Abstract
RATIONALE
Tandem-ion mobility spectrometry/mass spectrometry methods have recently gained traction for the structural characterization of proteins and protein complexes. However, ion activation techniques currently coupled with tandem-ion mobility spectrometry/mass spectrometry methods are limited in their ability to characterize structures of proteins and protein complexes.
METHODS
Here, we describe the coupling of the separation capabilities of tandem-trapped ion mobility spectrometry/mass spectrometry (tTIMS/MS) with the dissociation capabilities of ultraviolet photo-dissociation (UVPD) for protein structure analysis.
RESULTS
We establish feasibility of dissociating intact proteins by UV irradiation at 213 nm between the two TIMS devices in tTIMS/MS and at pressure conditions compatible with ion mobility spectrometry (2–3 mbar). We validate that the fragments produced by UVPD under these conditions arise from a radical-based mechanism in accord with prior literature on UVPD. The data suggest stabilization of fragment ions produced from UVPD by collisional cooling due to the elevated pressures used here (“UVnoD2”), which otherwise do not survive to detection. The data account for a sequence coverage for the protein ubiquitin comparable to recent reports, demonstrating the analytical utility of our instrument in mobility-separating fragment ions produced from UVPD.
CONCLUSIONS
The data demonstrate that UVPD carried out at elevated pressures of 2–3 mbar yields extensive fragment ions rich in information about the protein, and that their exhaustive analysis requires IMS separation post-UVPD. Hence, because UVPD and tTIMS/MS each have been shown to be valuable techniques on their own merit in proteomics, our contribution here underscores the potential of combining tTIMS/MS with UVPD for structural proteomics.
Introduction
Ion activation methods have long been coupled with mass spectrometry to probe the structure of ions.1 Tandem mass spectrometry (MS/MS) separated the ionization event from the activation of the ions, which allowed coupling with a variety of activation methods.2 Early activation methods explored in MS/MS focused on increasing the internal energy of ions by energetic collisions with neutral collision gas particles. Collisional-activation of ions can be achieved in a variety of mass spectrometers, including quadrupole, time-of-flight, or ion cyclotron resonance mass analyzers.3,4 To this day, tandem-mass spectrometry by means of collision-induced dissociation (CID) remains a widely used approach to produce fragment ions of peptides in order to identify proteins in the field of proteomics.3
Collisional activation, however, is less efficient for sequence analysis of large biological ions, such as intact proteins or protein complexes. Top-down sequence and structure analysis of intact proteins and protein complexes is often more efficiently accomplished by alternative activation methods, such as surface-induced dissociation (SID),5 electron-based dissociation (electron transfer/capture dissociation),6,7 or ultraviolet photodissociation (UVPD).8–10 These methods, often used in conjunction with each other and/or CID to produce complementary fragment ions, have enabled top-down sequencing and structure characterization of proteins and protein complexes.11–13 Crucially, these methods often preserve post-translational modifications of peptides and proteins,8,14–16 which is ascribed to differences in the timescale and molecular mechanism of the dissociation reaction relative to CID. These activation methods thus allow characterization of differentially modified proteins (proteoforms). Moreover, these methods have also enabled the characterization of the subunit architecture and topology of protein complexes,5,7,17 which is challenging by collisional-activation methods alone.
UVPD in particular has proven to be a very versatile tool to study the primary, tertiary, and quaternary structures of proteins and protein complexes.9,10 UVPD begins when the ion absorbs UV light, producing an electronically excited state of the ion.18–20 The electronically excited ion then may undergo a variety of relaxation processes, or it may dissociate. The photodissociation mechanism of small peptides in the gas-phase has been extensively studied21–23 and UVPD has been comprehensively exploited analytically to identify sequences of peptides,8 glycopeptides,24 or carbohydrates,25 including glycosaminoglycans.26 Notably, UVPD can fragment the peptide backbone of glycopeptides without breaking the peptide-glycan bond.14,24 This ability to dissociate the protein backbone while preserving otherwise volatile post-translational modifications takes on increased relevance in the light of the ongoing pandemic because viral envelope proteins, such as the S protein of SARS-CoV-2, are typically heavily glycosylated.27,28
UVPD has previously been coupled with ion mobility spectrometry/mass spectrometry (IMS/MS).29–37 IMS/MS determines the momentum transfer cross section of an ion from measuring the velocity it acquires when traversing a gas-filled chamber under the influence of an electric field.38 The benefit of IMS is that the measured cross section is directly related to the atomic structure of the ion, thereby enabling structurally selective UVPD measurements. Various implementations have been reported; for example, UVPD was performed in quadrupolar ion traps/guides attached to the exit of drift tubes.34,35 Alternatively, UVPD was coupled with traveling wave IMS/MS instrumentation; here, UVPD was carried out in both the pre-IMS “trap” cell36,37 and also in the post-IMS “transfer” cell30–32 of the “triwave” of a Synapt G2. The benefit of performing ion mobility separation upstream of UVPD is that this enables conformationally-selective UVPD experiments. By contrast, the benefit of performing ion mobility separation downstream of UVPD is that fragment ions produced from UVPD are mobility-separated, which facilitates fragment identification by decongesting the resulting individual mass spectra.
In this Communication, we introduce coupling the separation capabilities of tandem-trapped ion mobility spectrometry / mass spectrometry (tTIMS/MS)39 with the dissociation capabilities of UVPD. Further, we validate feasibility of dissociating intact proteins at 2–3 mbar by irradiation with 213 nm UV light produced from a Nd:YAG laser. The construction of our novel, UVPD-capable tandem-TIMS (tTIMS/MS-UVPD) instrument was based upon experiences with our prior tandem-TIMS instrument obtained through its application to the study of peptide oligomers, and native-like structures of proteins and their complexes.39–43 The tTIMS/MS-UVPD instrument described here combines the benefit of conformer-selection of the precursor ion prior to UVPD with mobility-separation of the fragment ions produced by UVPD. Furthermore, the instrument described here is constructed from a commercial timsTOFPro instrument and therefore enables parallel accumulation/serial fragmentation (PASEF) workflows44 for the fragment ions generated from UVPD. Hence, this instrument appears highly promising for top-down analysis of proteins and protein complexes. In the following, we (1) describe the coupling of the separation capabilities of tandem-trapped ion mobility spectrometry/mass spectrometry (tTIMS/MS) with the dissociation capabilities of ultraviolet photodissociation (UVPD); (2) establish feasibility of dissociating intact proteins by UV irradiation at 213 nm and pressure conditions compatible with ion mobility spectrometry (2–3 mbar); (3) validate that the fragments produced by UVPD are produced from a radical-based mechanism in accord with prior literature on UVPD; (4) obtain a sequence coverage of approximately 40% for the protein ubiquitin which is comparable to recent reports coupling high resolution mass spectrometers with UVPD at 213 nm45,46 and (5) demonstrate the analytical potential of our instrument for top-down protein analysis by mobility-separation of fragment ions produced from UVPD.
Experimental Details
Materials and Sample preparation.
Ubiquitin from bovine erythrocytes (≥98 %) and water (LC/MS grade) were obtained from Sigma-Aldrich (St. Louis, MO). Methanol (LC/MS grade) and acetic acid (glacial) were obtained from Fisher Scientific (Pittsburgh, PA). High-concentration ESI tuning mix were obtained from Agilent (Santa Clara, CA). For trapping experiments, an aqueous ubiquitin solution with 1 v% acetic acid and ~10 μM was prepared. For UVPD and CID measurements, ubiquitin samples (~10 μM) were prepared in 1:1 methanol/water solution with 1 v % acetic acid. ESI tuning mix was used as obtained for ion mobility calibration.
Calibration and Data Analysis.
Collision cross sections in tTIMS are determined via a calibration procedure described elsewhere.47–49 Calibration was performed by using cross sections reported by Stow el at.50 for perfluorophosphazenes ions contained in the ESI tuning mix as described.48 All cross sections are reported for nitrogen gas. Data analysis of the mass and mobility spectra were performed using Compass DataAnalysis version 5.1 (Bruker Daltonics, Billerica, MA). We used two different approaches to identify fragment ions in the CID and UVPD spectra. First, we identified monoisotopic peaks of the fragment ions using the SNAP algorithm implemented in Compass DataAnalysis, followed by comparison of the obtained m/z to the theoretical m/z predicted by ProteinProspector (UCSF, San Francisco, CA). Assignments were made with an error tolerance of 10 ppm. Peaks were additionally annotated by manually comparing isotopic patterns observed in the experiment to isotopic patterns calculated for fragment ions predicted by ProteinProspector. All fragment ion types (a, b, c, x, y, z, including their +/− 1 and +/− 2 congeners, and d, v, w, ions due to side-chain losses) including their neutral loss and adduct ions were considered for fragment ion assignment.
