Abstract
PCR primers were patterned after chitinase genes in four γ-proteobacteria in the families Alteromonadaceae and Enterobacteriaceae (group I chitinases) and used to explore the occurrence and diversity of these chitinase genes in cultured and uncultured marine bacteria. The PCR results from 104 bacterial strains indicated that this type of chitinase gene occurs in two major groups of marine bacteria, α- and γ-proteobacteria, but not the Cytophaga-Flavobacter group. Group I chitinase genes also occur in some viruses infecting arthropods. Phylogenetic analysis indicated that similar group I chitinase genes occur in taxonomically related bacteria. However, the overall phylogeny of chitinase genes did not correspond to the phylogeny of 16S rRNA genes, possibly due to lateral transfer of chitinase genes between groups of bacteria, but other mechanisms, such as gene duplication, cannot be ruled out. Clone libraries of chitinase gene fragments amplified from coastal Pacific Ocean and estuarine Delaware Bay bacterioplankton revealed similarities and differences between cultured and uncultured bacteria. We had hypothesized that cultured and uncultured chitin-degrading bacteria would be very different, but in fact, clones having nucleotide sequences identical to those of chitinase genes of cultured α-proteobacteria dominated both libraries. The other clones were similar but not identical to genes in cultured γ-proteobacteria, including vibrios and alteromonads. Our results suggest that a closer examination of chitin degradation by α-proteobacteria will lead to a better understanding of chitin degradation in the ocean.
Surveys of microbial diversity without cultivation have discovered types of microbes not detected in culture-based studies largely because <1% of the microorganisms observable in nature can be cultivated using standard techniques (3). Furthermore, the few bacteria that can be cultured appear to be very different from uncultured bacteria, based on comparisons of 16S rRNA gene sequences. Consequently, the lack of closely related cultured representatives raises questions about the metabolic capacities of the uncultured bacteria and the role of these microbes in specific biogeochemical processes. Some physiological capacities are restricted to specific taxonomic groups of microbes, for example, oxygenic photoautotrophy in cyanobacteria, but in general, the relationship between the taxonomy of uncultured bacteria and many biogeochemically interesting capacities is not known. One approach for exploring the metabolic capacities of uncultured bacteria is to examine genes encoding enzymes involved in specific biogeochemical processes. This approach is a step toward identifying microbial groups driving those processes and determining whether the metabolism of cultured bacteria adequately represents the metabolic capacities of uncultured bacteria.
Previous studies have already examined several genes encoding enzymes mediating biogeochemical reactions in C, N, and S cycles and have compared these genes in cultured and uncultured bacteria in natural microbial communities. These enzymes, genes, and bacteria include the nitrogen-fixing enzyme (nifH) (42) in nitrogen-fixing bacteria, nitrite reductase (nirK and nirS) and nitrous oxide reductase (nosZ) in denitrifying bacteria (5, 18), particulate methane monooxygenase (pmoA) and methanol dehydrogenase (mxaF) found in bacteria using C1 and methylated compounds (30, 31), and dissimilatory bisulfite reductase (dsv) in sulfate-reducing bacteria (7). In most cases, the genes of cultured and uncultured bacteria were not identical, suggesting that cultured bacteria do not adequately model biogeochemical processes driven by uncultured bacteria.
The genes that have been examined to date in uncultured bacteria are essential to the microbes' survival, even if only under selected conditions (e.g., denitrification genes are essential only under anoxic conditions). In contrast, little is known about nonessential genes in uncultured bacteria. Variation and correlation of nonessential genes with rRNA gene phylogeny might differ from those of essential genes. Differences could arise from various mechanisms, including different rates of evolution, gene duplication, and lateral gene transfer (9). There seems to be less resistance to lateral transfer of nonessential genes than essential genes (9).
Genes encoding chitinases and other glycosyl hydrolases may be particularly interesting examples of nonessential genes in uncultured bacteria, since previous work has already suggested that the evolution of these enzymes has been impacted by lateral gene transfer (10). Chitinases are probably not essential for heterotrophic bacteria living in most environments, including the oceans, where chitin is very abundant (25), because many other organic carbon sources are available. Although perhaps nonessential to an individual bacterium, hydrolysis of chitin and other high-molecular-weight biopolymers by hydrolases is still an essential first step in the degradation of organic material in nature. Many types of cultured bacteria and archaea are known to hydrolyze chitin (17, 21), but the identity of uncultured bacteria degrading chitin in nature is unknown. Chitinase genes cloned directly from uncultured marine microorganisms suggested the presence of a large pool of uncultured chitin-degrading bacteria in aquatic systems (8).
Information on bacterial chitinase genes is largely restricted to cultured γ-proteobacteria and gram-positive bacteria. Since γ-proteobacteria are widespread in the ocean (11), comparing chitinase genes in cultured and uncultured bacteria in this phylogenetic group should be informative. In contrast, gram-positive bacteria are quite rare in seawater (11). To access chitinase genes in uncultured γ-proteobacteria and potentially in other bacteria, it may be possible to use a PCR-based approach with oligonucleotide primers patterned after conserved nucleotide sequences of chitinase genes in cultured bacteria. However, it is not possible to design a single pair of PCR primers that will amplify even all γ-proteobacterial chitinases because they are too different. Svitil and Kirchman (38) did identify 13 and 15 consensus amino acids in the catalytic and chitin-binding domains of bacterial chitinases, respectively. But since PCR primers require adjacent identical nucleotides or amino acids, the conserved regions in bacterial chitinases are insufficient for designing PCR primers that would amplify all bacterial chitinases. An alternative approach would be to target selected subsets of bacterial chitinases. One such subset is group I, which has the highest average percent similarity of the five chitinase groups classified by Svitil and Kirchman (38).
