Abstract
The denitrifying strain T1, identified as Thauera aromatica, is able to grow with toluene serving as its sole carbon source. Previous work identified two genes, tutD and tutE, that are involved in toluene metabolism. Two small open reading frames, tutF and tutG, which may also play a role in toluene metabolism, were also identified. The present work examines the transcriptional organization and regulation of these toluene utilization genes. Northern analysis indicates that the four genes are organized into two operons, tutE and tutFDG, and that both operons are regulated in response to toluene. Primer extension analysis has identified major transcriptional start sites located 177 bp upstream of the tutE translational start and 76 bp upstream of the tutF translational start. Furthermore, a fifth gene, tutH, has been identified immediately downstream of tutG. It is transcribed from the same start site as tutFDG and is predicted to code for a 286-amino-acid protein with a calculated molecular mass of about 31,800 Da. The TutH protein is predicted to have an ATP/GTP binding domain and is similar to the NorQ/NirQ family of proteins.
Toluene is a hazardous substance that poses health risks to humans. A number of microorganisms that are able to metabolize this aromatic hydrocarbon under denitrifying conditions have been isolated and include Thauera aromatica K172 (27), Azoarcus sp. strain T (11, 17), and T. aromatica T1 (formerly known as strain T1) (12, 28). Biochemical studies with cell extracts of T. aromatica K172 and Azoarcus sp. strain T have shown that the first step in anaerobic toluene metabolism in these two organisms is the enzymatic formation of benzylsuccinate from toluene and fumarate (4, 6). This is a highly stereospecific reaction carried out by benzylsuccinate synthase, an enzyme recently isolated from T. aromatica K172 (4–6, 19). This purified enzyme has been shown to be an α2β2γ2 complex consisting of two subunits each of the BssA, BssB, and BssC proteins (19).
The genes coding for the benzylsuccinate synthase protein subunits have been cloned from T. aromatica K172 and designated bssCAB (19). Highly similar genes cloned from T. aromatica T1 and designated tutFDG (Fig. 1) most likely also code for a benzylsuccinate synthase (8). Based on similarities of the BssA and TutD proteins with pyruvate formate-lyase, and based on the reported mechanism for pyruvate formate-lyase (23, 24, 29), it has been proposed that these enzymes function by formation of a glycine free radical (8, 19). Biochemical work with the benzylsuccinate synthase enzyme from T. aromatica K172 and mutagenesis studies of the tutD gene of T. aromatica T1 support this mechanism of action (8, 19).
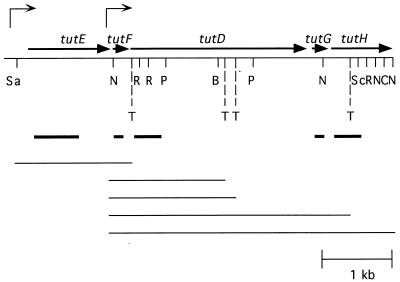
FIG. 1.
Restriction map of the region of cosmid clone 13-6-4 that contains the tutE tutFDGH genes. The five identified open reading frames are indicated with thick arrows. Thin arrows above the map indicate the major transcriptional start sites identified. T, potential terminator site. The probes used for Northern analysis are also indicated below the map with thick lines. Potential transcripts (from the starts to the terminators) are shown below the probes with thin lines. Restriction site abbreviations: B, BamHI; C, ClaI; N, NcoI; P, PstI; R, EcoRI; Sa, SacII; Sc, SacI. Sites blocked by methylation are omitted.
In addition to the bssA and tutD genes encoding benzylsuccinate synthase and its likely homologue, genes coding for a proposed benzylsuccinate synthase-activating enzyme have also been cloned from T. aromatica K172 and T. aromatica T1 and designated bssD and tutE, respectively (8, 19). The proposed function is based on the similarities of these gene products with pyruvate formate-lyase-activating enzymes (8, 19). The bssD and tutE genes are located upstream of and transcribed in the same direction as bssA and tutD (8, 19) (Fig. 1). It has been shown that in T. aromatica K172, the bss genes are grouped into an operon and transcribed as a single unit (19). Additionally, genes involved in the regulation of toluene metabolism in T. aromatica K172 and T. aromatica T1 have been identified (9, 18).