Trapping of mobility selected native-like ions.
Ubiquitin ions were electrosprayed from an aqueous solution (~10 μM) with 1 v% acetic acid, and mobility separated in TIMS-1 by linear ramping of the potential at the entrance of the mobility analysis region from −217 V to −213 V while keeping the potential at the exit funnel at −100 V. Ubiquitin 7+ ions were mobility selected by timing the potential at the L2 lens; for details see elsewhere.39 Subsequently, mobility selected ions were trapped by applying −130 V at V3 and −120 V at V2 in the ion trap for 0, 90, 410, and 910 ms, respectively. Next, ions were eluted from the ion trap by lowering the potential on V3 and accumulated in the TIMS-2 analyzer for 100 ms. Ions were mobility analyzed in TIMS-2 by linear ramping of the potential at the entrance of the mobility analysis region from −110 V to −10 V at a rate of 1.0 V ms-1. RF peak-to-peak amplitudes in TIMS-1, ion trap, and in TIMS-2 were set to ~330 V, ~330 V, and ~220 V, respectively. RF frequencies of 464 kHz, 795 kHz, and 439 kHz were applied to TIMS-1, the ion trap, and TIMS-2, respectively. The pressures were set as follows; p1=3.54 mbar, p2=2.51 mbar, p3=2.17 mbar, p4=0.95 mbar.
Collision-induced dissociation.
CID was performed between the accumulation region and the mobility analysis region inside the TIMS-2 analyzer in accord with a recent report.51 We electrosprayed ubiquitin from a MeOH:H2O solution (see above). Gradually decreasing dc voltages were set throughout TIMS-1 and at L1/L2/L3 lenses to transmit ions into the ion trap. For direct comparison to UVPD experiments, ions were stored for ~450 ms in the ion trap. Subsequently, ions were eluted from the ion trap and accumulated in the TIMS-2 analyzer for 100 ms. We applied 150 V between the accumulation and mobility analysis regions to perform CID of the ions. Finally, the ions are mobility analyzed in TIMS-2 by linear ramping of the potential at the entrance of the mobility analysis region from −170 V to 20 V at a rate of 1.9 V ms-1. RF peak-to-peak voltages in TIMS-1, multipole ion trap, and in TIMS-2 were set to ~250 V, ~180 V, and ~180 V. RF frequencies and gas pressures used are identical to the trapping experiments described above.
Results and Discussion
In the following, we (1) describe the coupling of the separation capabilities of tandem-TIMS/MS with the dissociation capabilities of UVPD; (2) demonstrate feasibility of dissociating intact proteins by UV irradiation at 2–3 mbar; and (3) discuss the analytical potential of coupling tandem-TIMS with UVPD for structural proteomics.
In most prior applications, UVPD is carried out at comparatively high vacuum of the HCD or C-trap of an Orbitrap instrument or in an ion cyclotron resonance (ICR) cell. Even when coupling UVPD with ion mobility spectrometry, the UV irradiation event is most often carried out separately from the ion mobility device in a trap that operates at much lower pressure relative to the ion mobility separation.30–32,34–37 The pressure conditions likely significantly impact the photoactivation of ions. In the collision-less environment at high vacuum, the ions are expected to undergo sequential dissociations, as the ions lack a collisional relaxation pathway. One might thus expect that many fragile (radical) fragment ions or labile PTMs do not survive to detection. Conversely, at more elevated pressures compatible with ion mobility spectrometry, such fragile species could be stabilized due to efficient collisional cooling. At such pressure regimes, the ion-neutral collision frequency is at least ~2–4 orders of magnitude higher than under typical UVPD conditions in an Orbitrap or ICR cell. Hence, it can be expected that vibrational-translational energy transfer due to ion-neutral collisions deactivates the UV-photoactivated ions significantly more efficiently under these elevated-pressure conditions. While this may in theory offer benefits in terms of stabilizing labile groups on UVPD product ions, it remains unclear if UVPD carried out at these pressures yields complementary information to CID tandem mass spectra, and thus whether the technique is analytically useful. Indeed, while photoactivation of small molecules by irradiation of visible light at 488 nm was reported for a tandem-IMS instrument at ~10 mbar,33 the feasibility of carrying out UVPD experiments of intact proteins at these pressure regimes has not yet been established.
This raises the question what wavelengths and what light sources might be best suited for these types of experiments. Multiple wavelengths have been used for UVPD studies; wavelengths greater than 240 nm require specific chromophores to absorb the UV photons, for example the π- π* transitions αof aromatic rings for wavelengths of 266 nm.52 By contrast, wavelengths shorter than ~200–210 nm are largely absorbed by the amide backbone. It is generally agreed that the maximum absorbance for proteins in the condensed phase takes place around 190 nm, due to the strong σ-σ* absorbance of the peptide bond.53 Hence, UVPD carried out at 193 nm and 157 nm demonstrated extensive sequence coverage for top-down protein sequence analysis irrespective of precursor charge state.10 Nevertheless, both 157 and 193 nm UV photons are produced from Excimer lasers and require hazardous gases. Additionally, both wavelengths are generally referred to as vacuum-UV and requiring the entire laser system to be set up in vacuo to minimize power loss. It should be noted that the absorbance band of the peptide bond is rather broad, and thus for example 205 nm is often chosen as a more convenient absorption wavelength for concentration measurements in solution.54 Similarly, in gas-phase work, it has been shown that the absorption cross section at 210 nm is very appreciable.19 UVPD experiments of proteins at 213 nm thus combine the high absorption cross section of the protein backbone, while minimizing absorption by atmospheric gases, and are thus conducive to elevated pressure conditions compatible with ion mobility spectrometry. Further, 213 nm photons are typically produced by higher harmonics of solid state Nd:YAG lasers, facilitating implementation of UVPD at 213 nm.
Orthogonal tandem-TIMS instrument coupled with UVPD
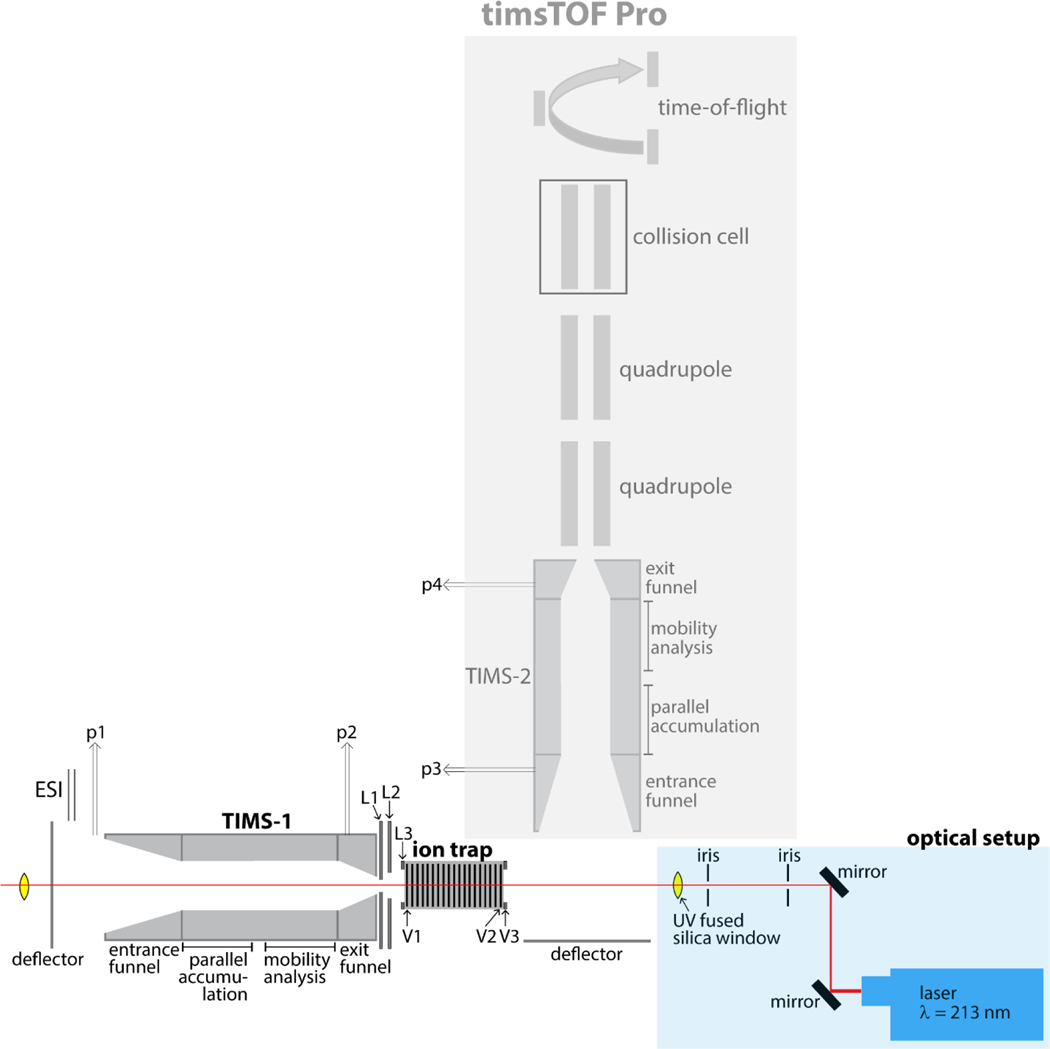
Figure 1 shows the schematics of our newly constructed orthogonal tandem-TIMS/MS instrument and the coupling with a UV laser. As shown in the Figure, we modified a commercially available timsTOF Pro instrument (Bruker Daltonics, Billerica, MA) by (1) incorporating an additional TIMS device between the electrospray capillary and the one already present in the timsTOF Pro; (2) inserting a linear ion trap operating at 2–3 mbar in-between the two TIMS devices; and (3) incorporating a 213 nm laser beam produced from the 5th harmonic of a Nd:YAG laser. Gas pressures are monitored by capacitance manometers. More details on the timsTOF pro,44,55 differential pumping, ion gating and ion activating between two TIMS devices can be found elsewhere.39,41,42
Figure 1. Schematics of the newly constructed tTIMS/MS instrument.
