Abstract
Cancer is a leading public health problem worldwide. Its treatment remains a daunting challenge, although significant progress has been made in existing treatments in recent years. A large concern is the poor therapeutic effect due to lack of specificity and low bioavailability. Gene therapy has recently emerged as a powerful tool for cancer therapy. However, delivery methods limit its therapeutic effects. Exosomes, a subset of extracellular vesicles secreted by most cells, have the characteristics of good biocompatibility, low toxicity and immunogenicity, and great designability. In the past decades, as therapeutic carriers and diagnostic markers, they have caught extensive attention. This review introduced the characteristics of exosomes, and focused on their applications as delivery carriers in DNA, messenger RNA (mRNA), microRNA (miRNA), small interfering RNA (siRNA), circular RNA (circRNA) and other nucleic acids. Meanwhile, their application in cancer therapy and exosome-based clinical trials were presented and discussed. Through systematic summarization and analysis, the recent advances and current challenges of exosome-mediated nucleic acid delivery for cancer therapy are introduced, which will provide a theoretical basis for the development of nucleic acid drugs.
Graphical Abstract
Keywords: Gene therapy, Exosome, Nucleic acid drug, Delivery, Cancer treatment
Introduction
Cancer remains one of the leading causes of death globally, with the prevalence of 410 million mortalities annually [1]. In 2021, there had been 19.29 million patients diagnosed with cancer, and nearly 10 million people died of cancer [2]. To fight cancer, various treatments such as surgical therapy, chemotherapy, and radiotherapy have been developed. These strategies have become more focused and personalized based on the type and stage of the disease, which has led to a decline in cancer-related mortality over the past decades [3]. Despite their undisputed contribution, these invasive and/or often lacking cancer cell-selective techniques lead to a wide range of harmful side-effects, such as high recurrence rates, enormous trauma, poor survival and impaired life quality, which often hamper therapy success. Consequently, there is an urgent demand to develop safe and efficacious therapeutic techniques for treating cancer.
Gene therapy is the therapeutic delivery of genetic material into cells to compensate for abnormal genes by either turning off genes that produce faulty proteins or introducing genes to make a beneficial protein to treat disease [4–6]. It is a safe and effictive method for treating a wide range of diseases, especially for cancer. The effect of gene therapy depends on the targeting of nucleic acids drugs , the delivery efficiency, and accuracy of delivery tools. The nucleic acids including specific DNA, messenger RNA (mRNA), microRNA (miRNA), small interfering RNA (siRNA), circular RNA (circRNA), which have been widely exploited for gene therapy. However, nucleic acids are negatively charged and hydrophilic, which cannot directly penetrate cell membranes and are vulnerable to enzymatic degradation, so they cannot be effectively transported to cells [4]. In this situation, delivery systems are necessary, which cannot only prevent the nucleic acids degrading in the bloodstream and being filtered out by the kidney, but can deliver them to desired locations.
Extracellular vesicles (EVs) are small membranous vesicles released from different cells to the extracellular matrix, which can participate intercellular communication between cells [7]. According to EVs’ size and origin, they are divided into three subgroups: (1) apoptotic bodies (500 nm–5 μm) released during programmed cell death, (2) microvesicles (150–500 nm) from the budding of the plasma membrane, and (3) exosomes (40–150 nm) from endosomes [8]. Owing to their nano size, exosomes are considered as the most promising drug delivery tools. Compared to conventional delivery systems such as lipid nanoparticles (LNPs), exosomes have the following advantages: (1) Exosomes are more stable in body fluid than LNPs, because LNPs can be easily removed by macrophages or reticuloendothelial cells [9]. (2) Due to their endogenous source and high biocompatibility, exosomes have relatively low cytotoxicity and immunogenicity [10]. (3) Exosomes can provide better drug protection during delivery, because drugs are within the double-layer exosomal membrane, while drugs appeared outside the LNPs, which are easier to degrade [11]. (4) Exosomes can deliver both hydrophobic and hydrophilic molecules. And they have effective homing ability to tumor sites, which may be attributed to their multivalent display of cell-derived surface moieties [10]. (5) Exosomes derived from tumor can escape the phagocytosis of mononuclear phagocyte system through the binding of CD47 on exosomal surface and signal regulatory protein alpha (SIRPα) on the face of macrophages and sending out “don’t eat me” signal [12]. (6) Exosome can cross the blood–brain barrier and reach the brain tissue owing to their small size and characteristics [13]. (7) Exosomes have high cellular uptake and are easily modified according to the target cells owing to membrane proteins such as tetraspanin and fibronectin [10].
Herein, we summarized exosomes’ characteristics and applications as various nucleic acid (DNA, mRNA, miRNA, siRNA and circRNA) delivery carriers for cancer therapy. Meanwhile, the challenges and the prospective in using exosome-mediated nucleic acids are also discussed.
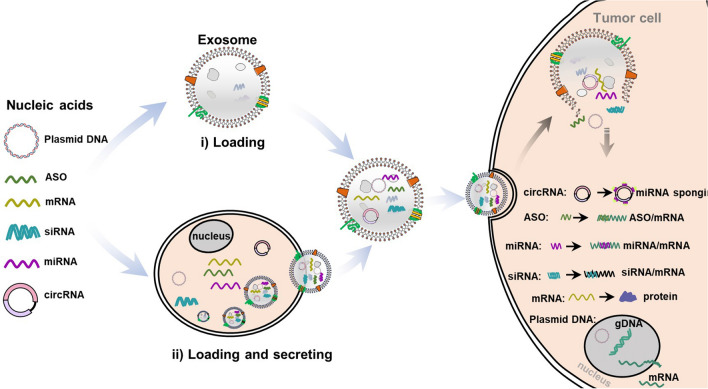
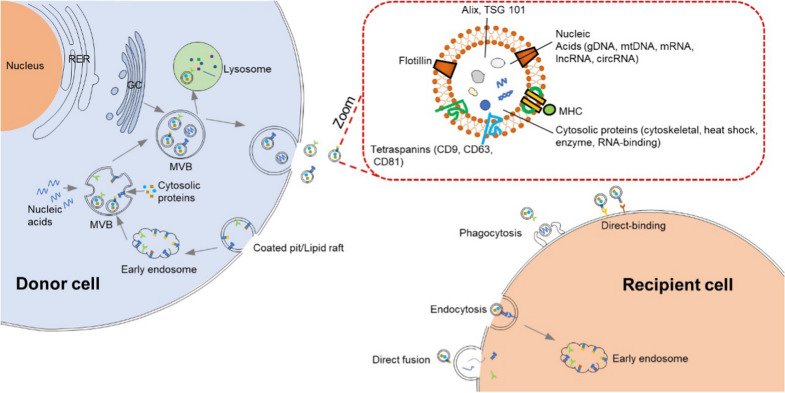
The biogenesis of exosome
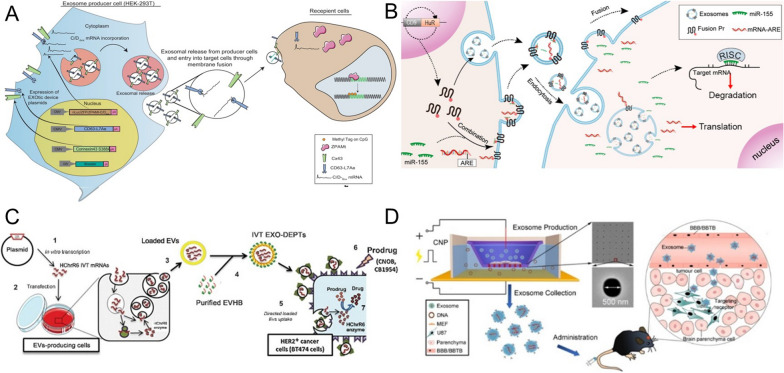
Exosomes are native nanovesicles with a diameter of 30–120 nm secreted from various cell types, including cancer cells, dendritic cells, B cells, T cells, mast cells and epithelial cells, and exist in different body fluids such as blood, urine, malignant effusions, bronchoalveolar lavage fluid and breast milk, etc. [14–17]. They were first found in the supernatant of sheep erythrocytes cultured in vitro in 1983 [18, 19]. At that time, exosomes were considered as the “Garbage Bags” for cells to eliminate unwanted products out of the cells. Subsequently, people found that they were formed by plasma membrane invagination, followed by acidification and maturation of mass exchange into the late endosomes. Late endosomes eventually form multi-vesicles, the membrane of which is sunken inward and sprouts to form intraluminal vesicles, which are exosomes. Finally, they are secreted out of the cell by fusing the plasma membrane (Fig. 1) [17]. The natural internal cargo of exosomes includes specific mRNAs, miRNAs, proteins, etc. (Fig. 1). Several proteins such as tetraspanins (CD9, CD63, CD81), heat shock proteins, and fusion proteins (flotillin) are identified on the surface of exosomes (Fig. 1). These tetraspanins could be used as a specific marker to isolate exosomes. Some studies revealed that exosomes’ target-homing capabilities depended on the surface proteins binding to receptor molecules on the target cell [20, 21]. In addition, many tumor cells secreted exosomes tenfold more than normal cells [21]. Furthermore, the presence of specific genetic information within exosomes derived from tumor cells offers opportunities to develop simple liquid biopsy-based approaches for cancer diagnosis or to monitor the effectiveness of cancer treatment [22].
Fig. 1.
The biogenesis, contents, and internalization of exosomes
The approaches and advances in exosome-mediated delivery
The effect of exosome-mediated therapy mainly depends on the source of exosomes, the loading methods of therapeutic molecules, the efficiency of cell uptake of exosomes. Exosomes derived from different cell types have diverse functions. For example, human embryonic kidney (HEK293) cells have been widely used in the field of biopharmaceutical manufacturing owing to the advantages of easy to growth, non-needing harsh culture conditions, and high transfection efficiency [23]. Moreover, HEK293 cells can accept various transfection methods and allow gene manipulation to modify the exosomal surface or load cargos during exosomal biogenesis [24]. And exosomes derived from HEK293 are immune inert and do not trigger inflammatory reactions in vivo [23]. In addition, cancer cells can secrete a large number of exosomes, because the overexpressed Rab27a and Rab27b proteins in cancer cells are involved in the process of exosome release [25]. Cancer cell-derived exosomes have a tropism toward cell origin due to their abundant biological components similar to their parent tumor cells, which can be used for cancer targeting [26]. Qiao et al. [26] isolated exosomes from two cancer cell lines (HT1080, human fibrosarcoma cells, and Hela, human cervical cancer cells) and observed that the uptake of HT1080 exosomes in HT1080 cells was twofold that of Hela exosomes. Furthermore, in vivo therapeutic experiments revealed that the inhibition rate of HT1080 exosomes loaded with a common chemotherapy drug Doxil was threefold higher than that of Hela exosomes with Doxil. However, there are some limitations, such as an unsatisfactory pharmacokinetic profile, being involved in tumor development and metastasis, and potential safety issues, which are expected to be improved to be better used in cancer treatment [10]. Besides, exosomes derived from immune cells have also been widely studied. For instance, monocytes- and macrophages-derived exosomes have been shown to evade immune phagocytosis [27]. Dendritic cell (DC)derived exosomes hold a significant advantage as they have been proven secure in different types of cancer [28]. And these exosomes loaded with tumor antigens have been effective against non-small cell lung cancer (NSCLC) [29]. As is shown in Table 1, exosomes from different sources have different advantages and disadvantages. Therefore, the purpose of good therapy can be achieved by selecting appropriate exosomes according to therapeutic requirements.
Table 1.
The advantages and disadvantages of different types of exosomes
| Types | Sources | Advantages | Disadvantages | References |
|---|---|---|---|---|
| Cell-secreted exosomes | Human embryonic kidney cells | Ease of growth; non-demanding maintenance conditions; high transfection efficiency; ideal host cells for membrane modification through gene manipulation | Immune inert | [23] |
| Cancer cells | Large secretion; targeting homotypic tumor | Have a less ideal pharmacokinetic profile; be involved in tumour development and metastasis; having potential safety issues | [30] | |
| Immune cells (e. g. macrophage cells, dendritic cells, natural killer cells) | Reduced immunogenicity; inducing potent cellular immune responses; containing killer proteins and cytotoxic molecules to inhibit tumour growth; penetrating the blood–brain barrier | Lack of understanding of mechanisms regarding how exosomal components interact with acceptor cells | [10] | |
| Stem cells (e. g. mesenchymal stem cells) | Immune regulation characteristics; low production cost; good homing and penetrating ability | The unclear cargo composition of exosomes and biological behavior mechanism | [31, 32] | |
| Blood-derived exosomes | Blood | Wide source and easy access; reduced unexpected mutations in cell culture; no occurring horizontal gene transfer; high transfection efficiency; natural brain targeting ability | Not determined | [33] |
| Food-derived exosomes | Milk-derived exosomes | Rich sources; crossing through the gastrointestinal tract via the neonatal Fc receptor; improving the oral bioavailability of drugs; improving the effectiveness and stability of drugs; improving human and mouse intestinal cells | Variation in shape, size, and cargo contents of exosomes; the unclear mechanism of the absorption, movement, and action | [34–36] |
| Plants-derived exosomes (e. g. grape, strawberry, lemon) | Rich sources; have the stability in the digestive environment | Less understanding of the ability in the process; the unclear mechanism of the absorption, movement, and action | [36, 37] |
Co-incubation, transfection, and electroporation are the frequently-used methods of therapeutic molecules loading into exosomes [38]. The aqueous core and bilayer lipid membrane of the exosomes make the loading of hydrophilic and hydrophobic drugs easier through co-incubation [39]. When hydrophilic molecules fail to spontaneously pass through the lipid bilayer, loading can be achieved by liposome transfection and electroporation to form transient pores on the exosomal membrane. Transfection-based approaches have been proved to have better loading efficiency and protein stability, but they are undesirable because of their toxicity and side effects of transfectants in altering cell gene expression [40]. Electroporation has been widely used as a safer method in therapeutic molecules loading into exosomes. Shtam et al. [40] provided sufficient evidence that the nucleic acids were more effectively introduced into exosomes from HeLa cells using electroporation than chemical treatment. However, not all cell-derived exosomes can be loaded by electroporation. For example, Ohno et al. [41] found that when using HEK293T cells as the source of exosomes, liposome transfection could load nucleic acids successfully while electroporation did not. Therefore, the method may need to be optimized for each exosome and cell type. In addition, in recent years, several new loading methods are emerging gradually. For instance, an active delivery modality exploits the HIV-1 TAR and RNA-TAT peptide interaction by swapping the wild type pre-miR loop with the TAR RNA loop. The modified pre-miR is designed to recognize the TAT peptide introduced into the exosomes using a Lamp2a fusion protein. The loading of the miRNA into exosomes was enhanced using this TAT-TAR interaction [42, 43]. The modified calcium chloride (CaCl2) method (CaCl2-heat shock) has successfully loaded nucleic acids into exosomes through forming CaCl2-nucleic acid complex, which was absorbed by exosomes under heat shock at 42 °C [44–46]. In addition, plasmid-mediated therapeutic molecule transfer has been gradually applied. A constructed plasmid containing therapeutic molecule genes is transfected into exosome-producing cells. After culture, the exosomes produced by donor cells contain therapeutic molecules [47, 48]. Based on the above, the correct selection of therapeutic molecules loading into the exosomes can achieve unexpected therapeutic effects.
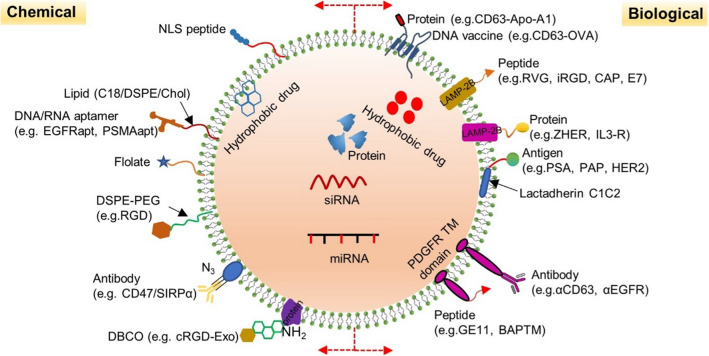
In addtion, exosomes loaded therapeutic molecules also face several challenges, including competition from endogenous exosomes, the internalization/clearance by the mononuclear phagocyte system, and targeting [49–51]. To solve these problems, it is essential for specific modification on exosome surfaces. These modification include chemical and biological modifications (Fig. 2). The former depends on the biological binding of targeted ligands to surface proteins, but surface protein inactivation or exosome aggregation may occur. The latter is an important strategy to display functional ligands on the exosome membrane, but it requires plasmid construction and overexpression of proteins in donor cells. Despite the defects, both methods have been successfully applied. For instance, Zhan et al. [52] constructed the the amphiphilic phosphatidylcholine (PC) exosome through inserting PC into the membrane lipid layer of the reticulocyte exosome from the blood. Compared with natural exosome, PC exosome increased the efficiency of tumor cell internalization by nearly twice. After loading therapeutic drugs, PC exosome significantly promoted the accumulation of drugs in tumor cells and showed enhanced antitumor activity in vitro. In addition, the surface of bovine serum‐derived exosomes is modified with α‐d‐mannose to facilitate interaction with mannose receptors on DCs and efficient delivery of immune stimulators to the DCs [53]. Zuo et al.[54]. added a potent adjuvant, high mobility group nucleosome-binding protein 1 (HMGN1) to tumor cell-derived exosomes, which enhanced the ability of DC to activate T cells and sustained protective immune response for about 9 weeks.
Fig. 2.
