Abstract
PINK1 and parkin constitute a mitochondrial quality control system mutated in Parkinson’s disease. PINK1, a kinase, phosphorylates ubiquitin to recruit parkin, an E3 ubiquitin ligase, to mitochondria. PINK1 controls both parkin localization and activity through phosphorylation of both ubiquitin and the ubiquitin‐like (Ubl) domain of parkin. Here, we observed that phospho‐ubiquitin can bind to two distinct sites on parkin, a high‐affinity site on RING1 that controls parkin localization and a low‐affinity site on RING0 that releases parkin autoinhibition. Surprisingly, ubiquitin vinyl sulfone assays, ITC, and NMR titrations showed that the RING0 site has higher affinity for phospho‐ubiquitin than phosphorylated Ubl in trans. We observed parkin activation by micromolar concentrations of tetra‐phospho‐ubiquitin chains that mimic mitochondria bearing multiple phosphorylated ubiquitins. A chimeric form of parkin with the Ubl domain replaced by ubiquitin was readily activated by PINK1 phosphorylation. In all cases, mutation of the binding site on RING0 abolished parkin activation. The feedforward mechanism of parkin activation confers robustness and rapidity to the PINK1‐parkin pathway and likely represents an intermediate step in its evolutionary development.
Keywords: autophagy, mitophagy, open‐loop control, Parkinson's disease, ubiquitin
Subject Categories: Organelles, Post-translational Modifications & Proteolysis, Structural Biology
Two distinct phospho‐ubiquitin binding sites differentially control mitochondrial recruitment and activation of the parkin E3 ligase.
Introduction
Parkinson’s disease is one of the most common neurodegenerative diseases. It causes motor symptoms due to the loss of dopaminergic neurons of the substantia nigra in the midbrain. Over 90% of the cases are sporadic and occur late in life. However, 5–10% of the cases are attributed to autosomal mutations that induce the disease in younger patients (Koros et al, 2017). Many of these mutations are found in PARK2 (Kitada et al, 1998) and PARK6 (Valente et al, 2004) genes and are responsible for the earliest onset cases. These genes encode respectively for parkin and PINK1 proteins, which work together in a mitochondrial quality control process consisting in tagging proteins of damaged mitochondria with ubiquitin molecules. The accumulation of ubiquitinated proteins at the mitochondrial surface triggers the degradation of either the whole damaged mitochondria through mitophagy (Pickles et al, 2018) or the excision of damaged portions through the formation of mitochondrial‐derived vesicles (Sugiura et al, 2014). Parkin and PINK1 are also involved in the suppression of mitochondrial antigen presentation (Matheoud et al, 2016) and activation of the STING pathway (Sliter et al, 2018), suggesting a role of inflammation in Parkinson’s disease.
Parkin is a cytosolic E3‐ubiquitin ligase composed of a N‐terminal ubiquitin‐like domain (Ubl) linked to a R0RBR module formed by four zinc‐binding domains: RING0, RING1, IBR, and RING2 (Figs 1A and EV1). As an RBR‐type E3 enzyme, parkin mediates the transfer of ubiquitin from an E2 enzyme onto a cysteine in the RING2 domain, followed by a second transfer onto the substrate protein (Wenzel et al, 2011). X‐ray structures of parkin have revealed that it adopts an autoinhibited conformation in basal cell conditions (Riley et al, 2013; Trempe et al, 2013; Wauer & Komander, 2013) in agreement with its previously reported autoinhibition (Chaugule et al, 2011). The active cysteine located on the RING2 domain is partially occluded by its RING0 domain, and the E2‐binding site on RING1 is blocked by the parkin Ubl domain and a short α‐helix referred to as the repressor element of parkin (REP). In cells, parkin needs to both translocate to mitochondria and undergo a conformational change to release the autoinhibition. Parkin recruitment to mitochondria depends on PINK1, a serine/threonine kinase, which acts as a sensor of mitochondrial defects (Geisler et al, 2010; Matsuda et al, 2010; Narendra et al, 2010; Vives‐Bauza et al, 2010). PINK1 accumulates at the surface of damaged mitochondria when mitochondrial protein import is impaired. There, it phosphorylates ubiquitin molecules present at mitochondrial surface. Parkin binds phosphorylated ubiquitin (pUb) with high affinity, which promotes its accumulation onto mitochondria (Kane et al, 2014; Kazlauskaite et al, 2014; Koyano et al, 2014; Ordureau et al, 2014). Parkin is then in turn phosphorylated by PINK1 to activate its ubiquitination activity (Kondapalli et al, 2012; Kazlauskaite et al, 2015; Kumar et al, 2015; Sauve et al, 2015; Wauer et al, 2015a). The most recent x‐ray structures of the complex of phosphorylated parkin (pParkin) and pUb have revealed that the phosphorylation of the parkin Ubl domain (pUbl) is responsible for the structural rearrangement within parkin that exposes the active cysteine (Gladkova et al, 2018; Sauve et al, 2018) (Fig EV1). Phosphoserine 65 of pUbl binds a phosphate‐binding site on RING0 formed by residues K161, R163, and K211. The relocation of the pUbl domain displaces the catalytic domain RING2 from RING0. The released RING2 domain is free to reposition next to the ubiquitin‐charged E2 for the transfer of ubiquitin to the parkin active cysteine and subsequently to target proteins. The addition of new ubiquitin molecules to the mitochondrial outer surface provides more substrates for PINK1, which leads to additional parkin recruitment to mitochondria (Seirafi et al, 2015). This positive feedback mechanism amplifies the signal for mitophagy and explains the observation that parkin activity is required for its recruitment (Lazarou et al, 2013; Ordureau et al, 2014; Shiba‐Fukushima et al, 2014).
Figure 1. Parkin recruitment and mitophagy without Ubl phosphorylation.
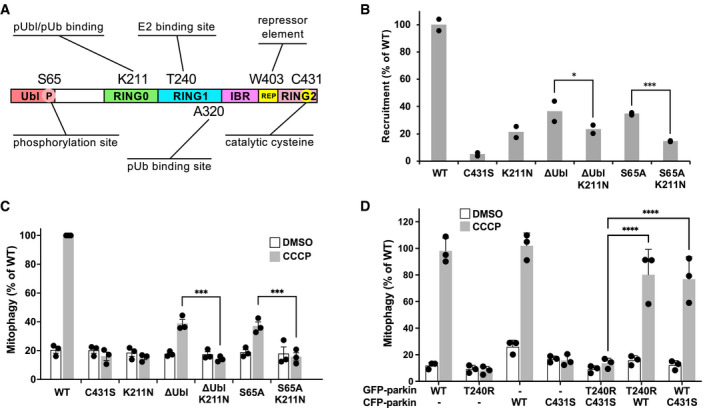
- Schematic representation of parkin highlighting key residues involved in parkin activation and activity.
- Partial recruitment to mitochondria of parkin mutants without the Ubl‐phosphorylation site. Graph shows recruitment of GFP‐parkin WT, K211N, C431S, and ΔUbl (deletion of residues 1–76), ΔUbl K211N, S65A, S65A K211N measured in U2OS cells 120 min after depolarization of mitochondria by 20 μM CCCP. Mutation K211N of the RING0 pUbl‐binding site decreased recruitment even in the absence of Ubl‐phosphorylation. Results are shown from two independent biological replicates with two‐way analysis of variance (ANOVA) with Bonferroni post‐test.
- Mitophagy in the absence of Ubl‐phosphorylation. Mitophagy was detected by fluorescence‐activated cell sorting (FACS) of untreated and CCCP‐treated (20 μM for 4 h) U2OS cells containing mitochondrially targeted mKeima (mt‐Keima) and transiently expressing GFP‐parkin. The K211N mutation was epistatic to the Ubl‐phosphorylation and decreased mitophagy to the levels seen in the parkin catalytically dead C431S mutant. For statistical analysis, a one‐way ANOVA with Tukey’s post‐test was performed on data from three biological replicates. Error bars indicate s.e.m.
- Lack of complementation between parkin mutants defective in E2‐binding and ubiquitin transfer. U2OS mt‐Keima cells were transfected with CFP or GFP alone (labeled: ‐) or as parkin fusion proteins and mitophagy measured by FACS for untreated and CCCP‐treated (20 μM for 12 h). For statistical analysis, a one‐way ANOVA with Tukey’s post‐test was performed on data from three biological replicates. Error bars indicate s.e.m.
Data information: *P < 0.05, ***P < 0.001, ****P < 0.0001.
Figure EV1. Conformational changes in parkin upon phosphorylation or pUb binding.
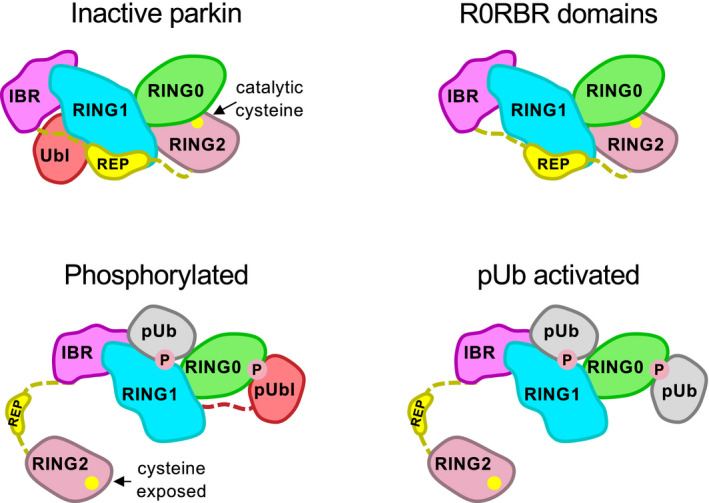
Full‐length parkin and the R0RBR module have low basal ubiquitin ligase activity due to masking of the RING2 catalytic site (cysteine 431) by the RING0 domain. Parkin is recruited to damaged mitochondria by a high‐affinity site on the RING1 domain that binds phospho‐ubiquitin (pUb). Upon phosphorylation of the Ubl domain or the binding of a second pUb, parkin becomes activated as the phosphorylated Ubl domain (pUbl) or pUb binds to a site on RING0 releasing and exposing the RING2 catalytic site.
