Abstract
Among the most urgent public health threats is the worldwide emergence of carbapenem-resistant Enterobacteriaceae1–4, which are resistant to the antibiotic class of ‘last resort’. In the United States and Europe, carbapenem-resistant strains of the Klebsiella pneumoniae ST258 (ref. 5) sequence type are dominant, endemic6–8 and associated with high mortality6,9,10. We report the global evolution of pathogenicity in carbapenem-resistant K. pneumoniae, resulting in the repeated convergence of virulence and carbapenem resistance in the United States and Europe, dating back to as early as 2009. We demonstrate that K. pneumoniae can enhance its pathogenicity by adopting two opposing infection programs through easily acquired gain- and loss-of-function mutations. Single-nucleotide polymorphisms in the capsule biosynthesis gene wzc lead to hypercapsule production, which confers phagocytosis resistance, enhanced dissemination and increased mortality in animal models. In contrast, mutations disrupting capsule biosynthesis genes impair capsule production, which enhances epithelial cell invasion, in vitro biofilm formation and persistence in urinary tract infections. These two types of capsule mutants have emerged repeatedly and independently in Europe and the United States, with hypercapsule mutants associated with bloodstream infections and capsule-deficient mutants associated with urinary tract infections. In the latter case, drug-tolerant K. pneumoniae can persist to yield potentially untreatable, persistent infection.
Carbapenem-resistant K. pneumoniae have recently acquired a virulence plasmid from hypervirulent K. pneumoniae strains in south-east Asia, resulting in the first report in 2018 of the feared convergence of hypervirulence with carbapenem resistance11. To date, that particular virulence plasmid (pLVPK) has not been detected in carbapenem-resistant K. pneumoniae strains of the ST258 sequence type12, which are endemic in the United States and Europe6–8, and only a single ST258 isolate has been reported to carry hypervirulence-associated siderophores12.
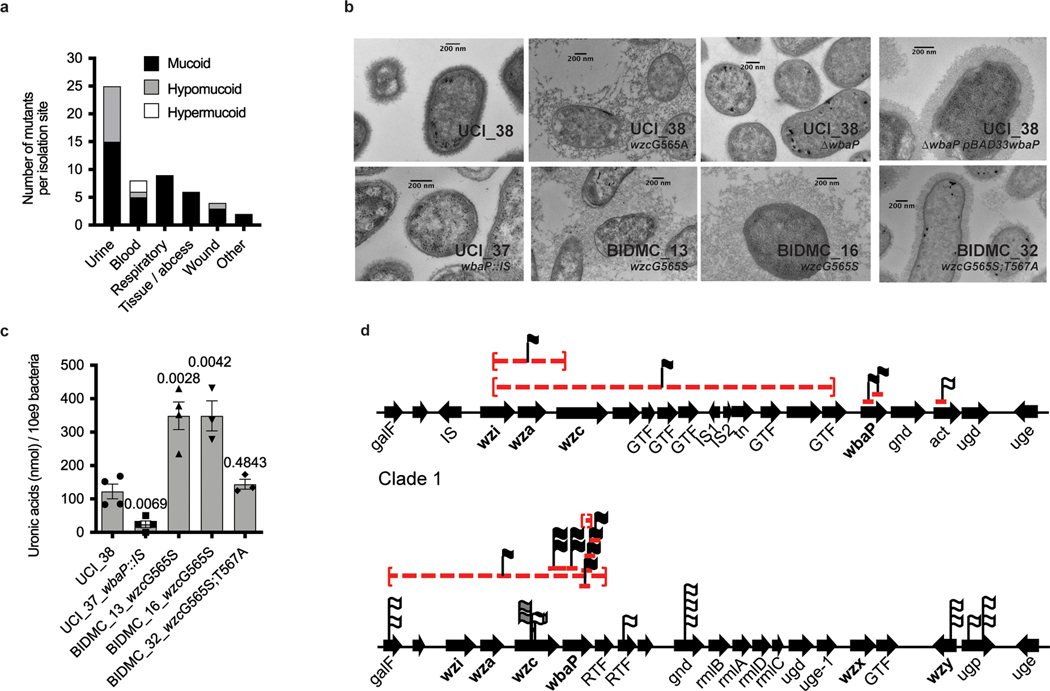
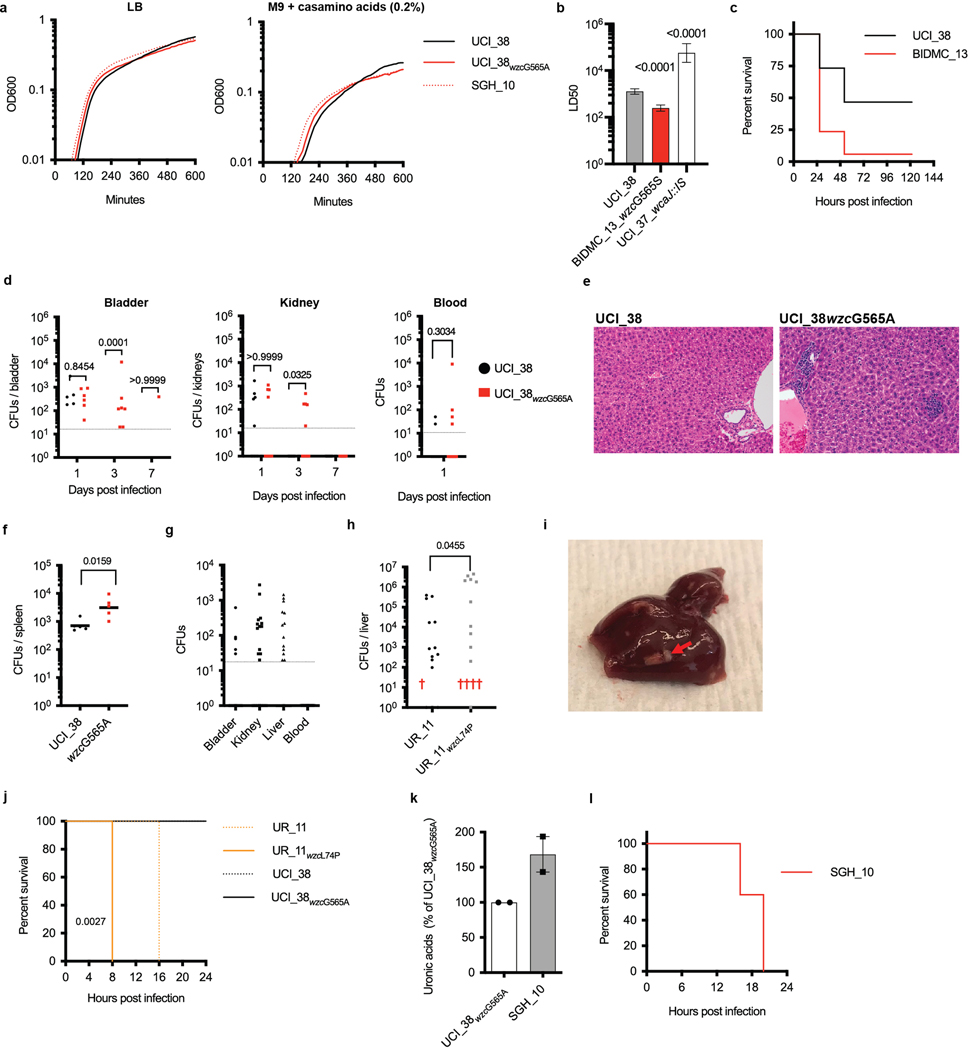
To explore whether the convergence of carbapenem resistance with virulence is more widespread, and could potentially involve alternative virulence mechanisms, we screened a previously reported collection of 54 K. pneumoniae ST258 strains13 collected from US patients for hypercapsule production, a previously recognized hallmark of hypervirulence14. Based on their ability to form hypermucoid colonies on blood agar plates (Extended Data Fig. 1a and Supplementary Table 1), we found that 74% (40/54) of the isolates were regular mucoid, 22% (12/54) were hypomucoid (Extended Data Fig. 1b) and 4% (2/54) were hypermucoid (Fig. 1a). Interestingly, the two hypermucoid isolates were isolated from bloodstream infections (P = 0.0196, Fisher’s exact test), whereas hypomucoidity was significantly associated with isolation from urinary tract infections (UTIs; P = 0.0069, Fisher’s exact test), which was the most frequent infection type from which ST258 strains were isolated (24/54, 46%) (Fig. 1a and Supplementary Table 1). Mucoidity has been associated with the state of the K. pneumoniae capsule, the polysaccharide layer coating the bacterium. We confirmed that hyper-and hypomucoidity of the ST258 clinical isolates indeed correlated with hypercapsule14 and capsule deficiency15, respectively, by imaging a representative set of isolates displaying the different colony phenotypes via electron microscopy (Fig. 1b) and quantifying the amount of uronic acids in each of the respective strain’s polysaccharide capsule (Fig. 1c)16.
Fig. 1 |. Capsule heterogeneity in screened collection of 54 clinical ST258 isolates.
a, Frequency of mucoidity phenotypes of strains isolated from different infection sites. Hypermucoid isolates are associated with bloodstream infections (P = 0.0196) and hypomucoid isolates with UTIs (P = 0.0069). b, Transmission electron microscopy of representative regular, hyper-and hypomucoid clinical isolates and isogenic strains: regular mucoid UCI_38, BIDMC_32 and UCI_38ΔwbaP(pBAD33wbaP); hypermucoid UCI_38wzcG565A, BIDMC_13 and BIDMC_16; and hypomucoid UCI_38ΔwbaP and UCI_37. Hypermucoid isolates display increased polymerization of the capsule, whereas capsule-deficient isolates do not produce a capsule, compared with regular mucoid UCI_38. One representative image from four images obtained from one section is shown. c, Capsule production measured by uronic acid content of isolates displaying altered mucoidity. Hypermucoid strains produce more capsule, compared with regularly mucoid isolates. The mean and s.e.m. of three independent experiments are shown. P values are displayed above the columns based on the comparison with UCI_38 (unpaired, two-tailed Student’s t-test). d, Mutations found in the capsule biosynthesis genes in the screened collection of ST258 isolates belonging to clade 1 and clade 2, which harbor different capsule operons. The position of the mutations is indicated. Red lines are indicative of deletions (dashed) and ISs. Flags lacking the red pedestal represent non-synonymous SNPs. Linked flags are indicative of multiple mutations found in an isolate. The mucoidity phenotypes associated with IS elements and SNPs are indicated by the color of the flag: white, mucoid; black, hypomucoid; gray, hypermucoid.
We first considered the capsule-deficient strains. As the capsule is thought to be essential for infection, we were surprised to identify capsule-deficient ST258 strains among the clinical urine isolates, because such strains had been assumed to be avirulent17. The 12 capsule-deficient ST258 isolates had 11 different mutations disrupting capsule-biosynthesis genes (Fig. 1d and Supplementary Table 1), with two closely related isolates carrying the same mutations (UCI_43 and UCI_44). The mutations included large deletions of several core capsule biosynthesis genes (wzi, wza, wzc) and, most commonly, insertion sequences (ISs) in wbaP, the homolog of which, in Escherichia coli, mediates the first step of capsule biosynthesis18. We engineered a mutant strain to confirm that deletion of wbaP in the normal capsule-producing clinical strain UCI_38 (UCI_38ΔwbaP) did indeed abolish capsule production, which could be restored by complementation with wbaP (Figs. 1b and 2a). Deletion of wbaP in two other clinical ST258 isolates from different ST258 (clade 2) subclades also abolished capsule production (BWH_36 (ref. 13) and BWH_45 (ref. 13); Extended Data Fig. 2a), demonstrating that wbaP plays a similar role in K. pneumoniae to that in E. coli.
The repeated acquisition of capsule-disrupting mutations implied strong selective pressure to inactivate capsule production in these urine isolates. We thus investigated the impact of capsule inactivation on phenotypes associated with UTIs, including biofilm formation and infection of bladder epithelial cells, features crucial for uropathogenic E. coli to establish and persist in UTIs19–21. The engineered deletion strain UCI_38ΔwbaP and the closely related hypocapsule clinical isolate UCI_37wbaP::IS (ref. 13) both formed more robust biofilms and invaded bladder epithelial cells more efficiently compared with wild-type UCI_38, a normal capsule strain and the complemented mutant (Fig. 2b,c and Extended Data Fig. 2b,c); these same results were recapitulated in wbaP deletion strains in two other ST258 strain backgrounds (BWH_36 and BWH_45; Extended Data Fig. 2c). Deletion of wbaP increased invasion and intracellular replication in LAMP1-positive vacuoles (Fig. 2d and Extended Data Fig. 2f–l), and resulted in a tenfold larger intracellular bacterial reservoir for UCI_38ΔwbaP mutants compared with the isogenic parent UCI_38, which persisted over the course of 48 h (Extended Data Fig. 2d). Importantly, intracellularly persisting K. pneumoniae can adopt a drug-tolerant phenotype, similar to intracellular uropathogenic E. coli22, as demonstrated by our finding that a capsule-deficient clinical isolate (BIDMC_54; Extended Data Figs. 1b and 2b) could not be eradicated (by colistin or meropenem/vaborbactam; Extended Data Fig. 2e), despite susceptibility to these antibiotics when the bacteria are grown extracellularly.
