Summary
A common theme across multiple successful immunotherapies for cancer is the recognition of tumor-specific mutations (neoantigens) by T cells. The rapid discovery of such antigen responses could lead to improved therapies through the adoptive transfer of T cells engineered to express neoantigen reactive T-cell receptors (TCR). Here, through CITE-seq (Cellular Indexing of Transcriptomes and Epitopes by Sequencing) and TCR-seq of non-small cell lung cancer (NSCLC) tumor-infiltrating lymphocytes (TIL), we develop a neoantigen-reactive T-cell signature based on clonotype frequency and CD39 protein and CXCL13 mRNA expression. Screening of TCRs selected by the signature allows us to identify neoantigen-reactive TCRs with a success rate of 45% for CD8+ and 66% for CD4+ T cells. Because of the small number of samples analyzed (4 patients), generalizability remains to be tested. However, this approach can enable the quick identification of neoantigen-reactive TCRs and expedite the engineering of personalized neoantigen-reactive T cells for therapy.
Graphical Abstract

eTOC blurb
The discovery of T-cell receptors (TCR) that recognize tumor-specific neoantigens can lead to improved therapies through the adoptive transfer of T cells engineered to express these TCRs. Here, Hanada et al. design a signature of neoantigen-reactive T cells that enables the rapid identification of both CD4 and CD8 neoantigen-reactive TCRs.
Introduction
Concerted efforts carried out over the past 30 years have led to the identification of a broad array of antigens targeted by patients’ tumor-reactive T cells (Bruggen et al., 1991; Kawakami and Rosenberg, 1997; Morgan et al., 2003) and studies carried out primarily over the past decade have indicated that the tumor-specific somatic mutations encode the dominant tumor-associated antigens associated with responses to immune checkpoint blockade (ICB) and adoptive T cell transfer (ACT). Some of the strongest evidence for this hypothesis has been the correlation observed between clinical benefit and tumor mutational load. This point is demonstrated most strikingly by the relatively high response rates to immune checkpoint therapy of multiple cancers with DNA repair defects (Gubin et al., 2014; Le et al., 2015; Rizvi et al., 2015; Rooij et al., 2013; Snyder et al., 2014). A study of the reactivity of tumor-infiltrating lymphocytes (TIL) from melanomas that induced objective responses in over half of patients treated was able to find T cells reactive with mutant gene products (Robbins et al., 2013). While they represent the safest class of immune targets for human cancer immunotherapy due to their lack of expression in normal tissues, the identification of neoantigen-reactive T cells in patients with cancer has been hampered by the complexities of these systems developed to identify these targets. In many studies, neoantigens are defined as tumor-specific mutant epitopes that are predicted by an algorithm to bind to one of the patient’s MHC alleles without verifying the ability of T cells to recognize the peptide candidates; however, in the largest study to date where T-cell responses were systematically investigated, only 1.6% of candidate neoantigens were documented to generate such a response (Parkhurst et al., 2019). The extensive analysis needed to validate neoantigen reactivities has led to attempts to develop more rapid methods for identifying tumor-reactive T-cells based on their expression of phenotypic markers. A significant enrichment of tumor-reactive CD8+ T cells was achieved by sorting T cells from fresh tumor (Gros et al., 2014; Inozume et al., 2010) or peripheral blood (Gros et al., 2016) that express the immune checkpoint cell surface marker PD-1. Populations of CD8+ T cells that express CD103, a marker of tissue-resident T cells up-regulated by chronic TCR stimulation and TGF-β (Duhen et al., 2018), appeared to be more highly activated by autologous tumor cells than the corresponding CD103 negative population (Djenidi et al., 2015). Expression of the T-cell exhaustion marker CD39 has also been found to enrich tumor-reactive CD8+ T cells from melanoma and head and neck cancer fresh tumor samples as measured by lysis of autologous tumor lines (Duhen et al., 2018). These studies were confined to CD8+ cells and did not molecularly define the antigens recognized by the T cells. CD39 also appeared to enrich tumor-reactive CD4+ T cells from HPV-induced fresh tumor specimens (Kortekaas et al., 2020), although CD39 is also expressed by regulatory T cells (Treg) (Mandapathil et al., 2009).
