ABSTRACT
Selection for Listeria monocytogenes strains that are tolerant to quaternary ammonium compounds (such as benzalkonium chloride [BC]) is a concern across the food industry, including in fresh produce processing environments. This study evaluated the ability of 67 strains of produce-associated L. monocytogenes and other Listeria spp. (“parent strains”) to show enhanced BC tolerance after serial passaging in increasing BC concentrations and to maintain this tolerance after substreaking in the absence of BC. After serial passaging in BC, 62/67 “BC passaged cultures” showed higher MICs (4 to 20 mg/L) than parent strains (2 to 6 mg/L). After the substreaking of two isolates from BC passaged cultures for each parent strain, 105/134 “adapted isolates” maintained MICs (4 to 6 mg/L) higher than parent strain MICs. These results suggested that adapted isolates acquired heritable adaptations that confer BC tolerance. Whole-genome sequencing and Sanger sequencing of fepR, a local repressor of the MATE family efflux pump FepA, identified nonsynonymous fepR mutations in 48/67 adapted isolates. The mean inactivation of adapted isolates after exposure to use-level concentrations of BC (300 mg/L) was 4.48 log, which was not significantly different from inactivation observed in parent strains. Serial passaging of cocultures of L. monocytogenes strains containing bcrABC or qacH did not yield adapted isolates that showed enhanced BC tolerance in comparison to that of monocultures. These results suggest that horizontal gene transfer either did not occur or did not yield isolates with enhanced BC tolerance. Overall, this study provides new insights into selection of BC tolerance among L. monocytogenes and other Listeria spp.
IMPORTANCE Listeria monocytogenes tolerance to quaternary ammonium compounds has been raised as a concern with regard to L. monocytogenes persistence in food processing environments, including in fresh produce packing and processing environments. Persistence of L. monocytogenes can increase the risk of product contamination, food recalls, and foodborne illness outbreaks. Our study shows that strains of L. monocytogenes and other Listeria spp. can acquire heritable adaptations that confer enhanced tolerance to low concentrations of benzalkonium chloride, but these adaptations do not increase survival of L. monocytogenes and other Listeria spp. when exposed to concentrations of benzalkonium chloride used for food contact surface sanitation (300 mg/L). Overall, these findings suggest that the emergence of benzalkonium chloride-tolerant Listeria strains in food processing environments is of limited concern, as even strains adapted to gain higher MICs in vitro maintain full sensitivity to the concentrations of benzalkonium chloride used for food contact surface sanitation.
KEYWORDS: Listeria monocytogenes, Listeria spp., adaptation, benzalkonium chloride, survival, tolerance
INTRODUCTION
Listeria monocytogenes is a foodborne pathogen that causes approximately 1,600 cases of foodborne illness and 260 deaths per year in the United States (1). L. monocytogenes persistence in built environments used for food production can provide a source of contamination in food products, which increases the risk of recalls and foodborne illness outbreaks (2, 3). While L. monocytogenes outbreaks and recalls have been linked to a diversity of foods, including dairy, seafood, and ready-to-eat deli meat (4), more recently (i.e., since 2009), L. monocytogenes has been implicated in several outbreaks related to environmental contamination in fresh produce packing or processing environments, including outbreaks associated with celery (5), cantaloupe (6), caramel apples (7), and packaged lettuce (8).
Listeria spp. (defined here as all species of Listeria excluding L. monocytogenes) are often referred to as index organisms for L. monocytogenes, and thus detection of Listeria spp. in a given processing facility location indicates conditions that would facilitate the presence and/or survival of L. monocytogenes in that same environment (9, 10). Throughout the world, regular monitoring of food production environments involves testing for all species of Listeria (including both Listeria spp. and L. monocytogenes), rather than L. monocytogenes alone (11). Therefore, it is important to understand how Listeria spp., in addition to L. monocytogenes, respond to interventions that seek to control L. monocytogenes.
Effective sanitation in food processing environments, including fresh produce packing and processing facilities, is essential for mitigating L. monocytogenes contamination of finished products. Quaternary ammonium compounds are sanitizers that are widely used for sanitation on both food contact and nonfood contact surfaces to control microorganisms such as L. monocytogenes and Listeria spp. (12). A commonly used class of quaternary ammonium compounds are alkyl-(C8-C18) dimethyl benzyl ammonium chlorides (also known as benzalkonium chlorides). Commercial formulations of sanitizers with benzalkonium chloride (BC) as an active ingredient are approved for use on food contact surfaces, utilized for nonorganic production, at concentrations between 150 and 400 mg/L (termed herein as use-level concentrations) in U.S. food packing, processing, and manufacturing environments (21 CFR 178.1010 [13]) (14). The efficacy of BC is measured by its ability to reduce target microorganism levels in a defined period (e.g., a 5-log reduction within 30 s) at use-level concentrations (15, 16). There is concern that frequent use of BC across the food industry, including in fresh produce packing and processing environments, may be contributing to the selection of L. monocytogenes and Listeria spp. that are less effectively inactivated by BC, which could increase the risk of persistence of L. monocytogenes and Listeria spp. in food facilities (17, 18).
Genetically encoded efflux pump systems represent an important mechanism of BC tolerance in L. monocytogenes and Listeria spp. (19). These efflux pumps can be encoded chromosomally, such as by the fepRA operon, which encodes the MATE family efflux pump FepA (20), or on mobile genetic elements, such as the three-gene cassette bcrABC, which is often found located on a pLM80-type plasmid, (21, 22), and qacH, which is located on the Tn6188 transposon (23). Both bcrABC and qacH have been frequently observed in strains of L. monocytogenes and Listeria spp. isolated from food production environments and are known to confer tolerance of L. monocytogenes and Listeria spp. to BC (17, 24–28). There is concern that horizontal gene transfer of these genes could facilitate widespread and enhanced tolerance of L. monocytogenes and Listeria spp. to BC in the processing environment (29). Recently, an experimental study showed horizontal gene transfer of a mobile genetic element containing bcrABC as well as cadmium resistance genes from Listeria spp. to a streptomycin-resistant L. monocytogenes (30). In these experiments, selection for streptomycin resistance and cadmium resistance was used to obtain transformants (30), which can be argued to not be representative of the type of selection that is expected to occur in food-associated environments. Therefore, there is a need for studies that evaluate the horizontal gene transfer of BC resistance genes, and subsequent impacts on BC tolerance, in L. monocytogenes and Listeria spp. under conditions that more closely simulate real-world conditions.
There is considerable debate surrounding the terminology used to describe the ability of L. monocytogenes and Listeria spp. to grow in the presence of sanitizers like BC. With respect to clinical antibiotics, the term resistance is generally referred to, in both scientific literature (31) and by government organizations such as the World Health Organization (https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance), as the ability of bacteria to maintain growth in therapeutic levels of an antibiotic agent. This definition is less relevant when applied to sanitizers such as BC, because use-level concentrations of BC are highly unlikely to support bacterial growth (32). In addition, in the context of food processing environments, improper development or implementation of sanitation standard operating procedures can result in the presence of BC at concentrations below use-level concentrations (18). Therefore, it is important to evaluate the growth of L. monocytogenes and Listeria spp. exposed to BC at concentrations below the recommended use-level concentrations. Recently, the term “tolerance” has been proposed as an alternative to the term “resistance” to describe the ability of microorganisms to show reduced susceptibility to sanitizers at concentrations below use-level concentrations (32). In this study, we sought to investigate the ability of L. monocytogenes and Listeria spp. to grow in low levels of BC (defined here as <20 mg/L), and we will henceforth use the term “tolerance” (as defined in reference 32) to refer to the capacity of strains of L. monocytogenes and Listeria spp. to acquire the ability to grow in low levels of BC to which they were once sensitive.
The specific purpose of this study was to investigate (i) the ability of monocultures of L. monocytogenes and Listeria spp. to acquire enhanced tolerance when exposed to sequentially increasing low levels of BC and (ii) the impact of this acquired enhanced BC tolerance on the ability of L. monocytogenes and Listeria spp. to survive exposure to use-level concentrations of BC. Additionally, we also examined (iii) whether L. monocytogenes cocultures that include strains carrying different BC resistance genes (i.e., bcrABC, qacH) could, after exposure to increasing BC concentrations, give rise to isolates with BC tolerance that exceeds tolerance observed in monocultures. Due to the recent emerging concerns about L. monocytogenes in fresh produce, we used produce-associated isolates for this study. However, as sanitizers such as BC are widely used for sanitation throughout food processing sectors, our findings should be applicable broadly to all food processing sectors.
RESULTS
Parent strains showed MICs to BC ranging from 1 to 6 mg/L, with strains carrying BC resistance genes showing significantly higher MICs.
Initial MIC experiments of the 67 strains of L. monocytogenes and Listeria spp. characterized here (Table 1) showed a narrow range of MICs, from 1 to 6 mg/L, with an estimated marginal mean of 2.30 mg/L (Fig. 1); these initial MICs are referred to here as “parent MICs.” Results from two-way analysis of variance (ANOVA) and post hoc tests (Table 2) showed that strains of L. monocytogenes and Listeria spp. that carry the BC resistance genes bcrABC (n = 10) or qacH (n = 1) showed significantly higher MICs (estimated marginal mean of 5.37 mg/L; range of 4 to 6 mg/L BC) than strains lacking these genes (estimated marginal mean MIC of 1.95 mg/L; range of 1 to 2 mg/L BC) (P < 0.05). bcrABC was present in six L. monocytogenes, two L. innocua, and two L. welshimeri strains; qacH was present in one L. monocytogenes strain. Parent MICs for L. monocytogenes and Listeria spp. (i.e., all L. innocua, L. ivanovii, L. marthii, L. seeligeri, and L. welshimeri strains examined in this study) were not significantly different from one another, with an estimated marginal mean MIC for L. monocytogenes of 2.59 mg/L (range of 2 to 6 mg/L) and an estimated marginal mean MIC for Listeria spp. of 2.12 mg/L (range of 1 to 6 mg/L) (P > 0.05).
TABLE 1.