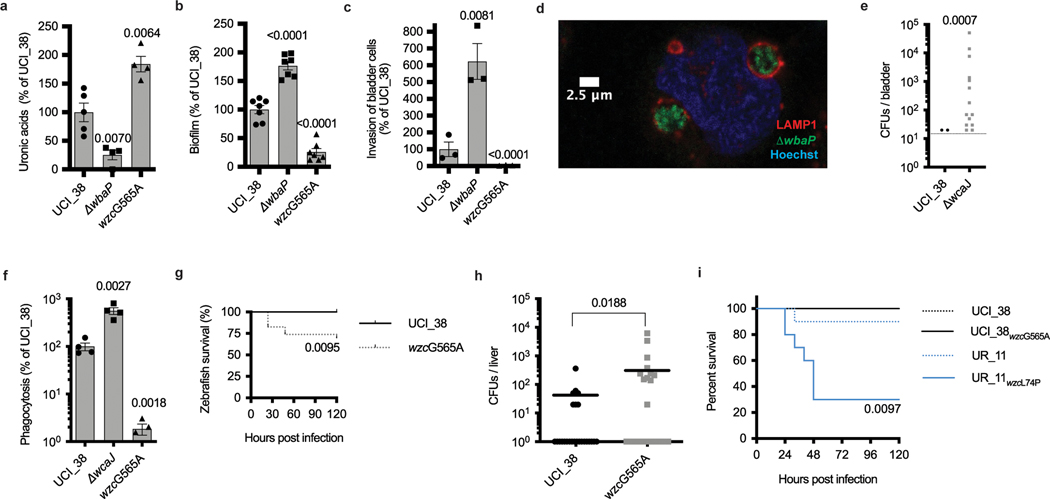
Fig. 2 |. Gain-and loss-of-function mutations increase the virulence or the persistence of K. pneumoniae ST258.
a, Capsule production of isogenic mutants of UCI_38 as measured by uronic acid content confirms that disruption of wbaP impairs capsule production, whereas an SNP at position 565 leads to hypercapsule production (n = 4 biologically independent experiments). b, Biofilm formation of isogenic mutants in polystyrene plates. The capsule-deficient mutant forms more robust biofilms (n = 7 biologically independent experiments). c, Impact of capsule production on the invasion of human bladder epithelial cells. The isogenic capsule-deficient mutant displays increased invasion efficiency (n = 3 biologically independent experiments). d, Intracellular K. pneumoniae persist in LAMP1-positive vacuoles: UCI_38ΔwbaP expressing chromosomally integrated mNeon shown 24 h post-infection (see Extended Data Fig. 2f–l for further analysis). The experiments were repeated three times with similar results. e, Persistence of the capsule-deficient isogenic mutant UCI_38ΔwbaP is greater than wild-type UCI_38 in a mouse model of chronic UTI. Bladders were plated 14 d post-infection. The data of two independent experiments are shown. A total of 30 mice were infected per strain. f, Phagocytosis by J774 macrophages. The hypercapsule mutant UCI_38wzcG565A is more phagocytosis resistant than the wild-type parent UCI_38 and corresponding capsule-deficient mutant (n = 4 biologically independent experiments). g, Bloodstream infection of zebrafish larvae with a non-lethal dose of UCI_38 (inoculum = 1,293 ± 733 c.f.u.; n = 19 infected zebrafish) results in lethality after infection with the isogenic hypercapsule wzc mutant UCI_38wzcG565A (inoculum 1,556 ± 820 c.f.u.; n = 23 infected zebrafish). h, Dissemination of UCI_38 and UCI_38wzcG565A from the bladder to the liver in murine UTIs (3 d post-infection; Extended Data Fig. 4d; n = 25 mice (C3H/HeN) infected per group). The hypercapsule mutant displays greater dissemination to the liver than the isogenic, wild-type parent, as demonstrated by bacterial numbers (c.f.u.) recovered in mouse livers (one mouse infected with the hypercapsule mutant died 2 d post-infection). The geometric mean is shown. i, The hypercapsule mutant from UR_11(UCI_11wzcL74P) is lethal in a mouse model of intraperitoneal infection. Mice were infected with an inoculum of 1–2 × 106 bacteria and mouse survival was followed over time (n = 10 BALB/c mice were infected per group). P values are displayed in the figures (hypercapsulated strains were compared with parent strains). The significance was calculated using an unpaired, two-tailed Student’s t-tests (a–c,f), a two-tailed log-rank test (g) and two-tailed Mann–Whitney U-tests (e,h,i). P values are displayed in the figures and are all based on the comparison with UCI_38 or UR_11. The limit of detection is indicated with a dashed line.
The impact of a single loss-of-function mutation resulting in persistence was also seen in an in vivo UTI model. The isogenic UCI_38ΔwbaP mutant, but not the parent UCI_38, was able to establish persistent infection over 14 d in a C3H/HeN mouse model of chronic UTI23 (Fig. 2e and Extended Data Fig. 2m). However, the UCI_38ΔwbaP mutant did not cause increased inflammation, as observed by histology of the bladder at 3 d post-infection, and was recovered less from the kidneys compared with the parent UCI_38, suggesting that loss of capsule enhanced its persistence in the bladder rather than its virulence and dissemination, which is consistent with the in vitro findings of increased epithelial cell invasion and biofilm formation (Extended Data Fig. 2m,n).
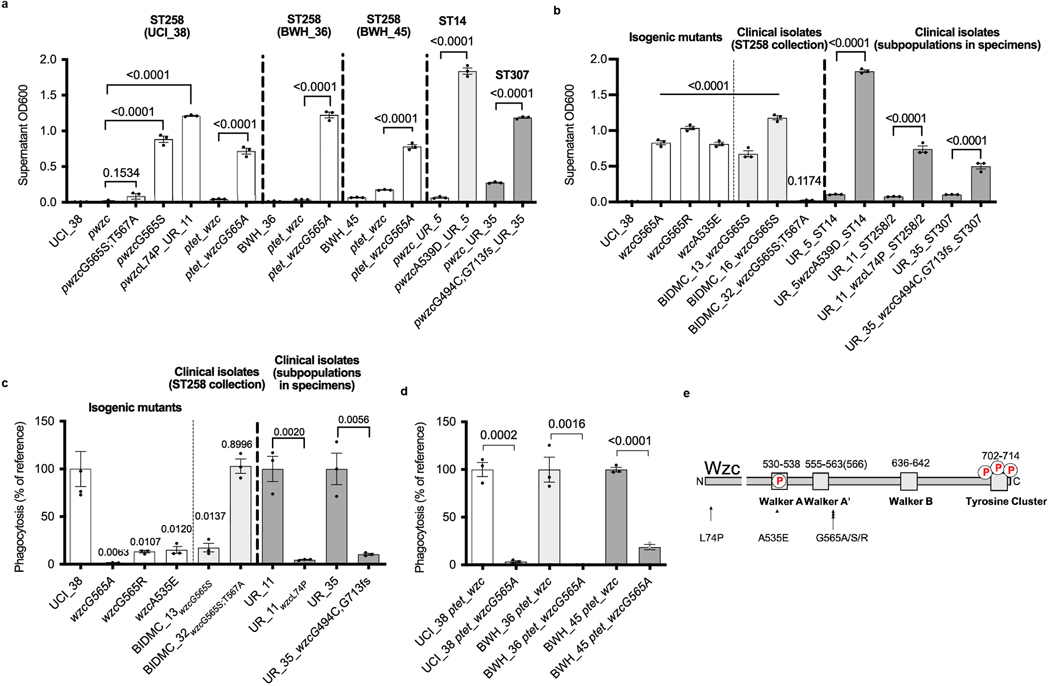
We next turned to understanding the genetic basis underlying the two hypercapsule blood isolates in the originally characterized collection of 54 strains. Sequence analysis of BIDMC_13 and BIDMC_16 revealed that they do not carry the capsule transcription factors rmpA/A2 and the siderophore aerobactin, which have been reported to be associated with hypervirulence in the Asian strain lineage24, in part by increasing capsule production and phagocytosis resistance25,26 (Supplementary Table 2). We also did not detect any mutations in regulators of capsule production (rcsAB, lon27). Instead, they carry a single missense mutation in the wzc gene, resulting in a glycine-to-serine substitution (Gly-565Ser; Supplementary Table 1). Wzc is required for high-level capsule polymerization in E. coli18, and mutations in the same region of the wzc gene of hypermucoid clinical Acinetobacter baumanii isolates result in a hypercapsule28 (Extended Data Fig. 3e). Episomal expression of the mutated wzc allele in UCI_38 confirmed that the Gly-565Ser substitution indeed conferred a hypercapsule, and that an additional Thr-567Ala mutation found in BIDMC_32 suppressed hypercapsule formation (Fig. 1b and Extended Data Fig. 3a,b). Although we were unable to engineer the point mutation corresponding to the Gly-565Ser substitution via two-step allelic exchange into the wild-type chromosomal copy of wzc in UCI_38, we did isolate a spontaneous single isogenic hypercapsule-producing mutant of UCI_38 harboring a Gly-565Ala substitution in wzc from a culture grown in vitro, based on its hypermucoid colony morphology (Extended Data Fig. 1c) and whole-genome sequencing, which did not reveal any additional mutations. In general, isolation of such wzc mutants was rare because otherwise we did not detect hypercapsule mutants from overnight cultures that were plated for single colonies, and the frequency of wzc mutants in overnight cultures was below the detection limit of 0.5%, as determined by deep sequencing. UCI_38wzcG565A, similar to the clinical isolate BIDMC_13wzcG565S, produced and polymerized significantly more capsule than its isogenic parent UCI_38 (Figs. 2a and 1b and Extended Data Fig. 3b), without any noticeable fitness cost in axenic culture (Extended Data Fig. 4a). Episomal expression of the mutated wzc allele containing the Gly-565Ala substitution in two other ST258 genetic backgrounds also resulted in hypercapsule production (BWH_36 and BWH_45; Extended Data Fig. 3d), thus confirming that a single point mutation in wzc causes hypercapsule production in K. pneumoniae ST258. Furthermore, the increased capsule indeed confers significant phagocytosis resistance to macrophages (Fig. 2f and Extended Data Fig. 3a–d)11,29, which has been reported previously as a contributing mechanism for increased virulence in hypervirulent strains25,26.
Hypercapsule production also resulted in increased virulence consistently across numerous animal models, including increased lethality in a bloodstream infection model of whole-organism zebrafish larvae (Fig. 2g and Extended Data Fig. 4b,c) and more effective dissemination in a murine UTI model—as evidenced by higher bacterial recovery in the kidneys and liver after inoculation with mutant rather than wild-type bacteria into the bladder (Fig. 2h and Extended Data Fig. 4d), increased inflammation on histological analysis of livers infected with the hypercapsule mutants (Extended Data Fig. 4e), and increased recovery of the hypercapsule mutant relative to a normal-capsule strain from the spleen in a mouse model of bloodstream infection (Extended Data Fig. 4f). In addition, after bladder inoculation with wild-type K. pneumoniae (UCI_38), we recovered phagocytosis-resistant, hypercapsule-producing wzc mutants (Gly-565Arg and Ala-535Glu) in abscesses of (toll-like receptor 4-deficient) mice, confirming the increased capacity of hypercapsulated ST258 strains to cause disseminated infection (Supplementary Table 3 and Extended Data Figs. 3a–c,e and 4g). Although we did not observe lethality with the in vitro-generated hypercapsule mutant (UCI_38wzcG565A) in mouse models of infection, this may be due to limitations in the mouse models because ST258 strains have been clearly documented as having high mortality in humans6,9,10 while being relatively avirulent in mice30. Thus, taken together, these results indicate that single mutations in wzc (wzcG565S/A/R, wzcA535E) confer phagocytosis resistance and consistently enhance the virulence of ST258 in multiple animal infection models.
