Highlights
-
•
Irradiation-induced USP7 leads to radioresistance in p53-mutated LSCC cells.
-
•
However, USP7 leads to radio-sensitivity in p53-wild type cells.
-
•
Targeting USP7 might improve radiotherapy's effects in LSCC and reduce side effects.
Keywords: Laryngeal, Radiotherapy, Head and neck cancer, Cancer genetic
Abstract
The relationship between ubiquitin specific protease 7 (USP7) and radio-sensitivity in laryngeal squamous cell carcinoma (LSCC) has not been reported yet. Using gene chip and Label-Free mass spectrometry, we found that USP7 was significantly increased both in radioresistant LSCC patients and LSCC cells receiving irradiation. Since p53 is the most important downstream gene of USP7 and is frequently mutated in LSCC, we investigated the effects of USP7 on radioresistance of LSCC cells with or without p53 mutation. We found that knockdown of USP7 increased the radio-sensitivity in p53-mutated LSCC cells, while inhibiting the radio-sensitivity in p53-wild type cells. Knockdown of USP7 significantly inhibited the expression of the p53 and p53 pathway. Overexpressing endogenous p53 by CRISPR/dCas9 could reverse the effects of USP7 on radiosensitivity both in vitro and in vivo. Our results demonstrated the irradiation-induced USP7 led to radioresistance in p53-mutated LSCC cells but radio-sensitivity in p53-wild type cells. Therefore, the clinical application of USP7 inhibitors may improve the effects of radiotherapy in LSCC with p53 mutations and reduce the side effects on surrounding normal tissues without p53 mutation.
Introduction
Laryngeal cancer is a high-incidence tumor of the head and neck, in which laryngeal squamous cell carcinoma (LSCC) is the most common pathological type [1]. Early LSCC can be treated with radical radiotherapy or surgery, while for advanced LSCC, a comprehensive treatment including radiation, surgery, and chemotherapy should be adopted, as well as multidisciplinary management of toxicities and follow-up strategies [2], [3], [4]. Therefore, improving the radiation effect is quite important for the treatment of LSCC [5].
Though radiation technology has improved greatly in recent years, the survival effect of LSCC is still not ideal due to the radioresistance. The current study finds that 6–15% and 20–31% of stage I\II LACC patients show radiation resistance. And the incidence of radioresistance in the advanced stage LSCC is even higher [6,7]. For these patients, the radiation dose may need to be increased along with the higher risk of damaging important surrounding organs, therefore, more toxic effects such as dysphagia [8].
The relationship between ubiquitin specific protease 7 (USP7) and radiation sensitivity in LSCC has not been reported yet. In this study, we found that USP7 regulated the radio-sensitivity of LSCC via the ubiquitin and degradation of p53. Our results showed that depending on the p53 mutation status, USP7 caused radioresistance of p53-mutated LSCC cells, while it promoted radio-sensitivity of p53-wild type LSCC cells. Since most LSCC samples harbor p53 mutations while normal tissues don't, agents inhibiting USP7 have the potential effects to improve the radiation sensitivity of LSCC and reduce radiation damage of surrounding normal tissues.
Methods
Specimens in clinic
The study was approved by the Ethics Committees of Eye and ENT Hospital, Fudan University (2020057–1), and performed in accordance with the principles of the Declaration of Helsinki. Informed consent from each patient or participant has been obtained.
We selected ten tumor tissue specimens from patients with stage III\IV of LSCC who received radiotherapy due to postoperative high-risk factors through retrospective analysis. The follow-up gene chip for high-throughput detection was performed by GMINIX Co (Shanghai, China). Among them, 5 cases were in the radiotherapy-resistance group, and the other 5 cases were in the radiotherapy-sensitive group. To reduce the heterogeneity between the two groups, the two groups of patients were matched one-to-one in terms of sex, age, tumor stage, primary tumor location, etc. To rule out the influence of chemotherapy on the judgment of the treatment effect, no patient included in this research received chemotherapy in the treatment process.
Cell culture
LSCC cell lines TU-212 (p53_P151S mutation), SNU-585 (p53_R273L mutation), SNU-899 (wild type p53), and SNU-1076 (wild type p53) were obtained from the Shanghai Academy of Life Sciences (Shanghai, China). Cells were cultured in RPMI 1640 medium containing 10% fetal bovine serum (FBS), 100 units/ml penicillin, and 100 μg/ml streptomycin antibiotics at 37 °C in an incubator of 5% CO2.
Cell transfection
To knockdown USP7, two different short hairpin RNAs (shRNAs) targeting USP7 and corresponding negative control were cloned into pLVX-IRES-Puro vectors by GeneChem Co., Ltd. (Shanghai, China). The CRISPR/Cas9-SAM system was used to overexpress endogenous p53 and constructed by GeneChem, too.
RNA extraction, RNA-Seq, and reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
All RNA from the four cell lines mentioned above was isolated using TRIzol (Thermo Fischer Scientific) reagent. RNA-Sequencing (RNA-Seq) of control and USP-knockdown TU-212 cells was performed by Oebiotech Co. (Shanghai, China). Gene expression was normalized to Fragments Per Kilobase of exon model per Million mapped fragments (FPKM) for subsequent analyses. Differentially expressed genes were identified using the parameters |logFC| > 0.5 and p < 0.05. Genes differentially expressed both in TU-212 SH1 and SH2 cells compared with control cells were further analyzed in the Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis as previously reported [9].
The mRNA levels were also detected by real-time PCR using the 7500 Real-time PCR System (Thermo Fischer). Relative fold changes in mRNA expression were calculated using the formula 2−ΔΔCq and β-actin as control. The primers were as follows: USP7 forward, 5′-GGAAGCGGGAGATACAGATGA-3′, USP7 reverse, 5′-AAGGACCGACTCACTCAGTCT-3′, p53 forward, 5′- CAGCACATGACGGAGGTTGT-3′, p53 reverse, 5′- TCATCCAAATACTCCACACGC-3′, β-actin forward, 5′-TGACGTGGACATCCGCAAAG-3′, β-actin reverse, 5′-CTGGAAGGTGGACAGCGAGG-3′.
