Graphical abstract

Keywords: Biophysics, Coronavirus, Molecular dynamics simulation, Machine learning
Abstract
SARS-CoV-2 main protease (Mpro) involved in COVID-19 is required for maturation of the virus and infection of host cells. The key question is how to block the activity of Mpro. By combining atomistic simulations with machine learning, we found that the enzyme regulates its own activity by a collective allosteric mechanism that involves dimerization and binding of a single substrate. At the core of the collective mechanism is the coupling between the catalytic site residues, H41 and C145, which direct the activity of Mpro dimer, and two salt bridges formed between R4 and E290 at the dimer interface. If these salt bridges are mutated, the activity of Mpro is blocked. The results suggest that dimerization of main proteases is a general mechanism to foster coronavirus proliferation, and propose a robust drug-based strategy that does not depend on the frequently mutating spike proteins at the viral envelope used to develop vaccines.
1. Introduction
COVID-19 is an ongoing pandemic threatening the lives and well-being of people across the globe [48]. A key means of curbing the pandemic is the development of vaccines. However, it is equally important to develop drugs to treat patients with the disease, and to prevent the proliferation of SARS-CoV-2 in the host. To determine a viable drug target, the key objective is to identify an indispensable mechanism in the viral replication cycle that can be targeted by potential therapeutic agents. From this perspective, the main protease (Mpro) of SARS-CoV-2 has been recently selected as a lucrative drug target due to three reasons [8]. First, its substrate specificity is unique [43]. Mpro cleaves the amide linkage in the viral polypeptide at 11 conserved locations defined by a fixed motif at a position after a conserved glutamine (Q) residue in the target sequence [52]. No human protease is known to have this brand of specificity [20]. Second, Mpro is essential for the maturation of the viral particles, thus inhibiting its action will severely hamper the virus’s ability to spread to a human host [15]. Third, given that viral proteases are commonly tested drug targets in viral diseases such as HIV and Hepatitis C [1], [13], [36], pre-existing drugs developed to block protease function, and whose safe use in humans has already been established, can be repurposed for speedy drug discovery [14], [23], [31], [35], [42], [43].
Mpro is a cysteine protease with a well-determined target sequence (-TSAVLQSGFRKM-) [7], [37], [45], [46], [50]. It has two main catalytic residues [40], [45], [46]: a cysteine residue (C145) and a histidine residue (H41) as illustrated in Fig. 1a and b. These residues reside within a substrate binding cleft facing each other. H41 forms a thiolate-imidazolium ion pair with C145 and extracts a proton from it. The deprotonated C145 then initiates a nucleophilic attack on the carbonyl carbon of a sessile peptide bond in the target substrate [40], [45], [46]. Thus, for the initiation of the chemical reaction mechanism, these two catalytic residues must reside close to each other at a distance less than 4 Å and in a configuration where the proton donor and acceptor face each other to form a stable hydrogen bond.
Fig. 1.
The structure of Mpro. a. The monomer is composed of three domains. b. The substrate-binding cleft region magnified. The monomeric enzyme (light blue) bound to the substrate analogue (red) with the catalytic residues H41 and C145 highlighted. c. Mpro in a dimeric form. The substrate-binding cleft and the substrate shown in red are depicted. d. The consensus sequence of the substrate with the cleavage site indicated. e. The dimer interface of Mpro shown in dark blue. The bound substrate is shown in red. The locations of all known mutations in the enzyme are shown in orange. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
The Mpro enzyme monomer is composed of three domains (Fig. 1a) [51]. Domains 1 and 2 contain the substrate-binding cleft, which contain the catalytic residues (Fig. 1a and b). Domains 2 and 3 are involved in the oligomerization of the enzyme. In this work, we refer to the apo and substrate-bound Mpro in the monomeric form as M− and M+, respectively. The dimeric form of Mpro can assume three different substrate-bound states: the apo state, the singly-bound state (in which only one of the protomers is bound to the substrate), or the doubly-bound state (in which each protomer is bound to one substrate). We refer to the apo Mpro dimer as D−D−, the singly-bound Mpro dimer as D+D−, and the doubly-bound dimer as D+D+. Following the same logic, D− denotes an apo protomer in a dimeric Mpro, and D+ denotes a protomer bound to a substrate in a dimeric enzyme. The enzyme has been suggested to be allosteric [11], [17], meaning that one chain of the dimer regulates the activation of the other chain. Importantly, Mpro can form a homodimer (Fig. 1c), which may be necessary for the activation of enzymatic activity of the protein [5], [6], [17], [29].
Although numerous crystal structures of Mpro are now available in the PDB database [PDB IDs: 6Y2E/F/G [51], 6WTK/M/J [44], 7BUY [24], 6WTT [32]], how the dimerization and substrate binding affect the enzymatic activity remain open questions. Does the substrate first bind a monomer, leading to dimerization? Or does dimerization take place first, followed by substrate binding? Most importantly, what exactly is the role of dimerization, and how would a decrease in dimerization help inhibit the functionality of Mpro?
In this work, we addressed these questions using extensive atomistic molecular dynamics (MD) simulations combined with machine learning (ML) techniques used to analyze ∼370 μs of all-atom MD simulation data. We simulated the Mpro enzyme in monomeric, dimeric, apo, and singly- and doubly-bound states (Table 1). Here, we focus specifically on two key aspects of the Mpro function: substrate binding and catalytic efficiency, which we elucidate through several intriguing structural features revealed by the ML analysis. The results suggest that the enzymatically active form of Mpro is dimeric due to its higher substrate-binding affinity and catalytic efficiency. Detailed analysis further unveiled a functional mechanistic pathway, in which substrate binding follows the formation of the Mpro dimer. The details of the simulation models, simulation approaches and ML-based analysis techniques are provided in Methods.
Table 1.
Simulation systems. The table lists the variants of Mpro systems. Nsim – the number of simulation repeats; tsim – the simulation length per repeat; ttot – total length of simulated trajectories. Double mutant has the R4A and E290A mutations, which were implemented to the enzyme structure [34], [38]. The descriptions 1A and 1B stand for one substrate per dimer bound to chain A or chain B, respectively. In all systems, the initial structure of the protein was taken from the PDB ID: 6LU7.
| System name | Protein state | Protein variant | Substrate | Nsim | tsim (µs) | ttot (µs) |
|---|---|---|---|---|---|---|
| Apo-monomer (M−) | Monomer | wild-type | – | 100 | 2 | 200 |
| Bound monomer (M+) | Monomer | wild-type | 1 | 20 | 2 | 40 |
| Apo-dimer (D−D−) | Dimer | wild-type | – | 20 | 2 | 40 |
| Doubly-bound dimer (D+D+) | Dimer | wild-type | 2 | 20 | 2 | 40 |
| Singly-bound dimer (chain A) (D+D−) | Dimer | wild-type | 1A | 10 | 2 | 20 |
| Singly-bound dimer (chain B) (D−D+) | Dimer | wild-type | 1B | 10 | 2 | 20 |
| Double mutant | Dimer | R4A, E290A | – | 5 | 2 | 10 |
Insights gained through this work suggest drug-based ways to prevent Mpro dimerization and propose that dimerization of main protease may be a more general mechanism used by viruses to foster viral proliferation. Further, in the specific context of COVID-19, vaccinations to prevent it are based on making fragments of the SARS-CoV-2 virus spike proteins, with the aim of triggering an immune response. However, given that continuous mutations in these spike proteins reduce the effectiveness of the vaccines, it is clear that a complementary strategy to prevent COVID-19 would be to inhibit the activity of the main protease, as it would not be dependent on mutations in the spike proteins.