The chemical and biological modification on exosome surfaces
Above all, to improve the efficiency and accuracy of drug delivery to achieve good therapeutic effect, the selection of exosomes, the loading mode of therapeutic molecules and surface engineering modification of exosomes are the main factors that should be considered comprehensively. Because exosomes from different cells have different functions, choosing the right exosomes as drug delivery tools can greatly improve the targeting of drug delivery. The loading capacity of therapeutic molecules can be significantly improved by appropriate loading method. Further, the engineering modification of exosome surface can achieve the loading efficiency and targeting of therapeutic molecules at the same time. Thus, integrating various advantages to deliver drug molecules through exosome can achieve the ideal therapeutic effect.
Exosomes-based nucleic acid delivery system for cancer treatment
Exosomes-based DNA delivery system
ASOs
In addition to the well-known genomic DNA, mitochondrial DNA and plasmid DNA, antisense oligonucleotides (ASOs) are another important DNA species which are single-stranded DNA molecules and usually consist of 12–25 nucleotides, can complementarity to target mRNA [55]. Following binding to the targeted RNA, the ASOs can regulate RNA function through several mechanisms. One is that ASOs can form RNA–DNA hybrid and serve as the substrate of RNase H-mediated cleavage, leading to the hydrolysis of a hybridized RNA strand [56, 57]. The formation of ASO-RNA heteroduplex also leads to splicing inhibition or exon skipping events by spatially blocking standard splicing sites [56]. Another is that ASO only plays a space-occupying role and does not directly degrade target RNA. For instance, ASOs can be designed to bind the miRNAs and block the targeted RNA, resulting in inhibition of translation of the RNA and increase the expression of a variety of proteins [57]. In addition, ASOs are designed to bind to a regulatory sequence in the 5-untranslated region of an mRNA that represses protein translation, such as an upstream open reading frame or stem-loop structure [57]. As a powerful molecular tool, ASOs are widely used in protein and RNA biology and are a highly selective therapeutic strategy for many diseases related to gene expression disorders. So far, over ten ASOs have been approved by Food and Drug Administration (FDA) [58]. Therefore, the successful loading of specific ASOs into cells play a key role in cancer therapy.
Loading methods for DNA into exosomes
As a common strategy, electroporation also allows DNA to be loaded into exosomes through creating pores on exosomal lipid bilayers. It has been shown that ASO4, ASO-210 or scramble ASO loaded into exosomes by electroporation could be delivered to recipient cells and knock down specific gene expression [59–61]. However, a major disadvantage of electroporation is the formation of nucleic acid and exosome aggregates during encapsulation, which will affect the function of nucleic acids [62]. Lamichhane et al. [59] reported that exosomes carrying plasmid DNA by electroporation delivered DNA to recipient cells; however, these DNAs were not functionally active. To solve these problems, it may be necessary to optimize electroporation parameters. In addition, some new loading methods have been developed. An exosomal liposome hybrid was formed through fusing the lipid bilayer of the exosomal membrane with liposomes, which could encapsulate and deliver large DNA molecules, such as CRISPR/Cas9 plasmid, and reduce the toxicity of liposomes [63]. Exosome-associated adeno-associated virus (exo-AAV) has also been proved to be a powerful system for DNA delivery. György et al. [64] cloned a mouse-codon-optimized gene encoding lipoma HMGIC fusion partner-like 5 (LHFPL5) with a hemagglutinin (HA) tag at the N terminus into an AAV vector, and then was transfected into HEK293T cells using the calcium phosphate and obtained exo-AAV1-HA-Lhfpl5, which could rescue hearing in a mouse model of hereditary deafness. Therefore, the successful loading will provide a basis for exosome-mediated DNA delivery for cancer treatment.
Delivery of therapeutic DNA
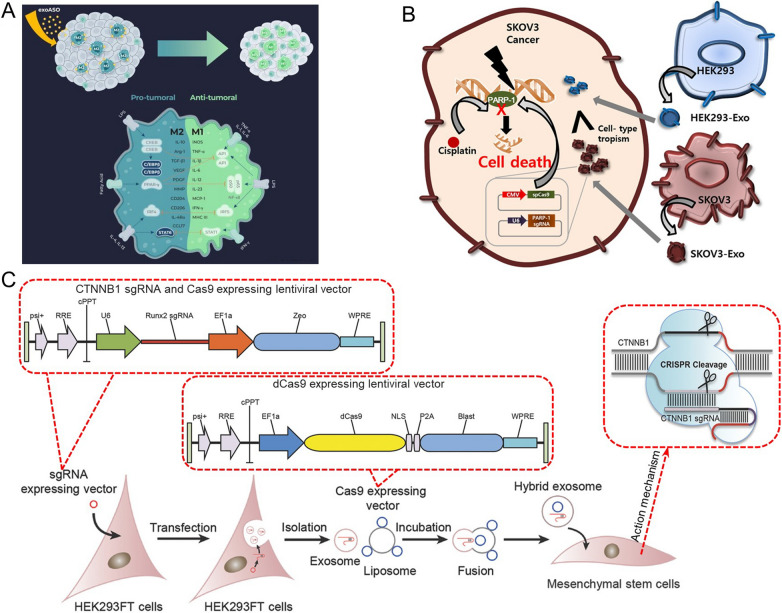
In recent years, it has been reported that exosome deliver various functionlized DNA into cells via the process of exosome-endocytosis to treat cancer [65–69]. However, due to their small size, the efficiency of packaging large DNA through exosomes is very low, which limit the application of the exosome-based drug delivery system. The relatively small ASO and plasmid DNA, or engineering modified exosomes are used to solve the problem. Codiak Biosciences [70] published the first preclinical data of engineered exosomes to deliver ASO (exoASO), demonstrating the potential of exoASO to M2 macrophages to target the expression of key immunosuppressive transcription factors STAT6 and C/EBP (Fig. 3A). The results revealed that the expression of TNF and IL-10 related to exoASO therapy increased up to 40-fold and 29-fold respectively, which was consistent with the repolarization from immunosuppressive M2 macrophages to immunostimulatory M1 macrophages, and exoASO-STAT6 significantly slowed tumor growth, and tumors in 50% of mice completely subsided. When exoASO-STAT6 was combined with anti-PD1 antibody, the tumor remission rate was further improved by 25%. It was exciting that FDA has recently approved the investigational new drug application of exoASO-STAT6. In addition, with the rise of CRISPR/Cas9-mediated genome editing, the delivery of Cas9-encoding plasmid through exosome has also been tried. For example, Kim et al. [71] reported that ovarian cancer-derived exosomes (SKOV3-Exo) could be efficiently electroporated with CRISPR/Cas9 plasmids in vivo to suppress the expression of poly (ADP-ribose) polymerase-1 (PARP-1). The results suggested that compared with SKOV3-Exo alone, the expression of PARP-1 was completely inhibited after treatment with CRISPR/Cas9-loaded SKOV3-Exo, and the tumor volume in the treatment group hardly changed within 20 days of intratumoral injection treatment, while that in the control group kept growing (Fig. 3B). Besides, Lin et al. [63] developed a kind of hybrid exosomes with liposomes to deliver the CRISPR/Cas9 expression plasmids into mesenchymal stem cell (MSC) target cells, and the results revealed that hybrid nanoparticles carried the large CRISPR/Cas9 expression plasmids could down-regulate the expression of gene Runx2 by twofold compared with the control group (only Runx2 guided CRISPR/dCas9 system) (Fig. 3C). Therefore, exosome mediated ASO or CRISPR/Cas9 plasmids into cells could correct or destroy oncogenes through regulating mRNA translation or therapeutic genome editing (gene destruction, gene correction, gene deletion, gene insert, etc.), respectively [72, 73].
Fig. 3.
A Engineered exosomes to deliver ASO to produce effective antitumor activity [70]. B Exosomes loaded with CRISPR/Cas9 targeting PARP-1 for cancer therapy [71]. C The hybrid exosomes successfully deliver CRISPR/dCas9 interference system [63]
Exosomes-based mRNA delivery system
mRNA
mRNA is an intermediate molecule that transmits the genetic code from DNA to the ribosome for protein expression. It has been considered as another promising tool for the treatment of a variety of diseases, especially cancer [74, 75]. Compared with DNA-based therapy, RNA-based therapy is more advantageous: (a) DNA transcription must precede translation and need to enter the nucleus. The efficiency is limited because less than 0.10% of cytoplasmic DNA enters the nucleus; In contrast, mRNA is directly translated when entering the cytosol, resulting in effective gene expression [76]; (b) mRNA has no risk of genome integration and will not cause insertion mutation [77]; (c) unlike DNA, mRNA is also translated in tumor dormant cells [78]. Based on the above advantages, the use of mRNA technology to develop vaccines for related diseases, including cancer, has gradually attracted extensive attention [79]. In particular, since the outbreak of COVID-19, a variety of mRNA vaccines have been rapidly developed using mRNA technology [80–82]. Nevertheless, mRNA is easy to be degraded by nuclease, easy to activate immune response, and large (104–106 Da) [83], which has become the main obstacle to the development of mRNA drugs. Exosomes, as a natural delivery carrier, can realize the effective delivery of mRNA.
Loading methods for mRNA into exosomes
As early as 2007, Valadi et al. [84] firstly found that exosomes were natural carriers of mRNA in mast cells. Subsequently, this phenomenon was also observed in many other cells [85–87]. However, the insertion of foreign mRNA into exosomes has been a challenge, because these electroporation- or chemical-based loading methods are not useful for packaging and delivering macromolecular mRNA via exosomes. Afterwards, Tsai et al. [88] reported that the exosome-liposome hybrid could efficiently transfect target cells with Antares2 mRNA. In the study, the purified mRNA was pre-incubated with polycationic lipid coating, and then mixed with equal amounts of purified exosomes. Based on this, the multiplexed mRNA COVID-19 vaccine was successfully developed. In addition, Kojima et al. [89] constructed an EXOsomal Transfer Into Cells (EXOtic) device, which transformed the way of sorting mRNA from natural but dynamic pathways to an engineering way. In this device, the archaea-derived L7Ae peptide (binding to the C/Dbox RNA structure) was fused with the exosomal marker protein CD63, recruiting those mRNAs containing C/Dbox into budding exosomes. And then exosomes carried these functional mRNAs into the cytoplasm of target cells (Fig. 4A). Recently, this method have also been adopted to package ZFP or ZPAMt mRNA with exosomes to inhibit HIV-1 transcription, inducing “blocking and locking” phenotypes in virusinfected cells [90]. In addition, Li et al. [91] fused the exosome membrane protein CD9 with RNA binding protein HuR to construct CD9-HuR functionalized exosomes, which has a strong ability to enrich specific RNAs (Fig. 4B). These functionalized exosomes were used to deliver dCas9 mRNA to target gene C/ebpα related to cell proliferation and differentiation in liver. And the expression of target gene was decreased by about 20-fold compared with free dCas9 mRNA. Furthermore, with the technical breakthrough of the exosome-loading mRNA method, this delivery system is gradually used to treat cancer.
Fig. 4.
A An EXOsomal Transfer Into Cells (EXOtic) device recruits those mRNAs containing C/Dbox into budding exosomes [90]. B CD9-HuR functionalized exosomes deliver dCas9 mRNA to target gene C/ebpα related to cell proliferation and differentiation in liver [91]. C Exosomes loaded with HChrR6 mRNA for breast cancer therapy [93]. D a cellular nano perforation technology for producing a large number of exosomes containing therapeutic mRNAs [94]
Delivery of therapeutic mRNA
Wang et al. [92] applied exosomes to deliver HChrR6-encoding mRNA (generated by transfection of cells with the XPort/HChrR6 encoding plasmid) to the HER2+ve human breast cancer cells, which caused nearly complete growth arrest of the breast cancer cells. This was the first time that exosome-mediated exogenous mRNA delivery has gained a therapeutic advantage. Subsequently, Forterre et al. [93] have successfully utilized similar methods to treat HER2+ve human breast cancer cells. In the study, exosomes from HEK293 cells delivered functional HChrR6 mRNA to HER2+breast cancer cells, and when administered systemically along with prodrug CB1954, they arrested the growth of HER2+human breast cancer xenografts in athymic mice by prodrug activation (Fig. 4C). In another work, Usman et al. [33] treated leukemia cells with exosome from human red blood cells (RBCs) loaded with Cas9 mRNA and gRNA targeting the human miR-125b-2 (an oncogenic miRNA in leukemia) locus. The results indicated the expression of miR-125a and miR-125b decreased by 90–98% after 2 days of treatment.
In addition to the delivery of exogenous mRNA, endogenous functional mRNA has also been caught attention. Yang et al. [94] reported a cellular nano perforation technology for producing a large number of exosomes containing therapeutic mRNAs. Firstly, the plasmid DNA was transfected into various sources cells, and then the cells were stimulated with focal and transient electrical stimulation to promote the release of exosomes carrying the transcribed mRNA. Based on this, PTEN and CDX (CD47 cloning targeted peptide) plasmids were transferred into glioma cells to obtain a large number of targeted functional exosomes, which enhanced cell uptake, restored the expression of PTEN protein, inhibited tumor growth, and prolonged survival with a median survival of 45 days, compared with 31 days for non-functional exosomes (Fig. 4D). Encouragingly, NeoCura (a Chinese company of RNA precision medicine based on artificial intelligence) and MDimune lnc (a Korean company based on extracellular vesicle drug delivery platform) recently jointly developed mRNA therapy for cancer vaccine delivery based on exosomes [95]. Based on the above, exosome mediated mRNA delivery has promising potential for cancer treatment (Fig. 4).
Exosomes-based miRNA delivery system
miRNA
miRNAs are a class of highly conserved single-stranded RNA with a length of 19–25 nucleotides, which are generally located in the non-coding region of the genome and do not encode proteins, but they play an important role in regulating gene expression [96, 97]. The specific sequence of the 3′ untranslated region (3′-UTR) of miRNA is completely or partially complementary to its targeted mRNA, leading to target degradation or translation inhibition, so as to negatively regulate the target protein expression. This process is also involved in the occurrence and development of tumors. For instance, several miRNAs, such as miR-149-5p, miR-29b and miR-34b, are poorly expressed in prostate cancer tissues [98]. There are some down-regulated miRNAs in bladder cancer, such as miR-145, miR-125b and miR-143, which even show anti-oncogenic properties; while some upregulated miRNAs, for instance, miR-17-5p, miR-20a, and miR-183, were oncogenic [99]. In NSCLC cells, miR-26, miR-21, miR-155 and miR-574-5p affected the progression of NSCLC through cell cycle regulation, escaping, apoptosis, metastasis regulation, etc. [100]. Besides endogenous miRNA, synthetic anti-miRNA oligonucleotides (AMOs) or miRNA mimics (miR mimics) have also been delivered into cells to suppress or enhance specific endogenous miRNAs’ function. Thus, regulating the expression of cancer-related genes through miRNA complementation is becoming a promising means of cancer treatment.
Loading methods for miRNA into exosomes
Recently, exosome based-miRNA therapy has developed more rapidly owing to its wide participation in gene regulation, small size, and easy to load. Electroporation is also used to load miRNA into exosomes. Studies have shown that each exosome was loaded with about 3000 miRNA molecules [101]. Table 2 also summarizes these studies of electroporation of miRNA into exosomes for therapy. In addition, miRNA could be loaded into exosomes by incubation at 37 °C [102]. However, the loading efficiency is not satisfactory, so this method is not often used. Besides, there are also commercial transfection reagents on the market, such as Exo-FectTM exosome transfection reagent, HiPerFect transfection reagent, Lipofectamine 2000 and 3000, which are used to load miRNA directly into exosomes (Table 2). Furthermore, in the case of heat shock, CaCl2 can mediate the transfection of miRNAs or their inhibitors into exosomes, and these RNAs have functional activity after transmission to recipient cells [44]. Additionally, another transfection method pre secretion of exosomes has also been proved to be effective. Trivedi et al. [103] introduced miRNA-125b into SK-LU-1 lung cancer cells using hyaluronic acid-polyethyleneimine (HA-PEI)/hyaluronic acid-polyethylene glycol (HA-PEG) combined nanoparticles as gene transfection agents, which successfully increased miRNA-125b expression in exosomes secreted by the lung cancer cells. Therefore, these available loading methods can be selected according to different requirments.
Table 2.
