Abstract
Laboratory-scale sequencing batch reactors (SBRs) as models for activated sludge processes were used to study enhanced biological phosphorus removal (EBPR) from wastewater. Enrichment for polyphosphate-accumulating organisms (PAOs) was achieved essentially by increasing the phosphorus concentration in the influent to the SBRs. Fluorescence in situ hybridization (FISH) using domain-, division-, and subdivision-level probes was used to assess the proportions of microorganisms in the sludges. The A sludge, a high-performance P-removing sludge containing 15.1% P in the biomass, was comprised of large clusters of polyphosphate-containing coccobacilli. By FISH, >80% of the A sludge bacteria were β-2 Proteobacteria arranged in clusters of coccobacilli, strongly suggesting that this group contains a PAO responsible for EBPR. The second dominant group in the A sludge was the Actinobacteria. Clone libraries of PCR-amplified bacterial 16S rRNA genes from three high-performance P-removing sludges were prepared, and clones belonging to the β-2 Proteobacteria were fully sequenced. A distinctive group of clones (sharing ≥98% sequence identity) related to Rhodocyclus spp. (94 to 97% identity) and Propionibacter pelophilus (95 to 96% identity) was identified as the most likely candidate PAOs. Three probes specific for the highly related candidate PAO group were designed from the sequence data. All three probes specifically bound to the morphologically distinctive clusters of PAOs in the A sludge, exactly coinciding with the β-2 Proteobacteria probe. Sequential FISH and polyphosphate staining of EBPR sludges clearly demonstrated that PAO probe-binding cells contained polyphosphate. Subsequent PAO probe analyses of a number of sludges with various P removal capacities indicated a strong positive correlation between P removal from the wastewater as determined by sludge P content and number of PAO probe-binding cells. We conclude therefore that an important group of PAOs in EBPR sludges are bacteria closely related to Rhodocyclus and Propionibacter.
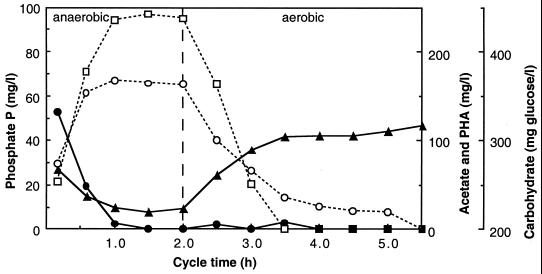
The removal of phosphorus (P) from wastewater can be achieved by chemical precipitation or by biological mechanisms in a process called enhanced biological phosphorus removal (EBPR). The basic configuration of an EBPR-activated sludge plant has the influent wastewater going into an anaerobic zone, where it is mixed with the returned microbial biomass from the secondary clarifier to form the so-called mixed liquor. This mixed liquor then flows into an aerobic zone, after which the biomass is separated from the treated wastewater in the secondary clarifier. Polyphosphate-accumulating organisms (PAOs) (40) are selectively enriched in these systems, and excessive phosphate accumulation occurs in the aerobic zone. Removal of a portion of the growing biomass (waste-activated sludge) results in the net removal of P from the wastewater. Figure 1 shows the profiles of chemical transformations relevant to EBPR.
FIG. 1.
Profiles of soluble extracellular phosphate-P (□), extracellular acetate (●), cellular PHA (○), and cellular carbohydrate (▴) during the anaerobic and aerobic reactor cycle stages of the P sludge of Bond et al. (8).
Empirical experience over the last 30 to 40 years of EBPR operation has permitted plant operators to more successfully conduct EBPR processes (K. J. Hartley and L. Sickerdick, presented at the Second Australian Conference on Biological Nutrient Removal from Wastewater, Albury, Victoria, Australia, 1994). However, the study of EBPR microbiology is important because the process does fail intermittently, PAOs have not been unambiguously identified, and the mechanisms of P removal are unknown. Researchers have constructed biochemical models that accommodate the gross chemical transformations observed in EBPR processes (15, 43).
There have been many investigations attempting to match the metabolic performance of bacterial isolates with the biochemical model suggested for EBPR. These have concentrated mostly on isolates of the genus Acinetobacter because members of this genus are easily isolated from EBPR sludges (21, 26, 42) and some isolates show some characteristics that may be important to EBPR (16, 36). However, evidence indicating that Acinetobacter may not be responsible for EBPR includes pure culture performances not correlating with biological models (7, 38) and analyses of EBPR bacterial communities indicating that Acinetobacter microorganisms are not present in high enough numbers to account for EBPR (7, 14, 24, 30, 41). Investigations of other EBPR-associated microorganisms are limited, although there has been some interest in gram-positive bacteria such as Microlunatus phosphovorus (33, 39), the gram-negative Lampropedia (35), and the Actinobacteria and α Proteobacteria (25). However, there is no general consensus that these bacteria are examples of PAOs, and indeed Mino et al. (31) concluded that rather than there being a single dominant PAO, several different bacterial groups may be important. The isolation of putative PAOs is hampered by the lack of an easy method of using the P removal phenotype in isolation strategies. It is evident that more knowledge of the microbial ecology of EBPR is required before research facilities commit to in-depth investigations of particular isolates. In this paper, we report the phylogenetic identification of a putative PAO in extremely efficient hyper-P-removing sludges containing over 15% PO4-P in the biomass, using 16S rRNA-directed probes.
MATERIALS AND METHODS
EBPR reactors.
Sequencing batch reactors (SBRs) with working volumes of 1 to 2 liters were operated under anaerobic/aerobic cycling conditions to achieve EBPR. SBR operation and monitoring were similar to that reported by Bond et al. (8), and operating data for three reactors are summarized in Table 1. Two reactors, A and B, were operated to highly enrich for PAOs. The performance of reactor GRC fluctuated over a 12-month period, and at regular stable operating times, the sludge was collected and used in the study.
TABLE 1.
