Abstract
The formation of viscous foams on aeration basins and secondary clarifiers of activated sludge plants is a common and widespread problem. Foam formation is often attributed to the presence of mycolic acid-containing actinomycetes (mycolata). In order to examine the relationship between the number of mycolata and foam, we developed a group-specific probe targeting the 16S rRNA of the mycolata, a protocol to permeabilize mycolata, and a statistically robust quantification method. Statistical analyses showed that a lipase-based permeabilization method was quantitatively superior to previously described methods (P << 0.05). When mixed liquor and foam samples were examined, most of the mycolata present were rods or cocci, although filamentous mycolata were also observed. A nested analysis of variance showed that virtually all of the measured variance occurred between fields of view and not between samples. On this basis we determined that as few as five fields of view could be used to give a statistically meaningful sample. Quantitative fluorescent in situ hybridization (FISH) was used to examine the relationship between foaming and the concentration of mycolata in a 20-m3 completely mixed activated sludge plant. Foaming occurred when the number of mycolata exceeded a certain threshold value. Baffling of the plant affected foaming without affecting the number of mycolata. We tentatively estimated that the threshold foaming concentration of mycolata was about 2 × 106 cells ml−1 or 4 × 1012 cells m−2. We concluded that quantitative use of FISH is feasible and that quantification is a prerequisite for rational investigation of foaming in activated sludge.
Microbe-based treatment problems occur in the plants of even the most sophisticated and experienced water utilities. One of the most intractable and widespread problems is the formation of viscous foams in activated sludge plants. Such foams appear as bacterial biomass floating on the surfaces of aeration basins and secondary sedimentation tanks. The presence of foam may lead to severe operational problems and may cause plants to fail effluent standards. The occurrence of foam is unpredictable, and there is uncertainty about the cause and mechanism of foaming; the control mechanisms used currently are empirical and frequently ineffective (45). It is thought that foams may form when gas bubbles are stabilized by the presence of hydrophobic particles and surfactants; the bubbles then rise to the surface and accumulate (45). The hydrophobic particles are assumed to be bacteria.
Microscopic examination of foams usually reveals the presence of large numbers of one or two filamentous bacterial morphotypes. It is assumed that the morphotypes that are branched are mycolic acid-containing actinomycetes (mycolata) (13, 48) and that unbranched filaments are Microthrix parvicella (45). Putative mycolata morphotypes are the most widely reported morphotypes (39, 43) and are often referred to as “nocardia.” However, a wide range of mycolata are now associated with foaming. Plate counting has revealed the presence of members of the genera Gordonia (23, 45), Skermania (44), Tsukamurella (22), Rhodococcus (36), and Nocardia (45). Micromanipulation techniques have been used to recover mycobacteria, rhodococci, nocardiae (46), and skermaniae (44) from foams. The mycolata have many phenotypic characteristics in common, form a distinct phyletic line, and are assigned to the genera Corynebacterium, Dietzia, Gordonia, Mycobacterium, Nocardia, Rhodococcus, Skermania, Tsukamurella, and Williamsia (21, 24).
The study of the relationships between foam formation and mycolata has suffered from severe methodological shortcomings. Morphological examination, the method used most commonly, is simply inadequate for identification or quantification of actinomycetes (9). Mycolata usually either form branched filaments or are pleomorphic in pure cultures (21, 24). However, culturing bacteria provides only a partial, selective assessment of biomass (5), and it provides no information about morphology in the environment. Antibody-based methods (26) are also unsatisfactory due to their inherent reliance on culturing, cross-reactivity (18), and uncertainty about the nature of the cell wall in the environment.
The difficulty of determining the number of mycolata in the environment has undermined attempts to study and understand foaming. Despite the extensive literature on the subject, there is still only circumstantial evidence that mycolata are involved in the process. There is a need for reliable quantitative techniques to enumerate mycolata and explore the relationships between the putative foam-causing organisms and foaming itself (45).
Using rRNA-targeted probes is a more promising technique because it does not require culturing and because the mycolata fall in a distinct phyletic line within the evolutionary radiation of the actinomycetes. Some workers have developed probes from clone sequences retrieved from activated sludge (41, 42), and other workers have taken a nested probe approach in which probes that target different phylogenetic levels are used (17, 19). When these probes were used in conjunction with quantitative membrane hybridization techniques, the results showed that mycolata were present in the foam, mixed liquor, return activated sludge, and anaerobic digester sludge of a variety of foaming plants in the United States; Gordonia amarae was found to be the dominant member of the mycolata in some, but not all, plants (19). de los Reyes et al. concluded that there was a need to use fluorescent in situ hybridization (FISH) in foaming studies and described using FISH to detect mycolata (17).
In subsequent studies a quantitative FISH method was compared with a method involving antibody probes and quantitative membrane hybridization (18). There appeared to be no relationship between the concentration of gordonia cells determined by membrane hybridization and the concentration determined by FISH. The counting method relied on a correlation between filament length and mycolata biomass that was determined by using exogenous cultures and was extrapolated for use with environmental samples. This strategy assumed that pure cultures were representative of organisms in situ and that only filamentous growth occurred in activated sludge. These assumptions may have confounded the comparison of the two methods.
Mycolata are not readily permeabilized by conventional FISH procedures (30). Paraformaldehyde (PFA) (17), HCl (30), and mutanolysin (42) have been used to permeabilize some members of the mycolata. However, no single procedure that permeabilizes representatives of all of the mycolata genera associated with foaming has been found. This could result in underestimation of the numbers of mycolata associated with foaming, particularly the nonfilamentous forms.
A second, more fundamental potential shortcoming of using FISH with activated sludge is the possibility that the technique is not readily amenable to statistical analysis (33, 49). While they did not explore statistical issues in detail, previous authors implied that the variability of FISH counts of bacteria in activated sludge was so great that valid statistical analyses were impractical. Other shortcomings of FISH may also have an impact on quantification (5, 17, 18), and many authors fail to indicate this in their results.
In this paper we describe a protocol for permeabilization of most representatives of mycolata genera. This protocol enabled us to use FISH to quantify mycolata in a statistically valid manner. We found that activated sludge foaming is a function of the number of mycolata cells and that there appears to be a plant-specific threshold mycolata concentration (in the mixed liquor) that is required in order for foaming to take place. We also found that filamentous and nonfilamentous organisms are involved in foaming in the plants which we studied and that the latter were prevalent in the mixed liquor during foaming.
MATERIALS AND METHODS
Organisms, culture media, and growth conditions.
