Abstract
The bacterial community structure of the activated sludge from a 25 million-gal-per-day industrial wastewater treatment plant was investigated using rRNA analysis. 16S ribosomal DNA (rDNA) libraries were created from three sludge samples taken on different dates. Partial rRNA gene sequences were obtained for 46 rDNA clones, and nearly complete 16S rRNA sequences were obtained for 18 clones. Seventeen of these clones were members of the beta subdivision, and their sequences showed high homology to sequences of known bacterial species as well as published 16S rDNA sequences from other activated sludge sources. Sixteen clones belonged to the alpha subdivision, 7 of which showed similarity to Hyphomicrobium species. This cluster was chosen for further studies due to earlier work on Hyphomicrobium sp. strain M3 isolated from this treatment plant. A nearly full-length 16S rDNA sequence was obtained from Hyphomicrobium sp. strain M3. Phylogenetic analysis revealed that Hyphomicrobium sp. strain M3 was 99% similar to Hyphomicrobium denitrificans DSM 1869T in Hyphomicrobium cluster II. Three of the cloned sequences from the activated sludge samples also grouped with those of Hyphomicrobium cluster II, with a 96% sequence similarity to that of Hyphomicrobium sp. strain M3. The other four cloned sequences from the activated sludge sample were more closely related to those of the Hyphomicrobium cluster I organisms (95 to 97% similarity). Whole-cell fluorescence hybridization of microorganisms in the activated sludge with genus-specific Hyphomicrobium probe S-G-Hypho-1241-a-A-19 enhanced the visualization of Hyphomicrobium and revealed that Hyphomicrobium appears to be abundant both on the outside of flocs and within the floc structure. Dot blot hybridization of activated sludge samples from 1995 with probes designed for Hyphomicrobium cluster I and Hyphomicrobium cluster II indicated that Hyphomicrobium cluster II-positive 16S rRNA dominated over Hyphomicrobium cluster I-positive 16S rRNA by 3- to 12-fold. Hyphomicrobium 16S rRNA comprised approximately 5% of the 16S rRNA in the activated sludge.
Activated sludge, a common biological treatment method for both municipal and industrial wastewater, represents a complex microbial community. Due to intricate interactions within the microbial community, process control of wastewater treatment plants can be difficult. Population shifts within the microbial community may result from changes in the plant operating conditions and cause sludge quality problems such as poor sludge settling, compaction, and dewatering (22). The application of molecular analysis to activated sludge is of considerable interest as a means for determining the microbial diversity and robustness, identifying populations associated with process upsets, and developing probes for diagnosing, monitoring, and controlling activated-sludge problems (5, 7, 8, 16, 27, 33, 37). Some operational problems with activated sludge can often be detected microscopically at the microorganism level. For example, poor sludge settling due to filamentous bulking is due to excessive growth of bacterial filaments (e.g., Sphaerotilus natans, Microthrix parvicella, Hyphomicrobium spp., Thiothrix nivea, etc.) (19). Diagnosis and correction of such activated-sludge problems require the correct identification of the responsible microbial population(s) and institution of appropriate process changes to select for or against specific organisms.
A potential for considerable variation between the activated sludge of industrial and that of municipal wastewater treatment exists due to the differences in chemical composition of the treated waste streams. In this study, the microbial community from an activated sludge system used to treat wastewater from chemical manufacturing processes was examined. This differs from municipal wastewater in that it is much higher in its total organic carbon load, comprising mostly simple organic acids and alcohols, with little fibrous or complex carbon sources present. One microorganism of particular interest routinely monitored in this wastewater treatment system is Hyphomicrobium. Hyphomicrobium spp. have been reported in both sewage treatment plants and adjacent waters (15, 17, 20). Recently, the 16S ribosomal DNA (rDNA) sequences for seven species of the genus Hyphomicrobium were published (30). These species fall into two distinct phylogenetic clusters indistinguishable by morphological characteristics alone. The maintenance of Hyphomicrobium in this industrial activated sludge is important due to its ability to degrade C-1 compounds such as methanol, which is found in the influent wastewater (18, 20). However, hypertrophic growth (i.e., hyphal elongation) of Hyphomicrobium can lead to poor sludge settling and compaction (A. J. Meyers and C. D. Meyers, Abstr. 86th Annu. Meet. Am. Soc. Microbiol. 1986, abstr. N-93, 1986). Therefore, the ability to differentiate and reliably monitor Hyphomicrobium levels in the flocs is fundamental for optimal control and operation of the wastewater treatment plant.
Other researchers (5, 33, 37) have proposed that a combination of approaches is needed to understand the basic microbial community structure of activated sludge. These methods include the construction and analysis of 16S rDNA libraries, hybridization with rRNA-targeted oligonucleotides, and comparison of those results with a specific focus on the presence of Hyphomicrobium-like strains.
MATERIALS AND METHODS
Samples.
Activated sludge samples were collected from the Eastman Chemical Company wastewater treatment plant (Tennessee Eastman Division, Kingsport, Tenn.). The influent wastewater contains primarily low-molecular-weight organic acids (e.g., acetic acid, propionic acid, n-butyric acid) and short-chain alcohols such as methanol, ethanol, and isopropanol (10). The plant includes four parallel trains, each with three aeration basins in series, and is operated in a modified step-feed flow configuration. The relevant operating characteristics for the 3 months in which samples were taken are provided in Table 1.
TABLE 1.
Performance summary for the wastewater treatment plant during the 3 months of sampling
| Process or parameter | Value for sample taken in:
|
||
|---|---|---|---|
| January 1995 | February 1995 | April 1995 | |
| Activated sludge process | |||
| Influent STOCb load (lb/day) | 121,927 | 128,465 | 149,909 |
| Influent methanol concn (mg/liter) | 101 | 124 | 108 |
| Influent acetic acid concn (mg/liter) | 484 | 422 | 516 |
| Flow split into 3 aeration basins (1/2/3) | 40/40/20 | 40/40/20 | 40/40/20 |
| Sludge age (days) | 12.1 | 15.4 | 11.8 |
| Solids inventory (tons) | 734 | 815 | 752 |
| Basin temp (°F) | 87.7 | 85.8 | 95.3 |
| Sludge volume index (g/ml) | 199 | 202 | 140 |
| Total filament rating (0–6)a | 3.2 | 3.3 | 4.0 |
| Hyphomicrobium rating (0–6)a | 2.4 | 2.0 | 4.6 |
| Clarification polymer concn (ppm) | 16.1 | 19.3 | 10.5 |
| Effluent total suspended solids (mg/liter) | 6.2 | 6.8 | 11.1 |
| Effluent STOC concn (mg/liter) | 24 | 24 | 27 |
| Aeration basin dissolved oxygen concn (mg/liter) | 1.5 | 1.8 | 1.7 |
| Sludge dewatering process | |||
| Waste activated-sludge concn (mg/liter) | 19,708 | 18,545 | 26,915 |
| Spin solids (% total solids) | 8.6 | 8.6 | 12.9 |
| Belt filter press cake solids (% total solids) | 10.5 | 10.3 | 13.3 |
| Cake production (dton/day) | 60.2 | 53.1 | 62.6 |
Microscopic rating scale based on that of Jenkins et al. (19): 0, none; 1, few; 2, some; 3, common; 4, very common; 5, abundant; and 6, excessive.