Isolates of L. monocytogenes and Listeria spp. selected for BC susceptibility experiments
| Parent strain FSL IDa | Adapted isolate A FSL IDb | Species | Isolation source (sample type) | Lineage | Clonal groupc | Resistance gened |
|---|---|---|---|---|---|---|
| FSL S11-0167 | FSL H9-0106 | L. innocua | Postharvest (environmental swab) | —e | — | bcrABC |
| FSL S10-3425 | FSL H9-0109 | L. innocua | Postharvest (environmental swab) | — | — | bcrABC |
| FSL S11-0456 | FSL H9-0107 | L. innocua | Postharvest (environmental swab) | — | — | NP |
| FSL S11-0003 | FSL H9-0108 | L. innocua | Postharvest (environmental swab) | — | — | NP |
| FSL S10-2287 | FSL H9-0110 | L. innocua | Preharvest (water) | — | — | NP |
| FSL S11-0176 | FSL H9-0111 | L. innocua | Postharvest (environmental swab) | — | — | NP |
| FSL S11-0315 | FSL H9-0112 | L. innocua | Postharvest (environmental swab) | — | — | NP |
| FSL S10-3544 | FSL H9-0113 | L. innocua | Postharvest (environmental swab) | — | — | NP |
| FSL S10-3605 | FSL H9-0114 | L. innocua | Postharvest (environmental swab) | — | — | NP |
| FSL S10-2131 | FSL H9-0115 | L. innocua | Preharvest (fecal) | — | — | NP |
| FSL S11-0426 | FSL H9-0116 | L. innocua | Postharvest (environmental swab) | — | — | NP |
| FSL R12-0030 | FSL H9-0117 | L. innocua | Postharvest (food) | — | — | NP |
| FSL S11-0073 | FSL H9-0118 | L. seeligeri | Postharvest (environmental swab) | — | — | NP |
| FSL S11-0115 | FSL H9-0119 | L. seeligeri | Postharvest (environmental swab) | — | — | NP |
| FSL S11-0119 | FSL H9-0120 | L. seeligeri | Postharvest (environmental swab) | — | — | NP |
| FSL S10-2573 | FSL H9-0121 | L. seeligeri | Preharvest (soil) | — | — | NP |
| FSL S10-2558 | FSL H9-0122 | L. seeligeri | Preharvest (soil) | — | — | NP |
| FSL S10-2630 | FSL H9-0123 | L. seeligeri | Postharvest (soil) | — | — | NP |
| FSL S11-0322 | FSL H9-0124 | L. seeligeri | Postharvest (environmental swab) | — | — | NP |
| FSL S11-3481 | FSL H9-0125 | L. seeligeri | Postharvest (environmental swab) | — | — | NP |
| FSL S11-0238 | FSL H9-0126 | L. seeligeri | Postharvest (environmental swab) | — | — | NP |
| FSL S11-0429 | FSL H9-0127 | L. seeligeri | Postharvest (environmental swab) | — | — | NP |
| FSL S11-0280 | FSL H9-0128 | L. seeligeri | Postharvest (environmental swab) | — | — | NP |
| FSL S11-0241 | FSL H9-0129 | L. seeligeri | Postharvest (environmental swab) | — | — | NP |
| FSL S11-0076 | FSL H9-0130 | L. seeligeri | Postharvest (environmental swab) | — | — | NP |
| FSL S11-0274 | FSL H9-0131 | L. seeligeri | Postharvest (environmental swab) | — | — | NP |
| FSL S10-3513 | FSL H9-0132 | L. marthii | Postharvest (environmental swab) | — | — | NP |
| FSL S10-3421 | FSL H9-0133 | L. marthii | Postharvest (environmental swab) | — | — | NP |
| FSL S10-3516 | FSL H9-0134 | L. marthii | Postharvest (environmental swab) | — | — | NP |
| FSL S11-0060 | FSL H9-0135 | L. welshimeri | Postharvest (environmental swab) | — | — | bcrABC |
| FSL S11-0106 | FSL H9-0136 | L. welshimeri | Postharvest (environmental swab) | — | — | NP |
| FSL R12-0129 | FSL H9-0137 | L. welshimeri | Postharvest (food) | — | — | NP |
| FSL R12-0130 | FSL H9-0138 | L. welshimeri | Postharvest (food) | — | — | NP |
| FSL S10-3574 | FSL H9-0139 | L. welshimeri | Postharvest (environmental swab) | — | — | NP |
| FSL S11-0256 | FSL H9-0140 | L. welshimeri | Postharvest (environmental swab) | — | — | bcrABC |
| FSL S10-2222 | FSL H9-0142 | L. welshimeri | Preharvest (soil) | — | — | NP |
| FSL S10-3496 | FSL H9-0143 | L. welshimeri | Postharvest (environmental swab) | — | — | NP |
| FSL F6-0674 | FSL H9-0144 | L. ivanovii | Postharvest (food) | — | — | NP |
| FSL R12-0049 | FSL H9-0145 | L. ivanovii | Postharvest (food) | — | — | NP |
| FSL R12-0334 | FSL H9-0078 | L. monocytogenes | Retail (environmental swab) | II | CC9 | NP |
| FSL R9-9908 | FSL H9-0079 | L. monocytogenes | Postharvest (environmental swab) | II | CC9 | bcrABC |
| FSL R12-0099f | FSL H9-0080 | L. monocytogenes | Postharvest (environmental swab) | II | CC9 | NP |
| FSL R12-0359 | FSL H9-0081 | L. monocytogenes | Retail (environmental swab) | I | CC5 | bcrABC |
| FSL R12-0180 | FSL H9-0082 | L. monocytogenes | Retail (environmental swab) | II | CC193 | NP |
| FSL R12-0326 | FSL H9-0083 | L. monocytogenes | Retail (environmental swab) | I | CC5 | bcrABC |
| FSL R12-0181 | FSL H9-0084 | L. monocytogenes | Retail (environmental swab) | II | CC29 | NP |
| FSL S10-1873 | FSL H9-0085 | L. monocytogenes | Preharvest (soil) | I | CC6 | bcrABC |
| FSL S11-0386 | FSL H9-0086 | L. monocytogenes | Postharvest (environmental swab) | I | CC6 | NP |
| FSL S11-0136 | FSL H9-0087 | L. monocytogenes | Postharvest (environmental swab) | I | CC4 | NP |
| FSL S11-0272 | FSL H9-0088 | L. monocytogenes | Postharvest (environmental swab) | I | CC4 | NP |
| FSL S10-1884 | FSL H9-0089 | L. monocytogenes | Preharvest (soil) | II | CC369 | NP |
| FSL S10-1977 | FSL H9-0090 | L. monocytogenes | Preharvest (food) | II | CC369 | NP |
| FSL S11-0146 | FSL H9-0091 | L. monocytogenes | Postharvest (environmental swab) | II | CC37 | NP |
| FSL R12-0133 | FSL H9-0092 | L. monocytogenes | Retail (environmental swab) | II | CC37 | NP |
| FSL S11-0216 | FSL H9-0093 | L. monocytogenes | Postharvest (environmental swab) | I | CC388 | NP |
| FSL S11-0432 | FSL H9-0094 | L. monocytogenes | Postharvest (environmental swab) | II | CC155 | bcrABC |
| FSL R12-0098 | FSL H9-0095 | L. monocytogenes | Postharvest (environmental swab) | II | CC155 | NP |
| FSL S10-3467 | FSL H9-0096 | L. monocytogenes | Postharvest (environmental swab) | III | CC1789 | NP |
| FSL R12-0260 | FSL H9-0097 | L. monocytogenes | Retail (environmental swab) | III | CC268 | NP |
| FSL S10-3558 | FSL H9-0098 | L. monocytogenes | Postharvest (environmental swab) | III | CC434 | NP |
| FSL R12-0093 | FSL H9-0099 | L. monocytogenes | Postharvest (environmental swab) | II | ST1861 | NP |
| FSL S10-2151 | FSL H9-0100 | L. monocytogenes | Preharvest (soil) | II | CC204 | NP |
| FSL R12-0085 | FSL H9-0101 | L. monocytogenes | Postharvest (environmental swab) | II | CC19 | NP |
| FSL S11-0027 | FSL H9-0102 | L. monocytogenes | Postharvest (environmental swab) | I | CC6 | NP |
| FSL R9-9884 | FSL H9-0103 | L. monocytogenes | Postharvest (environmental swab) | II | CC9 | bcrABC |
| FSL R12-0189 | FSL H9-0104 | L. monocytogenes | Retail (environmental swab) | I | CC5 | qacH |
| FSL R12-0324 | FSL H9-0105 | L. monocytogenes | Retail (environmental swab) | I | CC4 | NP |
Isolates from Cornell Food Safety Lab (FSL) culture collection. Isolate information can be found on the Food Microbe Tracker website, https://www.foodmicrobetracker.net/login/login.aspx.
Derivative isolates of parent strains that were obtained through serial passaging of parent strains in BC, followed by substreaking seven times in BC-free medium.
Includes both clonal complexes (CCs) and singleton sequencing types (STs).
These data indicate whether a parent strain possessed either bcrABC or qacH (genes associated with Listeria resistance to BC) or neither gene (NP, not present).
—, not applicable, as only L. monocytogenes can be classified to lineage and clonal group using a standardized nomenclature.
Strain FSL R12-0099 was derived from CFSAN007516 (SRA accession no. SRR1101447) which has a genome size of 3,023,201 bp and shows presence of the gene bcrABC. FSL R12-0099 in the Cornell FSL culture collection has a genome size of 2,947,467 bp and does not possess bcrABC (SRA accession no. SRR15829693). These two genomes differ in size by 75,734 bp, which is similar to the size of plasmid pLM80 (80,000 bp) on which the bcrABC cassette can be encoded (21). These results suggest evidence of the loss of bcrABC in FSL R12-0099.
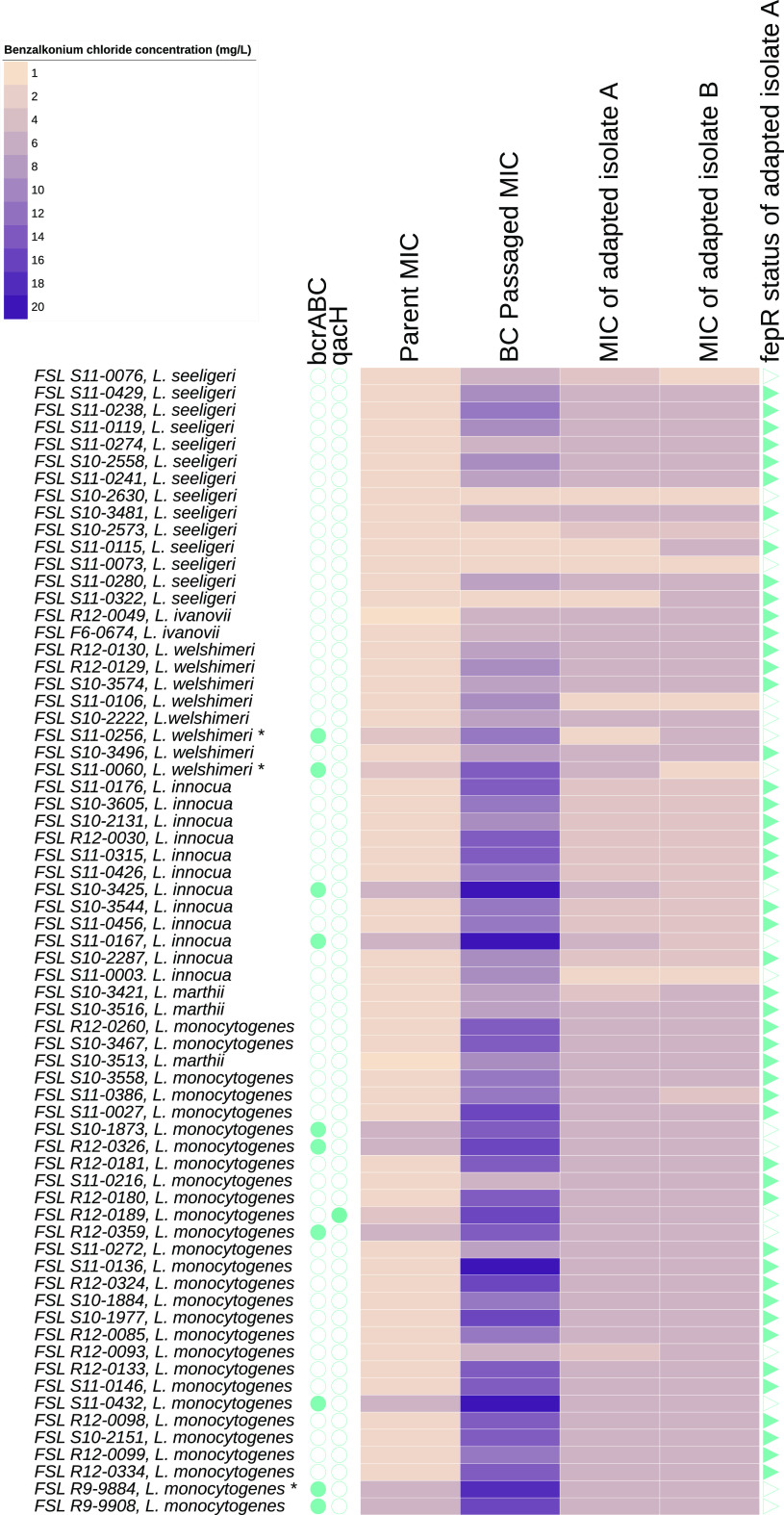
FIG 1.
Heat map of Listeria MICs to BC. Each row is specific to an individual parent strain of Listeria. For example, row 1 of the heat map corresponds to strain FSL S11-0076, which is a strain of L. seeligeri. “Parent MIC” indicates the initial MIC obtained for a given Listeria strain; “BC passaged MIC” indicates the MIC obtained through serial passaging of a strain in increasing concentrations of BC; “MIC of adapted isolate A” and “MIC of adapted isolate B” indicate the MIC obtained from two isolates obtained from serial passage experiments that were substreaked seven times onto BHI agar. Blue filled circles represent the presence of a BC resistance gene (bcrABC, qacH) in the Listeria parent strain. FSL S11-0256 and FSL S11-0060 showed loss of bcrABC in their corresponding adapted isolate A, and FSL S11-0256 and FSL R9-9884 showed loss of bcrABC in their corresponding adapted isolate B (an asterisk is used to identify these parent strains that showed loss of bcrABC in one of their respective adapted isolates). Blue filled triangles represent the presence of a nonsynonymous mutation in fepR in adapted isolate A.
TABLE 2.
Marginal mean estimates for MICs based on interaction models I and II
| Model and level | MIC estimate (mg/L) | SE | Lower CLa | Upper CL | Groupb |
|---|---|---|---|---|---|
| Interaction model Ic (fixed effects interaction of MIC type–Listeria type) | |||||
| Parent MIC–L. monocytogenes | 2.59 | 0.19 | 2.22 | 2.97 | A |
| Parent MIC–Listeria spp. | 2.12 | 0.13 | 1.86 | 2.37 | A |
| BC passaged MIC–L. monocytogenes | 13.38 | 0.98 | 11.45 | 15.31 | E |
| BC passaged MIC–Listeria spp. | 8.03 | 0.50 | 7.05 | 9.01 | D |
| Adapted MIC–L. monocytogenes | 5.91 | 0.34 | 5.25 | 6.58 | C |
| Adapted MIC–Listeria spp. | 4.41 | 0.21 | 3.99 | 4.83 | B |
| Interaction model IId (fixed effects interaction of MIC type–resistance gene) | |||||
| Parent MIC–present | 5.37 | 0.59 | 4.21 | 6.53 | B |
| Parent MIC–absent | 1.95 | 0.10 | 1.76 | 2.14 | A |
| BC passaged MIC–present | 16.14 | 1.77 | 12.66 | 19.62 | D |
| BC passaged MIC–absent | 9.04 | 0.44 | 8.17 | 9.90 | C |
| Adapted MIC–present | 5.23 | 0.46 | 4.32 | 6.14 | B |
| Adapted MIC–absent | 4.94 | 0.19 | 4.56 | 5.32 | B |
CL, confidence limit.