2. Methods
2.1. Atomistic molecular dynamics simulations
We used the GROMACS 2020.1 simulation package [30] for our atomistic molecular dynamics (MD) simulations. The simulation inputs were generated with CHARMM-GUI [25]. The simulations of Mpro were performed for both a dimer and a free monomer, whose coordinates were obtained from the PDB ID 6LU7. The simulations were carried out at 310 K with 150 mM KCl using a cubic box with initial dimensions 10×10×10 nm3, filled with ∼33000 water molecules. The force field used for the protein was Amber ff14SB [33] with compatible parameters for salt ions [26]. For water, we used the TIP3P [49] parameters. The Particle Mesh Ewald [9] technique was used to calculate electrostatic interactions. For short-range van der Waals interactions, we used a cut-off of 1.0 nm as parameterized for the Amber ff14SB [33]. The LINCS [19] algorithm was used to constrain the covalent bonds in the protein. Systems were equilibrated under NVT conditions and then simulated in production runs in the NpT ensemble at 1 atm using a timestep of 4 fs obtained by using heavy-hydrogens and reducing their oscillatory frequencies, thus slowing down the fastest degrees of freedom [21].
As to the substrate (Fig. 1d), we created a substrate analogue which we inserted in the binding site by replacing the drug N3 bound in the crystal structure. The initial coordinates (orientation and conformation) of the substrate in the binding site were obtained using structural alignment with the crystal structure of substrate-bound SARS-CoV Mpro (PDB ID: 2Q6G) using the canonical substrate sequence [7], [37] characteristic for SARS-CoV and SARS-CoV-2 Mpro. All variant forms of the monomer and the dimer bound to the substrate were explored. We thus simulated the monomer in apo (M−) and substrate-bound (M+) conditions. Similarly, the dimer was considered in its apo form (D−D−), doubly-bound state (D+D+; the substrate bound to both chain A and chain B), and the two singly-bound states where the substrate is bound to either chain A or chain B (D+D− and D−D+).
Each case was simulated for 2 μs through 10–120 independent replicas (Table 1). For the analysis of the simulation data, we discarded the first 250 ns of all trajectories and an equal number of data frames were selected from each state to avoid any statistical bias. The total simulation time was 370 μs. Protein structures were visualized with VMD [22].
2.2. Determinant of substrate binding
To have an objective measure of the substrate binding capacity of the enzyme, we created a model to represent the accessibility of the substrate-binding cleft in the enzyme binding site. From visual inspection of simulation trajectories, we chose five residues (T24, M49, N142, E166, N189) that flank the cleft. From the C-alpha atoms of these five residues (Fig. S1c) we built a polygon whose area acts as a proxy for the visibility of the substrate-binding cleft. This area was computed for both the monomer and the dimer in both apo and substrate-bound forms. Results for the polygon area were observed to be in very good agreement with visual identification of the tightly bound and loosely bound substrates in the active site (Fig. S1a and b).
2.3. Free energy calculations
We calculated the relative binding free energies of the Mpro-substrate complexes by the Molecular Mechanics-Poisson–Boltzmann Solvent-Accessible surface area (MMPBSA) method [3], using the g_mmpbsa tool [28]. The MMPBSA data were calculated from the substrate-bound systems (Table S1) at every 10 ns of each trajectory. The final free energy values were obtained by taking an average of all the simulation replicas in a given system. The MMPBSA method calculates the binding free energy of a protein-substrate complex, focusing on its enthalpic component, and assuming that the entropy of binding is similar and the differences in the ΔΔS ≈ 0. Consequently, the key importance of the results given by this method lies in the qualitative trends it observes rather than in quantitative figures. These free energy calculations indicated that the free energy of binding was the largest in systems where the binding cleft area was the smallest, and vice versa.
2.4. Machine learning: Background
We used two machine learning methods for characterization: the Gaussian Mixture Model (GMM) and the Partial Least Squares based Functional Mode Analysis (PLS-FMA) model. The GMM is used to indicate how different enzyme states are determined by the orientation of the catalytic residues. This allows us to classify these states with a quantitative and statistically robust parameter that can indicate their catalytic activation. Meanwhile, the PLS-FMA model is a robust technique to infer how the dynamics of the rest of the enzyme is specifically correlated to the catalytic residues.
2.5. Machine learning: Construction of the Gaussian Mixture model
As a direct indicator of the catalytic efficiency of the enzyme, we used the orientations of the catalytic residues C145 and H41. To simplify presentation, these complex variables were described by two distances: dCA as the distance between the C-alpha atoms of the two residues. and dSidechain as the distance between the CE1 atom of H41 and the S atom of C145 (Fig. 2c). To detect individual clusters in the distribution defined by these distances, we pooled all the MD simulation data (Table S1) together (Fig. 2a) to build a machine learning-based Gaussian Mixture Model (GMM) [12] to optimally separate the clusters. GMM is a probabilistic model that assumes data points to be generated from a mixture of Gaussian distributions. The choice of this model was motivated by observing the elliptically distributed data points in the raw representation of the coordinates selected for the clustering (Fig. 2a). For our GMMs, we used the Scikit-learn [2] package BayesianGaussianMixture. The model was initialized with centers selected by the k-means clustering technique.
Fig. 2.
Gaussian Mixture model (GMM) for estimating catalytic efficiency. a. Simulation data illustrated in terms of dCA (the distance between the C-alpha atoms of the catalytic residues) and dSidechain (the distance between the side chains of the catalytic residues). b. Cluster determination by the GMM. The colored circles on the clusters represent observed crystal structures currently available on the PDB. c. Representative conformations of the catalytic residues from each cluster.