Our goals were to compare the group I chitinase genes of cultured and uncultured bacteria and to examine the relationship between the phylogeny of these chitinase genes and the phylogeny of 16S rRNA genes. We hypothesized that PCR primers for group I chitinase genes would amplify chitinase genes of other cultured and uncultured γ-proteobacteria. It was unclear if more distantly related bacteria possess this type of chitinase as well. We also expected that chitinase genes of cultured and uncultured chitin-degrading bacteria would differ and that chitinase gene phylogeny would not follow the phylogeny of 16S rRNA genes. In fact, we found that a few chitinases from uncultured bacteria were very similar, and in some cases identical, to those in cultured bacteria, but overall, the phylogeny of chitinase genes differed from 16S rRNA phylogeny.
MATERIALS AND METHODS
Isolation and characterization of bacterial strains.
Surface seawater was collected from the Indian River Inlet on the Atlantic coast of Delaware and the University of Delaware dock at the Roosevelt Inlet, situated 8 km inside the Delaware Bay estuary. Bacterial strains were isolated on 1.5% agar prepared using unenriched seawater and seawater enriched with R2A nutrients, which include 0.5 g each of yeast extract, peptone, Casamino Acids, and dextrose per liter and 0.3 g of sodium pyruvate per liter (37). Plates were incubated at 20 to 25°C and inspected daily for growth. Strains were purified by two iterations of streaking on agar. Strains were analyzed by using a mixture of the restriction enzymes RsaI and HhaI (32) to digest 16S rRNA genes amplified using primers EubA and EubB (12). Strains having different restriction patterns were used in subsequent analyses. Bacterial strains were classified phylogenetically using fluorescent in situ hybridization of fluorochrome-labeled rRNA probes specific for α-proteobacteria, Alf1b (29), β-proteobacteria, Bet42a (29), γ-proteobacteria, Gam42a (29), the Cytophaga-Flavobacter cluster (28), and gram-positive bacteria having high DNA G+C content (35). Established hybridization conditions were used for each probe (41). One strain producing ambiguous results using fluorescent in situ hybridization was characterized by sequencing of the 16S rRNA gene.
Strains were screened for the ability to produce clearing zones on unenriched seawater agar and R2A-enriched seawater agar containing colloidal chitin. Cultures were also assayed for hydrolysis of the fluorogenic chitin analogue 4-methylumbelliferyl-β-d-N,N′-diacetylchitobioside. For this analysis, cultures were grown on R2A-enriched seawater broth and artificial seawater enriched with 50 mg of chitin oligomers (Vector Labs) per liter 10 mM NH3Cl, and 2 mM NaH2PO4.
Isolation of environmental DNA.
Coastal Pacific Ocean water was collected from a depth of 1 m at Station 5, located 60 km off the coast of Oregon (salinity equal to 28 ppt) in July 1997. Delaware Bay estuarine water was collected at a depth of 0.5 m from Station 16, located 80 km upstream from the mouth of the estuary (20) (salinity equal to 2.0 ppt) in September 1997. The coastal Pacific sample (10 liter) was filtered through a 1-μm-pore-size filter, and bacteria were collected on 0.2-μm Gelman Supor filters. The Delaware Bay sample (10 liters) was filtered through a 1-μm filter, and bacteria were collected using a Millipore Sterivex-GV filtration cartridge (0.22 μm). The samples were stored frozen at −80°C in a storage buffer (13). Frozen samples were thawed, and the cells were lysed using sodium dodecyl sulfate and proteinase K. The lysate was extracted sequentially with phenol-chloroform and chloroform. The nucleic acids were precipitated with ethanol and further purified using cetyltrimethylammonium bromide extraction.
PCR primer design.
We identified conserved amino acids in the chitinases encoded by the chiA genes of Alteromonas sp. (40), Aeromonas caviae (36), Serratia marcescens (22), and Enterobacter agglomerans (19). The conserved regions were in the putative catalytic and chitin-binding domains and a region with no known function (38). The forward primer was based on conserved amino acids in the catalytic region at amino acid 260 of ChiA in Alteromonas sp. strain 0-7. The reverse primer bound to a conserved region with no known function at amino acid 571 of chitinase A in the Alteromonas strain. Deoxyinosine residues in the third position of codons were used to accommodate every codon for all amino acids in the targeted regions without increasing degeneracy of the primers. This group of chitinase genes is called group I (38).
Clone library construction and screening.
Group I chitinase genes were amplified from DNA of coastal Pacific Ocean and Delaware Bay bacterioplankton using 25-μl PCR mixtures containing 4 ng of template DNA per μl, the four deoxynucleoside triphosphates (dTTP, dCTP, dGTP, and dATP) at 0.2 mM each, 1.5 mM MgCl2, 1 μM each primer, and 2.5 U of Taq DNA polymerase (Promega). Thermal cycling conditions included 1 min of denaturation at 94°C, 1 min of primer annealing at 50°C, and 3 min of primer extension at 72°C. This cycle was repeated 35 times. PCR products were cloned by using the TOPO-TA cloning kit with the pCR 2.1 vector (Invitrogen) following the manufacturer's protocol. Approximately 60 recombinant clones were screened for full-size inserts (approximately 900 bp) by transferring small aliquots of cells to PCR mixtures containing the group I chitinase gene primers and PCR amplified using the conditions described above. The PCR products were cut with a mixture of restriction enzymes HhaI and RsaI. The restriction fragments were separated by agarose gel electrophoresis using 2% Metaphore (FMC) agarose. Clones having identical restriction patterns were grouped together into clone families.
Nucleotide sequencing.
Nucleotide sequencing was performed using an ABI PRISM 310 (Perkin-Elmer) genetic analyzer and ABI PRISM Big Dye terminator cycle sequencing reagent. Double-stranded DNA templates were prepared using the manufacturer's alkaline-lysis procedure. M13 forward and reverse sequencing primers and internal sequencing primers were used to obtain the complete nucleotide sequences of both DNA strands of one clone from each clone family. Deduced amino acid sequences were analyzed by using the BLASTX (2) tool.
Phylogenetic analysis.