Operating data for three laboratory-scale EBPR reactors
| Sludge | Feed
|
Concn (mg/liter)
|
P% in sludged | ||||
|---|---|---|---|---|---|---|---|
| PO4-P (mg/liter) | Acetate (mg/liter) | CODa:P | MLSSb | PO4-Pc
|
|||
| End of anaerobic | Effluent | ||||||
| A | 57 | 309 | 9 | 3,692 | 144 | <0.05 | 15.1 |
| B | 53 | 425 | 4.3 | 1,160 | 110 | 28 | 17.2 |
| GRC | 28 | 389 | 18 | 3,070 | 77 | 6.7 | 6.7 |
COD, chemical oxygen demand.
MLSS, mixed liquor suspended solids (in milligrams per liter).
In both conditions, acetate was not detected.
P% = (PT − Pe/MLSS) × 100 (PT = total sludge phosphate in milligrams/liter; Pe = phosphate in the effluent in milligrams/liter; MLSS is in milligrams/liter).
Microbiological analyses. (i) Microscopy of EBPR mixed cultures.
Mixed cultures (sludges) from three SBRs (A, B, and GRC) were investigated by classical cell staining procedures and by fluorescence in situ hybridization (FISH) (4). Methylene blue staining (for polyphosphate) and Gram staining (8) were conducted with two sludges (A and GRC). For the B sludge, Neisser staining (for polyphosphate) was done as described in Eikelboom and van Buijsen (18), Gram staining by the modified Hucker method was from Jenkins et al. (23), and poly-β-hydroxyalkanoate (PHB) staining was as described by Murray (32). Light micrographs of Gram and methylene blue stains were captured on a Nikon Microphot FXA microscope via a charge-coupled device connected to a PC. Final images were prepared in Adobe Photoshop.
Samples were fixed and hybridized as reported by Bond et al. (8). Enumeration of α, β (including β-1 and β-2), and γ Proteobacteria, Actinobacteria, and Cytophaga-Flavobacterium (for probe details, see Table 2) were reported as proportions of all Bacteria (according to probe EUB338 [8]) for the A sludge. FISH preparations were viewed on both a Zeiss LSM510 and a Zeiss Axiophot. The Zeiss LSM510 confocal laser scanning microscope employed an Axiovert 100M SP inverted optical research microscope and a Plan-Neofluar 63×/1.25 numerical aperture objective. Scan time was 31.8 s per frame with 4.48-μs pixel dwell time. An argon laser 488-nm line and the HeNe 543-nm line were used for imaging. Frame size was 512 by 512 pixels. Images from the Zeiss Axiophot were collected by a cooled charge-coupled device and initially processed by Kontron software (KS200). Final images were prepared in Adobe Photoshop.
TABLE 2.
Information relevant to FISH oligonucleotides used in this study
| Probe | Sequence (5′–3′) | rRNA target sitea | Specificity | % Formamide | Reference |
|---|---|---|---|---|---|
| EUB338 | GCTGCCTCCCGTAGGAGT | 16S, 338–355 | Bacteria | 20 | 3 |
| ALF1b | CGTTCG(C/T)TCTGAGCCAG | 16S, 19–35 | α Proteobacteria | 20 | 29 |
| BET42a | GCCTTCCCACTTCGTTT | 23S, 1027–1043 | β Proteobacteria | 35 | 29 |
| BONE23a | GAATTCCATCCCCCTCT | 16S, 663–679 | β-1 Proteobacteria | 35 | 2 |
| BTWO23a | GAATTCCACCCCCCTCT | 16S, 663–679 | Competitor for BONE23a | 35 | 2 |
| GAM42a | GCCTTCCCACATCGTTT | 23S, 1027–1043 | γ Proteobacteria | 35 | 29 |
| HGC69a | TATAGTTACCACCGCCGT | 23S, 1901–1918 | Actinobacteria | 25 | 34 |
| CF319 | TGGTCCGTGTCTCAGTAC | 16S, 319–336 | Cytophaga-Flavobacterium | 35 | 29 |
| PAO462 | CCGTCATCTACWCAGGGTATTAAC | 16S, 462–485 | PAO clusterb | 35 | This study |
| PAO651 | CCCTCTGCCAAACTCCAG | 16S, 651–668 | PAO clusterb | 35 | This study |
| PAO846 | GTTAGCTACGGCACTAAAAGG | 16S, 846–866 | PAO clusterb | 35 | This study |
| Rc988 | AGGATTCCTGACATGTCAAGGG | 16S, 988–1009 | “Rhodocyclus group”b | NDc | This study |
Sequential FISH and methylene blue staining was carried out by first collecting images from FISH preparations. The coverslip was removed from the slide, the mounting fluid was removed by rapid washing and drying, and the slides were stained with methylene blue. The fields from which FISH images had been collected were located, and images of methylene blue stains were recorded.
(ii) Clone libraries.
Bacterial 16S rRNA gene (rDNA) clone libraries were prepared from genomic DNA extracted from frozen A, B, and P (8) sludges, and inserts from individual clones were amplified and grouped according to restriction fragment length polymorphism (RFLP) analysis using methods previously described (13). Briefly, primers 27f and 1492r (27) were used for PCR amplification of near-complete 16S rDNAs, and amplified genes were cloned using a TA cloning kit (Invitrogen, San Diego, Calif.). Clones of RFLP group representatives were partially sequenced using primer 530f (27) and phylogenetically analyzed (9, 13). A representative selection of clone inserts was fully sequenced with a range of primers (5). Phylogenetic analysis of the 16S rDNA sequences was performed as described previously (17). Briefly, sequences were compiled using the software package SeqEd (Applied Biosystems, Sydney, New South Wales, Australia). Each of the compiled sequences was compared to publicly available databases using the basic local alignment search tool (BLAST [1]) to determine approximate phylogenetic affiliations. All clone sequences were examined with the CHECK_CHIMERA program (28) to identify any chimeric sequences. The compiled sequences were aligned using the ARB software package and database (O. Strunk, O. Gross, M. Reichel, S. May, S. Herrmann, N. Stuckmann, B. Nonhoff, M. Lenke, A. Ginhart, A. Vilbig, T. Ludwig, A. Bode, K.-H. Schleifer, and W. Ludwig, http://www.mikro.biologie.tu-muenchen.de/), and alignments were refined manually. Phylogenetic trees were constructed by carrying out evolutionary distance analyses on the 16S rDNA alignments, using the appropriate tool in the ARB database. The robustness of the tree topology was tested by bootstrap analysis, using neighbor joining with the Kimura two-parameter and parsimony analysis (version 4.0b2a of PAUP* [37]).