We shuffled our pooled dataset of ∼400,000 points and randomly picked 10,000 points to build a GMM over this trial set (Fig. 2a). We used the Aikake Information Criterion (AIC) and the Bayesian Information Criterion (BIC) (Fig. S2b) [16] to choose the number of clusters that minimize the penalty score that indicates overfitting. Once this score can no longer be minimized, further addition of cluster centers leads to an overfitted model. Based on our judgment from the BIC and AIC curves, we chose a five-cluster model, since beyond this number there is little decrease in the penalty. To confirm that the five-cluster model was robust in prediction tasks, we repeated the model building with 10 non-overlapping training sets of 10,000 points each from the pooled data and built independent GMMs from each set. We then used these GMMs to predict labels for a reserved data set of 10,000 points (test set). The standard deviation of the prediction labels obtained for the test set from the 10 GMMs developed on the training sets were then used to judge the robustness of the models. Low standard deviations for all individual clusters implied that the clustering performed with the training sets leads to high-fidelity predictions in the test set, validating robustness (Fig. S2c).
2.6. Machine learning: Partial least squares based functional mode analysis (PLS-FMA)
We used machine learning (the PLS-FMA method [27]) to build a mechanistic model of protein dynamics associated with the coordinates of interest (function). The PLS-FMA model identifies a set of highly correlated collective atomic motions, i.e., a collective mode, that has a maximal linear correlation with a function of interest calculated from the simulation data. This collective mode can be composed of several components, each of which is highly correlated to the function of interest and orthogonal to each other. The cross_decomposition.PLSRegression module from scikit-learn was used to perform the PLS-FMA analysis. We used all the heavy atoms of the backbones and side chains to train the model.
The dSidechain and the binding cleft area coordinateas described above were used as the target functions on which the PLS-FMA model was trained. Only the monomeric simulations in the apo form (M−) were used to create this model. The model was validated by using only a half of the simulation data set to train the model, while reserving the other half for cross-validation. The coefficient of determination (R2) for the cross-validation set was used to check that the model was not overfitted. We found that the R2 value for the dSidechain coordinate was saturated with 15 components, indicating that adding more components would lead to overfitting. The R2 with 15 components was >0.8, thus leading to a robust model that was used for the analysis of the simulation data. For the binding cleft area coordinate, the model was found to reach an R2 value >0.8 with five components (Fig. S6). The collective modes for the dSidechain and the binding cleft area coordinates are illustrated in Movie S1 and S2, respectively.
3. Results
3.1. The dimer interface is strictly conserved
To get an idea of the evolutionary stress of the various parts of the enzyme, we combined all the reported mutations in the structure of Mpro to date [18]. These are shown on the enzyme surface as illustrated in Fig. 1e. All known mutations are distributed evenly on the enzyme surface including the area of the catalytic cleft but are absent from its dimerization interface. This indicates that the virus does not tolerate any modification at the dimerization interface of Mpro, highlighting that the residues responsible for dimerization are crucial for the function of Mpro and the maturation of SARS-CoV-2. This is the fundamental reason why, in this work, we focus on the dimer interface.
3.2. Active site is characterized by five states, one of them being active
The catalytic efficiency of Mpro was assessed based on the geometry of the catalytic residues H41 and C145. For efficient proton extraction from the thiol group of C145 by H41, these catalytic residues must be brought close to each other. Additionally, they must be in a correct orientation to form a hydrogen bond such that the side chains face each other to lead to a formation of a catalytic dyad that can participate in a proteolytic reaction mechanism. We studied the behavior of H41 and C145 using two figures of merit: the distance between the C-alpha atoms of these catalytic residues (dCA), and the distance between the sulfur atom of thiol in C145 and the Cε carbon atom of the imidazole ring of H41 (dSidechain) (Fig. 2). The coordinate dCA indicates how the backbone atoms of the catalytic residues are positioned, while dSidechain is an indicator of the proximity of the functional groups of the residues. The results based on the analysis of ∼370 μs atom-scale molecular dynamics (MD) simulation data of both Mpro monomers and dimers (Table 1) are depicted in Fig. 2a. The distribution of these coordinates in a variety of bound states is shown in Fig. S2a.
To analyze these data, we used a machine learning method known as the Gaussian Mixture Model (GMM, see Methods). In essence, we used the distributions of the different states (Fig. S2a) to build the GMM, which identified five clusters shown in Fig. 2b. Each of these clusters represents a distinct characteristic geometry of the catalytic residues. The structures closest to the centers of the GMM clusters are shown in Fig. 2c. Cluster 1 represents the activated configuration of the catalytic residues where the backbone atoms of the two residues are maximally separated, but the sidechains are in proximity. The orientation of the two head groups of these sidechains is ideally suited for the formation of the catalytic dyad, as the Nδ atom of H41 directly faces the thiol group of C145. In this configuration, the hydrogen bonding for the proton transfer is expected to be optimal. Analysis of the simulation data showed that this is the case: hydrogen bonding was found to be present in ∼80% of the cluster members. Although Cluster 2 has similar values for dCA and dSidechain as Cluster 1, the plane of H41 in Cluster 2 is turned away from the thiol group of C145. This makes the extraction of the proton by H41 difficult. This was expressed as a lower hydrogen bonding capacity, which decreased to 41% within these cluster members. Thus, Cluster 2 is not favorably suited to form the catalytic dyad. In Clusters 3 and 4, the H41 sidechain plane is far from the thiol group (Fig. 2c), which is also inefficient for dyad formation. Cluster 5 represents another inactive configuration, since the distance between the two catalytic residues is unsuitable for dyad formation.
Altogether, our results provide compelling evidence that the catalytic residues of Mpro have five different states, and only one of them (Cluster 1) is catalytically active.
3.3. The substrate binding cleft has two distinct states: Tightly and weakly bound
We used our atom-scale simulation data to analyze the substrate-binding capacity of Mpro, which in practice was implemented by representing the accessibility of the substrate-binding cleft with the area of a polygon that forms the cleft surface (Fig. S1c). In essence, a large area indicates that the cleft is open and the substrate can only bind loosely, while a small area indicates strong binding. This analysis revealed that the binding cleft (Fig. 1c) can bind the substrate either tightly (Fig. S1a) or weakly (Fig. S1b). We have discovered through free energy calculations that a smaller area of the binding cleft corresponds to tighter binding through a lower (more negative) value of binding free energy, and vice versa.
3.4. Monomeric Mpro has low substrate affinity
The apo-monomer (M−) has a very flexible binding cleft (Fig. 3a) characterized by large fluctuations of the cleft area. The substrate-bound monomer (M+; Fig. 3b), on the other hand, has a stable but large cleft area (∼75 Å2), where the flanking residues that enclose the binding site face away from the cleft. This informs that monomeric Mpro cannot bind the substrate tightly, since even when the substrate is bound to the monomeric Mpro, the substrate may slip out of its cleft. Free energy calculations also indicated that the monomeric Mpro has a much lower affinity for the substrate than the dimeric states.
Fig. 3.