Representatives of five species of mycolata, including Gordonia amarae DSM 43392T (T = type strains), Gordonia bronchialis DSM 43247T, Mycobacterium peregrinum DSM 43271T, Nocardia asteroides DSM 43005T, and Rhodococcus rhodochrous DSM 43241T, as well as strain N1171, which was tentatively called “Tsukamurella spumae” (22), were grown in glucose-yeast extract broth (25) at 30°C for 4 to 7 days (that is, until the stationary phase, as determined by measuring the optical density at 600 nm). In addition, Escherichia coli NCIMB 4174 was grown overnight in Luria-Bertani broth (34) at 30°C, and Corynebacterium glutamicum NCIMB 10025T was grown to the stationary phase in corynebacterium broth at 30°C.
Environmental samples.
Samples of activated sludge mixed liquor and foam were collected from a pilot plant and a full-scale plant at Stoke Bardolph Water Reclamation Works, Nottingham, United Kingdom. All of the samples except the samples treated as described by de los Reyes et al. (17) (see below) were immediately fixed in absolute ethanol (1:1, vol/vol), transported to the laboratory, and stored at −20°C until they were used.
(i) Pilot plant.
In order to observe the effect of sludge age on the population of mycolata, FISH counts were obtained for samples taken from a large pilot activated sludge plant situated at Stoke Bardolph Water Reclamation Works, at which foaming in a full-scale activated sludge plant occurs. The pilot plant consisted of a clarifier and a 20-m3 aeration basin that was 1.85 m deep and was seeded with sludge from a nonfoaming plant; this plant received primary settled waste from the full-scale plant. The sludge age was controlled by maintaining the concentration of mixed liquor suspended solids by removing sludge directly from the aeration basin. The plant was run for 2 weeks at each sludge age in order to ensure stability and was increased stepwise from about 5 to 11 days. Grab samples were collected at a range of sludge ages. Throughout operation of the pilot plant instantaneous sludge ages were calculated daily, and the values obtained were used in comparisons with FISH counts obtained for grab samples.
(ii) Full-scale plants.
There are two full-scale activated sludge plants on the same site as the pilot plant, and both receive the same wastewater. The mean depth of one of these plants (plant A) is 3.5 m; this plant is highly baffled and is aerated by mechanical mixers, and extensive foaming occurs at it. The depth of the other plant (plant B) is 5.5 m; this plant is unbaffled and is aerated by diffused air, and no foaming occurs at it.
Total counts.
Activated sludge total counts were obtained by using the universal DNA stain 4′,6-diamidino-2-phenylindole (DAPI). Samples were washed and then resuspended in phosphate-buffered saline (PBS) (8 g of NaCl per liter, 0.2 g of KCl per liter, 1.15 g of Na2HPO4 per liter, 0.2 g of KH2PO4 per liter; pH 7.3). Three 10-fold serial dilutions were prepared with MilliQ water containing DAPI at a final concentration of 3.3 μg ml−1 and were incubated for 12 min. Aliquots (30 μl) were filtered onto 0.2-μm-pore-size black polycarbonate filters (Millipore Corp.) and washed with MilliQ water. The filters were placed onto slides, and each filter was mounted with a drop of Citifluor (Citifluor Ltd., Canterbury, United Kingdom) antifadent. A coverslip was placed over the resultant preparation, and nail varnish was used as a sealant. Counts were obtained by using a model 14 standard microscope (Carl Zeiss, Göttingen, Germany) equipped with a 50-W high-pressure mercury lamp and the appropriate filter sets (Carl Zeiss). Dilutions that resulted in between about 30 and 300 cells per field of view were used, and 10 random fields of view were examined. Total cell counts were calculated by the method of Kepner and Pratt (28), checked for normality and, when appropriate, transformed (47).
Oligonucleotide probes.
An oligonucleotide probe designed to target the 16S rRNA of members of the mycolata was generated after we compared the aligned sequences of representative validly described mycolata with the sequences of other bacteria by using the AL16S program (12). The sequence of this probe, S-*-Myc-0657-a-A-16 (Myc657) (Table 1) (1), was verified by using the CHECK_PROBE program of the Ribosomal Database Project (31). In addition, bacterial probe S-D-Bact-0338-a-A-18 (Bact338) (6) and nonspecific probe S-D-Bact-0338-a-S-18 (an antisense Bact338 probe; Non338) (32) were also used. All probes were made, labelled, and obtained commercially (Genosys Biotech Ltd., Cambridge, United Kingdom).
TABLE 1.
Sequences of probe Myc657, its target, and the corresponding small-subunit rRNA of selected organisms
| Organism or sequence | Probe or target sequencea |
|---|---|
| Probe sequence | 3′-ATGAYGTCCCCTCTGA-5′ |
| Target sequence | 5′-UACURCAGGGGAGACU-3′ |
| Escherichia coli | 5′-.CUC.U..A..G.GG.-3′ |
| Bacillus subtilis | 5′-.G.A.A..A.....G.-3′ |
| Corynebacterium glutamicum | 5′-.G...U..........-3′ |
| Corynebacterium hoagii | 5′-................-3′ |
| Dietzia maris | 5′-................-3′ |
| Gordonia amarae | 5′-................-3′ |
| Mycobacterium fortuitum | 5′-................-3′ |
| Nocardia farcinica | 5′-................-3′ |
| Nocardia asteroides | 5′-....U...........-3′ |
| Rhodococcus erythropolis | 5′-................-3′ |
| Skermania piniformis | 5′-....C...........-3′ |
| “Tsukamurella spumae” | 5′-.....U..........-3′ |
| Tsukamurella wratislaviensis | 5′-................-3′ |
Only mismatching nucleotides are indicated.
Fixation and permeabilization procedures. (i) Screening possible permeabilization agents.
A number of possible permeabilization agents were screened by using pure cultures representing five mycolata genera (Table 2). Permeabilization and fixation were carried out in solution. Portions (100 μl) of cultures were centrifuged at 13,000 × g for 3 min, and each pellet was washed in 1 ml of PBS. Except for the cells fixed in paraformaldehyde (see below), the cells were serially dehydrated by using increasing concentrations of ethanol in water (60, 80, and 100% [vol/vol]; 3 min each). Test organisms were then subjected to the following permeabilization treatments.
TABLE 2.
Effectiveness of permeabilizing agents as determined by whole-cell hybridization with representative strains of mycolata
| Straina | No. of carbon atoms in mycolic acids | Relative fluorescence of hybridized cells withb:
|
||||||
|---|---|---|---|---|---|---|---|---|
| 4% PFA | Diethyl ether | Xylene | 1 M HCl
|
Lipase
|
||||
| 50 min | 60 min | 50 min | 60 min | |||||
| Gordonia bronchialis DSM 43247T | 48–66c | ± | ± | ± | ++ | + | + | + |
| Mycobacterium peregrinum DSM 43271T | 60–90d | − | − | − | + | − | ± | + |
| Nocardia asteroides DSM 43005T | 46–60e | − | ND | ND | + | ± | ND | + |
| Rhodococcus rhodochrous DSM 43241T | 36–48f | + | ND | ND | ++ | ± | ++ | − |
| “Tsukamurella spumae” N1171 | 48–76g | − | − | − | − | + | + | ++ |
DSM, Deutsche Sammlung von Mikroorganismen und Zellkulturen, Braunschweig, Germany.