(iii) Probe design from clone libraries, probe synthesis, and use with the A sludge.
PAO-specific probes (Table 2) were designed using the probe design tool in the ARB software package. Based on comparative analysis of all sequences in the ARB database comprised of publicly available sequences and our in-house clone sequences, the program selected specific regions within the putative PAO sequences which allowed their discrimination from all other reference sequences. Sequences were subsequently confirmed for specificity using BLAST (1). The designed oligonucleotides were synthesized and labeled at the 5′ end with the indocarbocyanine dye CY3 by Genset (Paris, France). These fluorescently labeled probes were evaluated with paraformaldehyde-fixed A sludge. The formamide concentration for optimum probe stringency was determined by performing a series of FISH experiments at 5% formamide increments starting at 0% formamide. Under all but the lowest-stringency conditions, the morphologically distinct clusters of methylene blue-positive coccobacilli were the only cells which bound the PAO probes. Therefore, the optimum formamide concentrations were determined by reference to the coccobacillus clusters. This was necessary because there are no pure cultures whose 16S rRNA would bind the PAO probes. A similar approach was used for “Microthrix parvicella” by Erhart et al. (19). Generally all three designed PAO probes were applied to any one individual sample spotted on the slide.
(iv) Use of designed probes with other sludges.
A range of sludges from laboratory-scale processes and full-scale EBPR plants was collected, fixed, and hybridized with the newly designed PAO probes after determining the formamide concentration for optimum probe stringency.
RESULTS
EBPR reactors.
Reactor operating data are summarized in Table 1. Our laboratory-scale systems exhibited carbon and phosphorus transformations like those found in full-scale EBPR processes and were therefore deemed good models for EBPR. Table 1 shows that all three SBRs were performing EBPR, since there was P release and acetate uptake by the biomass during the initial anaerobic stage. This can be appreciated by comparing PO4-P and acetate data in the feed and at the end of the anaerobic stage (Table 1). During the subsequent aerobic period, all sludges took up large amounts of P, as seen by comparing the PO4-P values at the end of the anaerobic stage with those from the effluent. A and B sludges were defined as hyper-P removing since they contained >15% PO4-P, which equates to ca. 50% inorganic polyphosphate. The GRC sludge was a good P-removing sludge; it removed >20 mg of PO4-P per liter from the wastewater (compare 28 mg of PO4-P/liter in the influent with 6.7 mg/liter in the effluent) and contained 6.7% P (Table 1).
Microscopy.
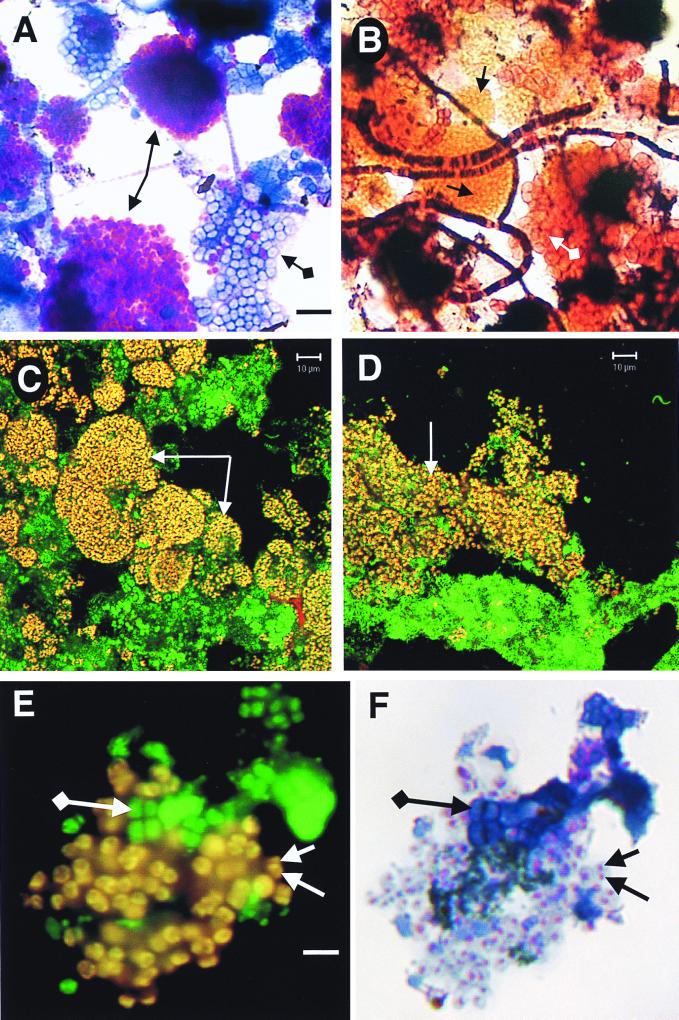
Large clusters comprising hundreds of gram-negative, polyphosphate-containing (either methylene blue- or Neisser-positive) coccobacilli overwhelmingly dominated the A and B sludges. Figure 2A demonstrates the purple polyphosphate-containing coccobacillus clusters in the GRC sludge, while Fig. 2B shows the typical microbial complexity of an EBPR sludge by Gram staining. Occasional tetrad-arranged cocci or “G” bacteria (6) were observed according to their characteristic arrangement in packets of four or eight cells. Table 3 reports the group hybridization results from the hyper-P-removing A sludge. β Proteobacteria, specifically of the subdivision β-2 (i.e., cells hybridizing to probe BTWO23a), dominated the A sludge community, suggesting that PAOs may be members of this bacterial subdivision.
