Abstract
Objectives
Slaughterhouse staff is occupationally exposed to antimicrobial resistant bacteria. Studies reported high antimicrobial resistance gene (ARG) abundances in slaughter pigs. This cross-sectional study investigated occupational exposure to tetracycline (tetW) and macrolide (ermB) resistance genes and assessed determinants for faecal tetW and ermB carriage among pig slaughterhouse workers.
Methods
During 2015–2016, 483 faecal samples and personal questionnaires were collected from workers in a Dutch pig abattoir, together with 60 pig faecal samples. Human dermal and respiratory exposure was assessed by examining 198 carcass, 326 gloves, and 33 air samples along the line, next to 198 packed pork chops to indicate potential consumer exposure. Samples were analyzed by qPCR (tetW, ermB). A job exposure matrix was created by calculating the percentage of tetW and ermB positive carcasses or gloves for each job position. Multiple linear regression models were used to link exposure to tetW and ermB carriage.
Results
Workers are exposed to tetracycline and macrolide resistance genes along the slaughter line. Tetw and ermB gradients were found for carcasses, gloves, and air filters. One packed pork chop contained tetW, ermB was non-detectable. Human faecal tetW and ermB concentrations were lower than in pig faeces. Associations were found between occupational tetW exposure and human faecal tetW carriage, yet, not after model adjustments. Sampling round, nationality, and smoking were determinants for ARG carriage.
Conclusion
We demonstrated clear environmental tetracycline and macrolide resistance gene exposure gradients along the slaughter line. No robust link was found between ARG exposure and human faecal ARG carriage.
Keywords: air, dermal exposure, faecal carriage, gloves, macrolide resistance, respiratory exposure, retail meat, tetracycline resistance
Introduction
Emerging antimicrobial resistance (AMR) poses a threat to global human and animal health (World Health Organization, 2014). Antimicrobial use (AMU) is the major driver for AMR in humans and animals in general (Holmes et al., 2016; Murphy et al., 2018). Human AMR carriage has been associated with direct and indirect contact with livestock (Bisdorff et al., 2012; Dorado-García et al., 2013; Dohmen et al., 2017a), while a high AMR prevalence has been documented in persons with regular occupational livestock contact including veterinarians (Wulf et al., 2008), farmers (Garcia-Graells et al., 2013; Geenen et al., 2013) and abattoir workers (van Cleef et al., 2010; Mulders et al., 2010; Gilbert et al., 2012; Dohmen et al., 2017b).
Contact with live animals was identified as the major risk factor for nasal carriage of (livestock-associated) methicillin-resistant Staphylococcus aureus [(LA-)MRSA] in pig and broiler slaughterhouse workers (van Cleef et al., 2010; Mulders et al., 2010; Gilbert et al., 2012). Similarly, a higher risk for faecal carriage of extended-spectrum β-lactamase-producing Enterobacteriaceae (ESBL-PE) was found for pig slaughterhouse employees working at earlier positions in the slaughter line (Dohmen et al., 2017b). Since AMR can also be indirectly transmitted to humans via the environment (Dorado-García et al., 2013; Dohmen et al., 2017a), it is necessary to quantify AMR in the air or on environmental reservoirs like carcasses or surfaces throughout the slaughter line.
Abovementioned studies focused their efforts on detection of specific resistant bacteria (MRSA, ESBL-PE). Our recent resistome studies demonstrated that antimicrobial resistance genes (ARGs) conferring resistance to tetracyclines and macrolides were among the most abundant ARGs in the resistome of European fattening pigs close to slaughter and were respectively associated with tetracycline and lincosamide/macrolide use in pigs (Munk et al., 2018; Van Gompel et al., 2019). These findings led us to adopt an in-depth study of the risk for ARG carriage in pig slaughterhouse workers by specifically studying faecal tetracycline (tetW) and macrolide (ermB) resistance genes. To assess occupational exposure to tetW and ermB, we simultaneously studied a variety of host (pig faeces) and environmental samples (pig carcasses, gloves worn by staff as a proxy for hand contamination, ambient air) at various positions along the pig slaughter line and assessed determinants for faecal tetW and ermB carriage among workers. In addition, we studied packed meat further down the chain as an indication for potential consumer exposure to ARGs.
Methods
Study design
Samples were collected within the largest Dutch pig slaughterhouse (Vion, Boxtel, the Netherlands), slaughtering daily ~18 000 Dutch fattening pigs. Sampling was performed in two 1-week sampling rounds (second week June 2015, first week July 2016). The human population from 2015 was previously largely tested for ESBL-PE (Dohmen et al., 2017b). Pig slaughter was divided into two main phases, slaughter in the ‘black area’ [successively: lairage, stunning, bleeding, scalding/dehairing, and carcass scorching (singeing)] and the ‘clean area’ [successively: evisceration, pluck removal (removal liver, lungs, heart, oesophagus, and tongue), carcass splitting, meat inspection, dressing, and carcass cooling)], followed by a third phase of cutting, deboning, and some further processing (Supplementary Figure S1, available at Annals of Work Exposures and Health online).
Respectively in 2015 and 2016, we collected in total (convenience sampling): 325 and 158 faecal samples from abattoir and other staff; 96 and 102 pooled carcass [skin (cork borer) and meat] samples; 16 and 17 air samples, 156 and 170 gloves worn by slaughterhouse staff, and 30 and 30 pig faecal samples. Carcass and glove sampling was divided over different time points to avoid bias based on daily bacterial build-up. Furthermore, 198 separate meat end-products (packed pork chops) were collected at the meat slicing and packing centre (Vion, Groenlo, the Netherlands) from June to September 2016.
Sample collection
Human faecal samples (N = 483) and gloves (N = 326)
Slaughterhouse employees were recruited by means of flyers, posters, and information provided on screens. Written information was translated into 11 languages to accommodate the 14 nationalities present and was distributed within all canteens together with stool sampling packages and questionnaires. Participants collected a stool sample and completed a questionnaire regarding personal details and occupational exposure (i.e. job position, AMU, animal contact, hospitalization, travelling, and meat consumption). A sample collection booth was installed at the main entrance of the slaughterhouse and was occupied by someone from the research team around all main breaks (from around 5 am to 5 pm). In 2015 and 2016, respectively 1781 (all slaughterhouse production workers) and 354 (production workers from lairage to cooling, to increase samples from earlier and scarcer slaughter line positions) employees were initially targeted to participate. Participants were compensated (25 euro) and gave written consent. The Medical Ethical Committee of the University Medical Centre Utrecht confirmed that the Dutch ‘Medical Research Involving Human Subjects Act’ did not apply for this study (Protocols 14–346/C, 14–403/C).
