Fan et al. quantitatively dissect the decisive regulation of migration paradigm (persistence and velocity) to the formation of the migrasome, a cellular organelle with vesicle-like morphology that mediates migracytosis. Their work adds up our current understanding of the role of cell behavior in migrasome studies.
Abstract
Migrasomes are recently discovered vesicle-like structures on retraction fibers of migrating cells that have been linked with transfer of cellular contents, shedding of unwanted materials, and information integration. However, whether and how the cell migration paradigm regulates migrasome formation is not clear. Here, we report that there are significantly fewer migrasomes in turning cells compared with straight persistently migrating cells. The major insight underlying this observation is that as the cells elongate, their rear ends become narrower, subsequently resulting in fewer retraction fibers during impersistent migration. In addition to migration persistence, we reveal that migration speed positively corelates with migrasome formation, owing to the derived length of retraction fibers. Substantiating our hypothesis, genetically removing vimentin compromises cell migration speed and persistence and leads to fewer migrasomes. Together, our data explicate the critical roles of two cell migration patterns, persistence and speed, in the control of migrasome formation by regulating retraction fibers.
Introduction
Migrasomes are recently discovered cellular organelles with vesicle-like morphology that mediate migracytosis, a cell migration–dependent mechanism for releasing cellular contents (Ma et al., 2015). It has been reported that migrasomes govern critical cellular processes including mitochondrial quality control (Jiao et al., 2021), cell–cell communication (Jiang et al., 2019), and lateral transfer of mRNA and proteins (Zhu et al., 2021). When cells crawl on extracellular substrates, retraction fibers (RFs) are pulled out of the plasma membrane from the cells’ rear ends (Taylor and Robbins., 1963). Migrasomes subsequently emerge on the RFs. When cells migrate away, the RFs break, and migrasomes are left behind (Ma et al., 2015; Wu et al., 2017).
Directed migration is an important behavior of cell motility, which can be characterized quantitatively by their velocity and persistence, describing the rate of migration and the continuity in directional migration, respectively (Petrie et al., 2009; Gail and Boone, 1970). In addition to external regulation, such as chemotaxis in response to soluble cues (Andrew and Insall, 2007), durotaxis in response to mechanical signals (Lo et al., 2000), and electrotaxis in response to electric fields (Zhao, 2009), the directional cell migration can also be regulated by intrinsic clues, the fundamental mechanisms of which are associated with the Rho family of small GTPases (Raftopoulou and Hall, 2004) and the ECM receptor–integrin family (Caswell and Norman, 2006).
Previous studies revealed that several factors contributed to migrasome formation. Tetraspanin 4 (TSPAN4), abundant in the membrane of migrasomes (Zhao et al., 2019; Ma et al., 2015), together with cholesterol, are necessary and sufficient for migrasome formation (Huang et al., 2019). Correct pairing of integrins with ECM proteins determines the migrasome formation (Wu et al., 2017). A chemical screen has identified Rho-associated coiled coil–containing protein kinase 1 (ROCK1) as a regulator for migrasome formation (Lu et al., 2020). Notably, migrasome formation strictly depends on cell migration. Blocking migration using the myosin II inhibitor blebbistatin or by modulating cell adhesions inhibits migrasome formation (Ma et al., 2015). However, how exactly distinct cell migration patterns contribute to migrasome formation has barely been addressed.
Vimentin is one of the most well-studied components of intermediate filaments (IFs; Ivaska et al., 2007). Recent studies have revealed that vimentin IFs interact and regulate cytoskeletal dynamics that drive cell motility. Vimentin controls actin stress fiber through GEF-H1 and RhoA (Jiu et al., 2017) and/or modulates lamellipodia formation (Helfand et al., 2011) to facilitate cell migration. Vimentin can also template microtubule networks (Gan et al., 2016) and/or strengthen focal adhesions (Bhattacharya et al., 2009) to enhance cell migration. Nevertheless, its correlation with biogenesis and function of migrasomes has not been previously reported.
In this study, we witness that cells form fewer migrasomes when making turns, and reveal that the reason is the narrower cell rear ends and fewer derived RFs during turning. In addition to persistence, cell migration speed is a critical aspect to confine migrasome formation by controlling the length of derived RFs. Depletion of vimentin IFs cause cell migration deficiency and consequently leads to fewer migrasomes, which verified our hypothesis, that migrasome formation is directly correlated with cell migration persistence and speed.
Results and discussion
There are fewer migrasomes formed when a migrating cell turns
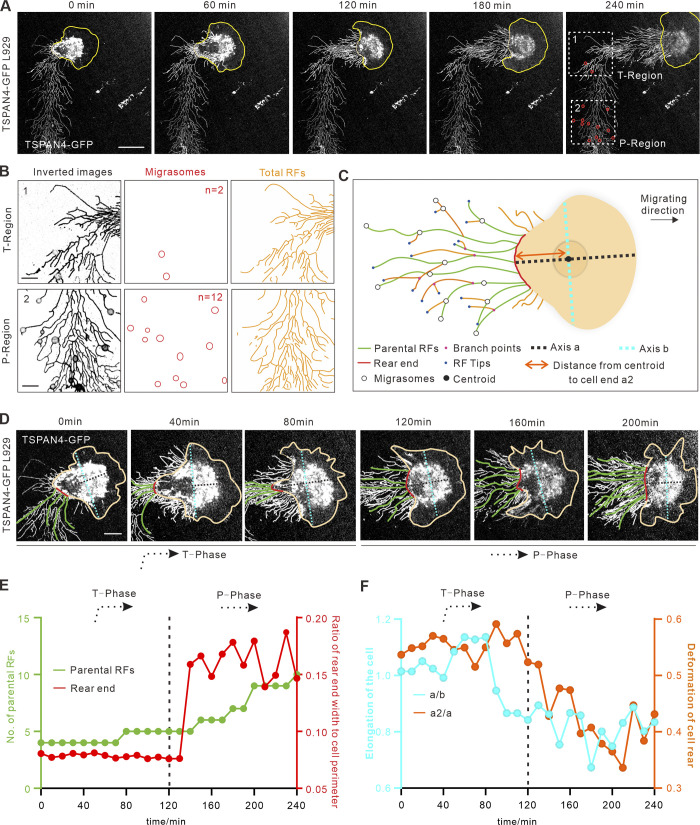
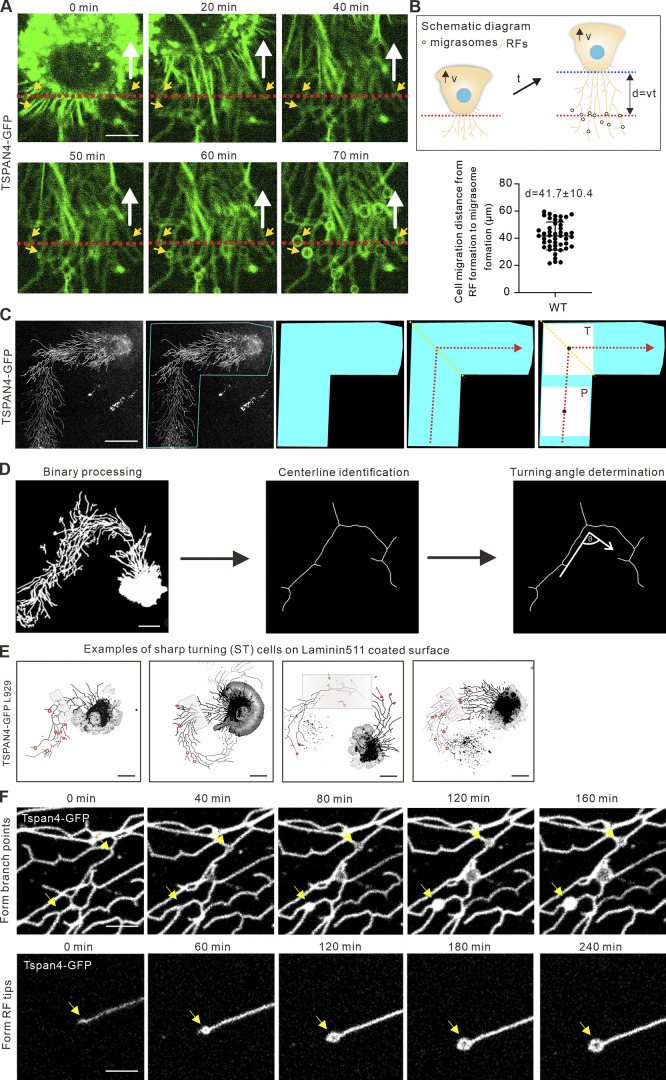
Random migration occurs when a cell receives a nondirectional motogenic signal in the absence of any external guiding factor (Stoker and Gherardi, 1991); thus cells are not always going straight during random migration but quite often change direction by different degrees. To experimentally explore whether persistence is a key factor for cell migration–mediated migrasome formation, we started live-cell imaging experiment by examining mouse fibroblast L929 cells with stable expression of TSPAN4-GFP to label migrasomes (Fig. 1 A; and Video 1). Because migrasomes were found to grow on RFs hours after the initiation of RF formation (Wu et al., 2017), we quantified the average migrating distance of the cell (41.7 ± 10.4 µm; mean ± SD) from the emergence of RFs to the appearance of migrasomes (Fig. S1, A and B; and Video 2), and decided to take regions 45 µm away from the cell rear body for measurements to ensure that migrasomes have enough time to form.
Figure 1.