In this study, we initially screened in vitro expanded NSCLC TIL to identify neoantigen-reactive T cells and sequenced their TCR variable regions that contain hypervariable complementarity determining region 3 (CDR3) sequences that are unique to individual T-cell clonotypes. In parallel, we performed CITE-seq combined with TCR-seq (TCR-CITE-seq) of the same patients’ fresh NSCLC TIL focusing on the clonotypes that contain the CDR3 regions from neoantigen-reactive T-cells. This analysis enabled us to develop a neoantigen-reactive T-cell signature in the fresh tumor and led to the identification of additional HLA class I and class II-restricted neoantigen-reactive TCRs. These results provide an impetus to further evaluate the application of this strategy for the development of streamlined cancer immunotherapies.
Results
Isolation of neoantigen-reactive T cells from NSCLC TIL
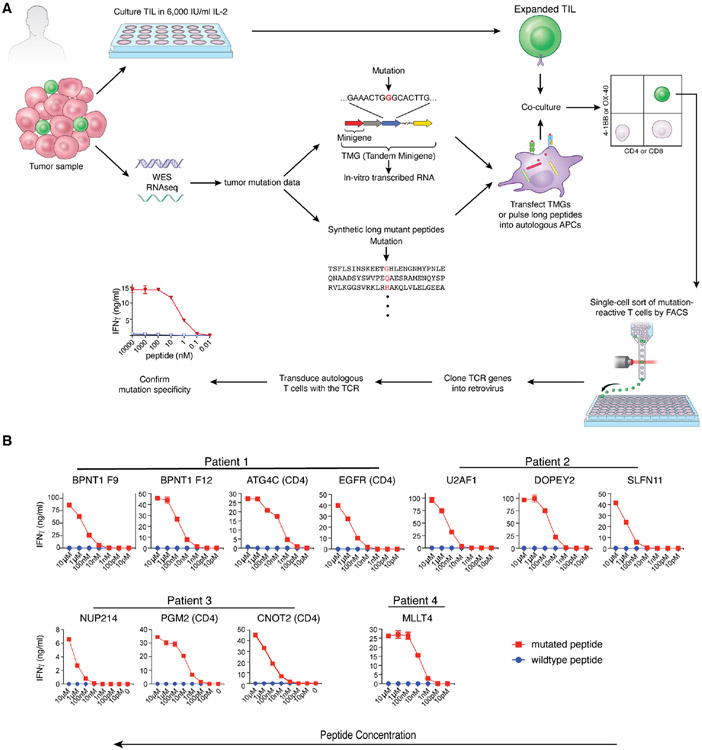
CITE-seq combined with TCR-seq (TCR-CITE-seq) is a method that can coordinately display cell surface proteins, transcriptome data, and TCR sequence information at the single-cell level. Hence, if TCR sequences from cells of interest are known, it is possible to know their transcriptome and cell surface protein expression. To identify TCR sequences expressed by neoantigen-reactive T cells, NSCLC TIL were initially expanded from four patient samples (Table S1) in vitro in the presence of interleukin-2. Whole exome sequencing (WES) analysis of patient fresh tumor and matched peripheral blood samples was carried out to identify non-synonymous somatic mutations that, in conjunction with the sequences that encoded the 12 upstream and downstream residues, were used to generate tandem minigenes (TMG) and peptide libraries, as previously described (Parkhurst et al., 2019; Robbins et al., 2013; Zacharakis et al., 2018) (Figure 1A). By screening a total of 514 mutated peptides and 39 tandem minigenes encoding mutations found by WES, we identified eleven mutation-reactive TCRs (seven from CD8+ and four from CD4+ T cells) and confirmed their neoantigen-specificity (versus wild type) by IFNγ secretion assay (Figure 1B). Patient 1 had a CD4 reactivity to a mutant epitope encoded by a common shared mutation of EGFR (p.E746_A750del) (Sharma et al., 2007) presented by HLA-DPA1*02:01, DPB1*01:01 and Patient 2 had an HLA-B*27:05-restricted CD8+ T cells that recognized a recurring mutation in the spliceosome gene U2AF1(p.S34F) described by others in NSCLC (Esfahani et al., 2019; Imielinski et al., 2012) and myelodysplastic syndrome (Wang et al., 2020). The additional 8 mutations that encoded identified HLA class I and class II neoantigens have not previously been identified and may represent passenger mutations that do not play an important role in tumorigenesis.
