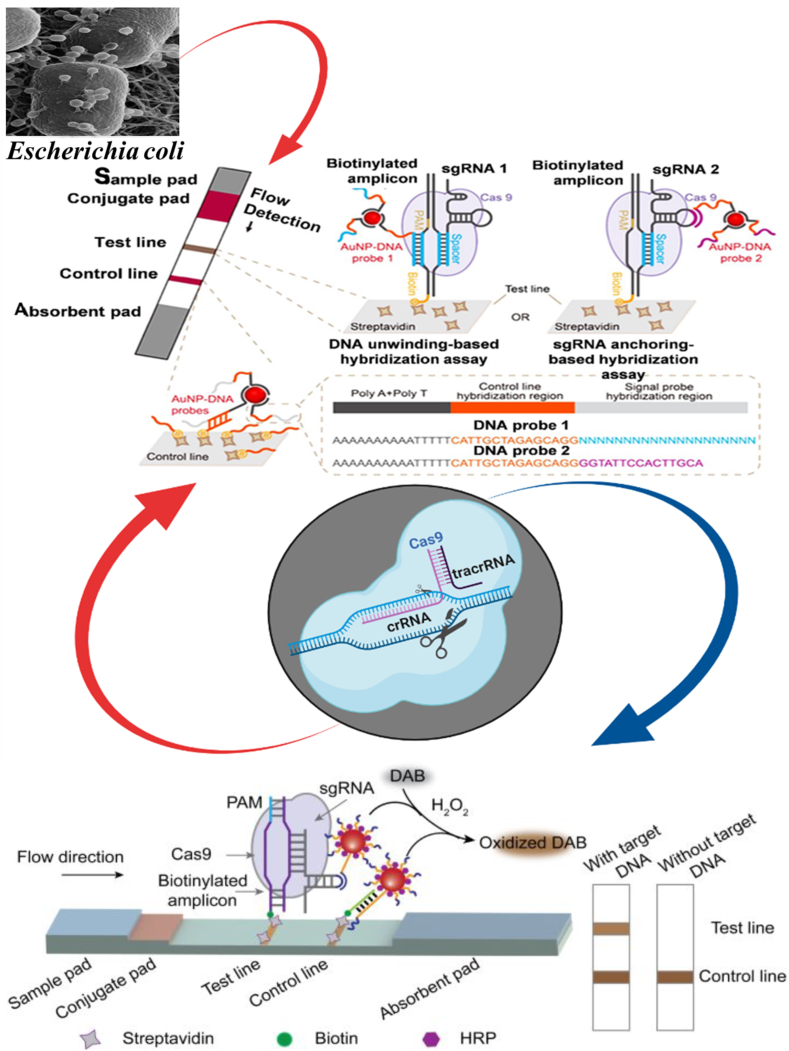
Fig. 2.
Detection mechanisms of using CRISPR-Cas against different food-borne pathogens. Schematic illustrating ASFV detection in suspected swine serum samples using the CASLFA technique along with, the CRISPR/Cas9-mediated test strip based on the DNA HRP-AuNP probes. DNA-HRP-AuNP probes are pre-embedded in conjugate pads. Cas9/sgRNA recognizes the biotinylated DNA product, is added dropwise to the sample pad and then flows through the conjugate pad and hybridizes with DNA-HRP-AuNP probes. The resulting complexes flow through the test line and are captured. Excess DNA-HRP-AuNP probes flow through the control line and are captured by precoated single-stranded DNA probes. When the target DNA is detected, the test strip will show two colored bands that form through a reaction catalyzed by HRP with the peroxidase substrate DAB, and only one control line band will appear when no target DNA is detected (Figures adapted and edited from Refs. [136,159]).
Escherichia coli O157:H7 is one of the most harmful foodborne pathogenic bacteria, causing an estimated 73,000 cases of infection in USA annually [39]. Escherichia coli belongs to the family of Gram-negative bacteria found as both pathogens and commensals in mammalian intestines and cause infections of urinary tract, neonatal meningitis, sepsis, pneumonia, surgical site infections through colonization [40]. The clonal dissemination of intestinal pathogenic E. coli O157:H7, primarily lead to contamination of food products [41]. The infection caused by food like diarrhoea in children is not only extremely contagious, but their transmission is unidentifiable [42]. Therefore, detecting the most common pathogens in foods and environmental sources would aid in better treatment. Standard culture-based approaches, qPCR, ELISA, have been conventionally applied to identify E. coli O157:H7 in the previous decades but are time-consuming, complex processes, and have low sensitivity [[43], [44], [45]]. Researchers have used several methodologies to detect E. coli O157:H7, including fluorescence, chemi-luminescence, and electrochemical detection in combination with various biosensors for quick analysis and great sensitivity [[46], [47], [48]]. The CRISPR Cas system has arisen as a new tool for the detection and antimicrobial of E. coli, supplementing the limits of conventional approaches due to its remarkable selectivity and genetic editing properties [49]. A genome-wide CRISPR-dCas9 screening by utilizing sgRNAs targeting random E. coli chromosome locations has been proved as an efficient method for diagnosis that utilizes CRISPR for the prediction of essential genes in the E. coli genome during in-vitro growth of functional capsids, encompassing the full phage cycle. Furthermore, the broad usage of CRISPR-dCas9 screening has made it a powerful detection tool in bacterial genomics [50]. A Cas9nAR-assisted lateral flow strip was used to identify two food-borne pathogens without the need for specialist analytical equipment, allowing it for the simultaneous amplification of two pathogens of interest in a mixed sample and immediate visualization of the detection in 3 h causing enhanced selectively and sensitively with a LOD (100 CFU/mL) [51].
Salmonella is the most common causes of foodborne infections posing a substantial health risk in United States with over 2600 different serovars, causing diarrhoea, fever, and stomach cramps post-infection. According to the WHO, about one million cases of reported salmonellosis are resulting in 20,000 hospitalizations and 400 morbidities annually [52,53]. Salmonella spp. strains are zoonotic MDR pathogens causing several outbreaks as reported by CDC. It acquires resistance in the animal host before being transmitted to humans via the food chain [54]. A variety of foods products including sprouts, chicken meat products, eggs, cucumbers, dry pepper, tuna, and tomatoes, have become increasingly linked to Salmonella outbreaks over the last decade. It can infect the food supply chain at any point throughout the manufacturing, processing, distribution, or marketing process. Because of the high frequency of Salmonella, effective methods for timely identification, detection, and monitoring are required. Detection of small numbers of microbial cells in food and clinical samples are incredibly useful, but it is still a major challenge [55].
Due to the use of culture-based techniques, current surveillance for this pathogen is limited to the detection of only the most abundant serovars. Thus, pathogenic serovars present in a small number of people remain undetected [53]. The CRISPR-based analysis aids rapid genotyping for future perspectives of infection identification and pathogen detection. The evolution of S. enterica sub-lineages is primarily driven by either the loss of coding regions with established metabolic responsibilities for functional decline or the acquirement of horizontally transmitted phage and plasmid DNA causing activation of virulence and resistance [56]. The CRISPR in S. enterica acts as a defence tool against invading plasmid as well as phage DNA, by altering the phenotype for CRISPR-mediated temporary immunity, regulating long-term sub-lineage development [56]. CRISPR analysis of spacer content of Salmonella was significantly connected to serovar specific multi-locus sequence type, independently verifying evolutionary patterns and emphasising the value of CRISPR-based genotyping [57]. CRISPR1 and CRISPR2 are two CRISPR loci in Salmonella sp, and the spacer content of these can be regarded as serovar-specific. The evolution of CRISPR-Cas systems for conservation of Cas gene, direct repeat organization, leader sequencing and protospacer pairing can be used as a detection tool for Salmonella identification [58]. The utilization of serovar-specific well-conserved CRISPR-Cas spacer content has become a unique way for genotyping, leading to easy and rapid detection. CRISPR-Sero Seq is an amplicon-based multiplexed sequencing technique for detecting several serovars in a single sample that was able to confidently detect a serovar constituting of 0.01% of the sample using mixed gDNA from two Salmonella serovars [53]. A detection technique (dubbed “APC-Cas”) has been developed utilizing a three-step amplification to produce robust fluorescence signals, identifying low quantities of a bacterial pathogen without isolation. APC-Cas utilizes a combination of allosteric probes based on nucleic acids and CRISPR-Cas13a with an aptamer domain for detection of S. enteritidis [59]. The efficiency of signal amplification of APC-Cas was successfully substantiated by recognizing low cross-reactivity of S. enteritidis with other pathogens. It is applied for the detection and quantification of Salmonella enteritidis cells (1–105 CFU) in a variety of samples, including milk, with sensitivity and accuracy comparable to or better than traditional real-time PCR [59].
The gram-positive bacterium Listeria monocytogenes is a common opportunistic food-borne MDR pathogen causing listeriosis in elderly, immune-compromised patients, foetuses, and infants, with a case of 30%fatality rate. The disease is spread mostly by the eating of contaminated food [60,61]. For precise detection of this infection rapid and sensitive technologies are necessary during food production. The detection of Listeria cells using CRISPR-Cas-assisted phage engineering via bioluminescence-reporter bacteriophages, through de novo genome assembly and activation using NLuc enzyme, has been incorporated into wide host range phage (A511) for enhanced sensitivity. The two serovar-specific Listeria phage responsible for reporter proteins are detected by NLuc-based phage within one day when present in samples such as milk, lettuce, cold cuts [62]. CRISPR-based subtyping has become feasible due to next-generation sequencing and new bioinformatics tools. CRISPR analyses saves time and is economical compared to WGS subtyping [63]. However, not all bacteria carry CRISPRs (approximately 48% of all bacteria), and hence are not considered as a good candidates for CRISPR-based subtyping [31].
Similarly, Shigella is a MDR food contaminated infectious pathogen, causing 600,000 morbidities worldwide annually in USA and China [[64], [65], [66]]. Even though single doses of norfloxacin and ciprofloxacin have been exhibited to be beneficial against shigellosis but are rendered ineffective owing to the emergence of AMR through point mutation, cellular impermeability, active efflux system, mutation(s) at target locations, and horizontal resistance gene transfer [67]. Shigella regulates the activity of the CRISPR-Cas system on its own, acquiring an external gene for survival. The insertion sequences cause mutations in bacterial genome by interfering expression of target sequences through transcription and translation alteration. Cas genes have the potential for detection of genetically transferred AMR strain, causing the alterations in the CRISPR-Cas machinery, based on the mechanism of CRISPR-Cas-mediated horizontal transfer of AMR genes in Shigella [68]. A quick and sensitive detection is required owing to the low infectious dosage of Shigella sp and thus introduction of Spa gene as a novel sequence for Shigella detection in any species of Shigella aids in its detection. An AuNPs-based optical geno-sensing system for the detection of four Shigella sp has been utilized wherein, AuNP-DNA probes were hybridized with the Spa gene sequence to the complementary target for inhibiting and detecting the Shigella sp from aggregating in an acidic environment with a LOD and LOQ (8.14 and 26.6 ng/mL) [69].
Overcoming the few limitations in the application of the food industry for the detection of food-borne pathogens, we are witnessing that the CRISPR-Cas tool has made a revolution in theragnosis with a variety of approaches for the most prevalent MDR food-borne pathogens including S. aureus, E. coli, Salmonella, Listeria, Shigella; making it the most reliable method over other easily available, less sensitive molecular and biochemical methods.
3. Anti-microbial activity using CRISPR-cas complex for food pathogens
CRISPR based antimicrobials are attractive substitute to antibiotics which possess their potentiality to target harmful and commensal pathogen indiscriminately. Which is due to their specificity and programmability. In beneficial bacteria, they are useful in minimizing the dissemination of antibiotic resistance or virulence factors over different strains and, controlling cell proliferation. In addition, they are beneficial for elimination of contaminants in the mixed food crops without any adverse effects on non-specific strains. Moreover, CRISPR-Cas are utilized to target harmful pathogens that causes spoilage in food materials [70]. CRISPR-Cas is often used as a pathogen clearing machinery of cells through the double-stranded cleavage of bacterial genomes, lacking the robust DNA repair pathways [71]. In spite of facing numerous challenges during modification genome, CRISPR-Cas enables antimicrobial to cause lethal damage to MDR food pathogens in a sequence-specific manner. CRISPR-Cas targets the bacterial genomes, showing the self-targeting lethal effect on host, which results in a non-reversible reduction for antimicrobial action as illustrated in Fig. 3.
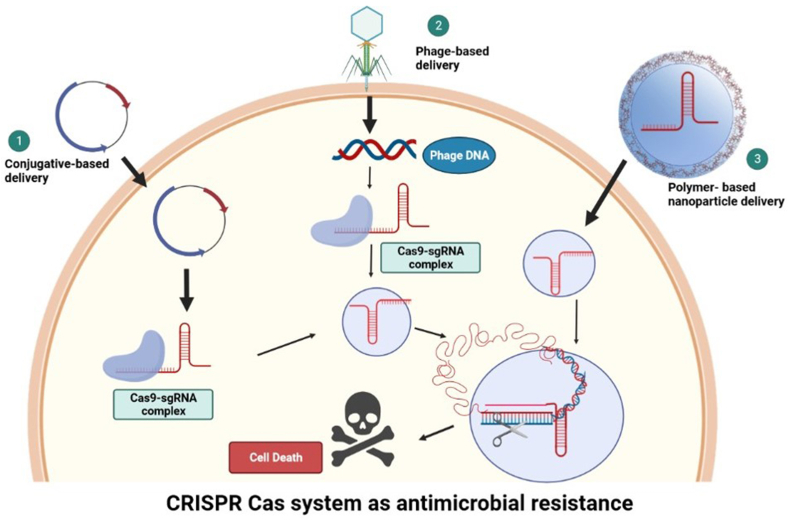
Fig. 3.
Graphical model demonstrating CRISPR-based delivery through three different mechanisms targeting antimicrobial activity 1. Conjugate-based delivery, 2. Plasmid-based delivery and 3. Polymer-based nanoparticle delivery.
Despite possessing difficulty in isolation of strains due to their similarity in physiological properties, mixed culturing offers a wide range of advantages and disadvantages [72]. However, traditional methods of isolation or control of a mixed population have only provided partial solutions, the native CRISPR-Cas systems function as an effective antimicrobial and capable of selectively eliminate targeted strains or control the mixed culture in a dose-dependent manner [73].