−, no fluorescence observed; ±, a few cells are poorly fluorescent; +, good fluorescence; ++, good to strong fluorescence; ND, not determined.
Data from reference 2.
Data from reference 35.
Data from reference 3.
Data from reference 15.
Data from reference 14.
(a) Standard PFA treatment.
Test cells were permeabilized and fixed by using 3 volumes of PFA (4%, wt/vol) for each volume of cell suspension, and the preparations were incubated overnight at 4°C prior to serial dehydration (see above) and hybridization.
(b) Diethyl ether.
Cells were incubated for 15 min at room temperature with 1 ml of diethyl ether (49) prior to fixation with ethanol-formaldehyde (90:10, vol/vol) for 30 min as described by Macnaughton et al. (30) and Braun-Howland et al. (11).
(c) Xylene.
Using a modification of the procedure described by Arnoldi et al. (7), we exposed cells to xylene for 10 min; the cells were then rehydrated and incubated with proteinase K (10 μg ml−1) for 30 min at 37°C prior to fixation with ethanol-formaldehyde as described above.
(d) Hydrochloric acid.
Test organisms were permeabilized as described by Macnaughton et al. (30) by incubating them with 1 M HCl at 37°C for 50 or 60 min prior to fixation with ethanol-formaldehyde.
(e) Lipase.
Cells were incubated with 300 μl of lipase (type VIII; final concentration, 0.2 mg ml−1; Sigma) at 37°C for 50 or 60 min. Subsequently, the cells were incubated with proteinase K (final concentration, 10 μg ml−1) at 37°C for 30 min and then washed three times in MilliQ water prior to fixation with ethanol-formaldehyde. After hybridization (see below), permeabilization was assessed qualitatively by using a conventional Carl Zeiss model 14 standard microscope equipped with a 50-W high-pressure mercury lamp and the appropriate filter sets, and the quality of fluorescence between treatments was compared and scored.
(ii) Quantitative comparison with recently described permeabilization techniques.
The lipase procedure was optimized, and the results were compared with the results obtained with other methods when the same activated sludge samples were used.
(a) Lipase.
A sample containing 2 × 108 cells was washed in 1 ml of PBS and dehydrated as described above. Following dehydration, the cells were incubated at 37°C with 75 μl of lipase (10 U μl−1 in PBS) for 60 min. The cells were subsequently incubated with proteinase K (final concentration, 10 μg ml−1) at 37°C for 30 min and then washed three times in 1 ml of MilliQ water and fixed in ethanol-formaldehyde for 30 min.
(b) PFA fixation.
Fresh portions of activated sludge mixed liquor containing 2 × 108 cells were fixed with 4% (wt/vol) PFA by using a modification of the method of de los Reyes et al. (17). In one treatment (treatment PFA1) samples were mixed with the fixative and left for 1 min, and in a second treatment (treatment PFA2) samples were mixed and immediately centrifuged at 13,000 × g for 3 min before hybridization.
(c) Mutanolysin.
A sample containing 2 × 108 cells was pretreated in solution with mutanolysin by using a modification of the method described by Schuppler et al. (42) prior to hybridization.
Hybridization.
During screening of possible agents in which pure cultures were used, permeabilization was followed by resuspension of the cells in 38 μl of simple hybridization buffer (0.9 M NaCl, 20 mM Tris-HCl [pH 7.2], 0.1% sodium dodecyl sulfate) containing 2 μl of tetramethyl rhodamine isothiocyanate (TRITC)-labelled oligonucleotide probe Bact338 (50 ng μl−1) (6). In addition, two negative controls were prepared; one of these controls was used to assess nonspecific binding (with TRITC-labelled probe Non338), and the other (lacking a probe) was used to monitor autofluorescence. All hybridizations were carried out at 48°C for 5 to 16 h. Following hybridization, the cells were washed twice in hybridization buffer at the hybridization temperature for 15 min before a final wash in MilliQ water. The cells were pelleted and resuspended in 300 μl of MilliQ water, and a 10-μl aliquot was placed on a gelatin-coated slide (4) and air dried. One drop of Citifluor antifadent (Citifluor Ltd.) was added to the sample, and a coverslip was applied to the preparation and sealed with nail polish before microscopy.
In other cases, using activated sludge samples, we performed hybridizations as described above, except that a more complex buffer containing 10× Denhardt's solution, poly(A) (0.5 mg ml−1) (4), and 30% formamide was used. A probe was added after 15 min of preincubation with the buffer at 37°C. Hybridizations were carried out overnight at the same temperature. The optimal hybridization conditions for probe Myc657 were determined empirically by increasing the formamide concentration in the hybridization buffer (32) and using different hybridization temperatures, as described previously (16).
Epifluorescent microscopy and image analysis.
Except as indicated, slides were viewed at a magnification of ×600 by using a confocal laser scanning microscope (CLSM) (model MRC 600; Bio-Rad, Hemel Hempstead, United Kingdom) which included a Nikon Optiphot 2 epifluorescent microscope. To analyze the success of various treatments, images were captured and probe-conferred fluorescence was measured by using the COMOS program (Bio-Rad). For environmental samples, extensively branched filaments were assumed to represent mycolata, and at least 20 sections of branched filaments per preparation were quantified.
Quantitative FISH procedure.
For every test sample we used two negative controls (see above) to determine autofluorescence and fluorescence due to nonspecific binding. In each case the levels of fluorescence of 20 cross sections of branched filaments were measured, and an average was calculated. The highest mean pixel intensity of the two controls was subtracted from the value for the test sample. We used randomly chosen fields of view for each test sample. Using the processed image, we counted only the cells that hybridized with both the TRITC-labelled mycolata-specific probe and the fluorescein isothiocyanate (FITC)-labelled eubacterial probe (that is, the cells that fluoresced a combination of red [TRITC] and green [FITC] [i.e., yellow]). The number of mycolata cells per milliliter was then determined from values for the area of the sample spot, the area of the field of view (FOV), the volume and dilution of the sample applied, and the original volume of the sample used for hybridization, as follows: total number of mycolata cells milliliter−1 = (number of mycolata cells per FOV × area of sample spot)/(area of FOV × volume applied × dilution × original sample volume).
Statistical analysis.
All quantitative data were checked for normality and, when appropriate, were transformed by using values determined by the Box-Cox transformation (10) before an analysis of variance (ANOVA) and multiple comparisons of means were carried out in most cases with the Minitab v11 program (Minitab Inc., State College, Pa.).