A commercially available timsTOF Pro instrument (Bruker Daltonics, Billerica, MA) is modified by (1) incorporating an additional TIMS device (TIMS-1) in-between the electrospray capillary and the TIMS device present in the timsTOF Pro (TIMS-2); the two TIMS devices are identical; (2) inserting a linear ion trap operating at 2–3 mbar in-between the two TIMS devices; and (3) incorporating a 213 nm laser beam produced from the 5th harmonic of a Nd:YAG laser. Gating of ions mobility-separated in TIMS-1 is controlled by timing voltages placed on L1 – L3. Storing and releasing ions in/from the linear ion trap is accomplished by timing voltages on V1 – V3.
The linear ion trap located in-between the TIMS devices is constructed from 75 printed circuit boards (PCBs) supporting segmented quadrupolar electrodes spaced 1.6mm apart. A quadrupolar electric field is generated between adjacent plates through the application of two radio frequency (rf) phases shifted by 180 degrees. All PCBs are driven via a single rf generator running at 795 kHz and up to 300 Vpp. The electrodes are resistively coupled and a direct-current (dc) electric field across the trap is created by placing an electric potential on the first (V1) and last (V2) electrodes of the trap. Trapping and ejecting of ions is controlled by an additional electrode (V3); ion trapping occurs for V2 < V3 whereas ions are ejected from the trap for V2 > V3.
To enable UVPD experiments, two UV fused silica glass windows with transmission between 185 nm – 2.1 μm were incorporated (VPCH42, Thorlabs, Newton, NJ); the laser beam enters the window proximal to the deflector region of TIMS-2 and exits through the window at the deflector region of TIMS-1 (see Figure 1). UV photons at 213 nm are created by means of the 5th harmonic of a diode-pumped solid state (DPSS) Q-switched nanosecond Nd:YAG laser (NL204, EKSPLA, Vilnius, Lithuania). The laser operates at a repetition rate of up to 1 kHz with an energy of up to 0.2 mJ per pulse. The beam width is approximately 0.7 mm in diameter with a beam divergence of <3 mrad. The beam path through the instrument is controlled via two dielectric coated mirrors for Nd:YAG laser use (213 nm, CVI Laser, Albuquerque, NM) as indicated in Figure 1. UVPD is performed on ions stored in the linear ion trap between TIMS-1 and TIMS-2. The tTIMS/MS settings used for UVPD experiments are identical to those used for CID described under Experimental Details (with the exception that 5 V was applied between the accumulation and mobility analysis region in TIMS-2 instead of 150 V used for CID). To demonstrate feasibility of conducting UVPD of intact proteins using this setup and the applied conditions, the trapped ubiquitin ions were irradiated with ~450 laser pulses (450 ms trap time) at a pulse energy of ~0.2 mJ prior to mobility-analysis in TIMS-2.
Softness of the ion trap enabling UVPD in the orthogonal tTIMS/MS
Our long-term goal is to further the ability to elucidate the solution structure of proteins and protein complexes by ion mobility spectrometry/mass spectrometry methods. Hence, for this purpose, the primary figure of merit of our instrument is the ability to retain native-like structures of proteins and protein complexes throughout the tandem-TIMS/MS-UVPD instrument. The UV photoactivation is carried out on proteins stored in the linear ion trap located in-between the two TIMS analyzers (see Figure 1). The purpose of the linear ion trap is to store the ions for a timescale sufficiently long so that UV irradiation produces sequence-informative fragment ions from the trapped proteins. Prior work coupling IMS/MS instruments with UVPD conducted trapping/UV irradiation on time-scales ranging up to 1 s.31,36 Hence, two aspects of the instrument are most critical for our purposes: 1) both TIMS devices must be capable of preserving native-like structures when operated sequentially; and 2) the trap must be able to store ions for time-scales conducive for UVPD measurements (i.e. up to ~1 s) without denaturing the native-like conformation due to ion heating. Thus, we assess the capability of our instrument in retaining native-like ubiquitin structures by mobility-separating ubiquitin ions in TIMS-1, followed by trapping of mobility-selected charge state 7+ (see above) in the linear ion trap between 0 ms (transmission) and 910 ms, followed by mobility-separation in TIMS-2.
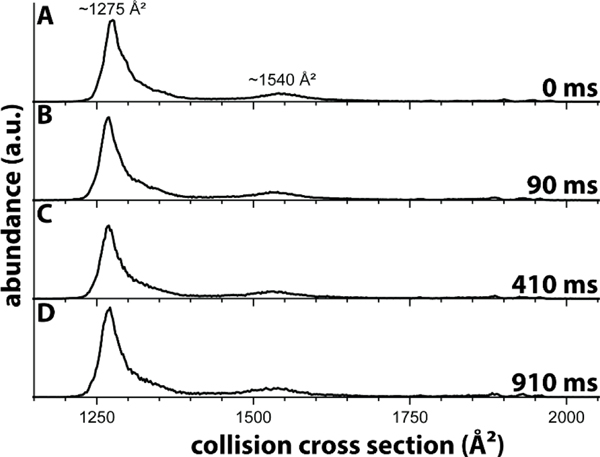
Figure 2 shows the ion mobility spectrum recorded for mobility-selected charge state 7+ after storage in the linear ion trap for up to 910 ms. Consistent with previously reported spectra for ubiquitin in nitrogen buffer gas,40,56–58 the spectrum recorded without trapping (Figure 2a) shows a main feature at ~1275 Å2 and a minor, broad feature around 1540 Å2. As discussed,40,57 the feature with a cross section centered at ~1275 Å2 corresponds to largely natively folded ubiquitin ions whereas the minor broad feature centered at ~1540 Å2 corresponds to partially unfolded ubiquitin ions. This observation underscores the “softness” of our orthogonal tandem-TIMS instrument based on the commercial TIMS platform. These data further demonstrate that timsTOFPro instruments are well-suited for structural studies of protein systems with instrument settings that minimize collisional activation of the ions.
Figure 2. Ion mobility spectrum for charge state 7+ of ubiquitin electrosprayed from a native solution.
The cross sections are consistent with presence of mainly natively folded ubiquitin ions. The data thus corroborate that the instrument described here is able to store proteins in the trap for up to at least ~1 s without significantly denaturing their structures.
