Abstract
A bacterial strain capable of growing on propachlor (2-chloro-N-isopropylacetanilide) was isolated from soil by using enrichment and isolation techniques. The strain isolated, designated GCH1, was classified as a member of the genus Pseudomonas. Washed-cell suspensions of strain GCH1 accumulated N-isopropylacetanilide, acetanilide, acetamide, and catechol. Pseudomonas strain GCH1 grew on propachlor with a generation time of 4.2 h and a rate of substrate utilization of 1.75 ± 0.15 μmol h−1. Gene expression did not require induction but was subject to catabolite expression. Acetanilide was a growth substrate with a yield of 0.56 ± 0.02 mg of protein μmol−1. GCH1 strain cells were immobilized by adsorption onto a ceramic support and were used as biocatalysts in an immobilized cell system. Propachlor elimination reached 98%, with a retention time of 3 h and an initial organic load of 0.5 mM propachlor. The viability of immobilized cells increased 34-fold after 120 days of bioreactor operation.
Propachlor (2-chloro-N-isopropylacetanilide) is an acylanilide herbicide widely used with corn, onion, cabbage, rose bushes, and ornamental plants. Microbial degradation (11, 15, 16, 19) is the primary mechanism of acylanilide dissipation from soil. We previously reported the isolation of Pseudomonas strain PEM1 (3, 10, 11), which metabolizes the herbicide propachlor, yielding N-isopropylacetanilide, acetanilide, and acetamide as intermediates, and the isolation of Acinetobacter strain BEM2, which follows a different pathway and yields N-isopropylaniline and isopropylamine as intermediates. Lee et al. (9) reported that N-isopropylaniline, N-isopropylacetanilide, N-(1-hydroxyisopropyl)acetanilide, and N-isopropyl-2-acetoxyacetanilide were formed in soil treated with propachlor. Villareal et al. (18) proposed a propachlor degradative pathway yielding 2-chloro-N-isopropylacetamide and catechol as intermediates.
Groundwater and soil contamination by herbicides has recently become of increasing concern (6, 7, 20). The relatively new concept of bioremediation provides a potentially cheap alternative to traditional disposal techniques, in addition to representing a genuine removal of contaminants by microbial degradation rather than the relocation of contaminants in such processes as landfilling. The problems presented by continuous fermentation processes could be resolved by the use of immobilized cells as biocatalysts (5, 7, 12, 21).
The aim of this study was to characterize the propachlor metabolism of a strain (GCH1) isolated from soil and to test a bioremediation system using immobilized cells in a reactor operating over a period of 150 days. The kinetics of the degradation and the viability of the immobilized cells are reported.
MATERIALS AND METHODS
Isolation of bacteria.
Ten soil samples (10) were collected from agricultural fields in Madrid, Spain, with a history of propachlor contamination. Minimal medium (MB) (4) supplemented with 45 mg of propachlor liter−1 was inoculated with 20 g of soil sample and incubated at 28°C without shaking. Aliquots were subcultured every 10 days for 40 days, and the final subculture was plated on MB agar plates with 1 mM propachlor as the carbon source. One of the isolates, designated strain GCH1, was selected for further analysis of substrate specificity and biochemical reactions (API 20NE kit; bioMérieux S.A., Marcy l'Etoile, France). Mole percent G+C content was estimated by the spectrometric method of Ulitzur (17) with DNA from Escherichia coli B as a standard. DNA was prepared with a Kristal DNA extraction kit (Cambridge Molecular Technologies, Cambridge, United Kingdom). As described by Widmer et al. (20), Pseudomonas-specific PCR primers Ps-for (0-mer; 5′-GGTCTGAGAGGATGATCAGT-3′) and Ps-rev (18-mer; 5′-TTAGCTCCACCTCGCGGC-3′) (Isogen Bioscience BV) were used for Pseudomonas taxonomic confirmation.
Media and growth conditions.
Cells were grown aerobically at 30°C in MB. Carbon sources were sterilized separately and added to give 0.1 to 2 mM propachlor, 10 mM glucose, 1 mM acetanilide, or 10 mM acetamide.
Cell immobilization.