The application of exosomes-based exogenous miRNA delivery system in cancer treatment in the last decade
| Exosome | The source of exosome | Therapeutic cargo | Loading method | Target gene | Mechanisms | Cancer types (cell lines) | Effects | References |
|---|---|---|---|---|---|---|---|---|
| Engineering exosome (Apo-A1-modified exosome) | HEK293T cells | miR-26a | Electroporation | CCNE2, CDK6, CCND2 | Down regulating of the expression levels of CCNE2, CCND2 and CDK6 | Liver cancer (HepG2) | Decreasing the rates of cell migration and proliferation | [21] |
| Engineering exosome (GE11 peptide or EGF-modified exosome) | HEK293T cells | let-7a | HiPerFect transfection reagent | Unidentified or uncharacterized genes | Breast cancer (HCC70 HCC1954, MCF-7) | Inhibiting tumor development | [41] | |
| Engineering exosome (magnetic molecules and L17E peptide- modified exosome) | Serum | dox, cholesterol-modified miR-21 inhibitor | Co-incubation | miR-21 | Interfering with nuclear DNA activity and down regulating the expression of oncogenes bcl-2, caspase-3 and p-akt | Human glioblastoma (U87), breast cancer (MDA-MB-231) | Inhibiting the growth of the tumors and alleviating side effects | [108] |
| Stem-cell-derived exosome | Human umbilical cord mesenchymal stromal cells | miR-145-5p | Exo-Fect™ exosome transfection reagent | Smad3 | Activating the TGF-β/Smad3 pathways | Pancreatic ductal adenocarcinoma (Capan-1, CFPAC-1, BxPC-3, Panc-1) | Inhibiting cell proliferation and invasion and increasing apoptosis and cell cycle arrest | [109] |
| Stem-cell-derived exosome | Bone marrow mesenchymal stem cells | LNA-antimiR-142-3p | Electroporation | APC, P2X7R | Decreasing the levels of miR-142-3p and miR-150, and increasing the targeted regulation of APC and P2X7R | Breast cancer (MCF-7) | Reducing cell clonogenicity and tumorigenicity | [110] |
| Stem-cell-derived exosome | Human umbilical cord mesenchymal stem cells | miR-6785-5p mimic | Lipofectamine 2000 transfection reagent | INHBA | Inhibiting the expression of INHBA | Gastric cancer (SGC7901, MGC803) | Suppressing cell angiogenesis and metastasis | [111] |
| Stem-cell-derived exosome | Human bone marrow mesenchymal stem cells | miR-205 mimic | Lipofectamine 2000 transfection reagent | RHPN2 | Inhibiting the expression of RHPN2 | Prostate cancer (LNCaP) | inhibiting cell proliferation, invasion, and migration and promoting cell apoptosis | [112] |
| Stem-cell-derived exosome | Human umbilical cord mesenchymal stem cells | miR-139-5p mimic | Lipofectamine 2000 transfection reagent | PRC1 | Inhibiting the expression of PRC1 | Bladder cancer (T24, J82, UMUC3, 5637) | Impeding the cell proliferation, migration, and invasion potentials | [113] |
| cancer-associated fibroblasts | Cancer-associated fibroblasts | miR-3188 mimic | Lipofectamine 2000 transfection reagent | BCL2 | Downregulating the expression of BCL2 | Head and neck cancer (HN4, HN30) | Inhibiting cell proliferation, colony formation ability and G1 to S cell cycle transition | [114] |
| Cancer-associated fibroblasts | Cancer-associated fibroblasts | miR-320a mimic | Lipofectamine 2000 transfection reagent | PBX3 | Suppressing the activation of the MAPK pathway | Hepatocellular carcinoma (MHCC97-H, SMMC-7721, Huh7) | Suppressing cell proliferation, migration and metastasis | [115] |
| Cancer-associated fibroblasts | Cancer-associated fibroblasts | miR-139 mimic | Lipofectamine 2000 transfection reagent | MMP11 | Decreasing the expression of MMP11 | Gastric cancer (N87, AGS) | Inhibiting cell growth and metastasis | [116] |
| Cancer-associated fibroblasts | Cancer-associated fibroblasts | miR-34 mimic | Lipofectamine 3000 transfection reagent | AR, CCL22, CCND1, CCNE2, CDK4, CDK6, c-Met, E2F3, E2F5, HMGA2, LETK3, MTA2, N-Myc, PAR2, SFRS2, SIRT1 | Downregulating the expression of target genes | Gastric cancer (AGS, AZ521, MKN1, NUGC3) | Inhibiting cell proliferation and invasion | [117] |
| Exosome-liposome hybrid | Human ovarian cancer cells | miR497, triptolide | Liposome | PI3K, AKT, mTOR | activatingpi3k/AKT/mTOR signaling pathway | Ovarian cancer (SKOV3) | Signifcantly enhancing tumor cell apoptosis and overcoming chemoresistant ovarian cancer | [118] |
| Engineering exosome (Her2-LAMP2-modified exosome) | HEK293T cells | miR-21 inhibitor, 5-FU | Electroporation | miR-21 | Downregulating miR-21 and rescuing PTEN and hMSH2 expressions | Colon cancer (HCT-1165FR) | Reversing drug resistance and significantly enhancing the cytotoxicity in 5-FU-resistant colon cancer cells | [107] |
| HEK293T cells | Let7c-5p | Lipofectamine RNAiMAX reagent | HMGA2, c-Myc | Downregulating the expression of HMGA2 and c-Myc | Breast cancer (MDA-MB-231) | Inhibiting cell proliferation and migration | [119] | |
| Stem-cell-derived exosome | Bone marrow-derived mesenchymal stem cells | LNA anti-miR-142-3p | Electroporation | APC, P2X7R | Suppressing the expression level of miR-142-3p and miR-150 and increasing the transcription of the regulatory target genes, APC and P2X7R | Breast cancer (4T1) | Decreasing cell proliferation | [120] |
| Stem-cell-derived exosome | Human umbilical cord mesenchymal stem cells | miR-375 mimic | Transfection | ENAH | Suppressing ENAH expression | Esophageal squamous cell carcinoma (KYSE70, ECA109, EC9706) | Inhibiting cell proliferation, invasion, migration, tumorsphere formation, and promoting apoptosis | [121] |
| Engineering exosome (TAT peptide-modified exosome) | A549 cells | miR-449a | TAT-TAR interaction | Bcl-2 | Inhibiting the expression of apoptosis inhibitor protein Bcl-2 | Non-small cell lung cancer (A549) | Inhibiting cell proliferation and promoting cell apoptosis | [43] |
| Engineering exosome (T7 peptide-modified exosome) | HEK293T cells | miR-21 antisense oligonucleotides | Electroporation | miR-21 | Reducing of miR-21 and inducing the expression of PDCD4 and PTEN | Glioblastoma (C6) | Resulting in a reduction of tumor sizes | [106] |
| Cancer-cell-derived exosome | HT-29 and SW480 cells | miR-375-3p mimic | Modified calcium chloride method | ZEB1 | Reducing the expression of β-catenin, vimentin, ZEB1, and snail significantly increasing the expression of E- cadherin in EMT process | Colon cancer (HT-29, SW480) | Reversing EMT process and inhibiting cell invasion and migration | [44] |
| FHC cells | miR-128-3p mimic | Electroporation | Bmi1, MRP5 | Suppressing Bmi1 and MRP5 expression | Oxaliplatin-resistant colorectal cancer (HCT116OxR, HT29OxR) | Enhancing cell chemosensitivity | [122] | |
| HEK293T cells | miRNA-497 mimic | Transfection | YAP1, HDGF, CCNE1, VEGF-A | Suppressing YAP1, HDGF, CCNE1, VEGF-A expression | Non-small cell lung cancer (A549) | Inhibiting the tumor growth and angiogenesis | [123] | |
| SCC084 cisplatin- resistant strain | miR-30a mimic | Lipofectamine RNAiMAX | Beclin1 | A concomitant decrease in Beclin1 and Bcl2 expression | Oral squamous cell carcinoma (SCC084) | Regaining sensitivity of the cisplatin-resistant OSCC cells | [124] | |
| Cancer-cell-derived exosome | Oral cancer patients and oral squamous cell carcinoma (OSCC) | miR-155 mimic | Lipofectamine RNAimax | FOXO3a | Modulation of EMT pathway and downregulation of FOXO3a | Oral cancer (SCC131) | Conferring cisplatin resistance in OSCC | [104] |
| Parental and cisplatin-resistant human OSCC cell lines | miR-155 inhibitor | Modified calcium chloride transfection method | FOXO3a | Upregulation of FOXO3a and induction of the mesenchymal-to-epithelial transition | Oral squamous cell carcinoma (UPCI-SCC-131) | Reversing chemoresistance in oral cancer | [46] | |
| Cisplatin-resistant OSCC cells | Anti-miR-21 | Lipofectamine 3000 | PTEN, PDCD4 | Downregulating the expression of PTEN and PDCD4 | Oral squamous cell carcinoma (HSC-3, SCC-9) | Inducing cisplatin resistance of OSCC cells | [125] | |
| HEK293T cells | Anti-miR-214 | Lipofectamine 2000 transfection reagent | miR-214 | Downregulation of miR-214 and overexpression of possible target proteins (PARP9, XRCC, LIN28B) | Gastric cancer (SGC7901) | REVERSING chemoresistance and repressing tumor growth | [126] | |
| HEK293T cells | miR-199a-3p mimic | Lipofectamine 2000 transfection reagent | ZEB1, MTOR, DNMT3A | Down-regulation of underlying target proteins (ZEB1, MTOR, DNMT3A) | Hepatocellular carcinoma (Huh-7) | Reversing chemoresistance to cisplatin in hepatocellular carcinoma | [127] | |
| Doxorubicin-resistant gastric cancer SGC7901/ADR cell | miR-501 inhibitor | Lipofectamine 2000 transfection reagent | miR-501 | Inducing downregulation of BLID, inactivating of caspase-9/-3 and phosphorylation of Akt | Gastric cancer (SGC7901) | Being sensitive to doxorubicin and attenuating proliferation, migration and invasion and increasing apoptosis | [105] |
Delivery of therapeutic miRNA
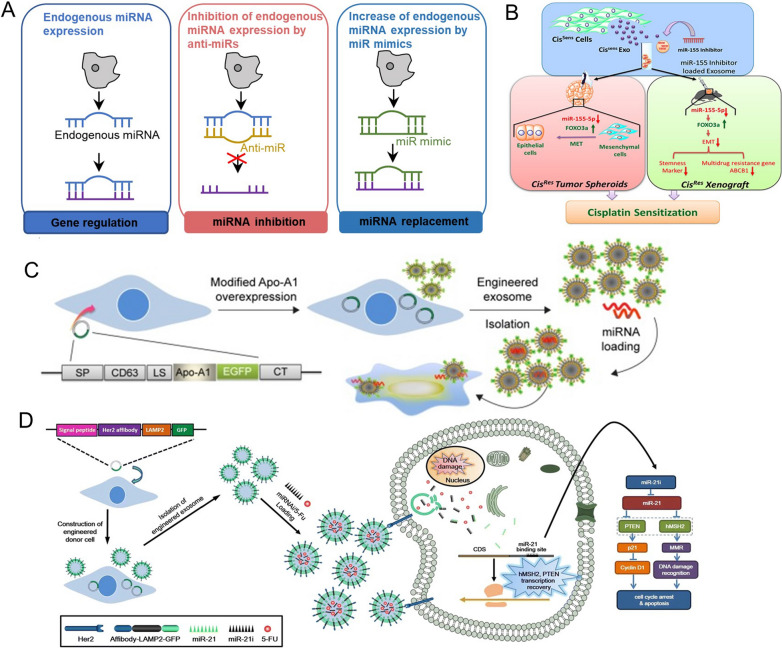
miRNA-based therapy is divided into two forms: miRNA replacement or inhibition (Fig. 5A). The former aims to introduce exogenous miRNAs (miR mimics) known to promote tumor inhibition. The latter provides specific miRNA inhibitor or AMOs to inhibit tumor promoting miRNA (oncomiR). Based on the different therapeutic requirements, a large number of successful cases have been reported. For example, as a tumor suppressor, miR-375 is negatively associated with epithelial-mesenchymal transition (EMT) in cancer patients. To increase its expression and reverse EMT process, Rezaei et al. [44] used tumor-derived exosomes to deliver miR-375 mimic, resulting in the inhibition of the migration and invasion abilities of colon cancer cells. In addition, miR-155 overexpression can enhance the invasive and chemoresistance of oral squamous cell carcinoma (OSCC) cells. To decrease miR-155 expression, Kirave et al. [104] introduced exosome as a carrier and miR-155 inhibitor as therapeutic agent to treat cisplatin-resistance OSCC, and the results revealed that exosomes loaded miR-155 inhibitor could reverse chemoresistance in oral cancer through upregulating the expression of FOXO3a and inducing the EMT transition (Fig. 5B). Similarly, exosome mediated miR-501 inhibitor delivery into doxorubicin (dox)-resistant gastric cancer cell, resulting in inhibiting the expression of miR-501 and makeing the cells sensitive to dox [105]. Although exosome based-miRNA has some effect in the tumor treatment, the therapeutic effect needs to be improved due to its targeting.
Fig. 5.
A Different therapeutic forms of miRNA. B Exosome mediated miR-155 inhibitor delivery to treat cisplatin-resistance oral squamous cell carcinoma [104]. C Apo-A1-modified exosomes loaded miR-26a selectively bound to HepG2 cells via the SR-B1 receptor-mediated endocytosis [21]. D Engineered exosome packaged miR-21i and chemotherapeutics 5-Fluorouracil (5-FU) to target 5-FU-resistant colorectal cancer cell [107]
Genetically modified exosome can target tumor cells through binding functional ligands modified on exosomal surface to overexpressed receptors on the tumor surface, so as to transfer more miRNA to tumor cells and further enhance the therapeutic effect. For example, Liang et al. [21] reported that the Apo-A1-modified exosomes loaded miR-26a (Apo-Exo/miR-26a) selectively bound to HepG2 cells via the SR-B1 receptor-mediated endocytosis. The results revealed that compared with the HepG2 cells incubated with exosome-loaded miR-26a, those with Apo-Exo/miR-26a could upregulate miR-26a expression about threefold, and downregulate key cyclins CCNE2 and CDK6 expression about onefold, and the inhibition of cell migration was twofold as high (Fig. 5C). Ohno et al. [41] revealed that modified exosomes with the GE11 peptide on their surfaces delivered let-7a miRNA specifically to xenograft breast cancer tissue in RAG2–/– mice, significantly inhibiting tumor development in vivo through upregulating the let-7a expression about 1000-fold and downregulating target gene HMGA2 about fivefold. Similarly, exosome surface were modified with target peptide transcriptional transactivator (TAT) protein and T7 respectively to deliver different miRNA to target tumor cells, which obviously inhibiting the tumor occurrence and development [43]43. Although the efficacy of this single gene therapy can be improved by strengthening the targeting of exosomes, due to the limitation of monotherapy, it is necessary to cooperate with other therapies to further improve the therapeutic effect.
Recently, it has been demonstrated that miRNAs and chemotherapeutics can be co encapsulated within engineered exosome and achieve more excellent anti-tumor effect. Liang et al. [107] integrated the fusion protein Her2-LAMP2 into the surface of exosomes to make exosome target EGFR receptor. And then engineered exosome packaged miR-21i and chemotherapeutics 5-Fluorouracil (5-FU) (THLG-EXO/5-FU/miR-21i) to target 5-FU-resistant HCT-116 colorectal cancer cell (HCT-1165−FR) through EGFR receptor-mediated endocytosis. The results revealed that the apoptotic proportion and proliferation inhibition rate of THLG-EXO/5-FU/miR-21i-treated HCT-1165FR cells increased by about 3.5-fold and fivefold respectively compared with that of THLG-EXO/miR-21i-treated cells (Fig. 5D). In addition, Zhan et al. [108] designed the exosome: (1) safe and sufficient blood exosomes; (2) binding the ligand-coupled superparamagnetic nanoparticles to the specific membrane proteins of exosome to achieve the separation, purification and tumor magnetic-targeting; (3) co-loading hydrophobic drugs dox and cholesterol-modified miR-21i to enhance the therapeutic effectiveness; (4) binding L17E peptide to promote the cytosolic release of encapsulated cargos. The engineered exosomes (D-Exos/miR21i-L17E) that met the above four requirements could be highly enriched in tumor targets. The results revealed that compared with Exo/miR21i-L17E groups, antitumor effect in vivo decreased two folds at the 18th day after administration in the D-Exos/miR21i-L17E group. Based on the above, the excellent antitumor could achive through combined therapy. The related research have been summarized in Table 2 in detail.
Exosomes-based siRNA delivery system
siRNA
siRNA is another class of double stranded DNA (dsRNA) with a length of about 25 bp, which could completely complementary to the targeted mRNA, resulting in gene silencing [128–131]. The mechanism is that endogenous dsRNA is recognized by ribonuclease protein Dicer, which cleaves the dsRNA into 21 to 23 bp with 2-nucleotide overhanging at 3′ ends. These cleavage products, named siRNAs, consist of a passenger and guide strands. After binding to the RNA-Induced Silencing Complex (RISC), the guide chain is guided to the target mRNA and cleaved into small fragments by the cleavage enzyme argonaute-2, which is located between bases 10 and 11 at the 5′ ends of the siRNA guide chain [132, 133]. Based on the above, siRNA has the potential to treat a variety of diseases by regulated the expression of target mRNA. Recently, FDA approved the Patisiran (siRNA is delivered to hepatocytes as a lipid complex) and Givosiran (siRNA is coupled to a GalNAc ligand that makes salivary glycoprotein receptors-mediated targeted delivery to hepatocytes) siRNA drugs, marking the beginning of the era of RNA interference (RNAi) therapy [134, 135].However, the successful therapy requires the safe and efficient delivery of siRNA into the cytoplasm to play an interference function.
Loading method for siRNA into exosomes
The concept of delivering siRNA using exosomes was first confirmed by Alvarez-Erviti et al. [136], who electroporating exogenous siRNA into exosomes for delivery both in vitro and in vivo, resulting in the knockdown of the specific gene BACE1. Similarly, some studies loaded siRNA into exosomes by electroporation (Table 3). Furthermore, Wahlgren et al. [137] delivered therapeutic siRNA into exosomes from peripheral blood by electroporation. And the effects of exosome concentration, siRNA concentration, and electroporation parameters on electroporation efficiency were studied. The results revealed that the changes of siRNA and capacitance had no effect on the electroporation efficiency, and when the concentration of exosomes was in the range of 0.25-1 mg/ml, the electroporation efficiency was the highest. In addition, like miRNA, there are also some commercial transfection reagents for transfecting siRNA into exosomes, such as Lipo2000, Lipo3000, Exo-fect Exosome Transfection Reagent, etc. (Table 3). Aqil et al. [138] loaded siRNA into milk exosomes through the Exo-fect Exosome Transfection Reagent, and the loading efficiency was about sixfold higher than that of electroporation. Besides, siRNA loaded into exosomes by sonication could be delivered to breast cancer cells, resulting in a 50% knockdown of an oncogene [139]. Exosomes by sonication induced less siRNA aggregation than electroporation [140]; However, the number of siRNAs entering recipient cells through exosomes is still limited. Therefore, sonication parameters need to be optimized to improve loading efficiency. As the loading methods mature, it is gradually applied to cancer therapy.