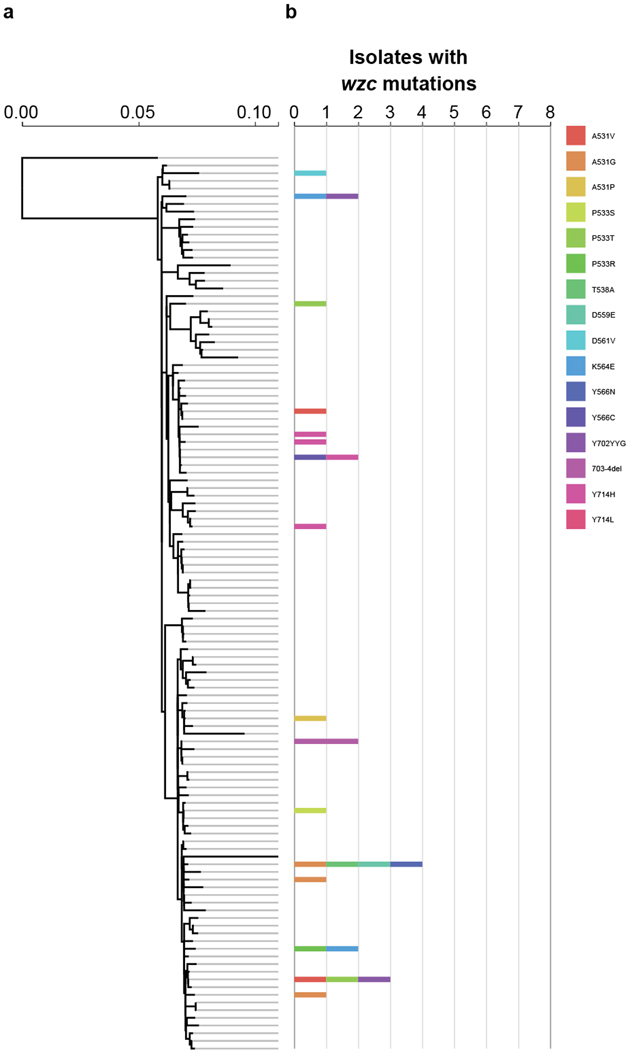
Given the ease with which ST258 strains could alter their infection programs through single mutational events, we suspected that the emergence of hypercapsule or capsule-deficient ST258 mutants was not limited to the characterized collection of 54 ST258 isolates. We thus searched all available ST258 clade 2 genomes in the National Center for Biotechnology Information (NCBI) RefSeq database for the presence of non-synonymous wzc mutations and wbaP disruptions. Of 966 genomes, 95 strains (10%) harbored mutations in the same wzc and wbaP capsule genes that we had observed in the original ST258 collection and from our in vitro and in vivo laboratory studies. These mutants appeared across the ST258 phylogeny from patient samples obtained from different parts of the world, including from two documented hospital outbreaks31,32 (Fig. 3, Extended Data Fig. 5 and Supplementary Tables 4 and 5). The excess of non-synonymous mutations versus synonymous mutations in wzc (86.5% versus 13.5%), as well as the absence of synonymous mutations in functional motifs of wzc, pointed to the repeated selection of wzc mutants. Indeed, we found 20 wzc mutations (2.1%; Fig. 3) that altered the same amino-acid positions we had previously identified as conferring hypercapsule production (positions 565 and 74; Supplementary Tables 1 and 3 and Extended Data Fig. 6), and 19 wzc mutations that were located next to hypercapsule-conferring mutations in wzc (2%) (Supplementary Table 4 and Extended Data Figs. 3e and 5); these findings resulted in an estimated prevalence of 4.1% of hypercapsule mutants among sequenced ST258 genomes. We also identified 56 wbaP disruptions, resulting in an estimated prevalence of 5.8% capsule deficiency among sequenced genomes (Fig. 3 and Supplementary Table 5), which is likely to be a low estimate given that disruptions in many other genes can interfere with capsule production. Thus, capsule-deficient and hypercapsulated mutants occur across the phylogeny of ST258, and can be induced with single mutations in different ST258 strain backgrounds (Extended Data Figs. 2a and 3a), to yield UTI-or virulence-related phenotypes (Extended Data Figs. 2c and 3d). These results suggest that a particular strain genetic background may not be required for the emergence of uropersistent (capsule-deficient) or increasingly virulent (hypercapsulated) K. pneumoniae (Fig. 4), and that these mutations probably benefit the pathogen during infection. Importantly, analysis of infection-related isolation sites of all sequenced ST258 isolates from RefSeq (infection site data available for 610 of 966 isolates) revealed significant associations of hypercapsule mutants with bloodstream infections (50% of all wzc mutants versus 30.45% of all wild-type strains are isolated from the blood, P = 0.0299) and capsule-deficient mutants with UTIs (62% of all wbaP mutants versus 39.8% of all wild-type strains are isolated from urine, P = 0.02) (Supplementary Table 6), thereby recapitulating the observations from the initial collection of 54 (ref. 13) ST258 isolates on a larger scale and pointing to the potential clinical impact of the capsule mutations in human infections.
Fig. 3 |. Hypercapsule and capsule-deficient mutants are widespread across the ST258 phylogeny.
a, ST258 clade 2 reference tree, comprising 117 reference genomes that represent the phylogenetic diversity of 966 ST258 clade 2 strains identified in RefSeq (Methods). b, Phylogenetic distribution of isolates in RefSeq carrying wzc mutations at positions 74 and 565, which were found to confer a hypercapsule. The count of strains carrying the designated mutation is mapped on to the phylogenetic reference tree, assigned to the strain in the reference tree that is most similar to the wzc mutation-carrying strain. Different mutations are indicated in different colors. Strains carrying wzc mutations from known hospital outbreaks are indicated: (1) Geneva (Switzerland)31 and (2) North Carolina (United States)32. c, Phylogenetic distribution of strains with inactivated wbaP. The count of strains carrying the designated wbaP mutation is mapped on to the phylogenetic reference tree and assigned to the strain in the reference tree that is most similar to the wbaP mutation-carrying strain. Colors indicate the length of the truncated WbaP protein relative to the consensus WbaP sequence of 475 amino acids. Mutations that occurred only once in our dataset are indicated in dark gray (other). Detailed information on the mutants is available in Supplementary Tables 4 and 5.
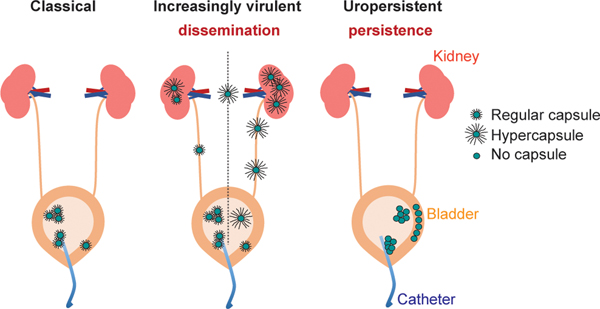
Fig. 4 |. A model for how capsule remodeling results in different infection phenotypes of K. pneumoniae.
Proposed model for the impact of capsule remodeling in catheter-associated UTIs. Classic K. pneumoniae strains are relatively resistant to phagocytosis during colonization of the bladder, by establishing biofilms on catheters and bladder epithelium and by invading bladder epithelial cells; the capsule, however, limits biofilm formation and invasion of bladder epithelial cells, thereby restricting infection to a relative degree. Increasingly virulent K. pneumoniae, which contain wzc mutations resulting in a phagocytosis resistance-conferring hypercapsule, can disseminate more effectively, but are unable to form a biofilm or invade bladder epithelial cells effectively. Uropersistent K. pneumoniae are impaired in capsule production and therefore are even more effective at forming a biofilm and invading bladder epithelial cells than classic strains. The increased invasion efficiency results in a larger intracellular reservoir, in which K. pneumoniae ST258 can persist in Klebsiella-containing vacuoles in the face of phagocytes, which leads to persistent UTIs and an untreatable intracellular reservoir.
The consistent detection of independently acquired capsule mutations in global genome sequence collections of colony-purified ST258 isolates suggested that the mutants had repeatedly undergone significant purifying selection, enabling their detection at relatively high frequency (10%), because only a single colony from plated patient specimens was picked for sequencing. We considered the possibility that such mutants could exist at even higher frequencies as subpopulations from patient samples that had not been subjected to a colony-purifying bottleneck. We collected 30 urine specimens prospectively from patients with K. pneumoniae UTIs (at the Brigham and Women’s Hospital, Boston (BWH)) and plated the specimens directly for single colonies to look for hypercapsule and capsule-deficient subpopulations, based on colony morphologies (example shown in Extended Data Fig. 1c) and string-test performance14. Indeed, we detected hypermucoid and hypomucoid subpopulations in 6 of 30 specimens (20%). We detected hypermucoid, phagocytosis-resistant, mutant subpopulations with hypercapsule-conferring SNPs in wzc (Extended Data Fig. 3a–c) in the urine of 3 of 30 patients (UR_5, UR_11, UR_35; Extended Data Fig. 6, visual detection limit: 0.2–0.5%); in one of these cases, we also detected a coexisting capsule-deficient subpopulation harboring a wbaP mutation (UR_5; Extended Data Fig. 6). (The limited time for bacterial growth after sample collection excluded the possibility that the mutants emerged in the specimen ex vivo (Supplementary Table 7).) We also detected homogeneous, capsule-deficient, biofilm-producing populations in 3 of 30 patient specimens, with 2 of them causing acute UTIs (based on symptoms and the presence of white blood cells in the urine), which included one patient with a history of recurrent UTIs (UR_15), confirming the ability of capsule-deficient mutants to persist in the host, with the capacity to cause acute (immunogenic) UTIs in patients, perhaps at a time of impaired mucosal defense (UR_15, UR_47, UR_53; Extended Data Fig. 2b and Supplementary Table 8).
Interestingly, the presence of hypercapsule mutant subpopulations in these urine specimens was significantly associated with multidrug resistance, occurring in 3 of the 6 multidrug-resistant (MDR) samples collected and in none of the 21 antibiotic-susceptible specimens (P = 0.0068; Supplementary Table 9). Importantly, all hypercapsule subpopulations were related to regular-capsule populations present in the same sample (Extended Data Fig. 6 and Supplementary Table 10), suggesting that they had probably been generated during the course of infection. Of note, the capsule mutants detected in these three patients spanned a number of multidrug-resistance-associated sequence types, and were not restricted solely to ST258 (including ST14 and ST307; Extended Data Fig. 6). Thus, the factors that contribute to the emergence and selection of MDR organisms in a patient probably also contribute to the evolution of virulent or persistent capsule mutants, perhaps affording the time for capsule mutations to be acquired9,10,33, as suggested by the multiple SNPs found in the capsule mutant subpopulations (Supplementary Table 10). Taken together, these results suggest that capsule mutants are frequently generated in MDR infections. They are prevalent (10–20% in all infections, 50% in drug-resistant infection) and can be missed by standard diagnostic procedures and whole-genome sequencing, which rely on colony purification of isolates from patient specimens. Although the analysis of available ST258 genomes shows that they are geographically diverse, this analysis of primary specimens, albeit a smaller set, suggests that potentially they also span a phylogenetic spectrum, with the potential to involve not only carbapenem-resistant ST258 strains but also MDR strains of other sequence types.
When we characterized the virulence of one of the hypercapsule ST258 mutants that had emerged in a patient UTI (UR_11wzcL74P, which did not harbor any known hypervirulence genes but did contain a wzc mutation), not only did we find that it was more virulent than its isogenic, normal-capsule parent, which is consistent with our previous findings with wzc mutants, but, notably, we also found that it was remarkably lethal in three different mouse infection models. In the mouse UTI model, compared with the normal-capsule parent (UR_11), the mutant killed 4 of 15 mice and caused pyogenic liver abscesses (which we had never previously observed in this infection model; Extended Data Fig. 4h,i). In a mouse model of bloodstream infection in which lethality is rarely observed with ST258 strains and only with high bacterial inocula (108) over long periods of time (days), the hypercapsule mutant UR_11wzcL74P displayed rapid and complete lethality by 8 h with an inoculum of 107 bacteria; although the normal-capsule mutant also displayed lethality, it did so with a significant delay in death compared with the hypercapsule mutant (Extended Data Fig. 4j). Finally, in a mouse intraperitoneal infection model, the hypercapsule mutant was lethal at an inoculum of 106 bacteria, compared with most previously described requirements of 108–109 ST258 bacteria for lethality in this model30,34 (Fig. 2i). Although the level of virulence of the hypercapsule mutant was less than that of the well-documented Asian hypervirulent ST23 strains29,35 (Extended Data Fig. 4k,l), the striking virulence of the hypercapsule mutant observed in all three mouse models, relative to the normal-capsule parent isolated from the same patient, demonstrates that hypercapsule production significantly increases the virulence of ST258, albeit with the absolute level of virulence depending on the strain background.