Parkin is also regulated through a feedforward (open‐cycle) mechanism, which does not require parkin phosphorylation. Multiple studies have shown that deletion of the Ubl domain or loss of serine 65 does not completely abolish parkin recruitment to mitochondria and mitophagy (Shiba‐Fukushima et al, 2012; Ordureau et al, 2014; Zhuang et al, 2016; Tang et al, 2017). While unphosphorylated parkin can be activated in vitro by pUb addition, the molecular mechanism has remained unexplored (Kazlauskaite et al, 2014). Here, we confirm the existence of a secondary mechanism for parkin activation that is independent of parkin phosphorylation but dependent on the RING0 pUbl‐binding site. We observe that pUb can bind to the pUbl‐binding site and, in fact, has higher affinity than the pUbl domain in trans. Experiments with phosphorylated polyUb chains reveal the avidity of the pUb and pUbl‐binding sites and mimic the effect of multiple immobilized pUb molecules in proximity to each other on the surface of mitochondria. Finally, experiments with parkin with the Ubl domain replaced by ubiquitin show the chimeric molecule is readily activated by phosphorylation. The feedforward mechanism increases the robustness of the pathway for the clearance of damaged mitochondria and likely represents an early feature in the evolutionary development of the PINK1/parkin pathway.
Results
Parkin recruitment and mitophagy in cells
To verify that parkin in cells could be activated in the absence of Ubl phosphorylation, we monitored the recruitment of different parkin variants (Fig 1A) from cytoplasm to mitochondria upon the addition of mitochondrial uncoupler carbonyl cyanide m‐chlorophenyl hydrazone (CCCP) (Fig 1B). WT parkin was actively recruited with over 80% of cells showing puncta of parkin on mitochondria after 2 h (Appendix Fig S1). The C431S mutant, which has no ligase activity, displayed the lowest recruitment as expected from its incapacity to ubiquitinate mitochondrial proteins and drive the PINK1/parkin feedback cycle. As observed by others, parkin bearing the S65A mutation or deletion of the Ubl domain (∆Ubl parkin) showed partial recruitment even though the mutations prevent parkin phosphorylation. The role of the RING0 pUbl‐binding site was evident as recruitment was further reduced by a second mutation (K211N) that disrupts the phosphoserine binding site (Wauer et al, 2015a; Gladkova et al, 2018; Sauve et al, 2018). These results demonstrate a mechanism of parkin activation that is independent of parkin phosphorylation but dependent on the pUbl‐binding site on RING0.
mt‐Keima assays were performed to assess the level of mitophagy in cells expressing parkin mutants. The assay measures mitophagy in cells expressing a mitochondrially targeted fluorophore that shifts its excitation spectrum when mitochondria enter the acidic environment of lysosomes. Addition of the protonophore CCCP to depolarize mitochondria induced mitophagy in cells expressing WT parkin (Fig 1C and Appendix Fig S2). Loss of the parkin active cysteine (C431S) or pUbl‐binding site (K211N) abolished the mitophagy response but cells expressing non‐phosphorylatable S65A parkin or ∆Ubl parkin showed partial mitophagy. The addition of the K211N mutation to inactivate the RING0 pUbl‐binding site eliminated the remaining mitophagic activity. These results demonstrate that an alternative mechanism, involving the pUbl‐binding site, can trigger mitophagy of depolarized mitochondria.
Parkin does not show complementation between inactivating mutations
A recurring model of parkin activity has posited that parkin forms dimers or oligomers to allow the catalytic RING2 domain of one molecule to contact the E2~Ub bound to another molecule (Gundogdu et al, 2021). We previously showed an absence of complementation in autoubiquitination assays (Sauve et al, 2018), but this left open the possibility of trans‐active parkin dimers on the surface of mitochondria. To test this, we looked for complementation in the mt‐Keima assay (Fig 1D) between the C431S mutant, which is catalytically dead, and the T240R mutant (Fig 1A), which is unable to bind E2 enzymes (Appendix Fig S3A). The mutants were tagged with GFP and CFP, so their expression could be measured and only cells expressing both proteins counted (Appendix Fig S3B and C). The mt‐Keima assay showed robust mitophagy in cells expressing WT parkin after 12 h of CCCP treatment. No mitophagy was detected in cells expressing the T240R and C431S mutants individually or together. Due to variations in the basal signal, the absence of complementation can best be seen in the individual experiments where cells expressing both mutants show no more mitophagy than cells expressing them separately with GFP or CFP (Fig 1D and Appendix Fig S3D). These results strongly argue against the existence of trans‐active parkin oligomers.
pUb binding activates parkin
We used two different in vitro assays to test whether pUb binding could activate parkin in the absence of Ubl phosphorylation. The first uses ubiquitin vinyl sulfone (UbVS), which is a chemically reactive derivative that crosslinks to the parkin active‐site cysteine (Borodovsky et al, 2001). The formation of the UbVS adduct is a sensitive measure of exposure of the parkin catalytic site and has been widely used to follow parkin activation (Riley et al, 2013; Ordureau et al, 2014; Wauer et al, 2015a). The advantages of the UbVS assay are its simplicity. It only measures accessibility of the parkin active‐site cysteine and does not require E1 and E2 enzymes or acyl transfer of ubiquitin to lysine residues. We used the R0RBR construct that is missing the Ubl domain but retains the catalytic RING2 domain and the pUb and pUbl‐binding sites on RING1 and RING0. Assays were done with R0RBR parkin purified as a complex with pUb.
UbVS assays showed that the addition of pUbl could promote the formation of R0RBR‐Ub crosslinks due to the release of the RING2 domain from RING0 (Fig 2A, upper left panel, and Appendix Fig S4, middle and lower panels). The activation of parkin could be more easily observed with the W403A mutation (Fig 2A, upper right panel) that partially derepresses parkin activity by destabilizing the REP helix (Trempe et al, 2013) (Fig 1A). Addition of 50 µM pUbl to the W403 mutant generated more than 50% R0RBR‐Ub crosslinks. The high concentration of pUbl required reflects the competition between RING2 and pUbl for binding RING0 and the fact that the RING2 domain is present in the same polypeptide chain at a high local concentration. We used the K211N mutation in the RING0 to confirm the essential role of the pUbl‐binding site. No UbVS crosslinks were observed, confirming that the release of RING2 was completely dependent on pUbl‐binding RING0 (Fig 2B).
Figure 2. Activation of parkin by pUbl or pUb binding.
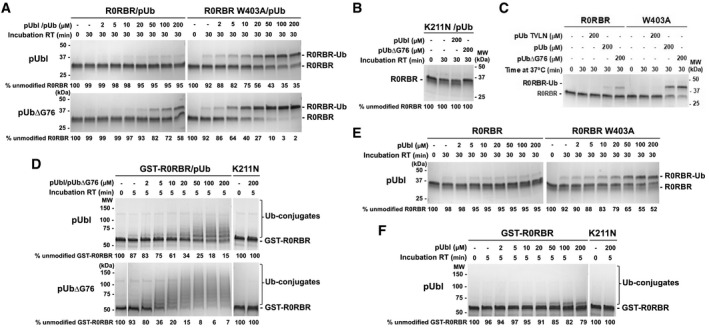
- Ubiquitin vinyl sulfone (UbVS) assays of RING2 release by addition of pUbl and pUb in trans to R0RBR parkin. Assays were performed with incubation of 2 μM wild‐type or W403A R0RBR purified in a 1:1 complex with pUb. Crosslinking was initiated by addition of 10 μM UbVS in the presence of pUbl or pUb∆G76.
- Inhibition of R0RBR activation by the K211N mutation.
- UbVS assays of RING2 release by various types of pUb.
- Autoubiquitination assays of parkin ligase activity induced by pUbl or pUb∆G76 in trans. 2 µM GST‐R0RBR in complex with pUb was incubated with 50 nM E1, 2 µM UbcH7, 100 µM ubiquitin, and 4 mM ATP.
- UbVS assays of RING2 release with R0RBR purified without pUb bound.
- Autoubiquitination assays of parkin ligase activity with GST‐R0RBR without pUb bound. The percentage of unmodified parkin band in each lane is indicated under the gel.
We next asked if pUb could similarly release the RING2 domain and generate UbVS crosslinks. The assays used pUb∆G76 (pUb without the C‐terminal glycine) for consistency with subsequent autoubiquitination assays where pUb interferes with the E2 discharging (Ordureau et al, 2014; Wauer et al, 2015b). Similar results were obtained with full‐length pUb (Fig 2C). Surprisingly, pUb∆G76 was much more efficient than pUbl in producing crosslinks, which suggests that it has a higher affinity for the RING0 pUbl‐binding site (Fig 2A, lower left panel). Addition of pUb∆G76 to wild‐type R0RBR led to detectable crosslinking at 20 µM and more than 40% at 200 µM. The R0RBR W403A mutant was again more sensitive with 50% crosslinking at 7 µM pUb∆G76 and nearly 100% at higher concentrations (Fig 2A, lower right panel). Assays with the K211N mutant confirmed that RING0 pUbl‐site was required for pUb‐induced crosslinks (Fig 2B). Only pUb adopting the canonical, major ubiquitin conformation (Wauer et al, 2015b) could bind RING0 pUbl‐binding site as shown by the absence of crosslinking in the presence of the pUb mutant TVLN that stabilizes the minor conformation (Gladkova et al, 2017) (Fig 2C).