Table 3.
The application of exosomes-based siRNA delivery system in cancer treatment in the last decade
| Exosome | The source of exosome | Therapeutic cargo | Loading method | Target gene | Mechanisms | Cancer types (cell lines) | Effects | References |
|---|---|---|---|---|---|---|---|---|
| Engineering exosome (tLyp-1-modified exosome) | HEK293T cells | siR1, siR2, siR3 | Electroporation | SOX2 | Knock-down the target gene expression | Non-small cell lung cancer (A549) | Reducing the stemness of cancer stem cells | [151] |
| Cancer-cell-derived exosome | Autologous breast cancer cells | siS100A4 | Incubation and extrusion method | S100A4 | Down-regulate the expression of S100A4 | Triple-negative breast cancer (4T1) | Inhibiting the growth of malignant breast cancer cells | [152] |
| HEK293T cells | si–c-Met | Lipofectamine 2000 transfection reagent | c-Met | Inhibiting the expression of c-Met | Gastric cancer (SGC7901) | Reversing the drug resistance of gastric cancer cells in vitro, and significantly inhibiting the tumor growth | [146] | |
| Engineering exosome (FA-displaying exosome) | HEK293T cells | Survivin siRNA | ExoFect exosomes transfection reagent | survivin | Knockdown the expression of survivin | Human oral epidermal carcinoma (KB) | Inhibiting tumor growth | [139] |
| Stem-cell-derived exosome | HEK293 cells, mesenchymal stem cell | PLK-1 siRNA | Electroporation | PLK-1 | Knockdown of PLK-1 mRNA and protein | Bladder cancer (UMUC3, SW780) | Inhibiting the bladder cancer cell proliferation | [142] |
| Engineering exosome (DARPin G3- modified exosome) | HEK293T cells | TPD52 siRNA | Electroporation | TPD52 | Binding specifically to HER2/Neu and siRNA molecules against TPD52 gene | Breast cancer (MDA-MB-231) | Inhibiting tumor growth | [153] |
| Normal fibroblast-like mesenchymal cells | KRAS siRNA | Electroporation | KRASG12D | Reducing KRASG12D mRNA levels and phosphorylated-ERK protein levels | Pancreatic cancer (MIA-PaCa-2, Capan-1) | Inhibiting tumor metastasis and increasing overall mouse survival | [154] | |
| HEK293T cells | HGF siRNA | Lipofectamine 2000 transfection reagent | HGF | Activating the HGF/c–Met signaling pathway | Gastric cancer (SGC‐7901) | Suppressing tumor growth and angiogenesis | [155] | |
| Cancer-cell-derived exosome | Cancer-associated fibroblasts | LINC00355 siRNA | Lipofectamine 2000 transfection reagent | LINC00355 | Decreasing the expression of LINC00355 | Bladder cancer (T24, 5367) | Repressing cell proliferation and invasion | [156] |
| Cancer-cell-derived exosome | Breast cancer | MALAT1 siRNA | Lipofectamine 2000 transfection reagent | MALAT1 | Down-regulating the expression of MALAT1 | Breast cancer (MCF-7, MDA-MB-231, MDA-MB-435S) | Suppressing cell proliferation | [157] |
| Cancer-cell-derived exosome | PANC-1 cells | PAK4 siRNA | Electroporation | PAK4 | Down-regulating the expression of PAK4 | Pancreatic cancer (PANC-1) | Inhibiting tumor growth and increasing mice survival | [158] |
| Human skin-derived fibroblasts(NB1RGB cells) | LCP1 siRNA | Electroporation | LCP1 | Suppressing LCP1 expression | Oral cancer (HSC-2, HSC-3, HSC-3-M3, HSC-4, Sa3, Ca9-22, KOSC-2, SAS, Ho-1-u-1, Ho-1-N-1, SAS-H1) | Suppressing the oncogenic activity of cancer cells | [159] | |
| HEK293T cells | TRPP2 siRNA | Incubation | TRPP2 | Suppressing TRPP2 protein expression levels | Human pharyngeal squamous cell carcinoma (FaDu) | Inhibiting migration, invasion and the EMT of cancer cells | [160] | |
| Cancer-cell-derived exosome | MCF-7, MCF-7/ADR cells | CD44 siRNA | Electroporation | CD44 | Suppressing CD44 expression | Breast cancer (MCF-7/ADR) | Reducing cell proliferation and enhancing susceptibility to DOX | [161] |
| MCF10A cells | CDK4 siRNA | Electroporation | CDK4 | Downregulating the CDK4 mRNA and protein expression | Breast cancer (MCF-7) | Inhibiting tumor growth | [162] | |
| HeLa cells | RAD51 siRNA, RAD52 siRNA | Electroporation | RAD51, RAD52 | Downregulating RAD51/RAD52 expression | Human cervical carcinoma (HT1080) | Resulting in apoptosis of the tumor cells | [40] | |
| Engineering exosome (iRGD peptide-modified exosome) | HEK293T cells | KRAS siRNA | Lipofectamine 2000 transfection reagent | KRAS | Silencing KRAS gene expression | Lung cancer (A549) | Inhibiting tumor growth | [163] |
| Stem-cell-derived exosome | Bone-marrow-derived mesenchymal stem cells | GRP78 siRNA | Lipofectamine 2000 transfection reagent | GRP78 | Inhibiting the expression of GRP78 | Hepatocellular carcinoma (HepG2, PLC) | Inhibiting the growth and invasion of the cancer cells | [145] |
| Stem-cell-derived exosome | Bone marrow mesenchymal stem cell | Galectin-9 siRNA | Electroporation | galectin-9 | tumor-suppressive macrophage polarization, cytotoxic T lymphocytes recruitment and Tregs downregulation | Pancreatic ductal adenocarcinoma (PANC-02) | Eliciting anti-tumor immunity | [100] |
| Engineering exosome (cRGD peptide-modified exosome) | RAW 264.7 cells | FGL1 siRNA, TGF-β1 siRNA | Exo-fect Exosome Transfection Reagent | FGL1, TGF-β1 | Blocking immune checkpoint FGL1 | Colorectal cancer(MC38) | An increased number of tumor infiltration CD8 + T cells, a decreased number of immunosuppressive cells, a significant anti-tumor effect | [150] |
| HEK293 cells | SCD-1 siRNA | Electroporation | SCD-1 | Regulating of fatty acids metabolism and increasing ROS level | Anaplastic thyroid carcinoma (Hth-7) | Inhibiting cellular proliferation and promoting cellular apoptosis | [164] | |
| Engineering exosome (iRGD peptide-modified exosome) | HEK-293 T cells | CPT1A siRNA | Lipofectamine 2000 transfection reagent | CPT1A | Regulating fatty acid oxidation | Colon cancer (HCT116, sw480) | Reversing oxaliplatin resistance and inhibiting tumor growth | [147] |
| Natural killer cells NK92MI | BCL-2 siRNA | Co-incubation | BCL-2 | Inhibiting the expression of BCL2 | Breast cancer (MCF-7, SKBR3, T-47D, MDA-MB-231) | Enhancing cancer cell’ intrinsic apoptosis | [141] | |
| Engineering exosome (E3 aptamer- modified exosome) | HEK293T cells | SIRT6 siRNA | Electroporation | SIRT6 | Inhibiting the expression of SIRT6 | Prostate cancer (C42B, DU145) | Inhibiting tumor growth and metastasis | [165] |
| Engineering exosome (EGFR RNA aptamer-modified exosome) | HEK293T cells | Survivin siRNA | ExoFect exosome transfection | survivin | Knockdown the expression of survivin | Non-small-cell lung cancer (A549) | Leading to potent gene knockdown, chemotherapy sensitization, and tumor regression | [166] |
| Engineering exosome (RNA nanotechnology- modified exosome) | HEK293T cells | Survivin siRNA | survivin | Knockdown the expression of survivin | Prostate cancer breast cancer colorectal cancer | Inhibiting tumor regression | [143] |
Delivery of therapeutic siRNA
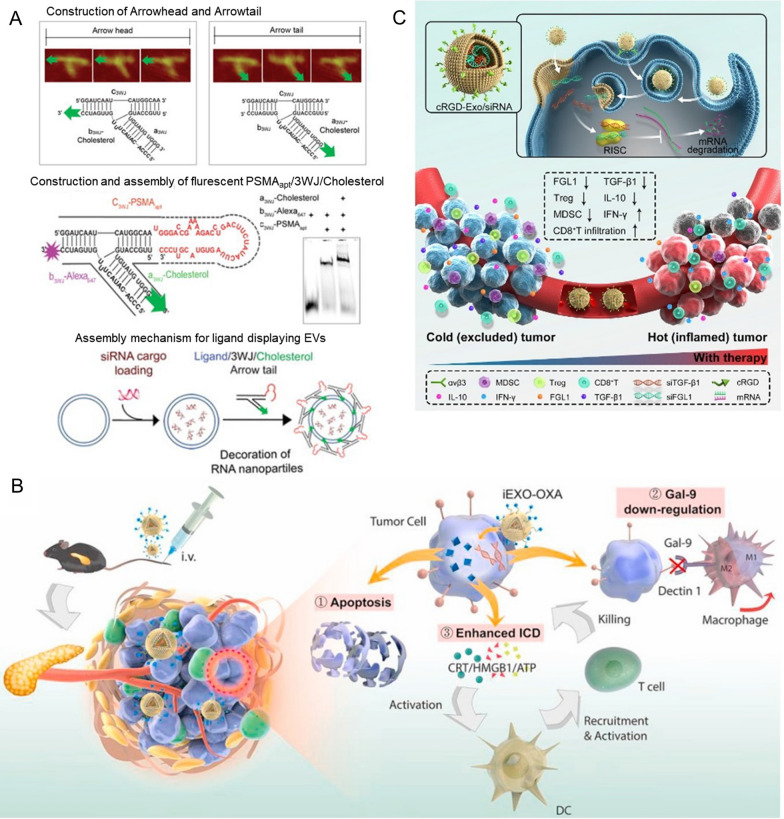
As we all know, cancer progression is related to the up-regulation of anti apoptotic protein such as BCL-2, PLK1, KRAS, survivin protein that initiates cell mitosis, and cell growth factor. siRNA exosomal therapy targeting tumor cells has been been committed to downregulating these oncogenes expression to inhibit cell proliferation and migration. Kaban et al. [141] loaded BCL-2 siRNA into natural killer (NK) cell-derived exosomes to treat ER + breast cancer, leading to enhanced apoptosis in breast cancer cells. Similarly, exosomes mediated the delivery of PLK-1 siRNA into bladder cancer cells, promoting cell apoptosis through silencing PLK-1 expression [142]. Additionally, Pi et al. [143] designed RNA nanoparticles-modified exosome to simultaneously target three cancer cells (breast cancer, prostatic cancer and colorectal cancer). In the structure of the RNA nanoparticle, the pRNA of phage phi29 (an RNA molecule with transport function) was extended into an arrow shape and connected with an RNA ligand (used to target and bind specific overexpressed receptors in tumor cells) and added the fluorescent dye alexa647 for imaging. The three RNA ligands were designed: prostate cancer specific membrane antigen RNA ligand, epidermal growth factor RNA ligand and folic acid ligand. And then this modified exosomes were used to deliver survivin siRNA. The results revealed that compared with the control (injection PBS), during the entire treatment period, the growth of breast and prostate cancer cells treated with the above exosome loaded with survivin siRNA was completely inhibited,the growth inhibition rate of colorectal cancer cell treated with the functional exosome increased onefold (Fig. 6A).
Fig. 6.
A RNA nanoparticle and RNA nanoparticles-modified exosome [143]. B Exosomes co-delivery chemotherapy drugs oxaliplatin (OXA) and nucleic acid drugs gal-9 siRNA to enhancing immunotherapy and reprogramming tumor microenvironment (TME) [100]. C cRGD-modified exosome with high siFGL1 and siTGF-β1 loading efficiency to realize the co-silence of FGL1 and TGF-β1 to to block immune checkpoints and simultaneously regulate TME [150]
As mentioned above, exosome mediated siRNA delivery can effectively inhibit the proliferation and migration of cancer cells. However, drug resistance is another major challenge in cancer treatment. Generally speaking, overexpression of chemotherapy resistance-associated proteins caused drug resistance in cancer cells [144]. Utilizing siRNA to overcome drug resistance has been widely reported. For example, Li et al. [145] used exosomes from bone marrow mesenchymal stem cells (BM-MSC) to deliver siRNA against Grp78 (overexpression in hepatocellular carcinoma and could promote the drug resistance to Sorafenib) in Sorafenib-sensitive hepatocellular carcinoma cells, leading to sorafenib-resistant cancer cells’ sensitivity sorafenib and the reversal of drug resistance. Similarly, Zhang et al. [146] reported that si-c-Met delivered by exosome showed a better inhibitory effect on the expression of c-Met (an essential role in drug resistance of various tumors) and significantly enhanced drug sensitivity. In addition, fatty acid oxidation (FAO) plays a crucial role in drug resistance of cancer cells. Carnitine palmitoyltransferase 1A (CPT1A), a key enzyme of FAO, is widely considered as an emerging therapeutic target. Lin et al. [147] utilized iRGD-modified exosomes to specifically deliver siCPT1A into colon cancer cells to suppress FAO, which have reversed the sensitivity of drug-resistant colon cancer cells to oxaliplatin. The above methods can effectively alleviate the drug resistance of cancer cells and provide new ideas for cancer treatment.
In addition, cancer immunotherapy utilizes the patient’s immune system to identify and destroy cancer cells, which is a specific protective strategy for cancer treatment [148]. Athough immune cells are common in the tumor microenvironment (TME), accounting for about 50% of the stromal cell components, only a few are anti-tumor effector cells, which may be responsible for the immune escape of tumor cells [149]. Thus, it is necessary for tumor immunotherapy to target TME and/or immune checkpoints. For example, Zhou et al. [100] designed a bio-platform targeting pancreatic ductal adenocarcinoma (PDAC) to enhance immunotherapy and reprogram TME. In this platform, exosomes derived from BM-MSCs as carrier co-delivered oxaliplatin (OXA) and gal-9 siRNA. Among them, OXA could both kill tumors and induce immunogenic cell apoptosis. siRNA interfered with the galectin-9 synthesis in tumor cells to reduce the transformation of macrophage M1. Compared with the chemotherapy or gene therapy alone, this combination treatment produced synergetic effects that affects cellular crosstalk in vivo, leading to overall change in TME, so as to further improve the antitumor efficacy, and the inhibition rate of cell growth was about twice higher (Fig. 6B). Furthermore, Pei et al. [150] established a cRGD-modified exosome with fibrinogen-like protein 1 (FGL1, an important immune checkpoint) siRNA and transforming growth factor-β (TGF-β1, an immunosuppressive cytokine in TME) siRNA (cRGD-Exo/siMix) to co-silence of FGL1 and TGF-β1. The results revealed that FGL1 expression was inhibited, which activated T cell recognition. Meanwhile, TGF-β1 expressiom was also silenced, which disaired the immunosuppressive microenvironment of tumor and promoted the infiltration of immune cells (Fig. 6C).
In general, exosome mediate the delivery of siRNA to silence the expression of key genes related to cell proliferation, drug resistance, immune checkpoints and TME, which will inhibit the development of tumor. Additionally, the simultaneous use of several siRNAs, and siRNA combined with chemotherapeutic drugs will achieve synergistic effects. The application of exosomes carrying siRNA for cancer treatment are summarized in Table 3 in detail. These methods will provide a reference for cancer treatment by using siRNA.
Exosomes-based circRNAs delivery system
circRNAs
circRNAs are covalently closed RNA molecules without 5′ caps and 3′ tails, generated by a process of back-splicing [167]. The length ranges from hundreds to thousands of nucleotides, and they are highly abundant in eukaryotes [168]. The circRNAs play important roles in the occurrence and development of human diseases, especially cancer, which can regulate multiple cancer-related biological processes. The main mechanisms are as follows [169]: (a) acting as miRNA proton sponges: circRNA competitively binds miRNA to regulate the expression of miRNA and its target genes; (b) regulating gene transcription: exon–intron circRNAs (EIciRNAs) interact with U1 small nuclear ribonucleoprotein (snRNP) to form the EIciRNAs-U1 snRNP complex, which binds to polymerase II (Pol II) to regulate the promoter region of host gene transcription; (c) binding with protein: circRNAs act as protein sponges or baits to regulate gene expression; (d) encoding small functional peptides: circRNAs have ribosome binding sites and stable open reading frame (ORF), which can encode corresponding peptides. For example, CiRS‐7, one of the most famous circRNAs, acts as more than 70 conventional miR-7-binding sites and modulates the expression of multiple cancer-related genes [170, 171]. circRNA_FoxO3 can be used as a protein scaffold of MDM2 and p53 to induce p53 degradation, which can induce cancer cell apoptosis [172]. circRNA_SHPRH encodes protein SHPRH-146aa, which function as a bait to protect SHPRH protein from ubiquitination through DTL mediated degradation, so as to inhibit glioma occurrence [173]. Although the circRNAs have been served as one of the most promising biomolecules for cancer therapy, their delivery efficiency is often limited by the selected delivery system.