Western blot
Cells were harvested at indicated time and lysed by using RIPA lysis buffer on ice. A total of 20 μg protein was loaded on 7%−15% sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The proteins were transferred to polyvinylidene difluoride membranes and incubated with the primary antibodies rabbit anti-USP7 (#4833, Cell Signaling Technology, CST, 1:1000), rabbit anti-p53 (#2527, CST, 1:1000), mouse anti-β-actin (#AA128, Beyotime Biotech., 1:3000) at 4 °C overnight. Then membranes were washed and incubated with corresponding secondary antibodies (Beyotime). Finally, the protein was visualized using an enhanced chemiluminescence detection kit (Beyotime). The density of the bands was analyzed, and the relative ratios of target genes and references were calculated.
Label-free liquid chromatography‑mass spectrometry (LC‑MS) analysis
Ten Gy radiation was used to irradiate the laryngeal squamous cell line TU-212, and total protein was extracted at 0, 2, 6, 12, 24, and 48 h after irradiation. The Label-Free high-throughput mass spectrometry method was performed by BANGFEI BIOSCIENCE Co. to detect the changes in the expression of the whole protein profile of the cells after irradiation as previously published [10].
Co-Immunoprecipitation (Co-IP)
Co-IP was performed using the Pierce Co-Immunoprecipitation (Co-IP) Kit (Thermo Fisher). Briefly, USP7 antibody (#4833, CST, 1:50) and Rabbit (DA1E) mAb IgG XP® Isotype Control (#3900, CST, 1:50) were cross-linked to protein A/G beads and then incubated with cell lysate. After thoroughly washed, proteins were eluted from the beads and then analyzed using western blots with rabbit anti-USP7 (#4833, CST, 1:1000), mouse anti-p53 (1C12) (#2524, CST, 1:1000), and corresponding secondary antibodies (Beyotime).
Cell colony formation assay
A total of 800 cells was seeded into the 6 cm plate with RPMI-1640 medium. Irradiation of 0 Gy, 2 Gy, 4 Gy, 6 Gy, 8 Gy, 10 Gy X-Ray was delivered 24 h later using an ONCOR™ linear accelerator. Cells were continued incubated for 14 days. Colonies were fixed with 4% methanol, stained with 1% crystal violet. The number of colonies with a diameter of ≥ 0.2 mm was analyzed using the surviving fractions [11].
Flow cytometric analysis
A total of 5 × 104 cells were harvested at 24 h after treatment with irradiation. Cell cycle distributions were examined. Briefly, The cells were fixed with 72% ethanol. The samples were left in the refrigerator for 2–5 min and centrifuged again at the same speed for 5 min. The cells were stained with 50 μg/mL propidium iodide for 15 min at room temperature, and then the specimens were run on a flow cytometer (BD Bioscience, USA). Cells were then analyzed by a FlowJo (v10.6.2).
Tumor formation in nude mice
Male BALB/c nude mice (5 weeks) were subcutaneously injected with 1 × 106 TU-212 and SNU-899 cells, respectively. After 7 days, the nude mice were irradiated with 6 Gy X-rays three times with an interval of 3 days. The mice were sacrificed 28 days after irradiation. The tumors were removed, and their weights were measured. Then, the tumors were fixed with formalin and embedded with paraffin for immunohistochemistry analysis.
Statistical analysis
GraphPad Prism 8 was used for plotting graphs and analyzing. Two tails of the Student T-test, one-way ANOVA, and Kolmogorov-Smirnov test were used to analyze statistical differences. P<0.05 was considered to indicate a statistical difference if not specified. The data were presented as mean ± standard error of the mean (SEM).
Results
The expression of USP7 in LSCC cells increased significantly after irradiation
The high-throughput detection of the gene chip revealed 150 differentially expressed coding genes and 82 differentially expressed non-coding genes between radiotherapy-sensitive and radiotherapy-resistant LSCC samples (p < 0.01). It is worth noting that USP7 was significantly up-regulated in the radiation-resistant group than in the sensitive group (Table 1).
Table 1.
Differences in gene expression of patients with laryngeal squamous carcinoma that are sensitive to and resistant to radiation.
| Gene name | p value* | RadiotherapySensitive group | RadiotherapyResistance group | multiple(Resistance/Sensitivity) | up/down |
|---|---|---|---|---|---|
| TMEM107 | 0.0006 | 0.32 | 5.95 | 18.6 | up |
| MYH2 | 0.0011 | 0.05 | 0.55 | 11.0 | up |
| SMAP2 | 0.0067 | 0.08 | 0.73 | 9.13 | up |
| BTG2 | 0.0043 | 0.51 | 4.59 | 9.07 | up |
| JAK3 | 0.0095 | 0.09 | 0.74 | 8.22 | up |
| USP7 | 0.0036 | 1.06 | 5.59 | 5.27 | up |
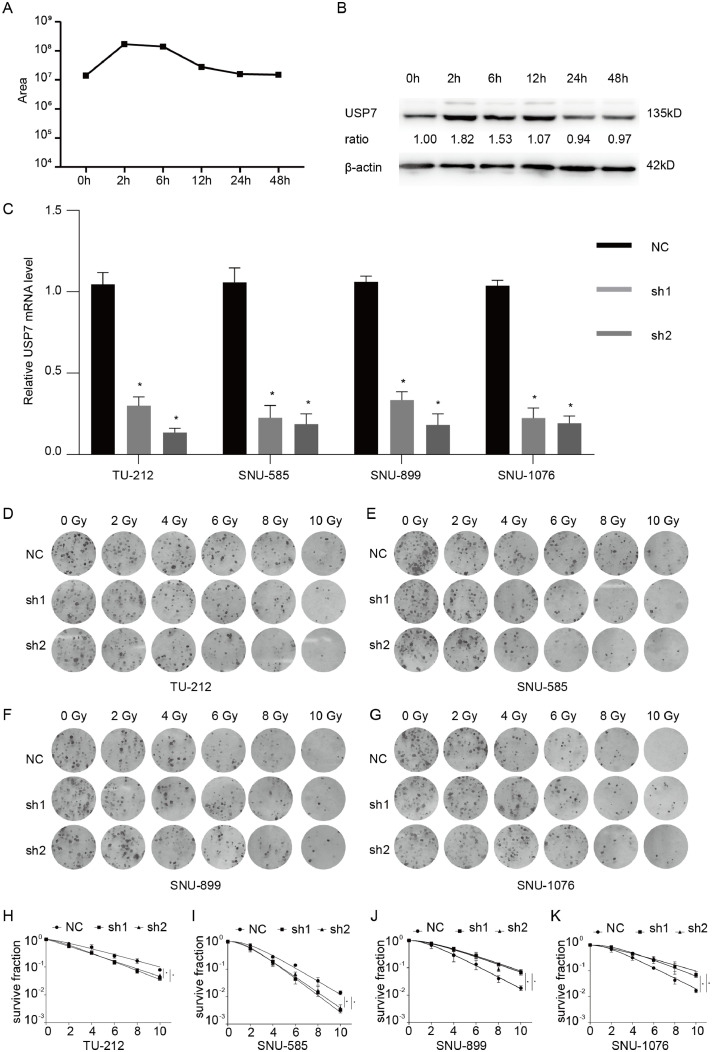
To further explore the mechanism related to the effect of radiotherapy for LSCC, we used 10 Gy radiation to irradiate the LSCC cell line TU-212, and extract total protein at 0, 2, 6, 12, 24, and 48 h after irradiation. The Label-Free high-throughput mass spectrometry method was used to detect the changes in the expression of the whole protein profile of the cells after irradiation. By comparing the results of mass spectrometry and gene chip, we found that USP7 was 21.2 times higher than the control group after 2 h of radiation exposure, and maintained at a higher level at 2–6 h. The expression decreased after 12 h. The western blot results are consistent with the above results (Fig. 1A, B).