This report focuses on the transcriptional analysis of the tutEFDG gene cluster of T. aromatica T1. Since the transcription of the bssDCAB genes of T. aromatica K172 is induced by toluene (19) and anaerobic toluene metabolism is inducible in T. aromatica T1 (15), RNA analysis of the tutEFDG gene cluster of T. aromatica T1 was undertaken to determine if their regulation also occurs at the level of mRNA abundance. In addition, the transcriptional organization of these genes was examined and transcriptional start sites were identified by primer extension and nuclease protection assays. Finally, the identification and characterization of tutH, a new gene that codes for a putative ATP/GTP binding protein, are presented.
MATERIALS AND METHODS
Strains and DNA manipulation.
Isolation, characterization, and identification of T. aromatica T1, a gram-negative peritrichous denitrifying organism, have been reported previously (12, 28). The Escherichia coli strain XL-1 Blue (Stratagene, La Jolla, Calif.) was used to propagate DNA. The isolation and characterization of cosmid 13-6-4 were described previously (8, 9). To isolate DNA for sequencing, large-scale preparations were performed using the Qiagen (Santa Clarita, Calif.) maxiprep according to the manufacturer's instructions. DNA manipulations were carried out as described by Ausubel et al. (2).
RNA preparation.
Wild-type T. aromatica T1 cells were grown under denitrifying conditions on a mineral salts medium (13) (vitamins and yeast extract omitted) with either pyruvate or toluene serving as the carbon source. When the density of the culture reached about 4 × 107 cells/ml, 35 ml of the culture was processed using the RNeasy Mini kit from Qiagen according to the manufacturer's instructions. Samples were run on a gel to confirm that there was no RNA degradation.
Northern gel analysis.
About 0.25 μg of total RNA was run on a 0.8% agarose gel containing 1% formaldehyde (2). Ethidium bromide was added to each RNA sample to a final concentration of 31 μg/ml before denaturation and loading to allow visualization of the RNA without affecting the efficiency of RNA transfer to the membrane (22). After electrophoresis, the gels were treated with 0.05 N NaOH for 30 min, 0.1 M Tris (pH 7.5) for 30 min, and 10× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) for 30 min. RNA was transferred to a Hybond-N Membrane (Boehringer Mannheim, Indianapolis, Ind.) by capillary blotting overnight. The RNA was cross-linked to the membrane by baking at 80°C for 2 h in a vacuum oven. Antisense, digoxigenin-labeled, gene-specific DNA probes spanning nucleotides 97 to 398 of the predicted tutD coding region (302 nucleotides), 106 to 792 of the predicted tutE coding region (687 nucleotides), 14 to 152 of the predicted tutF coding region (139 nucleotides), 36 to 241 of the predicted tutG coding region (206 nucleotides), and 59 to 470 of the predicted tutH coding region (412 nucleotides) were made by PCR (20) and are indicated in Fig. 1. Prehybridization was performed at 42°C for at least 1 h in DIG Easy Hyb solution (Boehringer Mannheim). The probe was heated to 95°C and then added to the prehybridization mix at a final concentration of about 50 ng/ml. Hybridization was continued overnight at 42°C. The blots were washed twice with 2× SSC–0.1% sodium dodecyl sulfate (5 min, room temperature) and twice with 0.5× SSC–0.1% sodium dodecyl sulfate (5 min, 65°C). The probes were visualized on BioMax ML film (Eastman Kodak, Rochester, N.Y.) using the DIG High Prime DNA Labeling and Detection Starter Kit II (Boehringer Mannheim) according to the manufacturer's instructions with the chemiluminescence substrate CSPD. Digoxigenin-labeled RNA (Boehringer Mannheim) was also loaded on the gel to serve as a size marker.