Figure 1. Isolation of neoantigen-reactive T cell receptors by the conventional method.
(A) Procedure for isolating neoantigen-reactive TCRs. Resected tumor samples were used for two purposes. One was for growing TIL and the other was for performing Whole Exome Sequencing (WES) and RNA-seq to identify tumor-specific mutations. Based on the mutation data, Tandem Minigenes (TMGs), containing a string of genes that encode 25 amino acids surrounding the mutation, were created for the presentation on class I MHC. TMGs are transcribed in vitro, and the RNA was introduced into the autologous dendritic cells by electroporation. For the class II MHC pathway presentation, 25-mer peptides, the center of which were mutated amino acids, were synthesized and were exogenously pulsed onto dendritic cells. These dendritic cells served as surrogate tumor cells and were co-cultured with expanded TIL. When TIL recognized a mutation, they up-regulated activation markers such as 4-1BB and OX-40. These activated T-cells were single-cell sorted by FACS, and the TCR genes were amplified by a PCR-based method. After cloning into a retrovirus vector, TCR genes were retrovirally transduced to the autologous T-cells. Their specificity was confirmed by co-culturing with DC presenting the mutated or the wildtype peptide. T-cell activation was measured by IFNγ ELISA. (B) Cognate peptides (mutated or wildtype) were pulsed onto autologous dendritic cells (CD8: 9-mer or 10-mer, 2 hours at RT, CD4: 25-mer, 3 hours at 37°C). After the incubation, dendritic cells were washed three times, and the TCR-transduced autologous T-cells were added. After 20-hour co-culture, IFNγ in the culture sup was measured by ELISA.
Populations of CD3+ cells were then isolated from cryopreserved vials of fresh tumor digests by fluorescence-activated cell sorting and subjected to TCR-CITE-seq analysis using 12 feature-barcoded (FBC) antibodies (Table S2). Cell surface protein analysis showed a typical CD4/8 ratio of roughly 2:1 and indicated that CD8 T cells were more terminally differentiated than CD4 T cells from the corresponding samples as judged by analysis of CD62L and CD45RA expression. FOXP3+ regulatory T cells constituted between 3.6 and 8.7% of CD4+ cells (Figure S1A). For all of the 4 evaluated patient samples, neoantigen-reactive CD8+ T cell clonotypes identified by T cell receptor alpha chain variable region (TRAV) and T cell receptor beta chain variable region (TRBV) sequencing mapped predominantly to a single cluster generated by t-SNE analysis of cell surface protein expression by FBC antibodies (Figure S1B). While neoantigen-reactive cells also predominantly mapped to a single transcriptome-based cluster of T cells from patient 3 and 2 clusters of T cells for patient 2, this method of analysis did not result in tight clustering of reactive T cells from patients 1 and 4.
CD39 and CXCL13 are highly expressed on neoantigen-reactive CD8 T cells
Next, for each patient, we compared the differential expression of proteins and genes for all neoantigen-reactive CD8+ T cells versus all other CD8+ T cells excluding patient 3 since only 4 neoantigen-reactive T cells were identified among the 4,490 CD8 T cells analyzed from this patient. Differential expression [FDR≤0.01, (Fold)2≥4] was found in 673, 382, and 505 genes or proteins in patients 1, 2, and 4, respectively, and among these, 24 were common to all three patients (22 of which were up-regulated and 2 that were down-regulated) (Figure 2A, B).