In a second set of assays, we measured autoubiquitination of GST‐R0RBR parkin upon the addition of pUbl or pUb (Fig 2D). We used GST‐tagged parkin because it displays a higher activity than untagged parkin, most likely due to lysine residues in GST acting as additional sites for ubiquitination. In agreement with the UbVS assays, addition of pUbl in trans activated parkin ligase activity detected by loss of the unmodified GST‐R0RBR parkin and the formation of higher molecular weight bands. pUb (Fig 2D, lower panel) was again more efficient than pUbl (Fig 2D, upper panel) in releasing parkin autoinhibition and required 10‐fold lower concentrations for activation. To confirm the importance of the RING0 Ubl‐binding site for parkin activation, we again used a K211N mutant. No autoubiquitination of K211N GST‐R0RBR was observed even at 200 µM pUb (Fig 2D). To compare the efficiency of parkin activation in trans and in cis, we measured the autoubiquitination activity of 2 µM GST‐R0RBR/pUb with 2 µM pUbl (Fig 2D, upper panel) and phosphorylated full‐length parkin in the presence of pUb (Fig EV2A, left panel). Only 24% of unmodified parkin band was still present in the in cis reaction (Fig EV2A, left panel) compared to 83% for the in trans reaction (Fig 2D, upper panel). This shows that activation in cis is more efficient. To rule out possible artifacts due to the presence of the GST tag, we repeated the in trans assays with R0RBR without a tag and a longer incubation time to compensate for the reduced autoubiquitination activity of untagged parkin. The results with untagged R0RBR (Fig EV2B) showed the same trend than those with GST‐parkin (Fig 2D). Parkin was most efficiently activated by pUbl in cis (Fig EV2A, right panel); however, when added in trans, pUb (Fig EV2B, lower panel) was more efficient than pUbl (Fig EV2B, upper panel).
Figure EV2. Activation of parkin by pUbl or pUb binding.
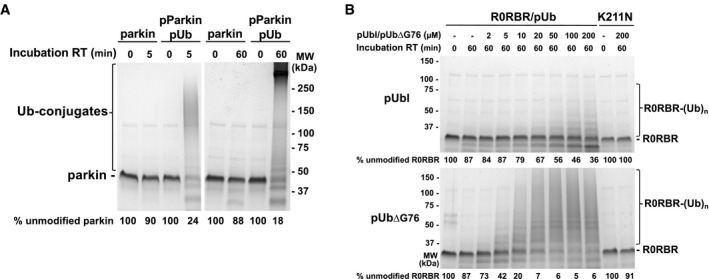
- Autoubiquitination assays of parkin ligase activity of inactive and activated full‐length parkin. Assays were performed with incubation of 2 μM of parkin or phosphorylated parkin purified in a 1:1 complex with pUb under the same conditions as autoubiquitination assays with GST‐R0RBR/pUb (Fig 2D) and R0RBR/pUb.
- Autoubiquitination assays of parkin ligase activity induced by pUbl or pUb∆G76 in trans. 2 µM R0RBR in complex with pUb was incubated with 50 nM E1, 2 µM UbcH7, 100 µM ubiquitin, and 4 mM ATP. The percentage of unmodified parkin band in each lane is indicated under the gel.
The UbVS and autoubiquitination assays used R0RBR parkin in complex with a stoichiometric amount of pUb bound to the RING1 site. To investigate possible coupling between the RING1 and RING0 sites, we repeated the pUbl titrations without pUb present. The UbVS assay showed a small decrease in the effectiveness of pUbl additions (Fig 2E). W403A R0RBR complexed with pUb required roughly 30 µM pUbl to achieve 50% crosslinking (Fig 2A, upper right panel) while the sample without pUb required 100 µM pUbl (Fig 2E, right panel). Autoubiquitination assays showed a much larger difference. Approximately, 10‐fold more pUbl was required to activate GST‐R0RBR parkin in the absence of pUb (Fig 2F) than in its presence (Fig 2D, upper panel).
Increased local concentration enhances parkin activation by pUb
To compare activation by pUb and pUbl in the context of intact parkin, we designed a chimeric molecule in which the Ubl domain has been replaced by ubiquitin (Fig 3A). To prevent pUb binding to the high‐affinity site on RING1, we introduced the A320R mutation that disrupts the RING1 site (Wauer et al, 2015a). The chimeric protein could be phosphorylated by PINK1 (Fig 3B) and crosslinked to UbVS as efficiently as wild‐type parkin (Fig 3C). The RING0 binding site was essential as the K211N mutation prevented crosslinking. Autoubiquitination assays showed that the chimera and wild‐type parkin had identical ubiquitination activity when phosphorylated, and activity was again completely blocked by K211N mutation in the pUbl‐binding site (Fig 3D). These results demonstrate that ubiquitin can fully replace the Ubl domain of parkin and, when phosphorylated, binds to RING0 to release the catalytic RING2 domain.
Figure 3. Ubiquitination activity of Ub‐R0RBR chimera.
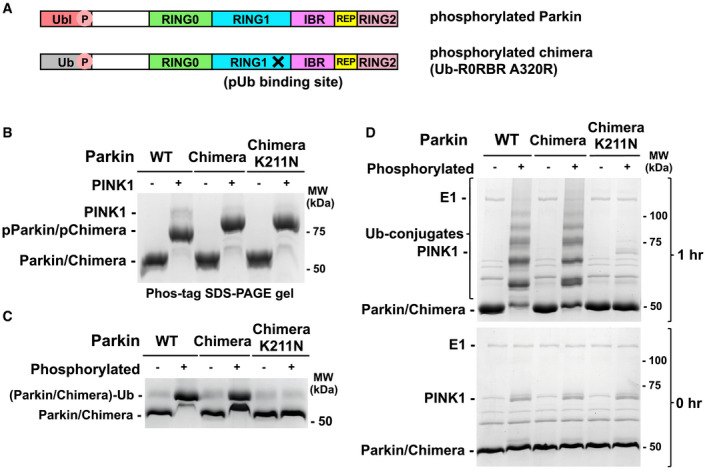
- Schematic representations of phosphorylated parkin (pParkin) and the phosphorylated chimera (pUb‐R0RBR) with the A320R mutation.
- Phosphorylation of wild‐type (WT) parkin and chimeric Ub‐R0RBR by PINK1. Reactions were analyzed on a Phos‐tag SDS–PAGE gel to assess the level of phosphorylation.
- Release of RING2 by Ub‐R0RBR chimera. Ubiquitin vinyl sulfone assays (10 μM UbVS) were performed with 3 μM phosphorylated and non‐phosphorylated full‐length parkin and Ub‐R0RBR.
- Ubiquitination activity of Ub‐R0RBR chimera. Autoubiquitination assays of phosphorylated full‐length parkin and Ub‐R0RBR (3.3 µM with 3 µm UbcH7 and 75 µM S65A ubiquitin) were performed to assess the impact of pUbl substitution by pUb on parkin activity.
On damaged mitochondria, PINK1 phosphorylation of ubiquitin molecules likely leads to an elevated local concentration of pUb. Mono‐ubiquitin and polyubiquitin chains can be phosphorylated by PINK1 in vitro, and polyphosphorylated ubiquitin chains typically phosphorylated on their terminal Ub molecules have been detected in cells following mitochondrial depolarization (Ordureau et al, 2014; Wauer et al, 2015b; Swatek et al, 2019). Here, we used phosphorylated polyubiquitin chains as a tool to mimic the mitochondrial surface modified with multiple pUb molecules. The existence of two pUb‐binding sites on parkin should lead to an increased affinity from cooperation between the sites. Binding of one end of a poly‐pUb chain to the high‐affinity RING1 site should bring a second pUb molecule in proximity to the low‐affinity RING0 site. Fitting of commercially available tetra‐pUb chains onto structures of activated parkin showed that they could bridge both pUb‐binding sites (Fig 4A).
Figure 4. Parkin activation by pUb chains.
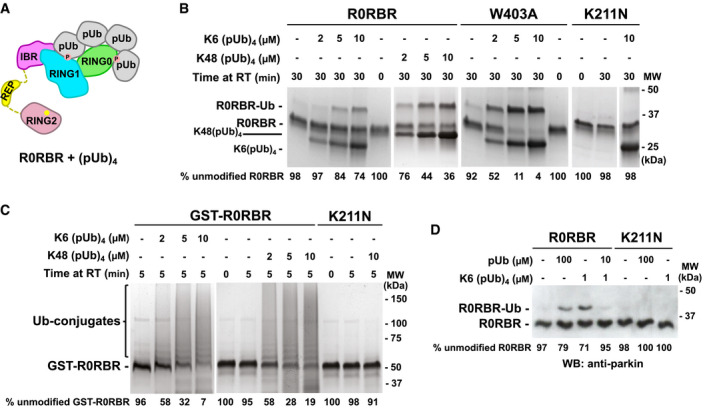
- Model of (pUb)4 chain binding to R0RBR.