Primer extension analysis.
The Primer Extension System-AMV Reverse Transcriptase kit was purchased from Promega (Madison, Wis.) and used according to the manufacturer's instructions. About 2.5 μg of total RNA was used for each reaction. Primers F-PE1 (5′ CTG CTT GCA TGT GGT GGT TC 3′), binding from 4 to 23 bp downstream of the translational start of tutF, and E-PE3 (5′ GAT CCA CCA CGA CCA TAG AAG 3′), binding 5 bp upstream to 15 bp downstream of the translational start of tutE, were labeled with T4 polynucleotide kinase (New England Biolabs, Beverly, Mass.) and [γ-32P]ATP (New England Nuclear, Boston, Mass.). The labeled primers were used for both the primer extension reaction and the sequencing ladder. The primer extension reaction products and the sequencing ladder were run on a standard 8 M urea–5% polyacrylamide sequencing gel.
Nuclease protection assay.
The Multi-NPA RNA/DNA/Oligo probe protection assay kit was purchased from Ambion (Austin, Tex.), and the manufacturer's standard procedure was followed. About 5 μg of total RNA was used for each reaction. Antisense gene-specific DNA probes of 354 bases (for tutE) or 623 bases (for tutF) spanning both the predicted transcriptional and translational start sites were synthesized by PCR (20) and labeled with T4 polynucleotide kinase (New England Biolabs) and [γ-32P]ATP (New England Nuclear). About 3 × 105 cpm of the probe was added to the assay mix. After completion of the reaction, the products were run on an 8 M urea–5% polyacrylamide gel.
DNA sequence analysis.
DNA was sequenced (both strands) by the dideoxy method of Sanger et al. (26) with [α-35S]dATP (New England Nuclear) serving as the label. Sequenase enzyme (modified T7 polymerase) and reagents were obtained in a Sequenase kit from Amersham Life Science (Arlington Heights, Ill.). The Bluescript vector and some primers used for sequence analysis were obtained from Stratagene. Synthetic oligonucleotide primers for the sequencing reactions were purchased from Life Technologies (Grand Island, N.Y.).
Computer analysis.
Searches for protein sequence similarity were carried out against the nonredundant GenBank protein database using the BLAST 2.0.2 program (1). The Motif program (21) was used to identify patterns in the protein sequences that could have a functional role. Potential factor-independent transcriptional terminator sites were identified with the Terminator program (7). Multiple sequence alignments were performed with the Lasergene software package from DNASTAR (Madison, Wis.).
Nucleotide sequence accession number.
The nucleotide sequence reported here has been submitted to the GenBank database and assigned accession number AF113168.
RESULTS AND DISCUSSION
Transcriptional regulation of the toluene utilization genes.
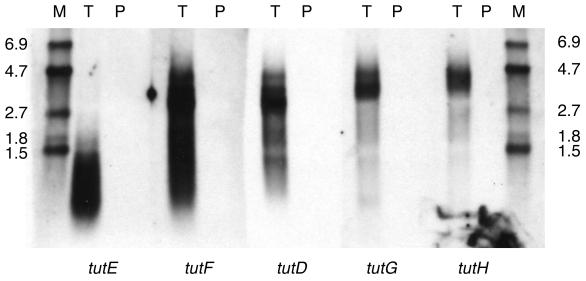
Northern analysis was used to examine the regulation of the toluene utilization genes of T. aromatica T1. Intense bands were detected when tutD, tutE, tutG (formerly open reading frame [ORF] 4) (8), and tutF (formerly ORF 2) (8) gene-specific probes were hybridized to aliquots of RNA isolated from toluene-grown cells (Fig. 2, lanes T). In contrast, no bands were detected by any of the tut gene-specific probes when aliquots of RNA isolated from pyruvate-grown cells were used (Fig. 2, lanes P). Since equal amounts of total RNA were loaded in the T and P lanes (as confirmed by ethidium bromide visualization of the 16S and 23S rRNA bands before transfer [data not shown]), these results indicate that the tut genes are induced by toluene. The bssDCAB genes of T. aromatica K172, which are similar to tutE tutFDG, are also regulated in response to toluene (19).