A ceramic material, granular sepiolite, was chosen as the support; the immobilization method used has been previously described (3, 5, 12). This porous material was placed into MB (10 g of sepiolite/20 ml of MB) and autoclaved; after 24 h, the mineral medium was replaced by a new sterile medium. Cells grown in 0.5 mM propachlor were harvested at the exponential phase and used as the inoculum. Immobilized cells were examined by scanning electron microscopy as previously reported (3, 12).
Use and mineralization of propachlor.
Kinetic parameters for the mineralization of propachlor were determined as previously reported (11). Metabolism was determined by measuring 14CO2 released from [ring-U-14C]propachlor (12 μCi/μmol). Cells pregrown in MB and glucose or propachlor were washed and resuspended in 10 ml of phosphate buffer (pH 7.2); 5 ml of the phosphate buffer was added to 50-ml flasks containing 1 μCi of [ring-U-14C]propachlor. Different amounts of unlabeled propachlor were added to the flasks to achieve final concentrations ranging from 0.1 to 2 mM. To initiate mineralization assays, media were inoculated with 106 washed cells of an early-stationary-phase culture of glucose- or propachlor-grown cells. 14CO2 formed from the mineralization was trapped on a 1 N NaOH solution located on the top of the bottles. Radioactivity was measured in a Packard model 2500 TR scintillation spectrometer. Total initial activity (and concentration) was determined by averaging counts of 1-ml aliquots sampled before, during, and after dispensation.
Experiments related to the regulation of propachlor metabolism required the use of chloramphenicol to block protein synthesis (13). Chloramphenicol at 110 μg ml−1 blocked growth within 35 min in exponentially growing batch cultures. The uptake (1) of l-[14C]phenylalanine was blocked by 110 μg ml−1 during 35 min of incubation (data not shown). Experiments were carried out with cells harvested at exponential phase, washed, and resuspended in MB containing 10 mM glucose; 10-ml aliquots of the cell suspension were dispensed into 50-ml serum flasks, and some of these suspensions received 110 μg of chloramphenicol ml−1. Flasks were preincubated 35 min at 30°C before the addition of 0.5 mM propachlor. Samples (1 ml) were removed at 45-min intervals and centrifuged, and the supernatant was analyzed by high-pressure liquid chromatography (HPLC) as described below.
Analytical methods.
The gas chromatography-mass spectrometry (GC-MS) analyses were performed with a Hewlett-Packard model 5890 series II gas chromatograph and an HP-5971 mass detector. The gas chromatograph was equipped with a methyl silicone capillary column (20 m; 0.22-mm inside diameter) programmed from 70 to 220°C (4°C/min), and the injector and detector interface temperatures were 170 and 300°C, respectively. HPLC analysis was performed by using a Waters model 616PDA996 equipped with a data analysis Millennium 20/10. Separation was achieved on a Novapack C-18 (3.9 by 150 mm) column, using a mobile phase consisting of 40% acetonitrile in water at a flow rate of 0.5 ml/min and measuring at 214 nm. The injection volume was 10 μl.
Characterization of propachlor degradation intermediates.
Intermediates were identified by experiments using nongrowing cells. Cultures of propachlor-grown cells were centrifuged at 10,000 × g for 10 min at 4°C; pellets were washed twice with 10 mM phosphate buffer (pH 7.2) and resuspended in the same buffer. Substrates were added to the cell suspensions and incubated at 30°C. Propachlor and the resulting intermediates in its degradation were analyzed by HPLC and GC-MS (10, 11). Samples for HPLC were evaporated to dryness under a nitrogen stream and redissolved in ethanol. For GC-MS, samples from the experimental cultures were extracted 1:1 with ethyl acetate, and 2-μl aliquots of the ethyl acetate extracts were injected into the column. Metabolites were identified by comparison of their electron impact-MS spectra with those obtained for standard and by coelution in HPLC and GC.
Laser scanning confocal microscopy analysis.
A Bio-Rad MRC 1024 confocal laser scanning microscopy was set up with the standard configuration. Immobilized cell samples were incubated with two fluorescent dyes, SYTO-13 and propidium iodide, during 15 min. Samples were washed twice in phosphate-buffered saline to eliminate the remaining dyes. The green emission from SYTO-13 was collected through 509- and 514-nm band-pass filters. Red emission from propidium iodide was collected at 610 nm.
Data analysis.