To determine occupational exposure to ARGs through hand contact, we examined the surface of disposable gloves worn by abattoir staff. At two time points (morning, afternoon) of one single day (for locations refer to Supplementary Figure S1, available at Annals of Work Exposures and Health online), we collected gloves (inside-out, chosen from the hand with most meat contact) in a stomacher bag (BagLight PolySilk®, Interscience, 400 ml, UK).
Pig carcass (N = 198) and faecal samples (N = 60), meat-end products (N = 198)
Carcass samples were collected on two consecutive days. The first day, sampling was divided over four time points (early morning, late morning, early afternoon, and late afternoon) (Supplementary Figure S1, available at Annals of Work Exposures and Health online). Each carcass sample consisted of four cork borer samples from four pigs closely succeeding in line, taken from the pigs’ shoulder (Ø 25 mm), subsequently pooled and collected in a stomacher bag (BagPage®+, with separation membrane, Interscience, 400 ml, UK). In parallel, at each time point, 14–16 pig faecal colon samples were collected at the veterinary inspection platform. The second day, we collected and similarly pooled meat samples in the morning (carcass cooling), and the morning and afternoon (cutting and deboning) from pigs slaughtered the previous day. Finally, packed meat end-products, intended for retail, were collected on six different mornings (pork chops, unconnected to previous sampling, N = 198) after packing under a modified atmosphere (~30% CO2, ~70% O2) at Vion Groenlo.
Air samples (N = 33)
Personal inhalable air samples were collected over the course of one shift (~8 h) by abattoir staff along the slaughter line (Supplementary Figure S1, available at Annals of Work Exposures and Health online) by means of a GilAir-5 pump [flow rate of 3.5 l/min, GSP conical sampler (JS holdings, UK)] containing a 37 mm Teflon (PTFE) SKC filter (2.0 µm, pore size, Ø 37 mm, SKC, Inc., USA). We only included successful 4–6 h measurements, excluding breaks, for measurement consistency.
Environmental samples laboratory preparation
Laboratory preparation steps including overall sample handling are described in detail in the Supplementary Material (available at Annals of Work Exposures and Health online). Briefly, Ringer’s solution was applied to the outside of each glove (by filling the inside-out turned gloves within a stomacher bag) and the stomacher bags containing the carcass samples or meat-end products, followed by further processing using a stomacher and centrifuge. Pellets were subsequently obtained and stored at −80°C until DNA extraction. Air filters were sterilely transferred to Greiner tubes which were then filled with extraction fluid and centrifuged. The fluid was freeze-dried and the resulting lyophilizate was stored at −20°C until DNA extraction.
DNA extraction and qPCR analysis
Samples were thawed just before DNA extraction. DNA from human and pig faeces was extracted by the modified QIAmp Fast DNA stool mini kit (Qiagen, cat. no. 51604) as described before (Knudsen et al., 2016). Glove DNA was extracted by the NucleoSpin®96 Food kit (Macherey-Nagel), while meat and air DNA was respectively extracted by the modified Nucleospin® Food kit (Macherey-Nagel) behind NucleoSpin® 8 Plant II kit. Following DNA extraction, tetW (Walsh et al., 2011) and ermB (Koike et al., 2010) genes were targeted by qPCR (Supplementary Table S1, available at Annals of Work Exposures and Health online). Additionally, qPCR was performed to target 16S rRNA (Fierer et al., 2005), a general molecular marker for microbial communities, used for normalization of tetW and ermB gene copies per total bacterial load. Refer to the Supplementary Material (available at Annals of Work Exposures and Health online) for more details regarding DNA extraction and qPCR.
Data preparation and statistical analysis
Questionnaires and field forms were entered and checked for consistency in EpiData 3.1 before importing into ‘R’ v.3.5.1 for data cleaning and statistical analysis (R Core Team, 2018). Potential determinants from the questionnaire were used in the analysis when at least 95% of the questions were answered. Due to potential difficulties in identifying antibiotics by its users, antibiotic usage was recorded after evaluating two questions from the questionnaire: (i) The question regarding the usage of any antibiotics or medication in the past year and (ii) In case medication was used, a question had to be answered with regard to the characteristics of the drugs used (name, indication, duration of usage, administration method). In case of doubt with regard to the type of medication used, the answer was marked as ‘potential antibiotic usage’. Participants were asked to choose up to three of the presented slaughter line positions where they worked the most and to describe their role in their own words. If workers indicated more than one job position, staff was assigned to the last, presumably cleaner, position in the slaughter line favouring specificity over sensitivity (Le Moual et al., 2018). If other positions were mentioned, they were assigned to the category ‘other’ unless the position clearly fitted in one of the slaughter line categories.
An ARG job exposure matrix (ARG-JEM) for glove and carcass contamination was created by calculating the percentage of detects (tetW, ermB) in carcass and glove samples for seven job area groups for which both carcass and gloves samples were available (Table 1, model population N = 327). These percentages were then allocated to workers in these positions for each combination of gene (tetW/ermB) and sample type (carcass/glove). Other environmental samples were only used for descriptive analysis due to localized sampling (pig faeces, meat-end products) or a limited number of samples (air).
Table 1.
General characteristics of the human slaughterhouse population and the human population included in the models.
| Characteristics | Full populationa (N = 482) N/total (%) | Population included in the modelsa (N = 327) N/total (%) |
|---|---|---|
| Gender (female) | 76/476 (16.0) | 47/322 (14.6) |
| Smoking (no) | 256/470 (54.5) | 177/319 (55.5) |
| Antibiotic use (no) | 427/475 (89.9) | 291/324 (89.8) |
| Mean age (10th–90th percentile) | 39.0 (24.0–54.1) N = 480b | 38.7 (25.0–54.0) N = 325b |
| Job positionc | ||
| Black area: | ||
| Lairage, bleeding | 23/482 (4.8) | 23/327 (7.0) |
| Scalding to singeing | 26/482 (5.4) | 26/327 (8.0) |
| Clean area: | ||
| After singeing | 8/482 (1.7) | 8/327 (2.5) |
| Evisceration, pluck removal | 52/482 (10.8) | 52/327 (15.9) |
| Inspection, dressing | 79/482 (16.4) | 79/327 (24.2) |
| Cutting and deboning: | ||
| Cutting room | 60/482 (12.5) | 60/327 (18.4) |
| Deboning room | 79/482 (16.4) | 79/327 (24.2) |
| Other areas: | ||
| Cooling | 18/482 (3.7) | n.a. |
| Organ area | 42/482 (8.7) | n.a. |
| Other (e.g. office, facility, cleaning) | 95/482 (19.7) | n.a. |
| Nationalities | ||
| Dutch | 77/482 (16.0) | 46/327 (14.1) |
| Polish | 186/482 (38.6) | 124/327 (37.9) |
| Romanian | 72/482 (15.0) | 55/327 (16.8) |
| Otherd | 124/482 (25.7) | 39/327 (11.9) |
| Unknown (missing) | 23/482 (4.8) | 13/327 (4.0) |
a N of the full (N in 2015 = 325, N in 2016 = 157) and model population (N in 2015 = 211, N in 2016 = 116) is an approximation of the size of the datasets used for the tetW and ermB (log10 copies) analyses before outlier removal. The tetW and ermB full and model datasets have 97.3% (full population) and 96.6% (model population) of all observations in common. Missings were excluded in the summary figures per variable.