STOC, soluble total organic carbon.
Isolation and growth of Hyphomicrobium strains.
Hyphomicrobium sp. strain M3 was previously isolated and identified from this activated sludge during an episode of poor settling due to the abundance of its elongated hyphae protruding from the flocs (Meyers and Meyers, Abstr. 86th Annu. Meet. Am. Soc. Microbiol. 1986). Isolation of the strain was on a liquid medium consisting of 1.36 g of KH2PO4, 0.5 g of (NH4)2SO4, 0.4 g of KNO3, 0.01 g of CaCl2 · 2H2O, 0.0031 g of MnSO4 · 4H2O, 2.13 g of Na2HPO4, 0.2 g of MgSO4 · 7H2O, 0.005 g of FeSO4 · 7H2O, and 0.0025 g of Na2MoO4 · 2H2O/liter with a final pH of 7.2 (1) and supplemented with 10 ml of methanol/liter under anoxic conditions at 30°C. After sequential enrichment of the strain on media with methanol, the strain was routinely cultivated aerobically at 30°C using the above-described medium without KNO3 and supplemented with 10 g of methylamine hydrochloride/liter. Isolation in pure culture was accomplished on this medium solidified with 15 g of agar/liter. Hyphomicrobium sp. strain M3 has been deposited in the American Type Culture Collection as ATCC 202122.
Strain M3 was identified as Hyphomicrobium based on morphological and physiological characteristics. In pure culture, this gram-negative, oval-shaped bacterium showed the diagnostic “mother cell,” with its monopolar budding giving rise to a “daughter cell” at the terminus of a thin, thread-like hypha (prostheca) (18). Hyphomicrobium sp. strain M3 is mesophilic (growth temperature range of 10 to 35°C; optimum, 30°C), is relatively slow growing (doubling time on methanol at 30°C is 9.1 h), is facultatively anaerobic, and has hyphal lengths of 3 to 5 μm (Meyers and Meyers, Abstr. 86th Annu. Meet. Am. Soc. Microbiol. 1986). This particular strain can utilize several C1 compounds (e.g., methanol, methylamine HCl, formic acid, methylurea, dimethylamine, and formamide) but not others (e.g., methane, formaldehyde, dimethyl ether, and dimethyl sulfoxide), qualifying it as a restrictive type of methyltroph. Selective C2 compounds such as acetic acid, ethanol, and ethylamine support its growth, while higher alcohols (e.g., n-isopropanol and n-butanol) and organic acids (e.g., propionic, n-butyric, succinic, and citric), sugars (e.g., glucose and sucrose), and other compounds (e.g., ethylene glycol and acetone) fail to support growth. A rather wide range of nitrogen sources are employed by Hyphomicrobium sp. strain M3; these include ammonium chloride, sodium nitrate, sodium nitrite, sodium azide, oxamic acid, and various amines (e.g., methylamine, dimethylamine, and glucosamine) and amides (e.g., formamide, acetamide, and N,N-dimethylformamide). Amino acids do not serve as either N or C sources for the organism. As with other Hyphomicrobium isolates, strain M3 accumulates intracellular reserves of poly-β-hydroxybutyric acid, engages in rosette formation, and produces a pellicle in quiescent liquid culture.
Extraction of DNA from sludge.
DNA was extracted from this sludge by following a modified method of Ogram et al. (28). Sludge samples (volumes ranging from 20 to 150 ml) were centrifuged at 5,500 × g at 10°C for 10 min. The supernatant fractions were discarded, and the pellets were resuspended in 50 ml of 0.12 M Na2PO4 (pH 8.0)–2.5 ml of 5% (wt/vol) sodium dodecyl sulfate. These samples were incubated at 70°C for 1 h with periodic inversions by hand. Five grams of 0.1-mm-diameter glass beads was added to the samples, and the samples were blended for two 2-min bursts separated by a 1-min rest period. The samples were recovered and centrifuged again at 5,500 × g for 25 min at 10°C; the resulting supernatants were collected and stored at 4°C. The pellets were resuspended in 25 ml of 0.12 M Na2PO4 and incubated at 70°C for 20 min with periodic inversions by hand. After centrifugation at 5,500 × g for 25 min at 10°C, the supernatants were pooled with the previously collected supernatants. Precipitation for >2 h at −20°C was performed by the addition of 0.1 volume of 2 M sodium acetate (NaOAc) solution and 0.8 volumes of isopropanol. The precipitates were then centrifuged at 11,500 × g at 4°C for 30 min and dried under vacuum at −100°C. Excess salts were eliminated by dialysis using Spectra dialysis tubing (molecular weight cutoff of 6,000 to 8,000) overnight against 10 mM Tris-HCl–1 mM EDTA buffer (pH 8.0) (TE buffer). Dialyzed samples were extracted with Tris-saturated phenol followed by extraction with chloroform-isoamyl alcohol (24:1 [vol/vol]). The recovered aqueous phases were precipitated at −20°C for 2 h by the addition of 0.1 volume of 2 M NaOAc and 2 volumes of absolute ethanol. The final precipitation product was centrifuged at 11,500 × g for 30 min at 4°C and dried under vacuum at −100°C. The samples were resuspended in 1 ml of sterile TE buffer and stored at −20°C. RNA was removed from the samples by treatment with 5 μl of DNAse-free RNase (Boehringer Mannheim Corp., Indianapolis, Ind.) at 37°C for 1 h.
PCR amplification and cloning.