The parameters of the logistic-type curve fitted to the growth data were estimated using NLIN, the nonlinear procedure of the statistical package SAS, as previously reported (11). Kinetic parameters of propachlor metabolism were calculated from Hanes plots of nonsaturating propachlor concentrations (2).
Bioreactor experiments.
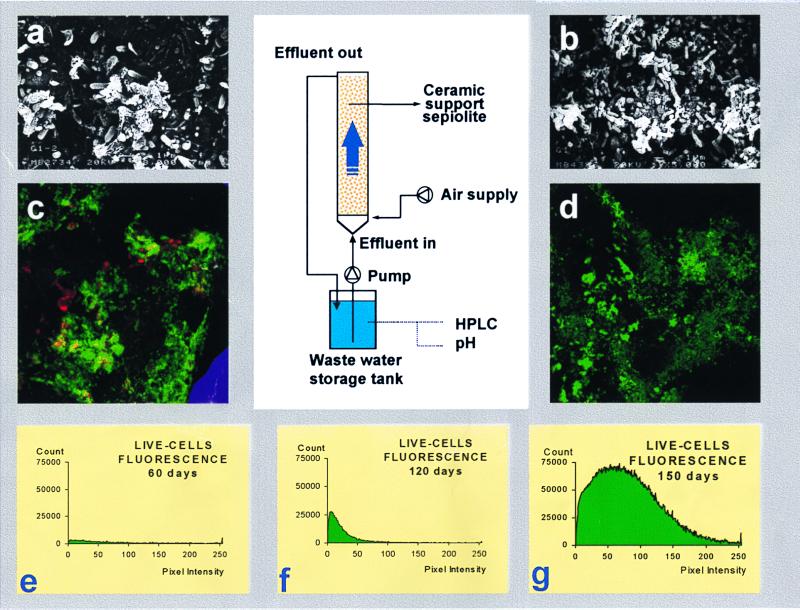
A schematic representation of the reactor setup is given in Fig. 1. The glass reactor had a working volume of 2.25 liters. The internal diameter of the column was 8 cm, the total height was 150 cm filled with 4.2 kg of ceramic support (sepiolite), and Pseudomonas strain GCH1 cells were used as the inoculum. The experiments were performed at room temperature (20 ± 2°C). Air was introduced by a fine bubble aerator in the bottom of the column. The reactor was operated with a cycle length of 15 days. A synthetic wastewater composed of 0.04 g of K2HPO4, 0.01 g of KH2PO4, 0.025 g of KNO3, 0.05 g of (NH4)2SO4, 0.025 g of CaSO4 · H2O, 0.025 g of MgSO4 · H2O, and 0.002 g of Fe(SO4) · 6H2O per liter (pH 7.2) was used. The final input concentration of propachlor was 105 mg/liter. Sampling was done periodically to monitor the fate of chlorinated herbicide and microorganisms.
FIG. 1.
Schematic of reactor for immobilized cell in ceramic support. (a and b) Scanning electron micrographs of immobilized GCH1 cells adsorbed onto the surface of the ceramic support, 48 h (a) and 360 h (b) after inoculation. (c and d) Laser scanning confocal micrographs showing viable cells after 60 days (c) and 120 days (d) of reactor operation. (e to g) Viability cell evolution during 150 days of reactor operation.
Chemicals.
Propachlor and [14C]propachlor were obtained from Monsanto España S.A. (Madrid, Spain). Acetamide and acetanilide were purchased from Aldrich (Milwaukee, Wis.). All chemical compounds were of the highest purity commercially available.
RESULTS
Identification of the isolated strain.
Enrichment cultures to obtain organisms that could utilize propachlor as the sole carbon source were prepared (10, 11). Samples were plated on Luria-Bertani (LB) medium, and resulting isolates were tested for the capability to grow on propachlor as the sole carbon source. By this technique, a pure culture, designated strain GCH1, which gave complete utilization of propachlor was isolated. The organism was a bacterium, on the basis of its morphological and biochemical properties. Strain GCH1 exhibited the following characteristics: oxidase positive, obligatorily aerobic, and 0.7 to 0.9 μm in diameter, and 1.4 to 2.1 μm in length. Electron microscopy of thin sections of cells showed a cell wall ultrastructure that is typical of gram-negative bacteria. The G+C content of the DNA was 63.2% ± 1.1%. The organism was not able to reduce nitrate to nitrite or to grow at 40°C; growth did not require the addition of vitamins to the growth medium. To further refine the taxonomic identification, purified DNAs from strain GCH1, Burkholderia cepacia ATCC 17759, and Pseudomonas anguilliseptica ATCC 33660 were tested with PCR performed at an annealing temperature of 55°C (20). The analyses revealed that P. anguilliseptica and GCH1 cultures tested positive by this PCR protocol. Thus, strain GCH1 could be considered a Pseudomonas sp.