CRISPR-Cas system become a unique antimicrobial with native CRISPR by providing a cell spacer which is transmitted through a plasmid with a self-targeting spacer [73]. The two sub-strains of E. coli share 99% sequential homology and have identical cellular processes that could effectively be separated by using the CRISPR-Cas system [73]. Since, the spacer sequence for the strain of interest can be developed by focusing variable genomic characteristics, the specific nature of CRISPR antimicrobials is incomparable in terms of point of specificity. CRISPR-Cas is a potent antimicrobial system to control the food pathogens or spoilage organisms, such as E. coli, Enterobacteriaceae, and S. aureus through the splitting of the genomes of target organism. Thus can be proved as a potent tool for pathogen clearance [[74], [75], [76]]. Interestingly, CRISPR mediated antimicrobials have been tested for the determination of strains having an absence of native CRISPR-Cas systems [75,77]. The characteristics of the CRISPR antimicrobials have been confirmed by the analysis of the differentiation potential between two strains in a single base pair [75]. Antimicrobials are also manipulated to handle targeted unwanted antibiotic resistance or virulence genes for the removal of strains [75]. A genome-wide library of guide RNAs constituting specific five-nucleotide seed sequences was used to drive catalytic dCas9 to stop gene transcription to deficits severe fitness or even kill E. coli regardless of their fifteen nucleotides in the guide sequence. These consequences are intensified by high dCas9 concentrations and mitigated by fine-tuning dCas9 expression while retaining robust on-target suppression. The antimicrobial role of the CRISPR-Cas complex was illustrated for off-targets with nine nucleotides of similarity to the guide RNA that can significantly suppress gene expression [78]. The Cas proteins genes in combination with crRNAs, and CRISPR array promoters degrade and penetrate foreign DNA in Escherichia coli. A promoter incorporated CRISPR directs the transcription of precursor crRNA, whereas the other promoter directs transcription of Cas gene upstream of the protein-coding genes. H-NS, the DNA-binding protein is involved in expression of Cas by suppressing the CRISPR-Cas promoters, causing anti-microbial activity using bacterial own defence systems [79]. Thus, CRISPR-Cas mediated antimicrobials deliver a rapid and comprehensive method for selection of certain strains without any effect on other closely connected strains. Additionally, they can be used for various purposes, like further research in identification of a strain, desirable elimination of contaminant from a media, to screen undesirable genes, and culture handling for optimal product quality. However, these application faces several obstacles. For example, the requirement of a suitable delivery system for the maintenance of populations of strain, particularly for those lacking native CRISPR-Cas systems. Small host range limits the usage of phage mediated delivery [73]. Targeting CRISPR is not always 100% effective because sequences in both the host and target mutate to escape CRISPR-Cas cleavage [71]. Further research focused to explore the causes of the CRISPR escape is required for the improvement of effectiveness of CRISPR antimicrobial.
4. Phage resistance of food pathogens using CRISPR-cas complex
Phage is primarily responsible for slow fermentation in the food industry because it causes contamination of starter cultures and are resistant to physiological treatments, such as, thermal processing, pasteurization, high pressure, and radiation technology [80]. Food industries have developed several ways, including changes in factory design and sanitation to minimize any economic losses associated with phage infection. The pathogens aiding in fermentation utilize CRISPR-Cas against invasive phage. The manifestation of CRISPR-Cas system is very much predictable in fermentation micro-organisms given the predominance of phage during fermentation processes. The adaptive and interacting nature of CRISPR-Cas indicates its capability for acquiring a spacer which encounters to be immune to this phage. The immunity-providing effect of native CRISPR-Cas systems is relatively simple and does not require the necessary engineering to promote the system to adapt to new spacers [81]. CRISPR-Cas system from S. thermophilus was transplanted into E. coli against phage infection by heterological protection. The sequence-specific CRISPR-Cas based interference causes alterations in the proto-spacer adjacent motif, evading CRISPR-encoded immunity in a McrA/HNH- and RuvC/RNaseH-motifs dependent manner. The transmission of active CRISPR-Cas systems across distant genera is the reason of heterologous interference generation against invading nucleic acids [82]. The available published outcome from the researcher across the globe is comprehending that not only for thero-diagnostics but CRISPR-Cas is also contributing to limiting food contamination and fermentation failure by phages, which were previously supposed to be attenuated by resistant or mutant microbes, those which have the potentiality to cause food borne diseases. Hence CRISPR-Cas is playing a significant role in the dual application of preventing phage-induced contamination as well as inhibiting the application of resistant pathogens for preventing phage contamination.
5. Plasmid vaccination of food pathogens using CRISPR-cas complex
CRISPR-Cas systems depict the intake of plasmids being prevented by dividing the DNA [83,84]. When a cell possessing CRISPR-Cas based immunity containing plasmid encounters, it causes the protospacer to be vaccinated against the uptake of that plasmid, resulting in evolution of strain through CRISPR-mediated vaccination. Another advantage gained from plasmid vaccination is the hindrance of unwanted DNA elements responsible for pathogenicity islands or antibiotic resistance genes, which are transmitted via plasmids [[85], [86], [87]]. The spread of AMR is a particular concern for the food industry and the occurrence of CRISPR-Cas was associated with an absence of AMR markers in Enterococci sp [87]. Moreover, phage resistance is engineered for CRISPR-Cas targeting plasmid-contained antibiotics-resistant genes in strains containing native CRISPR-Cas systems. This was successfully performed in S. thermophilus by selection of a strain without plasmid presented by a AMR gene containing plasmid [82].
6. Strain typing of food pathogens using CRISPR-cas complex
CRISPR-Cas is a potent technique not only for the detection of strains but also understanding the stress difference in microbial ecology investigation and development of environmental complex. Thus, providing epidemiological insights and understanding of populations of the genotypes [88,89]. Strain typing, a novel and reliable tool for sequencing CRISPR repeat-spacer arrays, has limitation of its application due to the occurrence of CRISPR-Cas in the strains of interest. However, industry related to starter culture is particularly favourable to the use of CRISPR-Cas in characterisation of strains because of the availability of CRISPR-Cas in many microorganisms related to fermentation industry [90].
CRISPR-Cas is favourable for strain-typing applications in microorganisms of fermentation process because of its high occurrence in food pathogens [90]. The utilization of repeat spacers for strain typing in S. thermophilus in the production of cheese and yogurts provides insight into the connectivity of different strains and their ecology [91]. The repeat-spacers of Cas genes were studied for the identification of Lactobacillus casei and Lactobacillus paracasei as probiotics [92]. Lactobacillus rhamnosus and Enterococcus faecalis based CRISPR genotyping could distinguish between related strains in handcraft and industrial dairy and meat products [93]. Sequence analysis of well-known commensal and probiotic microorganism Bifidobacterium genus and Lactobacillus gasseri exhibit genetic divergence while CRISPR-Cas based strain typing monitors the existence of spoilage microorganisms, in many fermentation processes [94,95]. Moreover, Salmonella sp, Lactobacillus buchneri, Campylobacter jejuni, Clostridium difficile, Corynebacterium diphtheriae, E. coli, Vibrio parahaemolyticus have also been genotyped via CRISPR-cas complex for stain typing and their application in food industry [96]. Thus, the CRISPR-Cas strain typing system is utilized to ensure the safety and quality of the food product, regardless of whether spoilage could be tracked or food pathogens are identified. Hence, with all reference to these progressive and prosperous applications, further research outcomes are expected with the data diminishing the limitation of the application of the CRISPR-Cas system in strain typing while enhancing its usage for preventing pathogenic outbreaks by available sequence identification.
7. Genome editing of food pathogens using CRISPR-cas complex
CRISPR-Cas are used as a tool for genome editing by Type II systems owing to their complex interference, simplicity in application, and the sequence-specific way of targeting DNA as illustrated in Fig. 4 [97]. For genome editing through CRISPR-Cas, four elements need to be established which includes the Cas9 protein effectors and the crRNA-tracrRNA complex. The crRNA-tracrRNA complex interacts with the target sequence of DNA and DNA repair mechanism within the host organism to repair the damage done by CRISPR-Cas for incorporation of the desired genomic changes [98]. Even though the pathogens contain a native CRISPR-Cas-type II, it probably contains no spacer that can focus precisely to the editing point. sgRNA has revolutionized the genome-editing field since sgRNA can be easily be developed for effectively targeting of genome of interest for Cas-9 delivery system. sgRNA simplified the two-component system of Cas9 and sgRNA which was easy to plan, package and transfer the cells of interest [97]. The vast bulk of genome editing of CRISPR in eukaryotes occurs when the cell performs its damage repairing machinery through endogenous DNA repairing mechanism like homology-orientated repairing, resulting into mutations on the site of cleavage [99]. However, CRISPR-Cas are used to modify the genome due to the lack of robust DNA repairing mechanisms in the bacterium, hence physiology and homeostasis mechanisms such as cleavage of double-strands of bacterial genomes causes cell death [98].
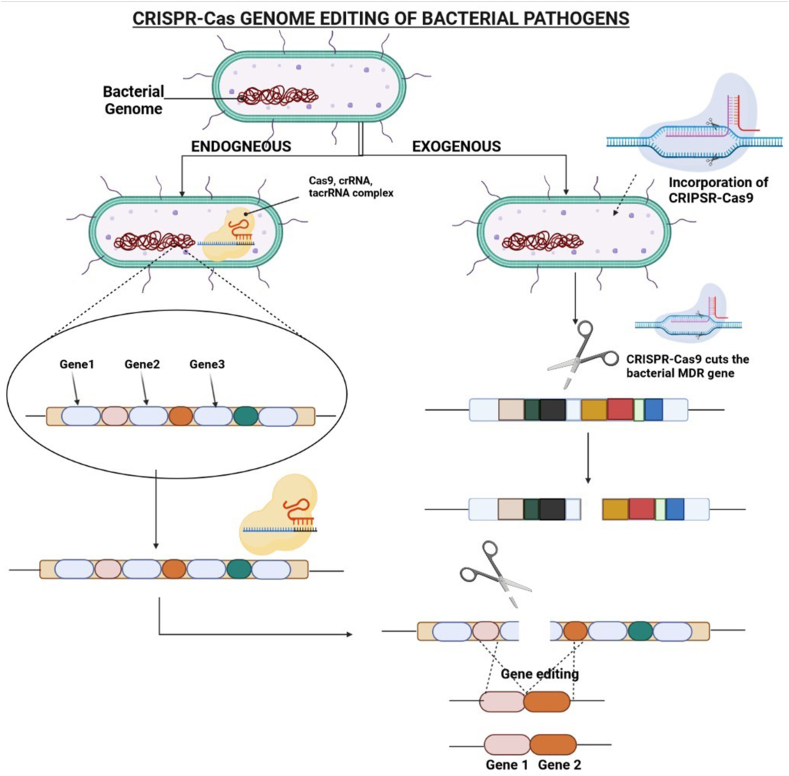
Fig. 4.
Genome editing for inactivation of MDR pathogens via CRISPR-Cas system. (1) Endogenous mechanism - pathogens containing a native CRISPR-Cas-type II without any spacer focuses on genome editing at desired location. crRNA-tracrRNA present inside bacterial cells forms a complex when come in contact with Cas9 protein. This complex further goes and bind to the bacterial gene at the site where DNA repairing is must and interrupt this repair mechanism, resulting into mutation at cleavage site. (2) Exogenous mechanism - Pathogens which do not have native CRISPR are incorporated with CRISPR (with the help of nano-carriers), where it acts as a scissors to cut the MDR gene, resulting into antibacterial sensitivity.
S. thermophilus in fermentation industry and Lactobacillus reuteri in probiotics exploits CRISPR-Cas to select certain genotypes [71,100]. CRISPR-Cas was utilized in S. thermophilus for selection of heterogeneous populations based on naturally occurring genotypes [71]. Using self-targeting of CRISPR-Cas, the individual genotypes for genomic islands of interest was screened. A combinatorial platform was developed with CRISPR-Cas to use ssDNA recombinant engineering, and enhance recoverable recombinants in L. reuteri to select mutants that were recombined with ssDNA [100]. CRISPR-Cas9 endonuclease complexed with dual-RNAs are used for the introduction of mutations in the genome of pathogens, eliminating the requirement for selectable markers and counter-selection systems through dual-RNA-Cas9 directed cleavage at the targeted genomic nucleic acids for unmutated pathogenic clearance [101]. The sequence of crRNA was modified to perform single-nucleotide and multi-nucleotide editing to reprogram dual-RNA-Cas9 specificity. Multiplex mutagenesis occurs when two crRNAs are used simultaneously and transformable cells are selected and a minor manipulation at the target sequences by dual-RNA- Cas9 cleavage that aids gene editing in S. pneumoniae and E. coli for the selection against non-recombinant cells [101].
The combination of ssDNA recombination and CRISPR-Cas possesses the ability to recover low efficiency and target codon mutagenesis. Even though ssDNA engineering is an impactful mechanism, its application is challenging, as it must be optimized for each strain. The primary focus is on the application of CRISPR-Cas for homologous recombination. Nevertheless, the utilization of CRISPR-Cas in genome editing increases its application in the food industry against MDR food pathogens. Furthermore, characterization to implement minor and significant mutations of bacterial genomes is required. Moreover, CRISPR-Cas is used for the natural selection of genotypes by exploiting their functionality from industry insights.
8. Nanomaterials(NM) as an antimicrobial and detection tool against food pathogens
The applications of nanotechnology have gained quite an interest in the food industry due to their high physical, and chemical stability, magnetic properties, and large constant lattice values [102,103]. These features are mainly used in the food industry to enhance the overall quality, shelf life, taste, flavor, processing capacity of the food, and increased specific surface area for antibacterial activity [104] (Fig. 5). These applications can primarily be summarized into two main groups which include the nano-structured food matrixes and nano-sensor for food. Nano-structured food constituents covering the broad range of processing and packaging industries are also applied as food preservatives, smart carriers, anti-caking age and antimicrobial agents, and fillers to strengthen the mechanical properties of packing material. Furthermore, nano-sensors can be used to improve food quality and safety assessment [18]. Food-borne infections have emerged as a major public health concern and thus causing the need for both quick and sensitive testing of various food-borne pathogens. Several metal-based NPs (MNPs) and organic NPs (ONPs) have been used directly or upon functionalization for enhanced binding affinity with the pathogen for improving the surface selectivity, leading to pathogen detection with accuracy [105]. Herein, we have highlighted the clinical significance of NPs as an antimicrobial and detection tool against food pathogens as illustrated in Fig. 6, Fig. 7.