Group refers to significant differences based on post hoc multiple-comparison adjustment with Tukey’s honestly significant difference (HSD) test. Within each interaction model (I or II), groups that do not share a given letter are significantly different (P < 0.05).
Interaction model I tests the interaction of fixed effects of (i) Listeria type (either L. monocytogenes or Listeria spp. [L. innocua, L. ivanovii, L. marthii, L. seeligeri, L welshimeri]) and (ii) MIC type (either parent MIC, BC passaged MIC, or adapted MIC) on MIC to BC. Parent MIC, the initial MIC for a strain of L. monocytogenes or Listeria spp.; BC passaged MIC, the MIC obtained through serial passaging a strain in increasing concentrations of BC; adapted MIC, the MIC obtained from two isolates obtained from serial passage experiments that were substreaked seven times onto BHI agar.
Interaction model II tests the interaction of fixed effects of (i) resistance gene present (strains of L. monocytogenes or Listeria spp. that carry bcrABC or qacH) or absent (strains of L. monocytogenes or Listeria spp. that do not carry bcrABC or qacH) and (ii) MIC type (either parent MIC, BC passaged MIC, or adapted MIC) on MICs for BC.
Serial passaging experiments showed that 62/67 parent strains were able to grow in BC concentrations above their parent MIC.
To assess the ability of L. monocytogenes and Listeria spp. to acquire enhanced tolerance to BC, monocultures of all 67 parent strains of L. monocytogenes and Listeria spp. were subjected to serial passaging in increasing concentrations of BC. The highest concentration of BC in which a given strain failed to show growth after 48 h of incubation was designated the “BC passaged MIC.” The estimated marginal mean BC passaged MIC for all 67 strains was 9.94 mg/L, which was significantly higher than the estimated marginal mean parent MIC of 2.30 mg/L (P < 0.05). Overall, 62/67 strains were able to grow to BC concentrations above their parent MICs. For these strains, BC passaged MICs ranged from 4 to 20 mg/L. The five strains that did not grow above parent MICs all belonged to the species L. seeligeri. Results from two-way ANOVA and post hoc tests showed that the BC passaged MICs for L. monocytogenes (estimated marginal mean of 13.38 mg/L; range of 6 to 20 mg/L) were significantly higher (P < 0.05) than the BC passaged MICs for Listeria spp. (estimated marginal mean of 8.03 mg/L; range of 2 to 20 mg/L) (Table 2).
To further characterize the impact of serial passages in the presence of BC on MICs, we also calculated the MIC fold change for all strains (Table 3). The fold change of BC passaged MIC/parent MIC for all strains ranged from 1 to 10. L. monocytogenes showed a slightly higher fold change (estimated marginal mean fold change of 4.74) than Listeria spp. (estimated marginal mean fold change of 4.03), but this difference was not significant (P > 0.05). The fold change of the BC passaged MIC relative to the parent MIC (BC passaged MIC/parent MIC) for strains of L. monocytogenes and Listeria spp. that carried BC resistance genes bcrABC or qacH was significantly lower (estimated marginal mean fold change of 2.25) than the fold change observed in strains that did not carry these BC resistance genes (estimated marginal mean fold change of 4.90) (P < 0.05).
TABLE 3.
Estimates of marginal means for fold change of MICs based on additive models I and II
| Model | Fixed effects | Level | Estimated fold change | SE | Lower CLa | Upper CL | Groupb |
|---|---|---|---|---|---|---|---|
| Ic | Fold change type–Listeria type | BC passaged MIC/parent MIC–L. monocytogenes | 4.74 | 0.43 | 3.89 | 5.60 | B |
| BC passaged MIC/parent MIC–Listeria spp. | 4.03 | 0.32 | 3.40 | 4.66 | B | ||
| Adapted MIC/parent MIC–L. monocytogenes | 2.38 | 0.21 | 1.97 | 2.79 | A | ||
| Adapted MIC/parent MIC–Listeria spp. | 2.02 | 0.15 | 1.73 | 2.32 | A | ||
| IId | Fold change type–resistance gene | BC passaged MIC/parent MIC–present | 2.25 | 0.25 | 1.76 | 2.74 | B |
| BC passaged MIC/parent MIC–absent | 4.90 | 0.28 | 4.35 | 5.46 | C | ||
| Adapted MIC/parent MIC–present | 1.13 | 0.12 | 0.89 | 1.36 | A | ||
| Adapted MIC/parent MIC–absent | 2.46 | 0.12 | 2.22 | 2.70 | B |
CL, confidence limit.
Group refers to significant differences based on post hoc multiple-comparison adjustment with Tukey’s honestly significant difference (HSD) test. Within each additive model (I or II), different numbers denote significant differences (P < 0.05).
Additive model I tests the fixed effects of (i) Listeria type (either L. monocytogenes or Listeria spp. [L. innocua, L. ivanovii, L. marthii, L. seeligeri, L welshimeri]) and (ii) MICs being compared (BC passaged MIC/parent MIC, adapted MIC/parent MIC) on the level of fold change. Parent MIC, the initial MIC for a strain of L. monocytogenes or Listeria spp.; BC passaged MIC, the MIC obtained through serial passaging a strain in increasing concentrations of BC; adapted MIC, the MIC obtained from two isolates obtained from serial passage experiments that were substreaked seven times onto BHI agar.
Additive model II tests the fixed effects of (i) resistance gene present (strains of L. monocytogenes or Listeria spp. that carry bcrABC or qacH) or absent (strains of L. monocytogenes or Listeria spp. that do not carry bcrABC or qacH) and (ii) MICs being compared (BC passaged MIC/parent MIC, adapted MIC/parent MIC) on the level of fold change.
L. monocytogenes and Listeria spp. maintain increased MICs after seven rounds of substreaking in the absence of BC.
Following serial passaging experiments, the culture that represented the concentration of brain heart infusion (BHI) broth supplemented with BC (BHI-BC) in which a given strain of L. monocytogenes or Listeria spp. failed to show growth was plated onto BC-free BHI agar (BHIA) and two individual colonies (adapted isolate A and adapted isolate B) were selected for further characterization. All colonies were substreaked for seven rounds on BC-free BHIA to determine whether the tolerance of L. monocytogenes and Listeria spp. to BC was maintained in the absence of selective pressure. These data were used to determine whether increased MICs acquired during serial passaging were due to transient or inherited tolerance. After seven rounds of substreaking, all isolates were characterized through MIC experiments, and the resulting MICs were designated “MIC of adapted isolate A” and “MIC of adapted isolate B” (Fig. 1). The estimated marginal mean adapted MIC for all L. monocytogenes and Listeria spp. was 4.99 mg/L (range of 2 to 6 mg/L), which was significantly higher than the estimated marginal mean parent MIC (2.30 mg/L) but also significantly lower than the estimated marginal mean BC passaged MIC (9.94 mg/L) (P < 0.05).
The majority of adapted isolates showed higher adapted MICs than their parent MICs (105/134) but lower adapted MICs than their BC passaged MICs (113/134). Results from two-way ANOVA and post hoc tests showed that L. monocytogenes adapted MICs were significantly higher (estimated marginal mean of 5.91 mg/L, range of 4 to 6 mg/L) than adapted MICs of Listeria spp. (estimated marginal mean of 4.41 mg/L, range of 2 to 6 mg/L) (P < 0.05). The presence (estimated marginal mean of 5.23 mg/L, range of 2 to 6 mg/L) or absence (estimated marginal mean of 4.94 mg/L, range of 2 to 6 mg/L) of BC resistance genes bcrABC or qacH was not associated with a significant difference in adapted MICs (P > 0.05) (Table 2).
Further analysis of the fold change of adapted MIC/parent MIC (Table 3) showed that the fold change of L. monocytogenes (estimated marginal mean fold change of 2.38) did not differ significantly from the fold change of Listeria spp. (estimated marginal mean fold change of 2.02) (P > 0.05), suggesting no difference in the abilities of L. monocytogenes and Listeria spp. to adapt to BC. The estimated marginal mean fold change of adapted MIC/parent MIC for L. monocytogenes and Listeria spp. that carried either bcrABC or qacH was significantly lower (estimated marginal mean fold change of 1.13) than the fold change of isolates that did not carry bcrABC or qacH (estimated marginal mean fold change of 2.46) (P < 0.05), suggesting that isolates carrying bcrABC or qacH showed limited adaptation to BC.
The 67 strains in this study were selected from a larger culture collection, where they were previously classified as the top 10% tolerant, top 10% sensitive, or average in their sensitivity (representing the 11th to 89th percentile) to use-level concentrations of BC (based on log reduction data collected in a previous study) (33). Hence, we examined whether these previous classifications for the strains of L. monocytogenes and Listeria spp. we selected were associated with the MICs for these strains. Our results found that the previous classification of strains of L. monocytogenes and Listeria spp. as tolerant, average, or sensitive to use levels of BC was not significantly associated with a difference in parent MICs, BC passaged MICs, or adapted MICs (P > 0.05).
Mutations in fepR are associated with enhanced tolerance of L. monocytogenes and Listeria spp. to low levels of BC.
To investigate putative mutations responsible for the observed enhanced tolerance in adapted isolates, we performed whole-genome sequencing (WGS) on a subset of adapted isolate A isolates (n = 16) and compared their genomes to the genomes of their respective parent strains using high-quality single nucleotide polymorphism (hqSNP) analysis. In all 16 adapted isolates sequenced, we detected mutations in fepR (lmo2088), a gene encoding a TetR family transcriptional regulator (Table 4). Mutations detected through hqSNP analysis included (i) nonsense mutations (i.e., SNPs that lead to premature stop codons in the coding sequence of fepR) in 8/16 isolates and (ii) missense mutations (i.e., SNPs that lead to an amino acid change in the fepR coding sequence) in 5/16 isolates. In addition, single nucleotide deletions leading to frameshift mutations in fepR were detected in 3/16 isolates; these deletions were detected by aligning genome assemblies of parent strains and adapted isolates to an annotated fepR sequence. Other mutations detected in adapted isolates included a missense mutation in an internalin-like protein in adapted isolate FSL H9-0100 and a nonsense mutation detected in the putative membrane protein YdfK in adapted isolate FSL H9-0112.
TABLE 4.
Mutations detected in fepR for adapted isolates of L. monocytogenes or Listeria spp.a
| Adapted isolate ID | Species | Total no. of SNPs in adapted isolate | Mutation detected in fepRb | Mutation typec in fepR | BioProject accession no. or SRA ID of parent straind |
|---|---|---|---|---|---|
| FSL H9-0080 | L. monocytogenes | 0 | Deletion | Frameshift | PRJNA761983 |
| FSL H9-0082 | L. monocytogenes | 1 | SNP | Missense | SRR9019233 |
| FSL H9-0095 | L. monocytogenes | 2 | SNP | Nonsense | SRR1027706 |
| FSL H9-0098 | L. monocytogenes | 2 | SNP | Missense | PRJNA761983 |
| FSL H9-0100 | L. monocytogenes | 4 | SNP | Nonsense | SRR12696480 |
| FSL H9-0097 | L. monocytogenes | 1 | SNP | Nonsense | SRR9019214 |
| FSL H9-0105 | L. monocytogenes | 1 | Deletion | Frameshift | SRR9019309 |
| FSL H9-0111 | L. innocua | 2 | SNP | Nonsense | PRJNA761983 |
| FSL H9-0112 | L. innocua | 4 | SNP | Nonsense | PRJNA761983 |
| FSL H9-0122 | L. seeligeri | 1 | SNP | Missense | PRJNA761983 |
| FSL H9-0131 | L. seeligeri | 2 | SNP | Nonsense | PRJNA761983 |
| FSL H9-0132 | L. marthii | 2 | SNP | Missense | PRJNA761983 |
| FSL H9-0134 | L. marthii | 1 | SNP | Missense | PRJNA761983 |
| FSL H9-0137 | L. welshimeri | 0 | Deletion | Frameshift | PRJNA761983 |
| FSL H9-0144 | L. ivanovii | 1 | SNP | Nonsense | PRJNA761983 |
| FSL H9-0145 | L. ivanovii | 2 | SNP | Nonsense | PRJNA761983 |
Mutations were identified by comparison of the whole-genome sequences of parent strains and adapted isolates.
fepR encodes a transcriptional regulator in the TetR family of transcriptional regulators (GenBank accession no. WP_010991061.1). SNP, single nucleotide polymorphism detected in fepR; deletion, deletion of a single nucleotide in fepR.