Propachlor metabolism by Pseudomonas strain GCH1.
In batch cultures, strain GCH1 showed a growth yield of 0.96 mg of protein μmol−1, and mean generation time during growth on 0.5 mM propachlor at 30°C was 4.2 h during the early exponential phase. The specific rate of propachlor metabolism was estimated to be 10.42 ± 0.9 nmol consumed mg of protein−1 h−1.
Preincubation in the presence of glucose and 110 μg of chloramphenicol of ml−1 did not affect the ability to degrade propachlor. Glucose-grown cells metabolized 68.7% ± 2.6% of the initial propachlor concentration (0.5 mM); when these cells were previously incubated in the presence of chloramphenicol, 67.6% ± 2.4% of the initial propachlor was metabolized. Propachlor-grown cells metabolized 72.5% ± 2.3% and 76.4% ± 2.1%, respectively, in the same conditions.
Cells pregrown on propachlor or glucose were used to study the kinetics of propachlor metabolism. Glucose-grown cells produced 14CO2 from [ring-U-14C]propachlor with a Ks of 0.18 ± 0.01 mM, and a Ks of 0.12 ± 0.01 mM was obtained when propachlor-grown whole cells were tested. The observed maximal specific rate of 0.33 ± 0.02 nmol μg of protein−1 h−1 for substrate mineralization by propachlor-grown GCH1 cells was higher than that obtained with glucose-grown cells [(3.6 ± 0.2) × 10−2 nmol of protein−1 h−1]. 14CO2 was measured as a product from the carbon atoms in the propachlor ring; no significant 14CO2 was released in control experiments without cells, and no counts were measured in controls without radioactivity. Thus, propachlor could be completely degraded by Pseudomonas strain GCH1, and the propachlor metabolism was constitutive.
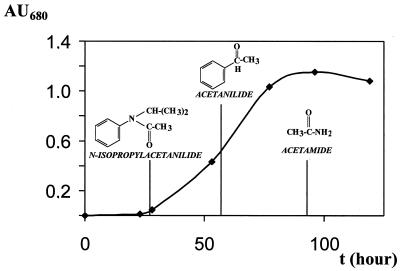
To characterize the metabolites formed during propachlor degradation, samples of the culture liquid were taken periodically. HPLC analyses of organic extracts from cultures revealed a number of products, and the most significant were identified by GC-MS. Propachlor disappeared during cell incubation, and simultaneously N-isopropylacetanilide appeared in the media as the first intermediate in the degradative pathway (Table 1). At the early exponential phase (Fig. 2), this compound was converted to acetanilide, which could be detected in the liquid medium during the exponential phase (25 to 60 h of incubation). When the culture had reached the stationary phase, acetamide was formed from the cleavage at the bond between the C atom of the aromatic ring and the N atom. Acetanilide (1 mM) and acetamide (10 mM) were tested as growth substrates. When strain GCH1 used acetanilide as sole carbon source and the growth yield obtained was 0.56 ± 0.02 mg of protein μmol−1, acetamide and catechol were detected in the supernatant by HPLC analysis. Acetamide also was found to be a growth substrate for strain GCH1 with a growth yield of 0.26 ± 0.03 mg of protein μmol−1. Thus, these results indicate that the propachlor catabolic pathway for strain GCH1 is the same that reported for Pseudomonas strain PEM1 (11).
TABLE 1.
Identification, by GC Rfs and MS properties, of intermediates detected in washed-cell incubations of Pseudomonas strain GCH1 with propachlor as the carbon source
| Compound | Rf | m/z of fragment ions (% relative intensity) |
|---|---|---|
| N-Isopropylacetanilide | 18.4 | 178(M+, 2), 162(M+ -CH3, 70), 120(M+ -COCH3-CH3, 100), 77(18) |
| Acetanilide | 9.7 | 136(M+, 2), 93(M+ -CO-CH3, 100), 66(15), 43(17) |
| Acetamide | 2.1 | Not detected |
FIG. 2.