b N = part of the population for whom a date of birth was available.
cIn total, around 24% of the workers worked in more than one area. Due to the targeting of workers from lairage to cooling in 2016, only four workers reported working in the cutting and deboning area (last position in the slaughter line) in 2016. n.a. = category not included in the models (gloves and carcass samples were not both available).
dE.g. Hungarian, Slovakian, and Portuguese nationalities.
All eligible questionnaire and ARG-JEM variables were separately linearly regressed against workers’ tetW or ermB carriage, expressed as log10 copies per gram faeces. Then, three separate multiple regression models were fitted per gene (tetW, ermB) including either the ARG-JEM derived carcass or gloves variable or the job position variable (with similar job positions included as in the JEM), next to associated covariates from the univariate analysis (P < 0.1) and potential confounders. Potential interactions and assumptions of the final models were checked (with and without potential outliers based on model diagnostics). Subsequently, geometric mean ratios (GMR) were computed by exponentiating regression coefficients and their 95% confidence intervals (CIs). Finally, a sensitivity analysis was performed including the 16S-normalized human faecal tetW and ermB log10 copies as the outcome variable.
Results
TetW and ermB were detectable in all human and pig faecal samples, whereas a smaller proportion of carcass (~20%), glove (~70%), and air (~30%) samples showed levels above the limit of detection (LOD, Supplementary Table S2, available at Annals of Work Exposures and Health online).
Human and pig samples
The human population (mean age 39 years, range 18–64) mostly consisted of men (84%), with 54.5% non-smokers (Table 1). Most common nationalities were Polish (38.6%), Dutch (16%), and Romanian (15%). Dutch, Polish, and Romanian staff are roughly proportionally represented within the black area (respectively, 26.5%, 24.5%, and 28.6% of all workers in the black area), while their majority is working at cutting and deboning (of all Dutch workers: 50.0%, of all Polish workers: 45.5%) or the clean area (of all Romanian workers: 57.4%). Most participants working at fixed positions along the slaughter line worked at inspection and dressing and within the deboning room (both 16.4%). Furthermore, the majority of staff (89.8%) reported not to have used any form of antibiotics 1 year before sampling.
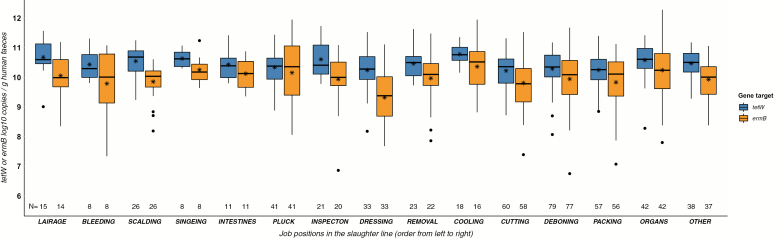
All human samples were found positive for tetW and ermB. TetW and ermB concentrations found in human faeces are lower compared to pig faeces. In both human and pig faeces, we observed higher concentrations of tetW than ermB. Human ARG concentrations varied across slaughter steps (Fig. 1).
Figure 1.
Prevalence of tetW and ermB log10 copies per gram stool from slaughterhouse staff along the slaughter line. The slaughter line runs from left to right: categories ‘organs’ and ‘other’ are technically not part of the slaughter line. Stars (*) depict the mean tetW or ermB log10 concentrations at a specific job location along the slaughter line. A box represents the 25th (Q1) to 75th (Q3) percentile, the centreline depicts the median (Q2). Dots represent values larger or smaller than Q1 − 1.5 or Q3 + 1.5 times the interquartile range. x-axis: N (numbers displayed above the x-axis): The number of human faecal samples eligible for analysis after qPCR quality control per slaughter position for each gene target (tetW, ermB). x-axis labels: The following job positions are included per slaughter step: Lairage: stables, stunning area. Bleeding: hanging and bleeding of stunned pigs. Scalding: positions after carcass scalding and carcass dehairing, before carcass singeing. Singeing: positions after carcass singeing, before evisceration. Intestines: removal of intestines (evisceration). Pluck: removal of liver, lungs, heart, oesophagus, and tongue. Inspection: veterinary inspection platform. Dressing: dressing of carcasses, removal of fat, and diaphragm. Removal: removal of heads and spinal cords. Cooling: cooling, includes carcass cooling but also several other cooling units. Cutting: separate carcass cutting room (within the same building as the main abattoir). Deboning: separate deboning area (within the same building as the main abattoir). Packing: packing and preparing meat for transport. Organs: multiple separate ‘warm’ and ‘cold organ units’: e.g. working with hearts, kidneys, and/or livers in separate organ areas. Other: includes office, facility and cleaning staff, and all other staff not working at a fixed position along the slaughter line.