- UbVS assays of release of RING2 by K6‐ or K48‐linked (pUb)4 chains. 10 μM UbVS was added to 2 μM R0RBR in the presence of increasing amount of K6(pUb)4 or K48(pUb)4 chains.
- Autoubiquitination assays of parkin activation by K6(pUb)4 or K48(pUb)4. Assays with 2 µM GST‐R0RBR, 50 nM E1, 2 µM UbcH7, and 100 µM ubiquitin were performed in the presence of increasing amount of K6(pUb)4 or K48(pUb)4 chains.
- Competition between K6(pUb)4 chains and monomeric pUb. UbVs (10 μM) was added to 20 nM of WT and K211N R0RBR in the presence of monomeric pUb and/or K6(pUb)4 chains. Reactions were resolved on SDS–PAGE and visualized with anti‐parkin antibody by western blotting.
Data information: The percentage of unmodified parkin band in each lane is indicated under the gel.
We conducted a UbVS assay of R0RBR with commercial K6‐ and K48‐linked tetra‐pUb chains (Fig 4B). Crosslinks were observed at 5 µM K6‐linked (pUb)4 and 2 µM K48‐linked (pUb)4, which is 10‐fold lower than with monomeric pUb (Fig 2A). K6(pUb)4 chains were also more efficient in crosslinking the W403A mutant although the ability of the assay to measure activation by low concentrations was limited by the need for a stoichiometric amount of pUb chains. No crosslinking was observed when RING0 pUbl‐binding site was mutated (Fig 4B). We also tested the ability of K6‐ and K48‐(pUb)4 chains to promote the autoubiquitination activity of GST‐R0RBR parkin (Fig 4C) and R0RBR parkin (Fig EV3). Robust autoubiquitination was observed at low concentrations of K6(pUb)4 and K48(pUb)4 chains and was dependent on the RING0 pUbl‐binding site. No ubiquitination activity was observed with the K211N mutant (Figs 4C and EV3).
Figure EV3. Autoubiquitination assays of parkin activation by K6(pUb)4 or K48(pUb)4 .
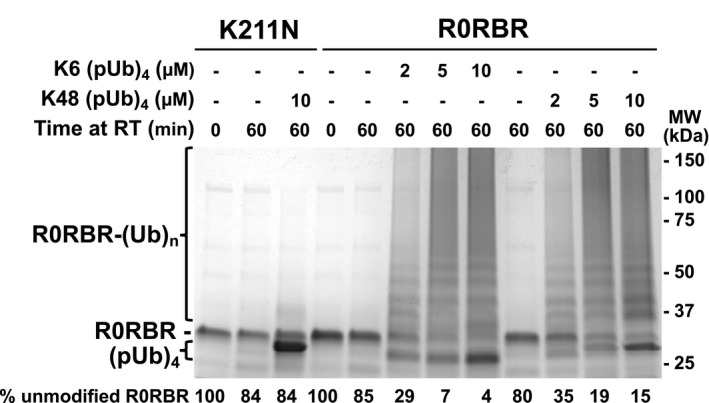
Assays with 2 µM R0RBR, 50 nM E1, 2 µM UbcH7, and 100 µM ubiquitin were performed in the presence of increasing amount of K6(pUb)4 or K48(pUb)4 chains. The percentage of unmodified parkin band in each lane is indicated under the gel.
To confirm that the activation capacity of tetra‐pUb chain was due to binding to RING1‐pUb‐binding site, we conducted a competition UbVS assay (Fig 4D). We switched to western blot detection, which allowed us to use 100‐fold less R0RBR (20 nM) in the assay and avoid the stoichiometry issue. The crosslinks observed with 1 µM K6(pUb)4 were comparable to those with 100 µM free pUb. When pUb was added at an intermediate concentration (10 µM) that would bind RING1 but not RING0, we observed a reduction in the intensity of the crosslinks induced by 1 µM K6(pUb)4. The 10‐fold excess of pUb outcompeted K6(pUb)4 binding to RING1 and eliminated the cooperativity that promoted K6(pUb)4 binding to RING0.
Characterization of pUb binding to RING0 by NMR
We previously used NMR spectroscopy to show that pUbl domain could bind R0RBR/pUb complex in trans (Sauve et al, 2018). Experiments with 15N‐labeled pUbl showed a loss of signal intensity for phosphoserine 65 upon addition of the complex of R0RBR and pUb. We repeated those experiments with 15N‐labeled pUb and the R0RBR W403A mutant (Fig 5A). Addition of the R0RBR W403A/pUb complex led to selective broadening and loss of signals for pUb residues around the serine 65 phosphorylation site (Figs 5A, middle panel and EV4A). When repeated with the double mutant K211N, W403A, the 15N‐labeled pUb spectrum was unchanged confirming that the changes were the result of pUb binding to the pUbl‐site on RING0 (Fig 5A, right panel). Phosphorylated ubiquitin exists in two conformations due to a shift of a β‐strand in a second, minor form (Wauer et al, 2015b). We observed similar changes in signal intensity for both conformations (Fig 5B), which we attribute to exchange between the two conformations, since UbVS assays with pUb TVLN mutant clearly show the minor pUb form is incapable of activating parkin (Fig 2C).
Figure 5. pUb binds to the parkin RING0 domain.
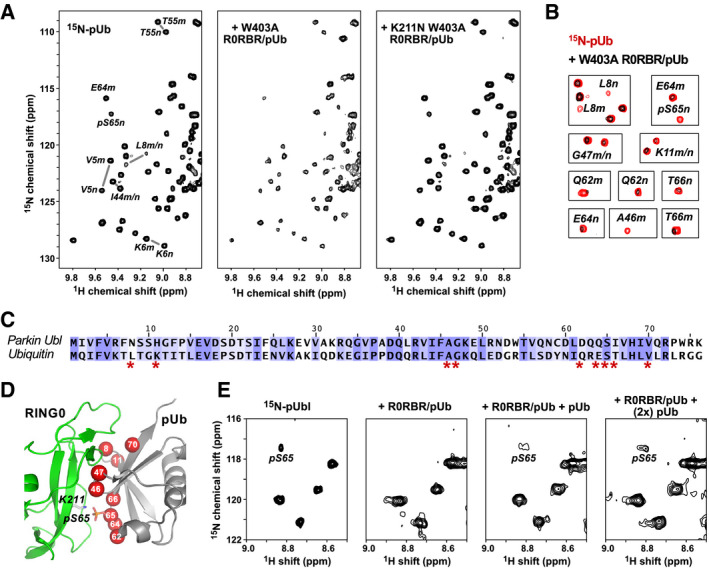
- 1H‐15N correlation spectra at 25°C of 50 µM 15N‐pUb alone (left) and in the presence of 100 µM complexes of pUb with W403A R0RBR (middle) or K211N W403A R0RBR parkin (right). Selected backbone amide signals are labeled according to residue number and the pUb conformation, major (m) or minor (n).
- Identification of 15N‐pUb signals that weaken or disappear when pUb is bound to W403A R0RBR.
- Sequence alignment of ubiquitin and the Ubl domain of human parkin colored according to similarity. Ubiquitin residues whose NMR signals change upon binding RING0 are marked by asterisks.
- Mapping of 15N‐pUb signals that weaken or disappear (red spheres) onto a model of pUb bound to parkin RING0.
- Competitive binding of pUbl and pUb to RING0. Portion of 1H‐15N NMR correlation spectra at 5°C of 60 µM 15N‐pUbl alone (left), in the presence of a 1:1 complex of rat parkin R0RBR and pUb (middle left), and in the presence of R0RBR/pUb supplemented with 60 μM pUb (middle right) and 120 μM pUb (right). The temperature was lower for this experiment to enhance detection of the weak binding by pUbl.
Figure EV4. pUb binds to the parkin RING0 domain.
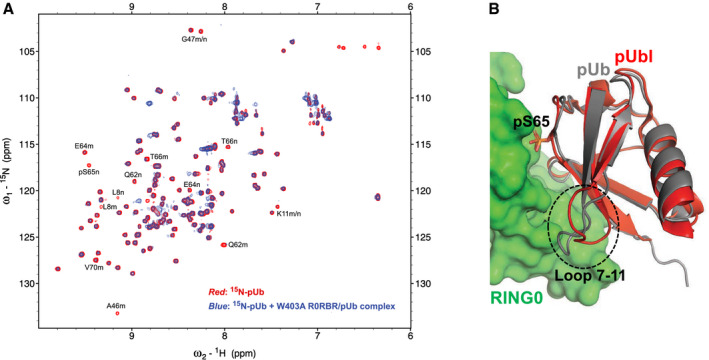
- The NMR correlation spectrum (red) of 15N‐pUb alone (50 μM) overlaid with the spectrum (blue) of 15N‐pUb (50 µM) in the presence of the parkin W403A R0RBR and pUb complex (100 μM). Signals that are strongly attenuated by binding are labeled by residue number and m for the major or n for minor pUb conformation.
- pUb was overlaid on pUbl in the pParkinΔREP‐RING2‐pUb structure (PDB 6GLC) (Gladkova et al, 2018). RING0 is displayed in green, pUbl in red, and pUb in gray.
We mapped the positions of the residues with the largest changes onto a model of pUb bound to RING0 based on the crystal structures of phosphorylated parkin. Ubiquitin and the parkin Ubl domain share 30% sequence identity and 71% similarity (Fig 5C), which allows for reliable modeling. The superimposition of pUb onto pUbl bound to RING0 shows identical conformations except for the loop formed by residues 7–11 (Fig EV4B). Comparison of the NMR changes with the model shows that the residues with the largest changes all reside at the RING0‐pUb interface (Fig 5D). This includes residues leucine 8 and lysine 11 that likely undergo a conformational rearrangement upon binding.