There has been tremendous interest in understanding the success of highly drug-resistant pathogens such as methicillin-resistant Staphylococcus aureus36 and now carbapenem-resistant K. pneumoniae. Although the previously reported, convergence event of carbapenem resistance with hypervirulence in Asia involved the acquisition of well-established hypervirulence factors rmpA2 and aerobactin into the ST11 sequence type11, we now demonstrate the globally occurring, parallel, within-host adaptive evolution of virulence in resistant ST258 infections by the acquisition of single hypercapsule-conferring mutations, resulting in classic virulence phenotypes, such as increased lethality and dissemination. Moreover, we show that capsule-deficient strains, which were previously believed to be completely avirulent17, actually have a fitness advantage in certain infections, because they display a greater persistence in mouse UTIs, while preserving the potential to cause acute infection, are drug tolerant and thus untreatable (Fig. 4 and Extended Data Fig. 2e), and can serve as a reservoir for antibiotic resistance genes in patients. Ecological stability stemming from chronic infection and the constant selection against immunogenicity appear to be driving the selection for uropersistent capsule-deficient mutants, in analogy to Pseudomonas aeruginosa mutants that emerge from cystic fibrosis patients, which are locked in a sessile lifestyle37. In addition, phagocytosis resistance, conferred by intracellular persistence and biofilm formation in the case of capsule-deficient mutants or hypercapsule production in the case of hypercapsule mutants, appears to be one contributing factor in the emergence of the two different infection programs38, which may include other factors such as cytokine responses39. Although the mutants have very clear advantages in certain infectious settings, in different or even fluctuating host environments encountered during different infection types, they may be disadvantageous, perhaps explaining why they have not completely taken over all strains.
The prevalence we have identified in Europe and the United States, and the evidence that the corresponding mutations are undergoing selection, suggest that these virulence and persistence mechanisms may contribute to the high mortality reported for carbapenem-resistant and MDR K. pneumoniae infections and to the spread of these strains; future studies will be revealing. Importantly, we found that capsule mutants are associated with and preferentially emerge in MDR infections, perhaps because of common features that may be shared by patients in whom drug-resistant bacteria and bacteria carrying these capsule mutations emerge. Such drug-resistant strains are occurring at an alarmingly high frequency in certain areas of the world2. At the same time, the detection of capsule mutants in drug-susceptible infections (albeit at a lower frequency compared with MDR infections) in our prospective collection of patient specimens suggests that the evolution of persistence and virulence could be a general phenomenon in K. pneumoniae infections, not restricted to drug-resistant infection. A better understanding of the evolutionary capacity of bacteria to become increasingly pathogenic will be necessary to detect, monitor and develop interventions that will minimize the potential impact of such pathogens on human health40.
Online content
Any methods, additional references, Nature Research reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at https://doi.org/10.1038/s41591-020-0825-4.
Methods
Strains and growth conditions.
Collection of K. pneumoniae ST258 isolates.
The collection of K. pneumoniae ST258 isolates is part of a recently characterized sequenced collection of carbapenem-resistant Enterobacteriaceae isolated from three Boston area hospitals (Beth Israel Deaconess Medical Center (BIDMC), Brigham and Women’s Hospital (BWH), Massachusetts General Hospital (MGH)) and one hospital in Irvine, California (University of California Irvine Medical Center (UCI))13. The collection includes 54 ST258 strains that were either prospectively collected from 2012 to 2013 over a period of 16 months or sporadically collected between 2006 and 2012 (historical), as indicated in Supplementary Table 1. Both ST258 clades (clade 1 and clade 2) are represented in the collection, which was found to be genetically diverse with only limited evidence of clonal spreading13. Most of the ST258 strains in the collection are carbapenem resistant and all strains are MDR8. The urine isolate UCI_38 was chosen for in-depth analysis of the impact of capsule mutations, because it did not harbor any mutations in capsule biosynthesis genes, could be transformed with common lab plasmids and was susceptible to chloramphenicol and gentamicin, which facilitated genetic manipulation and infection experiments. The primary isolates (single colonies on an agar plate stored at room temperature for 1–2 d) were streaked out on fresh agar plates, from which slants or glycerol stocks were prepared from overnight cultures and sent to the lab at BWH, which managed the collection of isolates. The isolates were streaked out one more time and overnight cultures were prepared from single colonies and frozen at −80 °C with Microbank beads. New glycerol stocks were created from the original Microbank stocks13 by streaking the existing stocks out for single colonies and preparing new glycerol stocks (final glycerol concentration: 25%) from overnight cultures started from a single colony. The glycerol stocks were stored at −80 °C and were used for all experiments described below. SGH_10 is a recently characterized ST23 isolate, which was isolated from a 35-year-old patient with liver abscesses and no underlying disease in Singapore in 2014, and was recently proposed as a reference strain for experimental and genomic studies of hypervirulent CG23 strains, because its genome was found to represent the predominant CG23-I sublineage and does not harbor atypical accessory genes24. All strains were cultivated in lysogeny broth (LB) medium at 37 °C under shaking conditions unless otherwise specified. Antibiotics were used at concentrations of 50 μg ml−1 of carbenicillin, 10 μg ml−1 of tetracycline and 25 μg ml−1 of chloramphenicol. The culture and storage conditions of strains from RefSeq are, in most cases, unclear.
Comparative assessment of colony mucoidity.
Isolates were grown overnight from glycerol stocks in 5 ml LB medium. Serial dilutions were prepared and plated for single, well-separated colonies on blood agar plates (tryptic soy agar with 5% defibrinated sheep blood, Remel). After 24 h incubation at 37 °C, colony mucoidity was determined (Extended Data Fig. 1a). UCI_38, a ST258 clade 2 isolate, which does not harbor mutations in capsule biosynthesis genes (Supplementary Table 1), and displays regular mucoidity, was used as a reference. Hypomucoid colonies could be easily identified by a grayish, flat appearance, and a translucent appearance on LB agar plates (Extended Data Fig. 1b), whereas hypermucoid colonies were obviously more mucoid and formed bigger colonies (Extended Data Fig. 1a,c). The significance of the association of mucoidity phenotypes with isolation sites was determined using Fisher’s exact test.
Generation of an isogenic wbaP deletion mutant in UCI_38, BWH_36 and BWH_45.
The deletion mutant wbaP was deleted via two-step allelic exchange using the suicide vector pDMS197 (ref. 41). Flanks of wbaP were amplified (724-base pair (bp) downstream, 635-bp downstream), cloned into the pir1-dependent and sacB-harboring pDMS197 vector via Gibson42 assembly, and electroporated into E. coli One Shot pir1. The resulting plasmid, pDMS197_wbaP, was electroporated into the donor strain MFDpir43. Matings of the donor strain harboring pDMS197_wbaP and K. pneumoniae UCI_38 were incubated for 4 h at 37 °C and then plated on LB agar plates supplemented with tetracycline (10 μg ml−1) and carbenicillin (50 μg ml−1) to select for the first crossover event resulting in chromosomally integrated pDMS197_wbaP (merodiploids). Merodiploid colonies were streaked over to non-salt-containing LB agar plates supplemented with 15% (w:v) sucrose to force a second crossover event (due to SacB-mediated sucrose sensitivity). The resulting deletion mutant was verified via PCR and whole-genome sequencing (see “Whole-genome sequencing of capsule mutant subpopulations from urine specimens”). Of note, the generation of merodiploids was very inefficient and therefore several attempts were needed to generate the isogenic deletion mutant. Multiple attempts at deleting wbaP via the lambda red system failed.
Cloning of wzc variants and expression in UCI_38, BWH_36 and BWH_45.
The wild-type and mutated wzc variants were amplified with the primers listed in Supplementary Table 11 and cloned into pBAD33 (ref. 44) or pBAD33tet downstream of an arabinose-inducible promoter via the Kpn1 and Sac1 restriction sites. Expression of wzc variants was induced with 50 mM L-arabinose.
Cloning of wbaP and expression in UCI_38.
The wbaP gene from UCI_38 was cloned with the primers listed in Supplementary Table 11 into pBAD33 (ref. 44) via the Kpn1 and Pst1 restriction sites. Expression of wbaP was induced with 50 mM L-arabinose. Induced overnight cultures were used in invasion assays and for electron microscopy.
Construction of pBAD33tet.
The chloramphenicol resistance gene in pBAD33 (ref. 44) was replaced with a tetracycline resistance gene from pACYC184 (ref. 45) via Gibson cloning42.
Integration of mNeon in UCI_38 and UCI_38ΔwbaP.
The integration of mNeon46 into the chromosome of UCI_38 and UCI_38ΔwbaP was carried out using a Tn7 transposon vector47. First, as a result of kanamycin resistance in UCI_38, the kanamycin-resistance cassette in the transposon vector pSTNSK47 was replaced with a tetracycline-resistance cassette from pACYC184 (ref. 45) via Gibson cloning42, resulting in pSTNST, which was introduced into UCI_38 and UCI_38ΔwbaP via electroporation. The mNeon was cloned in front the synthetic promoter proD48 and, with an optimized ribosome-binding site (biobrick part BB0030), into the mobilizable suicide vector pGP-Tn7-Cm47 between two Tn7 ends via Gibson cloning42. The construct was then used to transform UCI_38 pSTNST and UCI_38ΔwbaP pSTNST, leading to the integration of mNeon into the glmS site of UCI_38 (verified via PCR). The resulting strains were then cured of pSTNST, which harbors a temperature-sensitive origin of replication.
Detection of capsule mutants in patient samples.
Institutional review board approval was granted by the Partners Human Research Committee. Discarded, de-identified samples were collected from inpatients at the BWH, or from outpatients at BWH-affiliated locations, under study approval in 2018. Records on the specimen isolation time point, primary analysis time point, work shifts and time point of 4 °C storage enabled a worst-case-scenario estimate for the time that the cultures had been kept at room temperature after isolation from the patient and are displayed in Supplementary Table 7. Cultures that tested positive for only K. pneumoniae (usually within 24 h of specimen isolation), with a concentration of more than 100,000 bacteria per ml, were obtained and sent out for further analysis to MGH in a styrofoam coolbox with a cool pack, arriving within 1–4 h. The samples were immediately serially diluted in phosphate-buffered saline (PBS) and plated for colony-forming units (c.f.u.; without any culturing step) on LB agar plates, and 25% glycerol stocks with 750 μl urine were prepared for long-term storage. After incubation for 24 h at 37 °C 200–500 colonies were analyzed for variation of colony morphology, as an indicator of altered capsule production. LB agar plates were used rather than blood agar plates for the identification of capsule mutant subpopulations, because of the more consistent size and more characteristic texture of the colonies that resulted compared with colonies formed on blood agar plates, enabling the identification of hyper-and hypomucoid colonies by mucoidity (example shown in Extended Data Fig. 1c) and string-test. Hyper-and hypomucoid colonies were streaked out on LB and blood agar plates to confirm the mucoidity phenotype (hypomucoid: grayish appearance on blood agar plates, translucent appearance on LB agar plates; hypermucoid: positive string-test of single colonies on blood plates) and were then further analyzed for the presence of mutations using whole-genome sequencing (see “Whole-genome sequencing of capsule mutant subpopulations from urine specimens”).
Growth of isolates from specimens with capsule subpopulations.
Overnight cultures were washed in PBS and diluted to a final optical density (OD) of 0.02 in LB medium or urine from a healthy donor. The cultures were then transferred to a 96-well plate and the OD was measured every 10 min for up to 16 h at 24 °C (room temperature), which also included a short period of shaking before the OD was measured. The logarithmic growth phase was identified and two time points in the log(growth phase) were chosen to calculate the doubling time (OD2: late time point, OD1: early time point):
Biofilm formation.
Biofilm production was determined as described recently49. Shaking overnight cultures were back-diluted 1:1,000 in LB medium and a total volume of 200 μl was transferred to wells of untreated 96-well polystyrene plates. After 24 h incubation at 37 °C, the wells were washed four times with water and 100 μl of 0.1% crystal violet was added. After 10 min incubation crystal violet was removed and the wells were washed six times with water. Then, 200 μl of 80% ethanol/20% acetone was added and the plate was incubated for 10 min at room temperature before determining the OD590 with a microplate reader (Spectramax M5). At least six replicates were used per experiment.
Transmission electron microscopy.
Transmission electron microscopy was performed using the Electron Microscopy Facility of Harvard Medical School. A pellet of cells was fixed for at least 2 h at room temperature in 2.5% glutaraldehyde, 1.25% paraformaldehyde and 0.03% picric acid in 0.1 M sodium cacodylate buffer (pH 7.4), washed in 0.1 M cacodylate buffer and postfixed with 1% osmium tetroxide/1.5% potassium ferrocyanide for 1 h, washed twice in water, once in maleate buffer and incubated in 1% uranyl acetate in maleate buffer for 1 h, followed by two washes in water and subsequent dehydration in grades of alcohol (10 min each: 50%, 70%, 90%; 2 × 10 min: 100%). The samples were then incubated with propylene oxide for 1 h and infiltrated overnight in a 1:1 mixture of propylene oxide and Spurr’s low-viscosity resin (Electron Microscopy Sciences). The following day the samples were embedded in Spurr’s resin and polymerized at 60 °C for 48 h. Ultrathin sections (about 60 nm) were cut on a Reichert Ultracut-S microtome, transferred to copper grids stained with lead citrate and examined in a JEOL 1200EX transmission electron microscope, and images were recorded with an AMT 2k CCD camera.
Extraction and quantification of capsule.