Figure 2. Identification of genes and proteins expressed on neoantigen-reactive T cells isolated by the conventional TIL screening.
Comparative gene and protein expression analysis of neoantigen-reactive T-cell clonotypes and putative non-reactive CD8+ T cells. (A) Genes and proteins with (Fold)2≥4 differences and FDR≤0.01 were selected and shown in the Venn diagram by numbers. (B) 24 genes or proteins commonly up- or down-regulated among three patients. (C) Dot plots of cell surface proteins as analyzed by FBC antibodies. (Patient 1, blue dots: BPNT1-reactive clone F9, red dots: BPNT1-reactive clone F12, Patient 2, red dots: DOPEY2-reactive clone, orange dots: SLFN11-reactive clone, blue dots: U2AF1-reactive clone, patient 4, red dots: MLLT4-reactive clone, Black dots: other CD8 T-cells in all the patients.) See also related Figure S1.
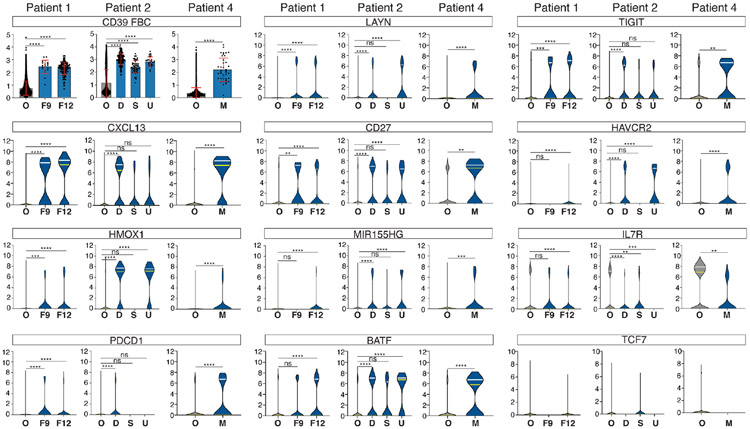
We examined the expression of these genes and proteins in each neoantigen-reactive T-cell clonotype. Two-dimensional plots of protein parameters (Figure 2C) showed that CD39+ and CD103+ represented the markers that were most significantly enriched in neoantigen reactive cells, markers that have previously been described as an identifier of tumor-reactive CD8+ T cells (Duhen et al., 2018). Another molecule known to enrich tumor-reactive cells is PD1 (Gros et al., 2014, 2016; Inozume et al., 2010). PD1 protein was highly expressed by neoantigen-reactive cells except in patient 4, the only patient with a history of anti-PD1 therapy. Since the tumor was resected 62 days after the last dose of pembrolizumab, it is possible that pembrolizumab, with a long in vivo half-life of 26 days (Dang et al., 2015), was still bound on T cells and blocked the binding of the feature-barcoded anti-PD1 antibody. High PDCD1 mRNA expression in these cells (Figure 3) supported this hypothesis. A study on melanoma TIL reported that CD39 is a key marker that separates all TIM3+ cells from TIM3− cells (Sade-Feldman et al., 2018). However, in our CITE-seq analysis on NSCLC, there were both CD39+TIM3+ and CD39+TIM3− neoantigen-reactive TIL. Accordingly, selecting just CD39+TIM3+ or PD1+TIM3+ cells from fresh NSCLC TIL may overlook a significant portion of neoantigen-reactive T cells. Very low CD45RA, intermediate CD45RO, and low CD62L was a consistent phenotype, but it did not cluster in a distinct population. RNA-seq analysis indicated that the majority of neoantigen-reactive T-cell clonotypes had an exhausted T-cell phenotype evidenced by up-regulating CXCL13 (Li et al., 2018; Sade-Feldman et al., 2018; Thommen et al., 2018; Tirosh et al., 2016; Zheng et al., 2017a), PDCD1, LAYN (Li et al., 2018; Thommen et al., 2018; Tirosh et al., 2016; Zheng et al., 2017a), CD27 (McLane et al., 2015; Thommen et al., 2018; Zheng et al., 2017a), BATF (Quigley et al., 2010), TIGIT, MIR155HG (Stelekati et al., 2018), and down-regulating IL7R (Figure 3). Within these parameters, CXCL13 stood out by its expression in the majority of neoantigen reactive clonotypes but was expressed by only a relatively small percentage of all other CD8+ T cells.