Migrasomes form less when a migrating cell turns. (A) Representative images from a time-lapse video of a L929 cell expressing TSPAN4-GFP. Yellow lines mark the outline of the cell, and white boxes indicate ROIs of T-region and P-region. Scale bar, 20 µm. (B) Analysis of TSPAN4-GFP signals in T-phase and P-phase depicted in A. Images were inverted, and migrasomes and RFs were masked on the right panel. n in middle panel represents the number of migrasomes. Scale bars, 10 µm. (C) Schematic diagram of a migrating cell. Different migration-associated components and indexes are defined in the diagram. (D) Enlarged time-lapse image of the cell shown in A, with rear end and parental RFs indicated as red and green lines, respectively. Axis a and axis b are indicated with cyan and black dashed lines, respectively. Scale bars, 10 µm. (E) Quantification of parental RFs (green lines) and the ratio of rear end width to cell perimeter (red lines) of the migrating cell in A. (F) Quantification of the length ratio of a to b (cyan) and a2 to a (orange) in each time point of the migrating cell in A. Black dashed line in E and F indicates the time cutoff point between T-phase and P-phase. n = 25 time points.
Video 1.
Time-lapse video of a turning TSPAN4-GFP L929 cell. Yellow lines indicate the outline of the cell. Duration of the video is 240 min, and the recording time interval is 10 min/frame. The display rate is 7 frames/s. Scale bar, 20 μm.
Figure S1.
Data analysis details. (A) Representative images from a time-lapse video indicate the time from RF derivation to migrasome formation in TSPAN4-GFP L929 cells. The white arrow indicates the direction of cell migration, the red dashed line denotes the position of cell rear at initial time point, and the yellow arrows indicated the position of migrasome appearance. Scale bars, 10 µm. (B) Schematic diagram and quantification of cell displacement from RF formation to migrasome appearance. Data are presented as mean ± SD; n = 48 cells. (C) Process diagram to determine the ROI of T-phase and P-phase in a sharp-turning TSPAN4-GFP L929 cell. Blue region marks the ROI of turning cell, yellow lines mark the position of turning, red lines mark the trajectory of migration, black dots mark the center of squares, and white squares mark the ROI of T-phase and P-phase. Scale bars, 20 µm. (D) Schematic diagram of the steps to define the turning angle. Scale bars, 20 µm. (E) Examples of sharp-turning TSPAN4-GFP L929 cells grown on laminin 511–coated surface. Gray boxes indicate the T-regions, and red circles indicate the migrasomes. Scale bars, 20 µm. (F) Representative images from time-lapse videos in TSPAN4-GFP L929 cells visualizing the migrasome formation process on branch points and tips of RFs. Yellow arrows indicate the position where migrasomes formed. Scale bars, 10 µm.
Video 2.
Time-lapse video of a migrating TSPAN4-GFP L929 cell, which indicates the distance from the emergence of RFs to the appearance of migrasomes. Duration of the video is 100 min, and the recording time interval is 10 min/frame. The display rate is 4 frames/s. Scale bar, 20 μm.
We next made an outline of this migrating cell and identified the central line of the migrating track at 240 min, when the cell completed turning and continued to migrate straight forward for >45 µm (Fig. S1 C). The turning region (T-region, 40 × 40–µm square) was subsequently defined (Fig. 1 A, region of interest [ROI] 1). A square of the same size, which was along the straight RFs before turning, is denoted as the persistent region (P-region), the center of which was 50 µm away from the center of T-region to avoid overlapping (Fig. 1 A, ROI 2). It appears that there were only two migrasomes in the T-region, compared with 12 in the P-region in this example. A clue for an intuitive causal factor of this effect is RFs, where migrasomes grow. By using the ridge detection plugin in ImageJ (Steger, 1998; Schindelin et al., 2012), we were able to calculate the total RFs by counting the linear segments between each intersection, and found that there are more RFs in the P-region than in the T-region (Fig. 1 B).
We next wondered how RFs directly derived from the cell body (defined as “parental RFs”) and the cell rear end (where the parental RFs grow) are reformed while the cell makes turns (Fig. 1 C). Thus, the turning phase (T-phase) was defined as cells migrating in the T-region (0–120 min), and the straightforward phase (P-phase) is the subsequent 120 min of rectilinear motion (Fig. 1 D). We identified that the cell rear end indeed became wider and parental RFs subsequently increased when the cell moved from T-phase to P-phase (Fig. 1 E), suggesting that a narrower rear end leads to fewer RFs derived from the cell body.
It is well known that cell shapes and polarity are constantly changing during migration (Bodor et al., 2020; Gómez-Moutón and Mañes, 2007; Zhong and Ji, 2013). To explain whether the morphology transformation could be the reason for the smaller rear end during turning, we defined three parameters: the distance along the migrating direction via centroid (Fig. 1 C, axis a), the distance perpendicularly to a via centroid (Fig. 1 C, axis b), and the distance from cell centroid to cell rear (Fig. 1 C, a2). Therefore, the elongation of the cell can be calculated by the ratio of a versus b, and the deformation of the cell rear can be calculated by the ratio of a2 versus a. Consistent with our speculation, this cell was more elongated, and the cell rear part became attenuated (longer and more narrow) while turning (Fig. 1 F). Together, the data from this turning cell indicate that the cell rear end and the RFs derived during cell turning are critical factors for migrasome formation.
Turning cells lead to narrower rear ends, fewer RFs, and consequently fewer migrasomes
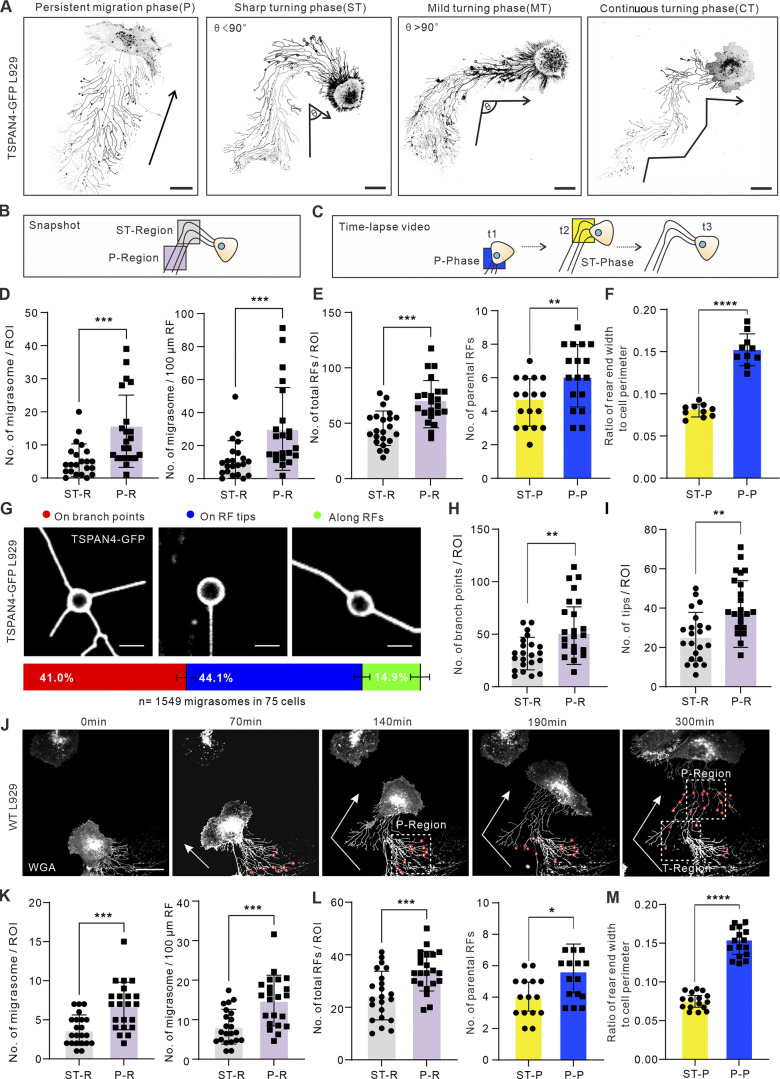
To evaluate whether there is indeed a tight correlation between cells turning and migrasome formation, we classified cell turnings into three categories: sharp turning (ST), mild turning, and continuous turning (Fig. 2 A). Among them, ST and mild turning were defined when the turning angles are less or more than 90°, respectively. To measure the turning angles, we first performed binary processing for the TSPAN4-GFP L929 cells, then identified the centerlines of the migration tracks; the turning angles were measured based on the centerlines (Fig. S1 D).
Figure 2.