Libraries of 16S rDNA were constructed by PCR amplification of the target genes from DNA extracted from sludge and Hyphomicrobium sp. strain M3, followed by cloning into pCRII (TA cloning kit; Invitrogen, Carlsbad, Calif.). PCR amplification of the 16S rDNA was performed using the eubacterial primers 27f and 1525r or 1492r (24). The PCR mixture consisted of 2 μl of DNA template, 2.5 μl of each primer (5 ng/μl), 10 μl of 10× PCR buffer (GeneAmp PCR reagent kit; Perkin-Elmer Corp., Norwalk, Conn.), 2 μl each of a 200 μM solution of dATP, dCTP, dGTP, and dTTP, and 75 μl of sterile water. Each reaction mixture was overlaid with filter-sterilized mineral oil. The reaction mixtures were heated to 100°C, followed by the addition of 0.5 μl of Taq polymerase (5 U/μl; Gibco BRL, Gaithersburg, Md.). PCR amplification was performed for 38 cycles using 94°C for 1 min, 60°C for 1 min, and 72°C for 1 min. For each set of PCR amplifications, a control reaction without template was performed to check the kit and solution purity.
Amplified DNA was cloned into pCRII plasmid, and the ligation mixture was transformed into OneShot competent Escherichia coli cells (Invitrogen Corp.) by following manufacturer protocols. Colonies containing plasmid inserts were identified by blue/white color selection on Luria-Bertani (LB) plates with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside). Alkaline lysis plasmid preparations were made from cultures grown in 100 ml of LB broth with either ampicillin (100 μg/ml) or kanamycin (50 μg/ml). Inserts were verified by restriction digestion of the plasmids with EcoRI.
DNA sequencing and analysis.
Plasmids containing 16S rDNA inserts were sequenced by Retrogen, Inc. (San Diego, Calif.), or the Molecular Biology Resource Facility at the University of Tennessee (Knoxville) using an ABI PRISM dye terminator cycle sequencing kit with AmpliTaq DNA polymerase (protocol P/N 402078, revision A) and an Applied Biosystems 373 DNA sequencer (Perkin-Elmer, Foster City, Calif.). Greater than 400 bp were sequenced for each insert using single primer extension with the 1492r primer or 27f primer (24). Nearly full-length 16S rDNA sequences were obtained for Hyphomicrobium sp. strain M3, and 18 16S rDNA clones from the sludge were obtained using the additional primers 27f, 530f, 907r, and 926f (24), M13f (−40), and M13r located on the pCRII plasmid.
All sequences were analyzed using the CHECK_CHIMERA and the SIMILARITY_RANK programs of the Ribosomal Database Project (26). The sequences were also analyzed using the BLAST program (3) (National Center for Biotechnology Information) to determine the closest available database sequences.
Selected DNA sequences were aligned using the Clustal W program (36). Published sequences were obtained from GenBank. A phylogenetic tree was constructed using Clustal W by distance matrix analysis and the neighbor-joining method (31). Bootstrap analysis was used to provide statistical confidence for the tree branch points. Phylogenetic trees were displayed using TREEVIEW (29).
Multiple pairwise alignments of the 16S rDNA sequences from April 1995 activated-sludge clones (those whose designations begin with 495) and Hyphomicrobium vulgare (X53182) were performed using the PILEUP program in the Genetics Computer Group data analysis package. Based on visual inspection of the sequence alignments, a probe for the region of Hyphomicrobium spp. corresponding to the 16S rRNA positions 1241 to 1260 in E. coli (9) was designed. The naming of the probe was based on the convention proposed by the Oligonucleotide Probe Database (2). Probe specificity was determined using the program PROBE_CHECK in the Ribosomal Database Project (26). Additional probes were designed to differentiate Hyphomicrobium clusters I and II based on alignments of full-length clone sequences and published sequences from seven Hyphomicrobium species. Hyphomicrobium probe sequences and target organisms are presented in Table 2.
TABLE 2.
Characteristics of oligonucleotide probes used to distinguish Hyphomicrobium spp.
| Target bacteria or probe | Oligonucleotide probe
|
16S rRNA location | No. of probe mismatches to 16S rDNA sequences from control strains and clones of:
|
||||
|---|---|---|---|---|---|---|---|
| Designation | Sequence | Cluster I | Cluster II | Z. ramigera | Bradyrhizobium japonicum | ||
| Hyphomicrobium genus | S-G-Hypho-1241-a-A-19 | GCTGC(G/C)CATTGTCACCGCC | 1241–1260 | 0 | 0 | 2 | 2 |
| Hyphomicrobium cluster Ia | S-S-HyphoC1-648-a-A-20 | CCTCTTCCGGACTCGAGACT | 648–667 | 0–1c | 2–3 | 4 | 5 |
| Hyphomicrobium cluster IIb | S-S-HyphoCII-654-a-A-18 | CCCACCTCTATCGGACTC | 654–672 | 2–5 | 0 | 6 | 4 |
| Universal probe 1390 | S-*-Univ-1390-a-A-18d | GACGGGCGGTGTGTAAA | 1390–1408 | 0 | 0 | 0 | 0 |
Cluster I strains: H. vulgare ATCC 25700 (Y14302), Hyphomicrobium-like sp. strain US-353 (U06473), Hyphomicrobium hollandicum IFAM KB-677 (Y14303), Hyphomicrobium aestuarii DSM 1564 (Y14304); cluster I clones: 1951, 1956, 49519, 49520.
Cluster II strains: Hyphomicrobium sp. strain M3, H. denitrificans DSM 1869 (Y14308), Hyphomicrobium methylovorum DSM 5458 (Y14307), Hyphomicrobium facilis sp. strain H-526 (Y14309), Hyphomicrobium facilis ATCC 27492 (Y14310); cluster II clones: 4953, 49512, 49518.
The cluster I probe has one mismatch in the last 3′ base to clones 49519 and 49520.
See reference 38.
Whole-cell hybridizations.