The capsule was extracted and uronic acids were quantified to assess capsule production. Overnight shaking cultures grown in LB medium were adjusted to an OD600 of 1.5 in LB medium and serial dilutions were plated for colony-forming units, to normalize the amount of uronic acids to 109 bacteria. The capsule was extracted as previously described16 in triplicate. Next, 500 μl culture, including LB medium controls, was mixed with 100 μl of 1% Zwittergent 3–14 detergent in 100 mM citric acid, pH 2.0, and heated for 30 min at 50 °C with occasional vortexing. The samples were centrifuged for 5 min at 16,000g and 300 μl of the supernatant was transferred to a new tube and precipitated with ethanol (final concentration 80%) at 4 °C for 30 min. The precipitate was centrifuged for 5 min at 16,000g and the supernatant was discarded, followed by drying and uptake of the precipitate in 250 μl of 100 mM HCL. The capsule was quantified as described previously50. The samples, including a series of tubes containing twofold serial dilutions of glucuronolactone standard, were cooled on ice and 20 μl of cooled 4 M ammonium sulfamate was added and mixed. Then, 1 ml of 25 mM sodium tetraborate in concentrated sulfuric acid was added to the samples, mixed and boiled at 100 °C for 15 min to hydrolyze capsule polysaccharides. The samples were cooled down to room temperate for approximately 30 min, before the addition of 40 μl of 0.15% m-hydroxybiphenyl and further incubation for 15 min to generate the chromophore. The samples were then transferred to disposable plastic cuvettes and the OD was determined at 520 nm. Uronic acids were quantified by comparing the OD with the glucuronolactone standard curve.
Centrifugation resistance as an indicator of excessive mucoidity.
Mucoidity was quantified as described recently with slight modifications51. Cultures were started in LB medium from glycerol stocks, which in the case of the cloned wzc variants was supplemented with 50 mM L-arabinose, and were incubated under shaking conditions at 37 °C for 18–20 h. Triplicate aliquots (2 ml) of the overnight cultures were centrifuged for 30 s at 16,000g, followed by immediate measurement of the OD of the supernatant (OD600).
Sequencing and identification of genes of interest.
Identification of capsule mutations (including IS elements).
The capsule biosynthesis genes of UCI_38 (clade 1) and BWH_22 (clade 2), which both display regular colony mucoidity, were compared with the corresponding capsule biosynthesis genes of other ST258 isolates belonging to the same clade (the genomes are available in the NCBI database; Supplementary Table 1). Each protein sequence in the K-locus (encompassing GalF and Ugd5) was used as a query in BLASTp to look for mutations in the capsule biosynthesis genes of clade 1 and clade 2 genomes from the ST258 collection. Protein truncations were further analyzed for deletions and the presence of ISs in the affected regions, which in the case of insertions led to the identification of annotated transposases. The nucleotide sequence of identified transposases including 100 bp of the flanking regions were further analyzed by searching the ISfinder database for corresponding, known ISs (https://isfinder.biotoul.fr) and are listed in Supplementary Table 1 (ref. 52).
Identification of rmpA/A2 and siderophores in the collection of ST258 genomes.
The presence or absence of rmpA/A2 and siderophores in the ST258 collection was determined using Kleborate v.0.30 (https://github.com/katholt/Kleborate)12,53.
Identification of mutations in RcsAB and Lon.
RcsA (EWD33475), RcsB (EWD32264) and Lon (EWD28182) from UCI_38 were used as queries in BLASTp to look for mutations in the collection of ST258 isolates13.
Sequencing of UCI_38ΔwbaP and UCI_38wzcG565A.
Illumina sequencing libraries were prepared using the Illumina Nextera XT Kit according to the manufacturer’s guidelines. Libraries were 100 bp, paired-end sequenced using the Hiseq2500 platform to achieve 100-fold coverage. The resulting paired reads were mapped to the UCI_38 genome and analyzed using Pilon v.1.23 (ref. 54) with default settings. All variants were manually examined and only confident calls with the expected coverage and good alignment quality were considered to pass. The wzcG565A and ΔwbaP mutations were the only mutations identified by Pilon across any of the strains.
Proximity of the capsule operon to the origin of replication.
The capsule operon of KPNIH33, GCA_000775375 (used as reference for the construction of the ST258 phylogenetic tree), was identified using Kaptive v.0.5.1 (ref. 55), whereas the origin of replication (oriC) was identified using BORIS56 (https://boris.mathematik.uni-marburg.de).
Deep sequencing of UCI_38 cultures.
An overnight culture (shaking, 5 ml LB medium) was started from a shortly outgrown single colony (grown overnight at room temperature followed by approximately 5 h of growth at 37 °C). Genomic DNA was isolated from 1 ml culture with the DNeasy Blood & Tissue Kit from Qiagen. The C-terminal part of wzc was amplified (5′-CAAGTCCAAATGCGGGTAAGAC and 5′-CCGGCTTTTGTTGAAAACACTTCTTTAC) with the high-fidelity Phusion polymerase (New England Biolabs). Libraries were 150 bp, paired-end sequenced by the MGH DNA core facility with a MiSeq platform to achieve 100,000×coverage. The reads can be found in the Sequence Read Archive (SRA) of the NCBI database under Bioproject, accession no. PRJNA506070, Biosamples SAMN10604457 and SAMN10604458. Deep coverage of wzc was achieved in two regions: basepairs 1647–1816 and 1884–2055/amino acids 549–605 and 628–685), covering the Walker A and Walker B boxes (Extended Data Fig. 3e). The raw reads were mapped to wzc with Bowtie 2 (v.2.3.2) and analyzed with Lofreq (http://csb5.github.io/lofreq, v.2.1.3.1 (ref. 57)) for the occurrence of SNPs with the -N option to disable the use of mapping quality from Bowtie 2 in the analysis, as recommended. Primer-binding regions were removed from the analysis (basepairs 1636–1656, 1965–1984), as recommended. Of note, Lofreq considered only small nucleotide variants (SNVs) with a quality score of 50 (error probability 1 in 100,000) and a minimum of 10 reads in its algorithm. Although the algorithm has been shown to reliably detect SNVs as low as 0.05% (ref. 57), the error rate of the Phusion High Fidelity polymerase is predicted to occur at a rate of 0.5% (based on a 4.4 × 10−7 error rate, stated by New England Biolabs, 30 cycles and PCR product of 350 bp), and therefore SNVs caused by polymerase errors cannot be distinguished from pre-existing SNVs at a frequency lower than 0.5%.
Identification of wzc and wbaP variants.
Some 6,312 K. pneumoniae assemblies were downloaded from RefSeq on 7 December 2018. Output from Kaptive v.0.5.1 (ref. 55), run with default settings, was used to identify 966 ST258 cps II/ST512 isolates based on capsule-type identification (KL107), and amino-acid substitutions and inactivating events in the Wzc and WbaP proteins, respectively. MUSCLE58 was used to align the amino-acid sequence from each isolate’s predicted Wzc amino-acid sequence to a Wzc consensus sequence. Amino-acid changes at positions found to confer hypercapsule production were identified in Wzc (Ala-74, Ala-535 and Ala-565), as well as amino-acid changes that occurred: (1) near known hypercapsule-associated mutations within the Walker box A (amino-acid positions 530–538), (2) within Walker box A (amino-acid positions 555–563), (3) next to Walker box A (amino-acid positions 564–566) and (4) within the tyrosine cluster (amino-acid positions 702–714; Extended Data Fig. 3e). To identify inactivating events in wbaP, Kaptive output was used to identify isolates lacking a wild-type copy of wbaP (that is, lacking a ‘very high confidence’ wbaP prediction). For these strains, Blastx was used to align the nucleotide sequence of the entire cps locus predicted by Kaptive for the WbaP reference amino-acid sequence chosen by Kaptive. Alignment length was determined by counting from the wild-type wbaP start codon (if present), up until a break in the alignment or a stop codon; strains lacking a start codon were predicted to have no intact wbaP. Mutations in wzc and wbaP were detected in high-quality genome sequences, including a selection of high-quality finished Illumina RefSeq assemblies, high-quality assemblies generated at the Broad Institute using a combination of Illumina data from both jump and fragment libraries13, as well as a high-accuracy assembly generated using a combination of Pacbio and Illumina data.
Construction of the ST258 clade 2 reference tree.
To generate a reference tree for mapping the placement of isolates carrying wzc and wbaP variants of interest, capsule-type KL107 isolate assemblies were filtered to include those that were generated using Illumina data, had read sets available at the SRA and were sequenced at the Broad Institute or submitted to the NCBI before 2016. Each read set was aligned to the ST258 clade 2 reference genome, K. pneumoniae strain KPNIH33 (GCA_000775375), using BWA mem v.0.7.12 (ref. 59), and SNPs were detected using Pilon v.1.23 with default settings54. An SNP matrix was constructed from all Pilon results. The matrix was then clustered using hierarchical agglomerative clustering to identify highly related groups of isolates. A single representative from each cluster with the most complete assembly was selected for downstream analysis (Supplementary Table 12). FastTree v.2.1.8 was used to build a phylogenetic tree from an SNP-based alignment of all variable positions passing Pilon’s default thresholds for coverage, ambiguity and SNP quality. Any alignment positions containing more than ten Ns was removed before tree building.
Phylogenetic placement of wzc and wbaP mutants within a cps II reference tree.
To place the wzc and wbaP mutant strains into phylogenetic context, we compared the k-mer set from each isolate assembly to the k-mer sets of all assemblies used to create the cps II reference tree. K-mer set comparisons were performed using a measure similar to the Jaccard distance: the size of the intersection of the reference k-mer set and the sample k-mer set was divided by the size of the smallest of the two k-mer sets, rather than that of the union, which reduces artifacts due to large insertions. Each strain was mapped to the reference isolate having the highest k-mer-based similarity score. In cases where there were multiple isolates mapping to the same reference, an average nucleotide identity value was calculated using the average nucleotide identity calculator from the Kostas lab (http://enve-omics.ce.gatech.edu/ani), which implements the algorithm of Goris et al.60. Clusters of SNPs were not masked.
Identification of carbapenemase genes.
The Antimicrobial Resistance (AMR) Reference Gene Database of the National Database of Antibiotic Resistant Organisms (https://www.ncbi.nlm.nih.gov/pathogens/antimicrobial-resistance) was downloaded on 22 January 2018. BLAST was used to search each wzc or wbaP mutant for genes encoding carbapenemases from the AMR Reference Gene Database. Hits having an e-value of 10−10 or lower across at least 80% of the reference gene sequence were retained.
Whole-genome sequencing of capsule mutant subpopulations from urine specimens.
Illumina sequencing libraries were prepared with the Illumina Nextera XT Kit according to the manufacturer’s guidelines. Libraries were 37 bp, paired-end sequenced using either the Nextseq500 (UR_5, UR_11) or the MisSeq platform (UR_35) with 50-bp reads to achieve 50-to 100-fold coverage. The reads were submitted to the SRA of the NCBI database under Bioproject, accession no. PRJNA506070. SRST2 (ref. 61) was used to assign an ST classification to each dataset. Using an approach similar to the Jaccard distance (see “Phylogenetic placement of wzc and wbaP mutants within a cps II reference tree”) to compare each sample to >2,500 available K. pneumoniae genome sequences, the best matching reference with a finished genome was selected for variant detection (Supplementary Table 13). Reads from each sample were aligned to the corresponding reference using BWA mem v.0.7.12 (ref. 59) and Pilon v.1.22 (ref. 54) was used with default settings to call SNPs against the reference. SNP positions with mapping quality <10 were removed. To calculate the number of SNPs that differed between two strains, we considered only positions that had high-quality, passing variant calls in both strains. SNPs with the following properties were removed: SNPs in regions annotated as transposases and phages; SNPs in regions predicted to be phage using Phaster62; and SNPs within 20 bp of the ends of scaffolds and within 1,000 bp of each other. Kaptive v.0.5.1 (ref. 55) (https://github.com/katholt/kaptive) was used to identify the K-loci (capsule biosynthesis operon).
In vitro models of infection.
Infection of bladder epithelial cells.
Bladder epithelial cells were infected as previously described63 with slight modifications. Briefly, the human bladder epithelial cell line 5637 (American Type Culture Collection, HTB-9) was seeded in 24-well plates and grown to confluence in RPMI 1640 medium supplemented with 10% fetal bovine serum. Bacteria were grown statically under aerated conditions from freezer stocks for 24 h and then subcultured for another 24 h under static and aerated conditions. Bladder cells were infected at a multiplicity of infection of 100 bacteria per host cell. Infection was synchronized via centrifugation (5 min, 600g) and after a 2-h incubation period the medium was removed, and the wells were washed twice with PBS, followed by the addition of medium containing 300 μg ml−1 of gentamicin to kill all extracellular bacteria. After 2 h the wells were washed three times with PBS and the cells were lysed with 0.1% Triton-X100 (20-min incubation). Serial dilutions of the lysates were plated for colony-forming units to determine invasion efficiency. For long-term persistence in host cells, the wells were washed twice with PBS and the cells were incubated with medium containing 50 μg ml−1 of gentamicin, and intracellular bacteria were enumerated at later time points. The intracellular population of BIDMC_54 was treated with 50 μg ml−1 of gentamicin, 100 μg ml−1 of colistin and 200 μg ml−1 of meropenem/vaborbactam, which corresponded to at least 100× the minimal inhibitory concentration. The activity of gentamicin against extracellular K. pneumoniae was confirmed with cytochalasin D-pretreated cells (Supplementary Fig. 1). Three biological replicates were used per experiment.