Either missense (SNP confers amino acid shift in FepR protein), nonsense (either SNP confers a premature stop codon in FepR), or deletion (confers a frameshift in FepR).
Whole-genome sequences of all adapted isolates can be found under BioProject accession number PRJNA761983. The parent strain ID that corresponds to each adapted isolate ID is shown in Table 1.
On the basis of the high frequency of fepR mutations in adapted isolates, we PCR amplified the fepR sequence, followed by Sanger sequencing of PCR amplicons, for the 51 remaining adapted isolate A isolates that had not been characterized by WGS. A comparison of the fepR gene sequence in all 67 adapted isolates and parent strains revealed that 48/67 of the adapted isolates of L. monocytogenes and Listeria spp. in this study showed nonsynonymous mutations in fepR (24 missense mutations, 16 nonsense mutations, and 8 frameshift mutations) compared to their respective parent strains (Fig. 1). Among the frameshift mutations detected, seven were the result of single nucleotide deletions, and one was the result of a duplication of 10 nucleotides in fepR in adapted isolate FSL H9-0115 (Fig. 2). Among the missense mutations detected, 16/24 were localized in the N-terminal DNA binding domain of FepR between amino acid residues 1 and 43; five nonsense and two frameshift mutations were also detected in this region of FepR. None of the 11 parent strains that carried bcrABC or qacH acquired fepR mutations in their respective adapted isolates. ANOVA and post hoc tests revealed that the presence of a fepR mutation in adapted isolates was significantly associated with enhanced adapted MICs (estimated marginal mean MICs of 5.27 mg/L and 4.21 mg/L for isolates that possess fepR mutation and do not possess fepR mutation, respectively) (P < 0.05), while no significant differences in adapted MICs were detected across the three types of mutations detected in fepR (i.e., nonsense, missense, frameshift) (P > 0.05).
FIG 2.
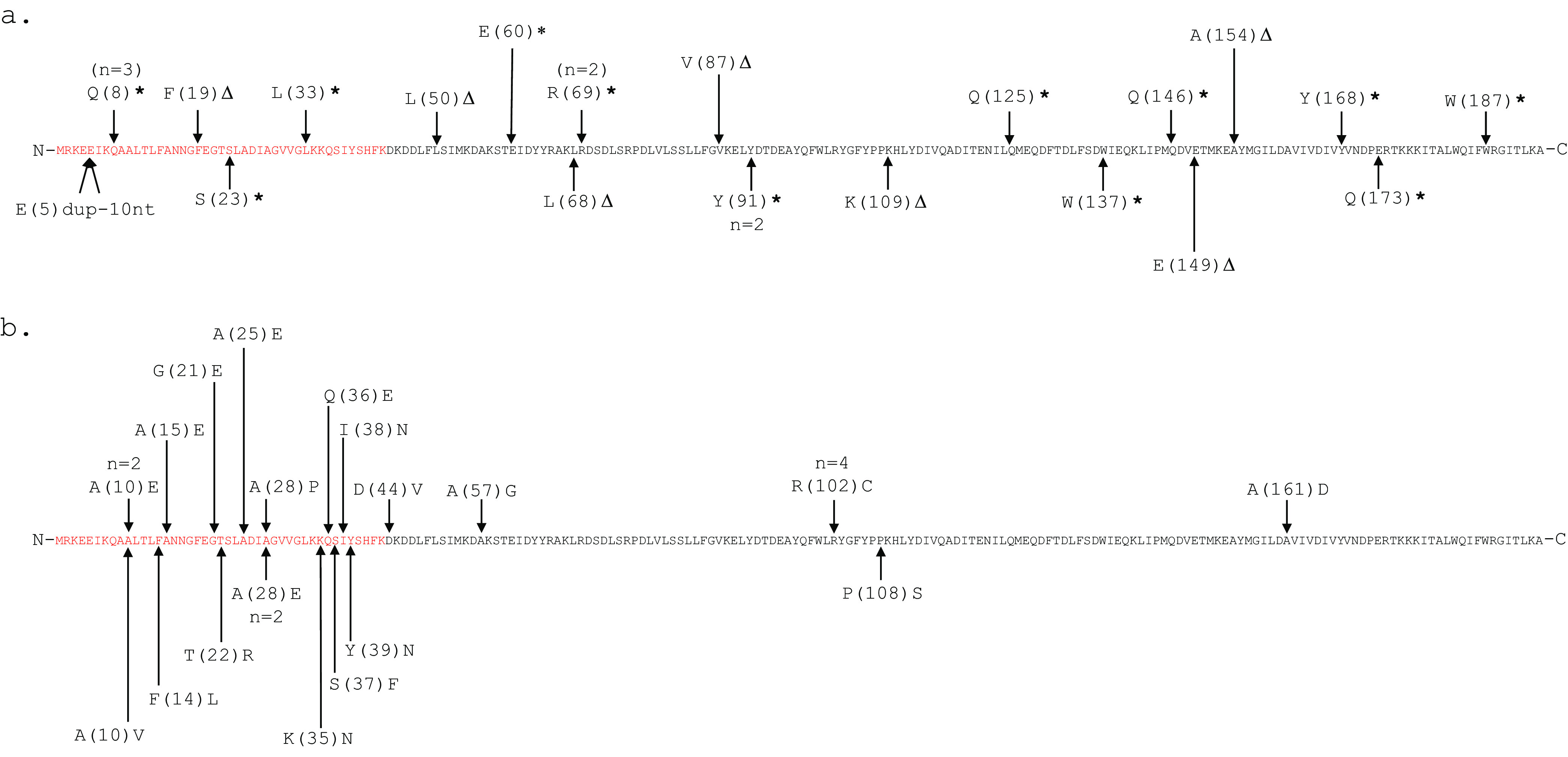
Location of nonsynonymous mutations in FepR in adapted isolates of L. monocytogenes and Listeria spp. from this study. Arrows indicate amino acid residues in which adapted isolates acquired unique mutations, and n is the number of isolates in which the unique mutation was detected. Amino acids colored in red represent the FepR DNA binding domain (45). (a) Nonsynonymous mutations that result in a nonsense mutation are denoted by an asterisk, single nucleotide deletions resulting in frameshift mutations are denoted by Δ, and a duplication resulting in a frameshift mutation (denoted by “dup-number of nucleotides” in length of duplication) (n = 24) are annotated on the 194-amino-acid FepR sequence (GenBank no. WP_010991061.1). (b) Nonsynonymous mutations that result in a missense mutation (n = 24) are annotated on the 194-amino-acid FepR sequence (GenBank no. WP_010991061.1).
While some adapted isolates show loss of bcrABC, this does not always result in reduced MICs.
PCR detection of bcrABC for adapted isolates revealed that four adapted isolates showed loss of bcrABC compared to their parent strains (including adapted isolates A and B derived from parent strain FSL S11-0256, adapted isolate A derived from parent strain FSL S11-0060, and adapted isolate B derived from parent strain FSL R9-9884) (Fig. 1). While two of these adapted isolates showed lower MICs than their respective parent strains, the other two adapted isolates (adapted isolate B from parent strain FSL S11-0256 and adapted isolate B from parent strain FSL R9-9884) both showed the same MICs as their respective parent strains (6 mg/L). In addition, both adapted isolate B from parent strain FSL S11-0256 and adapted isolate B from parent strain FSL R9-9884 did not show fepR mutations. These findings suggest possible additional mechanisms of adaptation to BC that could be further explored in future studies.
Tolerance of L. monocytogenes and Listeria spp. to low levels of BC is not associated with increased survival at use-level concentrations of BC.
In addition to investigating the tolerance of L. monocytogenes and Listeria spp. to low levels of BC, all parent strains and adapted isolate A isolates were assessed for their survival when exposed to a use-level concentration of BC (300 mg/L) for 30 s. Initial populations of parent strains (mean of 8.46 ± 0.02 log CFU/mL) and adapted isolates (mean of 8.45 ± 0.02 log CFU/mL) were reduced by 1.7 to 6.7 log (mean of 4.56 ± 0.09) and 1.8 to 6.6 log (mean of 4.48 ± 0.09), respectively, after treatment with BC (Fig. 3). An unpaired t test showed that log reductions of parent strains and adapted isolates were not significantly different (P = 0.57). Furthermore, the 11 parent strains that contained bcrABC or qacH and the 56 parent strains that did not contain bcrABC or qacH also did not differ significantly (P = 0.65) in observed log reductions in the presence of BC (mean log reductions of 4.47 ± 0.21 and 4.57 ± 0.10, respectively) (raw data deposited in GitHub).
FIG 3.
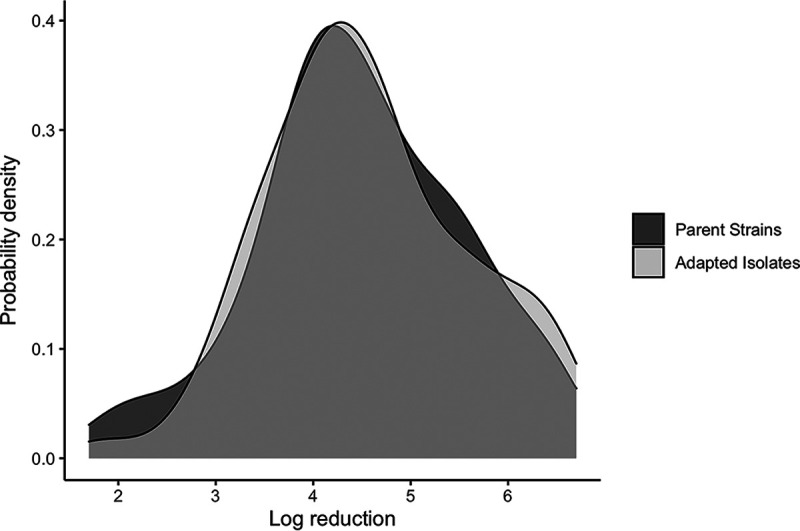
Density plot comparing log reductions of parent strains and adapted isolates after exposure to a use-level concentration (300 mg/L) of BC.
Cocultures of L. monocytogenes strains did not yield adapted isolates that show higher BC tolerance than monocultures exposed to BC.
Cocultures of L. monocytogenes containing either bcrABC or qacH, and strains containing neither genetic resistance determinant, were initially cocultured on a 0.45-μm filter in the absence of BC and subsequently serially passaged in increasing concentrations of BC to select for BC tolerance. This was followed by substreaking in the absence of BC to select for adapted isolates with heritable enhanced BC tolerance. The goal of these experiments was to determine whether the combination of conditions that facilitate horizontal gene transfer and select for BC tolerance could give rise to adapted isolates that showed enhanced BC tolerance in comparison to adapted isolates obtained from monocultures of L. monocytogenes. MICs of cocultures and monoculture controls immediately after filter mating on filter plates ranged from 8 to 10 mg/L, except for the monoculture of strain FSL R12-0334 (the only monoculture strain that contained neither bcrABC nor qacH), which showed an MIC of 4 mg/L (Table 5). After serial passaging, all monocultures and cocultures achieved BC passaged MICs in the range of 10 to 14 mg/L; cocultures did not show significantly higher BC passaged MICs than monocultures (P > 0.05). Similarly, MIC experiments performed on adapted isolates obtained after seven rounds of substreaking from monoculture and coculture serial passage experiments revealed that the adapted MIC across all adapted isolates obtained from cocultures and monocultures was 6 mg/L. Thus, L. monocytogenes cocultures did not yield adapted isolates that showed enhanced tolerance to BC compared to monocultures. These findings suggest that no rearrangements of BC resistance genes (bcrABC, qacH) occurred that resulted in enhanced BC tolerance in adapted isolates. As none of the adapted isolates from L. monocytogenes cocultures showed higher tolerance than adapted isolates from L. monocytogenes monocultures, no further characterization (e.g., subtyping to determine the identity of the adapted isolates) was performed.
TABLE 5.