Cell turning leads to narrower rear ends and consequently fewer RFs. (A) Examples of different migration patterns of TSPAN4-GFP L929 cells. Black arrows indicate the migration direction. Ө indicates the turning angle. Scale bars, 20 µm. (B) Schematic diagram of the analyzed region for quantifying the number of migrasomes (per ROI and per 100 μm) and total RFs. ST-region, sharp turning region. (C) Schematic diagram of the time point for calculating the number of parental RFs and rear ends. t1, t2, and t3 are ordinal chronological points of the cell. (D) Quantification of the number of migrasomes per ROI (T-region or P-region) or per 100 μm of RFs within the ROI in ST cells. n = 22 cells. (E) Quantification of total RFs (n = 22) and parental RFs (n = 17) within the ROI in T-phase and P-phase of the ST cell. (F) Quantification of the ratio of rear end width to cell perimeter within the ROIs in T-phase and P-phase of the ST cells. n = 10 cells. (G) Three types of migrasome localizations in RF networks and their corresponding proportions in TSPAN4-GFP L929 cells. Scale bars, 5 µm. Quantification is from n = 1,549 migrasomes in 75 cells. (H) Quantification of the branch points within the defined ROIs in T-phase and P-phase of the ST cells. n = 22 cells. (I) Quantification of the tips within the defined ROIs in T-phase and P-phase of the ST cells. n = 22 cells. (J) Representative images from a time-lapse video of WT L929 cell. White arrows indicate the migration direction; red circles indicate migrasomes, and white boxes indicate ROIs of T-region and P-region. Scale bar, 20 µm. (K) Quantification of the number of migrasomes per ROI (T-region or P-region), or per 100 μm of RFs within the ROI in ST cells. n = 22 cells. (L) Quantification of the number of total RFs and parental RFs within the ROI in T-phase and P-phase of the ST cell. n = 22 cells in left panel and n = 15 cells in right panel. (M) Quantification of the ratio of rear end width to cell perimeter within the ROIs in T-phase and P-phase of the ST cells. n = 16 cells. The data for quantification in D–F, H, I, and K–M are from n = 3 independent experiments. The data are presented as mean ± SD (t test). *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
To boost the discrepancy of migrasomes between straightforward migrating cells and turning cells, we decided to choose ST cells for further analysis and used the same strategy in Fig. 1 to define the T-region and P-region (Fig. 2, B and C). The results from numbers of ST cells further supported the conclusion that the reduced number of migrasomes results from the narrower rear ends and subsequently fewer RFs while cells change directions (Fig. 2, D–F). To further validate this finding, we observed cells cultured on a surface coated with laminin 511 (Humphries et al., 2006; Wu et al., 2017), another ECM protein, and found that there were fewer migrasomes at the T-region in ST cells (Fig. S1 E). It was noted that RFs on laminin 511 appeared not to extend as well as on fibronectin, which may due to the lower pairing of this ECM protein to specific integrins (Wu et al., 2017).
How do migration directionality and RFs regulate migrasome formation? As we observed, the location of migrasomes on RFs could be divided into three categories: on the branch points, on the tips, and along the linearized RFs (Figs. 2 G and S1 F). We measured the proportion of each case during random cell migration, and the calculation indicates that >80% of migrasomes grow de novo either on the intersections or on the tips of RFs (Fig. 2 G and Video 3). We further tested whether cell turning affects the organization of the RF network by manually quantifying the branch points and RF tips and found that sharp turnings indeed reduced the numbers of branches and tips (Fig. 2, H and I).
Video 3.
Time-lapse videos of migrasome formation in TSPAN4-GFP L929 cell–derived RFs. Migrasomes tended to generate on branch points and tips. Duration of the video is 240 min, and the recording time interval is 10 min/frame. The display rate is 5 frames/s. Scale bar, 10 μm.
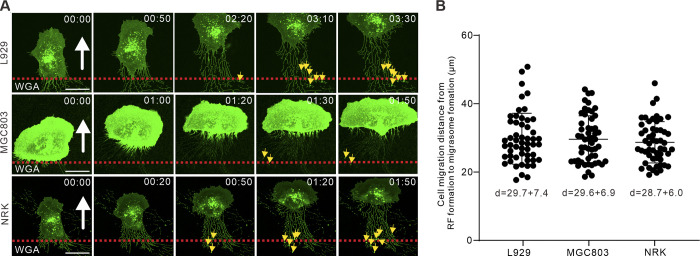
A recent study revealed that TSPAN4 promotes individual migrasome biogenesis (Huang et al., 2019). To exclude the effect of TSPAN4 on migrasomes, we repeated the experiments in WT L929 cells, which also form a fair amount of migrasomes. Fluorescently tagged wheat germ agglutinin was used to stain and track migrasome formation (Chen et al., 2019; Fig. 2 J and Video 4). The average migrating distance from the emergence of RFs to the appearance of migrasomes in WT L929 cells was measured (29.7 ± 7.4 µm; Fig. S2, A and B; and Video 5), and thus regions ≥30 µm away from the cell rear body were measured. Similar T- and P-regions were defined, and the same statistical tests were performed as in Fig. 2, D–F. Consistent with the results in TSPAN4-GFP L929 cells, migrasomes decreased during T-phase compared with straight migrating phase (Fig. 2 K), via reshaping RFs and rear ends (Fig. 2, L and M). Taken together, these data support the concept that cell migration persistence is a critical factor for migrasome formation via regulating widths of rear ends and numbers of RFs.
Video 4.
Time-lapse video of a turning WT L929 cell. Duration of the video is 300 min, and the recording time interval is 10 min/frame. The display rate is 4 frames/s. Scale bar, 20 μm.
Figure S2.
Cell migration displacement from RF formation to migrasome appearance. (A) Representative images from time-lapse videos indicate the time from RF derivation to migrasome formation in WT L929, MGC803, and NRK cells. The white arrows indicated the direction of cell migration, the red dashed lines denote the position of cell rear at initial time point, and the yellow arrows indicate the position of migrasome appearance. Scale bars, 20 µm. (B) Quantification of cell displacement from RF formation to migrasome appearance. Data are presented as mean ± SD; n = 54 cells in L929, 53 cells in MGC803 and 53 cells in NRK.
Video 5.
Time-lapse video of migrating L929, MGC803, and NRK cells, which indicate the distance from the emergence of RFs to the appearance of migrasomes. Duration of the videos are 200, 110, and 100 min, respectively. The recording time interval is 10 min/frame. The display rate is 4 frames/s. Scale bar, 20 μm.
There are more migrasomes formed when cells migrate more persistently and faster
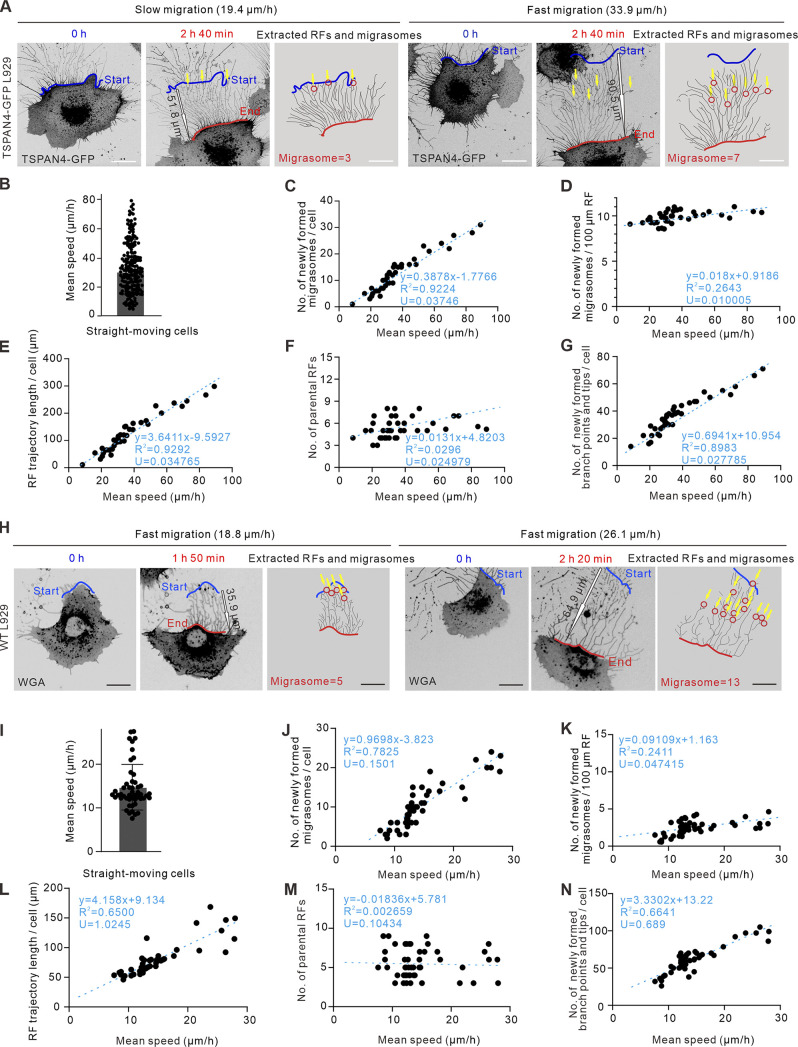
Cell migration speed is another critical aspect to confine migration behavior. We noticed that cell migrating speeds vary a lot even in the same Petri dish, and two straight-moving examples with slower and faster speed, respectively, are shown in Fig. 3 A (Video 6). Our measurement indicated that the speeds of straightforward-migrating TSAPN4-GFP L929 cells ranged from 15 to 80 µm/h, with an average speed of 30.5 µm/h (Fig. 3 B).
Figure 3.
Migrasomes form more when cells migrating faster regardless of their migration persistence. (A) Representative images from time-lapse videos of an L929 cell expressing TSPAN4-GFP. The left panel shows an example of a slow persistent migrating cell, and the right panel shows an example of a fast persistent migrating cell; red and blue lines indicate the start and end positions of the migrating cell, respectively; yellow arrows indicate the position of migrasome formation; and black outline arrows indicated the displacement of the migrating cell. Scale bar, 20 µm. (B) Quantification of average speed of straight-moving cells in the same culture condition. n = 200 cells. (C) Quantification of the number of newly formed migrasomes per cell in straight-moving cells. n = 39 cells. (D) Quantification of the number of newly formed migrasomes per 100-µm RF trajectory in straight-moving cells. n = 39 cells. (E) Quantification of the RF trajectory length per cell in straight-moving cells. n = 39 cells. (F) Quantification of the number of parental RFs per cell in straight-moving cells. n = 39 cells. (G) Quantification of the number of newly formed branch points and tips per cell in straight-moving cells. n = 39 cells. The fitting lines are indicated by the blue dashed line, and the fitting equations are listed in each panel from C to G, with goodness of fit R2 and uncertainty of the fitting slope U to define the correlation coefficient. (H) Representative images from time-lapse videos of WT L929 cells. The left panel shows an example of a slow persistent migrating cell, and the right panel shows an example of a fast persistent migrating cell; red and blue lines indicated the start and end position of the migrating cell, respectively; red circles indicate migrasomes; yellow arrows indicate the position of migrasome formation; and black outline arrows indicated the displacement of the migrating cell. Scale bar, 20 µm. (I) Quantification of average speed of straight-moving WT L929 cells in the same culture condition. n = 41 cells. (J) Quantification of the number of newly formed migrasomes per cell in straight-moving cells. n = 43 cells. (K) Quantification of the number of newly formed migrasomes per 100-µm RF trajectory in straight-moving cells. n = 44 cells. (L) Quantification of the RF trajectory length per cell in straight-moving cells. n = 39 cells. (M) Quantification of the number of parental RFs per cell in straight-moving cells. n = 41 cells. (N) Quantification of the number of newly formed branch points and tips per cell in straight-moving cells. n = 41 cells. The fitting lines are indicated by blue dashed line, and the fitting equations are listed in each panel from C–G and J–N with goodness of fit R2 and uncertainty of the fitting slope U to define the correlation coefficient. The data for quantification in C–G and J–N are from n = 3 independent experiments.