Fig. 1.
(A, B) USP7 expression of TU-212 cells at different time points after irradiation detected by LC-MS and western blot; (C) Knockdown effects of USP7 by siRNAs in LSCC cells; (D, E) Clone formation assays and single target multi-shot model curve fittings of USP7-knockdown LSCC cells with or without p53 mutations. Data are presented as mean ± SEM. *P < 0.05, **P < 0.01, and ***P < 0.001.
The alteration of radiosensitivity of p53-mutated or p53-wild type LSCC cell line were different after knockdown of USP7
To explore the role of USP7 in LSCC cells, we designed two different shRNAs targeting USP7, and transfected them into p53-mutated or p53-wild type LSCC cell lines. The results of qRT-PCR indicated that the expression of USP7 was significantly decreased in both p53-mutated and p53-wild type LSCC cell lines by USP7-shRNAs compared with USP7-NC (Fig. 1C).
However, we found that the effect of silencing of USP7 was different on the radiosensitivity of the p53-mutated and p53-wild type LSCC cell lines(Fig. 1D, E). The colony formation assay results demonstrated that knockdown of USP7 increased the radio-sensitivity compared with control in p53-mutated LSCC cell lines (TU-212, SNU-585). But knockdown of USP7 reduced the radio-sensitivity in p53-wild type LSCC cell lines (SNU-899, SNU-1076).
Knockdown of USP7 significantly changed the p53 pathway
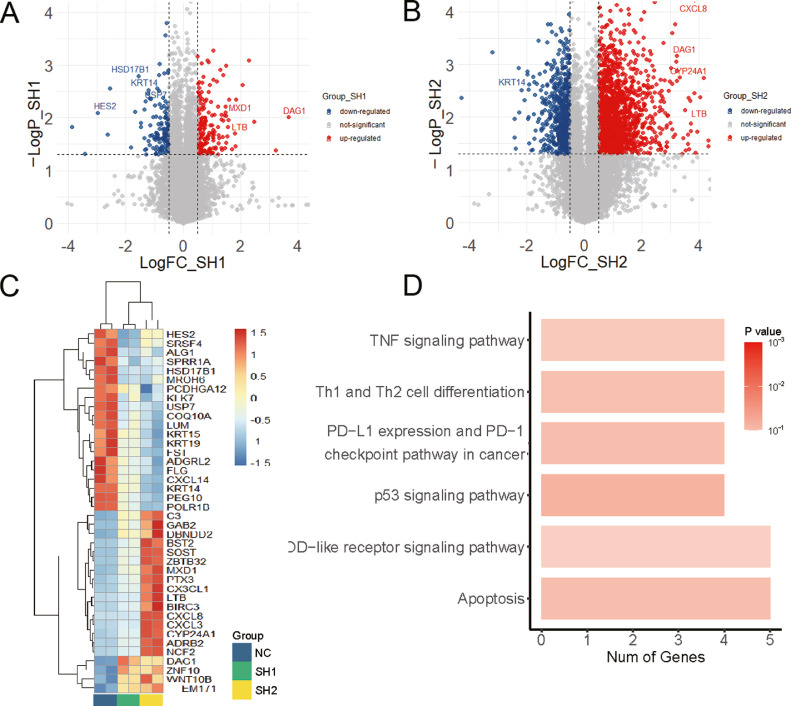
To future explore the role of USP7 on the radiosensitivity of p53-mutated or p53-wild type LSCC cell line, we performed RNA_Seq in control and USP7-knockdown TU-212 cells, and identified 80 genes (55 up-regulated and 25 downregulated) differentially expressed both in USP7-SH1 and USP7-SH2 cells compared with control TU-212 cells (Fig. 2A, B). The heatmap showed the top 40 genes with significant changes after USP7 knockdown, including USP7 itself (Fig. 2C), respectively. The differentially expressed genes both in USP7-SH1 and USP7-SH2 cells were analyzed using KEGG pathway, which indicated that p53 pathways ranked in the top 4 mapped pathways (Fig. 2D).
Fig. 2.
The volcano map (A, B), heatmap (C), and the KEGG analysis (D) of differentially expressed genes in TU-212 cells with USP7 knockdown.