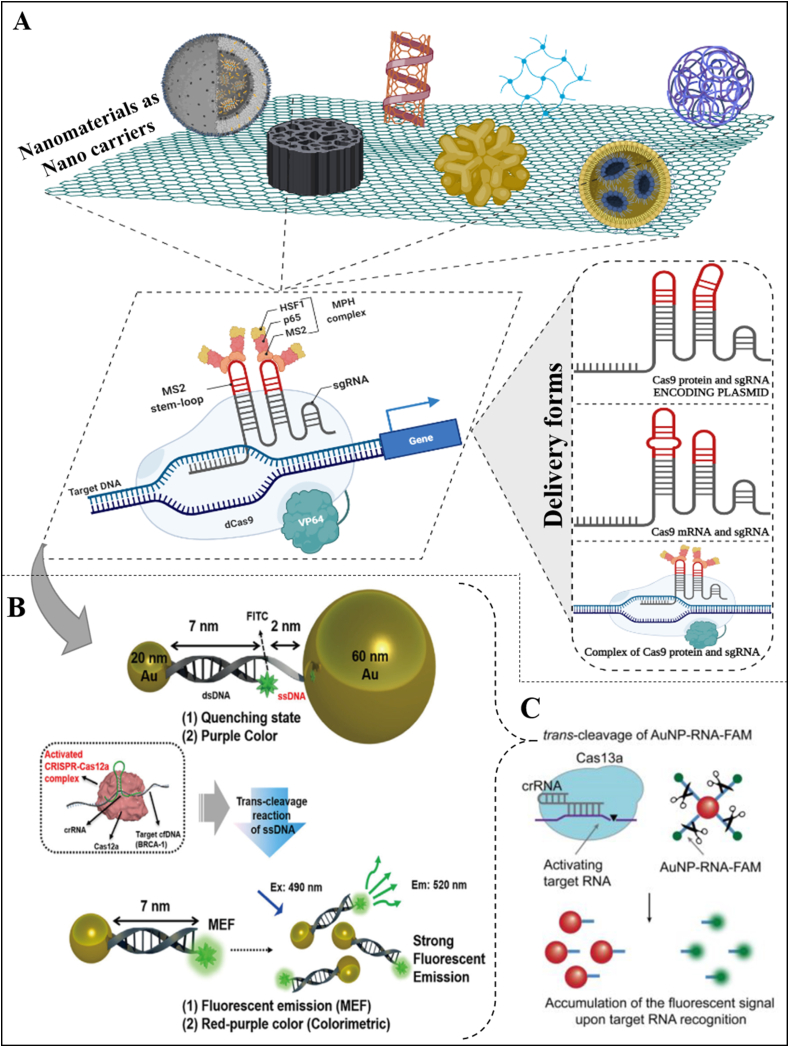
Fig. 5.
(A) Different types of nanoparticles as nano carriers used for detection and antimicrobial activity of food-borne pathogens. (B) DNA-functionalized gold nanoparticles combined with the trans-cleavage activity of the CRISPRCas12a would release the short-distance gold nanoparticles, greatly increasing the fluorescence intensity. (C) Schematic illstrating the CRISPR/Cas13a detection system based on the AuNP-RNA-FAM probes (Figures adapted and edited from Refs. [159,160]).
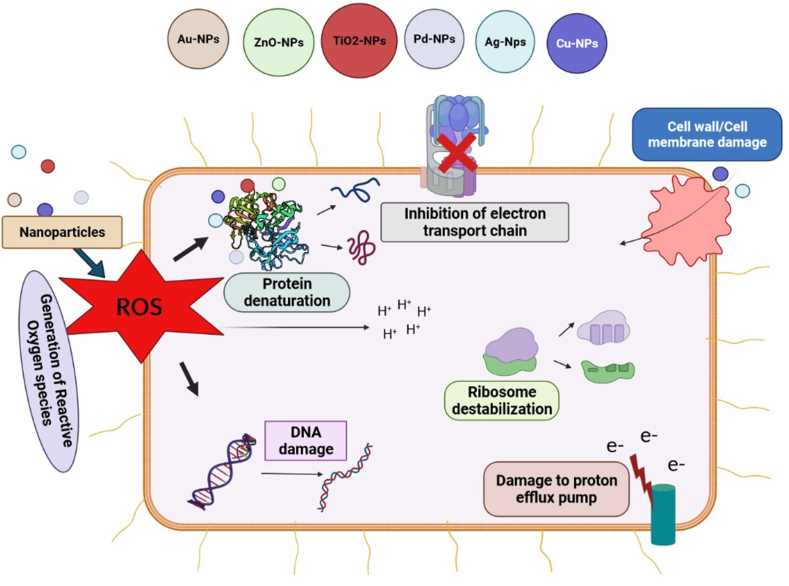
Fig. 6.
Mechanism of Antibacterial action of different nanoparticles. The naoparticles act agai.
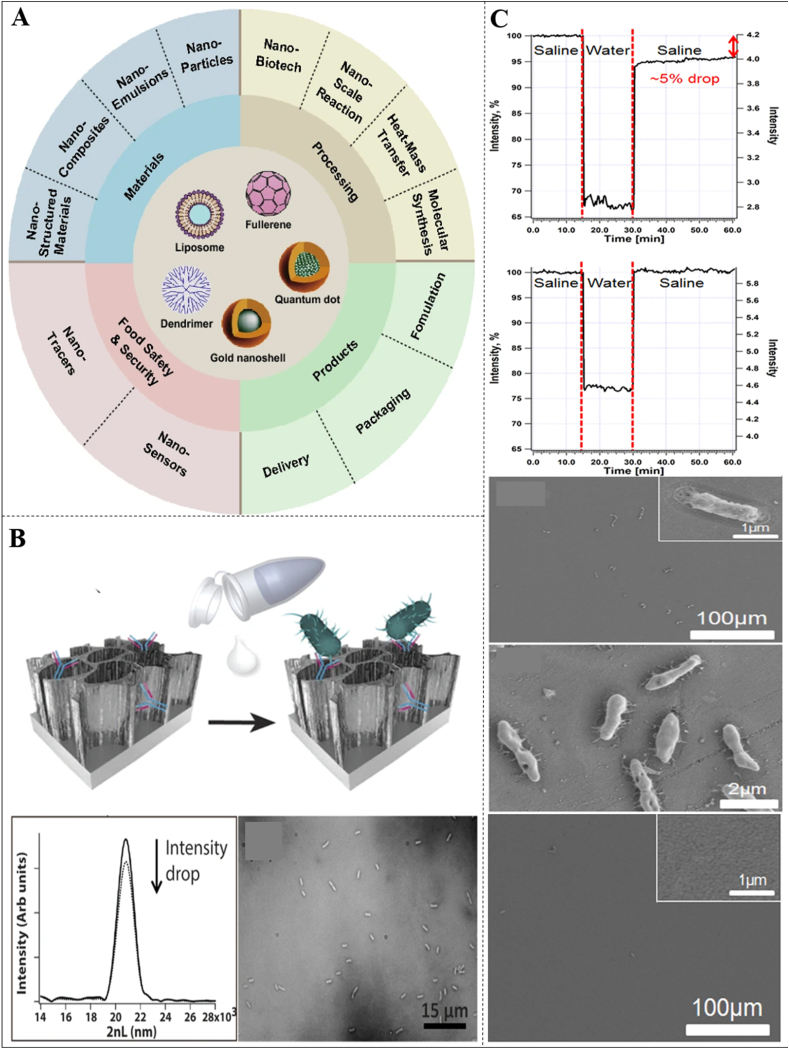
Fig. 7.
A. Nanotechnologies in Food Science: Applications, Recent Trends, and Future Perspectives. B. Specific capture probes (antibodies) were immobilized onto the porous surface to provide the active component of the biosensor. The biosensor was then exposed to the target bacteria to directly capture the bacteria cells onto the antibody-modified PSiO2 surface. A drop in the intensity of the thin-film optical interference spectrum of the biosensor results from bacteria capture. Microscopy tools (light microscope and HRSEM) and real-time PCR methods were used to confirm the presence of bacteria on the biosensor surface. C. Representative biosensing experiments and the corresponding HRSEM images of the biosensors immediately after the experiments. Spiked process water (104 cell/mL E. coli). Control - original process water (no E. coli). The corresponding HRSEM images (in two different magnifications) of the biosensor after a biosensing experiment with spiked process water, demonstrating bacteria capture. The inset presents enlargement of a captured bacterium on the biosensor surface. A corresponding HRSEM image of the biosensor after the control experiment (original process water, no E. coli) showing a negligible amount of cells. The inset presents enlargement of the biosensor surface (Figures adapted and edited from Refs. [161,162]).
The anti-bacterial properties exhibited by ZnO-NPs through cytoplasmic leakage caused by rupture of the cell membrane via abrasiveness and surface oxygen species for biocidal effects against E. coli DH5 and S. aureus owing to their stability under pressure through their interaction with pathogen [106,107]. The values of MIC and MBC suggest that nano-colloidal AgNPs possesses bacteriostatic effect but a weak bactericidal impact, against food-borne pathogens (Salmonella enterica, Serovar Typhi, Escherichia coli, Listeria monocytogenes, Bacillus cereus, Vibrio cholerae, Vibrio parahaemolyticus, and Staphylococcus aureus) and since they are inexpensive and exhibits great antibacterial efficacy, they could act as potential antimicrobial agents [108]. Alike chitosan, AgNPs impair bacterial cells causing the loss of the ability to permeate the membrane and impart anomalies in the regulation of membrane transport and denature nucleic acid [109]. The antibacterial activity of polyamines attached to the exterior surface of mesoporous silica particles for enhanced anti-bacterial efficacy. The enhancement is due to the covalent functionalization with N-(3-trimethoxysilylpropyl) diethylenetriamine against L. monocytogenes through interactions between the positive amine corona on the surface of NPs and the negatively charged bacteria membrane leading to the cell membrane disruption. NPs surface conjugated with polyamine is 100 times more potent than free polyamines for L. monocytogenes clearance [110]. A novel nano-carrier was self-assembled by coupling the magnetic properties of surface active maghemite NPs (core) with tannic acid (shell) in water to synthesize SAMN@TA hybrid against Listeria monocytogenes. The material is employed in the food sector as an affordable, efficient as well as eco-friendly anti-microbial nanomaterial [111]. Gold nanoparticles (AuNPs) of size range 7–40 nm and AgNPs of size range 10–40 nm were demonstrated to be highly potent against Gram-positive as well as Gram-negative bacteria [112] such as B. subtilis (AuNPs), P. aeruginosa (AgNPs), S. mutans (AgNPs), S. aureus (both AuNPs and AgNPs), E. coli and K. pneumoniae (both AuNPs and AgNPs) [[113], [114], [115]]. AuNPs cause reduced membrane potential by disrupting ETC and a decrease in ATP synthesis for irregular cell shape, membrane degradation, and the loss of its protective function [116,117]. The morphological characteristics of Au-NPs and Pt-NPs were assembled with Salmonella Enteritidis and Listeria monocytogenes as bacterial NPs vehicles for utilizing bacteria as a vehicle to transport a nano-conjugated drug. Au-NPs and Pt-NPs are encapsulated inside the bacteria via a self-organization method without the disruption of bacterial cell walls [118]. Due to the morphological interaction between Au-NPs and the pathogens (Salmonella enteritidis and Listeria monocytogenes), the aggregation of Au-NPs between flagella or biofilm networks did not permeate the bacterial cell. Pt-NPs entered Listeria monocytogenes cells to cause elimination via its bactericidal properties from the cells [119]. Anisotropic dumbbell-like Au–Pt NPs pose efficient catalytic and anti-bacterial activity using a DNA-programming approach for the detection and eradication of bacterial infections [120]. Furthermore, the oxygen reduction activity of octahedral Pt–Ni NPs is attributed to shape selectivity and tailored surface composition [121]. Rhamnolipids functionalized liposomes (rhamnosomes) were produced using nisin for the induction of innate antibacterial and antibiofilm properties to enhance its efficacy against the foodborne pathogens. RSNVs depicted synergy between nisin and rhamnolipids for improved antibiofilm activity [122]. Listerium monocytogees were investigated in intestinal epithelium cells upon pre-treated with a low or high dosage of TiO2 NPs for both adhesion and invasion. The increased invasion was exhibited due to cytoskeletal changes caused by NPs for upregulated levels of listeriolysin-O that protects L. monocytogenes from ROS for enhanced bacterial survival causing nano-toxicity. The potential risk of increased L. monocytogenes susceptibility related to the nanosized TiO2 intake at small dose should be considered [123].
The identification of E. coli in the samples of ground beef using MNCs functionalized with biotin-labelled polyclonal goat anti–E. coli antibodies immobilized over streptavidin-coated magnetic NPs for MNCs. MNCs presented a MCE of 94% for E. coli and the immunoreaction time was 15 min deprived of any enrichment [124]. Carbon-based nanotubes both single-walled (SWNT) and multi-walled (MWNT) possess unique properties as an advantage with enhanced aspect ratio, high surface area, and unique electrical and optical aspects. Chemical modification or functionalization of carbon nanotubes employed for in vitro and in vivo evaluations ensures homogeneous dispersion in aqueous settings for pathogen detection [125,126]. A carbon nanotube functions as a 1-D scaffold while being covalently connected with d-galactose molecules through ligand-receptor interaction by binding selectively with pathogenic Escherichia coli for cell agglutination [127]. For efficient detection of E. coli a micro-nanofluidic device functionalized with magnetic beads was utilized to capture the food pathogens through computational fluid dynamics (CFD), 3D-tomography, and machine learning to probe and bead stack the pathogen. Moreover, this nano-device is multiplexed, downsized, cost-effective, transparent, and easy to fabricate and operate which is making it ideal for pathogen separation in both laboratory and point-of-care (POC) settings [128].