Whole-cell hybridizations were performed using fluorescent 16S rRNA probes by following published methods (4, 6, 34) with the exceptions that Igepal CA-630 was substituted for Nonidet P-40 and an additional 5-min sonication step in an ice-water bath was used after fixation in paraformaldehyde. Prior to sonication, sludge samples were resuspended in 10 mM EDTA to enhance the permeability of the flocs and probe penetration. Hybridization experiments were performed with 5′-end-labeled fluorescein oligonucleotide probes obtained from Genosys (The Woodlands, Tex.). The control strains used to determine the concentration of formamide needed at 37°C to discriminate zero, one, and two mismatches using probe S-G-Hypho-1241-a-A-19 included H. vulgare ATCC 27500 (no mismatches), Hyphomicrobium sp. strain M3 (no mismatches), Zoogloea ramigera ATCC 19623 (two mismatches), and Nitrobacter winogradskyi ATCC 14123 (one mismatch). The optimum concentration of formamide for hybridization was determined using 20, 30, 40, and 60% (vol/vol) formamide concentrations. Both H. vulgare and Hyphomicrobium sp. strain M3 were positive with all formamide concentrations tested. Z. ramigera was positive at 20 and 30% (vol/vol) formamide and had weak fluorescence at 40 and 60% (vol/vol) formamide. N. winogradskyi was positive at 20 and 30% (vol/vol) formamide and negative at 40 and 60% (vol/vol) formamide. Based on these results, hybridization with this probe was carried out at 40% (vol/vol) formamide at 37°C. Slides were visualized using a Zeiss Axioskop with a Zeiss 100-W epifluorescence illuminator and Zeiss fluorescein isothiocyanate filter 31001. Pictures were obtained with a Zeiss (optronics) ZVS-3C75DE three-chip video camera and a TCX Frame Grabber board or Sony CVP-M3 video printer.
Extraction of RNA from sludge samples and slot blot hybridizations.
RNA was isolated from approximately 2 g of sludge solids or 100 ml from pure cultures by following previously published methods (14). Final RNA pellets were resuspended in 100 μl of diethyl pyrocarbonate-treated water and stored at −80°C. RNA dot blots were prepared as outlined by Sambrook et al. (32). RNA amounts on the dot blots were 1 μg and 500, 250, 100, 50, and 10 ng for control samples, and 0.1-ml aliquots were used for sludge samples. Samples were vacuum blotted onto 0.45-μm-pore-size Biotrans membranes (ICN, Irvine, Calif.) using a Bio-Rad dot blot apparatus and baked for 1 h at 80°C. The blots were pretreated for at least 2 h in a hybridization solution (11). Probes were end labeled with [δ-32P]ATP by following the protocol described by Life Technologies (Gaithersburg, Md.) and purified using Nuc-Trap push columns (Stratagene, La Jolla, Calif.). Stringency experiments were conducted with four control strains with zero to six probe mismatches as shown in Table 2. The optimal hybridization temperature was determined by hybridizing blots containing the control strains with the probes at temperatures from 42 to 65°C. Optimal hybridization was achieved at 50°C for S-S-HyphoC1-648-a-A-20, 52°C for S-S-HyphoCII-654-a-A-18, and 65°C for S-G-Hypho-1241-a-A-19. Hybridizations with universal probe 1390 were carried out at 45°C (38). Blots were hybridized overnight and were washed at the same hybridization temperature with 1× SSC (0.15 M NaCl plus 0.015 M sodium citrate)–0.1% sodium dodecyl sulfate solution two times for 15 min each. The RNA levels were quantified using a Storm 840 PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.). The number of nanograms of RNA hybridizing in sludge samples with each probe was determined by regression analysis of standard curves (nanograms of total 16S rRNA versus signal intensity) generated from probe hybridization with the positive-control strains. The percentage of the total RNA probing positive in each sample was calculated as [(nanograms of specific RNA)/(nanograms of universal 16S rRNA)] · 100. The percentage of the total 16S rRNA probing positive in sludge samples for each month, except January, was the average of results from three or four samples.
Nucleotide sequence accession numbers.
The 16S rDNA clone sequences were deposited in GenBank at the National Center for Biotechnology Information under accession no. AF097766 to AF097829, and the 16S rDNA sequence from Hyphomicrobium sp. strain M3 received accession no. AF098790.
RESULTS
Analysis of 16S rDNA sequences from activated-sludge samples.
Preliminary analysis based on sequences using the 1492r or 27f primers was performed on 67 16S rDNA clones from three different sludge samples taken from the same industrial wastewater treatment plant. Individual clones were given numbers based on the month and year of the sludge sample and an individual clone number. Clones from the January 1995 sample begin with 195, those from the February 1995 sample begin with 295, and those from the April 1995 sample begin with 495. All clones were subjected to CHIMERA_CHECK analysis through the Ribosomal Database Project. Three of the clones were considered potential chimeras and were not further analyzed. Preliminary work to classify the clones into known subdivisions was performed using the SIMILARITY_RANK program of the Ribosomal Database Project and BLAST analysis of all sequences in GenBank. None of the sequences were identical to previously isolated sequences. Sequences of the majority of strains exhibited 90 to 95% similarity to 16S rRNA sequences in GenBank. Four clones had sequences that were less than 90% similar to published 16S rRNA sequences in GenBank. The sequences of several of these clones showed high similarity to sequences obtained from a phosphate-removing sludge in a sequencing batch reactor (SBR) (8). These included clones 2951, 2952, 2958, and 29523 from the Holophaga/Acidobacterium cluster with 95% similarity to SBR1078 and SBR10103 and 1959 in an undescribed cluster of Cytophaga with 96% similarity to UNSBR1093. Seven clones also showed a 95 to 96% sequence homology with beta subdivision clones from municipal activated sludge (33). Based on these identifications, the cloned 16S rDNAs were assigned to Eubacteria groups and Proteobacteria subdivisions (Table 3). In this analysis, population shifts were seen among the 3 months, with alpha and beta type bacteria dominating in January and April. The February clones differed from the January and April clones in that alpha subdivision clones were absent and the majority of clones belonged to the Holophaga/Acidobacterium group.
TABLE 3.
Distribution of 16S rDNA cloned sequences from three different sludge samples
| Phylogenetic classification | % of clone sequences in:
|
||
|---|---|---|---|
| January | February | April | |
| Alpha subdivision of Proteobacteria | 30a | 0 | 46a |
| Beta subdivision of Proteobacteria | 50 | 11 | 21 |
| Gamma subdivision of Proteobacteria | 5 | 21 | 8 |
| Delta subdivision of Proteobacteria | 10 | 0 | 4 |
| Acidobacterium/Holophaga | 0 | 68 | 0 |
| Cytophaga | 0 | 0 | 16 |
| Firmicutes | 5 | 0 | 5 |
Includes Hyphomicrobium species.
Analysis of alpha subdivision clones.