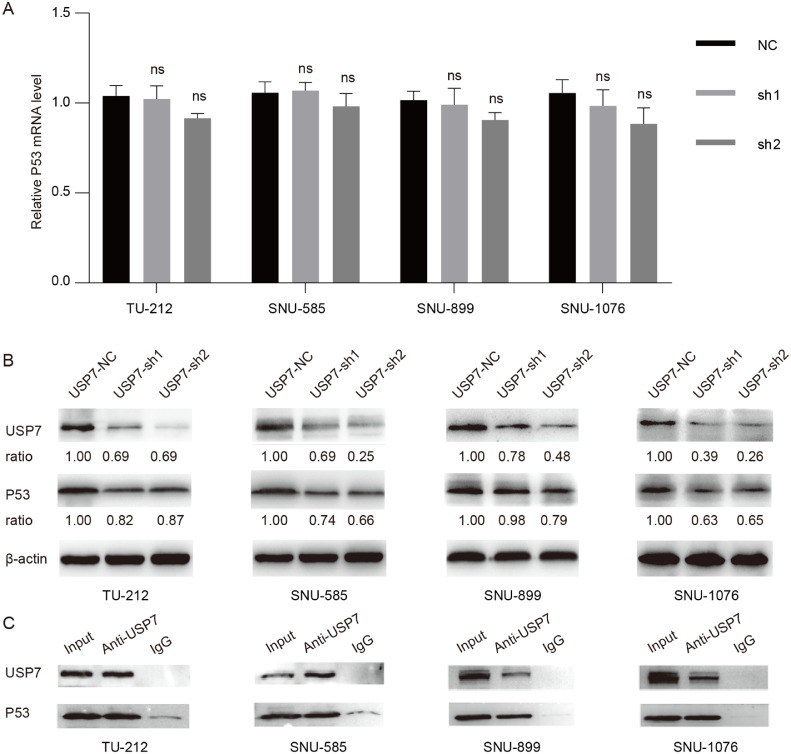
The results of qRT-PCR showed that the mRNA of p53 was not significantly changed after the knockdown of USP7 (Fig. 3A). However, the protein was significantly decreased after the knockdown of USP7 (Fig. 3B). Meanwhile, our co-IP assays showed that USP7 could directly bind to p53 in each LSCC cell line with or without p53 mutation (Fig. 3C). It was known that USP7 regulates the localization and stability of P53 by binding, deubiquitinating, and stabilizing p53 protein [12]. Our results also suggested that USP7 regulated the expression of p53 at the protein level, not the p53 mRNA.
Fig. 3.
The mRNA expression (A) and the protein expression of p53 (B) in LSCC cells with USP7 knockdown; (C) Co-IP results of USP7 and p53 using anti-USP7 antibody.
p53 mediated the regulation of USP7 on the radiosensitivity of LSCC cell line
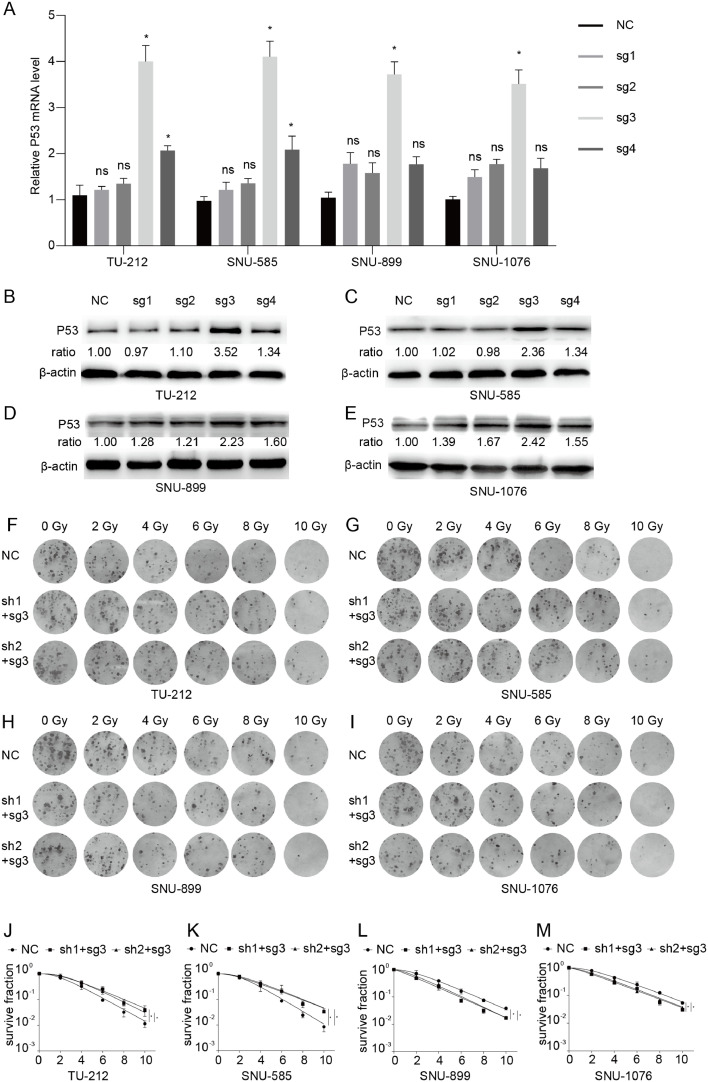
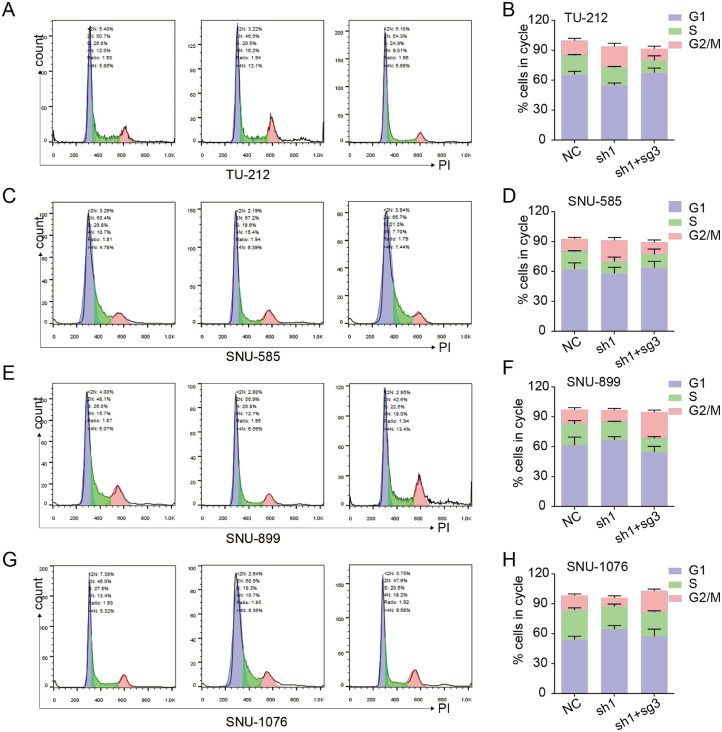
We overexpressed endogenous p53 in USP7 knockdown cells using CRISPR-Cas9 system. Among the four sgRNAs _USP7, sg3_ USP7 most significantly increased the expression of p53 both in p53-mutated and p53-wild type LSCC cell lines in the results of qRT-PCR and western blot (Fig. 4A, B). The colony formation assay results demonstrated that overexpression of p53 reversed the effects of USP7 on p53-mutated or p53-wild type LSCC cell line (Fig. 4C, D). To explore how did p53 mediate the regulation of USP7 on the radiosensitivity of LSCC cell line, flow cytometric analysis was performed. The results suggested that knockdown of USP7 increases the ratio of G2/M phase in the cell cycle in p53-mutated LSCC cell lines (Fig. 5A-D). However, the ratio of G2/M phase was reduced by the knockdown of USP7 in the cell cycle in p53-wild type LSCC cell line (Fig. 5E-H). Overexpressed endogenous p53 significantly reversed the effects of USP7 on the ratio of G2/M phase in the cell cycle both in the p53-mutated or p53-wild type LSCC cell line (Fig. 5A-H).