Video 6.
Time-lapse video of two persistent migrating (slow and fast) TSPAN4-GFP L929 cells. The recording time interval is 7.5 min/frame. The red line indicates the start position, and the blue line indicates the end position. Yellow arrows indicate the position of migrasome formation. The mean speed during the recording time is labeled above the images. The display rate is 6 frames/s. Scale bar, 20 μm.
We then quantified the number of migrasomes in these straight-moving cells. It is interesting to note that the number of migrasomes per cell scaled up with the speed of cell migration (Fig. 3 C), yet the amount of migrasomes remained at a similar level when quantified by migrasomes derived per 100-µm length of RFs (Fig. 3 D), indicating that the changes in the number of migrasomes are associated with the length of RFs. We then compared the correlation between cell migration speed and the length of RFs and found that the faster-migrating cells generated significantly longer RFs (Fig. 3 E). In contrast, the average number of parental RFs derived from the cell body showed no direct proportion with cell migration speed (Fig. 3 F), suggesting that the influence of cell migration velocity on migrasome formation is mainly due to the relative length of derived RFs. Moreover, more branch points and tips were accompanied by longer RFs generated with faster speed (Fig. 3 G). Not surprisingly, similar results were found in WT L929 cells (Fig. 3, H–N; and Video 7). Collectively, these data suggest that migration speed is another general mechanism that is critical for migrasome formation via regulating the length of derived RFs.
Video 7.
Time-lapse video of two persistent migrating (slow and fast) L929 cells. The recording time interval is 10 min/frame. The red line indicates the start position, and the blue line indicates the end position. Yellow arrows indicate the position of migrasome formation. The display rate is 4 frames/s. Scale bar, 20 μm.
Persistence and speed are critical cell migration parameters for migrasome formation
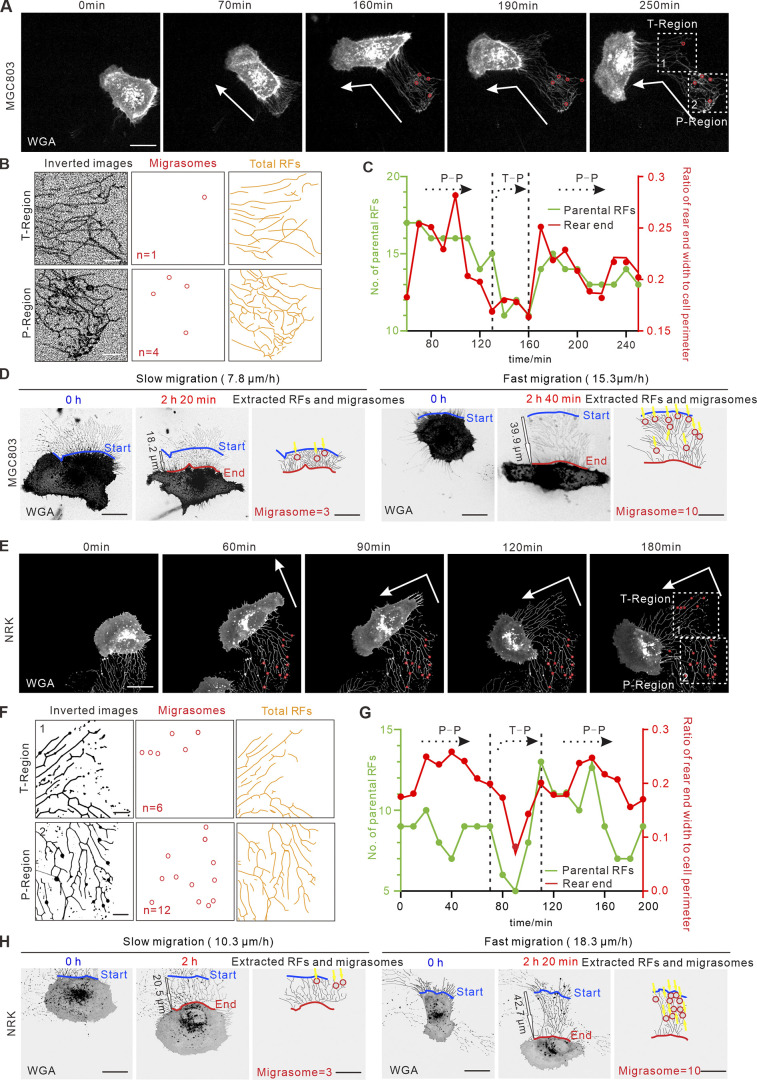
To verify that the persistence and speed of cell migration, in general, modulate the formation of migrasomes, we reproduced experiments in human gastric carcinoma cell line MGC803 and normal rat kidney cell line NRK. Migrasomes appear when cells migrate 29.7 ± 7.4 and 28.7 ± 6.0 µm away in MGC803 and NRK cells, respectively (Fig. S2, A and B; and Video 6). The T- and P-regions were thus defined 30 µm away from the cell rear body. Representative live-cell imaging revealed consistently that more migrasomes were formed when cells migrated more persistently and faster (Fig. 4, A, B, D, E, F, and H; and Videos 8 and 9), which is tightly associated with rear end width, number of parental RFs, and RF length (Fig. 4, C, D, G, and H). Together, results from WT L929, MGC803, and NRK provided more evidences to support the generalization of our findings.
Figure 4.
Persistence and speed of cell migration regulate migrasomes formation in MGC803 and NRK cells. (A and E) Representative images from a time-lapse video of WT MGC803 cell (A) and NRK cell (E). White arrows indicate the migration direction, red circles indicate migrasomes, and white boxes indicate ROIs of T-region and P-region. Scale bar, 20 µm. (B and F) Analysis of RFs and migrasomes in T-region and P-region depicted in A and E, respectively. Images were inverted, and migrasomes and RFs were masked on the right. n in middle panel represents the number of migrasomes. Scale bars, 10 µm. (C and G) Quantification of the number of parental RFs (green lines) and the ratio of rear end width to cell perimeter (red lines) of the migrating cell in A and E, respectively. (D and H) Representative images from time-lapse videos of WT MGC803 cells (D) and NRK cells (H). The left panel shows an example of a slow persistent migrating cell, and the right panel shows an example of a fast persistent migrating cell. The red and blue lines indicate the start and end position of the migrating cell, respectively; yellow arrows indicated the position of migrasome formation; and black outline arrows indicated the displacement of the migrating cell. Scale bar, 20 µm.
Video 8.
Time-lapse video of a turning MGC803 cell and a turning NRK cell. Duration of the video is 250 min and 190 min, respectively. The recording time interval is 10 min/frame. The display rate is 4 frames/s. Scale bar, 20 μm.
Video 9.
Time-lapse video of two persistent migrating (slow and fast) MGC803 cells and NRK cells. The recording time interval is 10 min/frame. The red line indicates the start position, and the blue line indicates the end position. Yellow arrows indicate the position of migrasome formation. The display rate is 4 frames/s. Scale bar, 20 μm.
Vimentin IFs control migrasome formation by regulating directed cell migration
The essential roles of cytoskeleton networks, including actin, microtubules, and IFs, in cell migration have been well characterized (Leduc and Etienne-Manneville, 2015; Gardel et al., 2010; Etienne-Manneville, 2013; Jiu et al., 2015). To regulate cell migration patterns by intrinsic factors and confirm whether they affect migrasome formation, we planned to manipulate the cytoskeleton system.