Figures 2b to 2d show the ion mobility spectra recorded for charge state 7+ after trapping for 90 ms to 910 ms. The plots reveal that 1) the number of features in the ubiquitin spectra remains constant; 2) abundances at around 2000 Å2, indicative of unfolded ubiquitin ions, are negligible; 3) the peaks remain centered at around 1275 Å2 and 1540 Å2; 4) the relative abundances of the two peaks remains approximately constant. Unfolding of the ubiquitin ions would have manifested itself by the emergence of features with cross sections centered at around 2000 Å2. Thus, taken together, the spectra plotted in Figure 2 show that we are operating our newly constructed orthogonal tTIMS/MS, including the ion trap, in a manner that largely retains the native structure of ubiquitin for up to at least ~1 s. This result is important, because a prior report coupling an ion trap with a drift tube ion mobility spectrometer observed unfolding of ubiquitin charge state 7+ after ~40 ms,59 probably due to rf heating in the trap.60
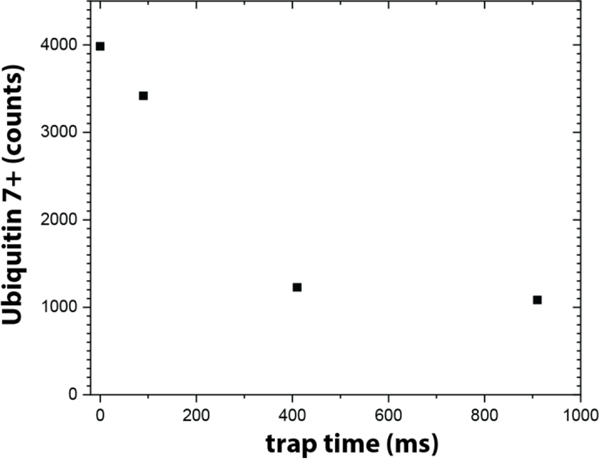
Figure 3 plots the abundances of ubiquitin ions detected when the ion trapping time is varied from ~0 ms to ~910 ms. Assuming that ion loss outside of the ion trap is negligible, these numbers correspond roughly to the number of ubiquitin ions that are retained in the linear ion trap for the duration of the trapping time. The data show that storage for ions up to approximately 1 s is possible but that the trapping efficiency decreases from ~85% after 100 ms to about 25%.
Figure 3. Storage efficiency of the ion trap.
Storage for intact protein ions up to approximately 1 second is possible in the current implementation of the ion trap. Note that ion storage at pressures >1 mbar is presently not yet well-understood and that we are currently optimizing the storage efficiency trapping of our trap by e.g. varying the RF amplitude and frequency.
While this may at first glance not appear to reflect adequate storage efficiency, it is important to consider the context. First, ion trapping at pressures >1 mbar is largely unexplored (typically, ion trapping is conducted at pressure regimes lower than 10−3 or 10−2 mbar).61 What is known is that the confining effective potential is strongly dampened at these pressures in a manner that depends both on the pressure and the mobility of the stored ions,62,63 in addition to the m/z, and the amplitude and frequency of applied RF potential.64 Hence, it is not yet fully understood how to most efficiently store ions at the 2–3 mbar regime compatible with our tandem-TIMS experiments. Second, while ion storage at ~5 mbar without significant loss for ~5 h was reported,61 this pertains to small organic compounds without consideration of ion heating or structural changes that might result therefrom in intact proteins. By contrast, our data here document the ability of storing intact ubiquitin 7+ at 2–3 mbar such that the structure of the protein does not change within ~1 s. Third, we are currently in the process of optimizing the performance of the trap by testing different amplitudes and frequencies of the applied rf voltage. Indeed, the fact that our current settings are sufficiently “soft” to retain largely the native ubiquitin structure for up to at least 910 ms (Figure 2) suggest that the RF amplitude and frequency used here (300 Vpp and 795 kHz, respectively) can still be varied to optimize the trapping efficiency without compromising “softness” (i.e. without denaturing the protein structure due to ion heating) for the 100 ms to 1 s time-scale of the UV irradiation. Moreover, for non-native top-down protein analysis, structural denaturation of the intact protein is not a concern and hence storage efficiency for extended periods of time can be straightforwardly improved by increasing the RF amplitude.
Feasibility of tTIMS/MS-UVPD of intact ubiquitin
We demonstrate proof-of-principle of conducting UVPD measurements of proteins trapped in the linear ion trap of tTIMS/MS by irradiation of 213 nm photons of the 8.6 kDa protein ubiquitin. To this end, we electrospray ubiquitin and compare the fragment ion spectrum obtained from UVPD in the ion trap to the fragment ion spectrum produced from CID.
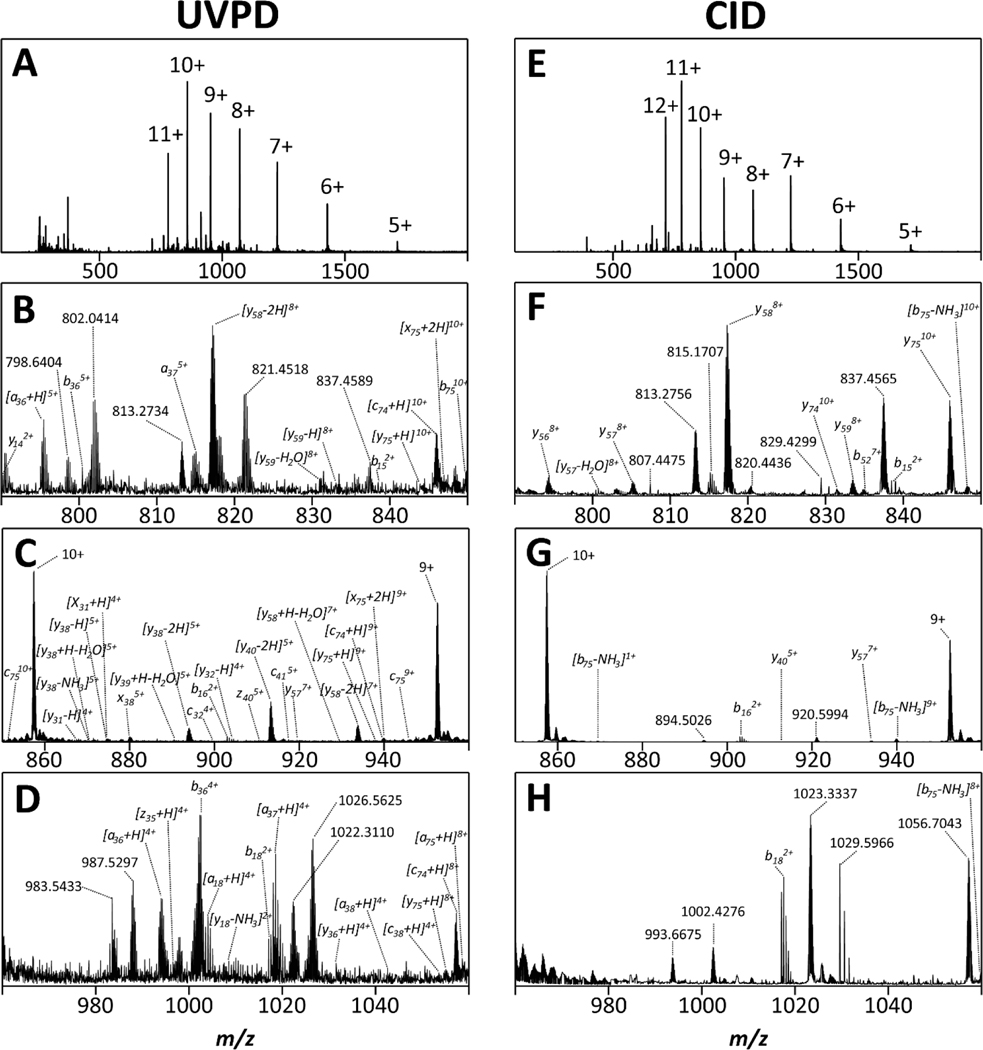
Figure 4 shows the mass spectrum recorded for ubiquitin after UV irradiation for ~450 ms in the ion trap (Figures 4a-d) and those produced from CID (Figures 4e-h). Figures 4a and 4e reveal that both fragmentation methods lead to substantial formation of fragment ions. We compare the nature of fragment ions produced by UVPD to those produced by CID in Figures 4b-d (UVPD) and Figures 4f-h (CID). We emphasize two observations:
Figure 4. Comparison of mass spectra produced by UVPD (A-D) and CID (E-H) of the protein ubiquitin.
The data reveal abundant formation of fragment ions after UV irradiation of ubiquitin ions stored in the trap, thereby demonstrating feasibility of conducting UVPD experiments at 2–3mbar using the instrument described in this work.