Fig. 4.
(A, B) Overexpression of endogenous p53 using different sgRNAs by CRISPR/Cas9-SAM; (C, D) Clone formation assays and single target multi-shot model curve fittings in USP7-knockdown LSCC cells with p53 overexpression. Data are presented as mean ± SEM. *P < 0.05, **P < 0.01, and ***P < 0.001.
Fig. 5.
Cell Cycle analyses of different LSCC cells with USP7 knockdown and p53 overexpression. (A, B) TU-212 cells; (C, D) SNU-585 cells; (E, F) SNU-899 cells; (G, H) SNU-1076 cells.
Knockdown of USP7 change the radiosensitivity of LSCC cell lines in vivo
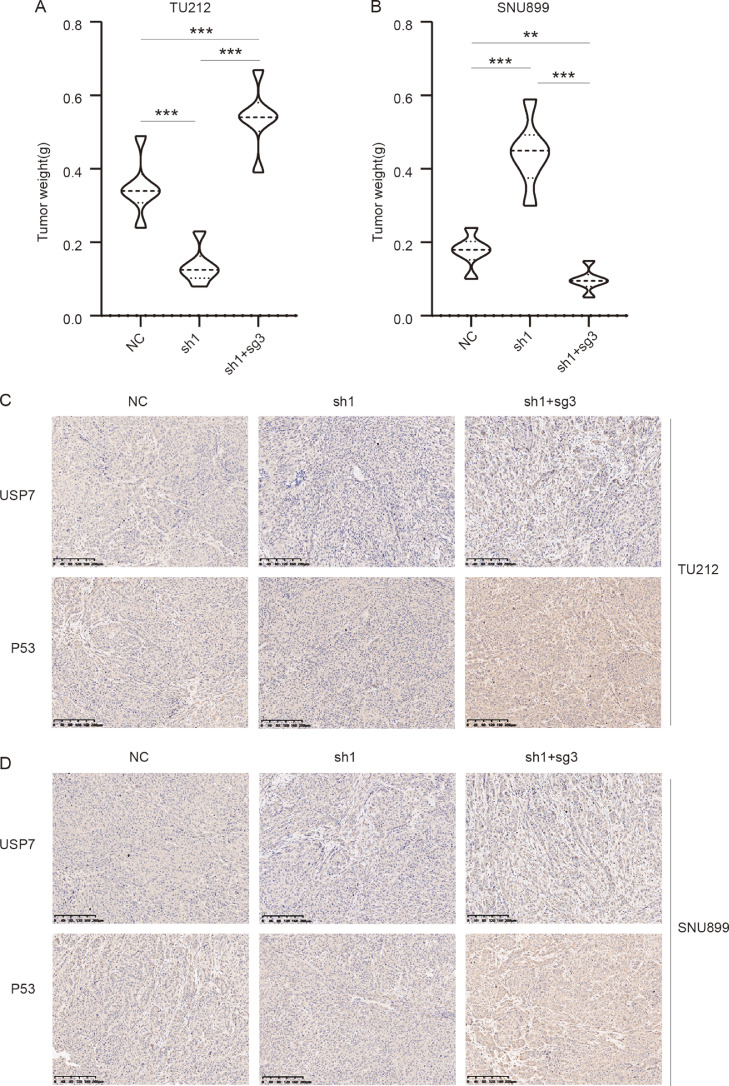
To verify the effect of USP7 on the radio-sensitivity in vivo, a subcutaneous tumor formation assay was performed. Knockdown of USP7 significantly increased the radio-sensitivity compared with control in p53-mutated TU-212 cell lines (Fig. 6A, C). But the radioresistance was significantly enhanced compared with control in p53-wild type SNU-899 cell lines. Overexpressed endogenous p53 significantly reversed the effects of USP7 on the radio-sensitivity both in the p53-mutated or p53-wild type LSCC cell line (Fig. 6B, D).
Fig. 6.
Tumor formation analyses in vivo. (A, B) Tumor weight of TU-212 and SNU-899 cells with USP7 knockdown and p53 overexpression; (C, D) Immunohistochemistry results of USP7 and p53 in TU-212 and SNU-899 cells with USP7 knockdown and p53 overexpression in vivo. Scale bar: 200 μm.
Discussion
LSCC is one of the most common tumors of head and neck malignant tumors. Radiotherapy is one of the main treatments for LSCC as well as surgery [13], [14], [15]. Improving the effects of radiotherapy, reducing the toxic effects, and preserving the organs' functions, such as voice rehabilitation, are quite important [16,17].