Meat, glove, and air samples
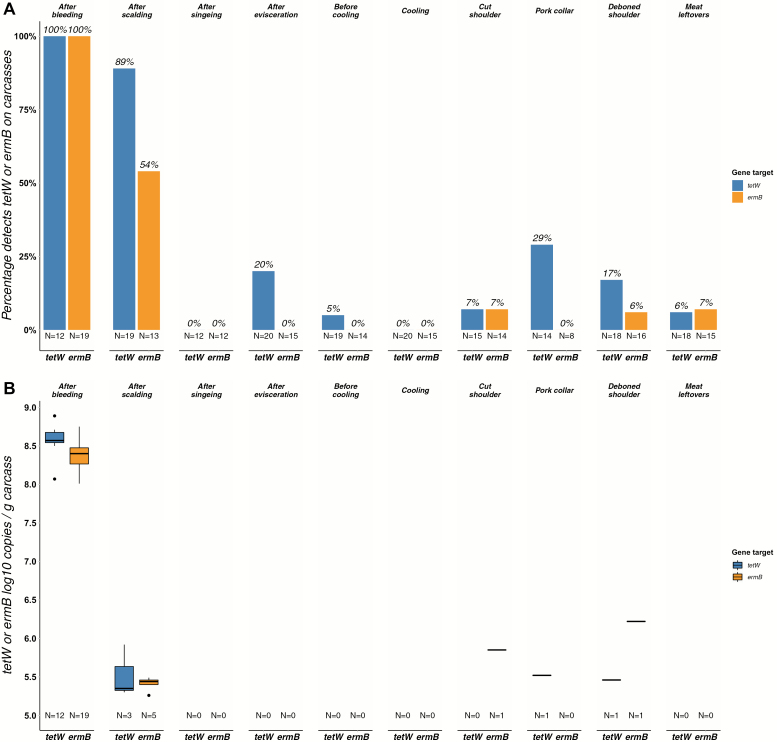
Carcass samples were predominantly tetW and ermB positive within the first slaughter steps (respectively, both 100% after bleeding, 89% and 54% after scalding, Fig. 2A). Remarkably, a rise in tetW and ermB positive samples was seen within cutting and deboning, although nearly all samples had gene concentrations below the limit of quantification (LOQ) (Fig. 2B). Additionally, we identified only one tetW (<LOQ, not originating from the sampled slaughterhouse) and no ermB positive packed pork chops. All pork chops originated from Dutch pigs, 21.3% came from the examined slaughterhouse.
Figure 2.
Bar plot (A): Percentage of tetW and ermB detects on pig carcasses and meat collected along the slaughter line (N = number of samples eligible for analysis after qPCR quality control). Boxplot (B): Total number of tetW and ermB log10 copies on carcasses along the slaughter line (N = number of samples > LOQ). A box represents the 25th (Q1) to 75th (Q3) percentile, the centreline depicts the median (Q2). Dots represent values larger or smaller than Q1 − 1.5 or Q3 + 1.5 times the interquartile range. The slaughter line runs from left to right.
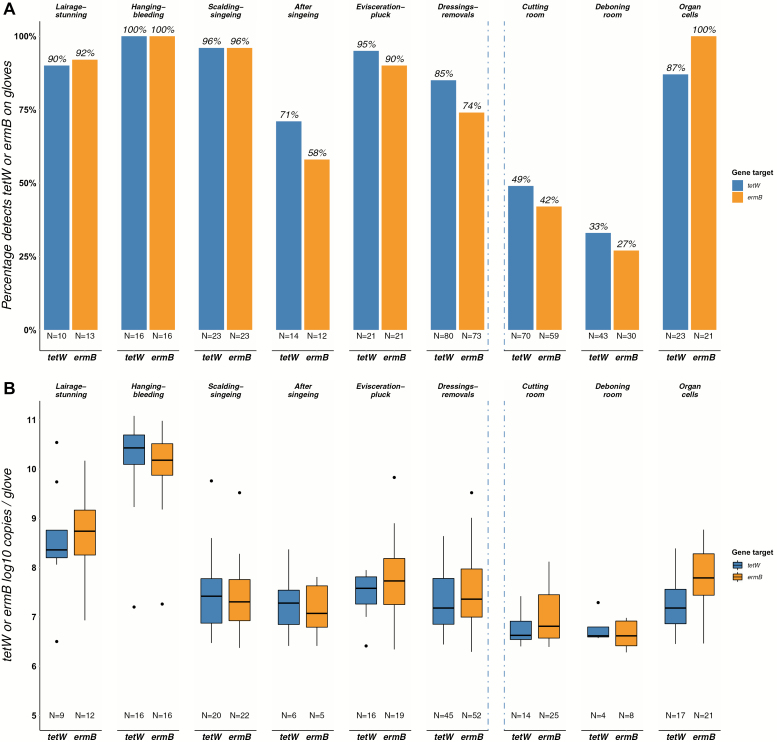
In comparison to carcass samples, gloves more frequently tested positive for tetW and ermB, and overall demonstrated higher median concentrations (samples >LOQ) (Fig. 3A,B). Additionally, a slight decrease was found after singeing, and an increasing-decreasing trend before and after evisceration and pluck removal. After cooling another decrease was visible, while slightly higher median tetW and ermB concentrations were found within gloves collected in the organ units. In gloves, tetW and ermB concentrations were also frequently low (of all positive samples respectively, 2–31% were <LOQ). After 16S-normalization, the same, but slightly more distinct trends were visible after ‘hanging and bleeding’ when examining ermB (Supplementary Figure S2, available at Annals of Work Exposures and Health online).
Figure 3.
Bar plot (A): Percentage of tetW and ermB detects on gloves worn by slaughterhouse staff along the slaughter line (N = number of samples eligible for analysis after qPCR quality control). Boxplot (B): Total number of tetW and ermB log10 copies on gloves worn by slaughterhouse staff along the slaughter line (N = number of samples > LOQ). A box represents the 25th (Q1) to 75th (Q3) percentile, the centreline depicts the median (Q2). Dots represent values larger or smaller than Q1 − 1.5 or Q3 + 1.5 times the interquartile range. Blue dotted lines represent the location of the cooling areas (including the carcass cooling) in the slaughter line. The slaughter line runs from left to right.
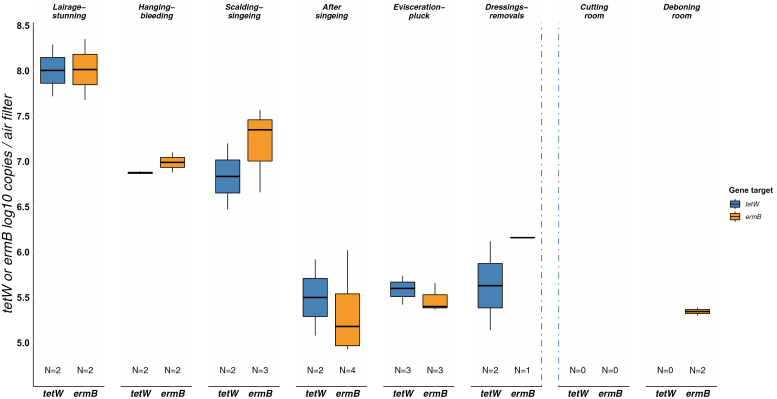
All air samples tested positive for tetW and ermB, except for samples from the cutting (tetW 0/3 and ermB 0/2 positive) and the deboning rooms (tetW 2/4 and ermB 2/3 positive). A clear declining gradient along the slaughter line was visible in samples >LOQ within the first steps of the slaughter line (Fig. 4). This overall trend was corroborated after 16S-normalization, apart from a seemingly lower median tetW and ermB concentration found at lairage and stunning (Supplementary Figure S3, available at Annals of Work Exposures and Health online).