FIG. 2.
Micrographs of mixed liquors from SBRs. (A and B) Bright-field micrographs of GRC sludge as operated according to data in Table 1. (A) Methylene blue stain. Standard-arrowed purple clusters of cells are those containing polyphosphate, while diamond-ended-arrowed blue cells do not contain polyphosphate. The bar is for both panels and is 6 μm. (B) Gram stain. Standard-arrowed orange clusters of cells match the morphology and size of the purple cells in panel A. Diamond-ended-arrowed pink cells match those of the blue cells in panel A. Morphologically identified “Nostocoida limicola” II can be seen as filaments of gram-positive and gram-negative cells. (C and D) Confocal laser scanning micrographs of sludges dual hybridized with EUB338 (25 ng, fluorescein labeled) and a mixture of all three PAO probes (Table 2; each 25 ng, CY3 labeled). Images were collected for fluorescein and CY3 channels, artificially colored, and superimposed. Arrowed yellow cells are the PAOs, since they are dual labeled with EUB338 (green) and PAO (red) probes. The bar for both panels C and D is 10 μm. (C) Mixed liquor from SBR A with operating data as given in Table 1. (D) Lightly sonicated mixed liquor from an EBPR SBR (ca. 10% P in the sludge) operating at 3.5% NaCl from a study of seafood-processing wastewater. Sludge kindly supplied by Nugul Intrasungkha. (E) Epifluorescence micrograph of GRC sludge (Table 1) dual hybridized with EUB338 (25 ng, fluorescein labeled) and PAO651 (Table 2; 25 ng, CY3 labeled). Separate images were collected for fluorescein and CY3 excitation, artificially colored, and superimposed. Standard-arrowed yellow cells are PAOs; diamond-ended-arrowed green-colored cells are other bacteria. The bar is for both panels and is 4 μm. (F) Methylene blue-stained image of the same field as in panel E. Standard-arrowed cells containing purple granules of polyphosphate are the same yellow cells in panel E. Diamond-ended-arrowed blue cells are the same green cells in panel E.
TABLE 3.
Proportions of major bacterial divisions in the A sludge by FISH and in the A, B, and P clone libraries as determined by RFLP analysis and sequencing of RFLP group representatives
| Bacterial division or subdivisiona | FISH of A sludge (% binding to probeb) | No. (% of total clones) in clone library determined by RFLP analysis
|
||
|---|---|---|---|---|
| A | B | P | ||
| α Proteobacteria | 12 | 38 (14) | 32 (13) | 5 (6) |
| β Proteobacteria (mostly Rhodocyclus relatives) | 80 (1 β-1; 81 β-2) | 13 (5) | 44 (18) | 15 (17) |
| Actinobacteria (mostly Terrabacter relatives) | 28 | 67 (24) | 22 (9) | 8 (9) |
| Cytophaga-Flavobacterium group | 14 | 83 (30) | 52 (21) | 45 (51) |
| Total clones in library analyzed by RFLP | NAc | 281 | 250 | 89 |
Comments in parentheses refer to clone library data only.
Percentage of cells in the A sludge binding the EUB338 probe for all Bacteria. β-1 and β-2 were determined by using BONE23a and BTWO23a probes. γ Proteobacteria comprised 1% of the Bacteria.
NA, not applicable.
Clone libraries.
A total of 281 bacterial 16S rDNA clones from the A sludge, 250 from the B sludge, and 89 from the P sludge (8) were evaluated by RFLP. These sludges were chosen to generate 16S rDNA sequences because they were high-performance EBPR systems (Table 1 and reference 8) and therefore a good source of PAO sequences from which specific FISH oligonucleotides could be designed. Group representatives were partially sequenced, and the overall results are reported in Table 3. Note that the relative proportions of phylogenetic groups in the A sludge clone library did not match those determined by FISH probing (Table 3). It is recognized that clone libraries may not provide quantitative data on the microbial community structure of the sample analyzed, but in this research, clone libraries were used simply as a mechanism to generate sequences for probe design.
Probe development.
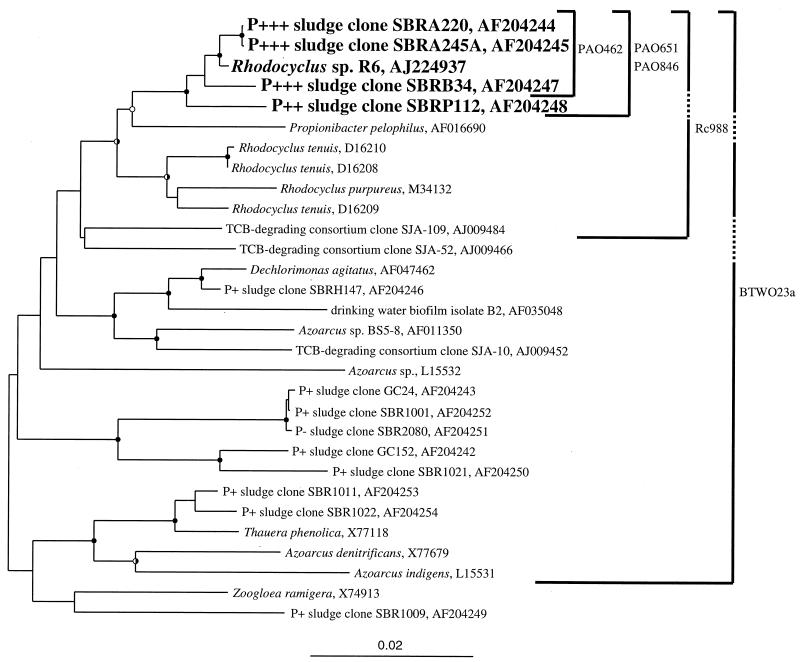
The putative PAOs were broadly highlighted as β-2 Proteobacteria by group hybridization experiments using the BTWO23a probe (Table 3 and reference 8). However, as the β-2 Proteobacteria probe (BTWO23a) was originally designed only as a competitor for the β-1 Proteobacteria probe (BONE23a [2]), its lack of specificity is not surprising. In addition to β-2 Proteobacteria, it also targets (with no mismatches) members of the β-3 Proteobacteria, some γ Proteobacteria, and a green nonsulfur division clone, OPB9. Despite this slight lack of specificity, probe BTWO23a helped refine the identity of the PAOs to a subset of the β Proteobacteria. However, additional more-specific probes are still required for the PAOs in the β-2 Proteobacteria group. To this end, all clones from the A, B, and P sludge libraries belonging to the β-2 Proteobacteria were fully sequenced (positions 28 through 1491, Escherichia coli numbering) in preparation for probe design. In addition, partially sequenced clones belonging to the β-2 Proteobacteria from two previously reported EBPR and non-EBPR clone libraries (9) and sludge clone SBRH147 from an unpublished library were fully sequenced. Figure 3 shows a phylogenetic tree of the completely sequenced (positions 28 through 1491, E. coli numbering) β-2 Proteobacteria clones from which the probes were designed and the specificity of the probes. Two main clusters of EBPR sludge clones were observed (SBRA220 cluster and GC24 cluster [Fig. 3]). However, only the SBRA220 cluster was comprised exclusively of clones from high-performance EBPR sludges. This became the focus group for probe design. Three PAO probes were designed to specifically target the SBRA220 cluster, and an additional probe of broader specificity called Rc988 (Fig. 3) was designed but not fully evaluated. All PAO probes are listed in Table 2 with their empirically determined optimum stringencies.