Delivery of therapeutic circRNA
circRNA naturally carried by exosomes has been widely developed for cancer treatment, which greatly improve its therapeutic effect owing to the high delivery efficiency. For example, Xue et al. [174] reported that exosomal circRNA_100284 acted as a sponge of miR-217, inhibiting cell proliferation by inducing a G2/M phase arrest in the cell cycle and targeting enhancer of zeste homolog (EZH) in various cancers. Chen et al. [175] introduced that exosomal circ-0051443 suppressed the hepatocellular carcinoma progression through competitive bounding to miR-331-3p. However, exosome-mediated exogenous circRNA delivery also faces some challenges. For one thing, the special circular structure of circRNA leads to the low circular efficiency. For another, macromolecular circRNAs also face the same problem as the large-size mRNA discussed above, which is difficult to load into the exosomes. To solve these problems, Yu et al. [176] constructed the target circRNA_DYM coding DNA into the lentivirus expression vector and then combined RVG-Lamp2b plasmid to transfected them into the HEK293T cells, and the engeneered exosome stably overexpressing the target circRNA_DYM (RVG-circDYM-EX) were secreted. This not only made the circRNA correctly and efficiently cyclized, but also could be easily loaded into the exosomes. The RVG-circDYM-EX was delivered to the brain to attenuate astrocyte disfunction induced by chronic unpredictable stress through binding to the transcription factor 1 (TAF1) and downregulating multiple downstream genes (Trpm6, Cyp39al). Similarly, Yang et al. [177] obtained the engeneered exosome modified with RVG-Lamp2b and loaded with circRNA_ SCMH1 and successfully transported them to the brain. The results revealed that the delivery system promoted functional recovery of rodent and non-human primate ischemic stroke models through binding to the methyl-CpG binding protein 2 and upregulating the expression of the target genes (Mobp, Igfbp3, Fxyd1 and Prodh).
In conclusion, the circular structure of circRNA can not only prevent being degraded and improve the expression time and amount of circRNA, but also be administered repeatedly, which makes it one of the emerging nucleic acid drugs. Some natural exosomal circRNA can play an important role in cancer therapy. And exogenous circular RNA can also be cyclized efficiently by constructing related lentivirus vectors, and can be loaded into exsome through transfecting the vectors into the target cells, which will provide references for the application of this system in cancer.
Exosomes-based other nucleic acids delivery system
Other nucleic acids
Other nucleic acid drugs, including long noncoding RNA (lncRNA), short hairpin RNA (shRNA), aptamer, etc., have been also introduced into cancer therapy. LncRNA, an RNA family with many members, has a length of over 200 bp and cannot be transformed into protein. Although it does not have the function of traditional RNA, it can regulate the activity of transcription factors [178, 179]. Moreover, some lncRNAs play a curical role in tumor proliferation, apoptosis, diffusion, and homeostasis maintenance [180–182]. For shRNA, structurally, it is more similar to miRNA, and both of them are local double-stranded RNA formed by hairpin structure [183]; Functionally, it is closer to siRNA, which is cleaved by the Dicer to form siRNA, and then performs interference through the siRNA pathway [184]. The shRNA is also a critical effector molecule in RNAi technology, and it could induce target mRNA degradation [185]. Another nucleic acid fragment, aptamer is a single-stranded DNA or RNA that can bind with different targets, such as chemical molecules, RNA, DNA or protein with high affinity and specificity to block protein–protein or receptor–ligands interactions. Pegaptanib (macugen), The first PEGylated RNA aptamer drug, pegaptanib (macugen), was approved by FDA in 2004, binding to extracellular VEGF165 with high specificity and affinity [186]. It can be seen that these RNAs will also play a key role in the cancer treatment.
Delivery of therapeutic other nucleic acids
Exosomes can also deliver these nucleic acids for cancer therapy. For instance, Zheng et al. [187] transfected the lncRNA PTENP1 lentiviral vector into HEK293A cells, and then secreted exosomes contained PTENP1. Eventually the exosomal PTENP1 protected PTEN by sponging miR-17 and inhibited the biomalignant behavior of bladder cancer. Similarly, Zheng et al. [188] obtained exosomal circLPAR1 by the same methods, which could suppress colorectal cancer cell growth through suppressing BRD4 expression via METTL3-eIF3h interaction. In addition, aptamers are often used to modify exosomes to enhance their ability to target tumors. Exosomes derived from HEK293T cells were modified by A9g (PSMA) aptamer and loaded with survivin siRNA, which could be specifically delivered to tumors and effectively block tumor growth [143]. All in all, these system are emerging, and its their successful delivery will also contribute to cancer therapy.
Exosome-based clinical applications for cancer treatment
As the discussed above, exosomes are a class of ideal drug delivery tool, which have also been performed in cancer clinical trials. The database www.ClinicalTrials.gov (accessed on April 2022), has been examined to assess the major exosomes’ clinical applications. 105 trials are registered within the study object “exosome” and “cancer”.
Table 4 summarizes the studies related to using “exosomes” for cancer therapy. Among them, immature DC-derived exosomes have been applied for melanoma and NSCLC with similar safety results. In addition, two clinical trials investigating plant-derived exosomes as cancer therapy are currently under way. At present, two clinical trials are on-going to study plant-derived exosomes for cancer treatment. In the first trial, grape-derived exosome-like nanoparticles are being tested for their effects on oral mucositis and related pain after radiotherapy and chemotherapy for head and neck cancer (NCT01668849). In the second study, plant-derived exosomes loaded with curcumin are being evaluated for their efficacy for treating colorectal cancer after oral administration (NCT01294072). The clinical studies on exosome-loaded nucleic acids for cancer treatment have been also “completed” or “ongoing”. For example, the phase I trial (NCT03608631) sponsored by the M.D. Anderson Cancer Center (Texas, USA) have investigated the use of MSC derived exosomes for the treatment of stage IV pancreatic cancer patients with KrasG12D mutation. The patients were injected with KrasG12D siRNA loaded into exosomes which targeted the oncogenic KRAS gene, reducing its expression in pancreatic tumors [154]. In addition, the clinical trials that tumor cell-derived exosome delivers ASO to treat malignant glioma of brain and neoplasms have been completed. In total, these exosome-based clinical applications for cancer treatment demonstrate the reliability of these delivery systems once again.
Table 4.
Exosome-based clinical applications for cancer treatment from clinical trials.com and references
| Cancer | Phase | Start year | Source of exosome | Therapeutic cargo | Status | Sponsor | Clinical trial number/Reference |
|---|---|---|---|---|---|---|---|
| Metastatic pancreas cancer with KrasG12D mutation | I | 2018 | Mesenchymal stromal cells | krasG12D siRNA | Ongoing | M.D. Anderson Cancer Center, Houston, Texas, United States | NCT03608631 |
| Non-small cell lung cancer | II | 2010 | Dendritic cells | Metronomic cyclophosphamide | Completed | Institute Gustave Roussy, Villejuif, France | NCT01159288 |
| Colon cancer | I | 2011 | Plant | Curcumin | Recruiting | University of Louisville Hospital, Louisville, Kentucky, United States | NCT01294072 |
| Head and neck cancer | I | 2012 | Grape | Lortab, fentanyl patch, mouthwash | Active, not recruiting | James Graham Brown Cancer Center, Louisville, Kentucky, United States | NCT01668849 |
| Malignant glioma of brain | I | 2012 | Tumor cells | IGF-1R antisense oligodeoxynucleotide | Completed | Thomas Jefferson University Hospital; Jefferson Hospital for Neurosciences, Philadelphia, Pennsylvania, United States | NCT01550523 |
| Malignant glioma neoplasms | I | 2015 | Tumor cells | IGF-1R antisense oligodeoxynucleotide | Completed | Thomas Jefferson University Hospital, Philadelphia, Pennsylvania, United States | NCT02507583 |
| Metastatic melanoma | I | 2000 | Autologous dendritic cell | Pulsed with MAGE 3 tumor peptides | Completed | Institute Curie, Paris, France | [189] |
| Non-small cell lung cancer | I | 2000 | Autologous dendritic cell | Pulsed with MAGE-A3, -A4, -A10, and MAGE-3DPO4 tumor peptides | Completed | Duke University Medical Center, Durham, NC, USA | [190] |
| Colorectal cancer | I | 2006 | Autologous ascites | The granulocyte–macrophage colony-stimulating factor | Completed | The Fourth Hospital Affiliated to Guangxi Medical University, Liuzhou, China | [191] |
Conclusions and future perspectives
Lower immunogenicity, lower toxicity, and better crossing biological barriers are the key advantages of exosomes over traditional nanocarriers. Based on these advantages, exosomes have shown great value in nucleic acid delivery, and can protect therapeutic substances from degradation and clearance by the host immune system. Additionally, the inherent targeting ability derived from their parental cells makes exosomes possess the potential of targeted delivery, enhancing the ability to penetrate the tumor vascular barrier and bioaccumulation at tumor sites, greatly improving their therapeutic efficacy. What’s more, therapeutic applications of exosomes as drug delivery vectors have been explored in numerous preclinical studies and several clinical trials. Thus, exosome-based delivery systems have unique advantages in cancer treatment. In this review, recent studies of using exosomes to deliver different nucleic acids (DNA, mRNA, miRNA, siRNA, circRNA, etc.) to treat various cancers are summarized.
Although significant progress has been made, some challenges hinder the exosomal therapeutic application. The first challenge is the large-scale production of exosomes for clinical trials. To increase the production of exosomes, bioreactors, 3D scaffolds, and microfluidic devices are adopted. For example, Haraszti et al. [192] applied 3D culture combined with tangential flow filtration (TFF) to increase the production of exosomes by 140-fold compared with 2D or 3D cultures or TFF. Another study found that using a hollow fiber bioreactor could increase the yield of exosomes by 40-fold [193]. Yang et al. [94] reported a cellular nanoporation (CNP) method to produce a large number of exosomes. The results revealed that compared with the traditional strategies (bulk electroporation and Lipo2000 transfection), CNP produced up to 50-fold more exosomes. The method of separation and purification of exosomes based on microfluidic devices also showed promising results [194–196]. Wang et al. [194] reported that a 3D nanostructured microfluidic chip could capture 90% of exosomes. In addition, some studies have shown that stress environments such as hypoxia, low pH and anticancer drugs could stimulate the production of more exosomes [197–199]. Also, food-derived exosomes, including bovine milk- and grape-derived exosomes, have shown promising results in preclinical studies [200, 201]. However, quality should be guaranteed while increasing output, especially considering the contamination or the size overlap between exosomes and other EVs.
The second challenge is to develop new methods for loading nucleic acids into exosomes to make up for defects of traditional methods. The low loading efficiency of current exosome-nucleic acid-loading strategies, including electroporation, incubation, and transfection, limits their application. For example, although electroporation is considered to be the best method for loading nucleic acids into exosomes, this method is easy to lead to the aggregation and degradation of nucleic acids and the change of exosome properties [202]. In addition, the simple incubation method is very limited in the type of the loaded cargo [203]. Transfection methods should further simplify the process and reduce the cost of mass production. To solve these problems, some potential new nucleic acid loading methods have been developed. For instance, Li et al. [91] constructed the fusion protein CD9-HuR through fusing the exosomal membrane protein CD9 with the RNA binding protein HuR, which would selectively enrich the target RNA into the exosomes. The results revealed that the specific RNA miRNA155 loaded into CD9-HuR modified exosomes was sevenfold higher than that of the control group. In addition, studies have shown that exosome-enriched RNAs shared three specific sequence motifs, including CAGUGAGC, UAAUCCCA, and ACCAGCCU, which played a role as cis-acting elements targeting to exosomes and helped us modify the exosomes to selectively enrich more candidate RNAs for therapeutic purposes [204]. Besides, it is necessary to develop new and efficient LNPs transfected with nucleic acid into exosomes. Wang et al. [205] developed a physical–chemical hybrid platform involving cationic LNPs exposed to cyclic stretch, which could effectively deliver siRNAs and plasmid DNAs. And the gene silencing efficiency was about 10% higher than that of commercial transfection reagents Lipo2000. Similarly, Hu et al. [206] developed thermostable ionizable lipid-like nanoparticles to deliver siRNA, which had good physical and chemical properties, thermal stability (not degraded at 40 °C for one week), and excellent siRNA transport efficiency (the same as that of Lipo2000). These techniques will provide new ideas for loading nucleic acid into exosomes.
The third challenge is to achieve personalized precision treatment of cancer. The heterogeneity of exosomes and complex in vivo environment limit the precise delivery and expected efficacy. To solve this problem, the cancer patient’s autologous exosomes can be the best choice of delivery carriers owing to the remarkable targeting ability against cancer cells. Among them, tumor cells can be obtained from minimally invasive and surgical samples and expanded in vitro under specific culture conditions. Gong et al. [207] domesticated tumor cells from gastric cancer patients under acidic conditions similar to the tumor tissue environment and found that the acidic environment would promote tumor cells to secrete more exosomes and improve the ability of these exosomes to be taken up by homologous gastric cancer cells. Futhermore, Wang et al. [208] obtained immune activated macrophage tumor hybrid cells by in vitro immune acclimation of patients’ own tumor cells and macrophages. The macrophage tumor chimeric exosomes secreted by these cells could inherit the functions of two kinds of source cells: (a) activating tumor specific immune response; (b) inheriting the homing ability of tumor cells and actively target tumor tissues. In addition, fluorescence, luminescence, PET-MRI and SPECT imaging techniques are used to track the biological distribution of exosomes and payloads, and obtain images with anatomical details, which will provide accurate and customized medical care for cancer patients [209].
Fourthly, considering that exosomes contain a group of discrete proteins and functional immune molecules, the application of exosomes may trigger a strong response of the host immune systems to a certain extent, resulting in the rapid disappearance of exosome-based drug delivery systems. Therefore, a comprehensive preclinical examination, including pharmacokinetics, toxicity characteristics and pharmacodynamics, should be performed to prevent potential side effects.
Abbreviations
- mRNA
Messenger RNA
- miRNA
MicroRNA
- siRNA
Small interfering RNA
- circRNA
Circular RNA
- EVs
Extracellular vesicles
- LNPs
Lipid nanoparticles
- SIRPα
Signal regulatory protein alpha
- HEK293
Human embryonic kidney
- DC
Dendritic cell
- NSCLC
Non-small cell lung cancer
- HEK
Human embryonic kidney
- CaCl2
Calcium chloride
- PC
Phosphatidylcholine
- HMGN1
High mobility group nucleosome-binding protein 1
- ASOs
Antisense oligonucleotides
- FDA
Food and drug administration
- exo-AAV
Exosome-associated adeno-associated virus
- LHFPL5
lipoma HMGIC fusion partner-like 5
- HA
hemagglutinin
- PARP-1
poly (ADP-ribose) polymerase-1
- MSC
Mesenchymal stem cell
- EXOtic
EXOsomal transfer into cells
- RBCs
Red blood cells
- 3′-UTR
3′ untranslated region
- AMOs
Anti-miRNA oligonucleotides
- miR mimics
miRNA mimics
- EMT
Epithelial–mesenchymal transition
- OSCC
Oral squamous cell carcinoma
- Dox
Doxorubicin
- 5-FU
5-Fluorouracil
- TAT
Transcriptional transactivator
- dsRNA
Double stranded DNA
- RISC
RNA-induced silencing complex
- RNAi
RNA interference
- RVG
Rabies virus glycoprotein
- NK
Natural killer
- BM-MSC
bone marrow mesenchymal stem cells (BM-MSC)
- FAO
Fatty acid oxidation
- CPT1A
Carnitine palmitoyltransferase 1A
- TME
Tumor microenvironment
- PDAC
Pancreatic ductal adenocarcinoma
- OXA
Oxaliplatin
- FGL1
Fibrinogen-like protein 1
- TGF-β1
Transforming growth factor-β
- EIciRNAs
Exon–intron circRNAs
- snRNP
Small nuclear ribonucleoprotein
- Pol II
Polymerase II
- ORF
Open reading frame
- EZH
Zeste homolog
- lncRNA
Long noncoding RNA
- shRNA
Short hairpin RNA
- TFF
Tangential flow filtration
- CNP
Cellular nanoporation
Author contributions
YZ wrote the manuscript. JL and CY revised the manuscript. QL, XZ, HH, ST, ZR, YC, XC and ML edited the different sections of the manuscript. All authors read and approved the final manuscript.
Funding
This work was granted by the National Natural Science Foundation of China (81900264), Natural Science Foundation of Guangdong Province (2019A1515012163, 2021A1515012159), Guangdong MedicalScience and Technology Research Funding(A2019359), the Shenzhen Science and Technology Innovation Fund (JCYJ20210324130609027), the Project of Administration of Traditional Chinese Medicine Guangdong Province (20211347), University Stable Support Research Funding of Shenzhen (20200813153346001), the Longgang Medical and Health Science and Technology Project (LGKCYLWS2020040 and LGKCYLWS2020044), SZU Top Ranking Project (86000000210).
Availability of data and materials
Not applicable.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jia Liu, Email: liujia870702@126.com.
Chengbin Yang, Email: cbyang@szu.edu.cn.