First, the data show that irradiation of ubiquitin ions in the trap of the tandem-TIMS instrument with UV photons clearly resulted in dissociation of the intact protein into fragment ions. This observation demonstrates that our newly constructed instrument enables UVPD of proteins stored in the ion trap operated at an elevated pressure of 2–3 mbar. Importantly, this shows that UVPD can be achieved at elevated pressures compatible with IMS.
Second, the data reveal that UVPD and CID produce fragment ions with different m/z. This observation corroborates that the dissociation mechanisms of CID and UVPD in our instrument differ. In accord with prior literature,63 we are unable to identify a large fraction of the fragment ions produced by UVPD despite considering all types of ions that are commonly observed (a, b, c, x, y, z, including the common +/− 1, +/− 2 congeners, and d, v, w, ions due to side-chain losses) including their neutral loss and adduct ions.
The analysis of our data suggests that the majority of fragment ions are y-type ions and that a, b, c-type ions are of similar frequency (Figure 5A). By contrast, prior studies conducted at lower pressures reported that a-type ions are typically more prominent in prior reports than y-type63 or b, c-type ions. This observation could point to differences in the dissociation mechanisms when UVPD is conducted at pressure regimes of 2–3 mbar instead of at the lower pressures prevalent in HCD/c-trap cells of an Orbitrap or in an ICR. Additionally, it appears plausible that the elevated pressure conditions used here would stabilize fragile (radical) species due to collisional cooling (“UVnoD2”) so that they survive to detection, whereas these fragile species would not survive to detection when UVPD is conducted under low-pressure conditions where no such collisional-cooling occurs. Nevertheless, the fragmentation pattern observed for ubiquitin (Figure 5B) shows that the assigned fragment ions account for a sequence coverage of 40% which is comparable to recent reports on high-resolution mass spectrometers coupled with UVPD at 213 nm.45,64
Figure 5. Counts for identified fragment ion types (A) and fragmentation map (B) obtained for the protein ubiquitin by UVPD at 213 nm.
(A) Number of assigned fragment ions classified according to their ion type; y-type ions predominate our assignments and a, b, c-type ions appear of similar frequency. (B) The analysis of the fragment ions account for a sequence coverage of approximately 40%, which is comparable to recent studies coupling high resolution mass spectrometers with UVPD at 213 nm.
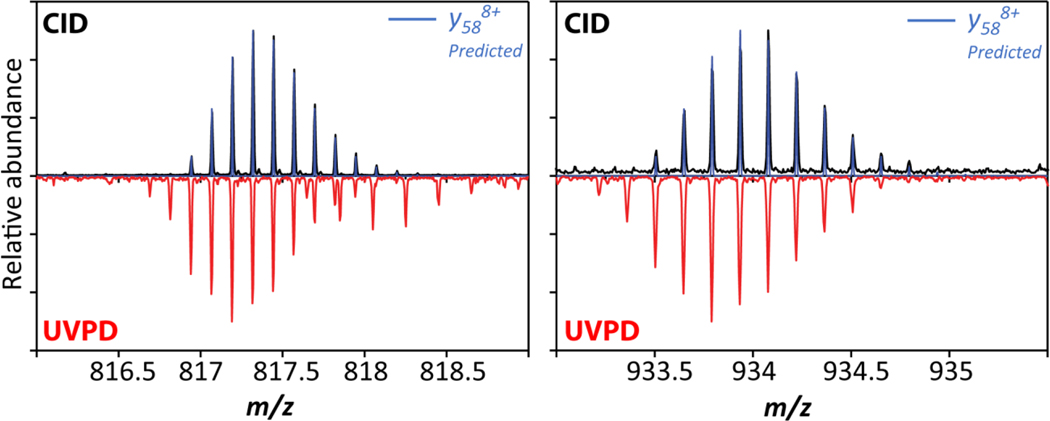
Figure 6 compares isotopic patterns for the fragment ions at m/z 817 and m/z 933 observed for both UVPD and CID in Figure 4. For the CID measurements, these ions are identified as y588+ and y587+ fragment ions, respectively, based on comparing the experimental and theoretical isotope patterns. These ions are produced from cleavage N-terminal to Pro19 and thus underscore the “proline-effect” in CID of protonated peptides and proteins.34,65,66 By contrast, the figure also shows that the isotopic patterns observed for the corresponding m/z species produced from UVPD differ from those observed for CID. Specifically, for the two cases plotted in Figure 6, the isotopic patterns for m/z 817 and m/z 933 produced from UVPD differ from those produced by CID by the loss of 1 or 2 hydrogen atoms and are thus assigned as [y58-1]+7 and [y58-1]+8, respectively. (Note that our data does not exclude presence of some fraction of [y58-2]+7 and [y58-2]+8, respectively.) This observation corroborates that hydrogen-transfer processes have occurred during the absorption/dissociation events in clear accord with prior reports of y-1 and y-2 ions produced from UVPD of peptides and proteins.7,67,68 Consequently, because these radical-driven processes are typical to UVPD and do not occur in CID of protonated peptides and proteins, the data shown in Figure 6 demonstrate that the reaction mechanism of UVPD carried out in our ion trap at pressures of 2–3 mbar differs mechanistically from that of CID.
Figure 6. Comparison of isotopic patterns for fragment ions observed at m/z 817 and m/z 933 produced from UVPD and CID.
The fragment ions produced from CID are assigned as y587+ and y588+ ions whereas those produced from UVPD correspond [y58−1]7+ / [y58-2]7+ and [y58-1]8+ / [y58-2]8+, respectively.
Taken together, the observations made in Figures 4, 5, and 6 show that (1) we succeeded in conducting UVPD of an intact protein at 2–3 mbar in the ion trap located in-between two TIMS devices; (2) performing UVPD as described here produces fragment ions through a radical-driven mechanism, therefore producing ions complementary to CID; (3) the sequence coverage obtained is comparable to recent reports coupling high-resolution mass spectrometers with UVPD conducted at high vacuum.45,64
Analytical potential for top-down structure analysis of proteins
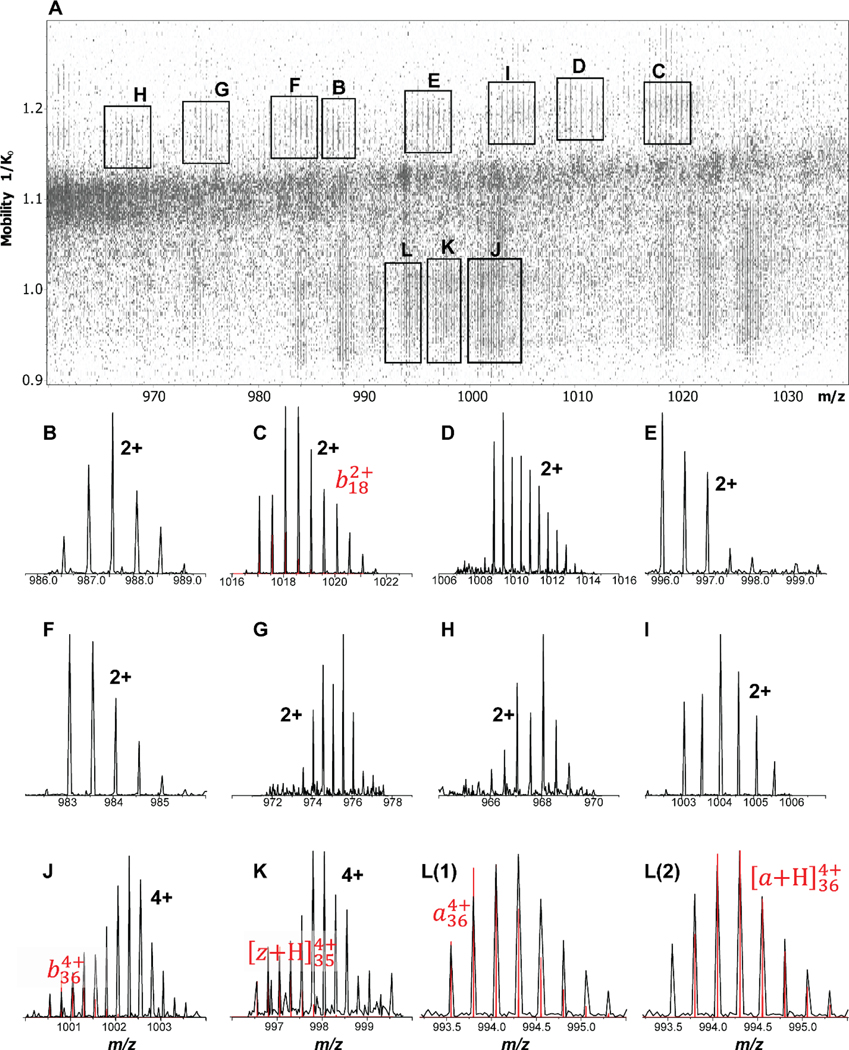
At first glance, the mass spectra shown in Figure 4 produced from UVPD appear somewhat noisier than the corresponding CID spectra and dominated by a small number of fragment ions. For example, the spectrum in Figure 4d appears to show 9 fragment ions at relatively poor signal/noise with the remainder of the m/z range appearing to be noise. The nested ion mobility-mass spectrum for this m/z range (see Figure 7), however, shows that what appears as noise in Figure 4d arises in fact from heavily congested fragment ion mass spectra. Specifically, the mobility-separation of the fragment ions produced from UVPD allows us to identify presence of a number of additional fragment ions that mass analysis alone is unable to reveal in Figure 4d. Indeed, the mobility-separation in TIMS-2 is capable of separating the various fragment ions to such an extent that their isotope patterns are essentially baseline resolved. We stress two facts: (1) our mobility-analysis is carried out at a scan rate of ~ 1.9 V/ms which is about 5–10 times higher than what is typically employed.39–42,69 This means that there is ample opportunity to improve the mobility separation of the ions depicted in Figure 7 by reducing the mobility scan rate, thereby improving the separation of fragment ions; (2) the data indicate presence of fragment ion species that differ in the presence of 1 or 2 hydrogen atoms; (3) The instrument discussed here enables PASEF44 workflows in which the fragment ions produced from UVPD are isolated and subsequently fragmented in the quadrupole/collision cell. Separately, PASEF has demonstrated great utility in bottom-up õmics experiments,70 and effective MS/MS/MS experiments carried out with tandem-TIMS42 and timsTOF Pro experiments51,55 demonstrated utility for protein top-down analysis. Hence, we anticipate that the instrument described here will prove particularly valuable for structural proteomics studies because it enables PASEF-assisted top-down analysis of proteins and protein complexes.