Similarity between chitinases (percent identical aligned amino acids) was determined from conceptual translations of open reading frames. Percentages of identical aligned nucleotides were compared for sequences having identical deduced amino acid sequences. Nucleotides were aligned using the corresponding amino acid alignment made using CLUSTAL in Sequence Navigator version 1.01 (Perkin-Elmer). Phylogenetic analysis was performed using SEQBOOT, DNADIST (Kimura 2-parameter model), NEIGHBOR, and CONSENSUS in PHYLIP version 3.527.
Nucleotide sequence accession numbers.
The nucleotide sequences described in this paper have been deposited in GenBank under accession no. AF193488 to AF193506.
RESULTS
Primer design.
Amino acid sequences of bacterial chitinase genes are very different, except for the catalytic and chitin-binding domains, but variation in even these two domains is too great for a single PCR primer set to match all bacterial chitinase genes. An alternative approach is to design PCR primers for sets of similar bacterial chitinases. The primers designed for this study were based on deduced amino acid sequences of chitinases in four γ-proteobacteria. This set of chitinases has been designated group I (38).
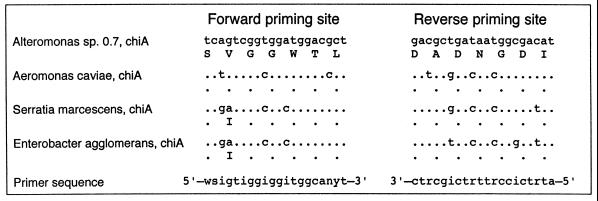
The forward PCR primer (IICRFORB) was patterned after conserved amino acids in the catalytic domain of group I chitinase genes, including the chiA genes in Alteromonas sp., A. caviae, S. marcescens, and E. agglomerans (Fig. 1). The chitin-binding domains were too variable for a PCR primer, but alignment of the complete deduced amino acid sequences of these four group I chitinase genes revealed 12 adjacent, conserved amino acids about 300 amino acids downstream from the forward priming site. A reverse primer (GRPI571AR) was patterned after seven of the conserved amino acids in this region (Fig. 1). The forward and reverse primers are degenerate 20-mer oligonucleotides incorporating every codon for the conserved amino acids. The oligonucleotides IICRFORB and GRPI571AR are referred to here as the group I primers.
FIG. 1.
Aligned nucleotide and deduced amino acid sequences used to design forward and reverse group I PCR primers IICRFORB and GRPI571AR, respectively. Dots indicate the same nucleotides or amino acids as in the Alteromanas sp. strain 0-7 sequence. The forward priming site is located in the hydrolytic domain, while the reverse priming site is approximately 900 bp downstream. The nucleotide sequences of the chiA genes of Alteromonas sp. (39), A. caviae (35), S. marcescens (22), and E. agglomerans (19) were obtained from GenBank.
Specificity of group I primers.
The group I primers yielded products having the predicted size (900 bp) from genomic DNA of 18 γ-proteobacteria out of the total of 38 strains tested, including the strains used to design the primers (Table 1). BLASTX analysis indicated that the deduced amino acid sequences of amplification products from γ-proteobacteria were >69% similar to known chitinase genes. Four of the 24 α-proteobacteria tested yielded 900-bp products having deduced amino acid sequences similar to those of chitinase genes (Table 1). All of the γ-proteobacteria amplifying with the group I primers were chitinolytic, but none of the four positive α-proteobacteria proved to be chitinolytic under the conditions tested. No amplification was obtained from the four chitinolytic α-proteobacteria.
TABLE 1.
Bacterial strains tested for amplification with PCR primers for group I chitinase genesa
| Classification | No. of strains tested
|
No. of strains amplifying
|
||
|---|---|---|---|---|
| Total | Chitinolytic | Chitinolytic | Nonchitinolytic | |
| α-Proteobacteria | 24 | 4 | 0 | 4 |
| β-Proteobacteria | 6 | 4 | 0 | 0 |
| γ-Proteobacteria | 38 | 28 | 18 | 0 |
| Cytophaga-Flavobacter | 30 | 17 | 0 | 0 |
| Gram positive | 6 | 4 | 0 | 0 |
Strains were scored as amplifying if they yielded 900-bp products having deduced amino acid sequences similar to those of known chitinases.
None of the 6 β-proteobacteria, 30 Cytophaga-Flavobacter bacteria, or 6 gram-positive bacteria we tested yielded 900-bp amplification products (Table 1), even though more than half of the strains in each group were chitinolytic. Less than 10% of all strains yielded nonspecific amplification products, i.e., a product smaller or larger than 900 bp. These nonspecific products had deduced amino acid sequences substantially different from those of chitinases.
Construction and composition of clone libraries.
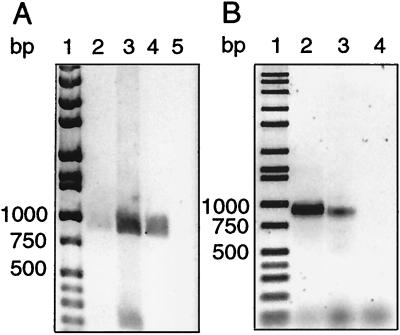
The occurrence of group I chitinase genes in naturally occurring bacteria was investigated using DNA extracted from bacterioplankton in the coastal Pacific Ocean and Delaware Bay. Bacterial community DNA from coastal and estuarine environments yielded the expected 900-bp products (Fig. 2). The Pacific sample required concentrating before cloning (Fig. 2A), while ample product was generated from the Delaware Bay sample without a concentrating step (Fig. 2B).
FIG. 2.
Ethidium bromide-stained agarose gel of PCR amplification products obtained with the group I primers. (A) Amplification of chitinase genes from microbial DNA collected from the coastal Pacific Ocean. Lanes: 1, molecular size markers; 2, PCR with bacterial community DNA; 3, PCR with bacterial community DNA concentrated by ultrafiltration; 4, PCR with Alteromonas sp. strain 0-7 DNA; 5, no-template control. (B) Amplification of chitinase genes from microbial DNA collected from the Delaware Bay estuary. Lanes: 1, molecular size markers; 2, PCR with bacterial community DNA; 3, PCR with Alteromonas sp. strain 0-7 DNA; 5, no-template control.