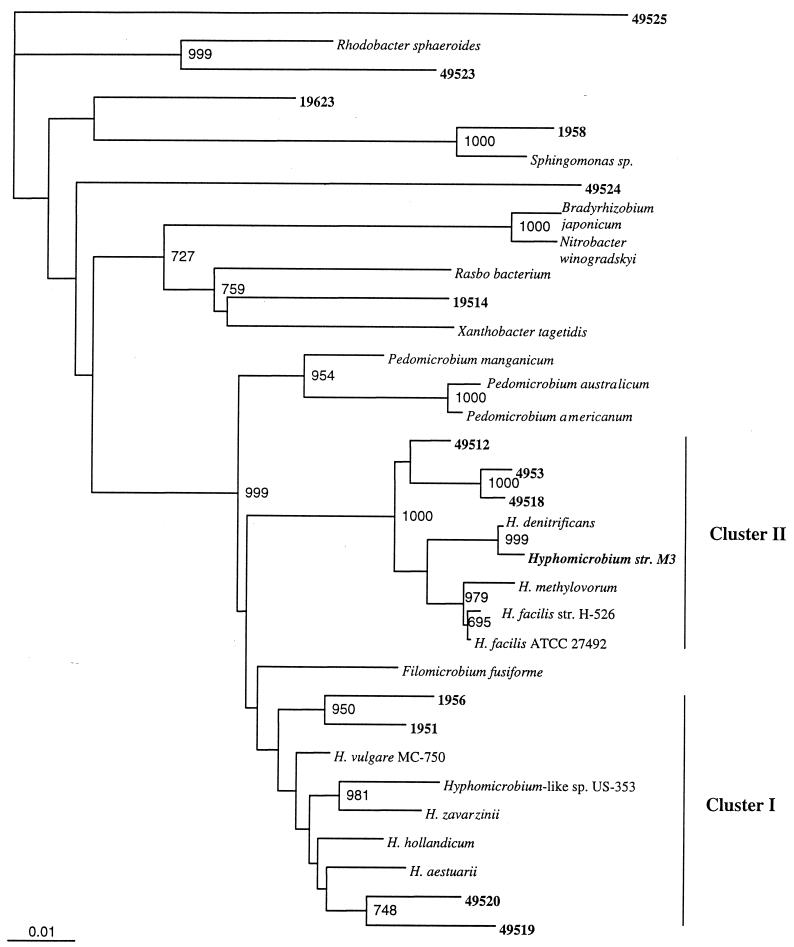
More than 1,300 bp of 16S rDNA sequences was determined for Hyphomicrobium sp. strain M3 and for 12 clones which fell into the alpha subdivision. A phylogenetic tree based on evolutionary distance and neighbor joining was constructed with 950 bp of 16S rDNA sequences from three Pedomicrobium spp., nine Hyphomicrobium spp., and three other members of the alpha subdivision (Fig. 1). In this phylogenetic analysis, known Hyphomicrobium spp. grouped into two clusters as described by Rainey et al. (30) and the Pedomicrobium spp. also fell into a distinctive phylogenetic group. The 16S rDNA sequence from Hyphomicrobium sp. strain M3 was 99.6% similar to that of Hyphomicrobium denitrificans DSM 1869T in Hyphomicrobium cluster II. Three of the clone sequences (4953, 49512, and 49518) also grouped with cluster II and were 96.2 to 96.5% similar to those of Hyphomicrobium sp. strain M3 and H. denitrificans DSM 1869T. Another two clones (1951 and 1956) were most closely related to Hyphomicrobium cluster I containing the H. vulgare species and were 96.6 to 96.8% similar to H. vulgare MC-750. Clones 49519 and 49520 also grouped most closely to the Hyphomicrobium cluster I (i.e., 94.9 to 95.5% similarity) but were only 94.3 and 95.1% similar to clones 1951 and 1956, respectively, and may represent a different cluster within the genus Hyphomicrobium.
FIG. 1.
Distance matrix tree showing the phylogenetic relationship between Hyphomicrobium sp. strain M3 and 16S rDNA clones from activated sludge libraries with the hyphal budding bacteria. Reference strains used were H. vulgare IFAM MC-750 (Y14302), Hyphomicrobium-like sp. strain US-353 (U06473), Hyphomicrobium hollandicum IFAM KB-677 (Y14303), Hyphomicrobium aestuarii DSM 1564 (Y14304), Hyphomicrobium zavarzinii ATCC 27495 (U59506), H. denitrificans DSM 1869 (Y14308), Hyphomicrobium methylovorum DSM 5458 (Y14307), Hyphomicrobium facilis sp. strain H-526 (Y14309), Hyphomicrobium facilis ATCC 27492 (Y14310), Filomicrobium fusiforme DSM 5304 (Y14313), Pedomicrobium austrailicum (X97694), Pedomicrobium americanum (X97692), Pedomicrobium manganicum (X97691), Rhodobacter sphaeroides (D16424), Rasbo bacterium (AF007948), Sphingomonas sp. strain B28161 (AJ001052), N. winogradskyi ATCC 14123 (L35507), Xanthobacter tagetidis (X99469), and Bradyrhizobium japonicum USDA110 (D13430). Bootstrap values per 1,000 bootstrap analyses are presented for values greater than 650. Clone sequences from the activated-sludge libraries from this study and Hyphomicrobium sp. strain M3 are in boldface.
Whole-cell hybridizations in activated sludge.
General Hyphomicrobium probe S-G-Hypho-1241-a-A-19 was designed to hybridize to all Hyphomicrobium strains based on sequence alignments between the 495 16S rDNA sequences and that of H. vulgare. This probe was intended to be degenerate in one position (G/C) to include H. vulgare, 49520, 49519, 4953, 49518, and 49512. Isolation and additional sequence analysis indicated that the probe would also hybridize to Hyphomicrobium sp. strain M3 and to the 16S rDNA cloned sequences 1951 and 1956. The specificity of the probe was initially checked using PROBE_CHECK from the Ribosomal Database Project. At that time the only strains which showed no mismatches were H. vulgare and Hyphomicrobium-like organism US-353. A few other alpha subdivision bacteria, including Rhizobium and Sphingomonas species, showed one or two mismatches to the probe. Most of the sludge 16S rDNA sequences from Hyphomicrobium-type clones were complementary to the probe with the G nucleotide, whereas sequences from H. vulgare and Hyphomicrobium-like sp. strain US-353 were complementary to the probe with the C nucleotide.