Figure 7. Analytical potential of mobility-separating UVPD fragment ions in our tandem-TIMS instrument.
(A) The nested ion mobility / mass spectrum for m/z 960 to 1040 shows a plethora of mobility-separated fragment ions that are not identified by mass analysis alone. (B) – (L) Extracted mass spectra for selected fragment ions observed in (A) with assigned ion or charge state annotated. The analysis shows that the mobility-separation in TIMS-2 is capable of separating the various fragment ions to such an extent that their isotope patterns are essentially baseline resolved. Notice that the experimental data show isotope patterns that indicate overlapping fragment ions.
Currently ongoing efforts in our laboratories aim to enable a broad community to conduct native top-down protein analysis on the basis of the instrument platform described above. Our data indicate that the most critical aspects toward that goal are (1) to optimize the ion trap to improve its storing efficiency while maintaining its “softness”; (2) further optimize the overlap between the trajectory of the ions stored in the trap and the laser beam; (3) to maximize sequence coverage by adjusting the laser power or irradiation time to the protein system of interest; and (4) to develop a PASEF method compatible with charge states and ion mobilities of fragment ions typical to top-down protein analysis. We will report on these ongoing efforts in due course.
Summary and Conclusions
We summarize:
We constructed a tandem-TIMS/MS (tTIMS/MS) in an orthogonal configuration on the basis of a commercial timsTOFPro instrument with a linear ion trap operating at 2–3 mbar inserted in-between the two TIMS devices.
We coupled this newly constructed tTIMS/MS with a 213 nm Nd:YAG laser to irradiate ions in the ion trap for UV photoactivation/photodissociation.
We demonstrated feasibility of conducting UVPD experiments of intact proteins trapped in the ion trap at 2–3 mbar.
We validated that the fragment ions produced by UVPD in our instrument are produced from a radical-based mechanism in accord with prior literature on UVPD.
Our data suggest stabilization of fragment ions produced from UVPD at the elevated pressures used here due to collisional-cooling (“UVnoD2”), which otherwise do not survive to detection.
The data account for a sequence coverage comparable to recent reports of high-resolution mass spectrometers coupled with UVPD.
We demonstrated the analytical potential of our instrument for top-down protein analysis by mobility-separation of fragment ions produced from UVPD.
In conclusion, our data demonstrate that UVPD carried out at elevated pressures of 2–3 mbar yields extensive fragment ions rich in information about the protein, and that their exhaustive analysis requires IMS separation post-UVPD. Hence, because UVPD and tTIMS/MS each have been shown to be valuable techniques on their own merit in proteomics, our discussion here underscores the potential of combining these two techniques for future structural proteomics studies.
Acknowledgements
The authors thank Jason E. Kuszynski, Geoffrey Strouse, and Lea Nienhaus (Department of Chemistry and Biochemistry, Florida State University) for assistance with the laser setup and alignment. This work was supported in part by the National Science Foundation and the National Institutes of Health under CHE-1654608 and R01GM135682 (C.B.)
References
- 1.Millard BJ Quantitative Mass Spectrometry. Heyden; 1979. [Google Scholar]
- 2.McLafferty FW Tandem Mass Spectrometry. Wiley; 1983. [Google Scholar]
- 3.Bayat P, Lesage D, Cole RB. Tutorial: Ion Activation In Taadem Mass Spectrometry using Ultra-High Resolution Instrumentation. Mass Spectrom Rev. 2020;39(5–6):680–702. doi: 10.1002/mas.21623 [DOI] [PubMed] [Google Scholar]
- 4.McLuckey SA, Goeringer DE. Slow heating methods in tandem mass spectrometry. J Mass Spectrom. 1997;32(5):461–474. [Google Scholar]
- 5.Sahasrabuddhe A, Hsia Y, Busch F, et al. Confirmation of intersubunit connectivity and topology of designed protein complexes by native MS. Proc Natl Acad Sci. 2018;115(6):1268–1273. doi: 10.1073/pnas.1713646115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qi Y, Volmer DA. Electron-based fragmentation methods in mass spectrometry: An overview: ExD IN MS. Mass Spectrom Rev. 2017;36(1):4–15. doi: 10.1002/mas.21482 [DOI] [PubMed] [Google Scholar]
- 7.Li H, Sheng Y, McGee W, Cammarata M, Holden D, Loo JA. Structural Characterization of Native Proteins and Protein Complexes by Electron Ionization Dissociation-Mass Spectrometry. Anal Chem. 2017;89(5):2731–2738. doi: 10.1021/acs.analchem.6b02377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang L, Reilly JP. Peptide Photodissociation with 157 nm Light in a Commercial Tandem Time-of-Flight Mass Spectrometer. Anal Chem. 2009;81(18):7829–7838. doi: 10.1021/ac9012557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brodbelt JS. Photodissociation mass spectrometry: new tools for characterization of biological molecules. Chem Soc Rev. 2014;43(8):2757–2783. doi: 10.1039/C3CS60444F [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brodbelt JS, Morrison LJ, Santos I. Ultraviolet Photodissociation Mass Spectrometry for Analysis of Biological Molecules. Chem Rev. 2020;120(7):3328–3380. doi: 10.1021/acs.chemrev.9b00440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li H, Nguyen HH, Ogorzalek Loo RR, Campuzano IDG, Loo JA. An integrated native mass spectrometry and top-down proteomics method that connects sequence to structure and function of macromolecular complexes. Nat Chem. 2018;10(2):139–148. doi: 10.1038/nchem.2908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O’Brien JP, Li W, Zhang Y, Brodbelt JS. Characterization of Native Protein Complexes Using Ultraviolet Photodissociation Mass Spectrometry. J Am Chem Soc. 2014;136(37):12920–12928. doi: 10.1021/ja505217w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Belov ME, Damoc E, Denisov E, et al. From Protein Complexes to Subunit Backbone Fragments: A Multi-stage Approach to Native Mass Spectrometry. Anal Chem. 2013;85(23):11163–11173. doi: 10.1021/ac4029328 [DOI] [PubMed] [Google Scholar]