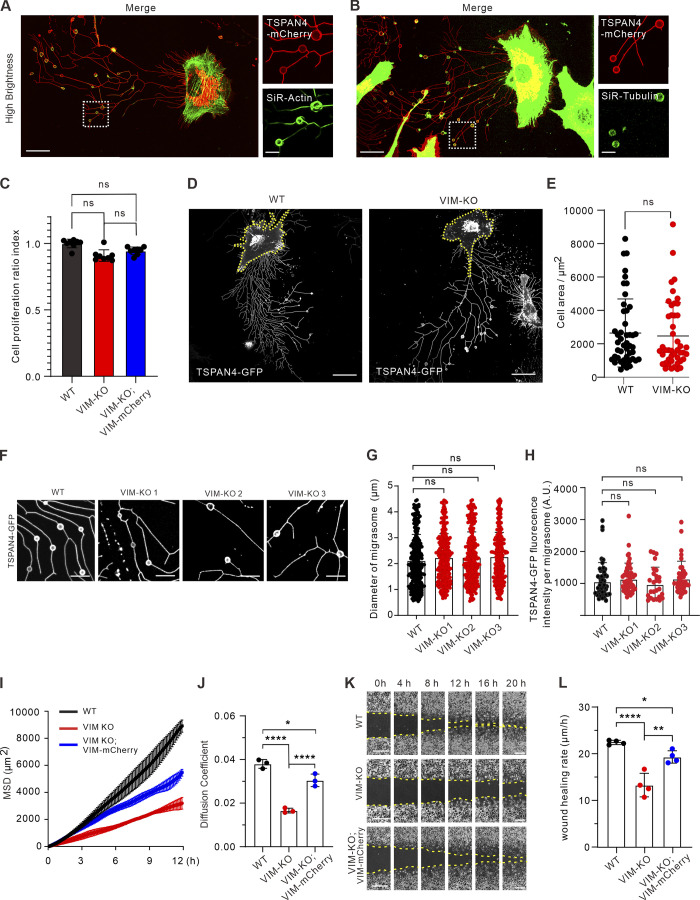
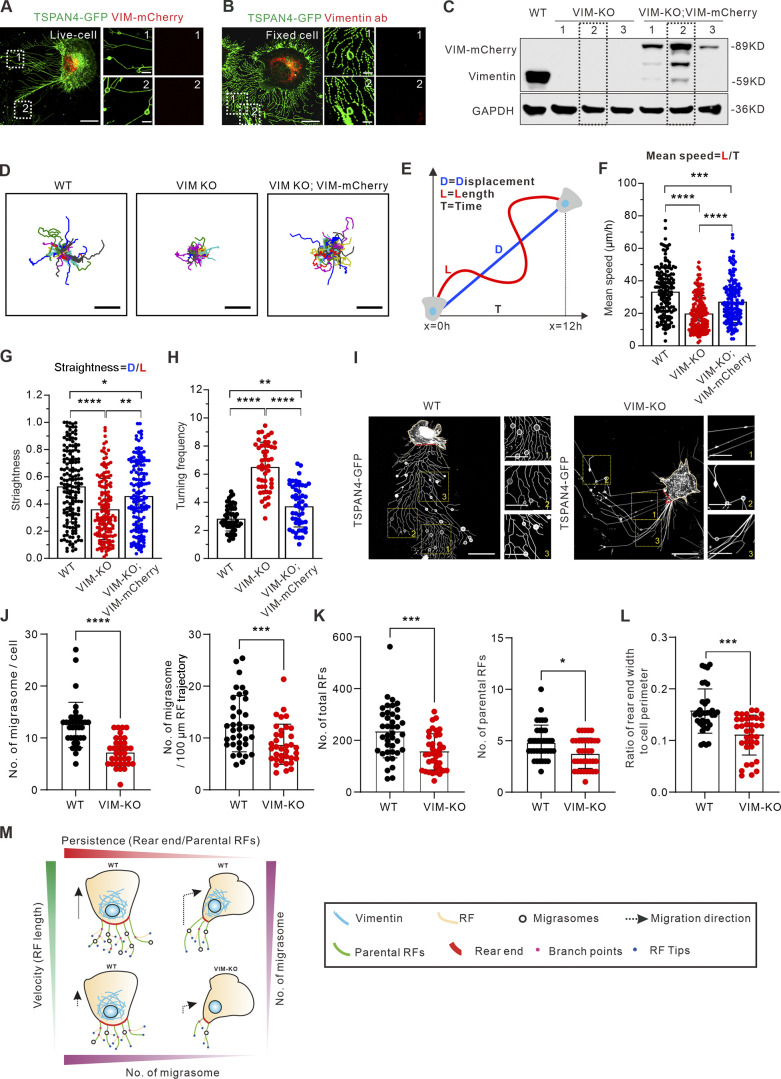
To find an optimal target, we first visualized the subcellular distribution of three cytoskeleton networks. It is not surprising that actin filaments are localized on both RFs and migrasomes, while microtubules are mainly on migrasomes (Fig. S3, A and B). We then visualized the localization of the major IF protein vimentin by exogenous expression of vimentin-mCherry (Fig. 5 A) and endogenous staining with antibody (Fig. 5 B). In both cases, vimentin showed no localization on RFs, and more importantly, no expression on migrasomes.
Figure S3.
Vimentin deficiency leads to defective cell migration and abnormal migrasome formation. (A) Representative images from a time-lapse video of TSPAN4-mCherry L929 cells incubated with SiR-actin. Magnified panels are the white box in the left panel. Scale bars, 20 µm. (B) Representative images from a time-lapse video of TSPAN4-mCherry L929 cells stained with SiR-tubulin. Magnified panels are the white box in the left panel. Scale bars, 20 µm. (C) Analysis of CCK8 assay in TSPAN4-GFP WT, VIM-KO, and VIM-KO; VIM-RES cells. (D) Examples of snapshots of live TSPAN4-GFP WT and VIM-KO L929 cells. Yellow dashed lines indicate the cell outlines. Scale bars, 20 µm. (E) Quantification of the cell area in TSPAN4-GFP WT and VIM-KO L929 cells. n = 47 cells. (F) Examples of RF-associated migrasomes in TSPAN4-GFP WT and VIM-KO L929 cells. Scale bars, 10 µm. (G) Quantification of the diameter of migrasomes in TSPAN4-GFP WT and VIM-KO L929 cells. n = 343 migrasomes in 23 cells. (H) Quantification of the TSPAN4-GFP intensity per migrasome in TSPAN4-GFP WT and VIM-KO L929 cells. n = 50 migrasomes in 11 cells. (I) Mean square displacement (MSD) analysis of representative trajectories per condition. n = 3 regions including 15 trajectories. (J) Diffusion coefficient (D, μm2/100 ms) was calculated from the slope of the fitted regression line derived by MSD analysis of H. n = 3 regions including 15 trajectories. (K) Examples of wound healing assay in TSPAN4-GFP WT, VIM-KO, and VIM-KO; VIM-RES cells. Scale bars, 100 µm. (L) Quantification of wound healing migration rate per condition. n = 4 independent experiments. The data for quantification in C, E, and G–L are from n = 3 independent experiments. Data are presented as mean ± SD (t test in E; one-way ANOVA in C, G, J, and K). *, P < 0.05; **, P < 0.01; ****, P < 0.0001.
Figure 5.
Vimentin regulates migrasome formation by modulating cell migration velocity and persistence. (A) Representative snapshots of live cells show the subcellular distribution of exogenously expressed vimentin-mCherry and TSPAN4-GFP in L929 cells. The white dotted outlines represent ROIs containing RFs and migrasomes. Scale bars, 10 and 2 µm in image and magnified view, respectively. (B) Representative immunofluorescence images show the subcellular distribution of endogenous vimentin with respect to TSPAN4-GFP. The white dotted outlines represent ROIs containing RFs and migrasomes. Scale bars, 10 and 2 µm in image and magnified view, respectively. (C) Western blot analysis of the extracts of WT, VIM-KO, and VIM-KO; VIM-mCherry TSPAN4-GFP expressing L929 cells. The dashed line indicates the cell line we chose for further experiments. (D) Cell random migration trajectory of WT, VIM-KO, and VIM-KO; VIM-mCherry TSPAN4-GFP–expressing cells within 12 h. n = 15 cells. Scale bars, 100 µm. (E) Schematic diagram for the quantification of cell migration speed and straightness. (F) Quantification of the mean speed of TSPAN4-GFP WT, VIM-KO, and VIM-KO; VIM-mCherry cells. n = 150 cells. (G) Quantification of the straightness of TSPAN4-GFP WT, VIM-KO, and VIM-KO; VIM-mCherry cells. n = 150 cells. (H) Quantification of the turning frequency of TSPAN4-GFP WT, VIM-KO, and VIM-KO; VIM-mCherry cells. n = 50 cells. (I) Snapshots of live TSPAN4-GFP–expressing WT and VIM-KO L929 cells. Yellow squares indicate the magnified fields, and red lines mark the rear end. Scale bars, 20 and 10 µm in image and magnified view, respectively. (J) Quantification of the number of migrasomes per cell and per 100-μm RF trajectory in TSPAN4-GFP WT and VIM-KO L929 cells. n = 36 cells. (K) The number of total RFs and parental RFs in TSPAN4-GFP–expressing WT and VIM-KO L929 cells. n = 36 cells. (L) Quantification of the ratio of rear end width to cell perimeter in TSPAN4-GFP WT and VIM-KO L929 cells. n = 36 cells. (M) Schematic model of cell migration pattern to orchestrate migrasome formation. The data for quantification in F–H and J–L are from n = 3 independent experiments. The data are presented as mean ± SD (t test in J–L; one-way ANOVA in I–K). *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. Source data are available for this figure: SourceData F5.
Next, three independent CRISPR/Cas9 vimentin knockout (KO) lines and accordingly three full-length rescue lines were established in TSPAN4-GFP L929 cells (Fig. 5 C), and one line from each condition was chosen for further analysis. Vimentin depletion did not affect cell size and proliferation according to our measurements (Fig. S3, C–E). Importantly, the size and TSPAN4 fluorescence intensity of individual migrasomes remain comparable among WT and three KO cell lines (Fig. S3, F–H), indicating that vimentin is not involved in migrasome biogenesis per se.
To confirm that vimentin depletion shows defects in the cell migration pattern, we carried out cell random migration and wound healing assays. Straightness is an index to generally describe the turning extent of cells and was measured by the ratio between cell migration displacement and track length (Gorelik and Gautreau, 2014; Fig. 5, D and E). Turning frequency was measured by quantifying turning events per 100-µm RFs. Depletion of vimentin significantly decreased straightness, turning frequency, and the speed of cell migration over long time lapses, which could be partially recovered with full-length vimentin (Fig. 5, D–H; Fig. S3, I–L; and Video 10). These results confirm that vimentin is involved in regulating cell migration patterns, as previously observed in other cells (Li et al., 2020; Ostrowska-Podhorodecka et al., 2021).
Video 10.