Macrophage phagocytosis assay.
J774 murine macrophages were seeded in 24-well plates and grown to confluence in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum. Bacteria were added at a multiplicity of infection of 10 bacteria per host cell and the inoculum was plated for colony-forming units. The plate was incubated for 30 min, followed by three washes with PBS and further incubation with 300 μg ml−1 of gentamicin (with exception of UR_35 isolates, which were treated with 300 μg ml−1 of amikacin due to gentamicin resistance) in growth medium to kill extracellular bacteria. After three washes the cells were lysed with 0.1% Triton-X100 for 20 min and serial dilutions of the lysate were plated for single colony-forming units on LB agar plates. The percentage of phagocytosed bacteria per inoculum was calculated and normalized to UCI_38. The activity of gentamicin or amikacin against extracellular K. pneumoniae was confirmed using cytochalasin D-pretreated cells (Supplementary Fig. 1). Three biological replicates per strain were used per experiment.
Confocal microscopy of intracellular K. pneumoniae.
Bladder epithelial cells (5637) were grown on glass coverslips for 2 d in 12-well plates and infected with UCI_38mNeon and UCI_38ΔwbaP_mNeon as described above, with the exception of a 1-min rather than a 5-min centrifugation step to synchronize infection, to more effectively wash off extracellular bacteria. The infected cells were fixed for 20 min with 4% paraformaldehyde in PBS and stained with a rabbit polyclonal anti-Lamp1 antibody (1:100, Abcam), followed by incubation with a secondary antibody conjugated to Alexa568 (donkey anti-rabbit immunoglobulin G, 1:200, Abcam), as described previously64. The nuclei were stained with Hoechst dye (1 μg ml−1, 20 min). All samples were mounted with Prolong Diamond antifade (Molecular Probes) and imaged using a Zeiss LSM 800 Airyscan confocal microscope. Images were analyzed using Image J. Three slides (six images per slide, corresponding to approximately 1,200–2,000 host cells per slide) were analyzed per condition.
Animal infection models.
Zebrafish husbandry and infections.
Wild-type AB and nacre lines were maintained in buffered reverse osmotic water systems. Fish were fed twice daily with dry feed and were exposed to a 14 h light:10 h dark cycle to maintain proper circadian conditions. Larvae (of undetermined sex given the early developmental stages used) were dechorionated manually and then anesthetized with 0.015% ethyl 3-aminobenzoate methanesulfonate before injection. Larvae were infected at 72 h postfertilization via the common cardinal vein, as visually ascertained under a stereomicroscope. The estimation of the inoculum size per experimental group (mean ± s.d.) was determined by immediate homogenization and plating of two fish at the beginning of the infection and two after the infection of each group. A coefficient of variation ≤30 in the inoculum size was established as a marker of quality for each experiment. Larvae were infected and returned to larval medium (E3), incubated at 29 °C and monitored for survival at regular intervals under a stereomicroscope. Dead larvae were defined by the absence of a heartbeat and circulating blood under a stereomicroscope. All vertebrate animal experiments were performed with the approval of MGH’s Institutional Animal Care and Use Committee.
Mouse model of UTI.
UTIs were established in C3H/HeN or C3H/HeJ (TLR4-deficient) mice as previously described23. Briefly, 8-week-old female C3H/HeN or C3H/HeJ mice (Charles River Laboratories) were inoculated by transurethral catheterization with 1–2 × 107 K. pneumoniae grown statically and under aerated conditions. The static cultures were started from a single, shortly outgrown colony in a 125-ml flask filled with 15 ml LB medium, and subcultured after 24 h for 18 h in a 250-ml flask filled with 25 ml. Approximately 500–1,000 c.f.u. of the inoculum were analyzed for the occurrence of capsule mutants (0.2% limit of detection). The mice were sacrificed at the indicated time points, and the bladders and kidneys were aseptically harvested, homogenized and plated for colony-forming units. The toll-like receptor 4-deficient mice (C3H/HeJ) were used as hosts because they prolonged infections caused by UCI_38 (Extended Data Fig. 4g). All vertebrate animal experiments were performed with the approval of MGH’s Institutional Animal Care and Use Committee.
Mouse models of bloodstream and intraperitoneal infection.
Female BALB/c or C3H/HeN mice aged 6–8 weeks were infected intraperitoneally (100 μl) or intravenously (50 μl) via retro-orbital vein inoculation with 106 or 107 K. pneumoniae harvested from the exponential growth phase and washed in PBS. Mice were monitored for clinical health-scoring criteria every 4 h for the first 16 h and then every 12 h. For mice displaying an elevated health score, monitoring was increased to every 4 h. Mice were euthanized when a ‘pre-lethal critical endpoint’ had been reached. The health-scoring criteria included evaluation of the activity, breath rate, posture, diarrhea, hair, coat, eyes, nose and weight. All vertebrate animal experiments were performed with the approval of MGH’s Institutional Animal Care and Use Committee.
Histological analysis of infected livers and bladders.
The bladders and livers of mice infected transurethrally with UCI_38 and UCI_38ΔwbaP were harvested 3 d post-infection and fixed in 10% formalin in PBS (pH 7) for 36 h. Slides were prepared, stained (with hematoxylin and eosin) and imaged by the Histopathology Research Core at MGH.
Statistics.
Prism 7 was used to calculate the significance with: unpaired, two-tailed, Student’s t-tests; log-rank tests; and two-tailed, Mann–Whitney U-tests. The type and number of replicates are indicated in the figure legends.
Reporting summary.
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Extended Data
Extended Data Fig. 1 |. Mucoidity phenotypes of clinical ST258 isolates.
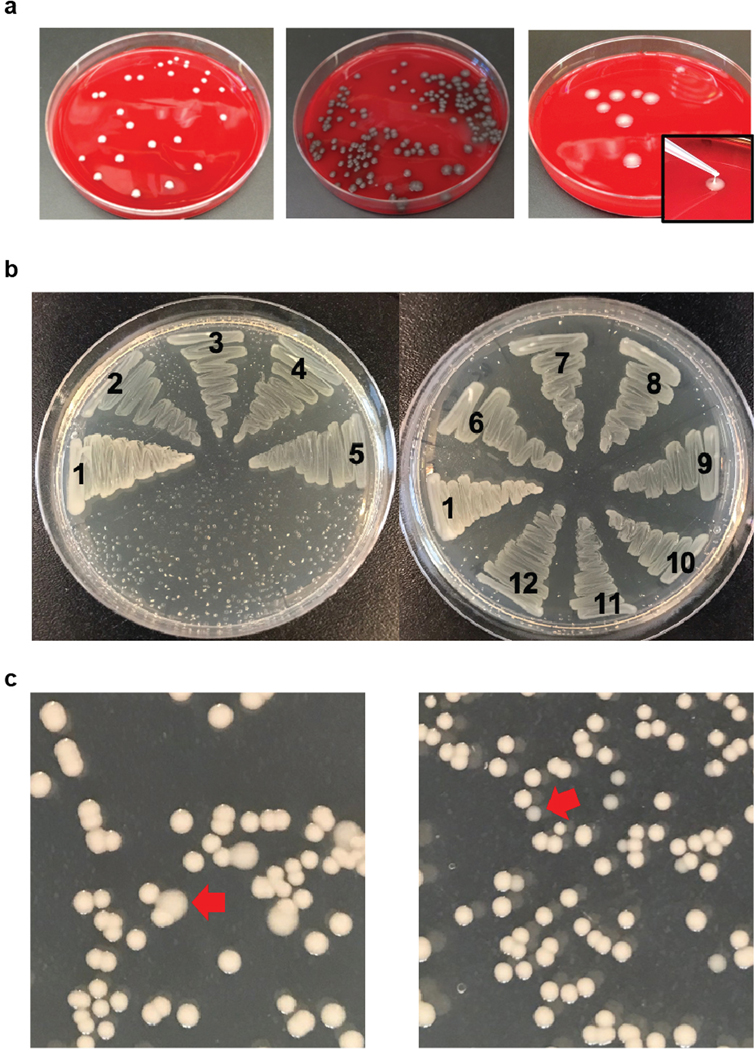
a, Representative colony phenotypes of clinical isolates plated on blood agar plates. The colony morphologies of UCI_38 (mucoid), UCI_37 (hypomucoid) and BIDMC_13 (hypermucoid) are shown. b, Hypomucoid clinical isolates display a translucent appearance on LB agar plates. All of the tested colony-purified ST258 isolates displayed the same colony mucoidity phenotype on blood or LB agar plates. 1, UCI_38 (control normal mucoid); 2, BIDMC_18A; 3, BIDMC_14; 4, BIDMC_54; 5, BIDMC_34; 6, UCI_37; 7, BWH_41; 8, UCI_43; 9, MGH_51; 10, MGH_71; 11, BIDMC_68; 12, MGH_73. c, Identification of capsule mutants on LB agar plates. In order to illustrate the feasibility of the identification of capsule mutant subpopulations on LB agar plates, UCI_38 was mixed with UCI_38wzcG565A (left) or UCI_38ΔwbaP (right) to achieve a concentration of mutants of approximately 5%. Hypermucoid colonies could be easily determined by the different colony morphology and bigger size (red arrow, left), while hypomucoid colonies could be easily identified by their translucent appearance and smaller size (red arrow, right). a-c were repeated 3 times independently with similar results.
Extended Data Fig. 2 |. Capsule-deficiency improves biofilm formation and invasion of bladder epithelial cells.