Additional evidence that pUb binds to the parkin pUbl‐binding site came from a competition experiment with 15N‐labeled pUbl (Fig 5E). As previously reported, the pUbl phosphoserine 65 peak disappears when bound to R0RBR/pUb (Sauve et al, 2018). Upon addition of excess of pUb, the pS65 peak reappeared confirming the competition for a single binding site.
Isothermal titration calorimetry (ITC) measurements
We conducted a series of ITC experiments to directly measure the affinity of pUb and pUbl to the RING0 binding site (Figs 6 and EV5). To decouple binding to RING0 from release of the RING2 domain, we used a truncated version of protein lacking the REP and RING2 domains, referred to as R0RB. The titration of R0RB with pUb revealed two binding sites with very distinct affinities: a high‐affinity site (2.9 nM) with positive enthalpy of binding and a low‐affinity site (365 nM) with negative enthalpy (Fig 6, top left). Based on previous studies of pUb binding to parkin, we attribute the high‐affinity site to binding of pUb to RING1 (Ordureau et al, 2014; Kazlauskaite et al, 2015; Sauve et al, 2015). The low‐affinity site corresponds to pUb binding to RING0. We carried out ITC measurements with two parkin mutants: A320R in RING1 and K211N in RING0. The mutation A320R weakened the high‐affinity site 500‐fold with essentially no effect on the low‐affinity site. Conversely, the K211N mutation eliminated the low‐affinity site with a small enhancement in high‐affinity site. We repeated the ITC experiments with pUbl (Fig 6, lower panels). The wild‐type R0BR fragment exhibited only one pUbl‐binding site (1.3 µM), which was assigned to the RING0 domain based on results with the mutant proteins. The RING1 A320R mutant bound pUbl with the same affinity as wild‐type, while the RING0 K211N mutant showed no detectable pUbl binding. Comparison of pUbl and pUb binding shows that pUbl binds RING0 with slightly weaker (fourfold) affinity in trans. This agrees well with the functional assays that show pUbl is roughly fourfold less effective in promoting UbVS crosslinking (Fig 2A).
Figure 6. Affinities of pUb and pUbl for the RING0 and RING1 binding sites measured by calorimetry.
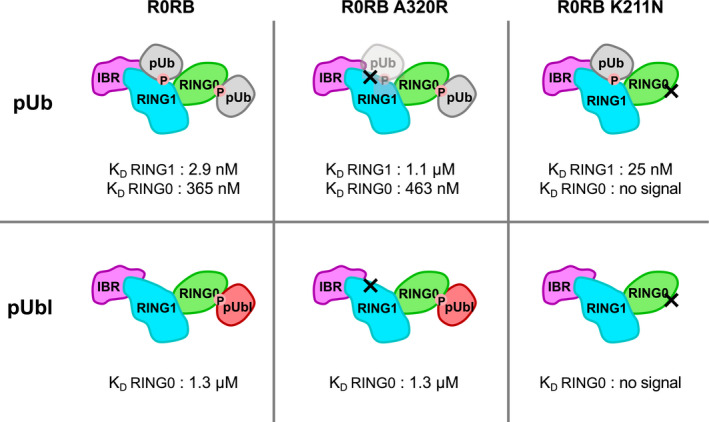
Models of complexes of the R0RB parkin fragment with either pUb or pUbl showing RING1 and RING0 binding sites. The affinities of complex formation were measured by ITC titrations and assigned to RING1 or RING0 based on the effects of mutations in either the RING1‐pUb‐binding site (A320R) or RING0‐pUbl‐binding site (K211N).
Figure EV5. Affinity of pUb and pUbl for RING0‐pUbl‐binding site.
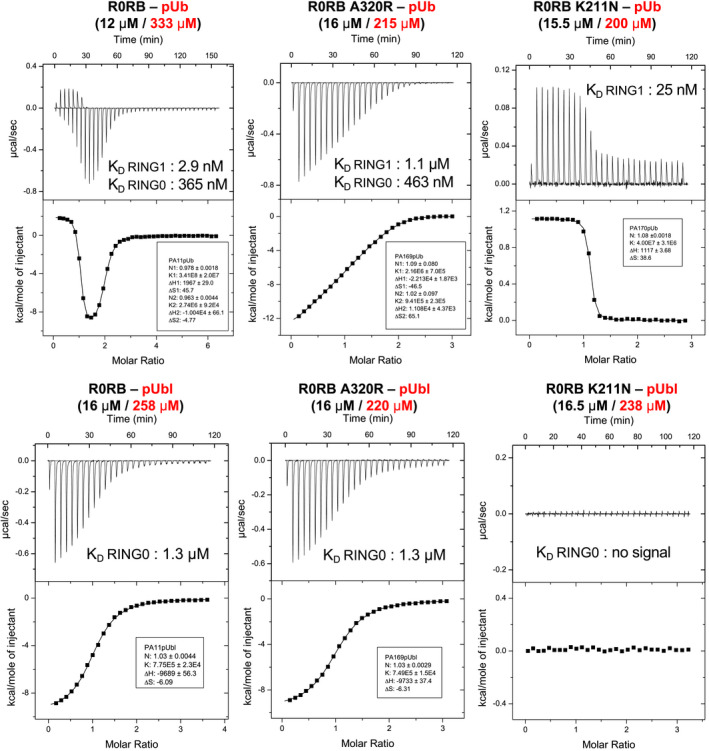
ITC data and fits for the titration of rat R0RB wild‐type, A320R or K211N with pUb and rat pUbl. Protein concentrations in the cell and the syringe and calculated affinities are indicated.
Discussion
Previous studies observed the existence of feedforward control in the PINK1/parkin pathway, but the molecular mechanism by which pUb directly activates parkin was not identified (Shiba‐Fukushima et al, 2012; Ordureau et al, 2014; Zhuang et al, 2016; Tang et al, 2017; Kazlauskaite et al, 2014). Here, we show that parkin can be activated without phosphorylation of its Ubl domain and that pUb binding is sufficient to activate ligase activity. pUb binds to the pUbl‐binding site allowing it to function as a signal for both recruitment of parkin to mitochondria and its activation (Fig 7).
Figure 7. Phosphorylation of ubiquitin on mitochondria acts on both recruitment and activation.
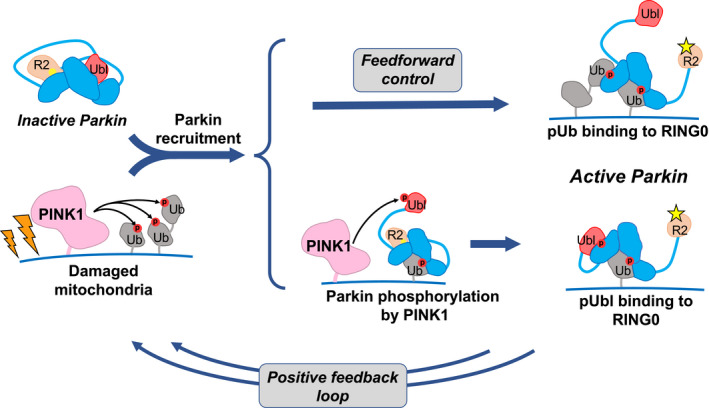
Accumulation of PINK1 leads to phosphorylation of ubiquitin on damaged mitochondria and recruitment of inactive parkin. Parkin can be activated through one of two ways. It can bind a second phosphorylated ubiquitin molecule or it can be phosphorylated on its Ubl domain. In the first case, the activation signal (pUb) is fixed and pre‐set, while in the second, it depends on the interaction between PINK1 and parkin. Both paths lead to a conformational change in which binding of pUb or pUbl to RING0 releases the catalytic RING2 domain (R2). Upon activation, parkin adds more ubiquitin molecules to the mitochondrial surface, which generates a positive feedback cycle of PINK1 activity and parkin recruitment. Phosphorylation sites are represented by red circles, and the parkin active‐site cysteine by yellow sphere when inactive and a star when active.
While the physiological importance of parkin activation by pUb in Parkinson's disease is unknown, there is a clear effect on mitochondrial recruitment and mitophagy in cultured cells. Loss of either the S65 phosphorylation site or the entire Ubl domain has less severe consequences than loss of the RING0 pUbl‐binding site (Fig 1B and C). The efficiency of mitophagy when the Ubl domain is absent is 20–40% compared with the full‐length protein (Fig 1C). Unfortunately, there is no way to perform the reciprocal experiment and selectively prevent pUb binding to quantify its contribution when both activation mechanisms, pUb‐ and pUbl‐binding, are present. The relative importance of the two activation mechanisms is probably affected by the level of ubiquitin present on the mitochondrial surface. The feedforward mechanism requires proximity of multiple pUb molecules, while parkin activation by phosphorylation requires proximity of PINK1 and pUb (Fig 7). Once parkin molecules in the pioneer round have been recruited and activated, the surface of the mitochondria will be covered with multiple pUb molecules, which would favor feedforward activation of subsequently recruited parkin. In future studies, it would be interesting to explore whether the two activation mechanisms are differentially sensitive to the basal level of mitochondrial ubiquitination. Feedforward activation of parkin likely provides a more robust response to PINK1 activity and accelerates the positive feedback responsible for the all‐or‐nothing, switch‐like behavior observed in cells.