FIG. 2.
Northern analysis of total RNA isolated from cells grown under denitrifying conditions with either toluene (T) or pyruvate (P) as the carbon source and visualized with gene-specific probes derived from tutE, tutF, tutD, tutG, or tutH. Samples of digoxigenin-labeled RNA were included to serve as size markers (M) and are labeled (in kilobases) on the sides.
It can also be seen from Fig. 2 that the banding pattern observed with the tutE gene-specific probe is distinct from the patterns observed with the tutF, tutD, and tutG gene-specific probes. The transcripts observed using the tutE probe are predominantly less than 1.5 kb in size, with a faint band observed at about 1.7 kb. Various sizes of mRNA transcripts are also observed with the other three probes, but the largest transcripts are approximately 4.5 kb in size. The banding patterns suggest that tutF, tutD, and tutG are part of one transcriptional unit and that tutE is a separate transcriptional unit.
Identification of a new toluene utilization gene.
Since the maximum size of the tutF, tutD, and tutG mRNA transcripts (about 4.5 kb) was significantly larger than needed to code for these genes (about 3.1 kb), an examination of the DNA downstream of tutG was undertaken. An additional open reading frame was identified and designated tutH. Figure 2 includes the results of a Northern analysis in which a tutH gene-specific probe was used to identify transcripts from toluene-grown cells. The pattern observed with RNA isolated from toluene grown cells with the tutH probe was similar to that seen with the tutF, tutD, and tutG probes (Fig. 2). In addition, the tutH probe did not detect any transcripts in RNA isolated from pyruvate-grown cells, indicating that tutH is also induced by toluene (Fig. 2).
Identification of the transcriptional start sites.
The Northern analysis described above suggested that the tutF, tutD, tutG, and tutH genes are likely to be contained within a single transcriptional unit, while the tutE gene is separate. Thus, primer extension and nuclease protection analyses were undertaken to identify the transcriptional start site(s) present for each gene.
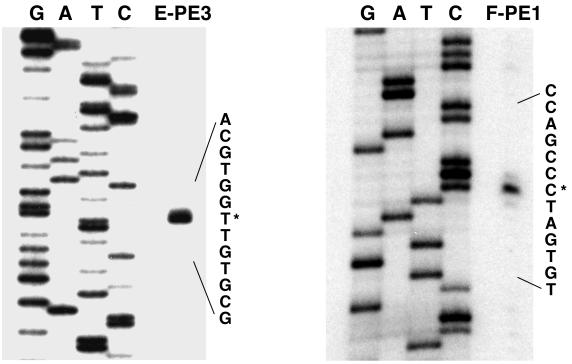
Figure 3 shows the results of a primer extension reaction using RNA isolated from toluene-grown cells and the E-PE3 primer, in which the predicted tutE translational start is contained. The major transcriptional start site is located 177 bp upstream of the tutE translational start. This same start site was also observed when a nuclease protection assay was carried out with a DNA probe spanning the tutE translational start (data not shown). Minor start sites (all of comparable intensity) were also observed 178 bp upstream (Fig. 3) and in the region 119 to 124 bp upstream of the tutE translational start (data not shown). None of these sites were observed when the analyses were performed with RNA isolated from pyruvate-grown cells (data not shown).
FIG. 3.
Primer extension analysis to map the transcriptional start sites of the tutE and tutF genes. End-labeled primer E-PE3 was used to identify the tutE start of transcription and end-labeled primer F-PE1 was used to identify the tutF start of transcription. The same primers were used to generate the sequencing ladder by the dideoxy method (lanes marked G, A, T, and C). The sequence encompassing the major transcriptional start (marked with an asterisk) is enlarged.