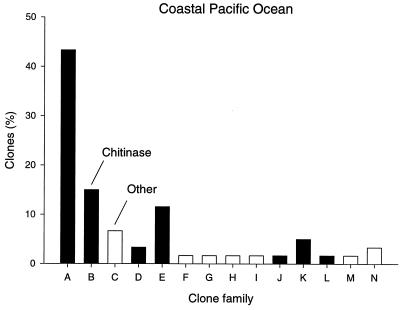
Sixty clones from the Pacific library treated with a mixture of restriction enzymes HhaI and RsaI yielded 14 different banding patterns. Clones sharing the same restriction pattern were assembled into clone families (Table 2). Twenty-six clones representing 43% of the Pacific library were assigned to clone family A (Table 2 and Fig. 3). Clone family B, comprised of nine clones, represented 15% of the library, while clone families C to N each contained seven or fewer clones. One clone from each clone family was completely sequenced and analyzed using BLASTX to determine its similarity to known chitinases. Seven of the 14 clone families in the Pacific library encoded proteins that were greater than 69% similar to known chitinases (Table 2 and Fig. 3). The seven chitinase clone families contained 82% of the clones in the library. Clone families not representing chitinase genes comprised half of the clone families but only 18% of the clones in the library.
TABLE 2.
Characteristics of cloned group I chitinase gene fragments amplified from microbial DNA from the coastal Pacific Ocean and Delaware Baya
| Clone family | No. of clones
|
Sequenced clone
|
% Amino acid similarity to chitinaseb | Deduced gene product | ||
|---|---|---|---|---|---|---|
| Pacific Ocean | Delaware Bay | Pacific Ocean | Delaware Bay | |||
| A | 26 | 30 | 5-8 | 16-2 | 70 | Chitinase |
| B | 9 | 17 | 5-27 | 16-1 | 69 | Chitinase |
| C | 4 | 2 | 5-28 | 16-10 | <1 | Other |
| D | 2 | 0 | 5-5 | NAc | 80 | Chitinase |
| E | 7 | 0 | 5-63 | NA | 86 | Chitinase |
| F | 1 | 0 | 5-2 | NA | <1 | Other |
| G | 1 | 0 | 5-11 | NA | <1 | Other |
| H | 1 | 0 | 5-14 | NA | <1 | Other |
| I | 1 | 0 | 5-19 | NA | <1 | Other |
| J | 1 | 0 | 5-26 | NA | 76 | Chitinase |
| K | 3 | 0 | 5-37 | NA | 78 | Chitinase |
| L | 1 | 0 | 5-40 | NA | 80 | Chitinase |
| M | 1 | 0 | 5-43 | NA | <1 | Other |
| N | 2 | 0 | 5-67 | NA | <1 | Other |
| O | 0 | 2 | NA | 16-12 | <1 | Other |
| P | 0 | 2 | NA | 16-15 | 78 | Chitinase |
| Q | 0 | 1 | NA | 16-27 | <1 | Other |
| R | 0 | 1 | NA | 16-85 | <1 | Other |
| S | 0 | 1 | NA | 16-23 | 78 | Chitinase |
| T | 0 | 1 | NA | 16-24 | 69 | Chitinase |
Sixty Pacific Ocean clones and 57 Delaware Bay clones were organized into families having identical restriction patterns produced by a mixture of HhaI and RsaI.
The deduced amino acid sequence was analyzed using BLASTX.
NA, not applicable.
FIG. 3.
Frequency distribution of clone families in the coastal Pacific Ocean library. The percentage of clones in each family was calculated relative to the total number of clones in the library.
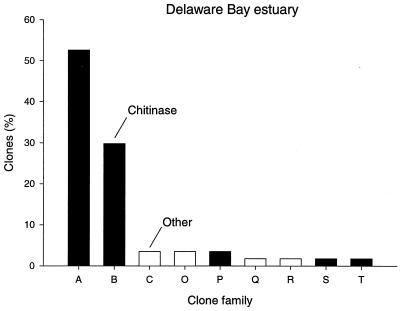
Digestion of 57 clones in the Delaware Bay library with a mixture of restriction enzymes HhaI and RsaI produced nine different restriction patterns. Clones having identical restriction patterns were segregated into nine clone families (Table 2), including three (A, B, and C) that occurred in the Pacific library as well. Clone families A and B, which dominated the Pacific library, also comprised the bulk of the Delaware Bay library (Table 2 and Fig. 4). Thirty clones representing 53% and 17 clones representing 30% of the Delaware Bay library were assigned to clone families A and B, respectively. Two clones were assigned to clone family C, which occurred in both libraries. Six clone families (O, P, Q, R, S, and T) represented by two or fewer clones were present only in the Delaware Bay library.
FIG. 4.
Frequency distribution of clone families in the Delaware Bay estuary library. The percentage of clones in each family was calculated relative to the total number of clones in the library.
Sequencing of one clone in each clone family of the Delaware Bay library revealed that 89% of the clones in the Delaware Bay library encoded proteins that were >69% similar to known chitinases (Table 2). Five of the nine clone families (A, B, P, S, and T) represented chitinase genes, whereas four clone families (C, O, Q, and R) did not encode chitinases.
Similarities among chitinase genes.
Chitinase genes in taxonomically related, cultured bacteria were similar, but chitinase phylogeny did not correlate completely with 16S rRNA phylogeny (Fig. 5). Chitinase genes in cultured Vibrio species were greater than 77% similar to each other, and phylogenetic analysis placed them in a clade separate from chitinases of other cultured γ-proteobacteria (Fig. 5). Chitinases in members of the families Alteromonadaceae and Enterobacteriaceae were less than 77 and 63% similar to Vibrio chitinases, respectively. Similarity was greater than 72% within clades of chitinases in Alteromonadaceae, Enterobacteriaceae, and α-proteobacteria. These assignments are supported by bootstrap values of 57 to 100 (Fig. 5).
FIG. 5.