The immunomagnetic MWNT is used as an alternative IMS platform for rapid and sensitive pathogen identification, owing to its ability to carry a large number of immuno-functionalities per unit weight of material while remaining stable in optically transparent liquids [126]. The use of IMS and MDA, preferentially concentrates Salmonella gDNA for improved detection of Salmonella-spiked raw chicken breast sample culture enrichment, allowing for serotyping and high accuracy SNP of food pathogens using shotgun sequencing has been implicated as a method of identification [129]. A specific, sensitive and accurate lateral flow immunoassay developed for the detection of S. typhi through QDNS, acting as signal reporters with visual LOD (5 × 103 CFU mL−1) for Salmonella typhimurium within 10 min [130]. Furthermore, non-Salmonella typhimurium bacteria were found in larger numbers depicts influence on Salmonella typhimurium detection [131]. m-AuNPs were characterized to selectively bind the mannose-specific adhesin FimH of type 1 pili by functionalizing with carbohydrate attachment as a labelling probe and multi-ligand carrier in biological systems. Thus, the robust specific binding of m-AuNP with bacterial type 1 pili paves a novel way for identifying particular proteins over the surface of the cell by using carbohydrate attached NPs [130]. A disposable immuno-sensor combining a magneto-immunoassay and AuNPs for electrochemical detection of Salmonella enterica by functionalizing a screen-printed carbon electrode (SPCE) with a permanent magnet. An anti-Salmonella magnetic beads (MBs-pSAb) were used to trap Salmonella-containing samples (skimmed milk), sandwiched with AuNPs-modified antibodies (sSAb-AuNPs) for detection using differential pulse voltammetry (DPV) causing 83% and 94% efficiency [128]. A label-free impedimetric aptamer-based bio-nano-sensor has been utilized based on electrochemically and chemical grafting of a diazonium-supporting layer onto SPEs which initiates immobilization of chemicals of aminated-aptamer, for capturing and detection of Salmonella. Furthermore, the cell systematic evolution of ligands by exponential enrichment was used to select DNA sandwich aptamers with excellent affinity and specificity for S. enteritidis detection. Sandwich capillary detection platforms utilized both aptamers crn-1 and crn-2 for recognition of target bacteria (S. enteritidis) precisely based on colour change with a detection limit (103 CFU/mL). Thus, this system is used to detect samples in food with a complicated matrix in a point-of-care naked eye setting [132]. A detecting system was established based on immune-magnetic NPs that incorporated magnetic and thermal properties of MNPs to allow efficient capturing, high sensitivity thermal sensor detection, and sterilization of Salmonella typhimurium. The spike in Salmonella typhimurium in drinking water can be detected with a detection limit of as low as 300 CFU/mL under ideal conditions [133].
A geno-sensor probe functionalized with a glassy carbon electrode was utilized for identification of Listeria monocytogenes in milk samples by modification of Pt-NPs dispersed in a chitosan matrix [134]. The DNA-based interfacial interaction between target DNA and Pt-NPs is immobilized with ssDNA and albumin to minimize non-specific DNA binding and increase signal sensitivity. It is a cost-effective, simple, user-friendly, and sensitive electroanalytical identification of gene sequences post hybridization with gDNA in the sample [134].
A highly sensitive fluorescence nano-bio sensor has been developed for the detection of Shigella species using two DNA probes for surface functionalization of AuNPs and MNPs. The target DNA was injected with the MNP-DNA probe and the DNA-AuNP-fluorescence DNA probe accompanied with a magnet used to separate the DNA probe complex while exhibiting enhanced sensitivity and specificity [135].
9. NM-CRISPR: A novel combinatorial therapeutic solution against food pathogens
Conventional diagnosis with limited stability and sensitivity necessitates the use of CRISPR as well as nanomaterials in the food industry to improve accuracy. Nanomaterials, primarily metal nanoparticles, offer a high degree of stability, as well as satisfactory optical and electrocatalytic properties, making them ideal for use as reporter molecules. CRISPR/Cas machinery on metal nanoparticles is being used in biosensor research to design a nanosensor with the motif of targeting a variety of pathogens [136,137]. Nanotechnology offers the potential to enhance the effectiveness and safety of CRISPR/Cas distribution, which has been known as a major limitation of its application. The combination of nanotechnology along with CRISPR/Cas technology holds a promising potential for next-generation thero-diagnostics of numerous food pathogens.
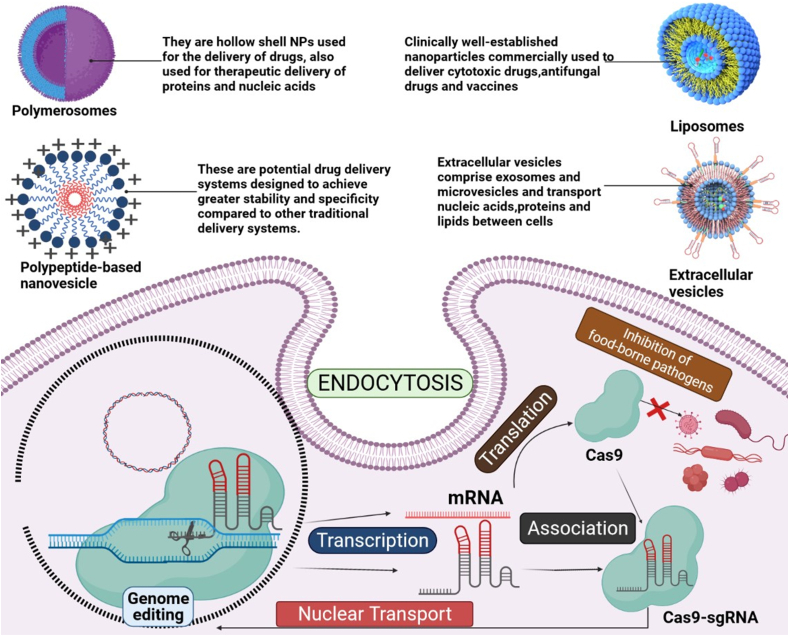
A spacer sequence containing crRNAs and tracrRNAs is produced by following transcription of CRISPR region and sgRNA processing. This sgRNA in conjugation with a protein complex possesses nucleotide degradation capabilities, allowing the DNA spacer sequence to be recognized by it [138]. The combination of sgRNAs and the CRISPR-Cas system triggered huge applications of genome editing for microbial detection and pathogen clearance. Despite great promises, the industrial application of the CRISPR-Cas system is inadequate due to lack of proper delivery. To overcome this aforementioned restriction several systems for delivering CRISPR-Cas and sgRNA to cells including viral and non-viral vectors. Nano-vesicles based on peptides, polymers, lipids, and extracellular vesicles are among the non-viral delivery systems. These nano-vesicles are designed to enhance the delivery by endosome escape and environmental factors such as light, pH, and environmental features [139]. Despite progress in CRISPR-Cas development, the widespread practical efficacy remains unchanged. The conventional adenovirus or retrovirus systems are highly transfection efficient, but their toxicological characteristics, together with immune-response initiation and viral genome introduction inside host chromosomes, restricted their use as a delivery system [140]. Non-viral delivery systems, are safer and not limited by genome editing components, but are relatively low in their delivery efficiency [141]. Cationic lipid NPs, and cationic liposomes are utilized for gene delivery as non-viral delivery systems, due to the electrostatic interaction of negatively charged nucleic acids and CRISPR-Cas9 RNP complexes with the positive charge of NPs. Although cationic lipid NPs offer promising systems to provide genome-editing components, strenuous exploitation of Cas-9 due to poor stabilization of cationic lipid NPs with plasmid DNA or mRNA is challenging. The lipid nano-shells encapsulating genome-editing cargo are used to overcome issues pertaining to stability caused by exposure of cationic lipoplexes to serum. Herein, we have elucidated the strategies that can be incorporated for delivery of CRISPR-Cas utilizing nanocarriers and their preclinical significance as illustrated in Fig. 8, Fig. 9 and Fig. 10.
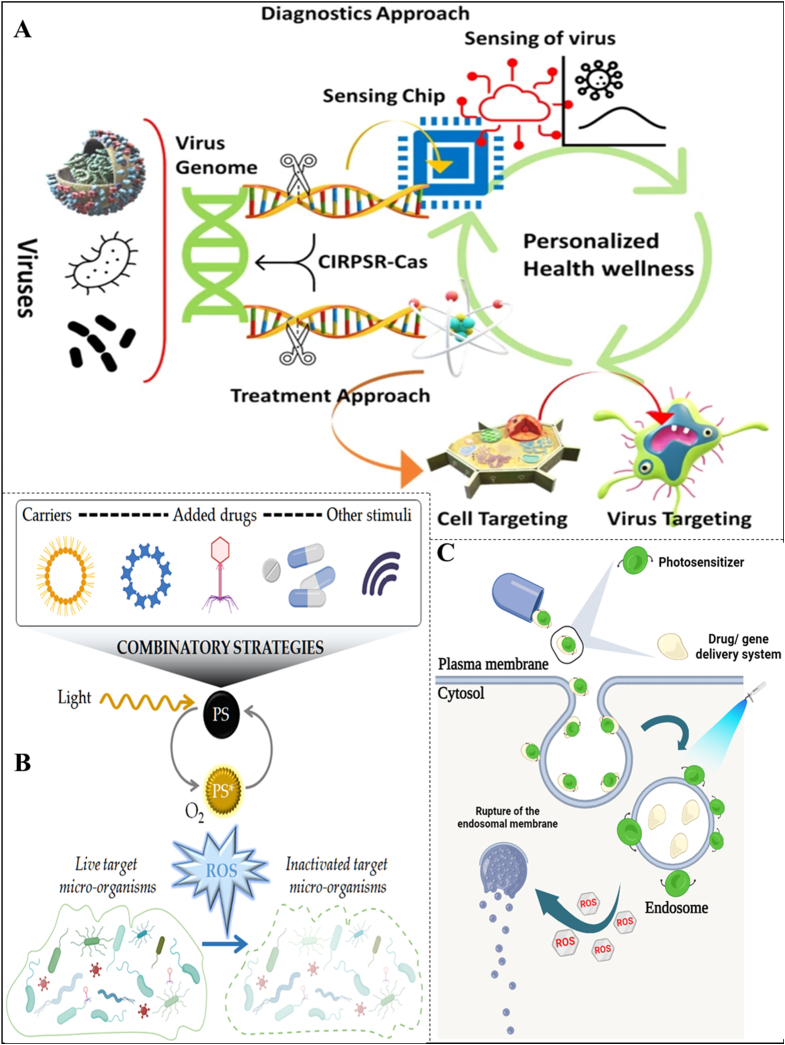
Fig. 8.
Different strategies of CRISPR-Cas complex delivery by nanomaterials as a therapeutic solution against food-borne pathogens. (A) The perspective of nanotechnology assisted CRISPR/Cas system for efficient diagnostics and treatment of a viral infection/diseases, overall, towards personalized health management. (B) Antimicrobial Photodynamic Therapy (pharmaceutics-13-01995-v3). (C) A pictorial representation illustrating the mechanism of photochemical internalization technology. The photosensitizer bind to the surface of the membrane. The gene/drug delivery system along with photosensitizer molecules internalize into the cells through endocytic pathway. The photosensitizers are integrated into the membranes of the endosomes and remain inactive. The photosensitizers becomess activated after light exposure and generates highly reactive singlet oxygen (1O2), causing rupture of the endosomal membranes. (Figures adapted and edited from Refs. [136,163]).
Fig. 9.
Mechanism of Nano-CRISPR synergy for pathogen clearance.
Fig. 10.
Modified Organic nanomaterials for their use as CRISPR delivery and their mechanistic application with detailed molecular understanding of genome editing upon delivery.
Most NPs enter cells through the endocytic pathways that include early fusion, maturation, and lysosomal degradation for efficient transmission of Cas-9 into the core to escape endosomes at the early stage. Liposome-mediated membrane fusion, cationic peptide-conducted membrane destabilization, PEI-mediated osmotic pressure, and pH-responsive polymer-mediated swelling are some of the endosomal escape strategies used to cause the fast release of nucleic or protein to the cytoplasm. A dendrimer encompassing glucuronyl-β-CD complex with Cas9-sgRNA is used to administer inside the cell, to induce gene editing efficiency at the injection site than ordinary Cas9-sgRNA conjugate [142]. A cationic diethylenetriamine movement for endosomal escape aids delivery of Cas9 mRNA and sgRNA by electrostatic interactions to the side chain of amphiphilic poly-aspartame derivates [143].
Phototherapy (PDT) represents another strategy for the induction or release of enclosed cargoes by laser-irradiated stimulation to deliver Cas9/sgRNA complex at the desired location and time. The light release via PDT aids in genomic modification at the target tissue while endosomal escape is facilitated, causing carriers to deliver their cargo. Photothermal as well as photodynamic activation studies have been conducted for intracellular delivery of Cas9-sgRNA plasmids [144]. A photothermal delivery system for plasmid DNA encoding Cas9 and sgRNA specific for the PLK1 gene has been developed, wherein a polymer derivative featuring semiconducting properties was grafted with PEI via thioester bond linkers. The transmission of dexamethasone from hydrophobic compartment of the polyplex contribute to nuclear-pore dilation, promoting the invasion of the Cas9-sgRNA plasmid [144].
A Glutathione-reducible synthetic lipid NPs was developed for the delivery of Cas9-sgRNA RNPs complexes [145]. Cas9-sgRNA RNPs can also be delivered through GSH-responsive disulfide bond-bearing cationic copolymers. Cas9-sgRNA-RNP complexes were also covered by the GSH-degradable cross-linker designed to supply Cas9-sgRNA, which were then polymerized by the formation of a disulfide bond for the NPs to cause degradation of GSH. Endosome escape occurs regardless of the cargo arrangement (DNA, mRNA, plasmids) encoding Cas9-sgRNA for delivering CRISPR via cell membrane penetration. The delivery system may become degraded by lysosomal enzymes once the endo-lysosome enters unless the endo-lysosome escapes via GSH delivery strategy [145].
A variety of pH-responsive polymers have also been introduced to make the endo-lysosomal release genome-editing materials. A derivative of polylysine-g-polyethylene glycol allows delivery of Cas9/sgRNA RNPs targeting STAT3 [146]. A variety of pH-sensitive amino lipids with modified amine head groups were synthesized for Cas9 plasmid delivery.