Figure 4.
Total number of tetW and ermB log10 copies on air filters collected along the slaughter line (N = number of samples > LOQ). A box represents the 25th (Q1) to 75th (Q3) percentile, the centreline depicts the median (Q2). Dots represent values larger or smaller than Q1 − 1.5 or Q3 + 1.5 times the interquartile range. Blue dotted lines represent the location of the cooling areas (including the carcass cooling) in the slaughter line. The slaughter line runs from left to right.
Determinants for tetW carriage in human stool
The percentage of tetW detected on carcasses and gloves was positively associated with the concentration of tetW in human stool (GMR for a 10% increase in detected tetW = 1.05, P = 0.052 for carcasses, and 1.07, P = 0.01 for gloves, Table 2). Adjustments for gender, age, smoking, AMU, and nationality mainly altered the carcass association (carcass: GMR = 1.03, P = 0.26; gloves: GMR = 1.08, P = 0.004). Additionally adjusting for sampling round yielded non-significant estimates for both carcasses and gloves (carcass: GMR = 1.004, P = 0.88, gloves: GMR = 1.01, P = 0.70; Table 2).
Table 2.
Determinants for tetW and ermB log10 copies per gram human stool.
| tetW | ermB | |||
|---|---|---|---|---|
| Risk factors | GMR [95% CI] | Adjusted GMR [95% CI] | GMR [95% CI] | Adjusted GMR [95% CI] |
| Environmental exposure variables | ||||
| Carcass (% detects per 10%) | 1.05 [0.9997, 1.10] | 1.004 [0.96, 1.05] | 0.99 [0.91, 1.08] | 1.01 [0.93, 1.09] |
| Gloves (% detects per 10%) | 1.07 [1.02, 1.13]a | 1.01 [0.95, 1.08] | 1.005 [0.92, 1.09] | 0.98 [0.89, 1.08] |
| Job position | ||||
| Black area | 1.91 [1.25, 2.92]a | 1.23 [0.76, 1.97] | 0.93 [0.46, 1.88] | 1.01 [0.48, 2.14] |
| Clean area | 1.41 [1.04, 1.90]a | 1.15 [0.81, 1.64] | 1.03 [0.63, 1.69] | 0.94 [0.54, 1.63] |
| Cutting and deboning | Reference | Reference | Reference | Reference |
| Potential covariates and confounders | ||||
| Gender (males versusbfemales) | 1.23 [0.83, 1.82] | 1.09 [0.75, 1.58] | 0.60 [0.32, 1.14] | 0.61 [0.34, 1.10] |
| Smoking (current versusbnon-smoker) | 0.59 [0.45, 0.78]a | 0.61 [0.47, 0.80]a | 0.71 [0.45, 1.12] | 0.59 [0.38, 0.89]a |
| Age | 1.02 [1.01, 1.03]a | 1.004 [0.99, 1.02] | 1.01 [0.99, 1.03] | 1.00 [0.98, 1.02] |
| Antibiotic use (use versusbnon-use)c | 1.12 [0.71, 1.76] | 1.17 [0.76, 1.80] | 0.87 [0.41, 1.86] | 1.11 [0.56, 2.20] |
| Nationalities | ||||
| Dutch | reference | reference | reference | reference |
| Polish | 0.44 [0.28, 0.67]a | 0.51 [0.32, 0.81]a | 2.23 [1.15, 4.32]a | 2.31 [1.12, 4.73]a |
| Romanian | 0.43 [0.26, 0.71]a | 0.47 [0.27, 0.81]a | 0.19 [0.09, 0.41]a | 0.19 [0.08, 0.44]a |
| Other | 0.33 [0.21, 0.52]a | 0.41 [0.25, 0.66]a | 0.85 [0.42, 1.71] | 0.88 [0.41, 1.88] |
| Unknown | 0.35 [0.15, 0.85]a | 0.48 [0.20, 1.15] | 3.41 [0.83, 13.96] | 4.69 [1.14, 19.27]a |
| Sampling year (2016 versusb2015) | 1.81 [1.36, 2.41]a | 1.86 [1.32, 2.63]a | 1.87 [1.17, 3.01]a | 2.24 [1.32, 3.80]a |
Unadjusted and adjusted regression coefficients are expressed as geometric mean ratios (GMRs). Three models were fitted per gene (tetW, ermB) including either carcass (JEM), gloves (JEM), or job position. These models were then adjusted for all potential covariates or confounders (gender, smoking, age, antibiotic use, nationality, sampling year). Adjusted estimates and CIs from the latter variables are shown from the model including gloves (JEM). No major differences were observed between these estimates and estimates from models including carcass (JEM) or job position.
a P values < 0.05. All numbers are rounded at two figures or the first integer behind the comma unless rounding would result in the misinterpretation of a non-significant effect.
bThis category was taken as the reference category in the regression analysis.
cAntibiotic use was recorded as (potential) antibiotic use in the past year before sampling. A sensitivity analysis showed no major differences between the inclusion or exclusion of potential antibiotic users. For each model (tetW/ermB), three outliers were removed based on the outcome of model diagnostics. A sensitivity analysis showed no major differences between the inclusion or exclusion of outliers.
In the univariate analysis, higher tetW concentrations were found within personnel working in the black (GMR = 1.91, P = 0.003) and clean area (GMR = 1.41, P = 0.03) compared with staff working within the cutting and deboning rooms. When the black, clean, and cutting-deboning areas were subdivided, only employees working at lairage and bleeding showed significantly different tetW concentrations (GMR = 1.94, P = 0.03) from staff working at deboning. Smoking was associated with lower tetW abundance (current versus non-smokers, GMR = 0.59, P = 0.0002), while age was positively associated. Additionally, faecal tetW abundances were significantly higher in Dutch staff compared with staff having other nationalities. After adjusting the models including either carcasses, gloves, or job position (for gender, age, smoking, antimicrobial use, nationality, and sampling round), only smoking, nationality and sampling round remained as determinants for tetW carriage (Table 2). Finally, a sensitivity analysis including the 16S-normalized human tetW outcome data, did not change our main conclusions (data not shown).