- 14.Ko BJ, Brodbelt JS. Comparison of glycopeptide fragmentation by collision induced dissociation and ultraviolet photodissociation. Int J Mass Spectrom. 2015;377:385–392. doi: 10.1016/j.ijms.2014.07.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Greer SM, Brodbelt JS. Top-Down Characterization of Heavily Modified Histones Using 193 nm Ultraviolet Photodissociation Mass Spectrometry. J Proteome Res. 2018;17(3):1138–1145. doi: 10.1021/acs.jproteome.7b00801 [DOI] [PubMed] [Google Scholar]
- 16.Fort KL, Dyachenko A, Potel CM, et al. Implementation of Ultraviolet Photodissociation on a Benchtop Q Exactive Mass Spectrometer and Its Application to Phosphoproteomics. Anal Chem. 2016;88(4):2303–2310. doi: 10.1021/acs.analchem.5b04162 [DOI] [PubMed] [Google Scholar]
- 17.Tamara S, Dyachenko A, Fort KL, Makarov AA, Scheltema RA, Heck AJR. Symmetry of Charge Partitioning in Collisional and UV Photon-Induced Dissociation of Protein Assemblies. J Am Chem Soc. 2016;138(34):10860–10868. doi: 10.1021/jacs.6b05147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zabuga AV, Kamrath MZ, Boyarkin OV, Rizzo TR. Fragmentation mechanism of UV-excited peptides in the gas phase. J Chem Phys. 2014;141(15):154309. doi: 10.1063/1.4897158 [DOI] [PubMed] [Google Scholar]
- 19.Hansen K, Skinnerup Byskov C, Nielsen SB. Energy flow in peptides after UV photoexcitation of backbone linkages. Phys Chem Chem Phys. 2017;19(30):19640–19645. doi: 10.1039/C7CP01768E [DOI] [PubMed] [Google Scholar]
- 20.Julian R R. The Mechanism Behind Top-Down UVPD Experiments: Making Sense of Apparent Contradictions. J Am Soc Mass Spectrom. 2017;28(9):1823–1826. doi: 10.1007/s13361-017-1721-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grégoire G, Jouvet C, Dedonder C, Sobolewski AL. Ab initio Study of the Excited-State Deactivation Pathways of Protonated Tryptophan and Tyrosine. J Am Chem Soc. 2007;129(19):6223–6231. doi: 10.1021/ja069050f [DOI] [PubMed] [Google Scholar]
- 22.Kang H, Jouvet C, Dedonder-Lardeux C, et al. Ultrafast deactivation mechanisms of protonated aromatic amino acids following UV excitation. Phys Chem Chem Phys. 2005;7(2):394–398. doi: 10.1039/B414986F [DOI] [PubMed] [Google Scholar]
- 23.Lepère V, Lucas B, Barat M, et al. Comprehensive characterization of the photodissociation pathways of protonated tryptophan. J Chem Phys. 2007;127(13):134313. doi: 10.1063/1.2770458 [DOI] [PubMed] [Google Scholar]
- 24.Zhang L, Reilly JP. Extracting Both Peptide Sequence and Glycan Structural Information by 157 nm Photodissociation of N-Linked Glycopeptides. J Proteome Res. 2009;8(2):734–742. doi: 10.1021/pr800766f [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Devakumar A, Thompson MS, Reilly JP. Fragmentation of oligosaccharide ions with 157 nm vacuum ultraviolet light. Rapid Commun Mass Spectrom. 2005;19(16):2313–2320. doi: 10.1002/rcm.2058 [DOI] [PubMed] [Google Scholar]
- 26.Klein DR, Leach FE, Amster IJ, Brodbelt JS. Structural Characterization of Glycosaminoglycan Carbohydrates Using Ultraviolet Photodissociation. Anal Chem. 2019;91(9):6019–6026. doi: 10.1021/acs.analchem.9b00521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Crispin M, Ward AB, Wilson IA. Structure and Immune Recognition of the HIV Glycan Shield. Annu Rev Biophys. 2018;47(1):499–523. doi: 10.1146/annurev-biophys-060414-034156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Watanabe Y, Allen JD, Wrapp D, McLellan JS, Crispin M. Site-specific glycan analysis of the SARS-CoV-2 spike. Science. 2020;369(6501):330–333. doi: 10.1126/science.abb9983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Adamson BD, Coughlan NJA, Markworth PB, Continetti RE, Bieske EJ. An ion mobility mass spectrometer for investigating photoisomerization and photodissociation of molecular ions. Rev Sci Instrum. 2014;85(12):123109. doi: 10.1063/1.4903753 [DOI] [PubMed] [Google Scholar]
- 30.Bellina B, Brown JM, Ujma J, et al. UV photodissociation of trapped ions following ion mobility separation in a Q-ToF mass spectrometer. The Analyst. 2014;139(24):6348–6351. doi: 10.1039/C4AN01656D [DOI] [PubMed] [Google Scholar]
- 31.Theisen A, Yan B, Brown JM, Morris M, Bellina B, Barran PE. Use of Ultraviolet Photodissociation Coupled with Ion Mobility Mass Spectrometry To Determine Structure and Sequence from Drift Time Selected Peptides and Proteins. Anal Chem. 2016;88(20):9964–9971. doi: 10.1021/acs.analchem.6b01705 [DOI] [PubMed] [Google Scholar]
- 32.Stiving AQ, Harvey SR, Jones BJ, et al. Coupling 193 nm Ultraviolet Photodissociation and Ion Mobility for Sequence Characterization of Conformationally-Selected Peptides. J Am Soc Mass Spectrom. 2020;31(11):2313–2320. doi: 10.1021/jasms.0c00259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Simon A-L, Chirot F, Choi CM, et al. Tandem ion mobility spectrometry coupled to laser excitation. Rev Sci Instrum. 2015;86(9):094101. doi: 10.1063/1.4930604 [DOI] [PubMed] [Google Scholar]
- 34.Warnke S, Baldauf C, Bowers MT, Pagel K, von Helden G. Photodissociation of Conformer-Selected Ubiquitin Ions Reveals Site-Specific Cis / Trans Isomerization of Proline Peptide Bonds. J Am Chem Soc. 2014;136(29):10308–10314. doi: 10.1021/ja502994b [DOI] [PubMed] [Google Scholar]
- 35.Zucker SM, Lee S, Webber N, Valentine SJ, Reilly JP, Clemmer DE. An Ion Mobility/Ion Trap/Photodissociation Instrument for Characterization of Ion Structure. J Am Soc Mass Spectrom. 2011;22(9):1477–1485. doi: 10.1007/s13361-011-0179-8 [DOI] [PubMed] [Google Scholar]
- 36.Mistarz UH, Bellina B, Jensen PF, Brown JM, Barran PE, Rand KD. UV Photodissociation Mass Spectrometry Accurately Localize Sites of Backbone Deuteration in Peptides. Anal Chem. 2018;90(2):1077–1080. doi: 10.1021/acs.analchem.7b04683 [DOI] [PubMed] [Google Scholar]
- 37.Theisen A, Black R, Corinti D, Brown JM, Bellina B, Barran PE. Initial Protein Unfolding Events in Ubiquitin, Cytochrome c and Myoglobin Are Revealed with the Use of 213 nm UVPD Coupled to IM-MS. J Am Soc Mass Spectrom. 2019;30(1):24–33. doi: 10.1007/s13361-018-1992-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mason EA, McDaniel EW. Transport Properties of Ions in Gases. Wiley-VCH; 1988.