References
- 1.Zaimy MA, Saffarzadeh N, Mohammadi A, et al. New methods in the diagnosis of cancer and gene therapy of cancer based on nanoparticles. Cancer Gene Ther. 2017;24:233–243. doi: 10.1038/cgt.2017.16. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Fuchs HE, et al. Cancer statistics, 2021. CA Cancer J Clin. 2021;71:7–33. doi: 10.3322/caac.21654. [DOI] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 4.Ramasamy T, Munusamy S, Ruttala HB, et al. Smart nanocarriers for the delivery of nucleic acid-based therapeutics: a comprehensive review. Biotechnol J. 2021;16:e1900408. doi: 10.1002/biot.201900408. [DOI] [PubMed] [Google Scholar]
- 5.Yang CB, Hu R, Anderson T, et al. Biodegradable nanoparticle-mediated K-ras down regulation for pancreatic cancer gene therapy. J Mater Chem B. 2015;3:2163–2172. doi: 10.1039/c4tb01623h. [DOI] [PubMed] [Google Scholar]
- 6.Zhang XM, Lin ZL, Yang JY, et al. Carbon dioxide-derived biodegradable and cationic polycarbonates as a new siRNA carrier for gene therapy in pancreatic cancer. Nanomaterials (Basel) 2021;11:2312. doi: 10.3390/nano11092312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alexandre B, Jeremy M, Rémy S, et al. Extracellular vesicles: future diagnostic and therapeutic tools for liver disease and regeneration. Liver Int. 2019;39:1801–1817. doi: 10.1111/liv.14189. [DOI] [PubMed] [Google Scholar]
- 8.Bunggulawa EJ, Wang W, Yin TY, et al. Recent advancements in the use of exosomes as drug delivery systems. J Nanobiotechnol. 2018;16:81. doi: 10.1186/s12951-018-0403-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Soltani F, Parhiz H, Mokhtarzadeh A, et al. Synthetic and biological vesicular nano-carriers designed for gene delivery. Curr Pharm Des. 2015;21:6214–6235. doi: 10.2174/1381612821666151027153410. [DOI] [PubMed] [Google Scholar]
- 10.Kim H, Jang H, Cho H, et al. Recent advances in exosome-based drug delivery for cancer therapy. Cancers. 2021;13:4435. doi: 10.3390/cancers13174435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim H, Kim EH, Kwak G, et al. Exosomes: cell-derived nanoplatforms for the delivery of cancer therapeutics. Int J Mol Sci. 2020;22:14. doi: 10.3390/ijms22010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pan SJ, Zhang YH, Huang M, et al. Urinary exosomes-based engineered nanovectors for homologously targeted chemo-chemodynamic prostate cancer therapy via abrogating EGFR/AKT/NF-kB/IkB signaling. Biomaterials. 2021;275:120946. doi: 10.1016/j.biomaterials.2021.120946. [DOI] [PubMed] [Google Scholar]
- 13.Li XM, Tsibouklis J, Weng TT, et al. Nano carriers for drug transport across the blood-brain barrier. J Drug Target. 2017;25:17–28. doi: 10.1080/1061186X.2016.1184272. [DOI] [PubMed] [Google Scholar]
- 14.Vlassov AV, Magdaleno S, Setterquist R, et al. Exosomes: current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials. Biochim Biophys Acta. 2012;1820:940–948. doi: 10.1016/j.bbagen.2012.03.017. [DOI] [PubMed] [Google Scholar]
- 15.Tan A, Rajadas J, Seifalian AM. Exosomes as nano-theranostic delivery platforms for gene therapy. Adv Drug Deliv Rev. 2013;65:357–367. doi: 10.1016/j.addr.2012.06.014. [DOI] [PubMed] [Google Scholar]
- 16.Azmi AS, Bao B, Sarkar FH. Exosomes in cancer development, metastasis, and drug resistance: a comprehensive review. Cancer Metastasis Rev. 2013;32:623–642. doi: 10.1007/s10555-013-9441-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mathivanan S, Ji H, Simpson RJ. Exosomes: extracellular organelles important in intercellular communication. J Proteomics. 2010;73:1907–1920. doi: 10.1016/j.jprot.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 18.Harding CSP. Transferrin recycling in reticulocytes: pH and iron are important determinants of ligand binding and processing. Biochem Biophys Res Commun. 1983;113:650–658. doi: 10.1016/0006-291x(83)91776-x. [DOI] [PubMed] [Google Scholar]
- 19.Pan BT, Johnstone RM. Fate of the transferrin receptor during maturation of sheep reticulocytes in vitro: selective externalization of the recepto. Cell. 1983;33:967–978. doi: 10.1016/0092-8674(83)90040-5. [DOI] [PubMed] [Google Scholar]
- 20.Fuhrmann G, Herrmann IK, Stevens MM. Cell-derived vesicles for drug therapy and diagnostics: opportunities and challenges. Nano Today. 2015;10:397–409. doi: 10.1016/j.nantod.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liang GF, Kan S, Zhu YL, et al. Engineered exosome-mediated delivery of functionally active miR-26a and its enhanced suppression effect in HepG2 cells. Int J Nanomedicine. 2018;13:585–599. doi: 10.2147/IJN.S154458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu HY, Chen L, Peng YJ, et al. Dendritic cells loaded with tumor derived exosomes for cancer immunotherapy. Oncotarget. 2018;9:2887–2894. doi: 10.18632/oncotarget.20812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu XH, Badawi M, Pomeroy S, et al. Comprehensive toxicity and immunogenicity studies reveal minimal effects in mice following sustained dosing of extracellular vesicles derived from HEK293T cells. J Extracell Vesicles. 2017;6:1324730. doi: 10.1080/20013078.2017.1324730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bellavia D, Raimondo S, Calabrese G, et al. Interleukin 3- receptor targeted exosomes inhibit in vitro and in vivo Chronic Myelogenous Leukemia cell growth. Theranostics. 2017;7:1333–1345. doi: 10.7150/thno.17092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ostrowski M, Carmo NB, Krumeich S, et al. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat Cell Biol. 2010;12:19–30. doi: 10.1038/ncb2000. [DOI] [PubMed] [Google Scholar]
- 26.Qiao L, Hu SQ, Huang K, et al. Tumor cell-derived exosomes home to their cells of origin and can be used as Trojan horses to deliver cancer drugs. Theranostics. 2020;10:3474–3487. doi: 10.7150/thno.39434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shenoda BB, Ajit SK. Modulation of immune responses by exosomes derived from antigen-presenting cells. Clin Med Insights Pathol. 2016;9:1–8. doi: 10.4137/CPath.S39925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pitt JM, Charrier M, Viaud S, et al. Dendritic cell-derived exosomes as immunotherapies in the fight against cancer. J Immunol. 2014;193:1006–1011. doi: 10.4049/jimmunol.1400703. [DOI] [PubMed] [Google Scholar]
- 29.Luan X, Sansanaphongpricha K, Myers I, et al. Engineering exosomes as refined biological nanoplatforms for drug delivery. Acta Pharmacol Sin. 2017;38:754–763. doi: 10.1038/aps.2017.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kok VC, Yu CC. Cancer-derived exosomes: their role in cancer biology and biomarker development. Int J Nanomed. 2020;15:8019–8036. doi: 10.2147/IJN.S272378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baek G, Choi H, Kim Y, et al. Mesenchymal stem cell-derived extracellular vesicles as therapeutics and as a drug delivery platform. Stem Cells Transl Med. 2019;8:880–886. doi: 10.1002/sctm.18-0226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yeo RW, Lai RC, Zhang B, et al. Mesenchymal stem cell: an efficient mass producer ofexosomes for drug delivery. Adv Drug Deliv Rev. 2013;65:336–341. doi: 10.1016/j.addr.2012.07.001. [DOI] [PubMed] [Google Scholar]
- 33.Usman WM, Pham TC, Kwok YY, et al. Efficient RNA drug delivery using red blood cell extracellular vesicles. Nat Commun. 2018;9:2359. doi: 10.1038/s41467-018-04791-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Munir J, Lee M, Ryu S. Exosomes in food: health benefits and clinical relevance in diseases. Adv Nutr. 2020;11:687–696. doi: 10.1093/advances/nmz123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen WY, Wang R, Li D. Comprehensive analysis of the glycome and glycoproteome of bovine milk-derived exosomes. J Agric Food Chem. 2020;68:12692–12701. doi: 10.1021/acs.jafc.0c04605. [DOI] [PubMed] [Google Scholar]
- 36.Adriano B, Cotto NM, Chauhan N, et al. Milk exosomes: Nature’s abundant nanoplatform for theranostic applications. Bioact Mater. 2021;6:2479–2490. doi: 10.1016/j.bioactmat.2021.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suharta S, Barlian A, Hidajah AC, et al. Plant-derived exosome-like nanoparticles: a concise review on its extraction methods, content, bioactivities, and potential as functional food ingredient. J Food Sci. 2021;86:2838–2850. doi: 10.1111/1750-3841.15787. [DOI] [PubMed] [Google Scholar]
- 38.Xi XM, Xia SJ, Lu R. Drug loading techniques for exosome-based drug delivery systems. Pharmazie. 2021;76:61–67. doi: 10.1691/ph.2021.0128. [DOI] [PubMed] [Google Scholar]
- 39.Das CK, Jena BC, Banerjee I, et al. Exosome as a novel shuttle for delivery of therapeutics across biological barriers. Mol Pharm. 2019;16:24–40. doi: 10.1021/acs.molpharmaceut.8b00901. [DOI] [PubMed] [Google Scholar]
- 40.Shtam TA, Kovalev RA, Varfolomeeva EY, et al. Exosomes are natural carriers of exogenous siRNA to human cells in vitro. Cell Commun Signal. 2013;11:88. doi: 10.1186/1478-811X-11-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ohno S, Takanashi M, Sudo K, et al. Systemically injected exosomes targeted to EGFR deliver antitumor microRNA to breast cancer cells. Mol Ther. 2013;21:185–191. doi: 10.1038/mt.2012.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sutaria DS, Jiang J, Elgamal OA, et al. Low active loading of cargo into engineered extracellular vesicles results in inefficient miRNA mimic delivery. J Extracell Vesicles. 2017;6:1333882. doi: 10.1080/20013078.2017.1333882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou W, Xu MM, Wang ZP, et al. Engineered exosomes loaded with miR-449a selectively inhibit the growth of homologous non-small cell lung cancer. Cancer Cell Int. 2021;21:485. doi: 10.1186/s12935-021-02157-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rezaei R, Baghaei K, Amani D, et al. Exosome-mediated delivery of functionally active miRNA-375-3p mimic regulate epithelial mesenchymal transition (EMT) of colon cancer cells. Life Sci. 2021;269:119035. doi: 10.1016/j.lfs.2021.119035. [DOI] [PubMed] [Google Scholar]
- 45.Zhang D, Lee H, Zhu ZW, et al. Enrichment of selective miRNAs in exosomes and delivery of exosomal miRNAs in vitro and in vivo. Am J Physiol Lung Cell Mol Physiol. 2017;312:1110–1121. doi: 10.1152/ajplung.00423.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sayyed AA, Gondaliya P, Mali M, et al. MiR-155 inhibitor-laden exosomes reverse resistance to cisplatin in a 3D tumor spheroid and xenograft model of oral cancer. Mol Pharm. 2021;18:3010–3025. doi: 10.1021/acs.molpharmaceut.1c00213. [DOI] [PubMed] [Google Scholar]
- 47.Katakowski M, Buller B, Zheng X, et al. Exosomes from marrow stromal cells expressing miR-146b inhibit glioma growth. Cancer Lett. 2013;335:201–204. doi: 10.1016/j.canlet.2013.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yao SR, Yin Y, Jin GY, et al. Exosome-mediated delivery of miR-204-5p inhibits tumor growth and chemoresistance. Cancer Med. 2020;9:5989–5998. doi: 10.1002/cam4.3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hu SQ, Wang XY, Li ZH, et al. Platelet membrane and stem cell exosome hybrid enhances cellular uptake and targeting to heart injury. Nano Today. 2021;39:101210. doi: 10.1016/j.nantod.2021.101210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang Y, Bi J, Huang J, et al. Exosome: a review of its classification, isolation techniques, storage, diagnostic and targeted therapy applications. Int J Nanomedicine. 2020;15:6917–6934. doi: 10.2147/IJN.S264498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Salunkhe S, Dheeraj Basak M, et al. Surface functionalization of exosomes for target-specific delivery and in vivo imaging & tracking: Strategies and significance. J Control Release. 2020;326:599–614. doi: 10.1016/j.jconrel.2020.07.042. [DOI] [PubMed] [Google Scholar]
- 52.Zhan Q, Yi KK, Li XP, et al. Phosphatidylcholine-engineered exosomes for enhanced tumor cell uptake and intracellular antitumor drug delivery. Macromol Biosci. 2021;21:e2100042. doi: 10.1002/mabi.202100042. [DOI] [PubMed] [Google Scholar]
- 53.Choi ES, Song J, Kang YY, et al. Mannose-modified serum exosomes for the elevated uptake to murine dendritic cells and lymphatic accumulation. Macromol Biosci. 2019;19:e1900042. doi: 10.1002/mabi.201900042. [DOI] [PubMed] [Google Scholar]
- 54.Zuo BF, Qi H, Lu Z, et al. Alarmin-painted exosomes elicit persistent antitumor immunity in large established tumors in mice. Nat Commun. 2020;11:1790. doi: 10.1038/s41467-020-15569-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Quemener AM, Bachelot L, Forestier A, et al. The powerful world of antisense oligonucleotides: from bench to bedside. Wiley Interdiscip Rev RNA. 2020;11:e1594. doi: 10.1002/wrna.1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Song ZX, Jia RR, Tang MF, et al. Antisense oligonucleotide technology can be used to investigate a circular but not linear RNA-mediated function for its encoded gene locus. Sci China Life Sci. 2021;64:784–794. doi: 10.1007/s11427-020-1743-8. [DOI] [PubMed] [Google Scholar]
- 57.Bennett CF. Therapeutic antisense oligonucleotides are coming of age. Annu Rev Med. 2019;70:307–321. doi: 10.1146/annurev-med-041217-010829. [DOI] [PubMed] [Google Scholar]
- 58.Winkle M, El-Daly SM, Fabbri M, et al. Noncoding RNA therapeutics-challenges and potential solutions. Nat Rev Drug Discov. 2021;20:629–651. doi: 10.1038/s41573-021-00219-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lamichhane TN, Raiker RS, Jay SM. Exogenous DNA loading into extracellular vesicles via electroporation is size-dependent and enables limited gene delivery. Mol Pharm. 2015;12:3650–3657. doi: 10.1021/acs.molpharmaceut.5b00364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang JL, Luo SL, Zhang JC, et al. Exosome-mediated delivery of antisense oligonucleotides targeting α-synuclein ameliorates the pathology in a mouse model of Parkinson’s disease. Neurobiol Dis. 2021;148:105218. doi: 10.1016/j.nbd.2020.105218. [DOI] [PubMed] [Google Scholar]
- 61.Zhang WX, Lin JX, Shi PL, et al. Small extracellular vesicles derived from MSCs have immunomodulatory effects to enhance delivery of ASO-210 for psoriasis treatment. Front Cell Dev Biol. 2022;10:842813. doi: 10.3389/fcell.2022.842813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang J, Chen D, Ho EA. Challenges in the development and establishment of exosome-based drug delivery systems. J Control Release. 2021;329:894–906. doi: 10.1016/j.jconrel.2020.10.020. [DOI] [PubMed] [Google Scholar]
- 63.Lin Y, Wu JH, Gu WH, et al. Exosome-liposome hybrid nanoparticles deliver CRISPR/Cas9 system in MSCs. Adv Sci (Weinh) 2018;5:1700611. doi: 10.1002/advs.201700611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.György B, Sage C, Indzhykulian AA, et al. Rescue of hearing by gene delivery to inner-ear hair cells using exosome-associated AAV. Mol Ther. 2017;25:379–391. doi: 10.1016/j.ymthe.2016.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Guescini M, Guidolin D, Vallorani L, et al. C2C12 myoblasts release micro-vesicles containing mtDNA and proteins involved in signal transduction. Exp Cell Res. 2010;316:1977–1984. doi: 10.1016/j.yexcr.2010.04.006. [DOI] [PubMed] [Google Scholar]
- 66.Balaj L, Lessard R, Dai L, et al. Tumour microvesicles contain retrotransposon elements and amplified oncogene sequences. Nat Commun. 2011;2:180. doi: 10.1038/ncomms1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kahlert C, Melo SA, Protopopov A, et al. Identification of double-stranded genomic DNA spanning all chromosomes with mutated KRAS and p53 DNA in the serum exosomes of patients with pancreatic cancer. J Biol Chem. 2014;289:3869–3875. doi: 10.1074/jbc.C113.532267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yokoi A, Villar-Prados A, Oliphint PA, et al. Mechanisms of nuclear content loading to exosomes. Sci Adv. 2019;5:eaax8849. doi: 10.1126/sciadv.aax8849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Andreeva OE, Shchegolev YY, Scherbakov AM, et al. Secretion of mutant DNA and mRNA by the exosomes of breast cancer cells. Molecules. 2021;26:2499. doi: 10.3390/molecules26092499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kamerkar S, Burzyn D, Leng C, et al. Abstract A50: Reprogramming of tumor-associated M2 macrophages with antisense oligonucleotide-loaded exosomes results in potent single-agent antitumor activity. Paper presented at the abstracts: AACR special conference on tumor immunology and immunotherapy; November 17–20, 2019; Boston, MA. 2020.