In order to assess the number of samples required at any one time and the number of random fields of view required to count any one sample, activated sludge mixed liquor was collected from two sides of the pilot plant and immediately fixed as described above. The two samples were subdivided into two parts, and counts for 10 random fields of view were obtained. The variance within samples and the variance between samples were determined by using nested analysis of variance and the Minitab v11 program (Minitab Inc.).
RESULTS
Comparison of permeabilization methods.
A comparison of permeabilization agents showed that the most effective permeabilizing agents were hydrochloric acid and lipase (Table 2). These agents were able to permeabilize all of the strains belonging to representative mycolata genera tested. The standard procedure in which PFA was used was found to be weakly effective, permeabilizing only two representative strains, G. bronchialis DSM 43247T and R. rhodochrous DSM 43241T. Treatment with diethyl ether or xylene was ineffective. The lipase method was investigated further as permeabilization with a strong acid was thought to be harsh. Indeed, prolonged use of hydrochloric acid resulted in reduced fluorescence, as shown previously (30). Optimization with environmental samples showed that incubation for 60 min with 750 U of lipase was required for effective permeabilization of extensively branched filaments if it was assumed that these morphotypes represented mycolata.
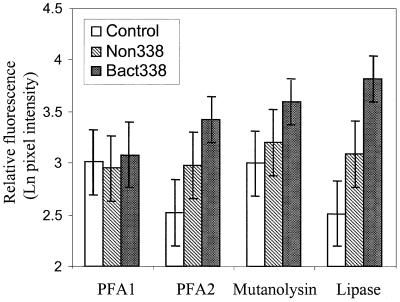
The effective use of FISH requires that labelled cells be distinguished from autofluorescing cells and from cells fluorescing due to nonspecific labelling. Four permeabilization protocols were compared by using the same sample of activated sludge in test preparations hybridized with Bact338 and appropriate controls. As determined by an ANOVA, the four treatments were significantly different (Fig. 1) (P < 0.001). Multiple comparisons of means (GT2 method pairwise comparisons [47]; family error, 0.0500; individual error, 0.00121) showed that the fluorescence of the positive cells permeabilized by the lipase method was greater than the fluorescence of the cells treated with either PFA (treatment PFA1 or PFA2) or mutanolysin. In addition, the lipase method distinguished between positive cells and the controls. The levels of fluorescence of the negative controls, including the autofluorescence control, were significant in all cases. The mutanolysin method gave particularly high values for autofluorescence and, along with treatments PFA1 and PFA2, appeared not to distinguish between labelled cells and controls when multiple comparisons were made. However, making overall multiple comparisons is very conservative since a low individual error rate is required to give an experimental error rate of 0.05 for all possible pairs of means (47).
FIG. 1.
Comparison of four permeabilization methods (treatments PFA1 and PFA2, mutanolysin, and lipase) by FISH in which the same sample of activated sludge (Bact338) was used. The results are expressed as the mean relative fluorescence of branched filaments in test and control samples for autofluorescence (Control) and nonspecific binding (Non338). The error bars represent 95% confidence as determined by the GT2 method, and nonoverlapping limits indicate that a difference was statistically significant.
When the mycolata cells in an activated sludge sample were enumerated by using the different treatments and dual hybridization with probes Myc657 and Bact338, the following counts were obtained: lipase treatment, 2.94 × 106 cells ml−1; PFA2 treatment, 1.14 × 106 cells ml−1; and mutanolysin treatment, 1.63 × 105 cells ml−1. These values were statistically significantly different (P < 0.001), and the value obtained with the lipase-treatment preparation was significantly greater than the other two values. The values obtained when the latter two methods were used were not statistically distinguishable at a P level of 0.05 (Tukey's pairwise comparison; individual error, 0.0205).
Probe design and optimization.
A probe targeting a region of the 16S rRNA specific for all mycolata based on a comparative analysis of aligned sequences was designed by using the AL16S program, recently available sequences, and the CHECK_PROBE program of the Ribosomal Database Project. The resultant probe, Myc657, is shown in Table 1 along with its target sequence and the sequences of representative target actinomycetes and reference organisms exhibiting one or more mismatches. The probe sequence matched the sequences of all but one Gordonia species and most species of the genera Nocardia, Mycobacterium, and Rhodococcus. The probe sequence exhibited one mismatch with the sequences of five Corynebacterium spp. (G:U at position 662), one Gordonia sp. (C:C at position 667), five Mycobacterium spp. (T:G and G:U at positions 658 and 662, respectively), seven Nocardia spp. (R:U or R:C at position 661), Skermania piniformis (R:C at position 661), and three Tsukamurella spp. (G:U at position 662) and two mismatches with the sequences of 30 Corynebacterium spp., one Mycobacterium sp., one Nocardia sp., two Rhodococcus spp. and Turicella otitidis, the only phylogenetically related organism belonging to the group that does not contain mycolic acids. In addition, the probe sequence exhibited three mismatches with the sequences of four Corynebacterium spp. and Rhodococcus fascians DSM 43241. Nontarget organism sequences exhibited at least two mismatches with the probe sequence. The optimal hybridization conditions determined for the probe were 30% formamide and 37°C. The stringency was such that target organisms included mycolata whose sequences exhibited only one mismatch with the probe sequence. The sequences of nontarget organisms exhibited two or more mismatches with the probe sequence. Under the conditions used the mean relative fluorescence intensity of the target organism, G. amarae DSM 43392T, was significantly different (P << 0.001) from the relative fluorescence intensity of a nontarget organism that had two mismatches, C. glutamicum NCIMB 10025T. In addition, the probe hybridized successfully with two other organisms that had the most common single mismatches with the probe, N. asteroides DSM 43005T (R:T mismatch at position 661) and “T. spumae” N1171 (G:T mismatch at position 662).
Sampling regimen.
The values for the number of mycolata cells in the subdivided samples were subjected to a nested ANOVA. The ANOVA showed that the variation between fields of view accounted for nearly all of the variation in the samples that had mean counts between 5 and 26 cells per field of view; the error was 69, and the coefficient of variation was 64%.