Substrate affinity and catalytic potential of the Mpro monomer. a. Distribution of the area of the substrate-binding cleft of the apo-monomer (M−). b. Distribution of the area of the substrate-binding cleft in the substrate-bound monomer (M+). The red dashed line illustrates the maximum of the distribution of the bound monomer. Shown in the inset is the structure of the substrate-binding cleft of M+ at an area of the substrate binding cleft matching the maximum of the distribution. The polygon formed from the C-alpha atoms of the residues surrounding the cleft is shown with a black dashed line. c,d. The cluster populations of M− and M+. Cluster numbering corresponds to Fig. 2. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Cluster 1, which represents the activated configuration of the catalytic residues, occupies ∼30% of the population of the apo-monomer (M−, Fig. 3c). In the substrate-bound monomer (M+), Cluster 1 occupies ∼75% of the population (Fig. 3d), indicating that the catalytic residues are oriented ideally for forming a dyad. However, given that the affinity of the substrate for the binding cleft of monomeric Mpro is low, the catalytic potential of the enzyme in the monomeric state is expected to be low, too.
3.5. Dimeric Mpro is specialized for substrate binding
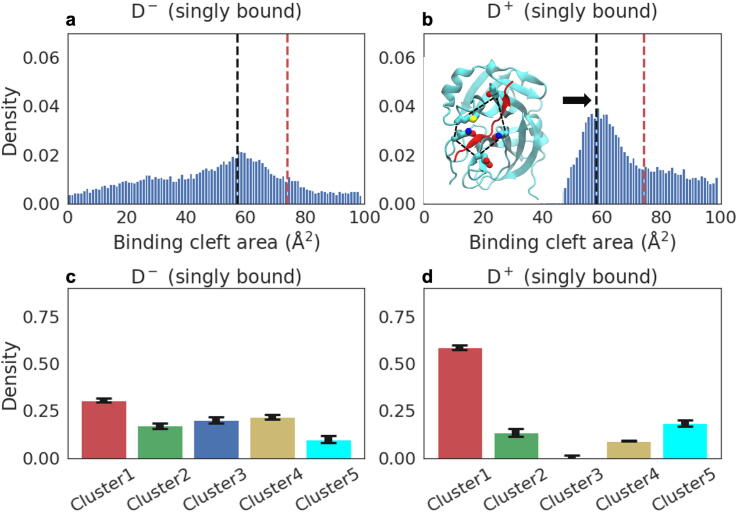
To resolve how dimerization affects substrate affinity, we studied various substrate-bound states of the Mpro dimer, including the apo-dimer (D−D−), as well as the singly- (D+D−) and doubly-bound (D+D+) forms (Table 1). The D−D− chains have a flexible substrate-binding cleft, like that of the apo-monomer (M−; Fig. S3a and b; cf. Fig. 3a). In D−D−, the catalytic residues are distributed across the five clusters. The population of either chain being in Cluster 1 is around ∼25%, like in the apo-monomer (Fig. S3c and d; cf. Fig. 3c). The doubly-bound dimer (D+D+) has a rigid substrate binding cleft (Figs. S4a and b), and unlike the substrate-bound monomer (M+; Fig. 3b), the cleft is narrower with an area of ∼ 60 Å2 (Figs. S4a and b). Cluster 1 has the highest population (∼40%) for each chain (Figs. S4c and d). For D+D−, we find that the apo protomer (D−) (Fig. 4a) behaves like the apo-monomer (M−) with respect to the area of the substrate-binding cleft (Fig. 3a). Meanwhile, the substrate-bound protomer (D+) has a narrow and rigid cleft (Fig. 4b). All residues that flank the cleft in D+ point into the cleft, blocking the substrate from escaping (Fig. 4b inset). D− in the singly-bound dimer has a population of ∼30% in Cluster 1 (Fig. 4c), while in the bound one it is ∼63% (Fig. 4d). This behavior does not depend on which chain the substrate is bound to, as expected.
Fig. 4.
Substrate affinity and catalytic potential of a singly-bound Mpro dimer. a. Distribution of the substrate-binding cleft area in a singly-bound dimer (D+D−): the chain not bound to the substrate (D−). b. As in panel A, but now for the chain bound to the substrate (D+). The black dashed line illustrates the maximum of the distribution for D+D−. For comparison, the red dashed line illustrates the maximum of the distribution of M+(see Fig. 3b). The inset describes the structure of the substrate-binding cleft of the substrate-bound chain at an area of the substrate binding cleft matching the maximum of the distribution. The polygon formed from the C-alpha atoms of the residues surrounding the cleft is depicted with a black dashed line. c,d. The cluster populations of D+D−: the unbound chain, and the chain bound to the substrate. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
These data indicate that only the singly-bound dimeric state (D+D−) is capable of both binding the substrate and maintaining an active dyad configuration of the catalytic residues.
Additionally, we found in all dimeric simulations that the C-terminus of one protomer can shield the substrate-binding cleft of the other protomer (Fig. S1d). To estimate this probability, we calculated a distribution for the distance from the C-terminus to the C-alpha atoms of the catalytic residue C145. To effectively block the exit of a substrate from the substrate-binding cleft, or to block entrance to this cleft, this distance should be 10 Å or less. We found that this condition is satisfied in ∼20% of the population in this distribution. This shielding is an additional barrier for the entry to (or the release of the substrate from) the active site, suggesting that this barrier could increase the residence time in the binding cleft to increase catalytic potential. Quantum-mechanical calculations would be needed to confirm this hypothesis.
3.6. Dimer forming interactions are coupled to the catalytic residues and the accessibility of the binding cleft
Our results revealed that the primary variable differentiating active Mpro enzymes from inactive ones are the dSidechain distance and the cleft area coordinate. We identified the intrinsic collective mode of the protein that is highly correlated with dSidechain and the cleft area using the PLS-FMA machine learning algorithm (see SI) to analyze the atom-scale simulation data. We found through cross-validation studies of our simulation data that for the dSidechain coordinate, the PLS-FMA model was maximally predictive with 15 components. The collective mode composed of these 15 components is visualized in Movie S1. For the cleft area coordinate, the PLS-FMA model was optimally predictive with 5 modes and is visualized in Movie S2. Analysis of the collective modes demonstrates that the dSidechain coordinate is tightly coupled to an inter-subunit salt-bridge pair formed between the residues R4 and E290 (Fig. 5a, Movie S1). We also observed that domain 3 of the substrate-bound Mpro monomer (M+) undergoes large-scale bending motions along the longitudinal axis of the enzyme, introducing the bending angle θ as a complementary key variable for this analysis (Fig. 5b). For decreasing values of θ, one observes the salt bridges to break (Fig. 5c and d), which indicates lower ability for formation of dimers. Our results indicate that the average bending angle for M+ is much smaller (about 175 degrees) than for the dimer (∼220 degrees), and for M− the bending angle (about 205 degrees) is more consistent with the dimer.