- 39.Liu FC, Ridgeway ME, Park MA, Bleiholder C. Tandem trapped ion mobility spectrometry. Analyst. 2018;143(10):2249–2258. doi: 10.1039/C7AN02054F [DOI] [PubMed] [Google Scholar]
- 40.Bleiholder C, Liu FC. Structure Relaxation Approximation (SRA) for Elucidation of Protein Structures from Ion Mobility Measurements. J Phys Chem B. 2019;123(13):2756–2769. doi: 10.1021/acs.jpcb.8b11818 [DOI] [PubMed] [Google Scholar]
- 41.Kirk SR, Liu FC, Cropley TC, Carlock HR, Bleiholder C. On the Preservation of Non-covalent Peptide Assemblies in a Tandem-Trapped Ion Mobility Spectrometer-Mass Spectrometer (TIMS-TIMS-MS). J Am Soc Mass Spectrom. 2019;30(7):1204–1212. doi: 10.1007/s13361-019-02200-y [DOI] [PubMed] [Google Scholar]
- 42.Liu FC, Cropley TC, Ridgeway ME, Park MA, Bleiholder C. Structural Analysis of the Glycoprotein Complex Avidin by Tandem-Trapped Ion Mobility Spectrometry–Mass Spectrometry (Tandem-TIMS/MS). Anal Chem. 2020;92(6):4459–4467. doi: 10.1021/acs.analchem.9b05481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bleiholder C, Liu FC, Chai M. Comment on Effective Temperature and Structural Rearrangement in Trapped Ion Mobility Spectrometry. Anal Chem. 2020;92(24):16329–16333. doi: 10.1021/acs.analchem.0c02052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meier F, Beck S, Grassl N, et al. Parallel Accumulation–Serial Fragmentation (PASEF): Multiplying Sequencing Speed and Sensitivity by Synchronized Scans in a Trapped Ion Mobility Device. J Proteome Res. 2015;14(12):5378–5387. doi: 10.1021/acs.jproteome.5b00932 [DOI] [PubMed] [Google Scholar]
- 45.Fornelli L, Srzentić K, Toby TK, et al. Thorough Performance Evaluation of 213 nm Ultraviolet Photodissociation for Top-down Proteomics. Mol Cell Proteomics. 2020;19(2):405–420. doi: 10.1074/mcp.TIR119.001638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Becher S, Wang H, Leeming MG, Donald WA, Heiles S. Influence of protein ion charge state on 213 nm top-down UVPD. The Analyst. 2021;146(12):3977–3987. doi: 10.1039/D1AN00571E [DOI] [PubMed] [Google Scholar]
- 47.Hernandez DR, DeBord JD, Ridgeway ME, Kaplan DA, Park MA, Fernandez-Lima F. Ion dynamics in a trapped ion mobility spectrometer. Analyst. 2014;139(8):1913–1921. doi: 10.1039/C3AN02174B [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chai M, Young MN, Liu FC, Bleiholder C. A Transferable, Sample-Independent Calibration Procedure for Trapped Ion Mobility Spectrometry (TIMS). Anal Chem. 2018;90(15):9040–9047. doi: 10.1021/acs.analchem.8b01326 [DOI] [PubMed] [Google Scholar]
- 49.Silveira JA, Ridgeway ME, Park MA. High Resolution Trapped Ion Mobility Spectrometery of Peptides. Anal Chem. 2014;86(12):5624–5627. doi: 10.1021/ac501261h [DOI] [PubMed] [Google Scholar]
- 50.Stow SM, Causon TJ, Zheng X, et al. An Interlaboratory Evaluation of Drift Tube Ion Mobility–Mass Spectrometry Collision Cross Section Measurements. Anal Chem. 2017;89(17):9048–9055. doi: 10.1021/acs.analchem.7b01729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Borotto NB, Graham KA. Fragmentation and Mobility Separation of Peptide and Protein Ions in a Trapped-Ion Mobility Device. Anal Chem. Published online July 14, 2021:acs.analchem.1c01188. doi: 10.1021/acs.analchem.1c01188 [DOI] [PubMed]
- 52.Ly T, Julian RR. Residue-Specific Radical-Directed Dissociation of Whole Proteins in the Gas Phase. J Am Chem Soc. 2008;130(1):351–358. doi: 10.1021/ja076535a [DOI] [PubMed] [Google Scholar]
- 53.Anthis NJ, Clore GM. Sequence-specific determination of protein and peptide concentrations by absorbance at 205 nm: Sequence-Specific Protein Concentration at 205 nm. Protein Sci. 2013;22(6):851–858. doi: 10.1002/pro.2253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Grimsley GR, Pace CN. Spectrophotometric Determination of Protein Concentration. Curr Protoc Protein Sci. 2003;33(1). doi: 10.1002/0471140864.ps0301s33 [DOI] [PubMed] [Google Scholar]
- 55.Larson EJ, Roberts DS, Melby JA, et al. High-Throughput Multi-attribute Analysis of Antibody-Drug Conjugates Enabled by Trapped Ion Mobility Spectrometry and Top-Down Mass Spectrometry. Anal Chem. Published online July 14, 2021:acs.analchem.1c00150. doi: 10.1021/acs.analchem.1c00150 [DOI] [PMC free article] [PubMed]
- 56.Liu FC, Kirk SR, Bleiholder C. On the structural denaturation of biological analytes in trapped ion mobility spectrometry – mass spectrometry. Analyst. 2016;141(12):3722–3730. doi: 10.1039/C5AN02399H [DOI] [PubMed] [Google Scholar]
- 57.Bleiholder C, Johnson NR, Contreras S, Wyttenbach T, Bowers MT. Molecular Structures and Ion Mobility Cross Sections: Analysis of the Effects of He and N 2 Buffer Gas. Anal Chem. 2015;87(14):7196–7203. doi: 10.1021/acs.analchem.5b01429 [DOI] [PubMed] [Google Scholar]
- 58.Bush MF, Campuzano IDG, Robinson CV. Ion Mobility Mass Spectrometry of Peptide Ions: Effects of Drift Gas and Calibration Strategies. Anal Chem. 2012;84(16):7124–7130. doi: 10.1021/ac3014498 [DOI] [PubMed] [Google Scholar]
- 59.Myung S, Badman ER, Lee YJ, Clemmer DE. Structural Transitions of Electrosprayed Ubiquitin Ions Stored in an Ion Trap over ∼10 ms to 30 s. J Phys Chem A. 2002;106(42):9976–9982. doi: 10.1021/jp0206368 [DOI] [Google Scholar]
- 60.Wyttenbach T, Bowers MT. Structural Stability from Solution to the Gas Phase: Native Solution Structure of Ubiquitin Survives Analysis in a Solvent-Free Ion Mobility–Mass Spectrometry Environment. J Phys Chem B. 2011;115(42):12266–12275. doi: 10.1021/jp206867a [DOI] [PubMed] [Google Scholar]
- 61.Zhang X, Garimella SVB, Prost SA, et al. Ion Trapping, Storage, and Ejection in Structures for Lossless Ion Manipulations. Anal Chem. 2015;87(12):6010–6016. doi: 10.1021/acs.analchem.5b00214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tolmachev AV, Vilkov AN, Bogdanov B, PĂsa-Tolić L, Masselon CD, Smith RD. Collisional activation of ions in RF ion traps and ion guides: The effective ion temperature treatment. J Am Soc Mass Spectrom. 2004;15(11):1616–1628. doi: 10.1016/j.jasms.2004.07.014 [DOI] [PubMed] [Google Scholar]
- 63.Tolmachev AV, Chernushevich IV, Dodonov AF, Standing KG. A collisional focusing ion guide for coupling an atmospheric pressure ion source to a mass spectrometer. Nucl Instrum Methods Phys Res Sect B Beam Interact Mater At. 1997;124(1):112–119. [Google Scholar]
- 64.Ng C-Y, Baer M, eds. Inhomogenous RF fields: a versatile tool for the study of processes with slow ions. In: State-Selected and State-to-State Ion-Molecule Reaction Dynamics. 1: Experiment. Vol LXXXII. Advances in Chemical Physics. Wiley; 1992. [Google Scholar]
- 65.Bleiholder C, Suhai S, Harrison AG, Paizs B. Towards Understanding the Tandem Mass Spectra of Protonated Oligopeptides. 2: The Proline Effect in Collision-Induced Dissociation of Protonated Ala-Ala-Xxx-Pro-Ala (Xxx = Ala, Ser, Leu, Val, Phe, and Trp). J Am Soc Mass Spectrom. 2011;22(6):1032–1039. doi: 10.1007/s13361-011-0092-1 [DOI] [PubMed] [Google Scholar]
- 66.Paizs B, Suhai S. Fragmentation pathways of protonated peptides. Mass Spectrom Rev. 2005;24(4):508–548. doi: 10.1002/mas.20024 [DOI] [PubMed] [Google Scholar]
- 67.Morrison LJ, Rosenberg JA, Singleton JP, Brodbelt JS. Statistical Examination of the a and a + 1 Fragment Ions from 193 nm Ultraviolet Photodissociation Reveals Local Hydrogen Bonding Interactions. J Am Soc Mass Spectrom. 2016;27(9):1443–1453. doi: 10.1007/s13361-016-1418-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cannon JR, Martinez-Fonts K, Robotham SA, Matouschek A, Brodbelt JS. Top-Down 193-nm Ultraviolet Photodissociation Mass Spectrometry for Simultaneous Determination of Polyubiquitin Chain Length and Topology. Anal Chem. 2015;87(3):1812–1820. doi: 10.1021/ac5038363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jeanne Dit Fouque K, Garabedian A, Leng F, et al. Trapped Ion Mobility Spectrometry of Native Macromolecular Assemblies. Anal Chem. 2021;93(5):2933–2941. doi: 10.1021/acs.analchem.0c04556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vasilopoulou CG, Sulek K, Brunner A-D, et al. Trapped ion mobility spectrometry and PASEF enable in-depth lipidomics from minimal sample amounts. Nat Commun. 2020;11(1):331. doi: 10.1038/s41467-019-14044-x [DOI] [PMC free article] [PubMed] [Google Scholar]