The crowding of multiple parkin molecules on the mitochondrial surface has led to the recurrent suggestion that parkin may function as a dimer (Gundogdu et al, 2021). This does not appear to be the case as no complementation was detected in cellular assays of mitophagy between RING1 mutants defective in E2‐binding and RING2 mutants defective in Ub‐transfer (Fig 1D). An identical result was observed in solution with parkin autoubiquitination assays (Sauve et al, 2018). While our results are limited to one pair of mutations, the lack of complementation strongly argues against parkin dimerization as an obligatory step in catalysis. If it does occur in cells, it makes a minor contribution to parkin activity.
The two phosphoserine binding sites in parkin have markedly different properties and function. The site on RING1 functions to recruit parkin to mitochondria. It binds pUb with nanomolar affinity and is extraordinarily selective. pUbl does not compete for binding and, in fact, parkin phosphorylation increases the affinity of pUb binding. This is due to allosteric coupling in the RING1 domain between pUb binding and release of the unphosphorylated Ubl domain (Kazlauskaite et al, 2015; Kumar et al, 2015; Sauve et al, 2015; Wauer et al, 2015a). This serves to promote parkin phosphorylation and to reduce the dissociation of parkin from mitochondria after activation. Independently, the Ubl domain undergoes a conformational change when phosphorylated and loses affinity for its site in the autoinhibited conformation of parkin. This promotes its binding to RING0 and increases the affinity of pUb binding.
The second phosphoserine binding site is on RING0 and functions to activate parkin through the release of the catalytic RING2 domain (Gladkova et al, 2018; Sauve et al, 2018). The site was thought to be low affinity and specific for pUbl; however, neither is true. The weak binding is a consequence of competition between pUbl and RING2 and does not reflect the true intrinsic affinity of the RING0 site for pUbl. As an intramolecular ligand, the RING2 domain has a high local concentration, and its displacement is disfavoured. pUbl must have higher intrinsic affinity than RING2 since it is able to displace RING2 from RING0 when parkin is activated. We can estimate the effective intramolecular concentration using the volume accessible to the tethered domain. If the linker restricts the binding partner to a sphere of diameter 70 Å, the effective concentration is roughly 10 mM. Further assuming that parkin has 1% basal activity (so RING2 is bound 99% of the time), we can estimate that the intrinsic affinity (Kd in trans) of RING2 for RING0 is roughly 100 µM. Conversely, if parkin is 99% active when the Ubl domain is phosphorylated, then pUbl must have 100‐fold higher affinity than RING2. This yields an intrinsic (trans) affinity of 1 µM for pUbl binding to RING0, which is remarkably close to the 1.3 µM affinity measured by ITC (Fig 6). Unlike the selective RING1 site, RING0 binds both pUb and pUbl with former exhibiting moderately higher affinity in functional assays (Fig 2), NMR experiments (Fig 5), and ITC titrations (Fig 6).
Several lines of evidence suggest that positive, cooperative coupling exists between the RING1 and RING0 sites. UbVS assays with the W403A mutant showed slightly more RING2 release when the RING1 site was occupied (Fig 2A compared with 2E). Similarly, NMR experiments showed stronger binding of 15N‐pUbl to the R0RBR/pUb complex than to R0RBR alone (Sauve et al, 2018). Previous hydrogen–deuterium exchange experiments with phosphorylated parkin detected less protection of RING2 when pUb was present than when it was not (Sauve et al, 2018). The largest synergistic effect was manifested in the autoubiquitination assays where pUbl addition to GST‐R0RBR in the absence of pUb was relatively ineffective in inducing polyubiquitination (Fig 2F compared with 2D). However, the interpretation is not as unambiguous as in the UbVS assays since it is possible that pUb binding stimulated other aspects of the autoubiquitination reaction such as E2~Ub binding.
Activation of parkin without its Ubl domain suggests that fusion of the Ubl and catalytic R0RBR core was a late event in evolution. In primordial parkin, a single pUb‐binding site would have sufficed to regulate its recruitment and activation. The addition of the RING0 domain to the catalytic RBR domains and the specialization of the two binding sites—one for recruitment and one for activation—would have provided greater control. Specialization likely preceded addition of the Ubl domain since specificity of the recruitment site is necessary for parkin localization. pUbl binding to the recruitment site would prevent parkin binding pUb on mitochondria. In contrast, the activation site remained nonspecific and the persistence of its ability to bind both pUbl and pUb argues for a conserved biological function of the feedforward mechanism.
The observation of pUb binding to RING0 may be relevant for understanding other possible functions of parkin. A variety of results suggest that parkin plays a role in membrane trafficking in nerve terminals. It associates with synaptic membranes in a phosphorylation‐dependent manner and can ubiquitinate the synaptic proteins: endophilin and synaptojanin (Trempe et al, 2009; Cao et al, 2014). Synaptojanin is mutated in an early‐onset form of Parkinson's disease, and endophilin is a risk locus for Parkinson's disease (Chang et al, 2017; Koros et al, 2017). Both are substrates of the Parkinson's disease‐associated kinase LRRK2 (Arranz et al, 2015; Pan et al, 2017). The unexpected plasticity of parkin for binding pUb and pUbl suggests that these or other phosphoproteins may bind the RING1 or RING0 sites to control parkin localization and activation. More broadly, these studies offer hope that it may be possible to find small molecules that modulate parkin for treating Parkinson's disease.
Material and Methods
Mitochondria GFP‐parkin recruitment time‐lapse microscopy
Human osteosarcoma U2OS cells stably expressing fusion proteins of GFP with human parkin (https://www.addgene.org/45875/) wild‐type, mutated, and/or deleted (∆Ubl (res. 77‐465)) were seeded on a 35‐mm Glass Bottom 4 Compartment Dish (Greiner Bio‐One) (Tang et al, 2017). After 18 h, cells were stained with MitoTracker DeepRed FM (ThermoFisher) at a final concentration of 50 nM following the manufacturer’s guidelines. Cells were then transferred to a heated stage maintained at 37°C and 5% CO2 using a Zeiss temperature controller and cell perfusion system. Cells were treated with CCCP at a final concentration of 20 µM. Microscopy was performed on a Zeiss AxioObserver.Z1 inverted fluorescent microscope. Fully automated multidimensional acquisition was controlled using the Zen Pro software (Zeiss). Fixed exposure times were as follows: GFP 150 ms; and MitoTracker 40 ms. Images were taken at 2‐min intervals for a total of 120 min. Parkin recruitment was visualized by the appearance of punctate GFP fluorescence overlapping Mitotracker fluorescence. The percentage of cells exhibiting parkin recruitment to the mitochondria was calculated at 2‐min intervals over 120 min. Approximately, 500 cells were examined per mutant over two separate experiments. The fluorescence intensity of GFP‐positive puncta was not considered in parkin recruitment analysis.
Mitophagy assays
Mitophagy was assessed using flow cytometry analysis of mitochondrially targeted mKeima (a gift from A. Miyawaki, Laboratory for Cell Function and Dynamics, Brain Science Institute, RIKEN, Japan). U2OS cells stably expressing an ecdysone‐inducible mt‐Keima were induced with 10 µM ponasterone A, transfected with GFP‐parkin for 20 h, and treated with 20 µM CCCP or an equivalent volume of DMSO for 4 h (Tang et al, 2017). Complementation experiments included co‐expression of cerulean fluorescent protein (CFP, https://www.addgene.org/54604/) fusions and treated with CCCP or DMSO for 12 h. For flow cytometry analysis, cells were trypsinized, washed, and resuspended in PBS prior to analysis on a Thermo Attune NxT cytometer (Thermo) equipped with 405, 488, and 561 nm lasers and 610/20, 620/15, 530/30, and 525/50 filters (NeuroEDDU Flow Cytometry Facility, McGill University). Measurement of lysosomal mitochondrially targeted mKeima was made using a dual‐excitation ratiometric pH measurement where pH 7 was detected using 610/20 filter and excitation at 405nm and pH 4 using 620/15 filter and excitation at 561 nm. For each sample, 100,000 events were collected and single GFP‐parkin‐positive cells were subsequently gated for mt‐Keima. Data were analyzed using FlowJo v10.7.2 (Tree Star). The percentage of cells with an increase in the 405:561nm ratio (i.e., cells within the upper gate) was quantified and normalized to the percentage observed in GFP‐WT‐parkin CCCP‐treated samples for each replicate. PRK8 antibody (Ab77924, AbCam) was used to detect parkin expression in U2OS cells.
Cloning, expression, and purification of recombinant proteins in E. coli
Single‐point mutations and deletions were generated using PCR mutagenesis (Agilent). Gibson assembly (NEB) was used to produce Ub‐R0RBR chimera: a PCR fragment containing ubiquitin gene (res. 1–76) and a PCR fragment containing rat parkin residues 77–465 were introduced into the BamHI/XhoI sites of pGEX‐6P1 (GE Healthcare). The serine (S77) following glycine 76 of ubiquitin was mutated in proline to prevent chimera cleavage after ubiquitin glycine 76 by E. coli endogenous proteins during protein expression. A320R mutation was introduced in the chimera to prevent pUb binding to the high‐affinity pUb‐binding site on RING1.
All protein expressions were done in BL21 (DE3) E. coli and purification of full‐length human parkin, human or rat R0RBR (res. 141‐465), rat R0RB (res. 141‐379), Ub‐R0RBR, pUb, human or rat pUbl (res. 1–76), His‐UbcH7, Tc‐PINK1, and human His‐E1 were purified as previously described (Berndsen & Wolberger, 2011; Sauve et al, 2015). The rat parkin fragments were used as a matter of convenience; for example, rat R0RB was used in ITC experiments because it is well expressed. Rat parkin was chosen for NMR titrations with 15N‐labeled pUbl, since the spectrum of the rat domain shows better separation of pS65 peak than the spectrum of the human Ubl domain.