Fig. 5.
Machine Learning model based on the dSidechain coordinate. a. Inter-chain salt-bridges between the residues R4 and E290. b. The definition of the bending angle to quantify the motion of domain 3 of Mpro. The C-terminus is shown as a curved dark green curve attached to domain 3. Color scheme used here is consistent with the color scheme in Fig. 1a. c. Distance between the side chains of the inter-subunit salt-bridge forming residues as a function of dSidechain. d. Bending angle as a function of dSidechain. e. Area of the binding site interface as a function of dSidechain. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
To understand how the salt-bridge pair as well as the substrate-binding cleft and the bending angle behave in the collective mode, we explored the behavior of all three quantities against dSidechain. Fig. 5c shows that the salt-bridge pairs break as the distance between the side chains of the catalytic residues increases. The bending angle decreases along the collective mode as dSidechain increases, retracting domain 3 away from the dimerization interface (Fig. 5d). The area of the substrate-binding cleft increases along the collective mode as dSidechain increases (Fig. 5e). As dSidechain increases linearly along the collective mode, the corresponding value of dSidechain is shown on the x-axis of Fig. 5e. Further, the bending angle was observed to be highly correlated with the potential ability of a monomer to form strong salt-bridges. The substrate-bound (M+) case demonstrated predominantly higher propensity towards broken inter-subunit salt-bridges at the dimeric interface (Fig. S5a). Its bending angle, as defined in Fig. 5b, is also smaller compared to the dimeric forms and fluctuates more than M− (Fig. S5b). Additional data (Fig. S5c and d) demonstrate the strong correlation of these inter-subunit salt-bridges with the bending angle in the two monomeric states. The dimeric state, on the other hand, shows little variation in the bending angle (Fig. S5b and d).
A complementary PLS-FMA model developed for the binding cleft area revealed concerted dynamics of inter-domain salt-bridges, shown in Fig. 6a, along its predicted collective mode. As the area of the cleft increases, one observes the formation of a salt-bridge between the residues R40 of domain 1 and D187 of domain 2. This is accompanied by the breaking of a salt-bridge between D153 and R298 of domains 2 and 3, respectively, while simultaneously forming a salt-bridge between the residues R131 and D289 of domains 2 and 3 (Fig. 6b). These rearrangements are correlated to the the dimer-forming interface, potentially strengthening the salt-bridge it can form with the E290 residue of the second chain (Fig. 6c). Interestingly, R40 sits directly in the neighborhood of the catalytic residue H41 and influences its rearrangement and formation of the catalytic dyad by affecting the dSidechain coordinate (Fig. 6d). Combined inference from Fig. 6c and d indicates that the formation of the dimeric interfacial salt-bridges assists the formation of the catalytic dyad by bringing the catalytic residues together. This collective mode (Movie S2) highlights the mechanism of transmission of information from the substrate binding site to the dimerization interface. We further verified the relevance of the salt-bridges identified in this collective mode by analysing the conservation of the residues involved in a variety of coronavirus orthologs (Fig. 6e). We found that the residues involved in the salt-bridge between domains 1 and 2 are uniformly conserved in all the sequences available from the PDB database for the Mpro enzyme. In essence, other salt-bridges also show substantial sequence conservation of the charges of the residues (>60%), indicating functional relevance.
Fig. 6.
Machine Learning model based on the binding cleft area coordinate. a. Intra-chain salt-bridges that rearrange along the collective mode. Anionic residues are shown in red and the cationic residues in blue. Domain colors for the protein correspond to the color scheme used in Fig. 1a. b. Rearrangement of the salt-bridge distance as a function of the binding cleft area. DI – DII, DII – DIII (1), DII – DIII (2), and DIII – DIII indicate a salt-bridge between the residues R40-D187 of domains 1 and 2, the residues R131-D289 of domains 2 and 3, the residues R298-D153 of domains 2 and 3, and the residues R298-D295 of domain 3, respectively. c. Distance between the side chains of the inter-subunit salt-bridge forming residues (R4 and E290) as a function of binding cleft area. d. dSidechain as a function of the binding cleft area. e. The salt-bridge forming residues of SARS-CoV-2 Mpro (R40, D187, D131, D197, D153, D295, D298) are aligned with analogous residues of other 15 coronavirus orthologs for which the structures of Mpro are available. The PDB IDs for these structures are shown in the first column and the corresponding strains are shown in the last column. Mpro orthologs were aligned with multiple sequence alignment [38]. Positively and negatively charged residues are highlighted with blue and orange colors, respectively. Identified intra-subunit salt bridges are marked using dashed lines. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
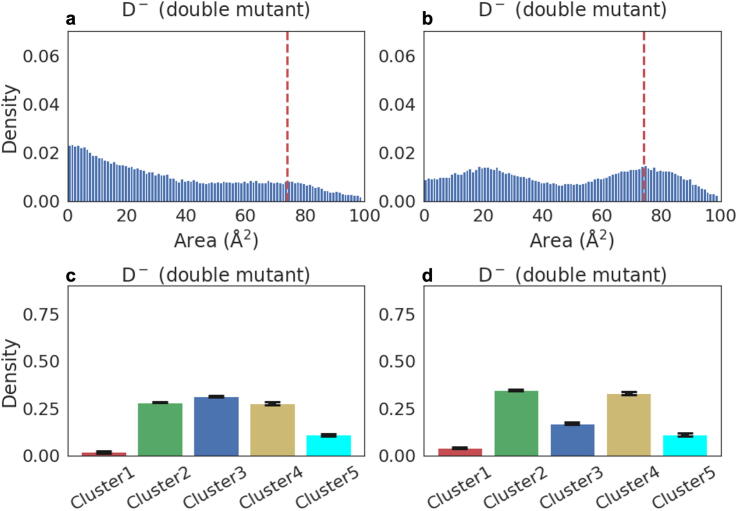
3.7. Mutations in the inter-chain salt-bridge inactivate the enzyme
The machine learning analysis of the simulation data predicts that the inter-chain R4-E290 salt-bridges are critical for the activation of the catalytic site and thereby Mpro function. To test the validity of this hypothesis, we performed additional atomistic simulations where we doubly mutated the salt-bridge forming residues to alanine in the apo dimer (D−D−). Fig. 7 shows the effects of these mutations on the cleft area and the distribution of the clusters. The cluster populations for the mutated dimer demonstrate that the population of Cluster 1, representing the active state, is drastically suppressed to 5–8%, which is the lowest value observed for any state simulated in this work. In brief, our results are consistent with the view that the impairment of the inter-chain R4-E290 salt-bridges alters the configuration of the catalytic residues and hinders the formation of the dyad structure necessary for catalysis. In silico mutations of the interfacial salt bridge residues confirm the allosteric coupling between the catalytic site and the dimer interface, as predicted by the machine learning model.