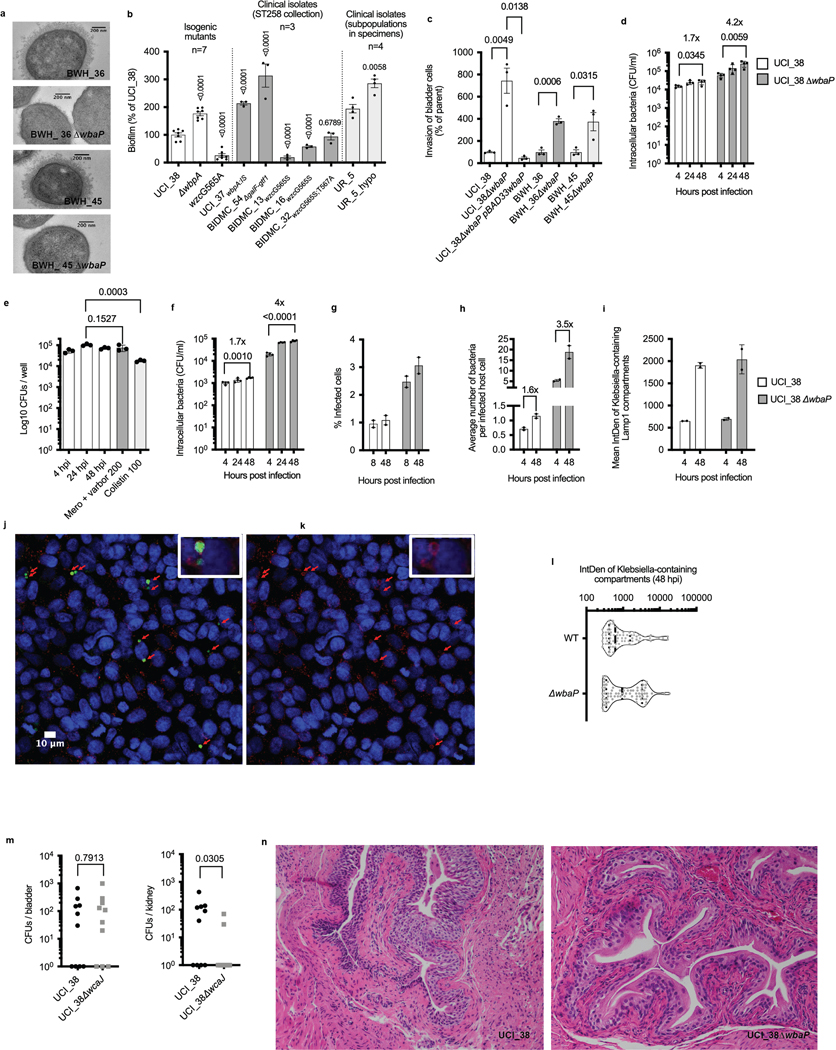
a, wbaP deletion in BWH_36 and BWH_45 abolishes capsule production. Transmission electron microscopy of BWH_36, BWH_45 and corresponding wbaP deletion mutants shown. For every isolate, one representative image from four images obtained from one section is shown. b, Impaired capsule production improves biofilm formation. The isogenic set of ST258 capsule mutants derived from UCI_38 is shown, as well as clinical ST258 capsule mutants and the regular and hypomucoid isolate from the patient specimen UR_5. UCI_37, BIDMC_54 and UR_5_hypo are hypomucoid; BIDMC_13 and BIDMC_16 are hypermucoid. Number of biologically independent experiments indicated. Mean and SEM of at least three independent experiments shown. Significance calculated with an unparalleled two-tailed t-test. c, Capsule production impairs the invasion of bladder epithelial cells. wbaP deletion in UCI_38, BWH_36 and BWH_45 increases the invasion of bladder epithelial cells, while wbaP expression in the isogenic wbaP deletion mutant of UCI_38 impairs bladder epithelial cell invasion. Mean and SEM shown. Significance calculated with an unparalleled two-tailed t-test. n = 3 biologically independent experiments. d, Impaired capsule production increases bladder epithelial cell invasion resulting in a larger intracellular reservoir of K. pneumoniae ST258. UCI_38 and the isogenic wbaP deletion mutant are shown. The net increase in CFUs over 48 h is indicated. Mean and standard deviation of n = 4 biologically independent experiments shown. Significance calculated with an unparalleled two-tailed t-test. e, Antibiotics of last resort are unable to clear intracellular carbapenem-resistant Klebsiella pneumoniae from bladder epithelial cells. Intracellular survival of the clinical capsule-deficient isolate BIDMC_54 shown. Mean and standard deviation of n = 3 biologically independent experiments shown. Significance calculated with an unparalleled two-tailed t-test. f, Intracellular survival of UCI_38_mNeon and UCI_38ΔwbaP_mNeon after 1-minute synchronization of infection (via centrifugation). The centrifugation time was reduced for confocal microscopy in order to more effectively wash off extracellular bacteria from host cells, which decreased invasion efficiency but did not affect the observed net increase in recovered CFUs over 48 h seen in Extended Data Fig. 2d. Mean and standard deviation of n = 4 biologically independent experiments shown. Significance calculated with an unparalleled two-tailed t-test. g, The number of host cells infected with the wild type and mutant is consistent over time, with the mutant displaying a 3fold higher frequency of infection compared to the wildtype. Percentage of infected cells at indicated timepoints, as determined by confocal microscopy shown. n = 2 biologically independent experiments. h, Average number of bacteria per infected cell, as determined by recovered CFUs (Extended Data Fig. 2f) and by the number of infected host cells (Extended Data Fig. 2g) at indicated timepoints. For the 4-hour time point, the number of infected cells determined at 8 hpi was used in order to better distinguish between live and dead bacteria. From 8 to 48 hours post infection, all identified intracellular fluorescent Klebsiella were in LAMP1-positive vacuoles. Mean and SEM shown. n = 2 biologically independent experiments. i, Intracellular Klebsiella grow in Klebsiella-containing LAMP1-positive vacuoles. The fluorescence intensity of mNeon-expressing Klebsiella in LAMP1-positive vacuoles determined via integrated density (IntDen) from confocal microscopy images taken under identical conditions. n = 2 biologically independent experiments. j, Representative image taken with a confocal microscope of bladder epithelial cells infected with mNeon expressing UCI_38ΔwbaP (green, 24 hpi), labeled with anti-LAMP1 antibodies (red) and Hoechst dye (blue). The experiments were repeated 3 times independently with similar results. k, The same image is shown without bacteria to visualize the LAMP1-positive compartments in which Klebsiella pneumoniae persist during infection. l, Heterogeneity of intracellular growth observed in Klebsiella-containing vacuoles. The capsule-deficient mutant displays growth more frequently resulting in 2-fold enhanced net-growth vs the wild type (Extended Data Fig. 2f,h). The integrated density of mNeon-expressing Klebsiella in LAMP1-positive vacuoles is shown (n = 70 UCI_38 vs n = 73 UCI_38ΔwbaP containing compartments, from n = 2 biologically independent experiments shown). Median indicated with a dashed line, quartiles indicated with dashed lines. m, Bacteria recovered from the bladder and the kidney of mice infected transurethrally with UCI_38 or UCI_38ΔwbaP, 3 days post infection. n = 10 mice per group. Significance was calculated with the Mann-Whitney two-tailed U test. P values indicated in the figures. n, The capsule-deficient mutant UCI_38ΔwbaP causes localized infection with no observed immunogenicity at 3 days post infection. Representative histological images of haematoxylin and eosin (H&E) stained liver tissue sections from mouse UTIs, 3 days post infection (n = 3 bladders analyzed). For f-i the mean and SEM is shown. P values are indicated in the figures.
Extended Data Fig. 3 |. SNPs in wzc confer phagocytosis resistance via hypercapsule production.
a, Engineered isogenic strains containing wzc mutations identified in the clinical ST258 isolates produce a hypercapsule. The wzc genes from normal and hypercapsule producing isolates and from the same patient samples (UR_5, UR_11, UR_35) were cloned into the expression plasmid pBAD33 or pBAD33tet (ptet, a tetracycline resistance conferring variant of pBAD33). The ST258 strain UCI_38 and other phylogenetically more distantly related ST258 strains BWH_36 and BWH_4513 were transformed with the resulting constructs and were tested for the ability to resist centrifugation as a measure of hypercapsule production51. After a brief centrifugation step the optical density of the supernatant was determined. b, Clinical isolates with wzc mutations produce a hypercapsule. Overnight cultures were tested for centrifugation resistance as indicator of excessive capsule production51. After a brief centrifugation step the optical density of the supernatant was determined. c, Hypercapsule producing clinical isolates are phagocytosis resistant. UCI_38 and isogenic hypercapsule producing wzc mutants, as well as other clinical isolates with wzc SNPs shown. UR_5 isolates and BIDMC_16 could not be tested for phagocytosis resistance due to extensive drug resistance. d, Mutated wzc induces a hypercapsule in distantly related ST258 strains (for a phylogenetic analysis of these isolates, see Cerqueira et al, 201713). For a-d the mean and SEM of n = 3 biologically independent experiments is shown. Significance calculated with an unparalleled two-tailed t-test. e, Hypercapsule conferring point mutations affect different regions of Wzc. The C-terminal domain of the Wzc protein from ST258 clade 2 and ST512 strains is shown. Wzc controls high level polymerization of capsule in E. coli18. Its activity depends on a C-terminal autokinase domain, which consists of Walker A and B boxes that are crucial for the phosphorylation of tyrosine residues located in the tyrosine cluster and which results in the negative regulation capsule polymerization28. Hypercapsule production has also been observed in clinical Acinetobacter baumanii isolates with point mutations in wzc and have only in some cases been found to affect autokinase activity28. The precise function of Wzc is unknown but it has been hypothesized that it regulates capsule biosynthesis via distinct interactions with other capsule biosynthesis proteins and may have a complex structural role in capsule assembly18. Outside of its autokinase activity, Wzc proteins are also known to form oligomers, independent of phosphorylation18. A535E is located in a Walker A box, while the G565 mutations are located just outside of the canonical Walker A’ box. However, additional conserved amino acids can be found in Walker A’ boxes of Enterobacteriaceae, potentially extending the Walker A’ box to amino acid 566. L74 is not located in a region of any known activity of Wzc but may affect oligomerization or other complex interactions.
Extended Data Fig. 4 |. Hypercapsule production increases ST258 virulence.
a, Hypercapsule production does not affect the growth rate of UCI_38 (ST258) or SGH_10 (ST23) in axenic culture. UCI_38 and the isogenic hypercapsule mutant UCI_38wzcG565A, as well as the hypervirulent isolate SGH_10 (ST23) were grown in LB medium or in M9 salts supplemented with 0.2% casamino acids. The experiments were repeated 3 times independently with similar results. b, The hypercapsule producing clinical isolate BIDMC_13 displays increased toxicity in a zebrafish bloodstream infection model compared to normal capsule strain UCI_38. The lethal dose (LD50) required to kill 50% of the zebrafish larvae population is indicated. Meanwhile, a capsule-deficient clinical isolate UCI_37 is more attenuated. Mean and SEM shown. n = 170 UCI_38, n = 111 BIDMC_13, n = 104 UCI_37. P-values indicated relative to the normal capsule UCI_38 strain. c, Survival of zebrafish larvae after bloodstream-infection with UCI_38 (n = 1773 + /-453 CFUs / fish) and the clinical hypercapsule mutant BIDMC_13 (n = 2213 + /−1113 CFUs / fish). P = 0.0024 (log-rank test). d, Hypercapsule facilitates the dissemination of murine UTIs (compare to Fig. 2h). C3H/HeN mice were infected transurethrally with UCI_38 and the isogenic hypercapsule mutant UCI_38wzcG565A. n = 10 mice were used per strain and timepoint. At day 3 post infection only the hypercapsule mutants were isolated from the bladder and the kidney. Significance was calculated with a two-tailed Mann-Whitney U test. The dashed line indicates the limit of detection. Dpi, days post infection. e, Representative histological images of haematoxylin and eosin (H&E) stained liver tissue sections from mouse UTIs 3 days post infection. The livers of n = 3 infected mice were analyzed per group. f, The hypercapsule mutant disseminates more effectively to the spleen in a mouse model of bloodstream infection. Recovered CFUs from homogenized spleens 3 days post infection shown. n = 5 mice were used per group. Significance was calculated with a two-tailed Mann-Whitney U test. g, Transurethral infection of TLR4-deficient C3H/HeJ mice with the normal capsule strain UCI_38 results in dissemination and persistent infection. Infections were established in 20 mice and CFU counts were determined 7 days post infection. The dashed line indicates the limit of detection. Hypercapsule mutants were isolated. h,i,j, A hypercapsulated ST258 mutant that emerged as a subpopulation in a patient displays enhanced virulence in mouse models of infection. h, Mouse UTI model. Recovered CFUs from the liver of mice infected transurethrally with normal (UC_11) and hyper-capsule producing (UCI_11wzcL74P) isolates from the urine specimen UR_11 (3 days post infection). The hypercapsule mutant killed 4 of 15 mice, while the parent isolate killed 1 of 15 mice, which is a degree of virulence that is usually not observed in mouse UTIs. Dead mice are indicated by the number of crosses. Livers from deceased mice are not included in the analysis. n = 15 mice per group. Significance calculated with a two-tailed Mann-Whitney U test. i, Pyogenic liver abscess caused by the hypercapsule mutant from UR_11 observed in 4 of 11 mouse UTIs. j, Mouse bloodstream model. The hypercapsule mutant from UR_11 (UCI_11wzcL74P) displays more rapid lethality in a mouse model of bloodstream infection compared to the normal capsule producing parent at an inoculum of 4×107. n = 5 C3H/HeN mice were infected per group. Mice infected with the normal capsule producing parent were sacrificed due to poor health scores at 16 hpi. Significance calculated with a log-rank test. k, Comparison of capsule production between the representative hypercapsulated ST23 isolate SGH_10 and the hypercapsulated ST258 strain UCI_38wzcG565A. The capsule was isolated and quantified by determining the amount of uronic acids. Mean and SEM of n = 2 independent experiments shown. l, The representative hypervirulent K. pneumoniae isolate SGH_10 displays rapid lethality in a mouse model of intraperitoneal infection. n = 5 BALB/c mice were infected with an inoculum of 1–2×106 CFUs and mouse survival was followed over time. SGH_10 is a representative Asian ST23 isolate harboring all known hypervirulence-associated genes.
Extended Data Fig. 5 |. identification of isolates with wzc mutations suspected to confer a hypercapsule.
a. ST258 clade 2 reference tree (including single locus ST512 variants), comprised of 117 reference genomes that represent the phylogenetic diversity of the 966 ST258 clade 2 strains identified in the Refseq database (see Methods). b. Phylogenetic distribution of wzc mutants mapped to the closest strain in the reference tree. Mutants harboring mutations close to the hypercapsule-conferring mutations positions that were found to confer a hypercapsule in this study are shown (Extended Data Fig. 3e).
Extended Data Fig. 6 |. Direct isolation of hypercapsule mutants from patients infected with multidrug-resistant K. pneumoniae.
The percentage of hypercapsule and capsule-deficient mutants (Hyper/Deficient) as determined by visual inspection of mucoidity and string-test positivity is shown, for 3 patient urine specimens plated and analyzed directly, as well as the identified wzc mutations (resulting amino acid substitutions shown) and sequence types (ST) for the hypercapsule population isolated from each specimen. The clonality of subpopulations isolated from patients was verified by whole genome sequencing. Point mutations in wzc were identified by comparing the wzc sequence of mutant isolates to regular capsule producing isolates in the same specimen or, in the case of the ST14 isolate, by comparing the sequence to the consensus ST14 sequence in NCBI. The ST307 hypercapsule mutant harbors an additional frameshift mutation (fs) in the tyrosine cluster of wzc (Extended Data Fig. 3e) that affects the last 7 codons (G713-Ter720) and extends the open reading frame by two codons.