MICs of BC for L. monocytogenes monocultures and cocultures obtained by three different experiments
| Isolate ID (abbreviation)a | Lineage, resistance gene | MIC (mg/L) obtained by indicated assayb |
||
|---|---|---|---|---|
| Filter plate MIC | BC passaged MIC | Adapted MIC | ||
| Monoculture FSL S11-0432 (S1) | II, bcrABC | 10 | 14 | 6 |
| Monoculture FSL S10-1873 (S2) | II, bcrABC | 10 | 14 | 6 |
| Monoculture FSL R9-9884 (S3) | II, bcrABC | 10 | 12 | 6 |
| Monoculture FSL R12-0334 (S4) | II, nonec | 4 | 10 | 6 |
| Monoculture FSL R12-0326 (S5) | I, bcrABC | 8 | 14 | 6 |
| Monoculture FSL R12-0189 (S6) | I, qacH | 8 | 12 | 6 |
| Monoculture FSL R12-0359 (S7) | I, bcrABC | 10 | 14 | 6 |
| Coculture I (S1, S3, S5, S6) | 10 | 12 | 6 | |
| Coculture II (S1, S5, S6, S7) | 10 | 12 | 6 | |
| Coculture III (S1, S4, S5, S6) | 8 | 10 | 6 | |
| Coculture IV (S2, S5, S6, S7) | 10 | 12 | 6 | |
| Coculture V (S2, S4, S5, S6) | 10 | 12 | 6 | |
| Coculture VI (S2, S3, S5, S6) | 10 | 14 | 6 | |
Monoculture, individual L. monocytogenes strains. Coculture, mixture of four L. monocytogenes strains combined and cultured together. Abbreviations in parentheses are isolate abbreviations used to describe the four isolates in each coculture.
Filter plate MIC, results from an MIC assay performed on cultures obtained after either four L. monocytogenes strains (i.e., cocultures), or individual L. monocytogenes strains (i.e., monocultures), were placed onto a 0.45-μm filter on BHI agar plates and allowed to incubate at 22°C for 48 h. BC passaged MIC, MIC obtained through serial passaging of L. monocytogenes cocultures I to VI or of monocultures (S1 to S7) in increasing concentrations of BC. Adapted MIC, the MIC for three adapted isolates that were taken from enumeration plates after serial passage experiments and substreaked for seven rounds onto BHI agar. All three adapted isolates (adapted isolate A, adapted isolate B, adapted isolate C) displayed the same adapted MIC.
No resistance gene (bcrABC or qacH) was identified in the genome.
DISCUSSION
In this study, we assessed the capacity of 67 diverse produce-associated strains of L. monocytogenes and Listeria spp. to acquire tolerance to low levels of BC, a sanitizer commonly used in produce packing and processing environments, and evaluated whether acquired tolerance to low levels of BC was associated with increased survival of L. monocytogenes and Listeria spp. when exposed to use-level concentrations of BC (i.e., 300 mg/L). While all strains of L. monocytogenes and Listeria spp. used in this study were isolated from environments associated with fresh produce production, since sanitizers such as BC are used widely throughout the food industry, our findings presented here are applicable beyond fresh produce packing and processing environments.
L. monocytogenes and Listeria spp. exposed to BC in serial passage experiments acquire inheritable and transient tolerance to low levels of BC.
In this study, we found that 105/134 adapted isolates of L. monocytogenes and Listeria spp. were able to acquire and maintain enhanced tolerance to BC through serial passage in increasing concentrations of BC and seven rounds of substreaking on BC-free media. By showing that this enhanced tolerance could be maintained in adapted isolates after selective pressure was removed, we have evidence to conclude that the acquired tolerance to BC in adapted isolates was due to inheritance of genetic mutations selected for during serial passaging. This phenomenon is referred to as tolerance due to genetic adaption (34). Additionally, we found that all species of Listeria represented in this study (L. innocua, L. ivanovii, L. marthii, L. monocytogenes, L. seeligeri, and L. welshimeri) could adapt to levels of BC up to 3-fold higher than their parent MICs. This is consistent with previous studies that have shown that BC-adapted L. monocytogenes strains can show 2- to 3-fold-higher MICs to BC than their wild-type counterparts (35, 36).
The majority of strains of L. monocytogenes and Listeria spp. were able to achieve higher BC passaged MICs than their parent MICs. However, for many strains of L. monocytogenes and Listeria spp., this acquired tolerance was not maintained or fully maintained after isolates were substreaked on BC-free medium. Thus, serial passage experiments also allowed L. monocytogenes and Listeria spp. to acquire transient tolerance to BC, which suggests that L. monocytogenes and Listeria spp. showed acclimation to BC (34) in addition to adaptation. Several studies have reported Gram-positive organisms acquiring transient tolerance to BC (37–39), even though some of these studies may not have referred to this observed phenomenon as “acclimation.” For example, in a study by Moore et al. (37), Staphylococcus haemolyticus was able to acquire tolerance to concentrations of BC that were 35-fold higher than its parental MIC (MIC value of 0.45 mg/L), but this tolerance was lost after passaging in the absence of BC for 7 days.
While horizonal gene transfer was previously reported to contribute to dispersal for bcrABC (30, 40), in our study, cocultures of L. monocytogenes strains carrying bcrABC, qacH, or neither genetic resistance determinant did not yield adapted isolates with enhanced tolerance to BC compared to the tolerance observed in adapted isolates obtained from monocultures. These findings could be because (i) selective pressure imposed by BC exposure in cocultures was not strong enough to select for transconjugants that did possess higher tolerance to BC or (ii) horizontal gene transfer of bcrABC and qacH did not occur within the coculture populations. This suggests that transfer of BC resistance genes to recipient isolates that already have resistance genes may be unlikely to occur or might not be strongly selected for, at least under the conditions used here. However, our study was limited to investigating the emergence of L. monocytogenes with enhanced BC tolerance among L. monocytogenes strains that were serially passaged in BC in planktonic cultures. As such, our findings may not be representative of interspecies and intergeneric transfer of BC resistance genes bcrABC and qacH and of selection that could occur in sessile cultures or multispecies biofilms. For example, interspecies transfer of bcrABC has been reported from Listeria spp. to L. monocytogenes (30), and intergeneric transfer of bcrABC has been reported from L. monocytogenes to Escherichia coli (40). As horizontal gene transfer continues to represent a potential mechanism for spread of BC tolerance in food processing environments (41, 42), future experiments using different approaches to select for BC-tolerant isolates should thus be performed to further probe whether horizontal gene transfer could give rise to transconjugants that do carry more than one BC resistance gene (i.e., bcrABC and qacH) and show an enhanced tolerance to BC compared to isolates that carry only one BC resistance gene.
Notably, our experiments revealed that L. monocytogenes and Listeria spp. (cocultures and monocultures) showed adaptation to a maximum MIC of 6 mg/L. Similar results were reported by Aase et al. (36), who showed that L. monocytogenes isolates with parent MICs to BC ranging from 1 to 7 mg/L were able to acquire inheritable tolerance to a maximum MIC of 7 mg/L BC. Results from Aase et al. (36) and our study both suggest that there is a biological barrier to the level of inheritable tolerance that L. monocytogenes and Listeria spp. can acquire to BC, with that level being ~6 to 7 mg/L. Most importantly, these findings support the view that there are limited concerns about L. monocytogenes and Listeria spp. acquiring tolerance to BC at levels that would impact the efficacy of quaternary ammonium compounds when they are used at recommended use-level concentrations.
Acquired tolerance of L. monocytogenes and Listeria spp. to BC is associated with nonsynonymous mutations in fepR.
The majority of strains of L. monocytogenes and Listeria spp. in our study acquired enhanced tolerance to BC, and this tolerance was significantly associated with the presence of nonsynonymous mutations (including missense, nonsense, and frameshift) in fepR. Importantly, adapted isolates representing all Listeria species included in this study showed evidence of mutations that would abolish FepR function. FepR has previously been shown to act as a local repressor for transcription of the operon fepRA (20). In addition to self-regulation, FepR also represses the transcription of fepA (lmo2087), which encodes an efflux pump that is part of the multidrug and toxic compound extrusion (MATE) family (43), which conceivably could remove BC compounds from the bacterial cytosol.
Consistent with our findings, Guérin et al. (20) reported that a single nonsense mutation in fepR in L. monocytogenes strain BM4716 was associated with a 2-fold-higher MIC to BC than that of L. monocytogenes parent strain BM4715. Moreover, strain BM4716 and a fepR deletion mutant of BM4715 (BM4715ΔfepR) both showed a 64-fold increase in gene expression of FepA compared to BM4715, demonstrating that loss of function of FepR results in overexpression of FepA. However, MIC assays conducted in the presence of the efflux pump inhibitor reserpine did not lead to the expected reduction in MIC for strain BM4716 (20), which may suggest that overexpression of efflux pump FepA may not be responsible for the enhanced BC tolerance associated with fepR mutations. Alternatively, reserpine added at a concentration of 10 mg/L may not have been used at the appropriate concentration to inhibit a highly expressed FepA or may not inhibit FepA as effectively as it does other efflux pumps. This is consistent with observations by Meier et al. (27), who found that 10 mg/L reserpine did not inhibit efflux activity in all eight of the L. monocytogenes strains that carried bcrABC in their study. Overall, data available to date strongly support that mutations in fepR resulting in truncation and loss of function in FepR are associated with and responsible for enhanced tolerance to low levels of BC.
In addition to mutations resulting in truncation of FepR (nonsense and frameshift mutations), we also observed missense mutations in 24 adapted isolates of L. monocytogenes and Listeria spp., and 22 of those isolates also showed the phenotype of acquired enhanced tolerance to BC. Similar to our findings, Bland et al. (44) showed that L. monocytogenes can acquire similar levels of tolerance to BC through both nonsense and missense mutations in fepR. In our study, we detected the majority (16/24) of missense mutations in the N-terminal DNA binding domain (NDB; helices α1 to α3) of FepR, which makes up the helix-turn-helix (HTH) motif that binds to the operator sequence of DNA on the fepRA operon (45). This NDB region of FepR is highly conserved with other TetR family transcriptional regulators (45); hence, previous findings in FepR homologues may be used to elucidate the likely effect of these missense mutations on FepR function. For example, TetR family regulators QacR and TetR both show a lack of water present at the NDB-operator DNA sequence binding interface, which facilitates tight binding of their NDB regions to DNA operator sequences (46, 47). Here, we saw 8/16 mutations in the NDB region of FepR that showed an amino acid change to glutamic acid (E) and 3/16 mutations that showed an amino acid change to asparagine (N), both of which have polar side chains that can participate in hydrogen bonding. We hypothesize that the incorporation of E and N residues in FepR’s NDB allows for the incorporation of water into the NDB-operator DNA sequence binding interface of fepRA, which can cause reduced or inhibited binding of FepR to the fepRA operator.
In addition, in our study, we also observed a handful of missense mutations located in regions localized outside of FepR’s NDB. One such mutation that was observed in L. innocua FSL H9-0107, an adapted isolate that showed 2-fold-enhanced tolerance to BC compared to its parent strain, resulted in an amino acid change from proline to serine at residue 108. Notably, this same amino acid change was reported by Bland et al. (44) in L. monocytogenes strain WRLP380, a strain which conferred 1.5-fold-enhanced tolerance to BC compared to its parent strain. Together, the findings from Bland et al. (44) and our study demonstrate that this particular amino acid change is associated with FepR loss of function, and further investigation is warranted to uncover the mechanism responsible for this observed phenotype. Additionally, these findings highlight the ability of L. monocytogenes and L. innocua to acquire BC tolerance through the same nonsynonymous mutation in fepR, further emphasizing the validity of L. innocua as an index organism for the presence of L. monocytogenes in food production environments where BC is used for sanitation (9). Interestingly, however, no fepR mutations were present in parent strains of L. monocytogenes and Listeria spp. in this study, suggesting selection against the loss-of-function mutation in fepR in more complex environments. Hence, the practical implications of fepR mutations in natural environments remains to be determined.
The BC tolerance phenotype is similar across L. monocytogenes and Listeria spp. with efflux mechanisms mediated by bcrABC and qacH or fepR mutations.
The presence of bcrABC and qacH in strains of L. monocytogenes and Listeria spp. from this study was associated with higher phenotypic tolerance to low levels of BC (parent MICs of 4 to 6 mg/L) than that of strains of L. monocytogenes and Listeria spp. that did not carry these genes (parent MICs of 1 to 2 mg/L). Similar findings have been reported for isolates carrying bcrABC and qacH obtained from a variety of food processing environments (17, 25, 27). For example, one study (17) reported that L. monocytogenes isolates carrying qacH showed MICs ranging from 5 to 12 mg/L (compared to strains without qacH that showed MICs of ≤ 5 mg/L), and Cooper et al. (25) showed that MICs for L. monocytogenes isolates carrying bcrABC were 2.5- to 3.5-fold higher than L. monocytogenes isolates that did not possess a BC resistance gene.
Notably, none of the adapted isolates in our study that were derived from parent strains carrying bcrABC or qacH acquired a mutation in fepR. Moreover, no L. monocytogenes or Listeria spp. with any of these three genotypes associated with BC tolerance (presence of bcrABC or qacH or nonsynonymous mutation in fepR) showed adapted MICs of >6 mg/L. Because there does not seem to be any phenotypic advantage in having one of these three genotypes over the others, we hypothesize that there was a lack of selective pressure for strains of L. monocytogenes and Listeria spp. already carrying either bcrABC or qacH to acquire inheritable tolerance through nonsynonymous mutations in fepR.