FIG. 3.
Phylogenetic tree of near-complete 16S rDNA sequences obtained from sludges A, B, P, SBRH, SBR1, SBR2, and GC (Gold Coast, Queensland, Australia) determined in this study and sequences from publicly accessible databases. Rubrivivax gelatinosus (D16213) was used as the outgroup in analyses but is not shown in the tree. Evolutionary distance and parsimonious analyses were carried out in PAUP∗ employing 2,000 bootstrap resamplings. Closed circles at nodes indicate >75% bootstrap support from both analyses; open circles indicate 50 to 75% from both analyses; half-shaded circles are for analyses where one algorithm gave >75% bootstrap support and the other gave 50 to 75%. The coding indicates that the clone came from a hyper-P-removing sludge (ca. 15% P in the sludge) (P+++), a good P-removing sludge (P++), a fair P-removing sludge (P+), or a non-P-removing sludge (P−). The specificity of the published β-2 Proteobacteria probe (BTWO23a) and those of probes designed in this work (PAO probes and Rc988) are shown by solid lines. Dashed lines indicate that the probe does not have 100% identity with the indicated sequence. For example, Rc988 has one mismatch (at position 1009) with the SBRP112 sequence. In addition to specifically targeting the sequences indicated in the tree, probe BTWO23a also targets (with no mismatches) members of the β-3 and γ Proteobacteria, Iodobacter, Chromobacterium, Chromatium spp., and a green nonsulfur division clone, OPB9. The scale indicates 0.02 nucleotide change per nucleotide position. TCB, trichlorobenzene.
It is worthwhile noting that probe PAO846 (Table 2) was originally designed to target positions 841 to 857 (E. coli numbering) and did not work in FISH. In light of a recent publication systematically documenting the in situ accessibility of FISH probes to E. coli 16S rRNA (20), the probe was redesigned to target positions 846 to 866, raising the putative probe accessibility from 8 to 42% to 42 to 52% (relative probe fluorescence). The redesigned probe worked well in FISH experiments, likely because E. coli and the Rhodocyclus-like PAOs have similar ribosomal higher-order structures (and therefore similar probe accessibility profiles) due to their relatively close phylogenetic relationship.
Actinobacteria was the second dominant group in the A sludge (Table 3), and design and evaluation of probes from these Actinobacteria clone sequences are under way.
Use of designed probes.
A series of fixed sludges including the A sludge, sludge from the GRC reactor at different operational periods, and the Loganholme sludge were evaluated with the designed PAO probes. In all full-scale and laboratory-scale EBPR sludges examined, the clusters of PAO probe-binding coccobacilli were distinct and uniform and resembled the yellow cells and clusters arrowed in Fig. 2C and D. Depending on EBPR performance, more or fewer clusters were present. For example, in the Loganholme sludge, a full-scale activated sludge plant treating domestic wastewater with an influent containing ca. 10 mg of PO4-P/liter, moderate numbers of clusters were observed. Large numbers of the clusters were observed in the hyper-P-removing A sludge (Fig. 2C). In the saline sludge (Fig. 2D) as in all sludges, the three PAO probes bound the same cells as bound the β-2 Proteobacteria probe (BTWO23a).
Figures 2E and F show results for FISH and methylene blue-stained sludge from the GRC reactor (Table 1). Yellow cells in Fig. 2E are dual hybridized with EUB338 (fluorescein) and PAO651 (CY3) probes and are therefore putative PAOs. In this preparation, the polyphosphate, which stains purple, appears as granules in the middle of PAOs (Fig. 2F). The typical appearance of PAOs by methylene blue staining is shown in Fig. 2A. Blue cells in Fig. 2F do not contain polyphosphate. The cells in Fig. 2E and F have been fixed in paraformaldehyde and undergone FISH, while those in Fig. 2A are simply dried onto microscope slides before staining. Therefore, either the paraformaldehyde or the chemicals used in FISH seem to have had an effect on the polyphosphate, making it appear as granules in the cells (Fig. 2F) rather than evenly distributed around the cells (Fig. 2A). Figure 2E and F clearly demonstrate that the putative PAOs according to FISH are those containing polyphosphate.