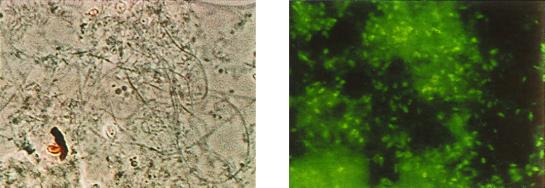
At this industrial wastewater treatment plant, detection and quantification of Hyphomicrobium levels in the sludge have been based on morphology and a rating system using conventional microscopic techniques. When phase-contrast illumination on either simply stained or Gram-stained slide preparations was used, individual Hyphomicrobium cells were indistinguishable from other microbial cells within the flocs; indeed, only those cells whose hyphae protruded from the floc periphery were readily identifiable as Hyphomicrobium. However, with the aid of the fluorescently labeled S-G-Hypho-1241-a-A-19 probe, clear visualization of Hyphomicrobium embedded within the floc structure by fluorescence microscopic examination was made possible (Fig. 2). Although the distinguishing hypha and tip structure are not seen on the inside of the floc as they are on the outside, the cells had the characteristic ovoid morphology seen at the tips of the cells on the outside of the flocs.
FIG. 2.
Fluorescence whole-cell hybridization of activated sludge samples with the probe S-G-Hypho-1241-a-A-19. Shown are photomicrographs comparing activated sludge under phase-contrast (left) and fluorescent (right) illumination. Magnification, ×920.
Characterization and quantification of Hyphomicrobium in activated sludge samples.
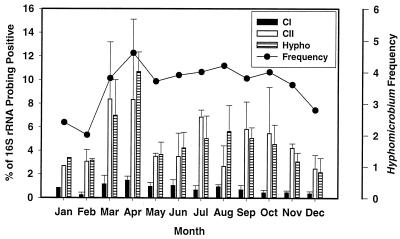
Based on the sequence alignments used to create the phylogenetic tree in Fig. 1, probes were designed for Hyphomicrobium cluster I and cluster II. H. vulgare MC-750 was used as the control strain for cluster I, and Hyphomicrobium sp. strain M3 was used as the control strain for cluster II (Table 2). These two probes also had higher numbers of mismatches to other known alpha subdivision sequences than S-G-Hypho-1241-a-A-19 and should allow better quantification of Hyphomicrobium populations (Table 2). RNA was extracted from 48 sludge samples collected from the wastewater treatment plant in 1995. The RNA was hybridized with the general Hyphomicrobium probe (S-G-Hypho-1241-a-A-19) and the cluster I (S-S-HyphoC1-648-a-A-20) and cluster II (S-S-HyphoCII-654-a-A-18) probes. The results from these analyses were arithmetically averaged by monthly periods (Fig. 3). The calculated values for total Hyphomicrobium-positive 16S rRNA using probe S-G-Hypho-1241-a-A-1 and for cluster II using probe S-S-HyphoCII-654-a-A-18 were similar for all months and ranged from 3 to 12% of the total 16S rRNA. The average percentage of Hyphomicrobium-positive 16S rRNA for the year was 4.8% ± 2.3%. The percentage of cluster I-positive 16S rRNA was lower and ranged from 0.2 to 1.5% of the total 16S rRNA. Hyphomicrobium-specific 16S rRNA was present throughout the year and peaked in March to April 1995. These results are in agreement with the visual microscopic ratings, which ranged from 2 (some) to 5 (abundant). The highest visual rating for Hyphomicrobium was in April, with an average score of 4.6.
FIG. 3.
Percentages of Hyphomicrobium-positive 16S rRNA (± standard deviations) in activated-sludge samples as determined by dot blot hybridization with the genus-specific Hyphomicrobium probe S-G-Hypho-1241-a-A-19 (Hypho) and the subgenus Hyphomicrobium probes S-S-HyphoC1-648-a-A-20 (cluster I [CI]) and S-S-HyphoCII-654-a-A-18 (cluster II [CII]).
DISCUSSION
Sequence analysis of 16S rRNA is a useful tool for studying the basic microbial community structure of activated sludge. For comparative purposes, ribosomal sequence data from a variety of activated sludge sources would be beneficial. In addition, to determine how operational parameters affect microbial communities, information on the plant operating conditions, as well as on sludge and effluent quality at the time of sampling is also needed. In this study, some plant operation data are provided (Table 1), although it is still unclear how process changes between January, February, and April affected the microbial community structure. The February sample was taken during a time period in which the sludge was experiencing poor dewatering due to zoogloeal infestation (23), whereas in April sludge dewatering had returned to normal conditions.
Libraries of 16S rDNA were constructed from three samples to relate potential shifts in the microbial populations to changes in operational parameters. While none of the clones were completely homologous to sequences found in GenBank, the majority of January (195) and April (495) clone sequences could be classified into well-known groups. Due to the increasing number of 16S rDNA sequences in GenBank, comparisons among diverse activated-sludge communities could be made. As found in other activated-sludge samples (8, 33, 37), the January and April samples had a large number of beta subdivision clones whose sequences had high similarity to published sequences. The microbial community in this same industrial sludge also had an abundance of alpha subdivision 16S rDNA sequences (16 of 64). Seven of these sequences were similar to the hyphal budding bacterium Hyphomicrobium or Pedomicrobium. Also of interest was the high similarity (96%) of the 49531 and 1959 clones to UNSBR1093 from a phosphate-removing bioreactor (8). These sequences have low similarity to other sequences in GenBank and may represent undescribed organisms in the activated sludge. Fourteen clones from February were most closely related to clones of soil bacteria (21, 25) which aggregate into cluster C of the recently described phylum Holophaga/Acidobacterium (25). Organisms belonging to this phylum appear to be widely distributed in soils, sediment, and activated sludge. The organisms in cluster C of this phylum have not been cultured, and their metabolic function in the activated sludge is unclear at this time.
The potential for biases in the construction of 16S rDNA libraries includes inefficient cell lysis, DNA recovery, and PCR amplification and cloning (13, 35). Therefore, the 16S rDNA sequences obtained in this present study may represent some of the members in the community but may not be all-inclusive or reflect the frequency of individuals in the community. For example, the February 16S rDNA sequences were very different from those sequences obtained from the January and April samples. It is unclear whether the 16S rDNA sequences obtained in the library from the February sample reflect changes in the microbial community during that time period or whether they are simply the result of bias in nucleic acid extraction efficiency and library formation. The fact that Hyphomicrobium type clones were not isolated in the February sludge library, even though the sludge samples probed positive with the general Hyphomicrobium probe (S-G-Hypho-1241-a-A-1) and the cluster II probe (S-S-HyphoCII-654-a-A-18) at approximately 3%, suggests that this library may be biased.