Fig. 7.
Substrate affinity and catalytic potential of the double mutant in the Mpro dimer. a,b. Distributions of the area of the substrate-binding cleft of the double mutant: chains A and B. The red dashed line depicts the maximum of the distribution of the bound monomer (Fig. 3b). c,d. The cluster populations of the double mutant: chains A and B. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
4. Discussion and conclusions
We combined extensive atomistic simulations with machine learning methods to investigate the dynamics of Mpro under conditions that describe substrate binding and dimerization. Using this approach, we developed a mechanistic model for its structure-function relationship. Our model revealed the activation mechanism of Mpro.
The analysis of structural Mpro data revealed that although a large number of different mutations have been observed for this enzyme, none of them are at the dimerization interface of Mpro, which strongly suggests that the functional state of Mpro is a dimer. The results of atomistic simulations support this view and depict a detailed picture of the mechanism of the enzyme’s function. The ML analysis unveiled that the affinity of the substrate for the substrate-binding cleft of monomeric Mpro is likely so low that the enzyme cannot use its high catalytic potential in the monomeric state. In essence, monomeric Mpro has low catalytic potential, as it cannot bind the substrate with high affinity. Meanwhile, in the dimeric structure a majority population of at least one chain exists in a form that is capable of both binding the substrate and maintaining a configuration of catalytic residues that can form an active dyad structure. Therefore, the functional state of Mpro is dimeric. The results revealed that even then the enzyme is active primarily in a state where the dimer is bound to only one substrate. This implies that the dimer is specialized for substrate retention, and the singly-bound dimeric (D+D−) form is ideally suited for catalysis.
Importantly, our results reveal that activation of Mpro requires dimerization, followed by substrate binding. Fig. 8 illustrates the model supported by our data. It postulates two main pathways for activation. In the first one, a monomer first binds to the substrate and then attempts to form a dimer. However, in this scenario, the inter-chain salt-bridges between R4 and E290 cannot be formed effectively due to large structural changes at the dimeric interface (Fig. 5a-d). Also, the bound monomer (M+) is trapped in a low-affinity state, even though it has a highly active configuration of the catalytic residues. In the second pathway, two monomers first form a dimer, after which the dimer is associated with either one or two substrates. Both monomers of the dimer have a similar high-affinity binding to the substrate as indicated by free energy calculations. However, the singly-bound dimer (D+D−) is catalytically more efficient, suggesting D+D− as the most active form.
Fig. 8.
Predicted pathways for the activation of Mpro. The primary mechanism of action is based on dimerization of Mpro monomers followed by the binding of a single substrate tightly bound to an enzyme, leading to a high-catalytic potential state (shown by green arrows). Alternatively, dimerization of Mpro monomers can be followed by the binding of two tightly bound substrates (shown by a blue arrow) but the catalytic potential of the enzyme is then less than 50% of the primary mechanism (occupancy of Cluster 1). In a different scenario, where a monomer first binds to the substrate and then tries to dimerize, there is a very active monomer that is weakly bound to the substrate (shown by a red arrow). However, in this case the salt bridges stabilizing the dimer are weakened, leading to an unstable dimer and hence low catalytic potential. The C-terminus is shown as a curved dark green curve attached to domain 3. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
These functional predictions can be tested in vitro. First, our results predict that monomeric Mpro has a low affinity for the substrate, which can be tested by, e.g., measuring the KM with Michaelis-Menten kinetics. Second, our results predict that the mutations E290A and R4A lead to an enzyme whose dimerization is low and catalysis is inefficient. On the other hand, based on experimental results already known, it can be concluded that the mechanism presented by our simulation results is justified. The E290A mutation in SARS Mpro (analogous to the E290 residue in SARS-CoV-2 Mpro) resulted in a complete loss of function [47]. In the same study, the R4A mutation (analogous to the R4 residue in Mpro of SARS-CoV-2) also leads to a significant reduction in enzymatic catalytic potential. Additionally, a drug known as x1187 was found to block the dimerization of Mpro and reduce its catalytic potential [11]. Taken together, there is strong empirical evidence that the mechanism suggested by our simulations and ML analysis can indeed translate into a therapeutic strategy.
Altogether, the simulations and the clustering analysis suggest that the catalytic potential of the enzyme is based on a collective mechanism in which the two chains of the enzyme modulate each other. This finding is in agreement with the fact that the enzyme is active only in the dimeric state and not in the monomeric state [39]. Dimerization affects the dynamics of the substrate binding sites, enhancing the affinity of the substrate for the enzyme. However, a catalytically active dimer is realized only when the substrate is bound to only one of the chains. Indeed, substrate binding to one of the chains is likely to prevent the binding of another substrate to the other chain by an anticooperative allosteric mechanism that involves blocking of the substrate-binding cleft of the latter with the C-terminus of the former.
Our model leads to an obvious question: why is the dimerization important from the point of view of the life cycle of the virus? We speculate that dimerization of Mpro acts as a switch, which allows the virus to regulate cell lysis. The virus cannot mature until it has enough monomers, and by rendering monomeric Mpro unsuitable for efficient catalysis, premature maturation is prevented. In other words, dimerization can only be enhanced when sufficient monomeric units have been synthesized from the viral mRNA, with the goal of regulating the time interval for the lysis of host cells. This model provides a general understanding of the maturation process across other viral species. Viral maturation that is too rapid will lead to a premature lysis of the host cells, leading to low viral particle numbers that are insufficient to propagate viral infection. This, in turn, can lead to activation of the host defense machinery, which can detect and effectively neutralize the few free viral particles. To prevent this, it is plausible that viruses use this dimerization mechanism to regulate the time scales of maturation for effective infection and proliferation. Preventing dimerization of the Mpro enzyme may, thus, be a means to hindering viral maturation, and can serve as a therapeutic target in drug-based methods [10] to combat COVID-19. This result has a concrete implication for the previously identified coronaviruses involved in Severe Acute Respiratory Syndrome (SARS), Middle East Respiratory Syndrome (MERS), and the closely related coronaviruses HKU5 and HKU7 [4], [41] on bat species. In all these four cases, there exists empirical evidence connecting the dimerization interface of the Mpro analogues of these enzymes with their activity, indicating a common mechanism. The strong preservation of a seemingly evolutionarily conserved mechanism of action thus forms a solid basis for future treatment of coronavirus-related diseases.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgments
We gratefully acknowledge CSC – IT Center for Science (Espoo, Finland) for providing ample computing resources.