Time-lapse video of cell random migration in WT, VIM-KO, and VIM-KO; VIM-mCherry TSPAN4-GFP–expressing cells within 12 h. Duration of the video is 720 min, and the recording time interval is 10 min/frame. Color-coded bar from blue to red indicates the speeds ranging from 0 to 80 μm/h. The display rate is 7 frames/s. Scale bar, 100 μm.
By snapshot quantification, there are far fewer migrasomes on the vimentin KO background regardless of whether the calculation was implemented by per cell or per 100-µm RFs (Fig. 5, I and J). Moreover, vimentin KO cells showed fewer RFs and defective cell rear ends (Fig. 5, K and L). Together, these results show that the effective contribution for migrasome formation by vimentin depends strongly on its effect on directed cell migration.
The complexity and heterogeneity of cell migration paradigms have frustrated efforts to understand how migrasome formation is related to cell migration, even though this interconnectedness is understood to play an essential role. In this regard, our work provides far-reaching effects on future study relevant to migrasome formation (Fig. 5 M). A migration cycle is accompanied by dramatic cytoskeletal and membrane rearrangement. Previous work revealed that rear retraction is controlled by membrane tension sensing and subcellular traction force redistribution, which leads to dramatic cell shape deformation to coordinate cell migration (Hetmanski et al., 2019; Zhong and Ji, 2013). Cells become elongated during turning in our results (Fig. 1, D and F), which lead to narrower rear ends, from which RFs grow. Therefore, RFs are tightly associated with migration pattern.
Migrasomes tend to generate on stiffer RFs where TSPAN4 and cholesterol accumulate (Huang et al., 2019). The numbers of branch points and tips are mildly reduced when cells make turns (Fig. 2, G–I). It is thus tempting to speculate that impersistent migration disrupts the topology-based rigidity distribution on RFs, and TSPAN4 and cholesterol on RFs are subsequently rearranged.
Vimentin has been reported to regulate cell migration in various cell types, such as fibroblasts (Vakhrusheva et al., 2019; Sliogeryte and Gavara, 2019), cancer cells (Xuan et al., 2020; Schoumacher et al., 2010), leukocytes (Nieminen et al., 2006), and astrocytes (De Pascalis et al., 2018). Importantly, vimentin localized on neither migrasomes nor RFs compared with actin and microtubule networks (Fig. 5, A and B). Unlike microtubules and actin filaments, vimentin filaments uniquely provide cells with a hyperelastic network that regulates cell stretchability and resilience (Hu et al., 2019). Thus, it is tempting to speculate that migrasomes are stiff because of the enrichment of TSPAN4 and cholesterol (Huang et al., 2019), whereas they may lack stretchability and resilience compared with the cell body because of the absence of vimentin IFs.
Cell migration plays a key role in organ formation, tissue regeneration, wound healing, cancer metastasis, and many diseases (Kurosaka and Kashina, 2008; Amini et al., 2019; Dekoninck and Blanpain, 2019; Villalobo and Berchtold, 2020). Persistent cell migration is beneficial to cell chemotaxis in tissue and organ formation, whereas impersistent cell migration allows cells, especially immune-associated cells, to better sense local environmental stimuli and signaling molecules (Worbs et al., 2017). That migration behavior controls migrasome formation potentiates a spatiotemporal dynamic regulatory function of migrasomes during various physiological and pathological processes. Our work has made a quantitative analysis of the association: that is, migration patterns affect the topology of RFs and thus affect migrasome formation, which is critical in current models of cell migration–generated migrasomes.
Materials and methods
Cell culture and transfection
Mouse fibroblast L929 cells, normal rat kidney epithelial cells (NRK cells), and human stomach (gastric) cancer MGC803 cells were kindly given by Dr. Li Yu (Tsinghua University, China). 293T cells were kindly given by Dr. Jing Zhong (Institute Pasteur of Shanghai, Chinese Academy of Sciences, China). All cells were maintained in high glucose (4.5 g/l) DMEM (06-1055-57-1A; Biological Industries) supplemented with 10% FBS (10270-106; Gibco), 100 U/ml penicillin, 100 µg/ml streptomycin, and 4 mM L-glutamine (complete DMEM) at 37°C in a humidified atmosphere with 5% CO2. Transient transfections were performed with JetPrime (Polyplus-transfection) according to the manufacturer’s instructions using a DNA:JetOPTIMUS ratios of 1:1.25 and 24-h incubation per assay.
Vimentin CRISPR/Cas9 KO cell line and rescue cell line generation
As previously described, vimentin KO cells were generated using CRISPR/Cas9 methods (Jiu et al., 2015) based on lentiCRISPRv2 vector (98290; Addgene) with two targets. Primers for vimentin target 1 were 5′-CACCGGAATGACTGCAGGGTGCTTT-3′ and 5′-AAACAAAGCACCCTGCAGTCATTCC-3′; primers for vimentin target 2 were 5′-CACCGAGCTCCGGTTGGAGCTGGGC-3′and 5′-AAACGCCCAGCTCCAACCGGAGCTC-3′. CRISPR clones were cultivated for 2 wk with 3 μg/ml puromycin before selecting clones with no discernible vimentin protein expression by Western blotting. Rescue cell lines were constructed by lentivirus. Briefly, lentivirus encoding vimentin-mCherry was used to establish L929 TSPAN4-GFP vimentin KO and vimentin-mCherry cell lines. 293T cells were cotransfected with the following vectors: pLVX-vimentin-mCherry, which was constructed by our lab using pLVX (125839; Addgene), HIV-1 packaging plasmid (psPAX2, 12260; Addgene), and a vesicular stomatitis virus glycoprotein expression vector (pMD2.G, 12259; Addgene). The rescue cell lines were selected by puromycin pressure.
Cell migration assay
Cells were collected from culture dishes using trypsin-EDTA and resuspended in complete DMEM at a concentration of 2 × 107 cells/ml. 20 µl cell suspension was added to the device inlet. The dish was pretreated for 3 h with 10 μg/ml fibronectin diluted in PBS at 37°C. Cells were allowed to adhere and spread overnight. Before imaging with an Olympus IX73 inverted microscope with an UplanFL 10×/0.3 objective (Olympus), cells were incubated with Hoechst 33342 for 10 min and washed twice with PBS, and medium was replaced with complete DMEM. Images were acquired by Olympus software CellSens Dimension v1.18. Average migration velocity and directional migration duration were quantified by tracking nucleus movement using Imaris Spots plugins (Bitplane). The recording was set as every 7.5 min for 12 h. Only cells that did not collide with one another were selected for measurements. 15-µm estimated xy diameter, 30-µm maximum distance, and 3-frame maximum gap sizes were set for analyzing in Imaris.
Wound healing assay
Cells were seeded in a fibronectin-coated (10 μg/ml) 6-well cell culture plate with a cell density of 25,000/cm2 and cultivated at 37°C in 5% CO2 overnight. Subsequently, the cell monolayers were scratched with a sterile 0.2-ml pipette tip to create linear wounds and washed with PBS to remove detached cells. Cells were incubated in complete DMEM and observed on an Olympus IX73 inverted microscope with an UplanFL 4×/0.13 objective (Olympus). Images were acquired by Olympus software CellSens Dimension v1.18. With ImageJ (National Institutes of Health), the wound areas between cells were measured and plotted at each time point. From the slope of the plot, a simple calculation yielded the cell migration rate in micrometers per hour (Jonkman et al., 2014). If we take A as the area of the gap and l as the length of the gap, the cell migration rate is given as
Western blot
All cell lysates were prepared by washing the cells once with PBS and scraping them into radioimmunoprecipitation assay lysis buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 1% Triton X-100, 1% sodium deoxycholate, and 0.1% SDS) supplemented with 1 mM PMSF, 10 mM DTT, 40 μg/ml DNase I, and 1 μg/ml of leupeptin, pepstatin, and aprotinin. All preparations were conducted at 4°C. Protein concentrations were determined with BCA Protein Assay kit (P0012; Beyotime) and equal amounts of total cell lysates were mixed with Laemmli sample buffer (LSB), boiled, and run on 12.5% SDS-PAGE gels. Proteins were transferred to nitrocellulose membrane with Trans-Blot Turbo transfer system (Bio-Rad) using Mini TGX gel transfer protocol. Membrane was blocked in 5% BSA for 1 h at RT. Primary and secondary antibodies were diluted into fresh blocking buffer overnight at 4°C and 1 h at RT, respectively. Proteins were detected from the membranes with SuperSigna West Femto Maximum Sensitivity Substrate (Thermo Fisher Scientific). Antibodies were used with the following dilutions in primary antibody dilution buffer (P0023A; Beyotime): vimentin rabbit monoclonal D21H3 antibody (dilution 1:1,000; 5741; Cell Signaling Technology), GAPDH rabbit polyclonal antibody (dilution 1:5,000 for WB; G9545; Sigma-Aldrich). HRP-linked anti-rabbit IgG antibody (dilution 1:5,000; 7074V; Cell Signaling Technology). Chemiluminescence was measured after resin with Western blotting ECL (180-501; Tanon).
Immunofluorescence microscopy
Cells were fixed with methanol for 5 min at −20°C, washed three times with PBS, and permeabilized with Saponin in PBS for 5 min. Cells were then blocked in PBS supplemented with 5% BSA. Vimentin rabbit monoclonal D21H3 antibody (dilution 1:200; 5741; Cell Signaling Technology) and Alexa Fluor 568 goat anti-rabbit IgG (H + L; dilution 1:1,000; A11011; Invitrogen) were applied to cells and incubated at RT for 2 h. All images were acquired by Olympus software CellSens Dimension v1.18 on an Olympus SpinSR10 Ixplore spinning disk confocal microscope equipped with an UplanApo 100×/1.5 oil objective (Olympus). The pixel size was optimized to achieve the maximum resolution, which was calculated to be 65 nm.