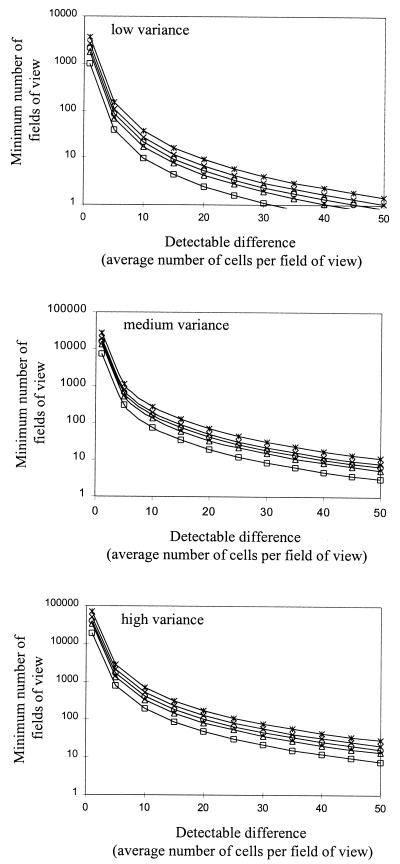
The variance in these samples was used to determine the minimum number of random fields of view required to detect a true difference of about 25 cells per field of view between two samples. This was done by rearranging the formula (47) φ = (nδ2/2as2)1/2 to give n = 2φ2 as2/δ2, where n is the minimum sample size, φ is a function of the F distribution (47), a is the group size, δ is the true difference between means, and s2 is the error. On this basis we calculated that a sample size of only five was required to give a 99% chance of detecting a difference between two samples at the 5% level of significance.
The number of fields of view required to detect a given difference at a given variance is a function of the number of cells per field of view, the number of samples compared, and the power required. However, it is the true difference that one wants to detect (i.e., the number of cells per field of view) that is the most important factor for any given variance (Fig. 2). In multiple comparisons of means the data exhibited heteroscadisticity (disproportionate variances), and transformations were required to perform parametric statistical analysis.
FIG. 2.
Influence of variance and the certainty (i.e., power) of a test on the minimum number of fields of view required to detect a given true difference (derived from an equation given in reference 47 for estimating sample size).
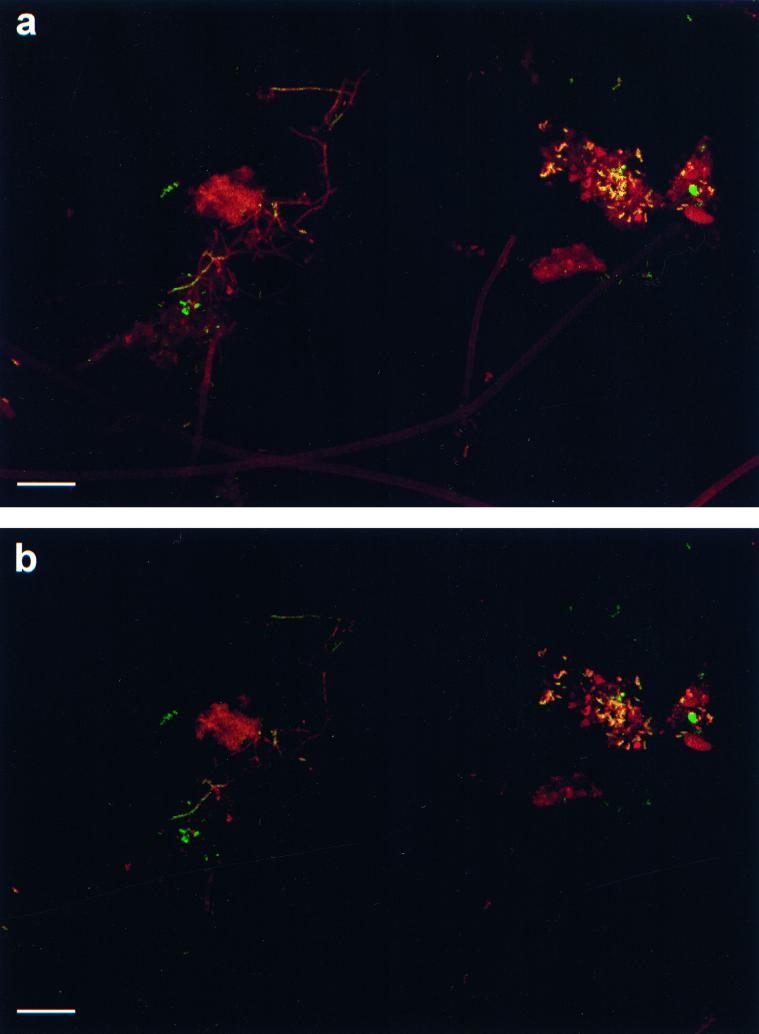
Cell morphology.
Samples of mixed liquor and foam from the pilot plant were examined by using FISH, and coccoidal, rod-shaped, and filamentous forms of mycolata were observed (Fig. 3). However, filaments were rarely observed in the mixed liquor in the samples, whereas in the foam filaments were seen more frequently. In both cases, however, rod-shaped and coccoid forms predominated. Many filaments exhibited low fluorescence; this was especially noticeable in the foam samples. In addition, the filamentous organisms observed exhibited the branching morphology characteristic of the organisms referred to as “Nocardia” spp. in typical morphological identification schemes (27). However, it should be noted that organisms with morphology characteristic of Mycobacterium parvicella were also observed.
FIG. 3.
CLSM images of an activated sludge sample hybridized with TRITC-labelled Myc657 and FITC-labelled Bact338. The sample was obtained from the Stoke Bardolph pilot plant. Bars = 10 μm. (a) Before subtraction of fluorescence intensities due to autofluorescence or nonspecific binding. (b) After subtraction of fluorescence intensities due to autofluorescence or nonspecific binding. Characteristic branched filaments are present, but a large number of rods and cocci are also present.
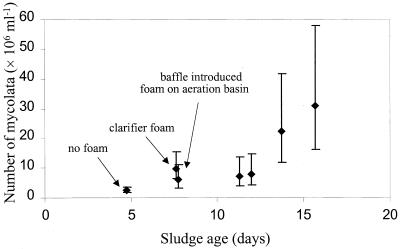
Pilot plant study.
A relationship between the concentration of mycolata and the presence of foam in the pilot plant was observed (Fig. 4). At the lowest sludge age mycolata were present at quite a high concentration (2.64 × 106 cells ml−1), but no foaming was observed. An increase in sludge age to 8 days resulted in a small but statistically significant increase in the number of mycolata associated with the appearance of foam in the secondary clarifier but not in the aeration tank. Addition of an inverted pipe to the overflow outlet of the aeration basin, simulating a baffle, led to accumulation of foam in this vessel without a statistically significant change in the concentration of mycolata in the mixed liquor. Furthermore, the foam persisted throughout the subsequent days of the study with one exception. At the greatest sludge age, foam disappeared from the surface of the aeration basin. This disappearance coincided with accidental loss of uniform aeration due to blockages in three of the four submerged air pipes.
FIG. 4.
Effect of sludge age on the number of mycolata determined by FISH by using a combination of probe Myc657 and probe Bact338 with mixed liquor samples obtained from the Stoke Bardolph pilot plant. An increase in sludge age to about 8 days resulted in a statistically significant increase in the number of mycolata and the appearance of foam on the clarifier. Introduction of a baffle resulted in the presence of foam on the surface of the aeration basin. The error bars indicate back-transformed 95% confidence intervals as determined by the GT2 method.