Funding
We thank the EU (Marie Curie Skłodowska fellowship scheme, no. 101033606), the Helsinki Institute of Life Science (HiLIFE) Fellow program, Human Frontier Science Program (HFSP, project no. RGP0059/2019), Sigrid Juselius Foundation, and Academy of Finland (project ID: 335527, 331349) for financial support.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2022.06.023.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Data Availability Statement
The authors declare no competing financial interest. The TPR files used for generating the data for analysis are available on Zenodo repositories with the following address: https://doi.org/10.5281/zenodo.5758986. The python code used for the analysis is available on request.
References
- 1.Agbowuro A.A., Huston W.M., Gamble A.B., Tyndall J.D.A. Proteases and protease inhibitors in infectious diseases. Med. Res. Rev. 2018;38:1295–1331. doi: 10.1002/med.21475. [DOI] [PubMed] [Google Scholar]
- 2.Aho K., Derryberry D., Peterson T. Model selection for ecologists: The worldviews of AIC and BIC. Ecology. 2014;95:631–636. doi: 10.1890/13-1452.1. [DOI] [PubMed] [Google Scholar]
- 3.Baker N.A., Sept D., Joseph S., Holst M.J., McCammon J.A. Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc. Natl. Acad. Sci. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barrila J., Bacha U., Freire E. Long-range cooperative interactions modulate dimerization in SARS 3CLpro. Biochemistry. 2006;45:14908–14916. doi: 10.1021/bi0616302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barrila J., Gabelli S.B., Bacha U., Mario Amzel L., Freire E. Mutation of Asn28 disrupts the dimerization and enzymatic activity of SARS 3CLpro. Biochemistry. 2010;49:4308–4317. doi: 10.1021/bi1002585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng S.-C., Chang G.-G., Chou C.-Y. Mutation of Glu-166 blocks the substrate-induced dimerization of SARS coronavirus main protease. Biophys. J. 2010;98:1327–1336. doi: 10.1016/j.bpj.2009.12.4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chuck C.-P., Chong L.-T., Chen C., Chow H.-F., Wan D.-C.-C., Wong K.-B. Profiling of substrate specificity of SARS-CoV 3CL. PLoS ONE. 2010;5:e13197. doi: 10.1371/journal.pone.0013197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dai W., Zhang B., Jiang X.-M., Su H., Li J., Zhao Y., et al. Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science. 2020;368:1331–1335. doi: 10.1126/science.abb4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Darden T., York D., Pedersen L. Particle mesh Ewald: An N⋅log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 10.Drayman N., DeMarco J.K., Jones K.A., Azizi S.-A., Froggatt H.M., Tan K., et al. Masitinib is a broad coronavirus 3CL inhibitor that blocks replication of SARS-CoV-2. Science. 2021;373:931–936. doi: 10.1126/science.abg5827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.El-Baba T.J., Lutomski C.A., Kantsadi A.L., Malla T.R., John T., Mikhailov V., et al. Allosteric inhibition of the SARS-CoV-2 main protease – insights from mass spectrometry-based assays. Angew. Chem. Int. Ed. Engl. 2020;59:23544–23548. doi: 10.1002/anie.202010316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fabian P., Varoquaux G., Gramfort A., Michel V., Thirion B., Grisel O., et al. Scikit-Learn: Machine learning in Python. J. Mach. Learn. Res. 2011;12:2825–2830. [Google Scholar]
- 13.Fernández-Montero J.V., Barreiro P., Soriano V. HIV protease inhibitors: recent clinical trials and recommendations on use. Expert Opin. Pharmacother. 2009;10:1615–1629. doi: 10.1517/14656560902980202. [DOI] [PubMed] [Google Scholar]
- 14.Fiorucci D., Milletti E., Orofino F., Brizzi A., Mugnaini C., Corelli F. Computational drug repurposing for the identification of SARS-CoV-2 main protease inhibitors. J. Biomol. Struct. Dyn. 2021;39:6242–6248. doi: 10.1080/07391102.2020.1796805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fu L., Ye F., Feng Y., Yu F., Wang Q., Wu Y., et al. Both Boceprevir and GC376 efficaciously inhibit SARS-CoV-2 by targeting its main protease. Nat. Commun. 2020;11:4417. doi: 10.1038/s41467-020-18233-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.GMM. (2022). Gaussian Mixture Models — Scikit-Learn 0.23.2 Documentation. https://scikit-learn.org/stable/modules/mixture.html.
- 17.Goyal B., Goyal D. Targeting the dimerization of the main protease of coronaviruses: A potential broad-spectrum therapeutic strategy. ACS Comb. Sci. 2020;22:297–305. doi: 10.1021/acscombsci.0c00058. [DOI] [PubMed] [Google Scholar]
- 18.Hadfield J., Megill C., Bell S.M., Huddleston J., Potter B., Callender C., et al. Nextstrain: Real-time tracking of pathogen evolution. Bioinformatics. 2018;34:4121–4123. doi: 10.1093/bioinformatics/bty407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hess B. P-LINCS: A parallel linear constraint solver for molecular simulation. J. Chem. Theory Comput. 2008;4:116–122. doi: 10.1021/ct700200b. [DOI] [PubMed] [Google Scholar]
- 20.Hilgenfeld R. From SARS to MERS: Crystallographic studies on coronaviral proteases enable antiviral drug design. FEBS J. 2014;281:4085–4096. doi: 10.1111/febs.12936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hopkins C.W., Le Grand S., Walker R.C., Roitberg A.E. Long-time-step molecular dynamics through hydrogen mass repartitioning. J. Chem. Theory Comput. 2015;2015(11):1864–1874. doi: 10.1021/ct5010406. [DOI] [PubMed] [Google Scholar]
- 22.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 23.Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., et al. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582:289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 24.Jin Z., Zhao Y., Sun Y., Zhang B., Wang H., Wu Y., et al. Structural basis for the inhibition of SARS-CoV-2 main protease by antineoplastic drug carmofur. Nat. Struct. Mol. Biol. 2020;27:529–532. doi: 10.1038/s41594-020-0440-6. [DOI] [PubMed] [Google Scholar]
- 25.Jo S., Kim T., Iyer V.G., Im W. CHARMM-GUI: A web-based graphical user interface for CHARMM. J. Comput. Chem. 2008;29:1859–1865. doi: 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- 26.Joung I.S., Cheatham T.E., 3rd Determination of alkali and halide monovalent ion parameters for use in explicitly solvated biomolecular simulations. J Phys Chem B. 2008;112(30):9020–9041. doi: 10.1021/jp8001614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krivobokova T., Briones R., Hub J.S., Munk A., de Groot B.L. Partial least-squares functional mode analysis: Application to the membrane proteins AQP1, Aqy1, and CLC-ec1. Biophys. J. 2012;103:786–796. doi: 10.1016/j.bpj.2012.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kumari R., Kumar R. Open source drug discovery consortium, & Lynn, A. g_mmpbsa – A GROMACS tool for high-throughput MM-PBSA calculations. J. Chem. Inf. Model. 2014;54:1951–1962. doi: 10.1021/ci500020m. [DOI] [PubMed] [Google Scholar]
- 29.Li C., Qi Y., Teng X., Yang Z., Wei P., Zhang C., et al. Maturation mechanism of severe acute respiratory syndrome (SARS) coronavirus 3C-like proteinase. J. Biol. Chem. 2010;285:28134–28140. doi: 10.1074/jbc.M109.095851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lindahl E., Abraham M.J., Hess B., van der Spoel D. GROMACS 2020 Source code. Zenodo. 2020 doi: 10.5281/zenodo.3562495. [DOI] [Google Scholar]
- 31.Liu S., Zheng Q., Wang Z. Potential covalent drugs targeting the main protease of the SARS-CoV-2 coronavirus. Bioinformatics. 2020;36:3295–3298. doi: 10.1093/bioinformatics/btaa224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma C., Sacco M.C., Hurst B., Townsend J.A., Hu Y., Szeto T., et al. Boceprevir, GC-376, and calpain inhibitors II, XII inhibit SARS-CoV-2 viral replication by targeting the viral main protease. Cell Res. 2020;30:678–692. doi: 10.1038/s41422-020-0356-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maier J.A., Martinez C., Kasavajhala K., Wickstrom L., Hauser K.E., Simmerling C. ff14SB: Improving the accuracy of protein side chain and backbone parameters from ff99SB. J Chem Theory Comput. 2015;11:3696–3713. doi: 10.1021/acs.jctc.5b00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.PyMOL. (2022). The PyMOL Molecular Graphics System, Version 1.2r3pre, Schrödinger, LLC.