As can also be seen in Fig. 3, results of a primer extension reaction using RNA isolated from toluene-grown cells and the F-PE1 primer (located just downstream of the predicted tutF translational start site) identified a major transcriptional start site 76 bp upstream of the tutF translational start. This site is located within the tutE coding region. This start site was also observed when a nuclease protection assay was carried out with a DNA probe spanning this region of the tutF translational start (data not shown). Minor transcriptional start sites (all of comparable intensity) were observed 75 and 77 bp upstream (Fig. 3) and in the region 125 to 129 bp upstream of the tutF translational start site (data not shown). None of these sites were observed when the analyses were performed with RNA isolated from pyruvate-grown cells (data not shown). These results are consistent with the results of the Northern analysis, indicating that the tutE transcript is separate from the tutFDGH transcript.
A primer extension reaction carried out with a primer located downstream of the predicted tutD translational start and a nuclease protection assay carried out with a DNA probe spanning the tutD translational start did not identify a transcriptional start immediately upstream of tutD. Instead, these reactions did identify the same start site located upstream of tutF (data not shown). Primer extension reactions carried out with primers located downstream of the predicted tutG and tutH translational starts and nuclease protection assays carried out with DNA probes spanning these translational start sites failed to identify transcriptional start sites immediately upstream of these genes (data not shown). The start site identified preceding tutF could not be verified for tutG and tutH due to its considerable distance from these genes (about 2.9 and 3.2 kb, respectively). However, the RNA analyses suggest that the tutF, tutD, tutG, and tutH genes are transcribed as a single unit from one start site. Thus, the variety of mRNA transcript sizes seen in the Northern analyses is likely not due to different transcriptional start sites but may be due to different transcriptional termination sites.
It is not surprising that the tutFDG genes, as well as the similar bssCAB genes of T. aromatica K172, which encode subunits of the benzylsuccinate synthase enzyme and are included in the bssDCAB gene cluster (19), are located in single operons. However, since both the tutE and bssD gene products are predicted to function as activators that enzymatically form a glycine free radical in the proteins encoded by the tutD and bssA genes, respectively, it might be expected that the activator proteins would be needed in smaller amounts than the activated proteins. Thus, organization into separate transcriptional units might be useful for regulatory purposes. Indeed, in the case of the pyruvate formate-lyase systems of E. coli, Haemophilus influenzae, and Clostridium pasteurianum, which show sequence similarities to the tutD and tutE genes and the bssA and bssD genes, the pyruvate formate-lyase-activating protein is located on a different transcriptional unit from the pyruvate formate-lyase (14, 24, 30). It is not clear why the tutE tutFDG and the bssDCAB gene clusters, coding for homologous proteins which are expected to carry out the same chemical reaction in two strains of the same bacterial species, would be organized in such different manners.
Computer analysis of terminator and promoter regions.
A search for potential factor-independent terminator-like sequences using the Terminator program (7) led to the identification of the four sites shown in Fig. 1. An additional terminator site is presumed to reside downstream of tutH. It is predicted that a transcript starting upstream of tutE and ending at the terminator past tutF would be about 1.5 kb and would be visualized by both the tutE and tutF gene-specific probes. Similarly, transcripts beginning at the start site upstream of tutF are predicted to be about 1.6, 1.8, 3.6, and greater than 4.3 kb, depending on which terminator is used. The tutF probe would be expected to identify all of these transcripts, while the tutG and tutH gene-specific probes would be expected to identify only the two largest. Indeed, results from the Northern analyses are consistent with the predicted locations of these putative terminators (Fig. 2). This computer analysis does not identify factor-dependent terminators that may also be present.
An examination of the regions immediately upstream of the defined transcriptional start sites failed to identify any consensus −35 or −10 sites. Additionally, a search failed to identify homology between the regions upstream of both tutE and tutF and any known bacterial promoter regions. A pairwise comparison of only the sequences upstream of tutE and tutF did identify a number of similar regions, ranging from 20 to 60 nucleotides in length. The significance (if any) of these sites is currently under investigation.