Live-cell imaging
For live-cell imaging, 35-mm glass-bottom dishes (MatTek Corp.) were coated with 10 µg/ml fibronectin (PHE0023; Gibco) in PBS for ≥3 h at 37°C, washed with PBS twice, and immersed in complete DMEM without phenol red (01-053-1A; Biological Industries) before seeding of cells. For labeling actin and microtubule cytoskeleton, cells were incubated with 0.2 µM SiR-Actin (CY-SC001; Cytoskeleton) and SiR-Tubulin (CY-SC002; Cytoskeleton) for 6 h, respectively.
Time-lapse images of cells with transient transfection were acquired with the Olympus CellSens Dimension system, consisting of an Olympus SpinSR10 Ixplore spinning disk confocal microscope and a Yokogawa CSU-W1 confocal scanner. Appropriate filters, heated sample environment (37°C), controlled 5% CO2, and UplanApo 100×/1.5 oil objective (Olympus Corp.) were used. The recording was set as every 10 min for 12 h, and one focal plane was recorded for all live-cell videos.
Statistical analysis
Statistical data analyses were performed with Excel (Microsoft) and Prism v8 (GraphPad Software). For the data with normal distribution, Student’s two-sample unpaired t test was used. If data did not follow a normal distribution, Mann–Whitney U test for two independent samples was conducted. One-way ANOVA followed by a Tukey’s post hoc test was used to evaluate differences between three or more groups. Statistical analysis was obtained with t test or U test for Fig. 2, D–F, H, I, K–M; Fig. 5, J–L; and Fig. S3 E and with one-way ANOVA for Fig. 5, F–H; and Fig. S3 C, G, H, J, and L. Significance is indicated by asterisks: *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
For analyzing the number of migrasomes, parental RFs, branch points and tips, we manually counted in the defined ROI views or cells. The position of centroid is the geometric center of the cell and equal to the arithmetic mean position of the uniformly distributed imaging pixel points within the spreading area of the cell. Therefore, the position of the centroid of the cell can be calculated with the following formula:
where are the coordinates of the centroid of the cell, n is the number of pixel points in the cell, and are the coordinates of the ith pixel within the cell.
For analyzing the number of migrasomes per 100 µm, migrasome number (Nmigrasome) and parental RFs number (NP–RF) per ROI in snapshot or per cell in live cells were manually counted. The average length of parental RF trajectories (L; µm) per ROI in snapshot or per cell in live cells were calculated by “line measurement” in ImageJ. This gave us the number of migrasomes per 100-µm RFs as
For analyzing the number of total RFs, “freehand selection” in ImageJ was used to identify the ROI of RFs, and appropriate line width was determined (5 pixels in this case) to make sure that all RFs were recognized. The numbers of recognized linear fragments between intersections of RFs were then calculated by “rigid detection” plugin in ImageJ (Steger, 1998) to obtain the number of total RFs.
For analyzing the ratio of rear end width versus cell perimeter, “free-hand line” of ImageJ was used to manually outline the rear ends and the cell perimeter, and the lengths were subsequently measured.
To estimate the linear regression in Fig. 3, C–G and J–N, we used “regression analysis” in Microsoft Excel (Office16) to fit the data with the linear equation: . a indicates the slope. The calculated fitting equation, goodness of fit (R2), and uncertainty of the fitting slope (U) are indicated in the figures. R2 tending to 1 indicates a better linear correlation between two sets of variables. The uncertainty of the slope was reflected by the range of slope fluctuation U, indicating the confidence interval for the slope (a ± U).
Online supplemental material
Fig. S1 shows details on the data analysis. Fig. S2 shows the quantification of cell displacement from RF formation to migrasome appearance in WT L929, MGC803, and NRK cells. Fig. S3 shows that vimentin deficiency leads to defective cell migration and abnormal migrasome formation. Video 1 shows a turning TSPAN4-GFP L929 cell. Video 2 shows a migrating TSPAN4-GFP L929 cell, which indicates the average time from the emergence of RFs to the appearance of migrasomes. Video 3 shows migrasome formation in TSPAN4-GFP L929 cell–derived RFs. Video 4 shows a turning WT L929 cell. Video 5 shows cell displacement from RF formation to migrasome appearance in WT L929, MGC803, and NRK cells. Video 6 shows two persistent migrating (slow and fast) TSPAN4-GFP L929 cells. Video 7 shows two persistent migrating (slow and fast) L929 cells. Video 8 shows a turning MGC803 cell and a turning NRK cell. Video 9 shows two persistent migrating (slow and fast) MGC803 and NRK cells, respectively. Video 10 shows cell random migration in WT, VIM-KO, and VIM-KO; VIM-mCherry TSPAN4-GFP expressing cells.
Supplementary Material
contains original blots for Fig. 5.
Acknowledgments
We thank Dr. Li Yu (Tsinghua University, China) for providing us with L929, NRK, and MGC803 cells and fruitful discussion of the project. We thank Dr. John E. Eriksson (Åbo Akademi University, Finland) for providing us with the vimentin plasmids.
This study was funded by National Natural Science Foundation of China (92054104, 31970660); CAS-VPST Silk Road Science Fund (GJHZ2021138); Key Research and Development Program, Ministry of Science and Technology of China (2021YFC2300204); Shanghai Municipal Science and Technology Major Project (2019SHZDZX02); and Key Laboratory of Molecular Virology & Immunology, Institut Pasteur of Shanghai (KLMVI-OP-202001).
The authors declare no competing financial interests.
Author contributions: C. Fan carried out the majority of the experiments and interpretation of the data. X. Shi carried out vimentin visualization experiments. H. Li and L. Wang analyzed the trajectory of the turning migration example. B. Ji and K. Zhao analyzed the centroid and migration direction. K. Shi and Y.-J. Liu helped with the revision. Y. Jiu conceived and designed the study and wrote the manuscript with contributions from all other authors.
References
- Amini, R., Labudina A.A., and Norden C.. 2019. Stochastic single cell migration leads to robust horizontal cell layer formation in the vertebrate retina. Development. 146:dev173450. 10.1242/dev.173450. [DOI] [PubMed] [Google Scholar]
- Andrew, N., and Insall R.H.. 2007. Chemotaxis in shallow gradients is mediated independently of PtdIns 3-kinase by biased choices between random protrusions. Nat. Cell Biol. 9:193–200. 10.1038/ncb1536 [DOI] [PubMed] [Google Scholar]
- Bhattacharya, R., Gonzalez A.M., Debiase P.J., Trejo H.E., Goldman R.D., Flitney F.W., and J.C.R. Jones. 2009. Recruitment of vimentin to the cell surface by beta3 integrin and plectin mediates adhesion strength. J. Cell Sci. 122:1390–1400. 10.1242/jcs.043042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodor, D.L., Pönisch W., Endres R.G., and Paluch E.K.. 2020. Of cell shapes and motion: The physical basis of animal cell migration. Dev. Cell. 52:550–562. 10.1016/j.devcel.2020.02.013 [DOI] [PubMed] [Google Scholar]
- Caswell, P.T., and Norman J.C.. 2006. Integrin trafficking and the control of cell migration. Traffic. 7:14–21. 10.1111/j.1600-0854.2005.00362.x [DOI] [PubMed] [Google Scholar]
- Chen, L., Ma L., and Yu L.. 2019. WGA is a probe for migrasomes. Cell Discov. 5:13. 10.1038/s41421-018-0078-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Pascalis, C., Pérez-González C., Seetharaman S., Boëda B., Vianay B., Burute M., Leduc C., Borghi N., Trepat X., and Etienne-Manneville S. 2018. Intermediate filaments control collective migration by restricting traction forces and sustaining cell–cell contacts. J. Cell Biol. 217:3031–3044. 10.1083/jcb.201801162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekoninck, S., and Blanpain C.. 2019. Stem cell dynamics, migration and plasticity during wound healing. Nat. Cell Biol. 21:18–24. 10.1038/s41556-018-0237-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etienne-Manneville, S. 2013. Microtubules in cell migration. Annu. Rev. Cell Dev. Biol. 29:471–499. 10.1146/annurev-cellbio-101011-155711 [DOI] [PubMed] [Google Scholar]
- Gail, M.H., and Boone C.W.. 1970. The locomotion of mouse fibroblasts in tissue culture. Biophys. J. 10:980–993. 10.1016/S0006-3495(70)86347-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardel, M.L., Schneider I.C., Aratyn-Schaus Y., and Waterman C.M.. 2010. Mechanical integration of actin and adhesion dynamics in cell migration. Annu. Rev. Cell Dev. Biol. 26:315–333. 10.1146/annurev.cellbio.011209.122036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan, Z., Ding L., Burckhardt C.J., Lowery J., Zaritsky A., Sitterley K., Mota A., Costigliola N., Starker C.G., Voytas D.F., et al. 2016. Vimentin intermediate filaments template microtubule networks to enhance persistence in cell polarity and directed migration. Cell Syst. 3:500–501. 10.1016/j.cels.2016.11.011 [DOI] [PubMed] [Google Scholar]