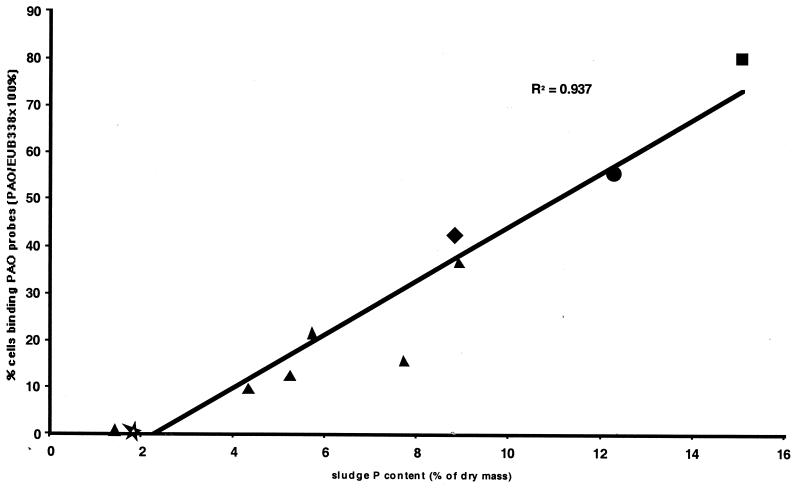
During various periods of stable operations of the GRC reactor, numbers of PAO probe-positive cells were counted and expressed as a proportion of EUB338-binding cells. The activity of the sludge from the GRC reactor was high, with the vast majority of cells binding the EUB338 probe. PAO probe-positive cells in other previously reported sludges like the Q, P, and S sludges (8, 10) were also counted. The proportion of PAO probe-binding cells was plotted versus the sludge percent P (Fig. 4). A positive correlation between these two parameters is evident.
FIG. 4.
Relationship between sludge P content (percentage of dry mass) and proportion of cells binding all three PAO probes as a percentage of the EUB338 probe-positive cells. Sludges evaluated included the Q (★), P (⧫), and S (●) sludges of Bond et al. (8, 10, 11), the GRC sludge at varying but stable P-removal efficiencies (▴), and the A sludge (■) (this study).
DISCUSSION
Our approach to discovering the identity of the PAOs was to selectively enrich for organisms with a P removal phenotype and to evaluate this enriched culture by non-culture-dependent methods like cloning and FISH. We present indirect and direct evidence of the identity of the PAOs and quantitation of the PAOs in sludges.
A total of 80% of the microbial biomass in the hyper-P-removing A sludge bound the β Proteobacteria probe (BET42a); these organisms were likely β-2 Proteobacteria since they bound the BTWO23a probe (Table 3). The numerical dominance of the BTWO23a-hybridizing group in the enriched sludge suggested that they played a major role in P removal. Additionally, clone library data of Bond et al. (9) had already associated bacteria of the Rhodocyclus group in the β-2 Proteobacteria with improved P removal. Probes for the putative PAOs (called PAO probes) were designed from a group of highly related β-2 Proteobacteria clone sequences (≥98% identical) retrieved from several enriched P-removing sludges (A, B, and P). These PAO probes were used with A sludge biomass, where they bound cell clusters whose morphology and arrangement resembled those binding the BTWO23a probe. The closest pure-cultured bacterial relatives to the β-2 Proteobacteria clone sequences (Fig. 3) are from Rhodocyclus (R. tenuis and R. purpureus) and Propionibacter pelophilus. A clone sequence from a recently published Swiss EBPR sludge (R6 [22]) was in the group containing the full clone inserts from the A, B, and P sludges (Fig. 3). Thus, by using this concerted hybridization approach (Fig. 3), we demonstrated that the designed probes were specific for the dominant β-2 Proteobacteria in the A sludge. In addition, the PAO probe-positive cells matched the morphology, size, arrangement, and abundance of those staining positive for polyphosphate by the methylene blue stain (Fig. 2A and C). When used with full-scale and other laboratory-scale EBPR sludges, the PAO probes and the β-2 Proteobacteria probe always bound the same cells whose morphology was similar to those staining positively for polyphosphate. Direct evidence that the PAO probes bound true PAOs came from sequential FISH and methylene blue-staining experiments. Cells containing polyphosphate also bound the probe PAO651 (Fig. 2E and F).
An indicative correlation between increasing P removal performance, as judged by percent P in the sludge, and levels of β Proteobacteria was observed when data for the P sludge (8.8% P; 45% β Proteobacteria [8]), the S sludge (12.3% P, 56% β Proteobacteria [10]), and the A sludge (15.1% P, 80% β Proteobacteria) were compared (plot not shown). Subsequent investigations with the PAO-specific probes on a range of laboratory-scale EBPR sludges demonstrated a positive correlation between percent P in the sludge and numbers of PAO probe-binding cells (Fig. 4). This quantitative data demonstrates that the designed PAO probes for particular β-2 Proteobacteria are very likely detecting a true PAO in the laboratory-scale processes.
Therefore, from Fig. 4 and data on the hybridization from the A sludge with PAO probes in concert with methylene blue stains, we have provided indirect, direct, and quantitative evidence that bacteria closely related to Rhodocyclus and P. pelophilus are examples of PAOs. However, other phylogenetically distinct PAOs may also exist in EBPR. The more rigorous validation of our probes in full-scale processes is now required.
ACKNOWLEDGMENTS
We thank the operators of the Logan City Loganholme Sewage Treatment Plant and the City of San Francisco Southeast Water Pollution Control Plant and Nugul Intrasungkha, operator of the saline reactor. Gene Tyson determined some of the RFLPs on the A and B sludge clone libraries, Paul Burrell provided the SBRH147 clone sequence from his clone library, Gavin Symonds of Carl Zeiss Australia assisted with preparation of the CSLM images, and Carl Zeiss Australia loaned the LSM510 for this purpose. We acknowledge all these for their valuable assistance.
Some of the work was funded by the CRC for Waste Management and Pollution Control Ltd., a center established and supported under the Australian Government's Cooperative Research Centres Program. Andrew Schuler was supported by a U.S. Environmental Protection Agency STAR graduate fellowship and by grant BES-9612640 from the National Science Foundation.