Sequence analysis of tutH.
Since the 4,905-bp SacII/EcoRI fragment of cosmid 13-6-4 (GenBank accession number AF036765) (8, 9) did not contain the complete sequence of the tutH gene, an additional 381 bp of this cosmid was sequenced on both strands. The 1,018-bp NcoI fragment (part of which is contained in the SacII/EcoRI fragment previously reported) containing the tutH sequence has been deposited in GenBank. Analysis of this sequence identified the complete tutH coding region, whose predicted protein product is 286 amino acids. The TutH protein has a calculated molecular mass of about 31,800 Da and a predicted pI of 5.4.
The BLAST program (1) was used to identify proteins similar to the predicted TutH protein. The four proteins with the highest degree of similarity were NorQ from Paracoccus halodenitrificans (25), Paracoccus denitrificans (10), and Rhodobacter sphaeroides (3) and NirQ from Pseudomonas stutzeri (16). The BLAST program calculated that these proteins are 27, 28, 27, and 22% identical (over nearly their entire sequence) to TutH, respectively.
The TutH protein sequence was also subjected to a Motif analysis (21). Amino acids 47 to 54 were identified as a putative ATP/GTP binding domain, a region that is conserved in the NorQ and NirQ proteins. This observation suggests that the NorQ-NirQ family of proteins and the TutH protein may use a similar mechanism involving ATP/GTP binding. It is possible that these proteins function in similar manners in their different systems and that this ATP/GTP binding is necessary for their function. The roles of NorQ and NirQ have not been elucidated, but studies of the NirQ protein from P. stutzeri suggest that it may posttranslationally regulate the activity of nitric oxide reductase (16). Based on results obtained with R. sphaeroides, it has been suggested that the NorQ protein may be involved in the assembly of the active nitric oxide reductase enzyme complex (3). The bssCAB gene products of T. aromatica K172, which are similar to those of the tutFDG genes, have been shown to form a complex (19). Based on this comparison, it is expected that the tutFDG gene products of T. aromatica T1 also form a complex. Thus, it can be speculated that the role of TutH is to posttranslationally modify one or more of the TutF, TutD, and TutG proteins and/or aid in the assembly of an active enzyme complex containing these proteins. Interestingly, a gene showing similarity to the tutH gene has not been observed in T. aromatica K172 (19). What role, if any, the tutH gene product plays in anaerobic toluene metabolism remains to be determined.
ACKNOWLEDGMENTS
This work was supported by National Science Foundation CAREER award MCB-9733210 and Ohio University Research Committee grant 97-12.
Bethany Henderson, Sarah Cunningham, and Karen Coschigano are acknowledged for their assistance with the Northern analysis, Olivia Harriott is acknowledged for assistance with the terminator analysis, and Bradley Bishop is acknowledged for technical assistance. Olivia Harriott, Anne Frazer, Lily Young, and Karen Coschigano are acknowledged for helpful discussion and comments on the manuscript.