The intracellular regulation of genome editing components is being actively regulated by physical strategies, including needle-based membrane pored formation which induces effective genome editing through incorporation of the sgRNAs-Cas9 complex inside the cell. The silicon-based nanoneedle ranges with 200 nm diameter nanoneedles where the protein complex of sgRNA and Cas9 were adsorbed on nano-needles hydrophobic surfaces [147]. Nano-wires have also been used for delivering genome editing tools [148]. An efficient, rapid delivery is possible via physical approaches [149]. Various stimuli (mechanical forces, electrical forces) are used to deform the cell membrane for successful delivery. Physical strategies pose unique benefits of induction for effective genome control in cells that are difficult to transfect, such as hematopoietic stem cells while delivering cargo like the CRISPR-Cas system.
Kang et al. used a nanoscale CRISPR complex containing a polymer-derivatized Cas-9 endonuclease and sgRNA to target the mecA gene through covalent modification with branched polyethyleneimine for bacterial genome editing efficiently causing MRSA inhibition [150]. In another investigation gold nanoclusters (AuNCs) were utilized along with CRISPR -Cas9 to transfer the Cas9 endonuclease (SpCas9) from Streptococcus pyogenes into the nucleus via a highly pH-dependent assembly process. Thus, utilizing this novel strategy of combinatorial therapy will pave a new platform against the food industry.
Though multiple studies are illustrating this combinatorial approach of nanotechnology and CRISPR, several researchers are on their way to shrinking the toxicological impact of nanocarrier for CRISPR delivery, for utilization of this novel strategy of combinatorial therapy to pave a new platform for the food industry in combating food-borne MDR pathogens.
10. Challenges and outlook
Despite its high specificity and sensitivity, CRISPR/Cas-based combinatorial detection has some shortcomings, including the requirement for pre-amplification procedures along with a compatible host having a suitable delivery system with enhanced efficacy for reduction of off-targeting. One of the major challenges in the transformation of genome editing tools is the non-target effects of inefficient genetic systems on both CRISPR and cellular tools. The genetics of the off-targeted effect has been developed for new variants of CRISPR tools. Nevertheless, the specificity of the delivery system mainly depends on off-target side effects. The non-specific uptake of Cas9 NPs generates unexpected mutations in the immune cell causing lethal toxicity by deleterious off-target mutations. So, future research focused to overcome these challenges, for preventing non-specific uptake by immune cells. Cas9/sgRNA delivery systems must be carefully considered for their route of administration in regular practice. Systemic administration allows particles in targeted tissue to be deposited deeply but causes unexpected gene-editing side effects due to their huge distribution volume in comparison with other routes of delivery.
Plasmid DNA expressing Cas9 and sgRNA have been exploited in some studies, but for these studies conducted successfully to the cytoplasm needs to contain the expression of Cas9 and sgRNA, and their complex need to invade into the nucleus for genetic modifications. The advantage of this single plasmid DNA-dispenser is easy, but its complex kinetics, complexity with sgRNA, and nuclear location reduce genome editing efficiency. The emerging platform for Cas9-sgRNA is natural vectors derived from extracellular vesicles. Although the biocompatibility and homeotropic-based targeting in these natural vesicles are superior, they pose several problems that should be addressed prior to their utilization for clinical practice. Based on the finding of the genome-editing machinery the detection of various pathogenic organisms has opened new doors to develop an effective, efficient and safe delivery system for pathogen detection.
Although several vectors for delivering CRISPR machinery have been studied in varied models but the field remains in its infancy. Additional modification shall be required for improving the limited efficiency of gene-editing, non-specific delivery, and unknown toxicity. Studies supporting regulatory perspectives are required for the approval of genome-editing components for future detection methods. Various strategies have been exploited for promoting editing efficiency and minimizing off-target genome modification. Upon addressing long-term safety problems and clinical problems with CRISPR delivery, these powerful tools could be used successfully for clinical practise. Reduction of off-target modification, consideration of the antimicrobial administration rout, and the search of a suitable vector following the development of nano-sensors and improvement of nanotechnological application by diminishing the toxicological effects should be focus of future research to overcome these challenges for making a promising revolutionary combinatorial theragnostic tool of MDR food pathogens.
11. Conclusion
Food-borne pathogens are one of the major causes of infection worldwide, and the rapid increase in food-borne diseases has been the major area of concern for healthcare worldwide. In summary we have highlighted the application of the CRISPR-Cas complex as well as nanoparticles for rapid detection and antimicrobial properties against food borne pathogens. CRISPR based detection of food pathogen is gaining recent interest in the last couple of years wherein CRISPR/Cas9 and CRISPR-Cas 13a system, have been successfully employed for detection of drug resistant S. aureus and E. coli respectively (Fig. 7). The serovar-specific CRISPR-Cas spacer system has emerged as a unique approach for genotyping and fast detection of Salmonella. Moreover, CRISPR based sub-typing is the basis of detection of Shigella and Listeria too. Apart from the utilization as a detection tool, CRISPR has come a long way to be used effectively as a powerful antimicrobial against severe MDR food pathogens by controlling the spoilage organisms and genetic editing tool. Not only as antimicrobial and detection tool but CRISPR-Cas system are also been used in case of fermentation industry for generating resistance to phages which are the main reason of slow fermentation. Another crucial application of CRISPR against food pathogens is plasmid vaccination, targeting phage resistance as well as antibiotic resistance. Strain typing and genome editing are some other efficient areas of applications of CRISPR-Cas system against severe food pathogens. Nano-compounds are also used in conjugation with some other assays (lateral flow immunoassay, immunomagnetic separation) as a successful detection method of food borne pathogens. Moreover, it is being utilized as an antimicrobial agent to inhibit the infection caused through contaminated food. Delivering the CRISPR system is the only limitation and utilization of nanocarrier nullified this problem. Herein we have presented with a novel combinatorial strategy of nano-CRISPR as a new potential therapeutic solution against food pathogens that elucidated the methodologies for the combinatorial therapy.
Data availability
All data included in this study are available upon request from the corresponding author.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We acknowledge infrastructure support available through DBT-BUILDER program (BT/INF/22/SP42155/2021) at KIIT UNIVERSITY.
Abbreviation
- MDR
Multidrug-resistant
- WHO
World Health Organization
- EFSA
European food Safety Authority
- CDC
Centers for Disease Control and Prevention
- AMR
Anti-microbial resistance
- RBP
Ribosome Binding Protein
- NPs
Nanoparticles
- ROS
Reactive Oxygen Species
- MRSA
Methicillin Resistant Staphylococcus aureus
- CCB
CRISPR-Cas13a based
- DEC
Diarrheagenic E. coli
- CFU
Colony Forming Unit
- NLuc
Nano-luciferase
- WGS
Whole Genome Sequencing
- LOD
Limit of Detection
- LOQ
Limit of Quantitation
- gRNA
Guide RNA
- crRNA
CRISPR RNA
- tracrRNA
Trans-activating CRISPR RNA
- H– NS
Histone-like nucleoid structuring
- MIC
Minimum Inhibitory Concentration
- MBC
Minimum Bactericidal Concentration
- AgNPs
Silver Nanoparticles
- AuNPs
Gold Nanoparticles
- Pt-NPs
Platimun Nanoparticles
- SAMN@TA
Surface active maghemite NPs with tannic acid
- RSNVs
Rhamnosome Nano-Vesicles
- MNCs
Magnetic nanoparticle-antibody conjugates
- MCE
Minimun Cpture Efficiency
- SWNT
Single-walled nanotube
- MWNT
Multi-walled nanotube
- IMS
Immune-magnetic separation
- MDA
Multiple displacement amplifcation
- SNP
Single nucleotide polymorphism
- SPCE
Screen-printed carbon electrode
- DPV
Differential pulse voltammetry
- MNPs
Magnetic Nanoparticles
- sgRNA
Single guide RNA
Contributor Information
Pritam Kumar Panda, Email: pritam.panda@physics.uu.se.
Yogendra Kumar Mishra, Email: mishra@mci.sdu.dk.
Suresh K. Verma, Email: sureshverma22@gmail.com, suresh.verma@physics.uu.se.
Mrutyunjay Suar, Email: msuar@kiitbiotech.ac.in.
References
- 1.Donkor E.S. Cockroaches and food-borne pathogens. Environ. Health Insights. 2020;14 doi: 10.1177/1178630220913365. 1178630220913365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Oliver S.P., Jayarao B.M., Almeida R.A. Foodborne pathogens in milk and the dairy farm environment: food safety and public health implications. Foodb. Pathog. Dis. 2005;2:115–129. doi: 10.1089/fpd.2005.2.115. [DOI] [PubMed] [Google Scholar]
- 3.WHO, World Health Organization WHO estimates of the global burden of foodborne diseases: foodborne disease burden epidemiology reference group 2007-2015. Encycl. Parasitol. 2015:1–265. [Google Scholar]
- 4.Tanwar J., Das S., Fatima Z., Hameed S. Multidrug resistance: an emerging crisis. Interdiscip. Perspect. Infect. Dis. 2014;2014 doi: 10.1155/2014/541340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lindahl J.F., Grace D. The consequences of human actions on risks for infectious diseases: a review. Infect. Ecol. Epidemiol. 2015;5:30048. doi: 10.3402/iee.v5.30048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pérez-Rodríguez F., Mercanoglu Taban B. A state-of-art review on multi-drug resistant pathogens in foods of animal origin: risk factors and mitigation strategies. Front. Microbiol. 2019;10 doi: 10.3389/fmicb.2019.02091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nikaido H. Multidrug resistance in bacteria. Annu. Rev. Biochem. 2010;78:119–146. doi: 10.1146/annurev.biochem.78.082907.145923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lund B.M. Microbiological food safety for vulnerable people. Int. J. Environ. Res. Publ. Health. 2015;12:10117–10132. doi: 10.3390/ijerph120810117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kumar R., Aadil K.R., Mondal K., Mishra Y.K., Oupicky D., Ramakrishna S., Kaushik A. Neurodegenerative disorders management: state-of-art and prospects of nano-biotechnology. Crit. Rev. Biotechnol. 2021 doi: 10.1080/07388551.2021.1993126. [DOI] [PubMed] [Google Scholar]
- 10.EFSA E. European food safety authority and European centre for disease prevention and control. EFSA J. 2015;13:4329. [Google Scholar]
- 11.Kunjachan S., Rychlik B., Storm G., Kiessling F., Lammers T. Multidrug resistance: physiological principles and nanomedical solutions. Adv. Drug Deliv. Rev. 2013;65:1852–1865. doi: 10.1016/j.addr.2013.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Adzitey F., Huda N., Ali G.R.R. Molecular techniques for detecting and typing of bacteria, advantages and application to foodborne pathogens isolated from ducks. 3 Biotech. 2013;3:97–107. doi: 10.1007/s13205-012-0074-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hille F., Charpentier E. CRISPR-cas: biology, mechanisms and relevance. Philos. Trans. R. Soc. B Biol. Sci. 2016;371 doi: 10.1098/rstb.2015.0496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Y., Li S., Wang J., Liu G. CRISPR/Cas systems towards next-generation biosensing. Trends Biotechnol. 2019;37:730–743. doi: 10.1016/j.tibtech.2018.12.005. [DOI] [PubMed] [Google Scholar]
- 15.Makarova K.S., Koonin E.V. Annotation and classification of CRISPR-Cas systems. Methods Mol. Biol. 2015;1311:47–75. doi: 10.1007/978-1-4939-2687-9_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tenaillon O., Barrick J.E., Ribeck N., Deatherage D.E., Blanchard J.L., Dasgupta A., Wu G.C., Wielgoss S., Cruveiller S., Médigue C., Schneider D., Lenski R.E. Tempo and mode of genome evolution in a 50,000-generation experiment. Nature. 2016;536:165–170. doi: 10.1038/nature18959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou J., Yin L., Dong Y., Peng L., Liu G., Man S., Ma L. CRISPR-Cas13a based bacterial detection platform: sensing pathogen Staphylococcus aureus in food samples. Anal. Chim. Acta. 2020;1127:225–233. doi: 10.1016/j.aca.2020.06.041. [DOI] [PubMed] [Google Scholar]