Figure 3. Analysis of gene and protein expression on neoantigen-reactive T cells at the clonotype level.
Clonotype-level comparisons of representative gene and protein expression that were selected in Figure 2. Protein expression (CD39 FBC) is shown by the dot and bar plot and gene expressions by violin plots. In dot and bar plots, the mean is shown by bars and ±1 SD by error bars. (O: other CD8 T cells, F9: BPNT1-reactive TIL clone F9, F12: BPNT1-reactive TIL clone F12, D: DOPEY2 reactive TIL clone, S: SLFN11-reactive TIL clone, U: U2AF1-reactive TIL clone, M: MLLT4-reactive TIL clone) Statistical analysis was done by one-way ANOVA and Dunnett’s multiple comparison test for patients 1 and 2 and by unpaired t-test for patient 4. In violin plots, statistical analysis was done for patients 1 and 2 by Kruskal-Wallis test and Dunn’s multiple comparison test and patient 4 by Mann-Whitney test. (ns P>0.05, * P≤0.05, ** P≤0.01, *** P≤0.001, **** P<0.0001)
Simple signature of ‘high-frequency TCR clonotype with CD39 protein and CXCL13 mRNA co-expression’ prospectively identifies neoantigen-reactive CD8+ T cells
In Patients 1, 2, and 3, cells expressing CD39 protein (Figure 4A, red dots) represented 23-34% of all CD8+ cells (Figure 4A, red and blue dots), but it was 5.7% in patient 4, the only patient who had received anti-PD1 antibody approximately 2 months prior to tumor resection. The frequency of CXCL13-expressing cells varied between 2.5 and 11.1% of CD8+ cells (Figure 4B). In general, CD39− cells had higher clonotypic frequencies than CD39+ cells in un-cultured TIL, as described previously (Simoni et al., 2018) (Figure 4C), and this trend was even more pronounced in peripheral blood lymphocytes (PBL), where some CD39− clonotypes were represented at 100-fold or higher frequencies than those of the majority of CD39+ clonotypes (Figure 4D). While the vast majority of CD39− T cells lacked expression of CXCL13, cells expressing high levels of CXCL13 predominated among cells expressing CD39 protein (Figure 4E). For all four patients, we selected the T-cell clonotypes with the highest frequency within the CD39+ plus CXCL13+ subset and high frequency clonotypes from the CD39− subset, generated retroviral constructs encoding the corresponding TCRs and evaluated their ability to confer transduced autologous PBL with the reactivity to candidate neoantigens identified by WES of the corresponding fresh tumor samples. Evaluation of TCRs identified from dominant clonotypes present in the 4 patient samples revealed that 15 of the 33 TCRs with unknown reactivity isolated from CD39+ T cells but none of the 21 TCRs isolated from CD39− T cells were neoantigen-reactive (Figure 4F). From patients 1, 3, and 4, we discovered additional TCRs reactive to the same neoantigens found by conventional testing of the expanded TIL (BPNT1 in patient 1, NUP214 in patient 3, and MLLT4 in patient 4). In patient 2, 8 out of 12 TCRs selected from CD39+ cells were neoantigen-reactive, five recognizing the previously identified mutant DOPEY-2 epitope. Importantly, the other three TCRs mediated recognition of previously unidentified