USP7 involve in a diverse array of cellular processes. Accumulating evidence indicates that USP7 plays critical role in cancers, neurological disorders, metabolic disorders, and immune dysfunction, making USP7 a potential target for cancer therapy [18], [19], [20], [21]. However, little is known about the relationship between USP7 and radiation. Our research not only explored and confirmed that USP7 significantly affected the radiotherapy of LSCC, but also revealed that USP7′s effects on radio-sensitivity depended on the mutation status of p53.
It is currently well known that USP7, one of the most studied deubiquitinating enzymes, can deubiquitinate p53 and activate the p53 signaling. USP7 was reported to bind to the carboxy-terminus of p53, while p53 specifically recognizes the N-terminal tumor necrosis factor-receptor associated factor (TRAF)-like domain of USP7 [22,23]. p53, as the most frequently mutated genes in LSCC, plays a pivotal role in tumorigenesis, cell growth rates, and even tumor response to chemotherapy and radiation [24], [25], [26]. The mechanism of how p53 status affects the cellular response to radiotherapy involves signaling pathways in cell survival, growth, apoptosis, ferroptosis, and DNA repair [27], [28], [29].
A comprehensive study on a systematic scale examined the role of p53 in radiosensitivity, including 39 human tumor cell lines from 9 histological types with different p53 statuses, and showed that cells that expressed p53-wild type exhibited more cell killing than cell lines that expressed mutant p53 [30]. It was also confirmed that cell lines from head and neck cancer harboring disruptive p53 mutations were more radioresistant than those with p53-wild type or non-disruptive p53 mutations [31]. However, many studies reported that mutant p53 had no effect or even enhanced cellular sensitivity to radiation [32]. It is quite important to be aware that p53 could present different types of mutations, and other p53 mutations do not have the same biological effects, which need to be explored furtherly. Okaichi K et al. [33] transferred different types of mutant p53 genes into human osteosarcoma Saos-2 cells (p53null), and found that cells with p53 mutations at codons 175, 244, 245, 273, and 282 were radioresistant, whereas those with mutations at codons 123, 195, 238, and 242 were radiosensitive. In this study, TU-212 cells with p53_P151S and SNU-585 cells with p53_ R273L were used. We supposed that the P151S mutation at the transactivation domain of p53 might disrupt its transcriptional activation function, and inhibit the transcript of its target genes through binding to their transcription initiation regions and blocking the binding of other factors with transcriptional activation activity. Meanwhile, the R273L mutation may lead to a gain-of-function (GoF) phenotype of p53 and the aggregation of p63 and p73, which are also important regulators of cell cycle as reported [34,35].
At present, the treatment plan of transfecting wild-type p53 into tumor cells by adenovirus has been tested in a number of clinical studies, which has a very good effect on tumor treatment and radiosensitivity [36]. However, it is still difficult to be applied to the clinic due to the specificity and safety of adenovirus. Our study inferred that targeting USP7 could increase the radiotherapy sensitivity of tumor cells with p53 mutations through the p53 pathway, effectively enhancing the killing effect of radiation on LSCC. A variety of specific USP7 inhibitors has been found these days, which may have significant application value in the radiation field [37,38]. Our research will provide a theoretical basis for the clinical application of USP7 inhibitors in radiotherapy.
Our study has several limitations. First, we couldn't validate the correlation between the outcome of radiotherapy and the expression of USP7 in a large cohort of patients due to the lack of enough specimens. Second, whether USP7 promotes the radio-sensitivity in normal laryngeal cells is still unsure. The effects of USP7 may be different between normal laryngeal cells and p53-wild type LSCC cells. Third, our study lacks in-depth exploration of p53′s downstream pathways in the radio-sensitivity, mainly due to the limited experimental conditions and funding. We hope those questions will be well addressed in the future study.
Conclusions
Radiotherapy induced the raised expression of USP7 in LSCC. USP7 inhibited the radio-sensitivity of p53-mutated LSCC cells, while it promoted radio-sensitivity of p53-wild type LSCC cells. Since most LSCC patients harbor p53 mutation, targeting USP7 has the potential effects to improve the radiation sensitivity of LSCC and reduce radiation damage of surrounding normal tissue.
CRediT authorship contribution statement
Hao Niu: Data curation, Formal analysis, Investigation, Validation, Writing – original draft. Yi Zhu: Data curation, Formal analysis, Investigation, Validation, Writing – original draft. Jie Wang: Conceptualization, Methodology, Project administration, Resources, Software, Visualization, Writing – original draft. Tian Wang: Conceptualization, Methodology, Project administration, Resources, Software, Visualization, Writing – original draft. Xiaosheng Wang: Conceptualization, Data curation, Project administration, Resources, Supervision, Visualization, Writing – review & editing. Li Yan: Conceptualization, Data curation, Funding acquisition, Project administration, Resources, Supervision, Visualization, Writing – review & editing.
Declaration of Competing Interest
None.
Acknowledgments
Acknowledgments
This work was supported by the National Natural Science Foundation of China (81703023), the Foundation of Shanghai Municipal Commission of Health and Family Planning (20184Y0204). We thank the International Science Editing Co. for language editing service.
Ethical approval statement
The study was approved by the Ethics Committees of Eye and ENT Hospital, Fudan University (2020057–1), and performed in accordance with the principles of the Declaration of Helsinki. Informed consent from each patient or participant has been obtained.
Data availability statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Contributor Information
Xiaosheng Wang, Email: xiaosheng.wang@fdeent.org.
Li Yan, Email: yanl13@fudan.edu.cn.