- 18.Ezhilarasi P.N., Karthik P., Chhanwal N., Anandharamakrishnan C. Nanoencapsulation techniques for food bioactive components: a review. Food Bioprocess Technol. 2013;6:628–647. doi: 10.1007/s11947-012-0944-0. [DOI] [Google Scholar]
- 19.Zorraquín-Peña I., Cueva C., Bartolomé B., Moreno-Arribas M.V. Silver nanoparticles against foodborne bacteria. Effects at intestinal level and health limitations. Microorganisms. 2020;8 doi: 10.3390/microorganisms8010132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shafiq M., Anjum S., Hano C., Anjum I., Abbasi B.H. An overview of the applications of nanomaterials and nanodevices in the food industry. Foods. 2020;9 doi: 10.3390/foods9020148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Steinvil A., Zhang Y.J., Lee S.Y., Pang S., Waksman R., Chen S.L., Garcia-Garcia H.M. Intravascular ultrasound-guided drug-eluting stent implantation: an updated meta-analysis of randomized control trials and observational studies. Int. J. Cardiol. 2016;216:133–139. doi: 10.1016/j.ijcard.2016.04.154. [DOI] [PubMed] [Google Scholar]
- 22.Husain S., Verma S.K., Hemlata, Azam M., Sardar M., Haq Q.M.R., Fatma T. Antibacterial efficacy of facile cyanobacterial silver nanoparticles inferred by antioxidant mechanism. Mater. Sci. Eng. C. 2021;122 doi: 10.1016/j.msec.2021.111888. [DOI] [PubMed] [Google Scholar]
- 23.Paul P., Verma S.K., Kumar Panda P., Jaiswal S., Sahu B.R., Suar M. Molecular insight to influential role of Hha–TomB toxin–antitoxin system for antibacterial activity of biogenic silver nanoparticles, Artif. Cells. Nanomed. Biotechnol. 2018;46:S572–S584. doi: 10.1080/21691401.2018.1503598. [DOI] [PubMed] [Google Scholar]
- 24.Kumari S., Kumari P., Panda P.K., Patel P., Jha E., Mallick M.A., Suar M., Verma S.K. Biocompatible biogenic silver nanoparticles interact with caspases on an atomic level to elicit apoptosis. Nanomedicine. 2020;15:2119–2132. doi: 10.2217/nnm-2020-0138. [DOI] [Google Scholar]
- 25.Verma S.K., Jha E., Kiran K.J., Bhat S., Suar M., Mohanty P.S. Synthesis and characterization of novel polymer-hybrid silver nanoparticles and its biomedical study. Mater. Today Proc. 2016;3:1949–1957. doi: 10.1016/j.matpr.2016.04.096. [DOI] [Google Scholar]
- 26.Tertis M., Cernat A., Mirel S., Cristea C. Nanodevices for pharmaceutical and biomedical applications. Anal. Lett. 2021;54:98–123. doi: 10.1080/00032719.2020.1728292. [DOI] [Google Scholar]
- 27.Verma S.K., Jha E., Panda P.K., Thirumurugan A., Suar M. 2019. Biological Effects of Green-Synthesized Metal Nanoparticles: A Mechanistic View of Antibacterial Activity and Cytotoxicity; pp. 145–171. [DOI] [Google Scholar]
- 28.Umesha S., Manukumar H.M. Advanced molecular diagnostic techniques for detection of food-borne pathogens: current applications and future challenges. Crit. Rev. Food Sci. Nutr. 2018;58:84–104. doi: 10.1080/10408398.2015.1126701. [DOI] [PubMed] [Google Scholar]
- 29.Grissa I., Vergnaud G., Pourcel C. The CRISPRdb database and tools to display CRISPRs and to generate dictionaries of spacers and repeats. BMC Bioinf. 2007;8 doi: 10.1186/1471-2105-8-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maji B., Gangopadhyay S.A., Lee M., Shi M., Wu P., Heler R., Mok B., Lim D., Siriwardena S.U., Paul B., Dančík V., Vetere A., Mesleh M.F., Marraffini L.A., Liu D.R., Clemons P.A., Wagner B.K., Choudhary A. A high-throughput platform to identify small-molecule inhibitors of CRISPR-cas9. Cell. 2019;177 doi: 10.1016/j.cell.2019.04.009. 1067-1079.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grissa I., Vergnaud G., Pourcel C. CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 2007;35 doi: 10.1093/nar/gkm360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sashital D.G. Pathogen detection in the CRISPR-Cas era. Genome Med. 2018;10 doi: 10.1186/s13073-018-0543-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Franklin Lowy.D. Staphylococcus aureus infection. N. Engl. J. Med. 2001;339 [Google Scholar]
- 34.Casewell M.W., Hill R.L.R. The carrier state: methicillin-resistant Staphylococcus aureus. J. Antimicrob. Chemother. 1986;18:1–12. doi: 10.1093/jac/18.supplement_a.1. [DOI] [PubMed] [Google Scholar]
- 35.Sanford M.D., Widmer A.F., Bale M.J., Jones R.N., Wenzel R.P. Efficient detection and long-term persistence of the carriage of methicillin-Resistant staphylococcus aureus. Clin. Infect. Dis. 1994;19:1123–1128. doi: 10.1093/clinids/19.6.1123. [DOI] [PubMed] [Google Scholar]
- 36.Tong S.Y.C., Davis J.S., Eichenberger E., Holland T.L., Fowler V.G. Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin. Microbiol. Rev. 2015;28:603–661. doi: 10.1128/CMR.00134-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lister J.L., Horswill A.R. Staphylococcus aureus biofilms: recent developments in biofilm dispersal. Front. Cell. Infect. Microbiol. 2014;4 doi: 10.3389/fcimb.2014.00178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kumar M. Multidrug-resistant Staphylococcus aureus, India, 2013–2015. emerg. Infect. Dis. 2016;22:1666–1667. doi: 10.3201/eid2209.160044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goode B., O'Reilly C., Dunn J., Fullerton K., Smith S., Ghneim G., Keen J., Durso L., Davies M., Montgomery S. Outbreak of escherichia coli O157 H7 infections after petting zoo visits, North Carolina state fair. Arch. Pediatr. Adolesc. Med. 2009;163:42–48. doi: 10.1001/archpediatrics.2008.525. [DOI] [PubMed] [Google Scholar]
- 40.Nataro J.P., Kaper J.B. Diarrheagenic Escherichia coli, clin. Microbiol. Rev. 1998;11:142–201. doi: 10.1128/CMR.11.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bélanger L., Garenaux A., Harel J., Boulianne M., Nadeau E., Dozois C.M. Escherichia colifrom animal reservoirs as a potential source of human extraintestinal pathogenic E. coli. FEMS Immunol. Med. Microbiol. 2011;62:1–10. doi: 10.1111/j.1574-695X.2011.00797.x. [DOI] [PubMed] [Google Scholar]
- 42.Aijuka M., Santiago A.E., Girón J.A., Nataro J.P., Buys E.M. Enteroaggregative Escherichia coli is the predominant diarrheagenic E. coli pathotype among irrigation water and food sources in South Africa. Int. J. Food Microbiol. 2018;278:44–51. doi: 10.1016/j.ijfoodmicro.2018.04.018. [DOI] [PubMed] [Google Scholar]
- 43.Fu Z., Rogelj S., Kieft T.L. Rapid detection of Escherichia coli O157:H7 by immunomagnetic separation and real-time PCR. Int. J. Food Microbiol. 2005;99:47–57. doi: 10.1016/j.ijfoodmicro.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 44.Mao X., Yang L., Su X.L., Li Y. A nanoparticle amplification based quartz crystal microbalance DNA sensor for detection of Escherichia coli O157:H7. Biosens. Bioelectron. 2006;21:1178–1185. doi: 10.1016/j.bios.2005.04.021. [DOI] [PubMed] [Google Scholar]
- 45.Yang L., Li Y. Simultaneous detection of Escherichia coli O157:H7 and Salmonella Typhimurium using quantum dots as fluorescence labels. Analyst. 2006;131:394–401. doi: 10.1039/b510888h. [DOI] [PubMed] [Google Scholar]
- 46.Guo Q., Han J.J., Shan S., Liu D.F., Wu S.S., Xiong Y.H., Lai W.H. DNA-based hybridization chain reaction and biotin–streptavidin signal amplification for sensitive detection of Escherichia coli O157:H7 through ELISA. Biosens. Bioelectron. 2016;86:990–995. doi: 10.1016/j.bios.2016.07.049. [DOI] [PubMed] [Google Scholar]
- 47.Ma F., Rehman A., Liu H., Zhang J., Zhu S., Zeng X. Glycosylation of quinone-fused polythiophene for reagentless and label-free detection of E. coli. Anal. Chem. 2015;87:1560–1568. doi: 10.1021/ac502712q. [DOI] [PubMed] [Google Scholar]
- 48.Ye R., Zhu C., Song Y., Song J., Fu S., Lu Q., Yang X., Zhu M.J., Du D., Li H., Lin Y. One-pot bioinspired synthesis of all-inclusive protein-protein nanoflowers for point-of-care bioassay: detection of: E. coli O157:H7 from milk. Nanoscale. 2016;8:18980–18986. doi: 10.1039/c6nr06870g. [DOI] [PubMed] [Google Scholar]
- 49.Sun X., Wang Y., Zhang L., Liu S., Zhang M., Wang J., Ning B., Peng Y., He J., Hu Y., Gao Z. CRISPR-Cas9 triggered two-step isothermal amplification method for E. coli O157:H7 detection based on a metal-organic framework platform. Anal. Chem. 2020;92:3032–3041. doi: 10.1021/acs.analchem.9b04162. [DOI] [PubMed] [Google Scholar]
- 50.Rousset F., Cui L., Siouve E., Becavin C., Depardieu F., Bikard D. Genome-wide CRISPR-dCas9 screens in E. coli identify essential genes and phage host factors. PLoS Genet. 2018;14 doi: 10.1371/journal.pgen.1007749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang L., Shen X., Wang T., Chen P., Qi N., Yin B.C., Ye B.C. A lateral flow strip combined with Cas9 nickase-triggered amplification reaction for dual food-borne pathogen detection. Biosens. Bioelectron. 2020;165 doi: 10.1016/j.bios.2020.112364. [DOI] [PubMed] [Google Scholar]
- 52.Scharff R.L. Economic burden from health losses due to foodborne illness in the United States. J. Food Protect. 2012;75:123–131. doi: 10.4315/0362-028X.JFP-11-058. [DOI] [PubMed] [Google Scholar]
- 53.Thompson C.P., Doak A.N., Amirani N., Schroeder E.A., Wright J., Kariyawasam S., Lamendella R., Shariat N.W. High-resolution identification of multiple Salmonella serovars in a single sample by using CRISPRSeroSeq. Appl. Environ. Microbiol. 2018;84 doi: 10.1128/AEM.01859-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Threlfall E.J. Antimicrobial drug resistance in Salmonella: problems and perspectives in food- and water-borne infections. FEMS Microbiol. Rev. 2002;26:141–148. doi: 10.1016/S0168-6445(02)00092-X. [DOI] [PubMed] [Google Scholar]
- 55.Shen Y., Xu L., Li Y. Biosensors for rapid detection of Salmonella in food: a review. Compr. Rev. Food Sci. Food Saf. 2021;20:149–197. doi: 10.1111/1541-4337.12662. [DOI] [PubMed] [Google Scholar]
- 56.Fricke W.F., Mammel M.K., McDermott P.F., Tartera C., White D.G., LeClerc J.E., Ravel J., Cebula T.A. Comparative genomics of 28 Salmonella enterica isolates: evidence for crispr-mediated adaptive sublineage evolution. J. Bacteriol. 2011;193:3556–3568. doi: 10.1128/JB.00297-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Monte D.F.M., Nethery M.A., Barrangou R., Landgraf M., Fedorka-Cray P.J. Whole-genome sequencing analysis and CRISPR genotyping of rare antibiotic-resistant Salmonella enterica serovars isolated from food and related sources. Food Microbiol. 2021;93 doi: 10.1016/j.fm.2020.103601. [DOI] [PubMed] [Google Scholar]
- 58.Shariat N., Timme R.E., Pettengill J.B., Barrangou R., Dudley E.G. Characterization and evolution of Salmonella CRISPR-Cas systems. Microbiol. 2015;161:374–386. doi: 10.1099/mic.0.000005. [DOI] [PubMed] [Google Scholar]
- 59.Shen J., Zhou X., Shan Y., Yue H., Huang R., Hu J., Xing D. Sensitive detection of a bacterial pathogen using allosteric probe-initiated catalysis and CRISPR-Cas13a amplification reaction. Nat. Commun. 2020;11 doi: 10.1038/s41467-019-14135-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Farber J.M., Losos J.Z. Listeria monocytogenes: a foodborne pathogen. CMAJ (Can. Med. Assoc. J.) 1988;138:413–418. doi: 10.1128/mr.55.4.752-752.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lomonaco S., Nucera D., Filipello V. The evolution and epidemiology of Listeria monocytogenes in Europe and the United States. Infect. Genet. Evol. 2015;35:172–183. doi: 10.1016/j.meegid.2015.08.008. [DOI] [PubMed] [Google Scholar]
- 62.Meile S., Sarbach A., Du J., Schuppler M., Saez C., Loessner M.J., Kilcher S. Engineered reporter phages for rapid bioluminescence-based detection and differentiation of viable Listeria cells. Appl. Environ. Microbiol. 2020;86 doi: 10.1128/AEM.00442-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shariat N., Dudley E.G. CRISPRs: molecular signatures used for pathogen subtyping. Appl. Environ. Microbiol. 2014;80:430–439. doi: 10.1128/AEM.02790-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mehata S., Duan G.C. Molecular mechanism of multi-drug resistance in Shigella isolates from rural China. Nepal Med. Coll. J. 2011;13:27–29. [PubMed] [Google Scholar]
- 65.Puzari M., Sharma M., Chetia P. Emergence of antibiotic resistant Shigella species: a matter of concern. J. Infect. Publ. Health. 2018;11:451–454. doi: 10.1016/j.jiph.2017.09.025. [DOI] [PubMed] [Google Scholar]
- 66.Sansonetti P.J. Shigellosis: an old disease in new clothes? PLoS Med. 2006;3:1465–1466. doi: 10.1371/journal.pmed.0030354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hussen S., Mulatu G., Yohannes Kassa Z. Prevalence of Shigella species and its drug resistance pattern in Ethiopia: a systematic review and meta-analysis. Ann. Clin. Microbiol. Antimicrob. 2019;18 doi: 10.1186/s12941-019-0321-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen S., Liu H., Liang W., Hong L. Insert sequences of CRISPR/Cas system regulate horizontal antibiotic gene transfer in Shigella. Int. J. Antimicrob. Agents. 2018 doi: 10.1016/j.ijantimicag.2018.09.020. https://www.sciencedirect.com/science/article/pii/S0924857918302796?dgcid=rss_sd_all. [DOI] [PubMed] [Google Scholar]
- 69.Elahi N., Baghersad M.H., Kamali M. Precise, direct, and rapid detection of Shigella Spa gene by a novel unmodified AuNPs-based optical genosensing system. J. Microbiol. Methods. 2019;162:42–49. doi: 10.1016/j.mimet.2019.05.007. [DOI] [PubMed] [Google Scholar]