Determinants for ermB carriage in human stool
No significant association was found between the percentage of ermB positive carcasses or gloves and ermB prevalence in workers’ stool (Table 2). Compared with Dutch staff, Polish staff carried higher and Romanian staff lower ermB abundances in their stool. Smoking had a negative effect (GMR = 0.59, P = 0.01) and sampling round was positively associated with ermB concentrations in stool (GMR = 2.24, P = 0.003). Similar observations were found in a sensitivity analysis with the 16S-normalized data, with the following differences in the adjusted model: females carried significantly higher ermB concentrations in their stool than males, while the effect of smoking was not significant (data not shown).
Discussion
Our findings indicate occupational exposure of slaughterhouse employees to tetW and to a lesser extent to ermB through dermal and inhalation exposure. Carcass and gloves contamination were analysed as a proxy of dermal exposure, potentially resulting in oral uptake via hand-mouth contact. We also found univariate associations between tetW in the slaughterhouse environment and human faecal tetW, which were attenuated in adjusted models. No such associations were found for faecal ermB abundance.
Our results show clear resistance gene gradients in carcasses along the slaughter line. TetW and ermB are mostly detected on carcasses after bleeding and partly after scalding (tetW > ermB) with only small numbers found after evisceration and pluck removal (tetW) and within the cutting and deboning area (tetW > ermB). Consistent with these findings, previous research in pigs has shown that scalding (apart from dehairing) and singeing consecutively reduce the number of resistant bacteria on carcasses (Hald et al., 2003; Wu et al., 2009; Vossenkuhl et al., 2014), while higher bacterial concentrations may be found after evisceration or pluck removal (Hald et al., 2003; Pearce et al., 2004). The increase in detection and abundance of tetW and ermB in carcasses after cooling suggests regrowth of resistant bacteria or increased cross-contamination from workers handling meat at the cutting and deboning rooms. Prior studies have reported conflicting effects of chilling (both increases and reductions) on bacterial contamination depending on the pathogen, or the chilling and detection method used (Pearce et al., 2004; Spescha et al., 2006; Vanantwerpen et al., 2016). Wu et al. (2009) showed a decreasing trend of culturable tetracycline resistant bacteria along the line, yet, in contrast to our study, respectively incalculable or no tetracycline resistant bacteria were found after cooling and cutting, probably related to the higher sensitivity of qPCR, which also detects non-viable bacteria. Rapidly decreasing ARG concentrations on carcasses in the slaughter line and nearly inexistent ARG levels in packed pork chops in our study are encouraging findings with respect to exposure to tetW and ermB resistance genes of abattoir staff working in a large slaughterhouse, but also of consumers of pork chops derived from Dutch reared and slaughtered pigs. Since 2007, Dutch antimicrobial sales in general and AMU in Dutch pigs have decreased drastically, possibly reducing the resistance levels entering the slaughterhouse and/or potential carcass contamination with resistant bacteria further down the line. However, ARG concentration reduction through scalding and singeing in the current study is likely to be much higher than resistance reduction previously seen in indicator bacteria isolated from Dutch pigs (Veldman and Mevius, 2018).
We also found decreasing tetW and ermB levels along the slaughter line in gloves and air samples. Similar gradients have previously been reported in an MRSA slaughterhouse study applying culture and qPCR (mecA, ST398, SCCmec) (Gilbert et al., 2012). Interestingly, despite no or few tetW and ermB positive carcasses after singeing, we found high percentages of positive gloves and air samples after singeing, including high numbers of samples >LOQ, especially on gloves. One way to explain this difference between gloves and carcasses is to consider tetW and ermB abundances on gloves a reflection of resistance found in the worker’s environment (e.g. contact with cutting boards, conveyor belts, scaffolds) including meat. Additionally, we could hypothesize that relatively more tetW and ermB positive gloves compared with carcasses could be caused by frequent hand-meat contact which might result in bacterial, and consequently ARG accumulation on gloves.
Human tetW and ermB concentrations are lower than in pigs and vary along the slaughter line, yet lack a decreasing gradient similar to the environmental samples. Univariate analysis revealed positive associations between the percentage of tetW positive carcasses or gloves and human tetW carriage, as well as a positive association between the percentage of tetW positive gloves and faecal tetW abundance after adjusting for gender, age, AMU, smoking, and nationality, but not after adjusting for sampling round. Also, no association with job position remained significant in the adjusted models. This contrasts with findings from pathogen-based studies where exposure to ESBL-PE and MRSA at certain slaughter positions (mainly live animal contact) was found to be a major determinant for, respectively, faecal ESBL and nasal MRSA carriage, although similar model adjustments as in our study were or could not always be applied (van Cleef et al., 2010; Mulders et al., 2010; Gilbert et al., 2012; Dohmen et al., 2017b).
Microbial composition is interrelated with the resistome (Pehrsson et al., 2016; Munk et al., 2018). Additionally, tetracycline and macrolide resistance genes (including tetW and ermB) are known to dominate the gastrointestinal resistome of healthy individuals (Pal et al., 2016). Prior research also identified inter-individual variation in relative resistance abundances or antibiotic resistance potentials in the resistome with respect to tetracycline and macrolide resistance gene classes, often dominated by significant country resistome differences (Forslund et al., 2013, 2014; Hu et al., 2013). These findings make it less likely to pick up additional differences resulting from AMR exposure along the slaughter line, especially in largely heterogeneous slaughterhouse populations potentially also characterized by different diets or other habits (e.g. diet can quickly influence the microbiome) (David et al., 2014). This is also reflected within the significant effects of nationality in our models. Part of the slaughterhouse population is originating from Eastern Europe and working temporarily in the Netherlands. Geographic differences in AMR prevalence have been previously established between Northern and Eastern-European countries, mostly suggesting a higher AMR prevalence in Eastern-European countries (European Centre for Disease Prevention and Control, 2017). Interestingly, Dutch nationality is associated with higher tetW concentrations compared to other staff, while Polish and Romanian staff, respectively, carry higher and lower ermB concentrations. An effect of international travelling (in our case interpreted as long-distance commuting) was previously described in AMR studies (van der Bij and Pitout, 2012), but its potential relationship with nationality and/or ARG carriage could not reliably be tested in our study due to too many missings. Finally, it must be noted that potential differences in the length of exposure to ARGs between employees (misclassification of long-term exposure due to temporary work) or short-term exposure in general, could also have contributed to the absence of a direct association between environmental exposure and human ARG carriage.