- 71.Kim SM, Yang Y, Oh SJ, et al. Cancer-derived exosomes as a delivery platform of CRISPR/Cas9 confer cancer cell tropism-dependent targeting. J Control Release. 2017;266:8–16. doi: 10.1016/j.jconrel.2017.09.013. [DOI] [PubMed] [Google Scholar]
- 72.Wang YY, Shahi PK, Xie RS, et al. A pH-responsive silica-metal-organic framework hybrid nanoparticle for the delivery of hydrophilic drugs, nucleic acids, and CRISPR-Cas9 genome-editing machineries. J Control Release. 2020;324:194–203. doi: 10.1016/j.jconrel.2020.04.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tong S, Moyo B, Lee CM, et al. Engineered materials for in vivo delivery of genome-editing machinery. Nat Rev Mater. 2019;4:726–737. doi: 10.1038/s41578-019-0145-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Thess A, Grund S, Mui BL, et al. Sequence-engineered mRNA without chemical nucleoside modifications enables an effective protein therapy in large animals. Mol Ther. 2015;23:1456–1464. doi: 10.1038/mt.2015.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liang XP, Li DP, Leng SL, et al. RNA-based pharmacotherapy for tumors: from bench to clinic and back. Biomed Pharmacother. 2020;125:109997. doi: 10.1016/j.biopha.2020.109997. [DOI] [PubMed] [Google Scholar]
- 76.Zou S, Scarfo K, Nantz MH, et al. Lipid-mediated delivery of RNA is more efficient than delivery of DNA in non-dividing cells. Int J Pharm. 2010;389:232–243. doi: 10.1016/j.ijpharm.2010.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wadhwa A, Aljabbari A, Lokras A, et al. Opportunities and challenges in the delivery of mRNA-based vaccines. Pharmaceutics. 2020;12:102. doi: 10.3390/pharmaceutics12020102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Marx V. How to pull the blanket off dormant cancer cells. Nat Methods. 2018;15:249–252. doi: 10.1038/nmeth.4640. [DOI] [PubMed] [Google Scholar]
- 79.Sahin U, Derhovanessian E, Miller M, et al. Personalized RNA mutanome vaccines mobilize poly-specific therapeutic immunity against cancer. Nature. 2017;547:222–226. doi: 10.1038/nature23003. [DOI] [PubMed] [Google Scholar]
- 80.Huang QR, Zeng JW, Yan JH. COVID-19 mRNA vaccines. J Genet Genomics. 2021;48:107–114. doi: 10.1016/j.jgg.2021.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Huang QR, Ji K, Tian SY, et al. A single-dose mRNA vaccine provides a long-term protection for hACE2 transgenic mice from SARS-CoV-2. Nat Commun. 2021;12:776. doi: 10.1038/s41467-021-21037-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bellos I, Karageorgiou V, Viskin D. Myocarditis following mRNA Covid-19 vaccination: a pooled analysis. Vaccine. 2022;40:1768–1774. doi: 10.1016/j.vaccine.2022.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li MY, Li Y, Li SQ, et al. The nano delivery systems and applications of mRNA. Eur J Med Chem. 2022;227:113910. doi: 10.1016/j.ejmech.2021.113910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Valadi H, Ekström K, Bossios A, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 85.Li YC, Zhao JJ, Yu SL, et al. Extracellular vesicles long RNA sequencing reveals abundant mRNA, circRNA, and lncRNA in human blood as potential biomarkers for cancer diagnosis. Clin Chem. 2019;65:798–808. doi: 10.1373/clinchem.2018.301291. [DOI] [PubMed] [Google Scholar]
- 86.Ji J, Chen R, Zhao L, et al. Circulating exosomal mRNA profiling identifies novel signatures for the detection of prostate cancer. Mol Cancer. 2021;20:58. doi: 10.1186/s12943-021-01349-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Matsuno Y, Kanke T, Maruyama N, et al. Characterization of mRNA profiles of the exosome-like vesicles in porcine follicular fluid. PLoS ONE. 2019;14:e0217760. doi: 10.1371/journal.pone.0217760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tsai SJ, Guo CX, Atai NA, et al. Exosome-mediated mRNA delivery for SARS-CoV-2 vaccination. BioRxiv. 2020 doi: 10.1016/j.jbc.2021.101266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kojima R, Bojar D, Rizzi G, et al. Designer exosomes produced by implanted cells intracerebrally deliver therapeutic cargo for Parkinson’s disease treatment. Nat Commun. 2018;9:1305. doi: 10.1038/s41467-018-03733-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Shrivastava S, Ray RM, Holguin L, et al. Exosome-mediated stable epigenetic repression of HIV-1. Nat Commun. 2021;12:5541. doi: 10.1038/s41467-021-25839-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Li ZL, Zhou XY, Wei MY, et al. In vitro and in vivo RNA inhibition by CD9-HuR functionalized exosomes encapsulated with miRNA or CRISPR/dCas9. Nano Lett. 2019;19:19–28. doi: 10.1021/acs.nanolett.8b02689. [DOI] [PubMed] [Google Scholar]
- 92.Wang JH, Forterre AV, Zhao J, et al. Anti-HER2 scFv-directed extracellular vesicle-mediated mRNA-based gene delivery inhibits growth of HER2-positive human breast tumor xenografts by prodrug activation. Mol Cancer Ther. 2018;17:1133–1142. doi: 10.1158/1535-7163.MCT-17-0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Forterre AV, Wang JH, Delcayre A, et al. Extracellular vesicle-mediated in vitro transcribed mRNA delivery for treatment of HER2(+) breast cancer xenografts in mice by prodrug CB1954 without general toxicity. Mol Cancer Ther. 2020;19:858–867. doi: 10.1158/1535-7163.MCT-19-0928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yang ZG, Shi JF, Xie J, et al. Large-scale generation of functional mRNA-encapsulating exosomes via cellular nanoporation. Nat Biomed Eng. 2020;4:69–83. doi: 10.1038/s41551-019-0485-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.https://www.biospace.com/article/neocura-and-mdimune-settle-cdvs-mediated-mrnacollaboration/
- 96.Lu TX, Rothenberg ME. MicroRNA. J Allergy Clin Immunol. 2018;141:1202–1207. doi: 10.1016/j.jaci.2017.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yang CB, Yin MJ, Xu GX, Lin, et al. Biodegradable polymers as a noncoding miRNA nanocarrier for multiple targeting therapy of human hepatocellular carcinoma. Adv Healthcare Mater. 2019;8:1801318. doi: 10.1002/adhm.201801318. [DOI] [PubMed] [Google Scholar]
- 98.Fu W, Hong ZM, You XJ, et al. Enhancement of anticancer activity of docetaxel by combination with Fuzheng Yiliu decoction in a mouse model of castration-resistant prostate cancer. Biomed Pharmacother. 2019;118:109374. doi: 10.1016/j.biopha.2019.109374. [DOI] [PubMed] [Google Scholar]
- 99.Su MJ, Aldawsari H, Amiji M. Pancreatic cancer cell exosome-mediated macrophage reprogramming and the role of microRNAs 155 and 125b2 transfection using nanoparticle delivery systems. Sci Rep. 2016;6:30110. doi: 10.1038/srep30110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhou WX, Zhou Y, Chen XL, et al. Pancreatic cancer-targeting exosomes for enhancing immunotherapy and reprogramming tumor microenvironment. Biomaterials. 2021;268:120546. doi: 10.1016/j.biomaterials.2020.120546. [DOI] [PubMed] [Google Scholar]
- 101.Batrakova EV, Kim MS. Using exosomes, naturally-equipped nanocarriers, for drug delivery. J Control Release. 2015;219:396–405. doi: 10.1016/j.jconrel.2015.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bryniarski K, Ptak W, Jayakumar A, et al. Antigen-specific, antibody-coated, exosome-like nanovesicles deliver suppressor T-cell microRNA-150 to effector T cells to inhibit contact sensitivity. J Allergy Clin Immunol. 2013;132:170–181. doi: 10.1016/j.jaci.2013.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Trivedi M, Talekar M, Shah P, et al. Modification of tumor cell exosome content by transfection with wt-p53 and microRNA-125b expressing plasmid DNA and its effect on macrophage polarization. Oncogenesis. 2016;5:e250. doi: 10.1038/oncsis.2016.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kirave P, Gondaliya P, Kulkarni B, et al. Exosome mediated miR-155 delivery confers cisplatin chemoresistance in oral cancer cells via epithelial-mesenchymal transition. Oncotarget. 2020;11:1157–1171. doi: 10.18632/oncotarget.27531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Liu X, Lu Y, Xu YC, et al. Exosomal transfer of miR-501 confers doxorubicin resistance and tumorigenesis via targeting of BLID in gastric cancer. Cancer Lett. 2019;459:122–134. doi: 10.1016/j.canlet.2019.05.035. [DOI] [PubMed] [Google Scholar]
- 106.Kim G, Kim M, Lee Y, et al. Systemic delivery of microRNA-21 antisense oligonucleotides to the brain using T7-peptide decorated exosomes. J Control Release. 2020;317:273–281. doi: 10.1016/j.jconrel.2019.11.009. [DOI] [PubMed] [Google Scholar]
- 107.Liang GF, Zhu YL, Ali DJ, et al. Engineered exosomes for targeted co-delivery of miR-21 inhibitor and chemotherapeutics to reverse drug resistance in colon cancer. J Nanobiotechnology. 2020;18:10. doi: 10.1186/s12951-019-0563-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zhan Q, Yi KK, Qi HZ, et al. Engineering blood exosomes for tumor-targeting efficient gene/chemo combination therapy. Theranostics. 2020;10:7889–7905. doi: 10.7150/thno.45028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ding YX, Cao F, Sun HC, et al. Exosomes derived from human umbilical cord mesenchymal stromal cells deliver exogenous miR-145-5p to inhibit pancreatic ductal adenocarcinoma progression. Cancer Lett. 2019;442:351–361. doi: 10.1016/j.canlet.2018.10.039. [DOI] [PubMed] [Google Scholar]
- 110.Naseri Z, Oskuee RK, Forouzandeh-Moghadam M, et al. Delivery of LNA-antimiR-142-3p by mesenchymal stem cells-derived exosomes to breast cancer stem cells reduces tumorigenicity. Stem Cell Rev Rep. 2020;16:541–556. doi: 10.1007/s12015-019-09944-w. [DOI] [PubMed] [Google Scholar]
- 111.Chen ZL, Xie Y, Chen WD, et al. microRNA-6785-5p-loaded human umbilical cord mesenchymal stem cells-derived exosomes suppress angiogenesis and metastasis in gastric cancer via INHBA. Life Sci. 2021;284:119222. doi: 10.1016/j.lfs.2021.119222. [DOI] [PubMed] [Google Scholar]
- 112.Jiang SJ, Mo CQ, Guo SJ, et al. Human bone marrow mesenchymal stem cells-derived microRNA-205-containing exosomes impede the progression of prostate cancer through suppression of RHPN2. J Exp Clin Cancer Res. 2019;38:495. doi: 10.1186/s13046-019-1488-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Jia YF, Ding XM, Zhou LH, et al. Mesenchymal stem cells-derived exosomal microRNA-139-5p restrains tumorigenesis in bladder cancer by targeting PRC1. Oncogene. 2021;40:246–261. doi: 10.1038/s41388-020-01486-7. [DOI] [PubMed] [Google Scholar]
- 114.Wang XN, Qin X, Yan M, et al. Loss of exosomal miR-3188 in cancer-associated fibroblasts contributes to HNC progression. J Exp Clin Cancer Res. 2019;38:151. doi: 10.1186/s13046-019-1144-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhang ZC, Li X, Sun W, et al. Loss of exosomal miR-320a from cancer-associated fibroblasts contributes to HCC proliferation and metastasis. Cancer Lett. 2017;397:33–42. doi: 10.1016/j.canlet.2017.03.004. [DOI] [PubMed] [Google Scholar]
- 116.Xu GF, Zhang B, Ye JH, et al. Exosomal miRNA-139 in cancer-associated fibroblasts inhibits gastric cancer progression by repressing MMP11 expression. Int J Biol Sci. 2019;15:2320–2329. doi: 10.7150/ijbs.33750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Shi L, Wang ZY, Geng XC, et al. Exosomal miRNA-34 from cancer-associated fibroblasts inhibits growth and invasion of gastric cancer cells in vitro and in vivo. Aging (Albany NY) 2020;12:8549–8564. doi: 10.18632/aging.103157. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 118.Li LX, He D, Guo QQ, et al. Exosome-liposome hybrid nanoparticle codelivery of TP and miR497 conspicuously overcomes chemoresistant ovarian cancer. J Nanobiotechnol. 2022;20:50. doi: 10.1186/s12951-022-01264-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kim H, Rhee WJ. Exosome-mediated Let7c-5p delivery for breast cancer therapeutic development. Biotechnol Bioproc Eng. 2020;25:513–520. [Google Scholar]
- 120.Naseri Z, Oskuee RK, Jaafari MR, et al. Exosome-mediated delivery of functionally active miRNA-142-3p inhibitor reduces tumorigenicity of breast cancer in vitro and in vivo. Int J Nanomedicine. 2018;13:7727–7747. doi: 10.2147/IJN.S182384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.He ZF, Li WH, Zheng TL, et al. Human umbilical cord mesenchymal stem cells-derived exosomes deliver microRNA-375 to downregulate ENAH and thus retard esophageal squamous cell carcinoma progression. J Exp Clin Cancer Res. 2020;39:140. doi: 10.1186/s13046-020-01631-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Liu T, Zhang X, Du LT, et al. Exosome-transmitted miR-128-3p increase chemosensitivity of oxaliplatin-resistant colorectal cancer. Mol Cancer. 2019;18:43. doi: 10.1186/s12943-019-0981-7. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 123.Jeong K, Yu YJ, You JY, et al. Exosome-mediated microRNA-497 delivery for anti-cancer therapy in a microfluidic 3D lung cancer model. Lab Chip. 2020;20:548–557. doi: 10.1039/c9lc00958b. [DOI] [PubMed] [Google Scholar]
- 124.Kulkarni B, Gondaliya P, Kirave P, et al. Exosome-mediated delivery of miR-30a sensitize cisplatin-resistant variant of oral squamous carcinoma cells via modulating Beclin1 and Bcl2. Oncotarget. 2020;11:1832–1845. doi: 10.18632/oncotarget.27557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Liu T, Chen G, Sun DW, et al. Exosomes containing miR-21 transfer the characteristic of cisplatin resistance by targeting PTEN and PDCD4 in oral squamous cell carcinoma. Acta Biochim Biophys Sin (Shanghai) 2017;49:808–816. doi: 10.1093/abbs/gmx078. [DOI] [PubMed] [Google Scholar]
- 126.Wang XY, Zhang HY, Bai M, et al. Exosomes serve as nanoparticles to deliver anti-miR-214 to reverse chemoresistance to cisplatin in gastric cancer. Mol Ther. 2018;26:774–783. doi: 10.1016/j.ymthe.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zhang K, Shao CX, Zhu JD, et al. Exosomes function as nanoparticles to transfer miR-199a-3p to reverse chemoresistance to cisplatin in hepatocellular carcinoma. Biosci Rep. 2020;40:BSR20194026. doi: 10.1042/BSR20194026. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 128.Irena M. RNA-based therapies. Nat Rev Drug Discov. 2007;6:863–864. [Google Scholar]
- 129.Watanabe T, Totoki Y, Toyoda A, et al. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008;453:539–543. doi: 10.1038/nature06908. [DOI] [PubMed] [Google Scholar]
- 130.Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 131.Yang CB, Chan KK, Lin WJ, et al. Biodegradable nanocarriers for small interfering ribonucleic acid (siRNA) co-delivery strategy increase the chemosensitivity of pancreatic cancer cells to gemcitabine. Nano Res. 2017;10:3049–3067. [Google Scholar]
- 132.Castanotto D, Rossi JJ. The promises and pitfalls of RNA-interference-based therapeutics. Nature. 2009;457:426–433. doi: 10.1038/nature07758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Tatiparti K, Sau S, Kashaw SK, et al. siRNA delivery strategies: a comprehensive review of recent developments. Nanomaterials (Basel) 2017;7:77. doi: 10.3390/nano7040077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Hoy SM. Patisiran: first global approval. Drugs. 2018;78:1625–1631. doi: 10.1007/s40265-018-0983-6. [DOI] [PubMed] [Google Scholar]
- 135.Scott LJ. Givosiran: first approval. Drugs. 2020;80:335–339. doi: 10.1007/s40265-020-01269-0. [DOI] [PubMed] [Google Scholar]
- 136.Alvarez-Erviti L, Seow Y, Yin HF, et al. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat Biotechnol. 2011;29:341–345. doi: 10.1038/nbt.1807. [DOI] [PubMed] [Google Scholar]
- 137.Wahlgren J, De LKT, Brisslert M, et al. Plasma exosomes can deliver exogenous short interfering RNA to monocytes and lymphocytes. Nucleic Acids Res. 2012;40:e130. doi: 10.1093/nar/gks463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Aqil F, Munagala R, Jeyabalan J, et al. Milk exosomes-Natural nanoparticles for siRNA delivery. Cancer Lett. 2019;449:186–195. doi: 10.1016/j.canlet.2019.02.011. [DOI] [PubMed] [Google Scholar]