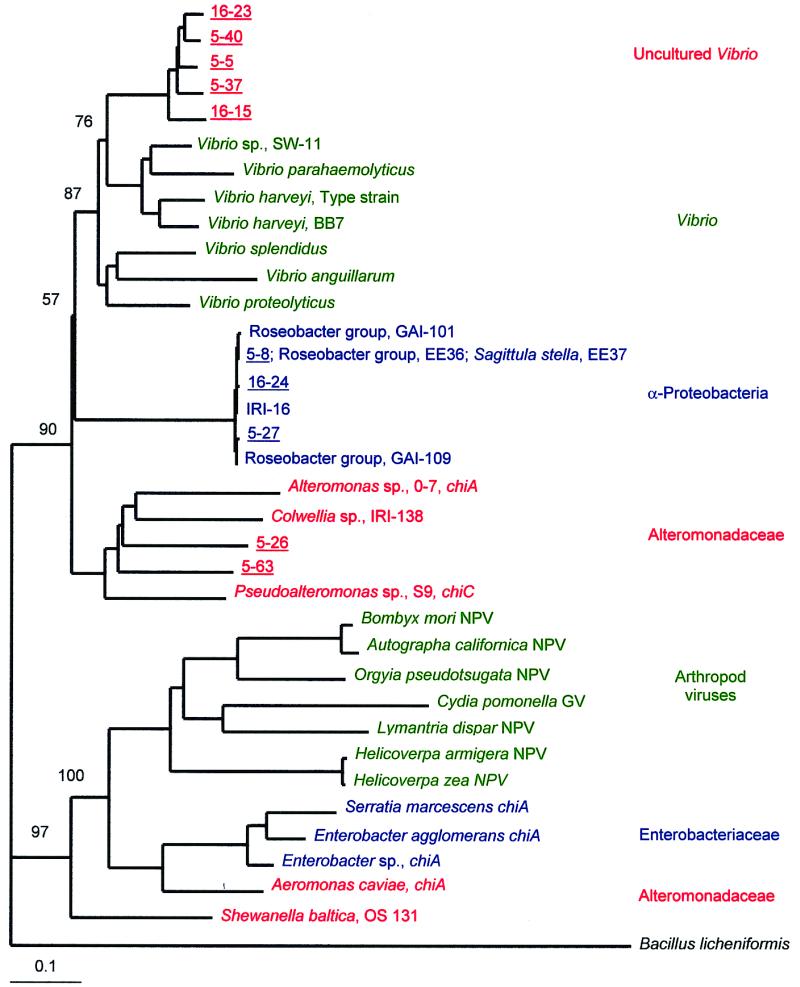
Additive phylogenetic tree of group I chitinase genes from cultured and uncultured bacteria and viruses of arthropods. The neighbor-joining analysis used a chitinase gene from B. licheniformis (GenBank accession no. U71214) as the outgroup. α-Proteobacteria (15, 16) and γ-proteobacteria were isolated from coastal and estuarine environments. Shewanella baltica was isolated from the Baltic Sea (42). The nucleotide sequences of the chitinase genes of reference strains of Pseudoalteromonas sp. (38), Enterobacter sp. (33), B. mori NPV (23), Autographa californica NPV (4), Orgyia pseudotsugata NPV (1), Helicoverpa zea NPV (26), Helicoverpa armigera NPV (GenBank accession no. AF114795), Lymantria dispar NPV (25), and Cydia pomonella granulovirus (GV) (24) were obtained from GenBank. Bootstrap values are indicated at the nodes separating the major groups. Genes from uncultured bacteria are underlined and are designated by a station and clone number (e.g., 5-40). The scale bar indicates the amount of genetic change measured as the number of nucleotide substitutions per site.
Group I chitinases occur in viruses infecting arthropods. The viral group I chitinases were more diverse than the bacterial groups revealed by our analysis and were 62 to 96% similar to each other. Viral chitinases were only 45 to 64% similar to group I chitinases from bacteria. However, these viral chitinases are more similar to bacterial group I chitinases than other bacterial chitinases are to bacterial group I chitinases. For example, bacterial group I chitinases are only 32% similar, on average, to the chitinase of Bacillus licheniformis (GenBank accession no. U71214).
Similarity between chitinases of uncultured and cultured bacteria ranged from 52 to 100% at the amino acid level (Fig. 5). At one extreme, clone 5-8 was identical at both the amino acid and nucleotide levels to the chitinases of α-proteobacteria including strain EE36 (14) and Sagittula stellata strain E37 (15) in the Roseobacter group. Clone 5-27 was also very (>97%) similar to the chitinase genes of α-proteobacteria. In contrast, the chitinases of uncultured bacteria represented by clones 5-5, 5-37, 5-40, 16-15, and 16-23 were most (84 to 89% at the amino acid level) similar to the chitinases of cultured Vibrio species but none were identical to that of a cultured strain. The node separating this clade of chitinase genes in uncultured bacteria from the clade of chitinases in cultured Vibrio species had a bootstrap value of 76 (Fig. 5). Finally, the chitinases of uncultured bacteria represented by clones 5-63 and 5-26 were most similar (76 to 83%) to chitinases of cultured bacteria including Colwellia sp., Alteromonas sp., and Pseudoalteromonas sp.
Group I chitinase genes in uncultured bacteria from the coastal Pacific and Delaware Bay were least similar to the chitinase genes of cultured enterobacteria (<64% similar) and viruses infecting arthropods (<58% similar). No genes of uncultured bacteria were in clades comprised of the family Enterobacteriaceae or viruses infecting arthropods (Fig. 5).