The presence of Hyphomicrobium in this industrial activated sludge was corroborated by its morphology using light microscopy, by isolation and identification of Hyphomicrobium sp. strain M3, and genetically by probe analysis. Hyphomicrobium sp. strain M3 was isolated from this sludge over 10 years, prior to the inception of this study (Meyers and Meyers, Abstr. 86th Annu. Meet. Am. Soc. Microbiol. 1986). Surprisingly, Hyphomicrobium sp. strain M3 showed a slightly higher sequence similarity to the cultured strain H. denitrificans DSM 1869T than to Hyphomicrobium clone isolates (99 versus 96%). This may represent a slight shift in the 16S rDNA sequence of the Hyphomicrobium cluster II population in this sludge or a bias that resulted from isolation and cultivation of Hyphomicrobium sp. strain M3. The placement of several cloned library sequences and strain M3 in the Hyphomicrobium cluster II was verified by phylogenetic analysis of 16S rDNA sequences. These analyses were in good agreement with the taxonomic placement of Pedomicrobium and Hyphomicrobium (12). Physiologically and morphologically, strain M3 belongs in the Hyphomicrobium genus because of its ability to utilize the C1 compounds (e.g., methanol and methylamine) and the formation of the characteristic mother cells with hyphae (18). Monopolar hyphal budding is the major morphological feature distinguishing Hyphomicrobium and Pedomicrobium (18). Hyphomicrobium sp. strain M3 has been shown to utilize monopolar budding as its mode of reproduction, based on light microscopic examination of pure cultures of the organism, thereby sustaining its genus identification.
A probe created to detect the Hyphomicrobium group was designed using the 16S rDNA library and was used to detect Hyphomicrobium spp. in activated-sludge samples. The distinctive morphology of Hyphomicrobium spp. allowed for the verification of the S-G-Hypho-1241-a-A-19 probe by whole-cell hybridization with activated sludge from the wastewater treatment plant. The fluorescein-labeled S-G-Hypho-1241-a-A-19 probe in conjunction with fluorescence microscopy provided a better resolution of Hyphomicrobium both within the structure and around the periphery of the flocs than that obtained with conventional light microscopy.
In this study, three cloned Hyphomicrobium sequences (4953, 49512, and 49518) fit into Hyphomicrobium cluster II with a high level of confidence. For purposes of probe creation, two other pairs of clones (1951 and 1956, and 49519 and 49520) were grouped with Hyphomicrobium cluster I although they were only 94 to 95% similar to each other and may represent different clusters within Hyphomicrobium. The grouping of these sequences together allowed H. vulgare MC-750 to be used as a control strain for hybridization studies. Hybridizations of samples taken from almost 1 year of operation indicated that the Hyphomicrobium cluster II organisms dominated over the Hyphomicrobium cluster I organisms by 3- to 12-fold. Holm et al. (17) suggest that morphologically and nutritionally similar isolates of Hyphomicrobium may show a high level of genetic diversity and that Hyphomicrobium populations may vary seasonally. In this wastewater treatment plant the Hyphomicrobium populations showed genetic diversity but did not vary seasonally. Gliesche and Fesefeldt (15) also did not see seasonal variations in Hyphomicrobium DNA/DNA hybridization group HG 27 in activated sludge. It would be interesting to determine whether plant upsets change the dominant Hyphomicrobium population.
In addition to validation of the molecular analysis methods by using more-conventional cultivation methods and microscopy, comparative information across a wide spectrum of activated sludge plants and operating conditions is needed. The isolation and cultivation of bacterial strains from sludge provide information on the roles and the niches of particular organisms in the community, whereas community analysis and probing methods are valuable in verifying that the isolated organisms are actually present in significant numbers within the community.
ACKNOWLEDGMENTS
This work was supported by a research grant from Eastman Chemical Company, Tennessee Eastman Division, Kingsport, and WMREI at the University of Tennessee.
We gratefully acknowledge the assistance of Claudia Werner for cultivation of strains and Neil Quigley at the Molecular Biology Resource Facility (UTK) for 16S rDNA sequencing. Thanks also to Janet Hensley (Eastman Chemical Company) for preparation of media and successful resuscitation of Hyphomicrobium sp. strain M3 cultures.
REFERENCES
- 1.Aaronson S. Experimental microbial ecology. New York, N.Y: Academic Press; 1970. Procedures for the enrichment and/or isolation of microorganisms: section J. Budding and stalked bacteria; pp. 128–130. [Google Scholar]
- 2.Alm E W, Oerther D B, Larsen N, Stahl D A, Raskin L. The oligonucleotide probe database. Appl Environ Microbiol. 1996;62:3557–3559. doi: 10.1128/aem.62.10.3557-3559.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 4.Amann R, Snaidr J, Wagner M, Ludwig W, Schleifer K-H. In situ visualization of high genetic diversity in a natural microbial community. J Bacteriol. 1996;178:3496–3500. doi: 10.1128/jb.178.12.3496-3500.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amann R, Lemmer H, Wagner M. Monitoring the community structure of wastewater treatment plants: a comparison of old and new techniques. FEMS Microbiol Ecol. 1998;25:205–215. [Google Scholar]
- 6.Amann R I, Krumholz L, Stahl D A. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J Bacteriol. 1990;172:762–770. doi: 10.1128/jb.172.2.762-770.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blackall L L, Burrell P C, Gwilliam H, Bradford D, Bond P C, Hugenholz P. The use of 16S rDNA clone libraries to describe the microbial diversity of activated sludge communities. Water Sci Technol. 1998;37:451–454. [Google Scholar]
- 8.Bond P L, Hugenholtz P, Keller J, Blackall L L. Bacterial community structures of phosphate-removing and non-phosphate-removing activated sludges from sequencing batch reactors. Appl Environ Microbiol. 1995;61:1910–1916. doi: 10.1128/aem.61.5.1910-1916.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brosius J, Palmer M L, Kennedy J P, Noller H F. Complete nucleotide sequence of a 16S ribosomal RNA gene from Escherichia coli. Proc Natl Acad Sci USA. 1978;75:4801–4805. doi: 10.1073/pnas.75.10.4801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bullard C M, Barber J B. Proceedings of the Water Environment Federation, 67th Annual Conference and Exposition. Alexandria, Va: Water Environment Federation; 1994. Improved operational performance using an extended sludge reaeration process. [Google Scholar]