The tolerance of L. monocytogenes and Listeria spp. to low levels of BC does not correlate with increased survival in use levels of BC.
Overall, the majority of isolates of L. monocytogenes and Listeria spp. in this study were able to acquire inherited tolerance to low levels of BC at a biological barrier of 6 mg/L. However, when we compared the survival of adapted isolates with acquired tolerance to low levels of BC to that of their respective parent strains at a use-level concentration of 300 mg/L BC, the adapted isolates did not show better survival than their parent strains. These findings are in accordance with those of Kastbjerg and Gram (48), who found that L. monocytogenes strains that were adapted to grow in 48 mg/L of BC did not show better survival in 125 mg/L BC than L. monocytogenes parent strains.
Additionally, the 11 parent strains carrying bcrABC and qacH also did not show increased survival after exposure to 300 mg/L BC compared to the other 56 parent strains in this study. In a study by Cooper et al. (25), the authors suggest that the presence of BC resistance genes, specifically bcrABC, can represent indicators for L. monocytogenes persistence in food processing environments. Based on our results, while the presence of bcrABC confers tolerance of L. monocytogenes and Listeria spp. to BC at levels below use-level concentrations, bcrABC does not confer increased survival of L. monocytogenes and Listeria spp. at use-level concentrations of BC. Therefore, based on our results, it is unlikely that the presence of bcrABC represents a plausible indicator of persistence of L. monocytogenes and Listeria spp. in food processing environments.
Conclusions.
Overall, our results demonstrate that nearly all strains of L. monocytogenes and Listeria spp. tested here showed the capacity to acquire inheritable tolerance to low levels of BC, presumably through nonsynonymous mutations in fepR. This level of tolerance associated with nonsynonymous mutations in fepR was comparable to the level of tolerance observed in L. monocytogenes and Listeria spp. that carry resistance genes bcrABC and qacH. In addition, our results showed that strains of L. monocytogenes and Listeria spp. that acquired tolerance to low levels of BC did not show significantly enhanced survival when exposed to concentrations of BC recommended for sanitation of food contact surfaces in food processing environments. These findings provide evidence to support that BC applied to food contact surfaces at typical use-level concentrations for quaternary ammonium compounds (e.g., those outlined in U.S. guidance documents, i.e., 21 CFR 178.1010 [13]) will have similar effectiveness at controlling L. monocytogenes and Listeria spp., regardless of whether the strains of L. monocytogenes and Listeria spp. are tolerant to low levels of BC. Additional studies are needed to explore the importance of horizontal gene transfer of BC resistance genes (e.g., bcrABC, qacH) in populations of L. monocytogenes and Listeria spp. under different conditions (e.g., in multispecies biofilms exposed to BC) to assess the emergence of strains of L. monocytogenes and Listeria spp. with enhanced survival to use-level concentrations of quaternary ammonium compounds.
MATERIALS AND METHODS
Bacterial isolates and culture preparation.
For this study, a diverse set of 67 strains of L. monocytogenes and Listeria spp. from different fresh produce-associated sources was assembled. The selected strains represent preharvest (n = 9), postharvest (n = 49), and retail (n = 9) sources associated with fresh produce, including environmental samples (e.g., soil, water, environmental swabs) as well as actual produce samples (Table 1). The 67 strains were selected from a larger collection of 588 produce strains (33), which had been characterized by whole-genome sequencing and initial screening for inactivation when exposed to use-level concentrations of three sanitizers, including BC. These data were used to select diverse BC-tolerant and -sensitive strains in this study. Strains were initially stratified into the 10% most BC-tolerant and -sensitive strains based on the preliminary data. For L. monocytogenes, we also selected nine of the most prevalent clonal complexes (CCs) within the entire isolate collection, as well as the three most common CCs associated with lineage III (to ensure representation of lineage III, which is less frequent than lineages I and II), yielding 12 common CCs. For these 12 CCs, we identified strains that represented the top 10% most BC-tolerant and -sensitive strains where available. For CCs that did not include strains that represented the 10% most BC-tolerant and -sensitive strains, we randomly selected strains for inclusion. In addition, the top three most tolerant isolates and top three most sensitive isolates from the entire culture collection were also selected if not already included in the strain set. Finally, any L. monocytogenes strains that represented unique genotypes with regard to presence/absence of bcrABC and qacH were also included (e.g., we included strain FSL R12-0189, which carried qacH but was not classified in the top 10% most tolerant or sensitive strains). With this strategy, the final L. monocytogenes set included 28 strains that represented lineages I, II, and III (10, 15, and 3 strains, respectively) and 16 different clonal groups, including 15 CCs (CC4, CC5, CC6, CC9, CC19, CC29, CC37, CC155, CC193, CC204, CC268, CC369, CC388, CC434, CC1789), and one singleton (ST1861); a singleton refers to a clonal group that has a sequence type (ST) that differs from all other existing STs by at least two alleles (49). In this set, 12 of the strains represented the 10% most BC-tolerant strains, 11 of the strains represented the 10% most BC-sensitive strains, and 5 of the strains did not fall into these categories but were selected because they represented either lineage III (n = 3) or a unique genotype with regard to the presence of bcrABC or qacH (n = 2). For species of Listeria other than L. monocytogenes (i.e., L. innocua, L. ivanovii, L. marthii, L. seeligeri, L. welshimeri), we picked the top 10% most BC-tolerant and -sensitive strains within each species. In addition, we included one L. welshimeri isolate that carried bcrABC and a single L. marthii isolate that contained the gene lmo1409, which represented a unique genotype for L. marthii strains from the original culture collection. The final strain set for non-monocytogenes Listeria spp. included 12 L. innocua, two L. ivanovii, three L. marthii, 14 L. seeligeri, and eight L. welshimeri strains (Table 1). More information about the strains, including available metadata, serotypes, and associated publications, can be found on the Food Microbe Tracker (https://www.foodmicrobetracker.net/login/login.aspx) under a strain’s isolate ID (e.g., FSL H9-0078) (Table 1) (50).
Strains were streaked from −80°C freezer stocks (frozen in brain heart infusion [BHI] broth with 15% glycerol) onto brain heart infusion agar (BHIA; BD, Franklin Lakes, NJ), followed by incubation at 37°C for 24 h. Plates were stored at 4°C for at least 24 h and up to 7 days for use in experiments. To prepare bacterial cultures, 5 mL of BHI broth was inoculated with a single colony, followed by static incubation at 22°C for 34 to 36 h. This culture was diluted 1:1,000 into fresh BHI broth, followed by static incubation at 22°C for 34 to 36 h to yield stationary-phase bacterial cultures (as determined through preliminary experiments; data not shown). Bacterial cultures grown to stationary phase consistently showed initial levels between 8.5 and 9.2 log CFU/mL.
MIC assay.
MICs of BC for individual strains were determined with the broth microdilution method, performed according to CLSI guidelines (CLSI standard M07-A9), with slight modifications in endpoint determinations (51). Briefly, BHI broth was supplemented with BC (n-alkyl dimethyl benzyl ammonium chloride; Sigma-Aldrich, St. Louis, MO; CAS no. 63449-41-2) to achieve appropriate BC target concentrations. For the MIC assays, bacterial cultures grown to stationary phase as described above were diluted 1:1,000 and inoculated into BHI broth supplemented with BC (BHI-BC) to yield final BC concentrations of 0.25, 0.5, 1, 2, 4, 6, 8, 10, and 12 mg/L and a final bacterial concentration of ~106 CFU/mL per well. Individual strains were also inoculated into BHI broth without BC to serve as positive growth controls, and all concentrations of BHI-BC without bacterial inoculation were included to serve as negative controls for each MIC assay. Absorbance (optical density) of cultures was measured at 600 nm (OD600) in a microplate reader (Biotek Instruments, Winooski, VT) before (T0) and after incubation for 24 h at 22°C (T24). Differences in absorbance before and after 24 h of incubation (ΔOD600) were calculated, and values of >0.100 indicated strain growth at a given BC concentration. This endpoint determination method, which deviates from the method recommended in the broth microdilution method (51), was used to account for any changes in absorbance that may occur immediately following inoculation of BHI-BC with cultures of L. monocytogenes or Listeria spp. at T0. No subsequent confirmation of growth of L. monocytogenes and Listeria spp. by using culture-based methods was performed, in accordance with the procedures outlined in the broth microdilution method (51).
Serial passage experiments.
Serial passage experiments were performed to assess the capacity of L. monocytogenes and Listeria spp. to acquire enhanced tolerance to BC. For serial passage experiments, each individual Listeria strain was serially passaged in increasing concentrations of BHI-BC in a 96-well microtiter plate with a 200-μL volume. Bacterial cultures in stationary phase were diluted to a final concentration of ~106 CFU/mL at an initial concentration of 0.25 mg/L BHI-BC, followed by static incubation for up to 48 h at 22°C to allow for growth. Growth was assessed by measuring ΔOD600 at 24 h, and cultures were incubated for another 24 h if they did not show growth after 24 h. When growth was detected, cultures were subcultured 1:10 in the next incremental concentration of BHI-BC (i.e., 0.5, 1, 2 mg/L, with subsequent incremental increases of 2 mg/L BHI-BC). When a strain failed to show growth in a given BC concentration, a 100-μL aliquot from the well in which the strain failed to show growth was diluted in phosphate-buffered saline (PBS). Dilutions were then plated on BHIA and incubated for 24 h at 37°C for enumeration. The BC concentration at which strains failed to show growth was recorded as the “BC passaged MIC.”
After serial passaging and plating onto BHIA, strains were assessed for the stability of their acquired tolerance to BC. Two individual colonies from enumeration plates (defined as adapted isolate A and adapted isolate B) from the serial passage experiment were substreaked onto BHIA in the absence of BC and incubated at 37°C for 24 h. This substreaking procedure was repeated for a total of seven times, and individual colonies of adapted isolate A and adapted isolate B from the seventh substreaked plate were assessed for their “adapted MIC” as detailed above. The same colonies of adapted isolate A and adapted isolate B used for MIC experiments were frozen in BHI broth with 15% glycerol and stored at −80°C.
WGS and bioinformatics analysis.
Whole-genome sequencing (WGS) was performed on 16 adapted isolates. The resulting sequences were compared to sequences of parent strains to evaluate the presence of genetic mutations. For selection of isolates for WGS, all adapted isolate A isolates were initially screened to include only isolates that displayed an adapted MIC at least 2-fold higher than that of their respective parent MIC. From this set, 16 adapted isolate A isolates were randomly selected for WGS characterization. Single colonies of adapted isolate A isolates were inoculated into 5 mL BHI broth and incubated for 15 to 18 h at 37°C, and then DNA from cultures was extracted using a QiAamp DNA minikit (Qiagen, Germantown, MD) in accordance with the manufacturer’s instructions. Genomic DNA was sequenced using the Illumina NextSeq 500 platform (Illumina, Inc., San Diego, CA) with 2 × 150 bp paired-end reads. The software Trimmomatic v0.39 was used to trim adapters and low-quality bases from raw sequencing data, and quality assessment was performed using FastQC v0.11.8 (https://www.bioinformatics.babraham.ac.uk/projects/fastqc) (52). Reads were assembled using SPAdes v3.15.2 using careful mode. Quality control of assemblies was performed using QUAST v5.0.2 (53), and average coverage was determined using SAMtools v1.11 (54). Genomes with an average coverage greater than 40× were included in genomic analysis. Contigs smaller than 500 bp were removed.
The genomes of adapted isolates were compared with their respective parental genomes through high-quality single nucleotide polymorphism analysis (hqSNP) using the CFSAN SNP Pipeline v2.2.1 (55). For each parent strain-adapted isolate pair, the CFSAN SNP Pipeline was run twice, once aligning the paired-end reads of the adapted isolate to the complete genome assembly of the parent strain and once aligning the paired-end reads of the parent strain to the complete genome assembly of the adapted isolate. SNPs that were identified in both analyses represented reliably identified SNP differences and were reported. If SNPs were located in an open reading frame, the sequence of the open reading frame was searched in NCBI against the nonredundant protein sequences (nr) database using the BLASTX program to determine gene identities and/or their putative associated protein functions (56). SNPs were classified as synonymous (i.e., silent mutations) or nonsynonymous (i.e., missense, nonsense, or frameshift mutations) in Geneious Prime v2021.1.1.
PCR and Sanger sequencing.