Surprisingly, although the same protocols were applied, higher tetW and ermB human faecal yields were found in samples from 2016 compared with 2015, even after 16S-normalization, less clearly identified within the environmental samples (data not shown). This potentially could be explained by study cohort differences, differences with regard to the temperature during sampling (resulting in different bacterial loads entering the slaughterhouse) or potential unidentified technical differences. A sensitivity analysis excluding data from 2016 confirmed the main significant associations detected in the adjusted models.
Smoking is reported to affect the gut microbiome (Lee et al., 2018). A negative effect of smoking on faecal tetracycline/macrolide resistance carriage is to the best of our knowledge not yet described before, although negative non-significant associations were reported in previous AMR slaughterhouse studies (including the ESBL-PE study using data from 2015) (Gilbert et al., 2012; Dohmen et al., 2017b).
Limitations of the study
We investigated exposure to and carriage of ARGs as a proxy for tetracycline and macrolide resistance, hence did not measure resistance expression in viable (pathogenic) bacteria, nor exposure to antimicrobial residues in pig faeces (Berendsen et al., 2015), and potentially on carcasses. Additionally, our study had a cross-sectional design and took place in only one, however large, slaughterhouse and was based on convenience sampling. Due to low ARG numbers (including samples <LOQ) on carcasses and gloves, we transformed these variables to the percentage of ARG detects. This reduces contrast and statistical power compared to the usage of average exposure per job area. Applying a JEM might also lead to less statistical power and misclassification, although generally, group-average exposure levels result in little bias of risk estimates (Armstrong, 1998). Other possible bias might have resulted from recalling e.g. past AMU or only sampling active staff (‘healthy worker effect’). Finally, we were not able to adjust our model for more covariates (e.g. animal contact, meat consumption) due to missing data.
Conclusion
Our study shows occupational dermal and respiratory exposure of slaughterhouse workers to tetracycline (tetW) and macrolide resistance (ermB) along the slaughter line (tetW > ermB). Exposure to tetW or ermB resistance through packed pork chops from Dutch reared and slaughtered pigs is low. Positive associations were found between tetW in the work environment and human faecal tetW carriage, yet, could not be upheld in adjusted models. No such associations were found for faecal ermB. Sampling round, nationality, and smoking are determinants for tetW and ermB carriage in slaughterhouse workers.
Supplementary Material
Acknowledgements
The authors would like to thank all Vion employees who participated in this study. We also specifically would like to thank all field workers, lab technicians, and students at the Institute for Risk Assessment Sciences, NL (Siegfried de Wind, Isabella van Schothorst, Erik van Deurssen, Gertie Bokken, Janieke van Veldhuizen, Christiaan van Dorsten, Siham Chaïbi, Jon Timmer, Daphne Beemsterboer, Zoë Morris, Remco Bergman), the Vion Food Group, NL (Eric Houdijk, Nina Wingens, Pim Sanders), the National Veterinary Research Institute (PIWet), PL (Ilona Samcik, Renata Kwit, Aleksandra Śmiałowska, Ewelina Kamińska), and the German Federal Institute for Risk Assessment (BfR), DE (Katharina Juraschek, Sead Hadziabdic) for their tremendous time and effort making this sampling and lab campaign a success. Lastly, the authors would like to thank Dik Mevius and Lützen Portengen for their advice and comments when preparing the article. Vion Food Group was not involved in the statistical analysis, the interpretation of the data, nor the decision to publish.
Funding
This work was part of the Ecology from Farm to Fork Of Microbial drug Resistance and Transmission (EFFORT) project (http://www.effort-against-amr.eu), co-funded by the European Commission, 7th Framework Programme for Research and Innovation (FP7-KBBE-2013–7, grant agreement: 613754). Research at the National Veterinary Research Institute (PIWet), Poland, was also supported by the donation of the Polish Ministry of Science: no. 3173/7PR/2014/2.
Data statement
Part of the human population included in this study (2015 cohort) is largely overlapping with the study population used in Dohmen et al. (2017b) in a study involving ESBL-PE testing.
References
- Armstrong BG. (1998) Effect of measurement error on epidemiological studies of environmental and occupational exposures. Occup Environ Med; 55: 651–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berendsen BJ, Wegh RS, Memelink Jet al. (2015) The analysis of animal faeces as a tool to monitor antibiotic usage. Talanta; 132: 258–68. [DOI] [PubMed] [Google Scholar]
- van der Bij AK, Pitout JD. (2012) The role of international travel in the worldwide spread of multiresistant Enterobacteriaceae. J Antimicrob Chemother; 67: 2090–100. [DOI] [PubMed] [Google Scholar]
- Bisdorff B, Scholhölter JL, Claußen Ket al. (2012) MRSA-ST398 in livestock farmers and neighbouring residents in a rural area in Germany. Epidemiol Infect; 140: 1800–8. [DOI] [PubMed] [Google Scholar]
- van Cleef BA, Broens EM, Voss Aet al. (2010) High prevalence of nasal MRSA carriage in slaughterhouse workers in contact with live pigs in The Netherlands. Epidemiol Infect; 138: 756–63. [DOI] [PubMed] [Google Scholar]
- David LA, Maurice CF, Carmody RNet al. (2014) Diet rapidly and reproducibly alters the human gut microbiome. Nature; 505: 559–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohmen W, Schmitt H, Bonten Met al. (2017a) Air exposure as a possible route for ESBL in pig farmers. Environ Res; 155: 359–64. [DOI] [PubMed] [Google Scholar]
- Dohmen W, Van Gompel L, Schmitt Het al. (2017b) ESBL carriage in pig slaughterhouse workers is associated with occupational exposure. Epidemiol Infect.; 145: 2003–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorado-García A, Bos ME, Graveland Het al. (2013) Risk factors for persistence of livestock-associated MRSA and environmental exposure in veal calf farmers and their family members: an observational longitudinal study. BMJ Open; 3: e003272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- European Centre for Disease Prevention and Control . (2017) Antimicrobial resistance surveillance in Europe 2015. Stockholm, Sweden: ECDC. pp. 1–120. [Google Scholar]