- 11.Church G M, Gilbert W. Genomic sequencing. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cox T L, Sly L I. Phylogenetic relationships and uncertain taxonomy of Pedomicrobium species. Int J Syst Bacteriol. 1997;47:377–380. doi: 10.1099/00207713-47-2-377. [DOI] [PubMed] [Google Scholar]
- 13.Farrelly V, Rainey F A, Stackebrandt E. Effect of genome size and rrn gene copy number on PCR amplification of 16S rRNA genes from a mixture of bacterial species. Appl Environ Microbiol. 1995;61:2798–2801. doi: 10.1128/aem.61.7.2798-2801.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fleming J T, Sanseverino J, Sayler G S. Quantitative relationship between naphthalene catabolic gene frequency and expression in predicting PAH degradation in soils at town gas manufacturing sites. Environ Sci Technol. 1993;27:1068–1074. [Google Scholar]
- 15.Gliesche C G, Fesefeldt A. Monitoring the denitrifying Hyphomicrobium DNA/DNA hybridization group HG27 in activated sludge and lake water using MPN cultivation and subsequence screening with the gene probe Hvu-1. Syst Appl Microbiol. 1998;21:315–320. doi: 10.1016/S0723-2020(98)80039-8. [DOI] [PubMed] [Google Scholar]
- 16.Godon J-J, Zumstein E, Dabert P, Habouzit F, Moletta R. Molecular microbial diversity of an anaerobic digestor as determined by small-subunit rDNA sequence analysis. Appl Environ Microbiol. 1997;63:2802–2813. doi: 10.1128/aem.63.7.2802-2813.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holm N C, Gliesche C G, Hirsch P. Diversity and structure of Hyphomicrobium populations in a sewage treatment plant and its adjacent receiving lake. Appl Environ Microbiol. 1996;62:522–528. doi: 10.1128/aem.62.2.522-528.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holt J G, Krieg N R, Sneath P H A, Stalely J T, Williams S T, editors. Bergey's manual of determinative bacteriology. 9th ed. Baltimore, Md: Williams and Wilkins; 1994. pp. 457–476. [Google Scholar]
- 19.Jenkins D, Richard M G, Daigger G T. Manual on the causes and control of activated sludge bulking and foaming. 2nd ed. Boca Raton, Fla: Lewis Publishers; 1993. [Google Scholar]
- 20.Kloos K, Fesefeldt A, Gliesche C G, Bothe H. DNA-probing indicates the occurrence of denitrification and nitrogen fixation genes in Hyphomicrobium: distribution of denitrifying and nitrogen fixing isolates of Hyphomicrobium in a sewage treatment plant. FEMS Microbiol Ecol. 1995;18:205–213. [Google Scholar]
- 21.Kuske C R, Barns S M, Busch J D. Diverse uncultivated bacterial groups from soils of the arid southwestern United States that are present in many geographic regions. Appl Environ Microbiol. 1997;63:3614–3621. doi: 10.1128/aem.63.9.3614-3621.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lajoie C A, Layton A C, Stapleton R D, Gregory I R, Meyers A J, Sayler G S. Molecular analysis and control of activated sludge. In: Sayler G S, Sanseverino J, Davis K L, editors. Biotechnology in the sustainable environment. New York, N.Y: Plenum Publishing Company; 1997. pp. 323–342. [Google Scholar]
- 23.Lajoie, C. A., A. J. Meyers, A. C. Layton, D. E. Taylor, I. R. Gregory, and G. S. Sayler. Zoogloeal clusters and biosolids dewatering potential in an industrial activated sludge wastewater treatment plant. Water Environ. Res., in press.
- 24.Lane D J. 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. New York, N.Y: John Wiley & Sons, Inc.; 1991. pp. 115–148. [Google Scholar]
- 25.Ludwig W, Bauer S H, Bauer M, Held I, Kirchhof G, Schulze R, Huber I, Spring S, Hartman A, Schleifer K H. Detection and in situ identification of representatives of a widely distributed new bacterial phylum. FEMS Microbiol Lett. 1997;153:181–190. doi: 10.1111/j.1574-6968.1997.tb10480.x. [DOI] [PubMed] [Google Scholar]
- 26.Maidak B L, Olsen G J, Larsen N, Overbeek R, McCaughey M J, Woese C R. The RDP (Ribosomal Database Project) Nucleic Acids Res. 1997;25:109–111. doi: 10.1093/nar/25.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Manz W, Wagner M, Amann R, Schleifer K-H. In situ characterization of the microbial consortia active in two wastewater treatment plants. Water Res. 1994;28:1715–1723. [Google Scholar]
- 28.Ogram A, Sayler G S, Barkay T. The extraction and purification of microbial DNA from sediments. J Microbiol Methods. 1987;7:57–66. [Google Scholar]
- 29.Page R D. TREEVIEW: an application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 30.Rainey F A, Ward-Rainey N, Gliesche C G, Stackebrandt E. Phylogenetic analysis and intrageneric structure of the genus Hyphomicrobium and the related genus Filomicrobium. Int J Syst Bacteriol. 1998;48:635–639. doi: 10.1099/00207713-48-3-635. [DOI] [PubMed] [Google Scholar]
- 31.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 32.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 33.Snaidr J, Amann R, Huber I, Ludwig W, Schleifer K-H. Phylogenetic analysis and in situ identification of bacteria in activated sludge. Appl Environ Microbiol. 1997;63:2884–2896. doi: 10.1128/aem.63.7.2884-2896.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stahl D A, Amann R. Development and application of nucleic acid probes. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. New York, N.Y: John Wiley & Sons, Inc.; 1991. pp. 205–248. [Google Scholar]
- 35.Suzuki M T, Giovannoni S J. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl Environ Microbiol. 1996;62:625–630. doi: 10.1128/aem.62.2.625-630.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thompson J D, Higgins D G, Gibson T J. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wagner M, Amann R, Lemmer H, Schleifer K-H. Probing activated sludge with oligonucleotides specific for Proteobacteria: inadequacy of culture-dependent methods for describing microbial community structure. Appl Environ Microbiol. 1993;59:1520–1525. doi: 10.1128/aem.59.5.1520-1525.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zheng D, Alm E W, Stahl D A, Raskin L. Characterization of universal small-subunit rRNA hybridization probes for quantitative molecular microbial ecology studies. Appl Environ Microbiol. 1996;62:4504–4513. doi: 10.1128/aem.62.12.4504-4513.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]