Mutations in fepR in adapted isolates were identified through PCR amplification, followed by Sanger sequencing of fepR. For lysate preparation, individual colonies of adapted isolate A isolates were resuspended in 100 μL distilled H2O, and DNA was extracted by heat lysis via incubation at 95°C for 15 min. After incubation, the suspension was centrifuged at 14,000 × g for 10 min, and supernatant was used as a template for PCR. Two conventional PCR assays were developed and performed using de novo-designed primers (Table 6) for amplification of 886-bp (for L. innocua, L. marthii, and L. monocytogenes isolates) and 819-bp (for L. seeligeri and L. welshimeri isolates) PCR products that contained the sequence of fepR (585 bp). PCR was conducted on an ABI 2720 thermal cycler (Thermo Fisher), using GoTaq green master mix (Promega) with an initial denaturation of 5 min at 95°C, followed by 30 cycles of denaturation for 20 s at 95°C, annealing for 30 s at 55°C, and extension for 1 min at 72°C, with a final extension of 10 min at 72°C. The remaining primers and deoxynucleoside triphosphates (dNTPs) were digested by adding exonuclease I (10 U; Thermo Fisher) and shrimp alkaline phosphatase (1 U; Thermo Fisher) to PCR products and incubating the samples at 37°C for 45 min, followed by 80°C for 15 min. Sanger sequencing was performed on PCR products by the Biotechnology Resource Center (Cornell University, Ithaca, NY). Alignments of fepR for each parent strain and its corresponding adapted isolate A were performed in Geneious Prime v2021.1.1.
TABLE 6.
Primers for fepR in Listeria strains used in this study
| Target organisms | Primer | Sequence (5′ to 3′)a | Reference |
|---|---|---|---|
| L. marthii, L. innocua, L. monocytogenes | fepR-mim-F | ACGAATTGATTAGCGARTTTTTAGAAGAAA | This study |
| fepR-mim-R | GCCKAGCTTATGCCCAACA | This study | |
| L. seeligeri, L. welshimeri | fepR-sw-F | ATMACTTACGAACGGTCGTCAGTT | This study |
| fepR-sw-R | GTACGGCWATATTTATCCCAGCAAG | This study |
Sequences contain degenerate sites to compensate for variability in target sequence. R means A or G, K means G or T, M means A or C, and W means A or T.
PCR amplification of bcrABC and qacH in parent strains and adapted isolates was performed using primers and conditions described previously (24).
Inactivation of parent strains and adapted isolates of L. monocytogenes and Listeria spp. at a use-level concentration of BC.
To determine whether adapted isolates showed increased survival (compared to parent strains) at use-level concentrations of BC, all 67 parent strains and all 67 adapted isolate A isolates were exposed to a use-level concentration of BC for 30 s. Prior to BC exposure, 1 mL of each bacterial culture was centrifuged at 10,000 × g for 2 min to pellet cells. Pellets were resuspended in 1 mL of phosphate-buffered saline adjusted to pH 8.0 (PBS-pH 8.0), and 200-μL aliquots of cell suspensions were subsequently transferred into individual wells of a 96-well deep-well plate. Then, 200 μL of 600 mg/L BC (dissolved in PBS-pH 8.0) was added to each cell suspension, yielding a final exposure level of 300 mg/L BC. Cells were exposed to this BC concentration for a total of 30 s, with pipetting up and down 10 times throughout the exposure period. Control cell suspensions were exposed to 200 μL PBS-pH 8.0 for 30 s. For neutralization, 400 μL of 1.43× Dey-Engley neutralizing broth (D/E broth; BD) was added by pipetting up and down 10 times, followed by incubation for 5 min at room temperature (~20 to 22°C). Neutralized cultures were serially diluted in D/E broth, followed by plating onto BHIA, incubation for 24 h at 37°C, and enumeration of colonies. Bacterial log reductions were calculated by subtracting the log CFU/mL of control cultures from log CFU/mL of BC-treated cultures.
Coculture experiments.
For coculture experiments, seven L. monocytogenes strains were selected (using no formal randomization procedures or prescreening criteria) from the set of 28 L. monocytogenes strains used in monoculture experiments. The focus of the selection was on strains with existing BC resistance genes to probe whether rearrangements of these genes could give rise to adapted isolates with enhanced tolerance to BC. The final L. monocytogenes strain set included five strains that carried bcrABC, one strain that carried qacH, and one strain that did not carry a BC resistance gene (Table 5). For each coculture experiment, four strains of L. monocytogenes were grown separately to stationary phase in BHI broth as described above, followed by combining 1 mL of the stationary-phase culture of each strain into a tube (yielding 4 mL of culture with about 9 log CFU/mL of each strain), thorough vortexing, and subsequent centrifugation for 10 min at 4,000 × g. Each coculture cell pellet was resuspended in 200 μL BHI broth, and the resuspension was transferred onto a 0.45-μm filter placed on a BHIA plate, followed by static incubation for 48 h at 22°C; this filter mating protocol has previously been detailed (30) and was used to facilitate horizontal transfer of resistance genes. As a control, monocultures of each of the seven individual strains were plated in a 200-μL volume on a 0.45-μm filter placed on BHIA and incubated for 48 h at 22°C. After incubation, each filter (including those for the monoculture controls) was removed with forceps, transferred into a 50-mL conical vial, and washed with 5 mL BHI broth by pulse vortexing. Filters were aseptically removed, and filter rinsates were diluted to achieve an OD600 of ~0.2. OD-adjusted cultures were used to perform a MIC experiment (“filter plate MIC”) and were serially passaged in BC as described above. After serial passaging, three individual colonies from enumeration plates were substreaked seven times, and each of the three isolates was used to perform adapted MIC experiments.
Statistical analysis.
Data were analyzed in R version 4.0.2 (R Core Team). MICs were log transformed to satisfy normality assumptions. Linear mixed-effects models were fit using the lme4 package to determine (i) the effect of the interaction between the type of MIC (i.e., parent, BC passaged, adapted), and whether an isolate was L. monocytogenes or Listeria spp. (i.e., L. innocua, L. ivanovii, L. marthii, L. seeligeri, or L. welshimeri), on the outcome of MIC value, (ii) the effect of the interaction between the type of MIC (i.e., parent, BC passaged, adapted), and whether an isolate carried a BC resistance gene (bcrABC or qacH), on the outcome of MIC value, (iii) the effect of whether an adapted isolate showed a mutation in fepR on the outcome of MIC value, and (iv) the effect of the type of MIC (i.e., filter plate, BC passaged, adapted), and whether L. monocytogenes strains were grown in cocultures or monocultures, on the outcome of MIC value. For these analyses, type of MIC, L. monocytogenes versus Listeria spp., and BC resistance gene (presence or absence) were considered fixed effects, and each individual isolate’s FSL ID was considered a random effect. Analysis of variance (ANOVA) was performed on linear mixed models, followed by post hoc analysis of estimated marginal means with Tukey’s adjustment using the emmeans package in R (57).
To assess the effect of exposure to use-level concentrations of BC on the log reductions of parent strains compared to adapted isolates, an unpaired t test was used to compare the mean log reductions of all parent strains to the mean log reductions of all adapted isolates. One-way ANOVA was performed to test for the effect of presence of a BC resistance gene on the outcome of log reduction following exposure to use-level concentrations of BC. P values of ≤0.05 were considered statistically significant. The detection limit for bacterial colony enumeration was 2 log CFU/mL. In cases where no colonies were observed, the value for the limit of detection (2 log CFU/mL) was used for statistical analyses.
Data availability.
Whole-genome sequence data related to this study are available in the NCBI Sequence Read Archive (SRA) under BioProject accession number PRJNA761983. All raw data files and R code used to carry out statistical analysis are available on GitHub (https://github.com/sjb375/Listeria_BC_adaptation).
ACKNOWLEDGMENTS
We thank Yi Chen at FDA-CFSAN for kindly providing isolates FSL R12-0098 (SRR1027706), FSL R12-0099 (SRR1101447), FSL R12-0085, and FSL R12-0093. We also thank Haley Oliver for kindly providing isolates FSL R12-0180 (SRR9019233), FSL R12-0260 (SRR9019214), FSL R12-0324 (SRR9019309), FSL R12-0133, FSL R12-0181, FSL R12-0189, FSL R12-0326, FSL R12-0334, and FSL R12-0359.
The work, based on a project entitled “Listeria develops reduced sanitizer sensitivity but not resistance at recommended sanitizer use levels,” was funded in whole or in part through a subrecipient grant awarded to the Center for Produce Safety through the Florida Department of Agriculture and Consumer Services Specialty Crop Block Grant Program.
Any opinions, findings, conclusions, or recommendations expressed in this publication are those of the authors and do not necessarily reflect the view of the Center for Produce Safety or the Florida Department of Agriculture and Consumer Services.
Contributor Information
Martin Wiedmann, Email: mw16@cornell.edu.
Johanna Björkroth, University of Helsinki.
REFERENCES
- 1.Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson M-A, Roy SL, Jones JL, Griffin PM. 2011. Foodborne illness acquired in the United States—major pathogens. Emerg Infect Dis 17:7–15. 10.3201/eid1701.p11101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ferreira V, Wiedmann M, Teixeira P, Stasiewicz MJ. 2014. Listeria monocytogenes persistence in food-associated environments: epidemiology, strain characteristics, and implications for public health. J Food Prot 77:150–170. 10.4315/0362-028X.JFP-13-150. [DOI] [PubMed] [Google Scholar]
- 3.Nüesch-Inderbinen M, Bloemberg GV, Müller A, Stevens MJA, Cernela N, Kollöffel B, Stephan R. 2021. Listeriosis caused by persistence of Listeria monocytogenes serotype 4b sequence type 6 in cheese production environment. Emerg Infect Dis 27:284–288. 10.3201/eid2701.203266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cartwright EJ, Jackson KA, Johnson SD, Graves LM, Silk BJ, Mahon BE. 2013. Listeriosis outbreaks and associated food vehicles, United States, 1998–2008. Emerg Infect Dis 19:1–9. 10.3201/eid1901.120393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gaul LK, Farag NH, Shim T, Kingsley MA, Silk BJ, Hyytia-Trees E. 2013. Hospital-acquired listeriosis outbreak caused by contaminated diced celery—Texas, 2010. Clin Infect Dis 56:20–26. 10.1093/cid/cis817. [DOI] [PubMed] [Google Scholar]
- 6.McCollum JT, Cronquist AB, Silk BJ, Jackson KA, O'Connor KA, Cosgrove S, Gossack JP, Parachini SS, Jain NS, Ettestad P, Ibraheem M, Cantu V, Joshi M, DuVernoy T, Fogg NW, Gorny JR, Mogen KM, Spires C, Teitell P, Joseph LA, Tarr CL, Imanishi M, Neil KP, Tauxe RV, Mahon BE. 2013. Multistate outbreak of listeriosis associated with cantaloupe. N Engl J Med 369:944–953. 10.1056/NEJMoa1215837. [DOI] [PubMed] [Google Scholar]
- 7.Angelo KM, Conrad AR, Saupe A, Dragoo H, West N, Sorenson A, Barnes A, Doyle M, Beal J, Jackson KA, Stroika S, Tarr C, Kucerova Z, Lance S, Gould LH, Wise M, Jackson BR. 2017. Multistate outbreak of Listeria monocytogenes infections linked to whole apples used in commercially produced, prepackaged caramel apples: United States, 2014–2015. Epidemiol Infect 145:848–856. 10.1017/S0950268816003083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Self JL, Conrad A, Stroika S, Jackson A, Whitlock L, Jackson KA, Beal J, Wellman A, Fatica MK, Bidol S, Huth PP, Hamel M, Franklin K, Tschetter L, Kopko C, Kirsch P, Wise ME, Basler C. 2019. Multistate outbreak of listeriosis associated with packaged leafy green salads, United States and Canada, 2015–2016. Emerg Infect Dis 25:1461–1468. 10.3201/eid2508.180761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chapin TK, Nightingale KK, Worobo RW, Wiedmann M, Strawn LK. 2014. Geographical and meteorological factors associated with isolation of Listeria species in New York State produce production and natural environments. J Food Prot 77:1919–1928. 10.4315/0362-028X.JFP-14-132. [DOI] [PubMed] [Google Scholar]