Growth of Pseudomonas strain GCH1 in batch culture with propachlor as the carbon source. Shown are the metabolic intermediates identified by GC-MS analysis as described in Materials and Methods.
Bioreactor operation.
The immobilized cell system was studied in a pilot-scale reactor (Fig. 1). The experiments were carried out by continuous recirculation of 10 liters of the synthetic water containing 0.5 mM propachlor, and the reactor was operated with a cycle length of 15 days. Initially the granular ceramic support was placed in the reactor and the propachlor sorption was studied. Synthetic wastewater containing 0.5 mM propachlor was passed through the column with a flow of 50 ml min−1. During the initial startup period, there was 39% propachlor sorption by the ceramic support; after 34 h, the reactor reached the steady state.
To study the biological degradation of propachlor by immobilized GCH1 cells, the reactor was inoculated with 5 × 106 cells g of support−1. This inoculum rapidly produced a biomass (Fig. 1) which effectively removed propachlor. Acclimation to utilize all 105 mg liter−1 in a water synthetic feed occurred within 48 h. Examination of the culture under the electron microscope (Fig. 1a) revealed that strain GCH1 cells were quickly adsorbed onto the surface of the ceramic support. The natural tendency of these microorganisms to adhere to solid surfaces could be observed during colonization of the support (Fig. 1b). Examination of these micrographs showed no apparent morphological changes upon immobilization.
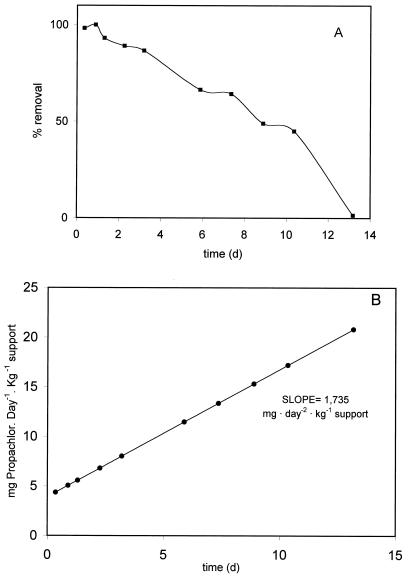
Figure 3 shows the results obtained from operation of the bioreactor treating the synthetic wastewater at a flow of 50 ml min−1 and hydraulic intensities of about 1 m h−1 during the 15 days of each experimental run. Samples were taken at different times (Fig. 3), and propachlor concentrations were measured by HPLC. About 50% of the initial propachlor was removed from the wastewater in 8 days (Fig. 3A), and the degradation was near 99% after 13 days. The retention time observed was 3 h in these experimental conditions. Propachlor degradation shows a first-order kinetic (Fig. 3B) with a specific rate of 1.73 mg of degraded propachlor kg of support−1 day−1. The pH 7.2 of the treated water was maintained during the cycle, and a very low degrading activity was detected when O2 was not supplied to the system. In samples of the effluent analyzed by HPLC, catechols were detected at a very high concentration, 16 ± 0.1 mg liter−1.
FIG. 3.
Results obtained from operation of the bioreactor treating synthetic wastewater. (A) Removal of propachlor from the wastewater expressed as percentage of initial propachlor concentration. (B) Kinetics of propachlor degradation. The results are the averages of a three-cycle operation run.
Immobilized cell samples from the operating reactor were analyzed by laser confocal microscopy; a combination of propidium iodide and SYTO 13 stain allowed monitoring cell viability after 60 days (Fig. 1c) and 120 days (Fig. 1d) of reactor operation. The viability of immobilized cells increased 2.75-fold after 120 days of operation and at the end of the process was 34-fold greater than at 60 days (Fig. 1e to g).