ADDENDUM
After the review process, a report by Hesselmann et al. (22) was published. Our paper essentially corroborates the data presented in that report. Hesselmann et al. (22) used methods similar to those used by us, including enrichment of hyper-P-removing mixed microbial communities in laboratory-scale SBRs, cell staining (Neisser and PHB), FISH with published group probes, cloning 16S rDNA from the enriched cultures, and probe design and application. An additional procedure used by Hesselmann et al. (22) was dot blot hybridization with extracted nucleic acids. Comparing the two reports, one finds differences in the cloning strategies, the number of clones sequenced and analyzed, and the number and specificity of probes designed. However, the major conclusion that Rhodocyclus-like bacteria, called Accumulibacter phosphatis by Hesselmann et al. (22), are genuine PAOs is common to both studies and suggests that these organisms may play a global role in EBPR.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Amann R, Snaidr J, Wagner M, Ludwig W, Schleifer K H. In situ visualization of high genetic diversity in a natural microbial community. J Bacteriol. 1996;178:3496–3500. doi: 10.1128/jb.178.12.3496-3500.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amann R I, Binder B J, Olson R J, Chisholm S W, Devereux R, Stahl D A. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol. 1990;56:1919–1925. doi: 10.1128/aem.56.6.1919-1925.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amann R I, Ludwig W, Schleifer K-H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blackall L L. Molecular identification of activated sludge foaming bacteria. Water Sci Technol. 1994;29(7):35–42. [Google Scholar]
- 6.Blackall L L, Rossetti S, Christenssen C, Cunningham M, Hartman P, Hugenholtz P, Tandoi V. The characterization and description of representatives of “G” bacteria from activated sludge plants. Lett Appl Microbiol. 1997;25:63–69. doi: 10.1046/j.1472-765x.1997.00176.x. [DOI] [PubMed] [Google Scholar]
- 7.Bond P L. Investigations of the microbial ecology of enhanced biological phosphorus removal in the activated sludge process. Ph.D thesis. Brisbane, Queensland, Australia: The University of Queensland; 1997. [Google Scholar]
- 8.Bond P L, Erhart R, Wagner M, Keller J, Blackall L L. The identification of some of the major groups of bacteria in efficient and nonefficient biological phosphorus removal activated sludge systems. Appl Environ Microbiol. 1999;65:4077–4084. doi: 10.1128/aem.65.9.4077-4084.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bond P L, Hugenholtz P, Keller J, Blackall L L. Bacterial community structures of phosphate-removing and non-phosphate-removing activated sludges from sequencing batch reactors. Appl Environ Microbiol. 1995;61:1910–1916. doi: 10.1128/aem.61.5.1910-1916.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bond P L, Keller J, Blackall L L. Bio-P and non-bio-P bacteria identification by a novel microbial approach. Water Sci Technol. 1999;39(6):13–20. [Google Scholar]
- 11.Bond P L, Keller J, Blackall L L. Characterisation of enhanced biological phosphorus removal activated sludges with dissimilar phosphorus removal performance. Water Sci Technol. 1998;37:567–571. [Google Scholar]
- 12.Brosius J, Dull T L, Steeter D D, Noller H F. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J Mol Biol. 1981;148:107–127. doi: 10.1016/0022-2836(81)90508-8. [DOI] [PubMed] [Google Scholar]
- 13.Burrell P C, Keller J, Blackall L L. Microbiology of a nitrite-oxidizing bioreactor. Appl Environ Microbiol. 1998;64:1878–1883. doi: 10.1128/aem.64.5.1878-1883.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cloete T E, Steyn P L. A combined fluorescent antibody-membrane filter technique for enumerating Acinetobacter in activated sludge. In: Ramadori R, editor. Biological phosphate removal from wastewaters. Oxford, England: Pergamon Press; 1987. pp. 335–338. [Google Scholar]
- 15.Comeau Y, Hall K J, Hancock R E W, Oldham W K. Biochemical models for enhanced biological phosphorus removal. Water Res. 1986;20:1511–1521. [Google Scholar]
- 16.Deinema M H, van Loosdrecht M C M, Scholten A. Some physiological characteristics of Acinetobacter spp. accumulating large amounts of phosphate. Water Sci Technol. 1985;17(12):119–125. [Google Scholar]
- 17.Dojka M A, Hugenholtz P, Haack S K, Pace N R. Microbial diversity in a hydrocarbon- and chlorinated-solvent-contaminated aquifer undergoing intrinsic bioremediation. Appl Environ Microbiol. 1998;64:3869–3877. doi: 10.1128/aem.64.10.3869-3877.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eikelboom D H, van Buijsen H J J. Microscopic sludge investigation manual. 1st ed. Delft, The Netherlands: TNO Research Institute for Environmental Hygiene; 1981. [Google Scholar]
- 19.Erhart R, Bradford D, Seviour R J, Amann R I, Blackall L L. Development and use of fluorescent in situ hybridization probes for the detection and identification of “Microthrix parvicella” in activated sludge. Syst Appl Microbiol. 1997;20:310–318. [Google Scholar]
- 20.Fuchs B M, Wallner G, Beisker W, Schwippl I, Ludwig W, Amann R. Flow cytometric analysis of the in situ accessibility of Escherichia coli 16S rRNA for fluorescently labeled oligonucleotide probes. Appl Environ Microbiol. 1998;64:4973–4982. doi: 10.1128/aem.64.12.4973-4982.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fuhs G W, Chen M. Microbiological basis of phosphate removal in the activated sludge process for the treatment of wastewater. Microb Ecol. 1975;2:119–138. doi: 10.1007/BF02010434. [DOI] [PubMed] [Google Scholar]