Supplementary Material
Acknowledgements
We thank L. Van Dijk for his help with the illustration of the phylogenetic analysis and Z. Ackermann-Bloom for advice on imaging. This work was supported by a generous gift from A. and J. Bekenstein and by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health (NIH) under award no. R01AI117043 to D.T.H and award number U19AI110818 to the Broad Institute. C.M.E. was supported by a Research Fellowship from the German Research Foundation (Deutsche Forschungsgemeinschaft) and by a Fund for Medical Discovery Postdoctoral Fellowship from MGH. C.A.R. was supported by the Mexican National Council for Science and Technology and Fundacion Mexico en Harvard. The Microscopy Core of the Program in Membrane Biology was partially supported by a Center for the Study of Inflammatory Bowel Disease grant (no. DK043351) and a Boston Area Diabetes and Endocrinology Research Center award (no. DK057521). The Zeiss LSM 800 Airyscan confocal microscope was purchased using an NIH Shared Instrumentation grant (no. 1S10OD021577-01).
Footnotes
Competing interests
The authors declare no competing interests.
Additional information
Extended data is available for this paper at https://doi.org/10.1038/s41591-020-0825-4.
Supplementary information is available for this paper at https://doi.org/10.1038/s41591-020-0825-4.
Peer review information Alison Farrell is the primary editor on this article, and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Data availability
Sequencing data from this study can be found in the SRA of the NCBI database under Bioproject accession no. PRJNA506070. All other datasets are available from the corresponding author on reasonable request.
References
- 1.Antibiotic Resistance Threats in the United States (Centers for Disease Control and Prevention, 2013). [Google Scholar]
- 2.Rapid Risk Assessment: Carbapenem-resistant Enterobacteriaceae—First update (European Centre for Disease Prevention and Control, 2018). [Google Scholar]
- 3.Pitout JD, Nordmann P. & Poirel L. Carbapenemase-producing Klebsiella pneumoniae, a key pathogen set for global nosocomial dominance. Antimicrob. Agents Chemother. 59, 5873–5884 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Antimicrobial Resistance—Global Report on Surveillance (World Health Organization, 2014). [Google Scholar]
- 5.Deleo FR et al. Molecular dissection of the evolution of carbapenem-resistant multilocus sequence type 258 Klebsiella pneumoniae. Proc. Natl Acad. Sci. USA 111, 4988–4993 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Munoz-Price LS et al. Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect. Dis. 13, 785–796 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee CR et al. Global dissemination of carbapenemase-producing Klebsiella pneumoniae: epidemiology, genetic context, treatment options, and detection methods. Front. Microbiol. 7, 895 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bowers JR et al. Genomic analysis of the emergence and rapid global dissemination of the clonal group 258 Klebsiella pneumoniae pandemic. PLoS ONE 10, e0133727 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dautzenberg MJ, Haverkate MR, Bonten MJ & Bootsma MC Epidemic potential of Escherichia coli ST131 and Klebsiella pneumoniae ST258: a systematic review and meta-analysis. BMJ Open 6, e009971 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Snitkin ES et al. Tracking a hospital outbreak of carbapenem-resistant Klebsiella pneumoniae with whole-genome sequencing. Sci. Transl. Med. 4, 148ra116 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gu D. et al. A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: a molecular epidemiological study. Lancet Infect. Dis. 18, 37–46 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Lam MMC et al. Tracking key virulence loci encoding aerobactin and salmochelin siderophore synthesis in Klebsiella pneumoniae. Genome Med. 10, 77 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cerqueira GC et al. Multi-institute analysis of carbapenem resistance reveals remarkable diversity, unexplained mechanisms, and limited clonal outbreaks. Proc. Natl Acad. Sci. USA 114, 1135–1140 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Siu LK, Yeh KM, Lin JC, Fung CP & Chang FY Klebsiella pneumoniae liver abscess: a new invasive syndrome. Lancet Infect. Dis. 12, 881–887 (2012). [DOI] [PubMed] [Google Scholar]
- 15.Sahly H. et al. Capsule impedes adhesion to and invasion of epithelial cells by Klebsiella pneumoniae. Infect. Immun. 68, 6744–6749 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Domenico P, Schwartz S. & Cunha BA Reduction of capsular polysaccharide production in Klebsiella pneumoniae by sodium salicylate. Infect. Immun. 57, 3778–3782 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Broberg CA, Palacios M. & Miller VL Klebsiella: a long way to go towards understanding this enigmatic jet-setter. F1000Prime Rep. 6, 64 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Whitfield C. Biosynthesis and assembly of capsular polysaccharides in Escherichia coli. Annu. Rev. Biochem. 75, 39–68 (2006). [DOI] [PubMed] [Google Scholar]
- 19.Hunstad DA & Justice SS Intracellular lifestyles and immune evasion strategies of uropathogenic Escherichia coli. Annu. Rev. Microbiol. 64, 203–221 (2010). [DOI] [PubMed] [Google Scholar]
- 20.Mysorekar IU & Hultgren SJ Mechanisms of uropathogenic Escherichia coli persistence and eradication from the urinary tract. Proc. Natl Acad. Sci. USA 103, 14170–14175 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Flores-Mireles AL, Walker JN, Caparon M. & Hultgren SJ Urinary tract infections: epidemiology, mechanisms of infection and treatment options. Nat. Rev. Microbiol. 13, 269–284 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blango MG & Mulvey MA Persistence of uropathogenic Escherichia coli in the face of multiple antibiotics. Antimicrob. Agents Chemother. 54, 1855–1863 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hung CS, Dodson KW & Hultgren SJ A murine model of urinary tract infection. Nat. Protoc. 4, 1230–1243 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lam MMC et al. Population genomics of hypervirulent Klebsiella pneumoniae clonal-group 23 reveals early emergence and rapid global dissemination. Nat. Commun. 9, 2703 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nassif X, Fournier JM, Arondel J. & Sansonetti PJ Mucoid phenotype of Klebsiella pneumoniae is a plasmid-encoded virulence factor. Infect. Immun. 57, 546–552 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Walker KA et al. A Klebsiella pneumoniae regulatory mutant has reduced capsule expression but retains hypermucoviscosity. MBio 10, e00089–19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gottesman S. & Stout V. Regulation of capsular polysaccharide synthesis in Escherichia coli K12. Mol. Microbiol. 5, 1599–1606 (1991). [DOI] [PubMed] [Google Scholar]
- 28.Geisinger E. & Isberg RR Antibiotic modulation of capsular exopolysaccharide and virulence in Acinetobacter baumannii. PLoS Pathog. 11, e1004691 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fang CT, Chuang YP, Shun CT, Chang SC & Wang JT A novel virulence gene in Klebsiella pneumoniae strains causing primary liver abscess and septic metastatic complications. J. Exp. Med. 199, 697–705 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wozniak JE et al. A nationwide screen of carbapenem-resistant Klebsiella pneumoniae reveals an isolate with enhanced virulence and clinically undetected colistin heteroresistance. Antimicrob. Agents Chemother. 63, e00107–19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ruppe E. et al. Clonal or not clonal? Investigating hospital outbreaks of KPC-producing Klebsiella pneumoniae with whole-genome sequencing. Clin. Microbiol. Infect. 23, 470–475 (2017). [DOI] [PubMed] [Google Scholar]
- 32.Spencer MD et al. Whole genome sequencing detects inter-facility transmission of carbapenem-resistant Klebsiella pneumoniae. J. Infect. 78, 187–199 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zilberberg MD, Nathanson BH, Sulham K, Fan W. & Shorr AF Carbapenem resistance, inappropriate empiric treatment and outcomes among patients hospitalized with Enterobacteriaceae urinary tract infection, pneumonia and sepsis. BMC Infect. Dis. 17, 279 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Skurnik D. et al. Extended-spectrum antibodies protective against carbapenemase-producing Enterobacteriaceae. J. Antimicrob. Chemother. 71, 927–935 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Russo TA & Marr CM Hypervirulent Klebsiella pneumoniae. Clin. Microbiol. Rev. 32, e00001–19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Uhlemann AC, Otto M, Lowy FD & DeLeo FR Evolution of community-and healthcare-associated methicillin-resistant Staphylococcus aureus. Infect. Genet. Evol. 21, 563–574 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Valentini M, Gonzalez D, Mavridou DA & Filloux A. Lifestyle transitions and adaptive pathogenesis of Pseudomonas aeruginosa. Curr. Opin. Microbiol. 41, 15–20 (2018). [DOI] [PubMed] [Google Scholar]
- 38.Kobayashi SD et al. Phagocytosis and killing of carbapenem-resistant ST258 Klebsiella pneumoniae by human neutrophils. J. Infect. Dis. 213, 1615–1622 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yoshida K. et al. Role of bacterial capsule in local and systemic inflammatory responses of mice during pulmonary infection with Klebsiella pneumoniae. J. Med. Microbiol. 49, 1003–1010 (2000). [DOI] [PubMed] [Google Scholar]
- 40.Kobayashi SD & DeLeo FR Re-evaluating the potential of immunoprophylaxis and/or immunotherapy for infections caused by multidrug resistant Klebsiella pneumoniae. Future Microbiol. 13, 1343–1346 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
References
- 41.Edwards RA, Keller LH & Schifferli DM Improved allelic exchange vectors and their use to analyze 987P fimbria gene expression. Gene 207, 149–157 (1998). [DOI] [PubMed] [Google Scholar]
- 42.Gibson DG et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345 (2009). [DOI] [PubMed] [Google Scholar]
- 43.Ferrieres L. et al. Silent mischief: bacteriophage Mu insertions contaminate products of Escherichia coli random mutagenesis performed using suicidal transposon delivery plasmids mobilized by broad-host-range RP4 conjugative machinery. J. Bacteriol. 192, 6418–6427 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guzman LM, Belin D, Carson MJ & Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177, 4121–4130 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chang AC & Cohen SN Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 134, 1141–1156 (1978). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shaner NC et al. A bright monomeric green fluorescent protein derived from Branchiostoma lanceolatum. Nat. Methods 10, 407–409 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Crepin S, Harel J. & Dozois CM Chromosomal complementation using Tn7 transposon vectors in Enterobacteriaceae. Appl. Environ. Microbiol. 78, 6001–6008 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Davis JH, Rubin AJ & Sauer RT Design, construction and characterization of a set of insulated bacterial promoters. Nucleic Acids Res. 39, 1131–1141 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Coffey BM & Anderson GG Biofilm formation in the 96-well microtiter plate. Methods Mol. Biol. 1149, 631–641 (2014). [DOI] [PubMed] [Google Scholar]
- 50.Blumenkrantz N. & Asboe-Hansen G. New method for quantitative determination of uronic acids. Anal. Biochem. 54, 484–489 (1973). [DOI] [PubMed] [Google Scholar]
- 51.Hsu CR, Lin TL, Chen YC, Chou HC & Wang JT The role of Klebsiella pneumoniae rmpA in capsular polysaccharide synthesis and virulence revisited. Microbiology 157, 3446–3457 (2011). [DOI] [PubMed] [Google Scholar]
- 52.Siguier P, Perochon J, Lestrade L, Mahillon J. & Chandler M. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 34, D32–D36 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lam MMC et al. Genetic diversity, mobilisation and spread of the yersiniabactin-encoding mobile element ICEKp in Klebsiella pneumoniae populations. Microb. Genom. 4, e000196 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Walker BJ et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 9, e112963 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wyres KL et al. Identification of Klebsiella capsule synthesis loci from whole genome data. Microb. Genom. 2, e000102 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sperlea T. et al. γBOriS: identification of origins of replication in gammaproteobacteria using motif-based machine learning. Preprint at bioRxiv 10.1101/597070 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wilm A. et al. LoFreq: a sequence-quality aware, ultra-sensitive variant caller for uncovering cell-population heterogeneity from high-throughput sequencing datasets. Nucleic Acids Res. 40, 11189–11201 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Edgar RC MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li H. & Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Goris J. et al. DNA–DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 57, 81–91 (2007). [DOI] [PubMed] [Google Scholar]
- 61.Inouye M. et al. SRST2: rapid genomic surveillance for public health and hospital microbiology labs. Genome Med. 6, 90 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Arndt D. et al. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 44, W16–W21 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mulvey MA, Schilling JD & Hultgren SJ Establishment of a persistent Escherichia coli reservoir during the acute phase of a bladder infection. Infect. Immun. 69, 4572–4579 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Eto DS, Sundsbak JL & Mulvey MA Actin-gated intracellular growth and resurgence of uropathogenic Escherichia coli. Cell Microbiol. 8, 704–717 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data from this study can be found in the SRA of the NCBI database under Bioproject accession no. PRJNA506070. All other datasets are available from the corresponding author on reasonable request.