- Fierer N, Jackson JA, Vilgalys Ret al. (2005) Assessment of soil microbial community structure by use of taxon-specific quantitative PCR assays. Appl Environ Microbiol; 71: 4117–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forslund K, Sunagawa S, Coelho LPet al. (2014) Metagenomic insights into the human gut resistome and the forces that shape it. Bioessays; 36: 316–29. [DOI] [PubMed] [Google Scholar]
- Forslund K, Sunagawa S, Kultima JRet al. (2013) Country-specific antibiotic use practices impact the human gut resistome. Genome Res; 23: 1163–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Graells C, van Cleef BA, Larsen Jet al. (2013) Dynamic of livestock-associated methicillin-resistant Staphylococcus aureus CC398 in pig farm households: a pilot study. PLoS One; 8: e65512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geenen PL, Graat EA, Haenen Aet al. (2013) Prevalence of livestock-associated MRSA on Dutch broiler farms and in people living and/or working on these farms. Epidemiol Infect; 141: 1099–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert MJ, Bos ME, Duim Bet al. (2012) Livestock-associated MRSA ST398 carriage in pig slaughterhouse workers related to quantitative environmental exposure. Occup Environ Med; 69: 472–8. [DOI] [PubMed] [Google Scholar]
- Hald T, Wingstrand A, Swanenburg Met al. (2003) The occurrence and epidemiology of Salmonella in European pig slaughterhouses. Epidemiol Infect; 131: 1187–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes AH, Moore LS, Sundsfjord Aet al. (2016) Understanding the mechanisms and drivers of antimicrobial resistance. Lancet; 387: 176–87. [DOI] [PubMed] [Google Scholar]
- Hu Y, Yang X, Qin Jet al. (2013) Metagenome-wide analysis of antibiotic resistance genes in a large cohort of human gut microbiota. Nat Commun; 4: 2151. [DOI] [PubMed] [Google Scholar]
- Knudsen BE, Bergmark L, Munk Pet al. (2016) Impact of sample type and DNA isolation procedure on genomic inference of microbiome composition. mSystems. 1(5): e00095-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike S, Aminov RI, Yannarell ACet al. (2010) Molecular ecology of macrolide-lincosamide-streptogramin B methylases in waste lagoons and subsurface waters associated with swine production. Microb Ecol; 59: 487–98. [DOI] [PubMed] [Google Scholar]
- Lee SH, Yun Y, Kim SJet al. (2018) Association between cigarette smoking status and composition of gut microbiota: population-based cross-sectional study. J Clin Med; 7: 282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Moual N, Zock JP, Dumas Oet al. (2018) Update of an occupational asthma-specific job exposure matrix to assess exposure to 30 specific agents. Occup Environ Med; 75: 507–14. [DOI] [PubMed] [Google Scholar]
- Mulders MN, Haenen AP, Geenen PLet al. (2010) Prevalence of livestock-associated MRSA in broiler flocks and risk factors for slaughterhouse personnel in The Netherlands. Epidemiol Infect; 138: 743–55. [DOI] [PubMed] [Google Scholar]
- Munk P, Knudsen BE, Lukjacenko Oet al. (2018) Abundance and diversity of the faecal resistome in slaughter pigs and broilers in nine European countries. Nat Microbiol.; 3: 898–908. [DOI] [PubMed] [Google Scholar]
- Murphy CP, Carson C, Smith BAet al. (2018) Factors potentially linked with the occurrence of antimicrobial resistance in selected bacteria from cattle, chickens and pigs: a scoping review of publications for use in modelling of antimicrobial resistance (IAM.AMR Project). Zoonoses Public Health; 65: 957–71. [DOI] [PubMed] [Google Scholar]
- Pal C, Bengtsson-Palme J, Kristiansson Eet al. (2016) The structure and diversity of human, animal and environmental resistomes. Microbiome; 4: 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce RA, Bolton DJ, Sheridan JJet al. (2004) Studies to determine the critical control points in pork slaughter hazard analysis and critical control point systems. Int J Food Microbiol; 90: 331–9. [DOI] [PubMed] [Google Scholar]
- Pehrsson EC, Tsukayama P, Patel Set al. (2016) Interconnected microbiomes and resistomes in low-income human habitats. Nature; 533: 212–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . (2018) R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. Available at https://www.r-project.org/. Accessed 12 December 2019. [Google Scholar]
- Spescha C, Stephan R, Zweifel C. (2006) Microbiological contamination of pig carcasses at different stages of slaughter in two European Union-approved abattoirs. J Food Prot; 69: 2568–75. [DOI] [PubMed] [Google Scholar]
- Vanantwerpen G, De Zutter L, Berkvens Det al. (2016) Impact of the sampling method and chilling on the Salmonella recovery from pig carcasses. Int J Food Microbiol; 232: 22–5. [DOI] [PubMed] [Google Scholar]
- Van Gompel L, Luiken REC, Sarrazin Set al. ; EFFORT Consortium. (2019) The antimicrobial resistome in relation to antimicrobial use and biosecurity in pig farming, a metagenome-wide association study in nine European countries. J Antimicrob Chemother; 74: 865–76. [DOI] [PubMed] [Google Scholar]
- Veldman KT, Mevius DJ (2018) Monitoring of antimicrobial resistance and antibiotic usage in animals in the Netherlands in 2017. Lelystad: Wageningen Bioveterinary Research. Available at: https://www.wur.nl/upload_mm/7/b/0/5e568649-c674-420e-a2ca-acc8ca56f016_Maran2018.pdf. Accessed 12 December 2019. [Google Scholar]
- Vossenkuhl B, Sharp H, Brandt Jet al. (2014) Modeling the transmission of livestock associated methicillin-resistant Staphylococcus aureus along the pig slaughter line. Food Control. 39: 17–24. [Google Scholar]
- Walsh F, Ingenfeld A, Zampicolli Met al. (2011) Real-time PCR methods for quantitative monitoring of streptomycin and tetracycline resistance genes in agricultural ecosystems. J Microbiol Methods; 86: 150–5. [DOI] [PubMed] [Google Scholar]
- World Health Organization . (2014) Antimicrobial resistance. Global report on surveillance. Geneva, Switzerland: World Health Organization. Available at: http://apps.who.int/iris/bitstream/10665/112642/1/9789241564748_eng.pdf. Accessed 12 December 2019. [Google Scholar]
- Wu S, Dalsgaard A, Vieira ARet al. (2009) Prevalence of tetracycline resistance and genotypic analysis of populations of Escherichia coli from animals, carcasses and cuts processed at a pig slaughterhouse. Int J Food Microbiol; 135: 254–9. [DOI] [PubMed] [Google Scholar]
- Wulf MWH, Sørum van Nes M, . et al. (2008) Prevalence of methicillin-resistant Staphylococcus aureus among veterinarians: an international study. Clin Microbiol Infect; 14: 29–34. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.