neoantigen, 2 being reactive with a mutant POLDIP2 epitope (#10 and #11) and 1 reactive with a mutant PNPLA6 epitope (#4), neither of which had been identified during standard screening with expanded TIL cultures. These results indicate that evaluation of the reactivity of TCRs isolated from CD8+ clonotypes expressing CD39 and CXCL13 in fresh TIL may represent a widely applicable strategy for identifying neoantigens and neoantigen-reactive TCRs that cannot readily be identified by screening in vitro expanded TIL. Overall, the frequency of neoantigen-reactive CD8+ T cells identified using this signature ranged between 0.17 and 1.52% of CD3+ cells in the 4 evaluated patient tumor samples. FBC dot plot analysis demonstrated that the majority of neoantigen-reactive CD8+ T cells expressed both CD39 and CD103 (Figure S2A). A substantial percentage of CD103+ T cells did not express CD39, none of which appeared to be neoantigen-reactive, demonstrating that CD103 alone does not represent a reliable marker for identifying tumor-reactive T cells. Uncultured TIL expressed widely varying levels of PD-1 protein, and in patient 2, higher levels of PD-1 expression were detected in many of the neoantigen-reactive clonotypes than in non-reactive clonotypes (Figure 4G), but this was not consistently observed for all patients. As was observed in the initial analysis (Figure 2C), TIM3 expression on neoantigen-reactive cells was not consistent (Figure S2B). Recent analysis on melanoma TIL suggested the strong correlation between the existence of tumor-reactive CD39−, CD69− T-cells in the adoptively transferred cells and clinical response (Krishna et al., 2020). However, by the single-cell analysis of T cells in the fresh NSCLC tumor, we were not able to identify CD39−, CD69− neoantigen-reactive T cells, likely due to their low frequency (Figure S2C and S3C).
Figure 4. Simple signature of expression of CD39 protein, CXCL13 transcript and high-frequency clonotype identifies CD8+ neoantigen-reactive T cells at high frequency.
(A and B) tSNE plots based on the cell surface protein expression data. Cell number and frequency in CD8+ T cells are shown in each box. (A) blue dots: CD8+, CD39−, red dots: CD8+, CD39+, gray dots: CD4+ T-cells (B) blue dots: CD8+, CXCL13−, red dots: CD8+, CXCL13+, gray dots: CD4+ T-cells (C) T-cell clonotype frequency in TIL as measured by single-cell TCR analysis (Red bars: neoantigen-reactive clonotypes, Blue bars: CD39− clonotypes, Gray bars: CD39+, CXCL13+ but not neoantigen-reactive) (D) T-cell clonotype frequency in PBL as measured by TCRβ deep sequencing. Total numbers of CDR3B were patient 1: 125,214, patient 2: 266,713, patient 3: 194,454, and patient 4: 318,838. (E) CXCL13 mRNA expression (yellow line: median, white lines: upper or lower quartile) (F) Test on neoantigen-specific reactivity by synthetic 9-mer peptides. Details on TCRs and peptides used for the assay are provided in the supplementary Table 3. The assay was performed in the same way as Figure 1B. (G) Identification of neoantigen-reactive TCRs in higher PD1 protein-expressing clonotypes. Red bars are neoantigen-reactive clones.