Mycolata in full-scale plants at the same site.
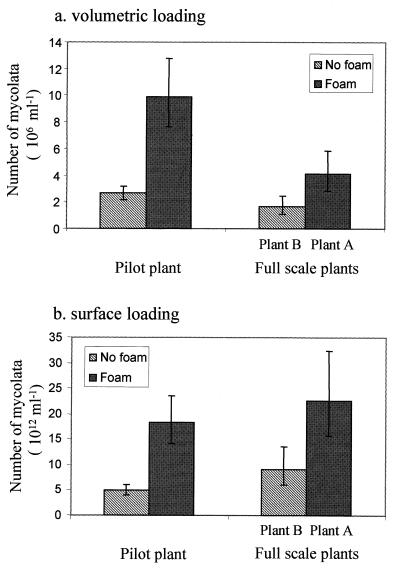
The number of mycolata per unit of volume in plant B (1.64 × 106 cells ml−1) was significantly lower (ANOVA; F-test; P = 0.003) than the concentration of mycolata in plant A (4.09 × 106 cells ml−1). However, to account for the difference in depth, the plants were compared on the basis of the number of mycolata per unit of surface area or surface load (27). The surface load at plant B (9.01 × 1012 cells m−2) was significantly lower than the surface load at plant A (2.25 × 1013 cells m−2) (P = 0.04).
In comparisons of mixed liquor samples with and without foam obtained at the unbaffled pilot plant, a volumetric loading analysis failed to distinguish between nonfoaming and foaming samples in all cases (Fig. 5a). However, when the results were analyzed on the basis of surface load, there was a statistically significant difference between the foam and the samples with no foam (Fig. 5b) (GT2 method; experimental error, 0.2). The surface loads at the pilot plant with and without foam were 1.83 × 1013 and 4.89 × 1012 cells m−2, respectively.
FIG. 5.
Comparisons of the numbers of mycolata determined by FISH under foaming and nonfoaming conditions and expressed on the basis of volumetric load (a) and surface load (b) at the pilot plant and at full-scale plants A and B at the Stoke Bardolph Water Reclamation Works. The GT2 method was used, and the error bars represent 80% confidence limits; nonoverlapping limits indicate that values were statistically significantly different. Intraplant differences between foaming and nonfoaming conditions were observed when the numbers of mycolata were expressed on the basis of both volumetric load and surface load. However, interplant differences were observed only when the numbers of mycolata were expressed on the basis of surface load.
DISCUSSION
The use of FISH for statistically valid quantification of the mycolata is practicable and is a prerequisite for rigorous studies of foaming in activated sludge. The simple detection of large numbers of mycolata cannot be considered a basis for studying or controlling foaming because the mere presence of mycolata does not cause foaming. Indeed, large numbers of mycolata were found in the absence of foam formation in the pilot plant and the full-scale plant.
Mycolata appeared to be associated with foam only when the concentration in the mixed liquor rose above a certain level or threshold, as previously speculated (19, 37, 38). This deduction is consistent with observations made in the pilot plant, in which modest but statistically significant increases in the concentration of mycolata were associated with foaming. Comparisons made at the full-scale plant also supported this conclusion, although other factors, such as differences in baffling, had an effect.
The threshold concept is consistent with the conceptual model of foam formation which requires the presence of rising air bubbles, hydrophobic particles, and surfactant. There would have to be a certain minimum number of hydrophobic particles in order for the foam to be stabilized. The foaming threshold appears to be a function of the physical layout of the plant, since foam on the aeration basin was induced by baffling with no associated change in mycolata concentrations. This is consistent with previous advice concerning minimizing baffling to avoid foaming (27). Consequently, we hypothesize that the greater the degree of baffling in a plant, the lower the foaming threshold mycolata concentration and vice versa. Plant-to-plant comparisons of foaming thresholds should take baffling into account.
It is important not only to define threshold values but also to define how they are expressed. Traditionally, the concentration of bacteria in a sample of activated sludge mixed liquor is expressed per unit of overall biomass (for example, per unit of suspended solids or per unit of small-subunit rRNA). However, foaming is a physical phenomenon, which is thought to be a function of the density of bacteria; therefore, the number of putatively foam-causing bacteria should be expressed per unit of volume. Moreover, the amount of foam that can be formed is a function of the number of bacteria per unit of area; therefore, this measure is more appropriate when units of different depths are compared.
Based on the presence of foam in the two baffled plants, a tentative estimate of the mycolata threshold foaming value for a baffled reactor is about 2 × 106 cells ml−1 or about 4 × 1012 cells m−2. The threshold value for an unbaffled reactor should be greater than this. Previous workers used other methods to suggest a range of actinomycete threshold values (ca. about 104 to 106 cells ml−1) (27, 37, 38). The wide range of values can be attributed to differences between, and the inadequacy of, the methods employed.
Our study provided evidence, but not definitive proof, of the role of mycolata in foaming. Filaments characteristic of M. parvicella were also observed in the mixed liquor, and the numbers of this organism and members of other groups of bacteria would have covaried with the number of mycolata and could have caused or at least contributed to foaming.
Using FISH ensures that all morphological forms are counted. Using filamentous morphology alone to identify bacteria associated with foam has led to speculation that filaments have a role in foam formation (27). However, the presence of large numbers of nonfilamentous mycolata highlight the inadequacy of morphological methods for identification of mycolata. Formation of filaments did not appear to be a prerequisite for foam formation since, as determined by FISH, comparatively few filaments were present in the mixed liquor during foaming. There is no fundamental physical reason why filaments should stabilize foam more effectively than nonfilamentous forms stabilize foam (8, 45). Foaming in the absence of filaments has been observed by workers using conventional methods (43). We found that casual microscopic observation led to overestimation of the relative numbers of filamentous bacteria, presumably because of the distinctive nature of their morphology. Nonfilamentous mycolata may be overlooked by workers who use FISH in a nonquantitative manner.
Permeabilization is very important for enumeration of mycolata. In our study we found that the lipase method was the most effective technique since it gave high counts and high specific fluorescence and permeabilized members of a wide variety of genera. It should be remembered that the cell wall composition of the bacteria in the environment may be different from the cell wall composition in pure culture and that there is almost certainly undiscovered diversity in activated sludge mycolata populations (22).