- 10.Townsend A, Strawn LK, Chapman BJ, Dunn LL. 2021. A systematic review of Listeria species and Listeria monocytogenes prevalence, persistence, and diversity throughout the fresh produce supply chain. Foods 10:1427. 10.3390/foods10061427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Farber JM, Zwietering M, Wiedmann M, Schaffner D, Hedberg CW, Harrison MA, Hartnett E, Chapman B, Donnelly CW, Goodburn KE, Gummalla S. 2021. Alternative approaches to the risk management of Listeria monocytogenes in low risk foods. Food Control 123:107601. 10.1016/j.foodcont.2020.107601. [DOI] [Google Scholar]
- 12.Merchel Piovesan Pereira B, Tagkopoulos I. 2019. Benzalkonium chlorides: uses, regulatory status, and microbial resistance. Appl Environ Microbiol 85:e00377-19. 10.1128/AEM.00377-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Code of Federal Regulations. 2011. 21 CFR 178.1010 - Sanitizing solutions. U.S. Food and Drug Administration, Washington, DC. 21 CFR 178.1010 - Sanitizing solutions. - Content Details - CFR-2011-title21-vol3-sec178-1010 (govinfo.gov). [Google Scholar]
- 14.U.S. Department of Agriculture. Allowed detergents and sanitizers for food contact surfaces and equipment in organic operations. National Organic Program, Agricultural Marketing Service, U.S. Department of Agriculture. [Google Scholar]
- 15.Martínez-Suárez JV, Ortiz S, López-Alonso V. 2016. Potential impact of the resistance to quaternary ammonium disinfectants on the persistence of Listeria monocytogenes in food processing environments. Front Microbiol 7:638. 10.3389/fmicb.2016.00638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Environmental Protection Agency. 2012. Product performance test guidelines: OCSPP 810.2300: sanitizers for use on hard surfaces—efficacy data recommendations. EPA 712-C-07-091. Office of Chemical Safety and Pollution Prevention, U.S. Environmental Protection Agency. [Google Scholar]
- 17.Møretrø T, Schirmer BCT, Heir E, Fagerlund A, Hjemli P, Langsrud S. 2017. Tolerance to quaternary ammonium compound disinfectants may enhance growth of Listeria monocytogenes in the food industry. Int J Food Microbiol 241:215–224. 10.1016/j.ijfoodmicro.2016.10.025. [DOI] [PubMed] [Google Scholar]
- 18.Duze ST, Marimani M, Patel M. 2021. Tolerance of Listeria monocytogenes to biocides used in food processing environments. Food Microbiol 97:103758. 10.1016/j.fm.2021.103758. [DOI] [PubMed] [Google Scholar]
- 19.Romanova NA, Wolffs PFG, Brovko LY, Griffiths MW. 2006. Role of efflux pumps in adaptation and resistance of Listeria monocytogenes to benzalkonium chloride. Appl Environ Microbiol 72:3498–3503. 10.1128/AEM.72.5.3498-3503.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guérin F, Galimand M, Tuambilangana F, Courvalin P, Cattoir V. 2014. Overexpression of the novel MATE fluoroquinolone efflux pump FepA in Listeria monocytogenes is driven by inactivation of its local repressor FepR. PLoS One 9:e106340. 10.1371/journal.pone.0106340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elhanafi D, Dutta V, Kathariou S. 2010. Genetic characterization of plasmid-associated benzalkonium chloride resistance determinants in a Listeria monocytogenes strain from the 1998-1999 outbreak. Appl Environ Microbiol 76:8231–8238. 10.1128/AEM.02056-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dutta V, Elhanafi D, Kathariou S. 2013. Conservation and distribution of the benzalkonium chloride resistance cassette bcrABC in Listeria monocytogenes. Appl Environ Microbiol 79:6067–6074. 10.1128/AEM.01751-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Müller A, Rychli K, Muhterem-Uyar M, Zaiser A, Stessl B, Guinane CM, Cotter PD, Wagner M, Schmitz-Esser S. 2013. Tn6188—a novel transposon in Listeria monocytogenes responsible for tolerance to benzalkonium chloride. PLoS One 8:e76835. 10.1371/journal.pone.0076835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harrand AS, Jagadeesan B, Baert L, Wiedmann M, Orsi RH. 2020. Evolution of Listeria monocytogenes in a food processing plant involves limited single-nucleotide substitutions but considerable diversification by gain and loss of prophages. Appl Environ Microbiol 86:e02493-19. 10.1128/AEM.02493-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cooper AL, Carrillo CD, Deschênes M, Blais BW. 2021. Genomic markers for quaternary ammonium compound resistance as a persistence indicator for Listeria monocytogenes contamination in food manufacturing environments. J Food Prot 84:389–398. 10.4315/JFP-20-328. [DOI] [PubMed] [Google Scholar]
- 26.Schmitz-Esser S, Anast JM, Cortes BW. 2021. A large-scale sequencing-based survey of plasmids in Listeria monocytogenes reveals global dissemination of plasmids. Front Microbiol 12:653155. 10.3389/fmicb.2021.653155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meier AB, Guldimann C, Markkula A, Pöntinen A, Korkeala H, Tasara T. 2017. Comparative phenotypic and genotypic analysis of Swiss and Finnish Listeria monocytogenes isolates with respect to benzalkonium chloride resistance. Front Microbiol 8:397. 10.3389/fmicb.2017.00397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Müller A, Rychli K, Zaiser A, Wieser C, Wagner M, Schmitz-Esser S. 2014. The Listeria monocytogenes transposon Tn6188 provides increased tolerance to various quaternary ammonium compounds and ethidium bromide. FEMS Microbiol Lett 361:166–173. 10.1111/1574-6968.12626. [DOI] [PubMed] [Google Scholar]
- 29.Rossi F, Rizzotti L, Felis GE, Torriani S. 2014. Horizontal gene transfer among microorganisms in food: current knowledge and future perspectives. Food Microbiol 42:232–243. 10.1016/j.fm.2014.04.004. [DOI] [PubMed] [Google Scholar]
- 30.Katharios-Lanwermeyer S, Rakic-Martinez M, Elhanafi D, Ratani S, Tiedje JM, Kathariou S. 2012. Coselection of cadmium and benzalkonium chloride resistance in conjugative transfers from nonpathogenic Listeria spp. to other Listeriae. Appl Environ Microbiol 78:7549–7556. 10.1128/AEM.02245-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brauner A, Fridman O, Gefen O, Balaban NQ. 2016. Distinguishing between resistance, tolerance and persistence to antibiotic treatment. Nat Rev Microbiol 14:320–330. 10.1038/nrmicro.2016.34. [DOI] [PubMed] [Google Scholar]
- 32.Bland R, Brown SRB, Waite-Cusic J, Kovacevic J. 2022. Probing antimicrobial resistance and sanitizer tolerance themes and their implications for the food industry through the Listeria monocytogenes lens. Compr Rev Food Sci Food Saf; 21:1777–1802. 10.1111/1541-4337.12910. [DOI] [PubMed] [Google Scholar]
- 33.Wiedmann M. 2020. Listeria develops reduced sanitizer sensitivity but not resistance at recommended sanitizer use levels. Center for Produce Safety, Woodland, CA. [Google Scholar]
- 34.Henderson LO. 2019. Did you just eat that? Novel approaches to control Listeria monocytogenes on ready-to-eat-foods. PhD dissertation. Cornell University, Ithaca, NY. [Google Scholar]
- 35.To MS, Favrin S, Romanova N, Griffiths MW. 2002. Postadaptational resistance to benzalkonium chloride and subsequent physicochemical modifications of Listeria monocytogenes. Appl Environ Microbiol 68:5258–5264. 10.1128/AEM.68.11.5258-5264.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aase B, Sundheim G, Langsrud S, Rørvik LM. 2000. Occurrence of and a possible mechanism for resistance to a quaternary ammonium compound in Listeria monocytogenes. Int J Food Microbiol 62:57–63. 10.1016/S0168-1605(00)00357-3. [DOI] [PubMed] [Google Scholar]
- 37.Moore LE, Ledder RG, Gilbert P, McBain AJ. 2008. In vitro study of the effect of cationic biocides on bacterial population dynamics and susceptibility. Appl Environ Microbiol 74:4825–4834. 10.1128/AEM.00573-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gadea R, Fernández Fuentes MÁ, Pérez Pulido R, Gálvez A, Ortega E. 2017. Effects of exposure to quaternary-ammonium-based biocides on antimicrobial susceptibility and tolerance to physical stresses in bacteria from organic foods. Food Microbiol 63:58–71. 10.1016/j.fm.2016.10.037. [DOI] [PubMed] [Google Scholar]
- 39.Kampf G. 2018. Adaptive microbial response to low-level benzalkonium chloride exposure. J Hosp Infect 100:e1–e22. 10.1016/j.jhin.2018.05.019. [DOI] [PubMed] [Google Scholar]
- 40.Xu D, Nie Q, Wang W, Shi L, Yan H. 2016. Characterization of a transferable bcrABC and cadAC genes-harboring plasmid in Listeria monocytogenes strain isolated from food products of animal origin. Int J Food Microbiol 217:117–122. 10.1016/j.ijfoodmicro.2015.10.021. [DOI] [PubMed] [Google Scholar]
- 41.Allen KJ, Wałecka-Zacharska E, Chen JC, Katarzyna K-P, Devlieghere F, Van Meervenne E, Osek J, Wieczorek K, Bania J. 2016. Listeria monocytogenes—an examination of food chain factors potentially contributing to antimicrobial resistance. Food Microbiol 54:178–189. 10.1016/j.fm.2014.08.006. [DOI] [Google Scholar]
- 42.Baquero F, Lanza V, Duval M, Coque TM. 2020. Ecogenetics of antibiotic resistance in Listeria monocytogenes. Mol Microbiol 113:570–579. 10.1111/mmi.14454. [DOI] [PubMed] [Google Scholar]
- 43.Jiang X, Yu T, Xu P, Xu X, Ji S, Gao W, Shi L. 2018. Role of efflux pumps in the in vitro development of ciprofloxacin resistance in Listeria monocytogenes. Front Microbiol 9:2350. 10.3389/fmicb.2018.02350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bland R, Waite-Cusic J, Weisberg AJ, Riutta ER, Chang JH, Kovacevic J. 2022. Adaptation to a commercial quaternary ammonium compound sanitizer leads to cross-resistance to select antibiotics in Listeria monocytogenes isolated from fresh produce environments. Front Microbiol 12:782920. 10.3389/fmicb.2021.782920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Samad A, Li Y, Zhang C, Chen F, Zeng W, Fan X, Jin T. 2019. X-ray crystal structure of putative transcription regulator lmo2088 from Listeria monocytogenes. Biochem Biophys Res Commun 520:434–440. 10.1016/j.bbrc.2019.10.033. [DOI] [PubMed] [Google Scholar]
- 46.Ramos JL, Martínez-Bueno M, Molina-Henares AJ, Terán W, Watanabe K, Zhang X, Gallegos MT, Brennan R, Tobes R. 2005. The TetR family of transcriptional repressors. Microbiol Mol Biol Rev 69:326–356. 10.1128/MMBR.69.2.326-356.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schumacher M, Miller MC, Grkovic S, Brown MH, Skurray RA, Brennan RG. 2002. Structural basis for cooperative DNA binding by two dimers of the multidrug-binding protein QacR. EMBO J 21:1210–1218. 10.1093/emboj/21.5.1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kastbjerg VG, Gram L. 2012. Industrial disinfectants do not select for resistance in Listeria monocytogenes following long term exposure. Int J Food Microbiol 160:11–15. 10.1016/j.ijfoodmicro.2012.09.009. [DOI] [PubMed] [Google Scholar]
- 49.Chen Y, Luo Y, Pettengill J, Timme R, Melka D, Doyle M, Jackson A, Parish M, Hammack TS, Allard MW, Brown EW, Strain EA. 2017. Singleton sequence type 382, an emerging clonal group of Listeria monocytogenes associated with three multistate outbreaks linked to contaminated stone fruit, caramel apples, and leafy green salad. J Clin Microbiol 55:931–941. 10.1128/JCM.02140-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vangay P, Fugett EB, Sun Q, Wiedmann M. 2013. Food Microbe Tracker: a web-based tool for storage and comparison of food-associated microbes. J Food Prot 76:283–294. 10.4315/0362-028X.JFP-12-276. [DOI] [PubMed] [Google Scholar]
- 51.Clinical and Laboratory Standards Institute. 2012. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically: approved standard , 9th ed. CLSI standard M07-A9. CLSI, Wayne, PA. [Google Scholar]
- 52.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gurevich A, Saveliev V, Vyahhi N, Tesler G. 2013. QUAST: quality assessment tool for genome assemblies. Bioinformatics 29:1072–1075. 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup . 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079. 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Davis S, Pettengill JB, Luo Y, Payne J, Shpuntoff A, Rand H, Strain E. 2015. CFSAN SNP Pipeline: an automated method for constructing SNP matrices from next-generation sequence data. PeerJ Comput Sci 1:e20. 10.7717/peerj-cs.20. [DOI] [Google Scholar]
- 56.States DJ, Gish W. 1994. Combined use of sequence similarity and codon bias for coding region identification. J Comput Biol 1:39–50. 10.1089/cmb.1994.1.39. [DOI] [PubMed] [Google Scholar]
- 57.Length R. 2019. Emmeans: estimated marginal means, aka least-squares means. R package version 1.3.4. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Whole-genome sequence data related to this study are available in the NCBI Sequence Read Archive (SRA) under BioProject accession number PRJNA761983. All raw data files and R code used to carry out statistical analysis are available on GitHub (https://github.com/sjb375/Listeria_BC_adaptation).