DISCUSSION
This report describes the isolation of a soil bacterium with the ability to degrade and mineralize propachlor. As reported above, Pseudomonas strain GCH1 is solely responsible for metabolism of the herbicide and can use it as a sole source of carbon and energy. The first step in the proposed degradative pathway (Fig. 2) involves dehalogenation; propachlor is attacked on the acetamide group with a chlorine as substituent (at C-2) to yield N-isopropylacetanilide. This dehalogenated intermediate is degraded by strain GCH1 with subsequent cleavage at the bond between the N atom and the C atom of the aromatic ring, the isopropyl side chain is removed, and acetanilide and acetamide are accumulated in the medium as products of the catabolism, which were identified by GC and HPLC analyses, respectively (Table 1). Recently, we reported (11) that Pseudomonas sp. strain PEM1 induced its own catabolism of propachlor following the same degradative pathway. Both Pseudomonas strain PEM1 and strain GCH1 mineralize propachlor, and CO2 was a product from the carbon atoms in the aromatic ring. This organism resembles Pseudomonas strain GCH1 in many respects but differs from it in that, in this isolate, the metabolism of propachlor is constitutive. The Ks of 0.18 mM found for the metabolism of propachlor by glucose-grown cells is similar to that found with propachlor-grown cells (0.12 mM); however, the maximal specific rate observed with propachlor-grown cells is higher than that obtained with glucose-grown cells.
The characterization of Pseudomonas strain GCH1 has several implications for the fate of acylanilide herbicides in the environment. When cells are immobilized onto a granular ceramic support, they maintain their viability (Fig. 1) and greatly increase the capacity to degrade propachlor. Morphological and physiological changes by the immobilization process (8, 21) have been reported to be a problem when this system is applied to biotechnological processes. Scanning electron microscopy and laser confocal microscopy analyses (Fig. 1) show that immobilized Pseudomonas strain GCH1 cells working in the reactor increased their viability 34-fold after 150 days, and examination of the micrographs shows no apparent morphological changes upon immobilization. The high level of biomass formation and the increase of viability are related to the increased rate of degradation observed in the immobilized cell system (Fig. 3B). Its implementation for the treatment of large volumes of wastewater to eliminate dilute organic chlorides provides a considerable advantage due to the use of native bacteria which biotransform the organic compounds in degrading biomass.
Immobilized cell systems have been applied for many biochemical processes (reviewed in references 3, 5, 8,;1 and and 12). Immobilization by adsorption, the gentlest fixation method, is mainly suitable for viable cells, compared to the immobilization of cells by entrapment into organic polymer (3). Previous studies demonstrated that the tolerance of various bacterial species to different organic compounds (3, 5, 12, 21) could be elevated by cell immobilization. Heipieper et al. (8) reported that phenol at sublethal concentrations is less inhibitory to immobilized microorganisms than to free microorganisms. We previously reported similar results (5, 10, 12), finding that strain DSZ1 degrades simazine or strain PEM1 degrades propachlor in an immobilized cell system or suspended cell system.
The results presented here indicate that biodegradation systems, using immobilized cells in the selected conditions, can be applied to removal of organic chemicals from water or soil. The high viability showed by strain GCH1 in the immobilized cell system is not surprising considering that if these bacteria are isolated from contaminated soils, then the ceramic material used as the support in the reactor improves the process implementation.
ACKNOWLEDGMENTS
This work was supported by grant AMB98-0501 from Comisión Interministerial de Ciencia y Tecnología and by grant CAM07M/0620/1997 from Comunidad Autónoma de Madrid.
We thank Jesús Sanz for the GC-MS analysis. We express our appreciation to Jaime Costa (Monsanto España S.A.) for providing propachlor and [14C]propachlor.