Briefly, proteins were purified by glutathione‐Sepharose (Cytiva) or Ni‐NTA agarose (Qiagen) affinity chromatography, followed by either 3C protease cleavage to remove the GST tag or Ulp protease cleavage to remove the His‐SUMO tag. Size‐exclusion chromatography was used as a last step. 15N‐labeled Ubl and Ub were produced in M9 minimal medium supplemented with 15NH4Cl. Phosphorylated Ub and Ubl were produced and purified according to published procedures (Ordureau et al, 2014; Wauer et al, 2015a). Purified proteins were verified using SDS–PAGE analysis. Protein concentrations were determined using UV absorbance. Tetra‐phospho‐ubiquitin chains (pUb)4 were obtained from Boston Biochem. Fiji was used for band quantification (Schindelin et al, 2012).
Ubiquitin vinyl sulfone assays
Ubiquitin vinyl sulfone assays (UbVS) were performed by incubating human full‐length, chimera Ub‐R0RBR or human R0RBR, WT or mutant with an excess of UbVS (Boston Biochem) in 50 mM Tris–HCl pH 8, 150 mM NaCl, 1 mM TCEP for 30 min at 37°C. Reactions were stopped by the addition of 5× SDS–PAGE loading buffer with 100 mM dithiothreitol (DTT) and analyzed on SDS–PAGE gel stained with Coomassie blue or by western blotting. For western blotting, the samples on SDS–PAGE gel were transferred to Immune‐Blot PVDF membrane. The membrane was first blocked with 5% BSA in TBS‐T (0.05% Tween 20) overnight at 4°C and then incubated with 1:2,000 dilution of rabbit anti‐Parkin antibody (Ab15954, AbCam) in the blocking solution. The membrane was washed with TBS‐T and incubated with 1:10,000 dilution of horseradish peroxidase (HRP)‐coupled goat anti‐rabbit IgG antibodies (Cell Signaling) in TBS‐T. After washing in TBS‐T, chemiluminescence was detected using ECL Prime (Cytiva) and bands were visualized with film.
Ubiquitination assays
The autoubiquitination assays of GST‐R0RBR (human) parkin were performed at 22°C for 5 min by adding 2 µM GST‐R0RBR WT, W403A or K211N, to 50 nM human His‐E1, 2 µM UbcH7, 100 µM ubiquitin in 50 mM Tris pH 8.0, 150 mM NaCl, 1 mM TCEP, 5 mM ATP, and 10 mM MgCl2 in the presence of increasing concentrations of pUbl or pUbΔG76. Autoubiquitination assays of R0RBR parkin were performed as for GST‐R0RBR except that the reactions were incubated at 22°C for 60 min. pUbΔG76 was used to prevent incorporation of pUb into the ubiquitination cascade by E1. Reactions were stopped by the addition of 5× SDS–PAGE loading buffer with 100 mM DTT, and the level of ubiquitination was analyzed on SDS–PAGE gels stained with Coomassie blue.
The autoubiquitination assays of full‐length human parkin and Ub‐R0RBR were done in two steps: first, the proteins were phosphorylated by TcPINK1, and then the autoubiquitination reaction was carried. The phosphorylation step was performed at 30°C for 3 h in 50 mM Tris pH 8.0, 100 mM NaCl, 1 mM TCEP, 0.1% CHAPS, 5 mM ATP, 10 mM MgCl2 with 0.1 µM GST‐PINK1 and 4 µM parkin. Protein phosphorylation was assessed using 7.5% SDS–PAGE gels containing 20 µM Phos‐tag (ApexBio) and 40 µM MnCl2. Autoubiquitination reactions were performed at 22°C for 1 hr by adding 3.3 µM phosphorylated or non‐phosphorylated parkin or Ub‐R0RBR to 50 nM human His‐E1, 3 µM UbcH7, 75 µM His‐ubiquitin S65A in 50 mM Tris–HCl pH 8.0, 150 mM NaCl, 1 mM TCEP, 4 mM ATP, and 8 mM MgCl2. The S65A mutation prevents ubiquitin phosphorylation by any PINK1 present. Reactions were stopped by the addition of 5× SDS–PAGE loading buffer, and the level of ubiquitination was analyzed on SDS–PAGE gels stained with Coomassie blue.
Isothermal titration calorimetry
ITC measurements were carried out at 20°C using VP‐ITC (Microcal) Samples were in 50 mM Tris–HCl, 150 mM NaCl, 1 mM TCEP, pH 7.4. The protein concentrations used for each titration are indicated in Fig EV5. WT and mutant R0RB were titrated with one injection of 5 μl followed by 28 injections of 10 μl of either pUb or pUbl. Data were modeled using either a single set of identical sites or two sets of identical sites using the Origin v7 software.
NMR experiments
Proteins used for NMR studies were buffer exchanged into 20 mM 4‐(2‐hydroxyethyl)‐1‐piperazineethanesulfonic acid (HEPES), 120 mM NaCl, 2 mM DTT, pH 7.4. For pUb titration, 100 μM unlabeled human R0RBR/pUb was added to 50 μM 15N‐labeled pUb. For pUbl titration, 120 μM unlabeled rat R0RBR/pUb was initially added to 60 μM 15N‐labeled pUbl. Then, 60 μM and 120 μM of pUb were added to the protein mixture. 1H‐15N heteronuclear single quantum coherence (HSQC) correlation spectra were acquired at field strengths of 600 MHz using Bruker spectrometers equipped with a triple‐resonance (1H, 13C, 15N) cryoprobe. Spectra were processed using NMRpipe and analyzed with SPARKY (Delaglio et al, 1995; Lee et al, 2015).
Author contributions
Véronique Sauvé: Conceptualization; Data curation; Formal analysis; Investigation; Visualization; Methodology; Writing—original draft; Writing—review & editing. George Sung: Conceptualization; Data curation; Formal analysis; Investigation; Visualization; Methodology. Emma J MacDougall: Conceptualization; Data curation; Formal analysis; Investigation; Visualization; Methodology. Guennadi Kozlov: Formal analysis; Investigation; Methodology. Anshu Saran: Investigation. Rayan Fakih: Investigation. Edward A Fon: Supervision; Funding acquisition; Project administration. Kalle Gehring: Conceptualization; Supervision; Visualization; Writing—original draft; Project administration; Writing—review & editing.
In addition to the CRediT author contributions listed above, the contributions in detail are:
VS performed construct design, protein purifications, UbVS and ubiquitination assays, and ITC experiments and wrote the manuscript. GS performed construct design, protein purifications, UbVS assays, and Western blots. EMD performed the recruitment to mitochondria and mt‐Keima assays and data analysis. GK performed NMR titrations and analysis. AS and RF performed protein purifications. EAF conceived experiments. KG conceived experiments, performed data analysis, and wrote the manuscript.
Disclosure and competing interests statement
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Acknowledgements
We thank Shafqat Rasool for the sample of ubiquitin TVLN and Kim A. Munro for advice for the ITC experiments. This work was supported by a Michael J. Fox Foundation grant (MJFF‐019029) to K.G., Canadian Institutes of Health Research grants (FDN 154301) to E.A.F and (FDN 159903) to K.G. and Canada Research Chairs (Tier 1) awards to E.A.F and K.G.
The EMBO Journal (2022) 41: e109460
Data availability
This study includes no data deposited in external repositories.