- 35.Riva L., Yuan S., Yin X., Martin-Sancho L., Matsunaga N., Pache L., et al. Discovery of SARS-CoV-2 antiviral drugs through large-scale compound repurposing. Nature. 2020;586:113–119. doi: 10.1038/s41586-020-2577-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rowe I.A., Mutimer D.J. Protease inhibitors for treatment of genotype 1 hepatitis C virus infection. BMJ. 2011;343:d6972–d. doi: 10.1136/bmj.d6972. [DOI] [PubMed] [Google Scholar]
- 37.Shi T.-H., Huang Y.-L., Chen C.-C., Pi W.-C., Hsu Y.-L., Lo L.-C., et al. Andrographolide and its fluorescent derivative inhibit the main proteases of 2019-nCoV and SARS-CoV through covalent linkage. Biochem. Biophys. Res. Commun. 2020;533:467–473. doi: 10.1016/j.bbrc.2020.08.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sievers F., Wilm A., Dineen D., Gibson T.J., Karplus K., Li W., et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Silvestrini L., Belhaj N., Comez L., Gerelli Y., Lauria A., Libera V., et al. The dimer-monomer equilibrium of SARS-CoV-2 main protease is affected by small molecule inhibitors. Sci. Rep. 2021;11:9283. doi: 10.1038/s41598-021-88630-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Świderek K., Moliner V. Revealing the molecular mechanisms of proteolysis of SARS-CoV-2 Mpro by QM/MM computational methods. Chem. Sci. 2020;11:10626–10630. doi: 10.1039/d0sc02823a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tomar S., Johnston M.L., St John S.E., Osswald H.L., Nyalapatla P.R., Paul L.N., et al. Ligand-induced dimerization of Middle East Respiratory Syndrome (MERS) coronavirus nsp5 protease (3CLpro): Implications for nsp5 regulation and the development of antivirals. J. Biol. Chem. 2015;290:19403–19422. doi: 10.1074/jbc.M115.651463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Trezza A., Iovinelli D., Santucci A., Prischi F., Spiga O. An integrated drug repurposing strategy for the rapid identification of potential SARS-CoV-2 viral inhibitors. Sci. Rep. 2020;10:13866. doi: 10.1038/s41598-020-70863-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ullrich S., Nitsche C. The SARS-CoV-2 main protease as drug target. Bioorg. Med. Chem. Lett. 2020;30 doi: 10.1016/j.bmcl.2020.127377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vuong W., Khan M.B., Fischer C., Arutyunova E., Lamer T., Shields J., et al. Feline coronavirus drug inhibits the main protease of SARS-CoV-2 and blocks virus replication. Nat. Commun. 2020;11:4282. doi: 10.1038/s41467-020-18096-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang H., He S., Deng W., Zhang Y., Li G., Sun J., et al. Comprehensive insights into the catalytic mechanism of middle east respiratory syndrome 3C-like protease and severe acute respiratory syndrome 3C-like protease. ACS Catal. 2020;10:5871–5890. doi: 10.1021/acscatal.0c00110. [DOI] [PubMed] [Google Scholar]
- 46.Wang Y.-C., Yang W.-H., Yang C.-S., Hou M.-H., Tsai C.-L., Chou Y.-Z., et al. Structural basis of SARS-CoV-2 main protease inhibition by a broad-spectrum anti-coronaviral drug. Am. J. Cancer Res. 2020;10:2535–2545. [PMC free article] [PubMed] [Google Scholar]
- 47.Wei P., Fan K., Chen H., Ma L., Huang C., Tan L., et al. The N-terminal octapeptide acts as a dimerization inhibitor of SARS coronavirus 3C-like proteinase. Biochem. Biophys. Res. Commun. 2006;339:865–872. doi: 10.1016/j.bbrc.2005.11.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.WHO coronavirus disease (COVID-19) dashboard (2022). https://covid19.who.int.
- 49.Jorgensen W.L., Chandrasekhar J., Madura J.D., Impey R.W., Klein M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 50.Xue, X., Yu, H., Yang., H., Xue, F., Wu., Z., Shen, W., Li, J., Zhou, Z., Ding, Y., Zhao, Q., Zhang, X. C., Liao, M., Bartlam, M., and Rao, Z. (2008). Structures of two coronavirus main proteases: implications for substrate binding and antiviral drug design. J. Virol. 82. 2515–2527. [DOI] [PMC free article] [PubMed]
- 51.Zhang L., Lin D., Kusov Y., Nian Y., Ma Q., Wang J., et al. α-ketoamides as broad-spectrum inhibitors of coronavirus and enterovirus replication: Structure-based design, synthesis, and activity assessment. J. Med. Chem. 2020;63:4562–4578. doi: 10.1021/acs.jmedchem.9b01828. [DOI] [PubMed] [Google Scholar]
- 52.Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., et al. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020;368:409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare no competing financial interest. The TPR files used for generating the data for analysis are available on Zenodo repositories with the following address: https://doi.org/10.5281/zenodo.5758986. The python code used for the analysis is available on request.