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Lipman W M D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Short protocols in molecular biology. New York, N.Y: Greene Publishing Associates and John Wiley & Sons; 1992. [Google Scholar]
- 3.Bartnikas T B, Tosques I E, Laratta W P, Shi J, Shapleigh J P. Characterization of the nitric oxide reductase-encoding region in Rhodobacter sphaeroides 2.4.3. J Bacteriol. 1997;179:3534–3540. doi: 10.1128/jb.179.11.3534-3540.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beller H R, Spormann A M. Anaerobic activation of toluene and o-xylene by addition to fumarate in denitrifying strain T. J Bacteriol. 1997;179:670–676. doi: 10.1128/jb.179.3.670-676.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beller H R, Spormann A M. Analysis of the novel benzylsuccinate synthase reaction for anaerobic toluene activation based on structural studies of the product. J Bacteriol. 1998;180:5454–5457. doi: 10.1128/jb.180.20.5454-5457.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Biegert T, Fuchs G, Heider J. Evidence that anaerobic oxidation of toluene in the denitrifying bacterium Thauera aromatica is initiated by formation of benzylsuccinate from toluene and fumarate. Eur J Biochem. 1996;238:661–668. doi: 10.1111/j.1432-1033.1996.0661w.x. [DOI] [PubMed] [Google Scholar]
- 7.Brendel V, Trifonov E N. A computer algorithm for testing potential prokaryotic terminators. Nucleic Acids Res. 1984;12:4411–4427. doi: 10.1093/nar/12.10.4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coschigano P W, Wehrman T S, Young L Y. Identification and analysis of genes involved in anaerobic toluene metabolism by strain T1: putative role of a glycine free radical. Appl Environ Microbiol. 1998;64:1650–1656. doi: 10.1128/aem.64.5.1650-1656.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coschigano P W, Young L Y. Identification and sequence analysis of two regulatory genes involved in anaerobic toluene metabolism by strain T1. Appl Environ Microbiol. 1997;63:652–660. doi: 10.1128/aem.63.2.652-660.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Boer A P N, van der Oost J, Reijnders W N M, Westerhoff H V, Stouthamer A H, van Spanning R J M. Mutational analysis of the nor gene cluster which encodes nitric-oxide reductase from Paracoccus denitrificans. Eur J Biochem. 1996;242:592–600. doi: 10.1111/j.1432-1033.1996.0592r.x. [DOI] [PubMed] [Google Scholar]
- 11.Dolfing J, Zeyer J, Binder-Eicher P, Schwarzenbach R P. Isolation and characterization of a bacterium that mineralizes toluene in the absence of molecular oxygen. Arch Microbiol. 1990;154:336–341. doi: 10.1007/BF00276528. [DOI] [PubMed] [Google Scholar]
- 12.Evans P J, Mang D T, Kim K S, Young L Y. Anaerobic degradation of toluene by a denitrifying bacterium. Appl Environ Microbiol. 1991;57:1139–1145. doi: 10.1128/aem.57.4.1139-1145.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Evans P J, Mang D T, Young L Y. Degradation of toluene and m-xylene and transformation of o-xylene by denitrifying enrichment cultures. Appl Environ Microbiol. 1991;57:450–454. doi: 10.1128/aem.57.2.450-454.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fleischmann, R. D., M. D. Adams, O. White, R. A. Clayton, E. F. Kirkness, A. R. Kerlavage, C. J. Bult, J.-F. Tomb, B. A. Dougherty, J. M. Merrick, K. McKenney, G. Sutton, W. FitzHugh, C. Fields, J. D. Gocayne, J. Scott, R. Shirley, L.-I. Liu, A. Glodek, J. M. Kelley, J. F. Weidman, C. A. Phillips, T. Spriggs, E. Hedblom, M. D. Cotton, T. R. Utterback, M. C. Hanna, D. T. Nguyen, D. M. Saudek, R. C. Brandon, L. D. Fine, J. L. F. J. L. Fuhrmann, N. S. M. Geoghagen, C. L. Gnehn, L. A. McDonald, K. V. Small, C. M. Fraser, H. O. Smith, and J. C. Venter. 1995. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science (Washington, D.C.) 269:496–512. [DOI] [PubMed]