- 139.Lamichhane TN, Jeyaram A, Patel DB, et al. Oncogene knockdown via active loading of small RNAs into extracellular vesicles by sonication. Cell Mol Bioeng. 2016;9:315–324. doi: 10.1007/s12195-016-0457-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Li SP, Lin ZX, Jiang XY, et al. Exosomal cargo-loading and synthetic exosome-mimics as potential therapeutic tools. Acta Pharmacol Sin. 2018;39:542–551. doi: 10.1038/aps.2017.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kaban K, Hinterleitner C, Zhou YJ, et al. Therapeutic silencing of BCL-2 using NK cell-derived exosomes as a novel therapeutic approach in breast cancer. Cancers (Basel) 2021;13:2397. doi: 10.3390/cancers13102397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Greco KA, Franzen CA, Foreman KE, et al. PLK-1 silencing in bladder cancer by siRNA delivered with exosomes. Urology. 2016;91(241):e241–e247. doi: 10.1016/j.urology.2016.01.028. [DOI] [PubMed] [Google Scholar]
- 143.Pi FM, Binzel DW, Lee TJ, et al. Nanoparticle orientation to control RNA loading and ligand display on extracellular vesicles for cancer regression. Nat Nanotechnol. 2018;13:82–89. doi: 10.1038/s41565-017-0012-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Martinez-Balibrea E, Martínez-Cardús A, Ginés A, et al. Tumor-related molecular mechanisms of oxaliplatin resistance. Mol Cancer Ther. 2015;14:1767–1776. doi: 10.1158/1535-7163.MCT-14-0636. [DOI] [PubMed] [Google Scholar]
- 145.Li HD, Yang C, Shi YJ, et al. Exosomes derived from siRNA against GRP78 modified bone-marrow-derived mesenchymal stem cells suppress Sorafenib resistance in hepatocellular carcinoma. J Nanobiotechnology. 2018;16:103. doi: 10.1186/s12951-018-0429-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Zhang QM, Zhang HY, Ning T, et al. Exosome-delivered c-Met siRNA could reverse chemoresistance to cisplatin in gastric cancer. Int J Nanomedicine. 2020;15:2323–2335. doi: 10.2147/IJN.S231214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Lin D, Zhang HQ, Liu R, et al. iRGD-modified exosomes effectively deliver CPT1A siRNA to colon cancer cells, reversing oxaliplatin resistance by regulating fatty acid oxidation. Mol Oncol. 2021;15:3430–3446. doi: 10.1002/1878-0261.13052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Marin-Acevedo JA, Soyano AE, Dholaria B, et al. Cancer immunotherapy beyond immune checkpoint inhibitors. J Hematol Oncol. 2018;11:8. doi: 10.1186/s13045-017-0552-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Clark CE, Hingorani SR, Mick R, et al. Dynamics of the immune reaction to pancreatic cancer from inception to invasion. Cancer Res. 2007;67:9518–9527. doi: 10.1158/0008-5472.CAN-07-0175. [DOI] [PubMed] [Google Scholar]
- 150.Pei X, Zhang XJ, Zhang L, et al. Targeted exosomes for co-delivery of siFGL1 and siTGF-β1 trigger combined cancer immunotherapy by remodeling immunosuppressive tumor microenvironment. Chem Eng J. 2021;421:129774. [Google Scholar]
- 151.Bai J, Duan JL, Liu R, et al. Engineered targeting tLyp-1 exosomes as gene therapy vectors for efficient delivery of siRNA into lung cancer cells. Asian J Pharm Sci. 2020;15:461–471. doi: 10.1016/j.ajps.2019.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Zhao LW, Gu CY, Gan Y, et al. Exosome-mediated siRNA delivery to suppress postoperative breast cancer metastasis. J Control Release. 2020;318:1–15. doi: 10.1016/j.jconrel.2019.12.005. [DOI] [PubMed] [Google Scholar]
- 153.Limoni SK, Moghadam MF, Moazzeni SM, et al. Engineered exosomes for targeted transfer of siRNA to HER2 positive breast cancer cells. Appl Biochem Biotechnol. 2019;187:352–364. doi: 10.1007/s12010-018-2813-4. [DOI] [PubMed] [Google Scholar]
- 154.Kamerkar S, LeBleu VS, Sugimoto H, et al. Exosomes facilitate therapeutic targeting of oncogenic KRAS in pancreatic cancer. Nature. 2017;546:498–503. doi: 10.1038/nature22341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Zhang HY, Wang Y, Bai M, et al. Exosomes serve as nanoparticles to suppress tumor growth and angiogenesis in gastric cancer by delivering hepatocyte growth factor siRNA. Cancer Sci. 2018;109:629–641. doi: 10.1111/cas.13488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Yan L, Wang PY, Fang WH, et al. Cancer-associated fibroblasts-derived exosomes-mediated transfer of LINC00355 regulates bladder cancer cell proliferation and invasion. Cell Biochem Funct. 2020;38:257–265. doi: 10.1002/cbf.3462. [DOI] [PubMed] [Google Scholar]
- 157.Zhang P, Zhou HX, Lu KF, et al. Exosome-mediated delivery of MALAT1 induces cell proliferation in breast cancer. Onco Targets Ther. 2018;11:291–299. doi: 10.2147/OTT.S155134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Xu LZ, Faruqu FN, Lim YM, et al. Exosome-mediated RNAi of PAK4 prolongs survival of pancreatic cancer mouse model after loco-regional treatment. Biomaterials. 2021;264:120369. doi: 10.1016/j.biomaterials.2020.120369. [DOI] [PubMed] [Google Scholar]
- 159.Kase Y, Uzawa K, Wagai S, et al. Engineered exosomes delivering specific tumor-suppressive RNAi attenuate oral cancer progression. Sci Rep. 2021;11:5897. doi: 10.1038/s41598-021-85242-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Wang CH, Chen L, Huang YY, et al. Exosome-delivered TRPP2 siRNA inhibits the epithelial-mesenchymal transition of FaDu cells. Oncol Lett. 2019;17:1953–1961. doi: 10.3892/ol.2018.9752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Wang XH, Cheng K, Zhang GQ, et al. Enrichment of CD44 in exosomes from breast cancer cells treated with doxorubicin promotes chemoresistance. Front Oncol. 2020;10:960. doi: 10.3389/fonc.2020.00960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Yang ZG, Xie J, Zhu J, et al. Functional exosome-mimic for delivery of siRNA to cancer: in vitro and in vivo evaluation. J Control Release. 2016;243:160–171. doi: 10.1016/j.jconrel.2016.10.008. [DOI] [PubMed] [Google Scholar]
- 163.Zhou Y, Yuan Y, Liu MH, et al. Tumor-specific delivery of KRAS siRNA with iRGD-exosomes efficiently inhibits tumor growth. ExRNA. 2019;1:28. [Google Scholar]
- 164.Wang MH, Ye Y, Zhang M, et al. Exosome-mediated delivery of SCD-1 siRNA promoted the death of anaplastic thyroid carcinoma cells via regulating ROS level. Clin Transl Oncol. 2021;24:288–296. doi: 10.1007/s12094-021-02682-x. [DOI] [PubMed] [Google Scholar]
- 165.Han Q, Xie QR, Li F, et al. Targeted inhibition of SIRT6 via engineered exosomes impairs tumorigenesis and metastasis in prostate cancer. Theranostics. 2021;11:6526–6541. doi: 10.7150/thno.53886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Li ZF, Yang LL, Wang HZ, et al. Non-small-cell lung cancer regression by siRNA delivered through exosomes that display EGFR RNA aptamer. Nucleic Acid Ther. 2021;31:364–374. doi: 10.1089/nat.2021.0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Chen L, Wang CL, Sun HY, et al. The bioinformatics toolbox for circRNA discovery and analysis. Brief Bioinform. 2021;22:1706–1728. doi: 10.1093/bib/bbaa001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Shi Y, Jia X, Xu J. The circRNA-microRNA code: emerging implications for cancer diagnosis and treatment. Mol Oncol. 2019;13:669–680. doi: 10.1002/1878-0261.12468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Wang CD, Tan SY, Li JW, et al. CircRNAs in lung cancer-Biogenesis, function and clinical implication. Cancer Lett. 2020;492:106–115. doi: 10.1016/j.canlet.2020.08.013. [DOI] [PubMed] [Google Scholar]
- 170.Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 171.Weng WH, Wei Q, Toden S, et al. Circular RNA ciRS-7-A promising prognostic biomarker and a potential therapeutic target in colorectal cancer. Clin Cancer Res. 2017;23:3918–3928. doi: 10.1158/1078-0432.CCR-16-2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Du WW, Fang L, Yang WN, et al. Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity. Cell Death Differ. 2017;24:357–370. doi: 10.1038/cdd.2016.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Zhang ML, Huang NN, Yang XS, et al. A novel protein encoded by the circular form of the SHPRH gene suppresses glioma tumorigenesis. Oncogene. 2018;37:1805–1814. doi: 10.1038/s41388-017-0019-9. [DOI] [PubMed] [Google Scholar]
- 174.Xue JC, Liu Y, Luo F, et al. Circ100284, via miR-217 regulation of EZH2, is involved in the arsenite-accelerated cell cycle of human keratinocytes in carcinogenesis. Biochim Biophys Acta Mol Basis Dis. 2017;1863:753–763. doi: 10.1016/j.bbadis.2016.12.018. [DOI] [PubMed] [Google Scholar]
- 175.Chen W, Quan YY, Fan SY, et al. Exosome-transmitted circular RNA has_circ_0051443 suppresses hepatocellular carcinoma progression. Cancer Lett. 2020;475:119–128. doi: 10.1016/j.canlet.2020.01.022. [DOI] [PubMed] [Google Scholar]
- 176.Yu XY, Bai Y, Han B, et al. Extracellular vesicle-mediated delivery of circDYM alleviates CUS-induced depressive-like behaviours. J Extracell Vesicles. 2022;11:e12185. doi: 10.1002/jev2.12185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Yang L, Han B, Zhang ZT, et al. Extracellular vesicle-mediated delivery of circular RNA SCMH1 promotes functional recovery in rodent and nonhuman primate ischemic stroke models. Circulation. 2020;142:556–574. doi: 10.1161/CIRCULATIONAHA.120.045765. [DOI] [PubMed] [Google Scholar]
- 178.Dykes IM, Emanueli C. Transcriptional and post-transcriptional gene regulation by long non-coding RNA. Genomics Proteomics Bioinformatics. 2017;15:177–186. doi: 10.1016/j.gpb.2016.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Sun QY, Hao QY, Prasanth KV. Nuclear long noncoding RNAs: key regulators of gene expression. Trends Genet. 2018;34:142–157. doi: 10.1016/j.tig.2017.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Wang JC, Su ZL, Lu SN, et al. LncRNA HOXA-AS2 and its molecular mechanisms in human cancer. Clin Chim Acta. 2018;485:229–233. doi: 10.1016/j.cca.2018.07.004. [DOI] [PubMed] [Google Scholar]
- 181.Kong XL, Duan Y, Sang YT, et al. LncRNA-CDC6 promotes breast cancer progression and function as ceRNA to target CDC6 by sponging microRNA-215. J Cell Physiol. 2019;234:9105–9117. doi: 10.1002/jcp.27587. [DOI] [PubMed] [Google Scholar]
- 182.Cao HL, Liu ZJ, Huang PL, et al. lncRNA-RMRP promotes proliferation, migration and invasion of bladder cancer via miR-206. Eur Rev Med Pharmacol Sci. 2019;23:1012–1021. doi: 10.26355/eurrev_201902_16988. [DOI] [PubMed] [Google Scholar]
- 183.Pushparaj PN, Aarthi JJ, Manikandan J, et al. siRNA, miRNA, and shRNA: in vivo applications. J Dent Res. 2008;87:992–1003. doi: 10.1177/154405910808701109. [DOI] [PubMed] [Google Scholar]
- 184.Rao DD, Vorhies JS, Senzer N, et al. siRNA vs. shRNA: similarities and differences. Adv Drug Deliv Rev. 2009;61:746–59. doi: 10.1016/j.addr.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 185.Snøve O, Rossi JJ. Expressing short hairpin RNAs in vivo. Nat Methods. 2006;3:689–695. doi: 10.1038/nmeth927. [DOI] [PubMed] [Google Scholar]
- 186.Zhou JH, Rossi J. Aptamers as targeted therapeutics: current potential and challenges. Nat Rev Drug Discov. 2017;16:440. doi: 10.1038/nrd.2017.86. [DOI] [PubMed] [Google Scholar]
- 187.Zheng R, Du ML, Wang XW, et al. Exosome-transmitted long non-coding RNA PTENP1 suppresses bladder cancer progression. Mol Cancer. 2018;17:143. doi: 10.1186/s12943-018-0880-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Zheng R, Zhang K, Tan SY, et al. Exosomal circLPAR1 functions in colorectal cancer diagnosis and tumorigenesis through suppressing BRD4 via METTL3-eIF3h interaction. Mol Cancer. 2022;21:49. doi: 10.1186/s12943-021-01471-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Escudier B, Dorval T, Chaput N, et al. Vaccination of metastatic melanoma patients with autologous dendritic cell (DC) derived-exosomes: results of thefirst phase I clinical trial. J Transl Med. 2005;3:10. doi: 10.1186/1479-5876-3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Morse MA, Garst J, Osada T, et al. A phase I study of dexosome immunotherapy in patients with advanced non-small cell lung cancer. J Transl Med. 2005;3:9. doi: 10.1186/1479-5876-3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Dai SM, Wei D, Wu Z, et al. Phase I clinical trial of autologous ascites-derived exosomes combined with GM-CSF for colorectal cancer. Mol Ther. 2008;16:782–790. doi: 10.1038/mt.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Haraszti RA, Miller R, Stoppato M, et al. Exosomes produced from 3D cultures of MSCs by tangential flow filtration show higher yield and improved activity. Mol Ther. 2018;26:2838–2847. doi: 10.1016/j.ymthe.2018.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Watson DC, Bayik D, Srivatsan A, et al. Efficient production and enhanced tumor delivery of engineered extracellular vesicles. Biomaterials. 2016;105:195–205. doi: 10.1016/j.biomaterials.2016.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Wang J, Li W, Zhang LC, et al. Chemically edited exosomes with dual ligand purified by microfluidic device for active targeted drug delivery to tumor cells. ACS Appl Mater Interfaces. 2017;9:27441–27452. doi: 10.1021/acsami.7b06464. [DOI] [PubMed] [Google Scholar]
- 195.Kanwar SS, Dunlay CJ, Simeone DM, et al. Microfluidic device (ExoChip) for on-chip isolation, quantification and characterization of circulating exosomes. Lab Chip. 2014;14:1891–1900. doi: 10.1039/c4lc00136b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Liga A, Vliegenthart ADB, Oosthuyzen W, et al. Exosome isolation: a microfluidic road-map. Lab Chip. 2015;15:2388–2394. doi: 10.1039/c5lc00240k. [DOI] [PubMed] [Google Scholar]
- 197.Harmati M, Tarnai Z, Decsi G, et al. Stressors alter intercellular communication and exosome profile of nasopharyngeal carcinoma cells. J Oral Pathol Med. 2017;46:259–266. doi: 10.1111/jop.12486. [DOI] [PubMed] [Google Scholar]
- 198.Kanemoto S, Nitani R, Murakami T, et al. Multivesicular body formation enhancement and exosome release during endoplasmic reticulum stress. Biochem Biophys Res Commun. 2016;480:166–172. doi: 10.1016/j.bbrc.2016.10.019. [DOI] [PubMed] [Google Scholar]
- 199.King HW, Michael MZ, Gleadle JM. Hypoxic enhancement of exosome release by breast cancer cells. BMC Cancer. 2012;12:421. doi: 10.1186/1471-2407-12-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Munagala R, Aqil F, Jeyabalan J, et al. Bovine milk-derived exosomes for drug delivery. Cancer Lett. 2016;371:48–61. doi: 10.1016/j.canlet.2015.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.www.ClinicalTrials.gov.
- 202.Johnsen KB, Gudbergsson JM, Skov MN, et al. Evaluation of electroporation-induced adverse effects on adipose-derived stem cell exosomes. Cytotechnology. 2016;68:2125–2138. doi: 10.1007/s10616-016-9952-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Ortega A, Martinez-Arroyo O, Forner MJ, et al. Exosomes as drug delivery systems: endogenous nanovehicles for treatment of systemic lupus erythematosus. Pharmaceutics. 2020;13:3. doi: 10.3390/pharmaceutics13010003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Batagov AO, Kuznetsov VA, Kurochkin IV, et al. Identification of nucleotide patterns enriched in secreted RNAs as putative cis-acting elements targeting them to exosome nano-vesicles. BMC Genomics. 2011;12(Suppl. 3):S18. doi: 10.1186/1471-2164-12-S3-S18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Wang MZ, Gu TW, Xu Y, et al. Mechanical stretching of cells and lipid nanoparticles for nucleic acid delivery. J Control Release. 2021;339:208–219. doi: 10.1016/j.jconrel.2021.09.021. [DOI] [PubMed] [Google Scholar]
- 206.Hu B, Li B, Li K, et al. Thermostable ionizable lipid-like nanoparticle (iLAND) for RNAi treatment of hyperlipidemia. Sci Adv. 2022;8:eabm1418. doi: 10.1126/sciadv.abm1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Gong CG, Zhang X, Shi M, et al. Tumor exosomes reprogrammed by low pH are efficient targeting vehicles for smart drug delivery and personalized therapy against their homologous tumor. Adv Sci (Weinh) 2021;8:2002787. doi: 10.1002/advs.202002787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Wang S, Li F, Ye T, et al. Macrophage-tumor chimeric exosomes accumulate in lymph node and tumor to activate the immune response and the tumor microenvironment. Sci Transl Med. 2021;13:eabb6981. doi: 10.1126/scitranslmed.abb6981. [DOI] [PubMed] [Google Scholar]
- 209.de Abreu RC, Fernandes H, da Costa Martins PA, et al. Native and bioengineered extracellular vesicles for cardiovascular therapeutics. Nat Rev Cardiol. 2020;17:685–697. doi: 10.1038/s41569-020-0389-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.