DISCUSSION
Although many types of cultured bacteria are known to degrade chitin and chitin degradation is widespread in nature, the relationship between uncultivated chitin-degrading bacteria and cultured strains is unknown. Since phylogenetic analysis of 16S rRNA genes indicates that cultured and uncultured bacteria differ greatly (37), we hypothesized that uncultivated chitin-degrading bacteria are probably different as well. We used a pair of PCR primers patterned after chitinase genes initially identified in γ-proteobacteria to explore the diversity of chitinase genes in uncultured bacteria from two contrasting marine environments. It was necessary to begin with primers based on chitinase genes in γ-proteobacteria because this is the only phylogenetic group of bacteria, aside from gram-positive bacteria, which are quite rare in the ocean (11), for which chitinase gene sequences are available. As predicted, most of the chitinase genes amplified from natural bacterial assemblages using the group I primers were different from the chitinase genes of cultured strains. However, some chitinase genes from uncultured bacteria were very similar and even identical to group I chitinase genes of α-proteobacteria. Phylogenetic analysis of group I chitinases of cultured strains revealed clusters of similar genes in taxonomically related bacteria. Still, the overall phylogeny of chitinase genes did not coincide with 16S rRNA phylogeny. Most prominently in the neighbor-joining analysis (Fig. 5), as well as in parsimony and maximum-likelihood analyses, the cluster of genes from α-proteobacteria formed a clade within the larger clade of genes from γ-proteobacteria, including alteromonads and vibrios.
Testing of the group I primers on various cultured strains revealed that group I chitinase genes occur, as expected, in many types of γ-proteobacteria, including Enterobacteriaceae, Aeromonas, Vibrio species, and various Alteromonadaceae, including strains of Shewanella and Pseudoalteromonas (Table 1). Unexpectedly, amplification products were obtained from strains in the Roseobacter group (Table 1), indicating that culturable marine α-proteobacteria possess group I chitinase genes. Of the 24 cultured α-proteobacteria strains we examined, only 1 produced a clearing zone on media containing colloidal chitin while 3 other strains hydrolyzed the fluorogenic chitin analogue 4-methylumbelliferyl-β-d-N,N′-diacetylchitobioside (D. L. Kirchman, M. T. Cottrell, L. Yu, and K. Dyit, unpublished data, 1999). However, none of the α-proteobacteria amplifying with the group I primers expressed a chitinolytic phenotype (Table 1). There are clearly aspects of chitin degradation by α-proteobacteria that are not understood. Chitin degradation in α-proteobacteria has not been extensively studied, but an examination of uncultured bacteria using fluorescence in situ hybridization of 16S rRNA-directed oligonucleotide probes combined with microautoradiography (6) suggested that α-proteobacteria are involved in chitin degradation in estuarine and coastal waters.
The most abundant type of group I chitinase gene amplified from estuarine and coastal Pacific Ocean bacterioplankton DNA was >98% similar to genes in cultured members of the Roseobacter group. In fact, the nucleotide sequences of clones in clone family A were identical to those of strain EE36 and S. stellata strain E37 in the Roseobacter group. More than half of the clones in the coastal Pacific and Delaware Bay libraries represent chitinase genes most similar to those in cultured α-proteobacteria, suggesting that chitin hydrolysis by culturable α-proteobacteria may adequately model hydrolysis by unculturable α-proteobacteria in the coastal Pacific Ocean and Delaware Bay. In contrast, none of the chitinase genes from uncultured bacteria were identical to those of γ-proteobacteria. These data suggest that chitin hydrolysis by these uncultured organisms in the ocean is not adequately represented by cultured bacteria in these groups.
Chitinase gene phylogeny only partially followed the phylogeny of 16S rRNA genes, which currently defines bacterial phylogeny, in particular, the various classes of proteobacteria. Our results show a closer relationship between chitinase genes of vibrios (γ-proteobacteria) and α-proteobacteria than between Vibrio chitinases and other γ-proteobacterial chitinases, such as those in some alteromonads. The deviation in chitinase gene phylogeny from 16S rRNA gene phylogeny may due to lateral gene transfer. Although 16S rRNA genes are presumed to be fairly resistant to lateral transfer, evidence is accumulating that transfer of nonessential genes, e.g., chitinase genes, between groups of bacteria is more prevalent than previously realized. In fact, microbial genomes appear to be patchworks of genes exchanged by lateral transfer (9). If this is so, the phylogeny of chitinase genes may not be particularly exceptional.
Lateral transfer of chitinase genes is an attractive hypothesis to explain the different phylogenetic relationships between group I chitinase and 16S rRNA genes. However, other mechanisms cannot be excluded. The phylogenetic relationships of similar chitinase genes could differ from the 16S rRNA tree if the chitinase genes are actually a mixture of different genes that arose from a gene duplication event (33). The 16S rRNA and chitinase gene trees would likely differ if the 16S rRNA gene reflects subsequent bacterial speciation decoupled from independent evolution of the chitinase genes. The 16S rRNA and chitinase gene trees would only match if all of the chitinase genes from bacteria having the corresponding 16S rRNA genes were compared. It is not possible to link definitively the chitinase and 16S rRNA genes in uncultured bacteria.
In the case of arthropod viruses, however, lateral gene transfer remains the most plausible explanation. The similarity of chitinases in bacteria and viruses alone provides a compelling argument for the idea that viruses obtained chitinase genes from bacteria. Furthermore, there is little evidence suggesting that these viruses acquired chitinase genes from their arthropod hosts. For example, the chitinase of Bombyx mori nuclear polyhedrosis virus (NPV) is 64% identical to ChiA of Enterobacter sp. strain G-1 at the amino acid level while the arthropod host and viral chitinase genes are not very similar, sharing only 24% identical aligned amino acids. In addition, the host gene lacks the priming sites for the group I chitinase primers. The viral group I chitinase genes probably originated in bacteria, not the arthropod host.
Group I chitinase genes provide an interesting perspective for examining the diversity of hydrolytic enzymes involved in organic matter degradation, the evolution of bacterial chitinase genes (38), and the relationship between chitinases of cultured and uncultured bacteria. Chitinase genes of uncultured microbes were both similar to and different from those of cultured bacteria. The most surprising similarity was between chitinase genes in uncultured bacteria and cultured α-proteobacteria. Culture-based studies give little indication that α-proteobacteria are important chitin degraders in aquatic systems, but our results suggest that exploration of the chitin-degrading capacities of uncultured α-proteobacteria will lead to a better understanding of chitin degradation in the ocean.