CD39 protein expression and high CXCL13 mRNA also identifies CD4+ neoantigen-reactive T-cells
Because of the small number of CD4+ neoantigen-reactive T cells identified by the traditional TIL screening method (three mutated ATG4C-reactive cells and seven mutated EGFR-reactive cells), we were not able to independently develop a phenotypic signature for CD4+ neoantigen reactive TIL. Extrapolating the methods used to find CD8+ neoantigen-reactive TIL, we evaluated the TIL that co-clustered in tSNE plots with these few CD4+ TIL of known neoantigen reactivity (Figure 5A). Comparison of the expression profiles of the 1,600 CD4+ T cells, the population where the previously identified TCRs mediating recognition of the ATG4C and EGFR HLA class II-restricted neoantigens were mapped (Figure 5A, orange dots), with all the 15,207 CD4+ T-cells that mapped outside of this cluster demonstrated that T cells expressing CD39 protein, TIGIT, and CXCL13 mRNA are highly enriched in the cluster (Figure 5B). The CD39+ population contained nearly all of the TIGIT+ T cells, and this CD39+, TIGIT+ population could be sub-divided into FOXP3+ Tregs and FOXP3−, CXCL13+ cells, the latter containing all the neoantigen reactive clonotypes (Figure 5A, C). To prospectively validate the CD39+ CXCL13+ signature, we evaluated reactivity of the nine most frequent clonotypes of unknown reactivity from the CD39+ CXCL13+ double-positive population in patient 1 against candidate 25-mer mutant peptides that were synthesized based upon WES of the corresponding fresh tumors (Figure 5D, E). Two of the nine evaluated TCRs recognized the previously identified EGFR neoepitope, 1 recognized the ATG4C neoantigen, while 2 were reactive with previously un-identified neoantigens, 1 recognizing a mutant BAG6 gene product and 1 recognizing a mutant SNX7 gene product (Figure 5F). In patients 2 and 3, the expression patterns of CD39, CXCL13, TIGIT, and FOXP3 were similar to patient 1 (Figure 5G and L). As was observed in CD8+ T cells, CD39− cells were detectable in both tumor and PBL, but CD39+ CXCL13+ cells existed almost exclusively in tumor (Figure 5H-J and M-O). From patients 2 and 3, nine CD39 and CXCL13 double negative TCRs failed to mediate neoantigen recognition, whereas 6 out of the 10 and 8 out of the 10 CD39+, CXCL13+ double positive TCRs were neoantigen-reactive (Figure 5K and P). It is noteworthy that in patient 2, we had not been able to find any neoantigen reactivity in CD4+ cells by the traditional screening method using expanded TIL but were successful using the phenotypic signature. The frequencies of the neoantigen-reactive clonotypes found was as low as 0.025% of total CD3+ cells in fresh tumor (Figure 5H), demonstrating the high sensitivity of the phenotype signature. These findings established that not only was the signature relevant to CD4+ neoantigen-reactive T cells, but they confirmed that it could detect additional neoantigens not used in the development of the signature. Cell surface protein analysis of neoantigen-reactive cells revealed that, although all the neoantigen-reactive clonotypes expressed CD39, a significantly lower percentage of CD4+ than CD8+ T cells expressed CD103, and neoantigen-reactive CD4+ clonotypes did not consistently express CD103 (Figure S3A) On the other hand, the population of neoantigen-reactive CD4+ T demonstrated consistent and high-level expression of PD1 to a greater extent than that was observed in neoantigen-reactive CD8+ neoantigen-reactive T cells, whereas TIM3 expression varied as it did in CD8+ cells (Figure S3B). Patient 4 was the only subject who had previously received anti-PD1 antibody (with no response). In this patient’s tumor, there were very few CD4+ CD39+ CXCL13+ clonotypes (Figure 5Q), in contrast to the CD8+ TIL from the same patient where the majority of CXCL13+ cells were CD39+ (Figure 4A and B). For completeness, we screened TCRs from 2 CD39dim, CXCL13+ clonotypes and 2 CD39−, CXCL13+ clonotypes (Figure 5R) but did not detect neoantigen reactivity.
Figure 5. CD39+, CXCL13+, high-frquency clonotype signature also identifies CD4+ neoantigen-reactive T cells.