- 70.Stout E., Klaenhammer T., Barrangou R. CRISPR-cas technologies and applications in food bacteria. Annu. Rev. Food Sci. Technol. 2017;8:413–437. doi: 10.1146/annurev-food-072816-024723. [DOI] [PubMed] [Google Scholar]
- 71.Selle K., Barrangou R. Harnessing CRISPR-Cas systems for bacterial genome editing. Trends Microbiol. 2015;23:225–232. doi: 10.1016/j.tim.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 72.Sieuwerts S., De Bok F.A.M., Hugenholtz J., Van Hylckama Vlieg J.E.T. Unraveling microbial interactions in food fermentations: from classical to genomics approaches. Appl. Environ. Microbiol. 2008;74:4997–5007. doi: 10.1128/AEM.00113-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gomaa A.A., Klumpe H.E., Luo M.L., Selle K., Barrangou R., Beisel C.L. Programmable removal of bacterial strains by use of genome- targeting CRISPR-cas systems. mBio. 2014;5 doi: 10.1128/mBio.00928-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bikard D., Euler C., Jiang W., Nussenzweig P.M., W G.G., Duportet X., Fischetti V.A., Marraffini L.A. Development of sequence-specific antimicrobials based on programmable CRISPR-Cas nucleases. Nat. Biotechnol. 2014;32:1146–1150. doi: 10.1038/nbt.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Citorik R.J., Mimee M., Lu T.K. Sequence-specific antimicrobials using efficiently delivered RNA-guided nucleases. Nat. Biotechnol. 2014;32:1141–1145. doi: 10.1038/nbt.3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Perera R.A., Wang P., Gomaa M.R., El-Shesheny R., Kandeil A., Bagato O., Siu L.Y., Shehata M.M., Kayed A.S., Moatasim Y., Li M., Poon L.L., Guan Y., Webby R.J., Ali M.A., Peiris J.S., Kayali G. Seroepidemiology for MERS coronavirus using microneutralisation and pseudoparticle virus neutralisation assays reveal a high prevalence of antibody in dromedary camels in Egypt, june 2013. Euro Surveill. 2013;18 doi: 10.2807/1560-7917.ES2013.18.36.20574. [DOI] [PubMed] [Google Scholar]
- 77.Bikard D., Euler C.W., Jiang W., Nussenzweig P.M., Goldberg G.W., Duportet X., Fischetti V.A., Marraffini L.A. Exploiting CRISPR-cas nucleases to produce sequence-specific antimicrobials. Nat. Biotechnol. 2014;32:1146–1150. doi: 10.1038/nbt.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cui L., Vigouroux A., Rousset F., Varet H., Khanna V., Bikard D. A CRISPRi screen in E. coli reveals sequence-specific toxicity of dCas9. Nat. Commun. 2018;9 doi: 10.1038/s41467-018-04209-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pul Ü., Wurm R., Arslan Z., Geißen R., Hofmann N., Wagner R. Identification and characterization of E. coli CRISPR-cas promoters and their silencing by H-NS. Mol. Microbiol. 2010;75:1495–1512. doi: 10.1111/j.1365-2958.2010.07073.x. [DOI] [PubMed] [Google Scholar]
- 80.Samson J.E., Moineau S. Bacteriophages in food fermentations: new frontiers in a continuous arms race. Annu. Rev. Food Sci. Technol. 2013;4:347–368. doi: 10.1146/annurev-food-030212-182541. [DOI] [PubMed] [Google Scholar]
- 81.Baltz R.H. Bacteriophage-resistant industrial fermentation strains: from the cradle to CRISPR/Cas9. J. Ind. Microbiol. Biotechnol. 2018;45:1003–1006. doi: 10.1007/s10295-018-2079-4. [DOI] [PubMed] [Google Scholar]
- 82.Rimantas S., Giedrius G., Christophe F., Rodolphe B., Philippe H., Virginijus S. The Streptococcus thermophilus CRISPR/Cas system provides immunity in Escherichia coli. Nucleic Acids Res. 2011;39:9275–9282. doi: 10.1093/nar/gkr606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Garneau Josiane E., Marie-Ève D., Manuela V., Romero Dennis A., Rodolphe B., Patrick B., Christophe F., Philippe H., Magadán Alfonso H., Sylvain M. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468:67. doi: 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- 84.Marraffini L.A., Sontheimer E.J. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science. 2008;322(80-):1843–1845. doi: 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Edgar R., Qimron U. The Escherichia coli CRISPR system protects from λ lysogenization, lysogens, and prophage induction. J. Bacteriol. 2010;192:6291–6294. doi: 10.1128/JB.00644-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nozawa T., Furukawa N., Aikawa C., Watanabe T., Haobam B., Kurokawa K., Maruyama F., Nakagawa I. CRISPR inhibition of prophage acquisition in streptococcus pyogenes. PLoS One. 2011;6 doi: 10.1371/journal.pone.0019543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Palmer K.L., Gilmore M.S. Multidrug-resistant enterococci lack CRISPR-cas. mBio. 2010;1 doi: 10.1128/mBio.00227-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Barrangou R., Dudley E.G. CRISPR-based typing and next-generation tracking technologies. Annu. Rev. Food Sci. Technol. 2016;7:395–411. doi: 10.1146/annurev-food-022814-015729. [DOI] [PubMed] [Google Scholar]
- 89.Barrangou R., Horvath P. CRISPR: new horizons in phage resistance and strain identification. Annu. Rev. Food Sci. Technol. 2012;3:143–162. doi: 10.1146/annurev-food-022811-101134. [DOI] [PubMed] [Google Scholar]
- 90.Gasiunas G., Barrangou R., Horvath P., Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc. Natl. Acad. Sci. U. S. A. 2012;109 doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Horvath P., Romero D.A., Cou A., Richards M., Moineau S., Boyaval P., Fremaux C., Barrangou R. Diversity , activity , and evolution of CRISPR loci in. J. Biotechnol. 2008;190:1401–1412. doi: 10.1128/JB.01415-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Smokvina T., Wels M., Polka J., Chervaux C., Brisse S., Boekhorst J., van Vlieg J.E.T.H., Siezen R.J. Lactobacillus paracasei comparative genomics: towards species pan-genome definition and exploitation of diversity. PLoS One. 2013;8 doi: 10.1371/journal.pone.0068731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Douillard F.P., Ribbera A., Kant R., Pietilä T.E., Järvinen H.M., Messing M., Randazzo C.L., Paulin L., Laine P., Ritari J., Caggia C., Lähteinen T., Brouns S.J.J., Satokari R., von Ossowski I., Reunanen J., Palva A., de Vos W.M. Comparative genomic and functional analysis of 100 Lactobacillus rhamnosus strains and their comparison with strain GG. PLoS Genet. 2013;9 doi: 10.1371/journal.pgen.1003683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Natsch A., Emter R., Badertscher R., Brunner G., Granier T., Kern S., Ellis G. Response to the Letter to the Editor Regarding Our Article (Natsch et al., 2015) Chem. Res. Toxicol. 2015;28:2082–2084. doi: 10.1021/acs.chemrestox.5b00423. [DOI] [PubMed] [Google Scholar]
- 95.Sanozky-Dawes R., Selle K., O'Flaherty S., Klaenhammer T., Barrangou R. Occurrence and activity of a type II CRISPR-Cas system in Lactobacillus gasseri. Microbiol. 2015;161:1752–1761. doi: 10.1099/mic.0.000129. [DOI] [PubMed] [Google Scholar]
- 96.Briner A.E., Barrangou R. Lactobacillus buchneri genotyping on the basis of clustered regularly interspaced short palindromic repeat (CRISPR) locus diversity. Appl. Environ. Microbiol. 2014;80:994–1001. doi: 10.1128/AEM.03015-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337(80-):816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Selle K., Barrangou R. CRISPR-based technologies and the future of food science. J. Food Sci. 2015;80:R2367–R2372. doi: 10.1111/1750-3841.13094. [DOI] [PubMed] [Google Scholar]
- 99.Barrangou R., van Pijkeren J.P. Exploiting CRISPR-Cas immune systems for genome editing in bacteria. Curr. Opin. Biotechnol. 2016;37:61–68. doi: 10.1016/j.copbio.2015.10.003. [DOI] [PubMed] [Google Scholar]
- 100.Oh J.H., Van Pijkeren J.P. CRISPR-Cas9-assisted recombineering in Lactobacillus reuteri. Nucleic Acids Res. 2014;42 doi: 10.1093/nar/gku623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wenyan Jiang L.A.M., Bikard David, Cox David, Zhang Feng. CRISPR-assisted editing of bacterial genomes. Nat. Biotechnol. 2013;31:233–239. doi: 10.1038/nbt.2508.CRISPR-assisted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kumari P., Panda P.K., Jha E., Pramanik N., Nisha K., Kumari K., Soni N., Mallick M.A., Verma S.K. Molecular insight to in vitro biocompatibility of phytofabricated copper oxide nanoparticles with human embryonic kidney cells. Nanomedicine. 2018;13:2415–2433. doi: 10.2217/nnm-2018-0175. [DOI] [PubMed] [Google Scholar]
- 103.Sheel R., Kumari P., Panda P.K., Jawed Ansari M.D., Patel P., Singh S., Kumari B., Sarkar B., Mallick M.A., Verma S.K. Molecular intrinsic proximal interaction infer oxidative stress and apoptosis modulated in vivo biocompatibility of P.niruri contrived antibacterial iron oxide nanoparticles with zebrafish. Environ. Pollut. 2020;267 doi: 10.1016/j.envpol.2020.115482. [DOI] [PubMed] [Google Scholar]
- 104.Umaraw P., Munekata P.E.S., Verma A.K., Barba F.J., Singh V.P., Kumar P., Lorenzo J.M. Edible films/coating with tailored properties for active packaging of meat, fish and derived products. Trends Food Sci. Technol. 2020;98:10–24. doi: 10.1016/j.tifs.2020.01.032. [DOI] [Google Scholar]
- 105.Sarkar B., Verma S.K., Akhtar J., Netam S.P., Gupta S.K., Panda P.K., Mukherjee K. Molecular aspect of silver nanoparticles regulated embryonic development in Zebrafish (Danio rerio) by Oct-4 expression. Chemosphere. 2018;206:560–567. doi: 10.1016/j.chemosphere.2018.05.018. [DOI] [PubMed] [Google Scholar]
- 106.Kaur P., Thakur R., Kumar S., Dilbaghi N. Interaction of ZnO nanoparticles with food borne pathogens Escherichia coli DH5α and Staphylococcus aureus 5021 & their bactericidal efficacy. AIP Conf. Proc. 2011;1393:153–154. doi: 10.1063/1.3653655. [DOI] [Google Scholar]
- 107.Das B.K., Verma S.K., Das T., Panda P.K., Parashar K., Suar M., Parashar S.K.S. Altered electrical properties with controlled copper doping in ZnO nanoparticles infers their cytotoxicity in macrophages by ROS induction and apoptosis. Chem. Biol. Interact. 2019;297:141–154. doi: 10.1016/j.cbi.2018.11.004. [DOI] [PubMed] [Google Scholar]
- 108.Petrus E.M., Tinakumari S., Chai L.C., Ubong A., Tunung R., Elexson N., Chai L.F., Son R. A study on the minimum inhibitory concentration and minimum bactericidal concentration of nano colloidal silver on food-borne pathogens. Int. Food Res. J. 2011;18:55–66. [Google Scholar]
- 109.Tamboli D.P., Lee D.S. Mechanistic antimicrobial approach of extracellularly synthesized silver nanoparticles against gram positive and gram negative bacteria. J. Hazard Mater. 2013;260:878–884. doi: 10.1016/j.jhazmat.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 110.Ruiz-Rico M., Pérez-Esteve É., de la Torre C., Jiménez-Belenguer A.I., Quiles A., Marcos M.D., Martínez-Máñez R., Barat J.M. Improving the antimicrobial power of low-effective antimicrobial molecules through nanotechnology. J. Food Sci. 2018;83:2140–2147. doi: 10.1111/1750-3841.14211. [DOI] [PubMed] [Google Scholar]
- 111.de Almeida Roger J., Magro M., Spagnolo S., Bonaiuto E., Baratella D., Fasolato L., Vianello F. Antimicrobial and magnetically removable tannic acid nanocarrier: a processing aid for Listeria monocytogenes treatment for food industry applications. Food Chem. 2018;267:430–436. doi: 10.1016/j.foodchem.2017.06.109. [DOI] [PubMed] [Google Scholar]
- 112.Kumari S., kumari P., Panda P.K., Pramanik N., Verma S.K., Mallick M.A. Molecular aspect of phytofabrication of gold nanoparticle from Andrographis peniculata photosystem II and their in vivo biological effect on embryonic zebrafish (Danio rerio) Environ. Nanotechnol. Monit. Manag. 2019;11 doi: 10.1016/j.enmm.2018.100201. [DOI] [Google Scholar]
- 113.Salomoni R., Léo P., Montemor A.F., Rinaldi B.G., Rodrigues M.F.A. Antibacterial effect of silver nanoparticles in Pseudomonas aeruginosa. Nanotechnol. Sci. Appl. 2017;10:115–121. doi: 10.2147/NSA.S133415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Shamaila S., Zafar N., Riaz S., Sharif R., Nazir J., Naseem S. Gold nanoparticles: an efficient antimicrobial agent against enteric bacterial human pathogen. Nanomaterials. 2016;6 doi: 10.3390/nano6040071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Qing Y., Cheng L., Li R., Liu G., Zhang Y., Tang X., Wang J., Liu H., Qin Y. Potential antibacterial mechanism of silver nanoparticles and the optimization of orthopedic implants by advanced modification technologies. Int. J. Nanomed. 2018;13:3311–3327. doi: 10.2147/IJN.S165125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Chlumsky O., Purkrtova S., Michova H., Svarcova V., Slepicka P., Fajstavr D., Ulbrich P., Demnerova K. The effect of gold and silver nanoparticles, chitosan and their combinations on bacterial biofilms of food-borne pathogens. Biofouling. 2020;36:222–233. doi: 10.1080/08927014.2020.1751132. [DOI] [PubMed] [Google Scholar]
- 117.Uma Suganya K.S., Govindaraju K., Ganesh Kumar V., Stalin Dhas T., Karthick V., Singaravelu G., Elanchezhiyan M. Blue green alga mediated synthesis of gold nanoparticles and its antibacterial efficacy against Gram positive organisms. Mater. Sci. Eng. C. 2015;47:351–356. doi: 10.1016/j.msec.2014.11.043. [DOI] [PubMed] [Google Scholar]