REFERENCES
- 1.Allende J L, Gibello A, Martín M, Garrido-Pertierra A. Transport of 4-hydroxyphenylacetic acid in Klebsiella pneumoniae. Arch Biochem Biophys. 1992;292:583–588. doi: 10.1016/0003-9861(92)90034-t. [DOI] [PubMed] [Google Scholar]
- 2.Cornish-Bowden A. Fundamentals of enzyme kinetics. London, England: Portland Press Ltd.; 1995. [Google Scholar]
- 3.Ferrer E, Blanco J, Alonso R, Martín M. Propachlor and alachlor degradation by immobilized and suspended Pseudomonas cells. In: Wijffels R M, et al., editors. Immobilized cells: basics and applications. Oxford, England: Elsevier Science; 1996. pp. 762–769. [Google Scholar]
- 4.Gerhardt P, Murray R G, Costilow R N, Nester E W, Wood W A, Krieg N R, Phillips G B, editors. Manual of methods for general and molecular bacteriology. Washington, D.C.: American Society for Microbiology; 1995. [Google Scholar]
- 5.Gibello A, Allende J L, Mengs G, Alonso R, Ferrer E, Martín M. Comparison of phenolic substrate utilization and growth kinetics between immobilized and suspended degradative-bacteria. Biocatal Biotransform. 1998;16:291–306. [Google Scholar]
- 6.Greer L E, Robinson J A, Shelton D R. Kinetic comparison of seven strains of 2,4-dichlorophenoxyacetic acid-degrading bacteria. Appl Environ Microbiol. 1992;58:1027–1030. doi: 10.1128/aem.58.3.1027-1030.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Greer L E, Shelton D R. Effect of inoculant strain and organic matter content on kinetics of 2,4-dichlorophenoxy-acetic acid degradation in soil. Appl Environ Microbiol. 1992;58:1459–1465. doi: 10.1128/aem.58.5.1459-1465.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heipieper H J, Keweloh H, Rehm H J. Influence of phenols on growth and membrane permeability of free and immobilized Escherichia coli. Appl Environ Microbiol. 1991;57:1213–1217. doi: 10.1128/aem.57.4.1213-1217.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee J K, Minard R D, Bollag J M. Microbial metabolism of propachlor (2-chloro-N-isopropylacetanilide) in soil suspension. Han'guk Nonghwahakhoe Chi. 1982;25:44–50. [Google Scholar]
- 10.Martin M, Fernández J, Ferrer E, Alonso R. Bioremediation of soil contaminated by Propachlor using native bacteria. Int Bioremed Biodeterior. 1995;35:213–225. [Google Scholar]
- 11.Martin M, Mengs G, Allende J L, Fernández J, Alonso R, Ferrer E. Characterization of two novel degradation pathways in two species of soil bacteria. Appl Environ Microbiol. 1999;65:802–806. doi: 10.1128/aem.65.2.802-806.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martín-Montalvo D, Mengs G, Ferrer E, Allende J L, Fernández J, Alonso R, Martin M. Simazine degradation by immobilized and suspended soil bacterium. Int Biodeterior Biodegrad. 1999;40:93–99. [Google Scholar]
- 13.Miller J M. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 14.Morgan P, Watkinson R. Microbiological methods for the clean-up of soils and ground water contaminated with halogenated organic compounds. FEMS Microbiol Res. 1989;63:277–300. doi: 10.1111/j.1574-6968.1989.tb03401.x. [DOI] [PubMed] [Google Scholar]
- 15.Novick N J, Alexander M. Cometabolism of low concentrations of propachlor, alachlor, and cycloate in sewage and lake water. Appl Environ Microbiol. 1985;49:737–743. doi: 10.1128/aem.49.4.737-743.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Novick N J, Mukherjee R, Alexander M. Metabolism of alachlor and propachlor in suspensions of pretreated soils and in samples from ground water aquifers. J Agric Food Chem. 1986;34:721–725. [Google Scholar]
- 17.Ulitzur S. Rapid determination of DNA base composition by ultraviolet spectroscopy. Biochim Biophys Acta. 1972;272:1–11. doi: 10.1016/0005-2787(72)90025-1. [DOI] [PubMed] [Google Scholar]
- 18.Villareal D T, Turco R F, Konopka A. Propachlor degradation by a soil bacterial community. Appl Environ Microbiol. 1991;57:2135–2140. doi: 10.1128/aem.57.8.2135-2140.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilber G G, Wang G. Biotransformation of herbicides in the presence of various electron acceptors. J Air Waste Manag Assoc. 1997;47:690–696. [Google Scholar]
- 20.Widmer F, Seidler R J, Gillevet P T, Watrud L S, Di Giovanni G D. A highly selective PCR protocol for detecting 16S rRNA genes of the genus Pseudomonas (sensu stricto) in environmental samples. Appl Environ Microbiol. 1998;64:2545–2553. doi: 10.1128/aem.64.7.2545-2553.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Willaert R G, Baron G V, De Backer L. Immobilised living cell systems modelling and experimental methods. J. Chichester, England: Wiley & Sons; 1996. [Google Scholar]