- 15.Frazer A C, Ling W, Young L Y. Substrate induction and metabolite accumulation during anaerobic toluene utilization by the denitrifying strain T1. Appl Environ Microbiol. 1993;59:3157–3160. doi: 10.1128/aem.59.9.3157-3160.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jüngst A, Zumft W G. Interdependence of respiratory NO reduction and nitrite reduction revealed by mutagenesis of nirQ, a novel gene in the denitrification gene cluster of Pseudomonas stutzeri. FEBS Lett. 1992;314:308–314. doi: 10.1016/0014-5793(92)81495-8. [DOI] [PubMed] [Google Scholar]
- 17.Krieger C J, Beller H R, Reinhard M, Spormann A M. Initial reactions in anaerobic oxidation of m-xylene by the denitrifying bacterium Azoarcus sp. strain T. J Bacteriol. 1999;181:6403–6410. doi: 10.1128/jb.181.20.6403-6410.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leuthner B, Heider J. A two-component system involved in regulation of anaerobic toluene metabolism in Thauera aromatica. FEMS Microbiol Lett. 1998;166:35–41. doi: 10.1111/j.1574-6968.1998.tb13180.x. [DOI] [PubMed] [Google Scholar]
- 19.Leuthner B, Leutwein C, Schulz H, Hörth P, Haehnel W, Schiltz E, Schägger H, Heider J. Biochemical and genetic characterization of benzylsuccinate synthase from Thauera aromatica: a new glycyl radical enzyme catalysing the first step in anaerobic toluene metabolism. Mol Microbiol. 1998;28:615–628. doi: 10.1046/j.1365-2958.1998.00826.x. [DOI] [PubMed] [Google Scholar]
- 20.Myerson D. Producing single-stranded DNA probes with the Taq DNA polymerase: a high yield protocol. BioTechniques. 1991;10:35–38. [PubMed] [Google Scholar]
- 21.Ogiwara A, Uchiyama I, Takagi T, Kanehisa M. Construction and analysis of a profile library characterizing groups of structurally known proteins. Protein Sci. 1996;5:1991–1999. doi: 10.1002/pro.5560051005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ogretmen B, Ratajczak H, Kats A, Stark B C, Gendel S M. Effects of staining of RNA with ethidium bromide before electrophoresis on performance of Northern blots. BioTechniques. 1993;14:932–935. [PubMed] [Google Scholar]
- 23.Plaga W, Frank R, Knappe J. Catalytic-site mapping of pyruvate formate lyase. Eur J Biochem. 1988;178:445–450. doi: 10.1111/j.1432-1033.1988.tb14468.x. [DOI] [PubMed] [Google Scholar]
- 24.Rödel W, Plaga W, Frank R, Knappe J. Primary structure of Escherichia coli pyruvate formate-lyase and pyruvate formate-lyase activating enzyme deduced from the DNA nucleotide sequences. Eur J Biochem. 1988;177:153–158. doi: 10.1111/j.1432-1033.1988.tb14356.x. [DOI] [PubMed] [Google Scholar]
- 25.Sakurai N, Sakurai T. Genomic DNA cloning of the region encoding nitric oxide reductase in Paracoccus halodenitrificans and a structure model relevant to cytochrome oxidase. Biochem Biophys Res Commun. 1998;243:400–406. doi: 10.1006/bbrc.1998.8106. [DOI] [PubMed] [Google Scholar]
- 26.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schocher R J, Seyfried B, Vazquez F, Zeyer J. Anaerobic degradation of toluene by pure cultures of denitrifying bacteria. Arch Microbiol. 1991;157:7–12. doi: 10.1007/BF00245327. [DOI] [PubMed] [Google Scholar]
- 28.Song B, Young L Y, Palleroni N J. Identification of denitrifier strain T1 as Thauera aromatica and proposal for emendation of the genus Thauera definition. Int J Syst Bacteriol. 1998;48:889–894. doi: 10.1099/00207713-48-3-889. [DOI] [PubMed] [Google Scholar]
- 29.Volker-Wagner A F, Frey M, Neugebauer F A, Schäfer W, Knappe J. The free radical in pyruvate formate-lyase is located on glycine-734. Proc Natl Acad Sci USA. 1992;89:996–1000. doi: 10.1073/pnas.89.3.996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weidner G, Sawers G. Molecular characterization of the genes encoding pyruvate formate-lyase and its activating enzyme of Clostridium pasteurianum. J Bacteriol. 1996;178:2440–2444. doi: 10.1128/jb.178.8.2440-2444.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]