The higher counts obtained when the lipase method was used were probably a function of the lower background counts, as well as the range of organisms permeabilized. It is notable that all methods produced some background fluorescence, which reinforces the need for a rigorous approach to distinguish between autofluorescence and labelling (17). This is particularly important with mycolata since unlabelled but morphologically distinctive cells could be easily mistaken for mycolata cells. We strongly recommend that in the future workers explicitly state how they distinguished between labelled and unlabelled mycolata. In addition, workers who used FISH previously (17, 18) may have been handicapped because they used epifluorescence microscopy in which fluorescence due to out-of-plane cells was problematic. The use of a CLSM prevents such problems.
An important drawback in the use of FISH is that the technique favors the detection of metabolically active, readily permeabilized cells. However, the ability of bacteria to cause foaming is a function of cell wall chemistry and not necessarily metabolic activity (19). Nevertheless, the observed quantitative relationship between FISH counts and foam validates the use of in situ hybridization in this context.
We believe that the statistically valid use of FISH to enumerate mycolata is important, not only because it enabled us to address a particular phenomenon in activated sludge but also because quantification is central to almost all ecology. If molecular microbial ecology is to make a substantial impact, then some form of quantification is required. Few workers have tried to use FISH quantitatively for studying specific bacterial populations. While some studies have highlighted the importance of distinguishing target populations from nontarget populations (18, 40), few workers have attempted to use FISH with statistical rigor (20, 29). Other workers have reported having considerable difficulty generating statistically valid data (33, 49). In such cases it may be necessary to use nonparametric tests (47). Microbial ecologists should strive to achieve the statistical standards of other areas of biology.
Nevertheless, quantitative FISH is laborious, and the method cannot be considered rapid. However, we are confident that studies of foaming and many other important intractable microbiological phenomena will benefit from the availability of reliable techniques for quantification.
ACKNOWLEDGMENTS
We thank Trevor Booth for guidance with the CSLM, Veronik Hermans for running the pilot plant, and John Upton for stimulating discussions.
This work was supported by Severn-Trent Water Ltd. and by an Engineering and Physical Sciences Research Council studentship.
REFERENCES
- 1.Alm E W, Oerther D B, Larsen N, Stahl D A, Raskin L. The oligonucleotide probe database. Appl Environ Microbiol. 1996;62:3557–3559. doi: 10.1128/aem.62.10.3557-3559.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alshamaony L, Goodfellow M, Minnikin D E. Free mycolic acids as a criteria in the classification of Nocardia and the ‘rhodochrous’ complex. J Gen Microbiol. 1976;92:188–199. doi: 10.1099/00221287-92-1-188. [DOI] [PubMed] [Google Scholar]
- 3.Alshamaony L, Goodfellow M, Minnikin D E, Mordarska H. Free mycolic acids as a criteria in the classification of Gordona and the ‘rhodochrous’ complex. J Gen Microbiol. 1976;92:183–187. doi: 10.1099/00221287-92-1-183. [DOI] [PubMed] [Google Scholar]
- 4.Amann R I, Krumholz L, Stahl D A. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J Bacteriol. 1990;172:762–770. doi: 10.1128/jb.172.2.762-770.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amann R I, Ludwig W, Schleifer K H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Amman R I, Binder B J, Olson R J, Chisholm S W, Devereux R, Stahl D A. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol. 1990;56:1919–1925. doi: 10.1128/aem.56.6.1919-1925.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arnoldi J, Schluter C, Duchrow M, Hubner L, Ernst M, Teske A, Flad H-D, Gerdes J, Bottger E C. Species-specific assessment of Mycobacterium leprae in skin biopsies by in situ hybridisation and polymerase chain reaction. Lab Investig. 1992;66:618–623. [PubMed] [Google Scholar]
- 8.Bickerman J J. Foams. New York, N.Y: Springer-Verlag; 1973. [Google Scholar]
- 9.Blackall L L. Molecular identification of activated-sludge foaming bacteria. Water Sci Technol. 1994;29:35–42. [Google Scholar]
- 10.Box G E P, Cox D R. An analysis of transformations. J R Stat Soc. 1964;B26:211–243. [Google Scholar]
- 11.Braun-Howland E B, Danielson S A, Nierzwicki-Bauer S A. Development of a rapid method for detecting cells in situ using 16S rRNA-targeted probes. BioTechniques. 1993;13:928–934. [PubMed] [Google Scholar]
- 12.Chun J. Computer assisted classification and identification of actinomycetes. Ph.D. thesis. Newcastle upon Tyne, United Kingdom: University of Newcastle-upon-Tyne; 1995. [Google Scholar]
- 13.Chun J, Kang S-O, Hah Y C, Goodfellow M. Phylogeny of mycolic acid-containing actinomycetes. J Ind Microbiol. 1996;17:205–213. [Google Scholar]
- 14.Collins M D, Jones D. Lipid composition of Corynebacterium paurometabolum (Steinhaus) FEMS Microbiol Lett. 1982;13:13–16. [Google Scholar]
- 15.Collins M D, Goodfellow M, Minnikin D E. A survey of the structures of mycolic acids in Corynebacterium and related taxa. J Gen Microbiol. 1982;128:129–149. doi: 10.1099/00221287-128-1-129. [DOI] [PubMed] [Google Scholar]
- 16.Davenport R J, Elliott J N, Curtis T P, Upton J. In situ detection of rhodococci associated with activated sludge foams. Antonie Leeuwenhoek. 1998;74:41–48. doi: 10.1023/a:1001795609982. [DOI] [PubMed] [Google Scholar]
- 17.de los Reyes F L, Ritter W, Raskin L. Group-specific small-subunit rRNA hybridization probes to characterize filamentous foaming in activated sludge systems. Appl Environ Microbiol. 1997;63:1107–1117. doi: 10.1128/aem.63.3.1107-1117.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de los Reyes F L, Oerther D B, de los Reyes M A, Hernandez M, Raskin L. Characterization of filamentous foaming in activated sludge systems using oligonucleotide hybridisation probes and antibody probes. Water Sci Technol. 1998;37:485–493. [Google Scholar]
- 19.de los Reyes M F, de los Reyes III F L, Hernandez M, Raskin L. Quantification of Gordona amarae strains in foaming activated sludge and anaerobic digester systems with oligonucleotide hybridization probes. Appl Environ Microbiol. 1998;64:2503–2512. doi: 10.1128/aem.64.7.2503-2512.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Franks H A, Harmsen H J M, Raangs G C, Jansen G J, Schut F, Welling G W. Variations in bacterial populations in human feces measured by fluorescent in situ hybridization with group-specific 16S rRNA-targeted oligonucleotide probes. Appl Environ Microbiol. 1998;64:3336–3345. doi: 10.1128/aem.64.9.3336-3345.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goodfellow M, Alderson G, Chun J. Rhodococcal systematics: problems and developments. Antonie Leeuwenhoek. 1998;74:3–20. doi: 10.1023/a:1001730725003. [DOI] [PubMed] [Google Scholar]
- 22.Goodfellow M, Stainsby F M, Davenport R, Chun J, Curtis T. Activated sludge foaming: the true extent of actinomycete diversity. Water Sci Technol. 1998;37:511–519. [Google Scholar]
- 23.Goodfellow M, Davenport R, Stainsby F M, Curtis T P. Actinomycete diversity associated with foaming in activated sludge plants. J Ind Microbiol. 1996;17:268–280. [Google Scholar]
- 24.Goodfellow, M., K. Isik, and E. Yates. Acta Nova Leopoldina, in press.