References
- 1.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.De Felice F., de Vincentiis M., Valentini V., Musio D., Mezi S., Lo M.L., Terenzi V., D'Aguanno V., Cassoni A., Di Brino M., et al. Follow-up program in head and neck cancer. Crit. Rev. Oncol. Hematol. 2017;113:151–155. doi: 10.1016/j.critrevonc.2017.03.012. [DOI] [PubMed] [Google Scholar]
- 3.Orlandi E., Iacovelli N.A., Tombolini V., Rancati T., Polimeni A., De Cecco L., Valdagni R., De Felice F. Potential role of microbiome in oncogenesis, outcome prediction and therapeutic targeting for head and neck cancer. Oral Oncol. 2019;99 doi: 10.1016/j.oraloncology.2019.104453. [DOI] [PubMed] [Google Scholar]
- 4.Saraniti C., Speciale R., Santangelo M., Massaro N., Maniaci A., Gallina S., Serra A., Cocuzza S. Functional outcomes after supracricoid modified partial laryngectomy. J. Biol. Regul. Homeost. Agents. 2019;33:1903–1907. doi: 10.23812/19-282-L. [DOI] [PubMed] [Google Scholar]
- 5.Steuer C.E., El-Deiry M., Parks J.R., Higgins K.A., Saba N.F. An update on larynx cancer. CA Cancer J. Clin. 2017;67:31–50. doi: 10.3322/caac.21386. [DOI] [PubMed] [Google Scholar]
- 6.Chera B.S., Amdur R.J., Morris C.G., Kirwan J.M., Mendenhall W.M. T1N0 to T2N0 squamous cell carcinoma of the glottic larynx treated with definitive radiotherapy. Int. J. Radiat. Oncol. Biol. Phys. 2010;78:461–466. doi: 10.1016/j.ijrobp.2009.08.066. [DOI] [PubMed] [Google Scholar]
- 7.Graboyes E.M., Zhan K.Y., Garrett-Mayer E., Lentsch E.J., Sharma A.K., Day T.A. Effect of postoperative radiotherapy on survival for surgically managed pT3N0 and pT4aN0 laryngeal cancer: analysis of the National Cancer Data Base. Cancer-Am. Cancer Soc. 2017;123:2248–2257. doi: 10.1002/cncr.30586. [DOI] [PubMed] [Google Scholar]
- 8.Guillen-Sola A., Soler N.B., Marco E., Pera-Cegarra O., Foro P. Effects of prophylactic swallowing exercises on dysphagia and quality of life in patients with head and neck cancer receiving (chemo) radiotherapy: the Redyor study, a protocol for a randomized clinical trial. Trials. 2019;20:503. doi: 10.1186/s13063-019-3587-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yan L., Hu C., Ji Y., Zou L., Zhao Y., Zhu Y., Wang X., Yu X. Identification of significant secreted or membrane-located proteins in laryngeal squamous cell carcinoma. J. Immunol. Res. 2022 doi: 10.1155/2022/9089397. 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Y., Zhan C., Chen G., Sun J. Labelfree quantitative proteomics and bioinformatics analyses of alcoholic liver disease in a chronic and binge mouse model. Mol. Med. Rep. 2018;18:2079–2087. doi: 10.3892/mmr.2018.9225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Niu H., Huang Y., Yan L., Zhang L., Zhao M., Lu T., Yang X., Chen Z., Zhan C., Shi Y., Wang Q. Knockdown of SMAD3 inhibits the growth and enhances the radiosensitivity of lung adenocarcinoma via p21 in vitro and in vivo. Int. J. Biol. Sci. 2020;16:1010–1022. doi: 10.7150/ijbs.40173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li M., Chen D., Shiloh A., Luo J., Nikolaev A.Y., Qin J., Gu W. Deubiquitination of p53 by HAUSP is an important pathway for p53 stabilization. Nature. 2002;416:648–653. doi: 10.1038/nature737. [DOI] [PubMed] [Google Scholar]
- 13.Shah N.K., Qureshi M.M., Dyer M.A., Patel S.A., Kim K., Everett P.C., Grillone G.A., Jalisi S.M., Truong M.T. Optimal sequencing of chemoradiotherapy for locally advanced laryngeal cancer. Laryngoscope. 2019;129:2313–2320. doi: 10.1002/lary.27771. [DOI] [PubMed] [Google Scholar]
- 14.Du Y., Shao S., Lv M., Zhu Y., Yan L., Qiao T. Radiotherapy Versus Surgery-Which Is Better for Patients With T1-2N0M0 Glottic Laryngeal Squamous Cell Carcinoma? Individualized Survival Prediction Based on Web-Based Nomograms. Front. Oncol. 2020;10:1669. doi: 10.3389/fonc.2020.01669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhan C., Yang X., Song X., Yan L. Radiotherapy vs surgery for T1-2N0M0 laryngeal squamous cell carcinoma: a population-based and propensity score matching study. Cancer Med. 2018;7:2837–2847. doi: 10.1002/cam4.1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grégoire V., Evans M., Le Q.T., Bourhis J., Budach V., Chen A., Eisbruch A., Feng M., Giralt J., Gupta T., et al. Delineation of the primary tumour Clinical Target Volumes (CTV-P) in laryngeal, hypopharyngeal, oropharyngeal and oral cavity squamous cell carcinoma: AIRO, CACA, DAHANCA, EORTC, GEORCC, GORTEC, HKNPCSG, HNCIG, IAG-KHT, LPRHHT, NCIC CTG, NCRI, NRG Oncology, PHNS, SBRT, SOMERA, SRO, SSHNO, TROG consensus guidelines. Radiother. Oncol. 2018;126:3–24. doi: 10.1016/j.radonc.2017.10.016. [DOI] [PubMed] [Google Scholar]
- 17.Serra A., Spinato G., Spinato R., Conti A., Licciardello L., Di Luca M., Campione G., Tonoli G., Politi D., Castro V., et al. Multicenter prospective crossover study on new prosthetic opportunities in post-laryngectomy voice rehabilitation. J. Biol. Regul. Homeost. Agents. 2017;31:803–809. [PubMed] [Google Scholar]
- 18.Qi S.M., Cheng G., Cheng X.D., Xu Z., Xu B., Zhang W.D., Qin J.J. Targeting USP7-Mediated Deubiquitination of MDM2/MDMX-p53 Pathway for Cancer Therapy: are We There Yet? Front. Cell Dev. Biol. 2020;8:233. doi: 10.3389/fcell.2020.00233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kisaï K., Koji S. Prognostic role of USP7 expression in cancer patients: a systematic review and meta-analysis. Pathol. Res. Pract. 2021;227 doi: 10.1016/j.prp.2021.153621. [DOI] [PubMed] [Google Scholar]