References
- Arranz AM, Delbroek L, Van Kolen K, Guimarães MR, Mandemakers W, Daneels G, Matta S, Calafate S, Shaban H, Baatsen P et al (2015) LRRK2 functions in synaptic vesicle endocytosis through a kinase‐dependent mechanism. J Cell Sci 128: 541–552 [DOI] [PubMed] [Google Scholar]
- Berndsen CE, Wolberger C (2011) A spectrophotometric assay for conjugation of ubiquitin and ubiquitin‐like proteins. Anal Biochem 418: 102–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borodovsky A, Kessler BM, Casagrande R, Overkleeft HS, Wilkinson KD, Ploegh HL (2001) A novel active site‐directed probe specific for deubiquitylating enzymes reveals proteasome association of USP14. EMBO J 20: 5187–5196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao M, Milosevic I, Giovedi S, De Camilli P (2014) Upregulation of Parkin in endophilin mutant mice. J Neurosci 34: 16544–16549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang D, Nalls MA, Hallgrimsdottir IB, Hunkapiller J, van der Brug M, Cai F, International Parkinson's Disease Genomics Consortium , 23andMe Research Team , Kerchner GA, Ayalon G et al (2017) A meta‐analysis of genome‐wide association studies identifies 17 new Parkinson's disease risk loci. Nat Genet 49: 1511–1516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaugule VK, Burchell L, Barber KR, Sidhu A, Leslie SJ, Shaw GS, Walden H (2011) Autoregulation of Parkin activity through its ubiquitin‐like domain. EMBO J 30: 2853–2867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6: 277–293 [DOI] [PubMed] [Google Scholar]
- Geisler S, Holmstrom KM, Skujat D, Fiesel FC, Rothfuss OC, Kahle PJ, Springer W (2010) PINK1/Parkin‐mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol 12: 119–131 [DOI] [PubMed] [Google Scholar]
- Gladkova C, Maslen SL, Skehel JM, Komander D (2018) Mechanism of parkin activation by PINK1. Nature 559: 410–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladkova C, Schubert AF, Wagstaff JL, Pruneda JN, Freund SM, Komander D (2017) An invisible ubiquitin conformation is required for efficient phosphorylation by PINK1. EMBO J 36: 3555–3572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundogdu M, Tadayon R, Salzano G, Shaw GS, Walden H (2021) A mechanistic review of Parkin activation. Biochim Biophys Acta Gen Subj 1865: 129894 [DOI] [PubMed] [Google Scholar]
- Kane LA, Lazarou M, Fogel AI, Li Y, Yamano K, Sarraf SA, Banerjee S, Youle RJ (2014) PINK1 phosphorylates ubiquitin to activate Parkin E3 ubiquitin ligase activity. J Cell Biol 205: 143–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazlauskaite A, Kondapalli C, Gourlay R, Campbell DG, Ritorto MS, Hofmann K, Alessi DR, Knebel A, Trost M, Muqit MM (2014) Parkin is activated by PINK1‐dependent phosphorylation of ubiquitin at Ser65. Biochem J 460: 127–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazlauskaite A, Martinez‐Torres RJ, Wilkie S, Kumar A, Peltier J, Gonzalez A, Johnson C, Zhang J, Hope AG, Peggie M et al (2015) Binding to serine 65‐phosphorylated ubiquitin primes Parkin for optimal PINK1‐dependent phosphorylation and activation. EMBO Rep 16: 939–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitada T, Asakawa S, Hattori N, Matsumine H, Yamamura Y, Minoshima S, Yokochi M, Mizuno Y, Shimizu N (1998) Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature 392: 605–608 [DOI] [PubMed] [Google Scholar]
- Kondapalli C, Kazlauskaite A, Zhang N, Woodroof HI, Campbell DG, Gourlay R, Burchell L, Walden H, Macartney TJ, Deak M et al (2012) PINK1 is activated by mitochondrial membrane potential depolarization and stimulates Parkin E3 ligase activity by phosphorylating Serine 65. Open Biol 2: 120080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koros C, Simitsi A, Stefanis L (2017) Genetics of Parkinson's disease: genotype‐phenotype correlations. Int Rev Neurobiol 132: 197–231 [DOI] [PubMed] [Google Scholar]
- Koyano F, Okatsu K, Kosako H, Tamura Y, Go E, Kimura M, Kimura Y, Tsuchiya H, Yoshihara H, Hirokawa T et al (2014) Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature 510: 162–166 [DOI] [PubMed] [Google Scholar]
- Kumar A, Aguirre JD, Condos TE, Martinez‐Torres RJ, Chaugule VK, Toth R, Sundaramoorthy R, Mercier P, Knebel A, Spratt DE et al (2015) Disruption of the autoinhibited state primes the E3 ligase parkin for activation and catalysis. EMBO J 34: 2506–2521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarou M, Narendra DP, Jin SM, Tekle E, Banerjee S, Youle RJ (2013) PINK1 drives Parkin self‐association and HECT‐like E3 activity upstream of mitochondrial binding. J Cell Biol 200: 163–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W, Tonelli M, Markley JL (2015) NMRFAM‐SPARKY: enhanced software for biomolecular NMR spectroscopy. Bioinformatics 31: 1325–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matheoud D, Sugiura A, Bellemare‐Pelletier A, Laplante A, Rondeau C, Chemali M, Fazel A, Bergeron JJ, Trudeau LE, Burelle Y et al (2016) Parkinson's disease‐related proteins PINK1 and Parkin repress mitochondrial antigen presentation. Cell 166: 314–327 [DOI] [PubMed] [Google Scholar]
- Matsuda N, Sato S, Shiba K, Okatsu K, Saisho K, Gautier CA, Sou YS, Saiki S, Kawajiri S, Sato F et al (2010) PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J Cell Biol 189: 211–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra DP, Jin SM, Tanaka A, Suen DF, Gautier CA, Shen J, Cookson MR, Youle RJ (2010) PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol 8: e1000298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ordureau A, Sarraf SA, Duda DM, Heo JM, Jedrychowski MP, Sviderskiy VO, Olszewski JL, Koerber JT, Xie T, Beausoleil SA et al (2014) Quantitative proteomics reveal a feedforward mechanism for mitochondrial PARKIN translocation and ubiquitin chain synthesis. Mol Cell 56: 360–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan PY, Li X, Wang J, Powell J, Wang Q, Zhang Y, Chen Z, Wicinski B, Hof P, Ryan TA et al (2017) Parkinson's disease‐associated LRRK2 hyperactive kinase mutant disrupts synaptic vesicle trafficking in ventral midbrain neurons. J Neurosci 37: 11366–11376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickles S, Vigie P, Youle RJ (2018) Mitophagy and quality control mechanisms in mitochondrial maintenance. Curr Biol 28: R170–R185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley BE, Lougheed JC, Callaway K, Velasquez M, Brecht E, Nguyen L, Shaler T, Walker D, Yang Y, Regnstrom K et al (2013) Structure and function of Parkin E3 ubiquitin ligase reveals aspects of RING and HECT ligases. Nat Commun 4: 1982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauve V, Lilov A, Seirafi M, Vranas M, Rasool S, Kozlov G, Sprules T, Wang J, Trempe JF, Gehring K (2015) A Ubl/ubiquitin switch in the activation of Parkin. EMBO J 34: 2492–2505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauve V, Sung G, Soya N, Kozlov G, Blaimschein N, Miotto LS, Trempe JF, Lukacs GL, Gehring K (2018) Mechanism of parkin activation by phosphorylation. Nat Struct Mol Biol 25: 623–630 [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda‐Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B et al (2012) Fiji: an open‐source platform for biological‐image analysis. Nat Methods 9: 676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seirafi M, Kozlov G, Gehring K (2015) Parkin structure and function. FEBS J 282: 2076–2088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiba‐Fukushima K, Arano T, Matsumoto G, Inoshita T, Yoshida S, Ishihama Y, Ryu KY, Nukina N, Hattori N, Imai Y (2014) Phosphorylation of mitochondrial polyubiquitin by PINK1 promotes Parkin mitochondrial tethering. PLoS Genet 10: e1004861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiba‐Fukushima K, Imai Y, Yoshida S, Ishihama Y, Kanao T, Sato S, Hattori N (2012) PINK1‐mediated phosphorylation of the Parkin ubiquitin‐like domain primes mitochondrial translocation of Parkin and regulates mitophagy. Sci Rep 2: 1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sliter DA, Martinez J, Hao L, Chen X, Sun N, Fischer TD, Burman JL, Li Y, Zhang Z, Narendra DP et al (2018) Parkin and PINK1 mitigate STING‐induced inflammation. Nature 561: 258–262 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Sugiura A, McLelland GL, Fon EA, McBride HM (2014) A new pathway for mitochondrial quality control: mitochondrial‐derived vesicles. EMBO J 33: 2142–2156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swatek KN, Usher JL, Kueck AF, Gladkova C, Mevissen TET, Pruneda JN, Skern T, Komander D (2019) Insights into ubiquitin chain architecture using Ub‐clipping. Nature 572: 533–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang MY, Vranas M, Krahn AI, Pundlik S, Trempe JF, Fon EA (2017) Structure‐guided mutagenesis reveals a hierarchical mechanism of Parkin activation. Nat Commun 8: 14697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trempe JF, Chen CX, Grenier K, Camacho EM, Kozlov G, McPherson PS, Gehring K, Fon EA (2009) SH3 domains from a subset of BAR proteins define a Ubl‐binding domain and implicate parkin in synaptic ubiquitination. Mol Cell 36: 1034–1047 [DOI] [PubMed] [Google Scholar]
- Trempe JF, Sauve V, Grenier K, Seirafi M, Tang MY, Menade M, Al‐Abdul‐Wahid S, Krett J, Wong K, Kozlov G et al (2013) Structure of parkin reveals mechanisms for ubiquitin ligase activation. Science 340: 1451–1455 [DOI] [PubMed] [Google Scholar]
- Valente EM, Abou‐Sleiman PM, Caputo V, Muqit MM, Harvey K, Gispert S, Ali Z, Del Turco D, Bentivoglio AR, Healy DG et al (2004) Hereditary early‐onset Parkinson's disease caused by mutations in PINK1. Science 304: 1158–1160 [DOI] [PubMed] [Google Scholar]
- Vives‐Bauza C, Zhou C, Huang Y, Cui M, de Vries RL, Kim J, May J, Tocilescu MA, Liu W, Ko HS et al (2010) PINK1‐dependent recruitment of Parkin to mitochondria in mitophagy. Proc Natl Acad Sci U S A 107: 378–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wauer T, Komander D (2013) Structure of the human Parkin ligase domain in an autoinhibited state. EMBO J 32: 2099–2112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wauer T, Simicek M, Schubert A, Komander D (2015a) Mechanism of phospho‐ubiquitin‐induced PARKIN activation. Nature 524: 370–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wauer T, Swatek KN, Wagstaff JL, Gladkova C, Pruneda JN, Michel MA, Gersch M, Johnson CM, Freund SM, Komander D (2015b) Ubiquitin Ser65 phosphorylation affects ubiquitin structure, chain assembly and hydrolysis. EMBO J 34: 307–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzel DM, Lissounov A, Brzovic PS, Klevit RE (2011) UBCH7 reactivity profile reveals parkin and HHARI to be RING/HECT hybrids. Nature 474: 105–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang N, Li L, Chen S, Wang T (2016) PINK1‐dependent phosphorylation of PINK1 and Parkin is essential for mitochondrial quality control. Cell Death Dis 7: e2501 [DOI] [PMC free article] [PubMed] [Google Scholar]