- 25.Gordon R E, Mihm J E. Identification of Nocardia caviae (Erikson) nov. comb. Ann N Y Acad Sci. 1962;98:628–636. [Google Scholar]
- 26.Hernandez M, Jenkins D, Beaman B L. Mass and viability estimations of Nocardia in activated sludge and anaerobic digesters using conventional stains and immunofluorescent methods. Water Sci Technol. 1994;29:249–259. [Google Scholar]
- 27.Jenkins D, Richard M G, Daigger G T. Manual on the causes and control of activated sludge bulking and foaming. 2nd ed. Boca Raton, Fla: Lewis Publishers; 1993. [Google Scholar]
- 28.Kepner R L, Pratt J R. Use of fluorochromes for direct enumeration of total bacteria in environmental samples: past and present. Microbiol Rev. 1994;58:603–615. doi: 10.1128/mr.58.4.603-615.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Langendijk P S, Schut F, Jansen G J, Raangs G C, Kamphuis G R, Wilkinson M H F, Welling G W. Quantitative fluorescence in situ hybridization of Bifidobacterium spp. with genus-specific 16S rRNA-targeted probes and its application in fecal samples. Appl Environ Microbiol. 1995;61:3069–3075. doi: 10.1128/aem.61.8.3069-3075.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Macnaughton S J, O'Donnell A G, Embley T M. Permeabilization of mycolic-acid-containing actinomycetes for in-situ hybridization with fluorescently labelled oligonucleotide probes. Microbiology. 1994;140:2859–2865. doi: 10.1099/00221287-140-10-2859. [DOI] [PubMed] [Google Scholar]
- 31.Maidak B L, Olsen G J, Larsen N, Overbeek O, McCaughey M J, Woese C R. The RDP (Ribosomal Database Project) Nucleic Acids Res. 1997;25:109–110. doi: 10.1093/nar/25.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Manz W, Amann R, Ludwig W, Wagner M, Schleifer K-H. Phylogenetic oligodeoxynucleotide probes for the major subclasses of proteobacteria: problems and solutions. Syst Appl Microbiol. 1992;15:593–600. [Google Scholar]
- 33.Manz W, Wagner M, Amann R, Schleifer K-H. In situ characterization of the microbial consortia active in two wastewater treatment plants. Water Res. 1994;28:1715–1723. [Google Scholar]
- 34.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 35.Minnikin D E, Minnikin S M, Hutchinson I G, Goodfellow M, Grange J M. Mycolic acid patterns of representative strains of Mycobacterium fortuitum, ‘Mycobacterium peregrinum’ and Mycobacterium smegmatis. J Gen Microbiol. 1984;130:363–367. doi: 10.1099/00221287-130-2-363. [DOI] [PubMed] [Google Scholar]
- 36.Mori T, Sakai Y, Honda K, Yano I, Hashimoto S. Stable abnormal foam in activated-sludge process produced by Rhodococcus with strong hydrophobic property. Environ Technol Lett. 1988;9:1041–1048. [Google Scholar]
- 37.Mori T, Itokazu K, Ishikura Y, Mishina F, Sakai Y, Koga M. Evaluation of control strategies for actinomycete scum in full-scale treatment plants. Water Sci Technol. 1992;25:231–237. [Google Scholar]
- 38.Pipes W O. Actinomycete scum production in activated sludge processes. Res J Water Pollut Control Fed. 1978;50:628–634. [Google Scholar]
- 39.Pujol R, Duchene P, Schetrite S, Canler J P. Biological foams in activated sludge plants: characterization and situation. Water Res. 1991;25:1399–1404. [Google Scholar]
- 40.Ramsing N B, Fossing H, Ferdelman T G, Andersen F, Thamdrup B. Distribution of bacterial populations in a stratified fjord (Mariager Fjord, Denmark) quantified by in situ hybridization and related to chemical gradients in the water column. Appl Environ Microbiol. 1996;62:1391–1404. doi: 10.1128/aem.62.4.1391-1404.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schuppler M, Mertens F, Schon G, Gobel U B. Molecular characterisation of nocardioform actinomycetes in activated sludge by 16S rRNA analysis. Microbiology. 1995;141:513–521. doi: 10.1099/13500872-141-2-513. [DOI] [PubMed] [Google Scholar]
- 42.Schuppler M, Wagner M, Schon G, Gobel U B. In situ identification of nocardioform actinomycetes in activated sludge using fluorescent rRNA-targeted oligonucleotide probes. Microbiology. 1998;144:249–259. doi: 10.1099/00221287-144-1-249. [DOI] [PubMed] [Google Scholar]
- 43.Seviour E M, Williams C J, Seviour R J, Soddell J A, Lindrea K C. A survey of filamentous bacterial populations from foaming activated sludge plants in eastern states of Australia. Water Res. 1990;24:493–498. [Google Scholar]
- 44.Soddell J A, Seviour R J. Numerical taxonomy of Skermania piniformis and related isolates from activated sludge. J Appl Microbiol. 1998;84:272–284. doi: 10.1046/j.1365-2672.1998.00341.x. [DOI] [PubMed] [Google Scholar]
- 45.Soddell J A, Seviour R J. Microbiology of foaming in activated sludge plants. J Appl Bacteriol. 1990;69:145–176. [Google Scholar]
- 46.Soddell J A, Knight G, Strachan W, Seviour R J. Nocardioforms not nocardia forms. Water Sci Technol. 1992;26:455–460. [Google Scholar]
- 47.Sokal R R, Rohlf F J. Biometry: the principles and practice of statistics in biological research. 3rd ed. New York, N.Y: W. H. Freeman and Co.; 1995. [Google Scholar]
- 48.Sutcliffe I C. Cell envelope composition and organisation in the genus Rhodococcus. Antonie Leeuwenhoek. 1998;74:49–58. doi: 10.1023/a:1001747726820. [DOI] [PubMed] [Google Scholar]
- 49.Wagner M, Erhart R, Manz W, Amann R, Lemmer H, Wedi D, Schleifer K-H. Development of an rRNA-targeted oligonucleotide probe specific for the genus Acinetobacter and its application for in situ monitoring in activated sludge. Appl Environ Microbiol. 1994;60:792–800. doi: 10.1128/aem.60.3.792-800.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]