ACKNOWLEDGMENTS
This research was supported by the U.S. Department of Energy. Collection of the samples was supported by the National Science Foundation.
We thank Mary Ann Moran, Åke Hagström, Wietse de Boer, Ingrid Brettar, and Manfred Höfle for donating bacterial strains.
REFERENCES
- 1.Ahrens C H, Russell R L Q, Funk C J, Evans J T, Harwood S H, Rohrmann G F. The sequence of the Orgyia pseudotsugata multinucleocapsid nuclear polyhedrosis virus genome. Virology. 1997;229:381–399. doi: 10.1006/viro.1997.8448. [DOI] [PubMed] [Google Scholar]
- 2.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 3.Amann R I, Ludwig W, Schleifer K-H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ayres M D, Howard S C, Kuzio J, Lopezferber M, Possee R D. The complete DNA sequence of Autographa californica nuclear polyhedrosis virus. Virology. 1994;202:586–605. doi: 10.1006/viro.1994.1380. [DOI] [PubMed] [Google Scholar]
- 5.Braker G, Fesefeldt A, Witzel K P. Development of PCR primer systems for amplification of nitrite reductase genes (nirK and nirS) to detect denitrifying bacteria in environmental samples. Appl Environ Microbiol. 1998;64:3769–3775. doi: 10.1128/aem.64.10.3769-3775.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cottrell, M. T., and D. L. Kirchman. Natural assemblages of marine proteobacteria and Cytophaga-Flavobacter consuming low- and high-molecular-weight dissolved organic matter. Appl. Environ. Microbiol., in press. [DOI] [PMC free article] [PubMed]
- 7.Cottrell M T, Cary S C. Diversity of dissimilatory bisulfite reductase genes of bacteria associated with the deep-sea hydrothermal vent polychaete annelid Alvinella pompejana. Appl Environ Microbiol. 1999;65:1127–1132. doi: 10.1128/aem.65.3.1127-1132.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cottrell M T, Moore J A, Kirchman D L. Chitinases from uncultured marine microorganisms. Appl Environ Microbiol. 1999;65:2553–2557. doi: 10.1128/aem.65.6.2553-2557.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doolittle W F. Phylogenetic classification and the universal tree. Science. 1999;284:2124–2128. doi: 10.1126/science.284.5423.2124. [DOI] [PubMed] [Google Scholar]
- 10.Garcia-Vallvé S, Palau J, Romeu A. Horizontal gene transfer in glycosyl hydrolases inferred from codon usage in Escherichia coli and Bacillus subtilis. Mol Biol Evol. 1999;16:1125–1134. doi: 10.1093/oxfordjournals.molbev.a026203. [DOI] [PubMed] [Google Scholar]
- 11.Giovannoni S, Rappé M. Evolution, diversity and molecular ecology of marine prokaryotes. In: Kirchman D L, editor. Microbial ecology of the oceans. New York, N.Y: John Wiley & Sons, Inc.; 2000. pp. 47–84. [Google Scholar]
- 12.Giovannoni S J. The polymerase chain reaction. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. New York, N.Y: Wiley; 1991. pp. 177–203. [Google Scholar]
- 13.Giovannoni S J, DeLong E F, Schmidt T M, Pace N R. Tangential flow filtration and preliminary phylogenetic analysis of marine picoplankton. Appl Environ Microbiol. 1990;56:2572–2575. doi: 10.1128/aem.56.8.2572-2575.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.González J M, Whitman W B, Hodson R E, Moran M A. Identifying numerically abundant culturable bacteria from complex communities: an example from a lignin enrichment culture. Appl Environ Microbiol. 1996;62:4433–4440. doi: 10.1128/aem.62.12.4433-4440.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.González J M, Mayer F, Moran M A, Hodson R E, Whitman W B. Sagittula stellata gen. nov., sp. nov., a lignin-transforming bacterium from a coastal environment. Int J Syst Bacteriol. 1997;47:773–780. doi: 10.1099/00207713-47-3-773. [DOI] [PubMed] [Google Scholar]
- 16.González J M, Moran M A. Numerical dominance of a group of marine bacteria in the α-subclass of the class Proteobacteria in coastal seawater. Appl Environ Microbiol. 1997;63:4237–4242. doi: 10.1128/aem.63.11.4237-4242.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gooday G W, editor. A survey of polysaccharase production: a search for phylogenetic implications. London, England: Academic Press; 1979. [Google Scholar]
- 18.Hallin S, Lindgren P E. PCR detection of genes encoding nitrite reductase in denitrifying bacteria. Appl Environ Microbiol. 1999;65:1652–1657. doi: 10.1128/aem.65.4.1652-1657.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harpster M H, Dunsmuir P. Nucleotide sequence of the chitinase B gene of Serratia marcescens Qmb1466. Nucleic Acids Res. 1989;17:5395. doi: 10.1093/nar/17.13.5395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoch M P, Kirchman D L. Seasonal and interannual variability in bacterial production and biomass in a temperate estuary. Mar Ecol Prog Ser. 1993;98:283–295. [Google Scholar]
- 21.Huber R, Stohr J, Hohenhaus S, Rachel R, Burggraf S, Jannasch H, Stetter K O. Thermococcus chitonophagus sp. nov., a novel, chitin-degrading, hyperthermophilic archaeum from a deep-sea hydrothermal vent environment. Arch Microbiol. 1995;164:255–264. [Google Scholar]
- 22.Jones J D G, Grady K L, Suslow T V, Bedbrook J R. Isolation and characterization of genes encoding 2 chitinase enzymes from Serratia marcescens. EMBO J. 1986;5:467–473. doi: 10.1002/j.1460-2075.1986.tb04235.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kamita S G, Maeda S. Sequencing of the putative DNA helicase-encoding gene of the Bombyx mori nuclear polyhedrosis virus and fine-mapping of a region involved in host range expansion. Gene. 1997;190:173–179. doi: 10.1016/s0378-1119(96)00671-3. [DOI] [PubMed] [Google Scholar]