- 118.Verma S.K., Jha E., Panda P.K., Kumari P., Pramanik N., Kumari S., Thirumurugan A. Molecular investigation to RNA and protein based interaction induced in vivo biocompatibility of phytofabricated AuNP with embryonic zebrafish, Artif. Cells. Nanomed. Biotechnol. 2018;46:S671–S684. doi: 10.1080/21691401.2018.1505746. [DOI] [PubMed] [Google Scholar]
- 119.Sawosz E., Chwalibog A., Szeliga J., Sawosz F., Grodzik M., Rupiewicz M., Niemiec T., Kacprzyk K. Visualization of gold and platinum nanoparticles interacting with Salmonella Enteritidis and Listeria monocytogenes. Int. J. Nanomed. 2010;5:631–637. doi: 10.2147/IJN.S12361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lu C., Tang L., Gao F., Li Y., Liu J., Zheng J. DNA-encoded bimetallic Au-Pt dumbbell nanozyme for high-performance detection and eradication of Escherichia coli O157:H7. Biosens. Bioelectron. 2021;187 doi: 10.1016/j.bios.2021.113327. [DOI] [PubMed] [Google Scholar]
- 121.Cui C., Gan L., Li H.H., Yu S.H., Heggen M., Strasser P. Octahedral PtNi nanoparticle catalysts: exceptional oxygen reduction activity by tuning the alloy particle surface composition. Nano Lett. 2012;12:5885–5889. doi: 10.1021/nl3032795. [DOI] [PubMed] [Google Scholar]
- 122.Niaz T., Shabbir S., Noor T., Imran M. Antimicrobial and antibiofilm potential of bacteriocin loaded nano-vesicles functionalized with rhamnolipids against foodborne pathogens. LWT (Lebensm.-Wiss. & Technol.) 2019;116 doi: 10.1016/j.lwt.2019.108583. [DOI] [Google Scholar]
- 123.Calderón-Gonzalez R., Terán-Navarro H., Frande-Cabanes E., Ferrández-Fernández E., Freire J., Penadés S., Marradi M., García I., Gomez-Román J., Yañez-Díaz S., Álvarez-Domínguez C. Pregnancy vaccination with gold glyco-nanoparticles carrying Listeria monocytogenes peptides protects against listeriosis and brain- and cutaneous-associated morbidities. Nanomaterials. 2016;6 doi: 10.3390/nano6080151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Varshney M., Yang L., Su X.L., Li Y. Magnetic nanoparticle-antibody conjugates for the separation of Escherichia coli O157:H7 in ground beef. J. Food Protect. 2005;68:1804–1811. doi: 10.4315/0362-028X-68.9.1804. [DOI] [PubMed] [Google Scholar]
- 125.Elkin T., Jiang X., Taylor S., Lin Y., Gu L., Yang H., Brown J., Collins S., Sun Y.P. Immuno-carbon nanotubes and recognition of pathogens. Chembiochem. 2005;6:640–643. doi: 10.1002/cbic.200400337. [DOI] [PubMed] [Google Scholar]
- 126.Lin Y., Jiang X., Elkin T., Fernando K.A.S., Gu L., Taylor S., Yang H., Jones E., Wang W., Sun Y.P. Carbon nanotubes for immunomagnetic separation of escherichia Coli O157:H7. J. Nanosci. Nanotechnol. 2006;6:868–871. doi: 10.1166/jnn.2006.130. [DOI] [PubMed] [Google Scholar]
- 127.Gu L., Elkin T., Jiang X., Li H., Lin Y., Qu L., Tzeng T.R.J., Joseph R., Sun Y.P. Single-walled carbon nanotubes displaying multivalent ligands for capturing pathogens. Chem. Commun. 2005:874–876. doi: 10.1039/b415015e. [DOI] [PubMed] [Google Scholar]
- 128.Chen X., Miller A., Cao S., Gan Y., Zhang J., He Q., Wang R.-Q., Yong X., Qin P., Lapizco-Encinas B.H., Du K. Rapid Escherichia coli trapping and retrieval from bodily fluids via a three-dimensional bead-stacked nanodevice. ACS Appl. Mater. Interfaces. 2020;12:7888–7896. doi: 10.1021/acsami.9b19311. [DOI] [PubMed] [Google Scholar]
- 129.Jarquin R., Hanning I., Ahn S., Ricke S.C. Development of rapid detection and genetic characterization of Salmonella in poultry breeder feeds. Sensors. 2009;9:5308–5323. doi: 10.3390/s90705308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Hu J., Tang F., Jiang Y.Z., Liu C. Rapid screening and quantitative detection of: Salmonella using a quantum dot nanobead-based biosensor. Analyst. 2020;145:2184–2190. doi: 10.1039/d0an00035c. [DOI] [PubMed] [Google Scholar]
- 131.Jiang W., Bikard D., Cox D., Zhang F., Marraffini L.A. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat. Biotechnol. 2013;31:233–239. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Bayraç C., Eyidoğan F., Avni Öktem H. DNA aptamer-based colorimetric detection platform for Salmonella Enteritidis. Biosens. Bioelectron. 2017;98:22–28. doi: 10.1016/j.bios.2017.06.029. [DOI] [PubMed] [Google Scholar]
- 133.Angelopoulou M., Tzialla K., Voulgari A., Dikeoulia M., Raptis I., Kakabakos S.E., Petrou P. Rapid detection of salmonella typhimurium in drinking water by a white light reflectance spectroscopy immunosensor. Sensors. 2021;21 doi: 10.3390/s21082683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Dong X., Wang P., Darby J.P., Tang Y., Overton C.M., Kathariou S., Sun Y.P., Yang L. Photoactivated carbon dots for inactivation of foodborne pathogens listeria and salmonella. Appl. Environ. Microbiol. 2021;87 doi: 10.1128/AEM.01042-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Elahi N., Kamali M., Baghersad M.H., Amini B. A fluorescence Nano-biosensors immobilization on Iron (MNPs) and gold (AuNPs) nanoparticles for detection of Shigella spp. Mater. Sci. Eng. C. 2019;105 doi: 10.1016/j.msec.2019.110113. [DOI] [PubMed] [Google Scholar]
- 136.Dubey A.K., Kumar Gupta V., Kujawska M., Orive G., Kim N.-Y., Li C., Kumar Mishra Y., Kaushik A. Exploring nano-enabled CRISPR-Cas-powered strategies for efficient diagnostics and treatment of infectious diseases. J. Nanostruct. Chem. 2022 doi: 10.1007/s40097-022-00472-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Phan Q.A., Truong L.B., Medina-Cruz D., Dincer C., Mostafavi E. CRISPR/Cas-powered nanobiosensors for diagnostics. Biosens. Bioelectron. 2022;197 doi: 10.1016/j.bios.2021.113732. [DOI] [PubMed] [Google Scholar]
- 138.Shalem O., Sanjana N.E., Zhang F. High-throughput functional genomics using CRISPR-Cas9. Nat. Rev. Genet. 2015;16:299–311. doi: 10.1038/nrg3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kim D., Le Q.V., Wu Y., Park J., Oh Y.K. Nanovesicle-mediated delivery systems for crispr/cas genome editing. Pharmaceutics. 2020;12:1–42. doi: 10.3390/pharmaceutics12121233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Wang Z., Lee Y.H., Wu B., Horst A., Kang Y., Tang Y.J., Chen D.R. Anti-microbial activities of aerosolized transition metal oxide nanoparticles. Chemosphere. 2010;80:525–529. doi: 10.1016/j.chemosphere.2010.04.047. [DOI] [PubMed] [Google Scholar]
- 141.Elmowafy E.M., Tiboni M., Soliman M.E. Biocompatibility, biodegradation and biomedical applications of poly(lactic acid)/poly(lactic-co-glycolic acid) micro and nanoparticles. J. Pharm. Invest. 2019;49:347–380. doi: 10.1007/s40005-019-00439-x. [DOI] [Google Scholar]
- 142.Taharabaru T., Yokoyama R., Higashi T., Mohammed A.F.A., Inoue M., Maeda Y., Niidome T., Onodera R., Motoyama K. Genome editing in a wide area of the brain using dendrimer-based ternary polyplexes of Cas9 ribonucleoprotein. ACS Appl. Mater. Interfaces. 2020;12:21386–21397. doi: 10.1021/acsami.9b21667. [DOI] [PubMed] [Google Scholar]
- 143.Kim H.J., Ogura S., Otabe T., Kamegawa R., Sato M., Kataoka K., Miyata K. Fine-tuning of hydrophobicity in amphiphilic polyaspartamide derivatives for rapid and transient expression of messenger RNA directed toward genome engineering in brain. ACS Cent. Sci. 2019;5:1866–1875. doi: 10.1021/acscentsci.9b00843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Huang H., Chai C., Li N., Rowe P., Minton N.P., Yang S., Jiang W., Gu Y. CRISPR/Cas9-Based efficient genome editing in Clostridium ljungdahlii, an autotrophic gas-fermenting bacterium. ACS Synth. Biol. 2016;5:1355–1361. doi: 10.1021/acssynbio.6b00044. [DOI] [PubMed] [Google Scholar]
- 145.Gong Y., Tian S., Xuan Y., Zhang S. Lipid and polymer mediated CRISPR/Cas9 gene editing. J. Mater. Chem. B. 2020;8:4369–4386. doi: 10.1039/d0tb00207k. [DOI] [PubMed] [Google Scholar]
- 146.Bai L., Zhou H., Xu R., Zhao Y., Chinnaswamy K., McEachern D., Chen J., Yang C.Y., Liu Z., Wang M., Liu L., Jiang H., Wen B., Kumar P., Meagher J.L., Sun D., Stuckey J.A., Wang S. A potent and selective small-molecule degrader of STAT3 achieves complete tumor regression in vivo. Cancer Cell. 2019;36 doi: 10.1016/j.ccell.2019.10.002. 498-511.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Yamagishi A., Matsumoto D., Kato Y., Honda Y., Morikawa M., Iwata F., Kobayashi T., Nakamura C. Direct delivery of Cas9-sgRNA ribonucleoproteins into cells using a nanoneedle array. Appl. Sci. 2019;9 doi: 10.3390/app9050965. [DOI] [Google Scholar]
- 148.Hansen-Bruhn M., de Ávila B.E., Beltrán-Gastélum M., Zhao J., Ramírez-Herrera D.E., Angsantikul P., Vesterager Gothelf K., Zhang L., Wang J. Active intracellular delivery of a cas9/sgRNA complex using ultrasound-propelled nanomotors. Angew. Chem. 2018;130:2687–2691. doi: 10.1002/ange.201713082. [DOI] [PubMed] [Google Scholar]
- 149.Du S., Liew S.S., Li L., Yao S.Q. Bypassing endocytosis: direct cytosolic delivery of proteins. J. Am. Chem. Soc. 2018;140:15986–15996. doi: 10.1021/jacs.8b06584. [DOI] [PubMed] [Google Scholar]
- 150.Kang Y.K., Kwon K., Ryu J.S., Lee H.N., Park C., Chung H.J. Nonviral genome editing based on a polymer-derivatized CRISPR nanocomplex for targeting bacterial pathogens and antibiotic resistance. Bioconjugate Chem. 2017;28:957–967. doi: 10.1021/acs.bioconjchem.6b00676. [DOI] [PubMed] [Google Scholar]
- 151.Chopade B., Ghosh Patil, Ahire Kitture, Jabgunde Kale, Pardesi Cameotra, Bellare Dhavale. Synthesis of silver nanoparticles using Dioscorea bulbifera tuber extract and evaluation of its synergistic potential in combination with antimicrobial agents. Int. J. Nanomed. 2012:483. doi: 10.2147/ijn.s24793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Verma S.K., Jha E., Panda P.K., Das J.K., Thirumurugan A., Suar M., Parashar S. Molecular aspects of core-shell intrinsic defect induced enhanced antibacterial activity of ZnO nanocrystals. Nanomedicine. 2017;13:43–68. doi: 10.2217/nnm-2017-0237. [DOI] [PubMed] [Google Scholar]
- 153.Tu Y., Lv M., Xiu P., Huynh T., Zhang M., Castelli M., Liu Z., Huang Q., Fan C., Fang H., Zhou R. Destructive extraction of phospholipids from Escherichia coli membranes by graphene nanosheets. Nat. Nanotechnol. 2013;8:594–601. doi: 10.1038/nnano.2013.125. [DOI] [PubMed] [Google Scholar]
- 154.Adams C.P., Walker K.A., Obare S.O., Docherty K.M. Size-dependent antimicrobial effects of novel palladium nanoparticles. PLoS One. 2014;9 doi: 10.1371/journal.pone.0085981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Hong X., Wen J., Xiong X., Hu Y. Shape effect on the antibacterial activity of silver nanoparticles synthesized via a microwave-assisted method. Environ. Sci. Pollut. Res. 2016;23:4489–4497. doi: 10.1007/s11356-015-5668-z. [DOI] [PubMed] [Google Scholar]
- 156.Abbaspour A., Norouz-Sarvestani F., Noori A., Soltani N. Aptamer-conjugated silver nanoparticles for electrochemical dual-aptamer-based sandwich detection of staphylococcus aureus. Biosens. Bioelectron. 2015;68:149–155. doi: 10.1016/j.bios.2014.12.040. [DOI] [PubMed] [Google Scholar]
- 157.Shen H., Wang J., Liu H., Li Z., Jiang F., Wang F.B., Yuan Q. Rapid and selective detection of pathogenic bacteria in bloodstream infections with aptamer-based recognition. ACS Appl. Mater. Interfaces. 2016;8:19371–19378. doi: 10.1021/acsami.6b06671. [DOI] [PubMed] [Google Scholar]
- 158.Kim D.M., Park J.S., Jung S.W., Yeom J., Yoo S.M. =Biosensing applications using nanostructure-based localized surface plasmon resonance sensors. Sensors. 2021;21 doi: 10.3390/s21093191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Hu M., Yuan C., Tian T., Wang X., Sun J., Xiong E., Zhou X. Single-step, salt-aging-free, and thiol-free freezing construction of AuNP-based bioprobes for advancing CRISPR-based diagnostics. J. Am. Chem. Soc. 2020;142:7506–7513. doi: 10.1021/jacs.0c00217. [DOI] [PubMed] [Google Scholar]
- 160.Choi J.H., Lim J., Shin M., Paek S.H., Choi J.W. CRISPR-Cas12a-Based nucleic acid amplification-free DNA biosensor via Au nanoparticle-assisted metal-enhanced fluorescence and colorimetric analysis. Nano Lett. 2021;21:693–699. doi: 10.1021/acs.nanolett.0c04303. [DOI] [PubMed] [Google Scholar]
- 161.Hu M., Yuan C., Tian T., Wang X., Sun J., Xiong E., Zhou X. Single-step, salt-aging-free, and thiol-free freezing construction of AuNP-based bioprobes for advancing CRISPR-based diagnostics. J. Am. Chem. Soc. 2020;142:7506–7513. doi: 10.1021/jacs.0c00217. [DOI] [PubMed] [Google Scholar]
- 162.Nile S.H., Baskar V., Selvaraj D., Nile A., Xiao J., Kai G. Nanotechnologies in food science: applications, recent Trends, and future perspectives. Nano-Micro Lett. 2020;12 doi: 10.1007/s40820-020-0383-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Ahmad A., Khan J.M., Haque S. Strategies in the design of endosomolytic agents for facilitating endosomal escape in nanoparticles. Biochimie. 2019;160:61–75. doi: 10.1016/j.biochi.2019.02.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data included in this study are available upon request from the corresponding author.