- 20.Nininahazwe L., Liu B., He C., Zhang H., Chen Z.S. The emerging nature of Ubiquitin-specific protease 7 (USP7): a new target in cancer therapy. Drug Discov. Today. 2021;26:490–502. doi: 10.1016/j.drudis.2020.10.028. [DOI] [PubMed] [Google Scholar]
- 21.Wu J., Kumar S., Wang F., Wang H., Chen L., Arsenault P., Mattern M., Weinstock J. Chemical approaches to intervening in Ubiquitin Specific Protease 7 (USP7) function for oncology and immune oncology therapies. J. Med. Chem. 2018;61:422–443. doi: 10.1021/acs.jmedchem.7b00498. [DOI] [PubMed] [Google Scholar]
- 22.Hu M., Gu L., Li M., Jeffrey P.D., Gu W., Shi Y. Structural basis of competitive recognition of p53 and MDM2 by HAUSP/USP7: implications for the regulation of the p53-MDM2 pathway. PLoS Biol. 2006;4:e27. doi: 10.1371/journal.pbio.0040027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ma J., Martin J.D., Xue Y., Lor L.A., Kennedy-Wilson K.M., Sinnamon R.H., Ho T.F., Zhang G., Schwartz B., Tummino P.J., Lai Z. C-terminal region of USP7/HAUSP is critical for deubiquitination activity and contains a second mdm2/p53 binding site. Arch. Biochem. Biophys. 2010;503:207–212. doi: 10.1016/j.abb.2010.08.020. [DOI] [PubMed] [Google Scholar]
- 24.Tchelebi L., Ashamalla H., Graves P.R. Mutant p53 and the response to chemotherapy and radiation. Subcell. Biochem. 2014;85:133–159. doi: 10.1007/978-94-017-9211-0_8. [DOI] [PubMed] [Google Scholar]
- 25.Kong X., Yu D., Wang Z., Li S. Relationship between p53 status and the bioeffect of ionizing radiation. Oncol. Lett. 2021;22:661. doi: 10.3892/ol.2021.12922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nathan C.A., Khandelwal A.R., Wolf G.T., Rodrigo J.P., Mäkitie A.A., Saba N.F., Forastiere A.A., Bradford C.R., Ferlito A. TP53 mutations in head and neck cancer. Mol. Carcinog. 2022;61:385–391. doi: 10.1002/mc.23385. [DOI] [PubMed] [Google Scholar]
- 27.Cheng G., Kong D., Hou X., Liang B., He M., Liang N., Ma S., Liu X. The tumor suppressor, p53, contributes to radiosensitivity of lung cancer cells by regulating autophagy and apoptosis. Cancer Biother. Radiopharm. 2013;28:153–159. doi: 10.1089/cbr.2012.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scian M.J., Stagliano K.E., Anderson M.A., Hassan S., Bowman M., Miles M.F., Deb S.P., Deb S. Tumor-derived p53 mutants induce NF-kappaB2 gene expression. Mol. Cell. Biol. 2005;25:10097–10110. doi: 10.1128/MCB.25.22.10097-10110.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang L., Kon N., Li T., Wang S.J., Su T., Hibshoosh H., Baer R., Gu W. Ferroptosis as a p53-mediated activity during tumour suppression. Nature. 2015;520:57–62. doi: 10.1038/nature14344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Williams J.R., Zhang Y., Zhou H., Gridley D.S., Koch C.J., Russell J., Slater J.S., Little J.B. A quantitative overview of radiosensitivity of human tumor cells across histological type and TP53 status. Int. J. Radiat. Biol. 2008;84:253–264. doi: 10.1080/09553000801953342. [DOI] [PubMed] [Google Scholar]
- 31.Skinner H.D., Sandulache V.C., Ow T.J., Meyn R.E., Yordy J.S., Beadle B.M., Fitzgerald A.L., Giri U., Ang K.K., Myers J.N. TP53 disruptive mutations lead to head and neck cancer treatment failure through inhibition of radiation-induced senescence. Clin. Cancer Res. 2012;18:290–300. doi: 10.1158/1078-0432.CCR-11-2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kong X., Yu D., Wang Z., Li S. Relationship between p53 status and the bioeffect of ionizing radiation. Oncol. Lett. 2021;22:661. doi: 10.3892/ol.2021.12922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Okaichi K., Ide-Kanematsu M., Izumi N., Morita N., Okumura Y., Ihara M. Variations in sensitivity to ionizing radiation in relation to p53 mutation point. Anticancer Res. 2008;28:2687–2690. [PubMed] [Google Scholar]
- 34.Li Y., Prives C. Are interactions with p63 and p73 involved in mutant p53 gain of oncogenic function? Oncogene. 2007;26:2220–2225. doi: 10.1038/sj.onc.1210311. [DOI] [PubMed] [Google Scholar]
- 35.Costa D.C., de Oliveira G.A., Cino E.A., Soares I.N., Rangel L.P., Silva J.L. Aggregation and Prion-Like Properties of Misfolded Tumor Suppressors: is Cancer a Prion Disease? Cold Spring Harb. Perspect. Biol. 2016;8 doi: 10.1101/cshperspect.a023614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang Z.X., Wang D., Wang G., Zhang Q.H., Liu J.M., Peng P., Liu X.H. Clinical study of recombinant adenovirus-p53 combined with fractionated stereotactic radiotherapy for hepatocellular carcinoma. J. Cancer Res. Clin. Oncol. 2010;136:625–630. doi: 10.1007/s00432-009-0701-6. [DOI] [PubMed] [Google Scholar]
- 37.An T., Gong Y., Li X., Kong L., Ma P., Gong L., Zhu H., Yu C., Liu J., Zhou H., et al. USP7 inhibitor P5091 inhibits Wnt signaling and colorectal tumor growth. Biochem. Pharmacol. 2017 doi: 10.1016/j.bcp.2017.02.011. [DOI] [PubMed] [Google Scholar]
- 38.Fan Y.H., Cheng J., Vasudevan S.A., Dou J., Zhang H., Patel R.H., Ma I.T., Rojas Y., Zhao Y., Yu Y., et al. USP7 inhibitor P22077 inhibits neuroblastoma growth via inducing p53-mediated apoptosis. Cell Death. Dis. 2013;4:e867. doi: 10.1038/cddis.2013.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.