- 24.Kang W, Tristem M, Maeda S, Crook N E, O'Reilly D R. Identification and characterization of the Cydia pomonella granulovirus cathepsin and chitinase genes. J Gen Virol. 1998;79:2283–2292. doi: 10.1099/0022-1317-79-9-2283. [DOI] [PubMed] [Google Scholar]
- 25.Kirchman D L, White J. Hydrolysis and mineralization of chitin in the Delaware Estuary. Aquat Microb Ecol. 1999;18:187–196. [Google Scholar]
- 26.Kuzio J, Pearson M N, Harwood S H, Funk C J, Evans J T, Slavicek J M, Rohrmann G F. Sequence and analysis of the genome of a baculovirus pathogenic for Lymantria dispar. Virology. 1999;253:17–34. doi: 10.1006/viro.1998.9469. [DOI] [PubMed] [Google Scholar]
- 27.Le T H, Wu T Q, Robertson A, Bulach D, Cowan P, Goodge K, Tribe D. Genetically variable triplet repeats in a RING-finger ORF of Helicoverpa species baculoviruses. Virus Res. 1997;49:67–77. doi: 10.1016/s0168-1702(97)01454-8. [DOI] [PubMed] [Google Scholar]
- 28.Manz W, Amann R, Ludwig W, Vancanneyt M, Schleifer K H. Application of a suite of 16S rRNA-specific oligonucleotide probes designed to investigate bacteria of the phylum Cytophaga-Flavobacter-Bacteroides in the natural environment. Microbiology (Reading) 1996;142:1097–1106. doi: 10.1099/13500872-142-5-1097. [DOI] [PubMed] [Google Scholar]
- 29.Manz W, Amann R, Ludwig W, Wagner M, Schleifer K-H. Phylogenetic oligodeoxynucleotide probes for the major subclasses of proteobacteria: problems and solutions. Syst Appl Microbiol. 1992;15:593–600. [Google Scholar]
- 30.McDonald I R, Murrell J C. The methanol dehydrogenase structural gene mxaF and its use as a functional gene probe for methanotrophs and methylotrophs. Appl Environ Microbiol. 1997;63:3218–3224. doi: 10.1128/aem.63.8.3218-3224.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McDonald I R, Murrell J C. The particulate methane monooxygenase gene pmoA and its use as a functional gene probe for methanotrophs. FEMS Microbiol Lett. 1997;156:205–210. doi: 10.1111/j.1574-6968.1997.tb12728.x. [DOI] [PubMed] [Google Scholar]
- 32.Moyer C L, Tiedje J M, Dobbs F C, Karl D M. A computer-simulated restriction fragment length polymorphism analysis of bacterial small-subunit rRNA genes: efficacy of selected tetrameric restriction enzymes for studies of microbial diversity in nature. Appl Environ Microbiol. 1996;62:2501–2507. doi: 10.1128/aem.62.7.2501-2507.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Page R D M, Holmes E C. Molecular evolution: a phylogenetic approach. Oxford, England: Blackwell Science; 1998. [Google Scholar]
- 34.Park J K, Okamoto T, Yamasaki Y, Tanaka K, Nakagawa T, Kawamukai M, Matsuda H. Molecular cloning, nucleotide sequencing, and regulation of the chiA gene encoding one of chitinases from Enterobacter sp. G-1. J Ferment Bioeng. 1997;84:493–501. [Google Scholar]
- 35.Roller C, Wagner M, Amann R, Ludwig W, Schleifer K H. In situ probing of gram-positive bacteria with high DNA G+C content using 23S ribosomal-RNA-targeted oligonucleotides. Microbiology (Reading) 1994;140:2849–2858. doi: 10.1099/00221287-140-10-2849. [DOI] [PubMed] [Google Scholar]
- 36.Sitrit Y, Vorgias C E, Chet I, Oppenheim A B. Cloning and primary structure of the chiA gene from Aeromonas caviae. J Bacteriol. 1995;177:4187–4189. doi: 10.1128/jb.177.14.4187-4189.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suzuki M T, Rappé M S, Haimberger Z W, Winfield H, Adair N, Strobel J, Giovannoni S J. Bacterial diversity among small-subunit rRNA gene clones and cellular isolates from the same seawater sample. Appl Environ Microbiol. 1997;63:983–989. doi: 10.1128/aem.63.3.983-989.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Svitil A L, Kirchman D L. A chitin-binding domain in a marine bacterial chitinase and other microbial chitinases: implications for the ecology and evolution of 1,4-beta-glycanases. Microbiology (Reading) 1998;144:1299–1308. doi: 10.1099/00221287-144-5-1299. [DOI] [PubMed] [Google Scholar]
- 39.Techkarnjanaruk S, Pongpattanakitshote S, Goodman A E. Use of a promoterless lacZ gene insertion to investigate chitinase gene expression in the marine bacterium Pseudoalteromonas sp. strain S9. Appl Environ Microbiol. 1997;63:2989–2996. doi: 10.1128/aem.63.8.2989-2996.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tsujibo H, Orikoshi H, Tanno H, Fujimoto K, Miyamoto K, Imada C, Okami Y, Inamori Y. Cloning, sequence, and expression of a chitinase gene from a marine bacterium, Altermonas sp. strain O-7. J Bacteriol. 1993;175:176–181. doi: 10.1128/jb.175.1.176-181.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zarda B, Hahn D, Chatzinotas A, Schonhuber W, Neef A, Amann R I, Zeyer J. Analysis of bacterial community structure in bulk soil by in situ hybridization. Arch Microbiol. 1997;168:185–192. [Google Scholar]
- 42.Zehr J P, Capone D G. Problems and promises of assaying the genetic potential for nitrogen fixation in the marine environment. Microb Ecol. 1996;32:263–281. doi: 10.1007/BF00183062. [DOI] [PubMed] [Google Scholar]
- 43.Ziemke F, Brettar I, Höfle M G. Stability and diversity of the genetic structure of a Shewanella putrefaciens population in the water column of the central Baltic. Aquat Microb Ecol. 1997;13:63–74. [Google Scholar]