- Gorelik, R., and Gautreau A. 2014. Quantitative and unbiased analysis of directional persistence in cell migration. Nat. Protoc. 9:1931–1943. 10.1038/nprot.2014.131 [DOI] [PubMed] [Google Scholar]
- Gómez-Moutón, C., and Mañes S. 2007. Establishment and maintenance of cell polarity during leukocyte chemotaxis. Cell Adh. Migr. 1:69–76. 10.4161/cam.1.2.4547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfand, B.T., Mendez M.G., Murthy S.N., Shumaker D.K., Grin B., Mahammad S., U. Aebi, T. Wedig, Y.I. Wu, K.M. Hahn, et al. 2011. Vimentin organization modulates the formation of lamellipodia. Mol. Biol. Cell. 22(8):1274–1289. 10.1091/mbc.E10-08-0699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetmanski, J.H.R., de Belly H., Busnelli I., Waring T., Nair R.V., Sokleva V., Dobre O., Cameron A., Gauthier N., Lamaze C., et al. 2019. Membrane tension orchestrates rear retraction in matrix-directed cell migration. Dev. Cell. 51:460–475.e10. 10.1016/j.devcel.2019.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, Y., Zucker B., Zhang S., Elias S., Zhu Y., Chen H., Ding T., Li Y., Sun Y., Lou J., et al. 2019. Migrasome formation is mediated by assembly of micron-scale tetraspanin macrodomains. Nat. Cell Biol. 21:991–1002. 10.1038/s41556-019-0367-5 [DOI] [PubMed] [Google Scholar]
- Humphries, J.D., Byron A., and Humphries M.J. 2006. Integrin ligands at a glance. J. Cell Sci. 119:3901–3903. 10.1242/jcs.03098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, J., Li Y., Hao Y., Zheng T., Gupta S., Parada G., Wu H., Lin S., Wang S., Zhao X., et al. 2019. High stretchability, strength, and toughness of living cells enabled by hyperelastic vimentin intermediate filaments. Proc. Natl. Acad. Sci. USA. 116:17175–17180. 10.1073/pnas.1903890116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivaska, J., Pallari H.M., Nevo J., and Eriksson J.E.. 2007. Novel functions of vimentin in cell adhesion, migration, and signaling. Exp. Cell Res. 313:2050–2062. 10.1016/j.yexcr.2007.03.040 [DOI] [PubMed] [Google Scholar]
- Jiang, D., Jiang Z., Lu D., Wang X., Liang H., Zhang J., Meng Y., Li Y., Wu D., Huang Y., et al. 2019. Migrasomes provide regional cues for organ morphogenesis during zebrafish gastrulation. Nat. Cell Biol. 21:966–977. 10.1038/s41556-019-0358-6 [DOI] [PubMed] [Google Scholar]
- Jiao, H., Jiang D., Hu X., Du W., Ji L., Yang Y., Li X., Sho T., Wang X., Li Y., et al. 2021. Mitocytosis, a migrasome-mediated mitochondrial quality-control process. Cell. 184:2896–2910.e13. 10.1016/j.cell.2021.04.027 [DOI] [PubMed] [Google Scholar]
- Jiu, Y., Peränen J., Schaible N., Cheng F., Eriksson J.E., Krishnan R., and Lappalainen P.. 2017. Vimentin intermediate filaments control actin stress fiber assembly through GEF-H1 and RhoA. J. Cell Sci. 130:892–902. 10.1242/jcs.196881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiu, Y., Lehtimäki J., Tojkander S., Cheng F., Jäälinoja H., Liu X., Vajosalo M., Eriksson J.E., and Lappalainen P.. 2015. Bidirectional interplay between vimentin intermediate filaments and contractile actin stress fibers. Cell Rep. 11:1511–1518. 10.1016/j.celrep.2015.05.008 [DOI] [PubMed] [Google Scholar]
- Jonkman, J.E.N., Cathcart J.A., Xu F., Bartolini M.E., Amon J.E., Stevens K.M., and P. Colarusso. 2014. An introduction to the wound healing assay using live-cell microscopy. Cell Adh. Migr. 8:440–451. 10.4161/cam.36224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurosaka, S., and Kashina A.. 2008. Cell biology of embryonic migration. Birth Defects Res. Part C Embryo Today. 84:102–122. 10.1002/bdrc.20125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leduc, C., and Etienne-Manneville S.. 2015. Intermediate filaments in cell migration and invasion: The unusual suspects. Curr. Opin. Cell Biol. 32:102–112. 10.1016/j.ceb.2015.01.005 [DOI] [PubMed] [Google Scholar]
- Li, C., Mcmanus F.P., Plutoni C., Pascariu C.M., Nelson T., Alberici Delsin L.E., Emery G., and Thibault P. 2020. Quantitative SUMO proteomics identifies PIAS1 substrates involved in cell migration and motility. Nat. Commun. 11:834. 10.1038/s41467-020-14581-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, P., Liu R., Lu D., Xu Y., Yang X., Jiang Z., Yang C., Yu L., Lei X., and Chen Y. 2020. Chemical screening identifies ROCK1 as a regulator of migrasome formation. Cell Discov. 6:51. 10.1038/s41421-020-0179-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo, C.M., Wang H.B., Dembo M., and Wang Y.L. 2000. Cell movement is guided by the rigidity of the substrate. Biophys. J. 79:144–152. 10.1016/S0006-3495(00)76279-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, L., Li Y., Peng J., Wu D., Zhao X., Cui Y., Chen L., Yan X., Du Y., and Yu L.. 2015. Discovery of the migrasome, an organelle mediating release of cytoplasmic contents during cell migration. Cell Res. 25:24–38. 10.1038/cr.2014.135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieminen, M., Henttinen T., Merinen M., Marttila-Ichihara F., Eriksson J.E., and Jalkanen S.. 2006. Vimentin function in lymphocyte adhesion and transcellular migration. Nat. Cell Biol. 8:156–162. 10.1038/ncb1355 [DOI] [PubMed] [Google Scholar]
- Ostrowska-Podhorodecka, Z., Ding I., Lee W., Tanic J., Abbasi S., Arora P.D., Liu R.S., Patteson A.E., Janmey P.A., and Mcculloch C.A.. 2021. Vimentin tunes cell migration on collagen by controlling beta1 integrin activation and clustering. J. Cell Sci. 134:jcs254359. [DOI] [PubMed] [Google Scholar]
- Petrie, R.J., Doyle A.D., and Yamada K.M.. 2009. Random versus directionally persistent cell migration. Nat. Rev. Mol. Cell Biol. 10:538–549. 10.1038/nrm2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raftopoulou, M., and Hall A.. 2004. Cell migration: Rho GTPases lead the way. Dev. Biol. 265:23–32. 10.1016/j.ydbio.2003.06.003 [DOI] [PubMed] [Google Scholar]
- Schindelin, J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. 2012. Fiji: An opensource platform for biological-image analysis. Nat. Methods. 9:676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoumacher, M., Goldman R.D., Louvard D., and Vignjevic D.M. 2010. Actin, microtubules, and vimentin intermediate filaments cooperate for elongation of invadopodia. J. Cell Biol. 189:541–556. 10.1083/jcb.200909113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sliogeryte, K., and Gavara N.. 2019. Vimentin plays a crucial role in fibroblast ageing by regulating biophysical properties and cell migration. Cells. 8:1164. 10.3390/cells8101164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steger, C. 1998. An unbiased detector of curvilinear structures. IEEE Transactions on Pattern Analysis and Machine Intelligence. Vol. 20. 113–125. [Google Scholar]
- Stoker, M., and Gherardi E.. 1991. Regulation of cell movement: The motogenic cytokines. Biochim. Biophys. Acta. 1072:81–102. 10.1016/0304-419x(91)90008-9 [DOI] [PubMed] [Google Scholar]
- Taylor, A.C, and Robbins E.. 1963. Observations on microextensions from the surface of isolated vertebrate cells. Dev. Biol. 6:660–673. 10.1016/0012-1606(63)90150-7 [DOI] [PubMed] [Google Scholar]
- Vakhrusheva, A., Endzhievskaya S., Zhuikov V., Nekrasova T., Parshina E., Ovsiannikova N., Popov V., Bagrov D., Minin A., and Sokolova O.S.. 2019. The role of vimentin in directional migration of rat fibroblasts. Cytoskeleton (Hoboken). 76:467–476. 10.1002/cm.21572 [DOI] [PubMed] [Google Scholar]
- Villalobo, A., and Berchtold M.W.. 2020. The role of calmodulin in tumor cell migration, invasiveness, and metastasis. Int. J. Mol. Sci. 21:765. 10.3390/ijms21030765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worbs, T., Hammerschmidt S.I., and Förster R.. 2017. Dendritic cell migration in health and disease. Nat. Rev. Immunol. 17:30–48. 10.1038/nri.2016.116 [DOI] [PubMed] [Google Scholar]
- Wu, D., Xu Y., Ding T., Zu Y., Yang C., and Yu L.. 2017. Pairing of integrins with ECM proteins determines migrasome formation. Cell Res. 27:1397–1400. 10.1038/cr.2017.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xuan, B., Ghosh D., Jiang J., Shao R., and Dawson M.R.. 2020. Vimentin filaments drive migratory persistence in polyploidal cancer cells. Proc. Natl. Acad. Sci. USA. 117:26756–26765. 10.1073/pnas.2011912117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, M. 2009. Electrical fields in wound healing-An overriding signal that directs cell migration. Semin. Cell Dev. Biol. 20:674–682. 10.1016/j.semcdb.2008.12.009 [DOI] [PubMed] [Google Scholar]
- Zhao, X., Lei Y., Zheng J., Peng J., Li Y., Yu L., and Chen Y.. 2019. Identification of markers for migrasome detection. Cell Discov. 5:27. 10.1038/s41421-019-0093-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, M., Zou Q., Huang R., Li Y., Xing X., Fang J., Ma L., Li L., Yang X., and Yu L.. 2021. Lateral transfer of mRNA and protein by migrasomes modifies the recipient cells. Cell Res. 31:237–240. 10.1038/s41422-020-00415-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, Y., and Ji B.. 2013. Impact of cell shape on cell migration behavior on elastic substrate. Biofabrication. 5:015011. 10.1088/1758-5082/5/1/015011 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
contains original blots for Fig. 5.