- 22.Hesselmann R P X, Werlen C, Hahn D, van der Meer J R, Zehnder A J B. Enrichment, phylogenetic analysis and detection of a bacterium that performs enhanced biological phosphate removal in activated sludge. Syst Appl Microbiol. 1999;22:454–465. doi: 10.1016/S0723-2020(99)80055-1. [DOI] [PubMed] [Google Scholar]
- 23.Jenkins D, Richard M G, Daigger G T. Manual on the causes and control of activated sludge bulking and foaming. New York, N.Y: Lewis Publishers; 1993. [Google Scholar]
- 24.Kämpfer P, Erhart R, Beimfohr C, Bohringer J, Wagner M, Amann R. Characterization of bacterial communities from activated sludge-culture-dependent numerical identification versus in situ identification using group- and genus-specific rRNA-targeted oligonucleotide probes. Microb Ecol. 1996;32:101–121. doi: 10.1007/BF00185883. [DOI] [PubMed] [Google Scholar]
- 25.Kawaharasaki M, Tanaka H, Kanagawa T, Nakamura K. In situ identification of polyphosphate-accumulating bacteria in activated sludge by dual staining with rRNA-targeted oligonucleotide probes and 4′,6-diamidino-2-phenylindol (DAPI) at a polyphosphate-probing concentration. Water Res. 1999;33:257–265. [Google Scholar]
- 26.Kerdachi D A, Healey K J. The reliability of cold perchloric acid extraction to assess metal-bound phosphates. In: Ramadori R, editor. Biological phosphate removal from wastewaters. Oxford, England: Pergamon Press; 1987. pp. 339–342. [Google Scholar]
- 27.Lane D J. 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. Chichester, England: Academic Press; 1991. pp. 115–175. [Google Scholar]
- 28.Maidak B L, Cole J R, Parker C T, Garrity G M, Larsen N, Li B, Lilburn T G, McCaughey M J, Olsen G J, Overbeek R, Pramanik S, Schmidt T M, Tiedje J M, Woese C R. A new version of the RDP (Ribosomal Database Project) Nucleic Acids Res. 1999;27:171–173. doi: 10.1093/nar/27.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Manz W, Amann R, Ludwig W, Wagner M, Schleifer K-H. Phylogenetic oligonucleotide probes for the major subclasses of Proteobacteria: problems and solutions. Syst Appl Microbiol. 1992;15:593–600. [Google Scholar]
- 30.Melasniemi H, Hernesmaa A, Pauli A S-L, Rantanen P, Salkinoja-Salonen M. Comparative analysis of biological phosphate removal (BPR) and non-BPR activated sludge bacterial communities with particular reference to Acinetobacter. J Ind Microbiol. 1999;21:300–306. [Google Scholar]
- 31.Mino T, van Loosdrecht M C M, Heijnen J J. Microbiology and biochemistry of the enhanced biological phosphate removal process. Water Res. 1998;32:3193–3207. [Google Scholar]
- 32.Murray R G E. Manual of methods for general bacteriology. Washington, D.C.: American Society for Microbiology; 1981. [Google Scholar]
- 33.Nakamura K, Hiraishi A, Yoshimi Y, Kawaharasaki M, Masuda K, Kamagata Y. Microlunatus phosphovorus gen. nov., sp. nov., a new gram-positive polyphosphate-accumulating bacterium isolated from activated sludge. Int J Syst Bacteriol. 1995;45:17–22. doi: 10.1099/00207713-45-1-17. [DOI] [PubMed] [Google Scholar]
- 34.Roller C, Wagner M, Amann R, Ludwig W, Schleifer K-H. In situ probing of Gram-positive bacteria with high DNA G+C content using 23S rRNA-targeted oligonucleotides. Microbiology. 1994;140:2849–2858. doi: 10.1099/00221287-140-10-2849. [DOI] [PubMed] [Google Scholar]
- 35.Stante L, Cellamare C M, Malaspina F, Bortone G, Tilche A. Biological phosphorus removal by pure culture of Lampropedia spp. Water Res. 1997;31:1317–1324. [Google Scholar]
- 36.Streichan M, Golecki J R, Schon G. Polyphosphate-accumulating bacteria from sewage treatment plants with different processes for biological phosphorus removal. FEMS Microbiol Ecol. 1990;73:113–124. [Google Scholar]
- 37.Swofford D L. PAUP*: Phylogenetic Analysis Using Parsimony (* and other methods), version 4.0b2a. Sunderland, Mass: Sinauer Associates; 1999. [Google Scholar]
- 38.Tandoi V, Majone M, May J, Ramadori R. The behaviour of polyphosphate accumulating Acinetobacter isolates in an anaerobic-aerobic chemostat. Water Res. 1998;32:2903–2912. [Google Scholar]
- 39.Ubukata Y. Some physiological characteristics of a phosphate removing bacterium isolated from anaerobic/aerobic activated sludge. Water Sci Technol. 1994;30(6):229–235. [Google Scholar]
- 40.van Loosdrecht M C M, Smolders G J, Kuba T, Heijnen J J. Metabolism of microorganisms responsible for enhanced biological phosphorus removal from wastewater. Antonie Leeuwenhoek. 1997;71:109–116. doi: 10.1023/a:1000150523030. [DOI] [PubMed] [Google Scholar]
- 41.Wagner M, Erhart R, Manz W, Amann R, Lemmer H, Wedi D, Schleifer K-H. Development of an rRNA-targeted oligonucleotide probe specific for the genus Acinetobacter and its application for in situ monitoring in activated sludge. Appl Environ Microbiol. 1994;60:792–800. doi: 10.1128/aem.60.3.792-800.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wentzel M C, Loewenthal R E, Ekama G A, Marais G V R. Enhanced polyphosphate organism cultures in activated sludge systems. Part 1. Enhanced culture development. Water South Africa. 1988;14:81–92. [Google Scholar]
- 43.Wentzel M C, Lötter L H, Ekama G A, Loewenthal R E, Marais G V R. Evaluation of biochemical models for biological excess phosphate removal. Water Sci Technol. 1991;23:567–576. [Google Scholar]