(A) Cell-surface protein-based tSNE plot of TIL from patient 1. Light blue dots indicate CD4+ T cells, red dots indicate EGFR-reactive CD4+ T cells, large blue dots indicate ATG4C-reactive CD4+ T cells. (B) Volcano plot showing the differentially expressed genes between the orange cluster and other CD4+ T-cells shown in (A). (C, G, L and Q) tSNE plots showing CD4+ cells by blue dots and CD39 protein-, CXCL13-, TIGIT-, and FOXP3-expressing cells by red dots. Frequency of cells in total CD4+ cells is shown by %. (D) Cell number of the candidate clonotypes within the orange cluster shown in (A), (red bars: neoantigen reactive, blue bars: non-reactive). (E, J and O) CXCL13 expressions in T-cell clones analyzed. (F, K and P) Neoantigen reactivities of TCR-transduced autologous T-cells were examined by co-culturing these cells with autologous dendritic cells pulsed with cognate mutated or wildtype 25-mer peptides. After a 20-hour co-culture, T-cell activation was examined by measuring IFNγ in the cell culture sup by ELISA. (H and M) clonotype frequency of TCRs analyzed within CD3+ TIL (light blue CD39−, red: CD39+ CXCL13+ and neoantigen-reactive, dark blue CD39+ CXCL13+ but not neoantigen-reactive). (I and N) Frequencies of candidate clonotypes in PBL measured by TRBV deep sequencing. (R) Expression of CD39 protein and CXCL13 in the four clonotypes tested for neoantigen-reactivity. All the tSNE plots in Figure 5 are cell surface protein-based.
Extended signature for neoantigen reactivity combining protein and mRNA expression
Through this study, we identified 25 clonotypes, 985 cells of CD8+ and 23 clonotypes, 303 cells of CD4+ neoantigen-reactive T cells that enabled us to analyze the characteristics of neoantigen-reactive T cells in more depth. First, among CD8+ cells, cell surface protein analysis showed higher expression of CD39, CD103, PD1, lower expression of CD4, CD8A, CD45RA, CD62L, CD134. and the expression of CD137 and TIM3 was variable depending on the clonotype (Figure S4). One unexpected finding from CITE-seq was that CD8+ T cells expressed a small amount of CD4 and on neoantigen-reactive T cells, both CD8A and CD4 were expressed less as compared with non-neoantigen-reactive CD8+ T cells (Figure S4). We also saw less CD8A expression on CD4+ neoantigen-reactive T cells as compared with all other CD4+ cells (Figure S5). For CD8 T cells, we created a neoantigen-reactive T-cell signature S=(CD39+, CD103+, PD1+, CD4low, CD8Alow, CD45RAlow, CD62Llow, CD134low) and analyzed its sensitivity and specificity by Receiver Operating Characteristic (ROC) curve analysis. For all the patients, ROC curves were excellent with Area Under the Curve (AUC) of higher than 0.9 (Figure 6A). In CD8+ T-cells, expression of CD39, CD103 and PD1 have been reported to be associated with tumor-reactivity (Duhen et al., 2018; Gros et al., 2014; Inozume et al., 2010). We compared our CD8+ T cell surface protein signature with S=(CD39+, CD103+) and S=(CD39+, CD103+, PD1+) and found our signature outperforms these in a statistically significant manner (Figure 6B and 6C). Both in CD4+ and CD8+ T cells, CD137 is known as an activation marker. However, very few CD4 neoantigen-reactive clonotypes showed the upregulation of CD137 and generally, trended lower than other cells (Figure 8B and S5). A signature for CD4+, (S=CD39+, PD1+, CD45RAlow, CD8Alow, CD137low), resulted in similarly good ROC curves with high AUC (Figure S5 and 6F).
Figure 6. Neoantigen-reactive T cell signatures designed by cell surface protein expression and gene expression show high sensitivity and specificity.
Receiver Operating Characteristic (ROC) curves created by protein and gene expression signature. For each ROC curve, Area Under the Curve (AUC) and the inflection point are shown in each box. A-C are ROC curves for CD8 cell surface protein signatures (A: Signature index = CD39+, PD1+, CD103+, CD4−, CD8A−, CD45RA−, C D62L−, CD134−, B: Signature index = CD39+, CD103+, C: Signature index = CD39+, CD103+, PD1+) For B and C, p-values as compared with the ROC of A are shown. (D) CD8 transcriptome signature (Signature index = CXCL13+, ENTPD1+, BATF+, GZMB+, CD27+, TIGIT+, PHLDA1+, CD74+, HLA-DMA+, HLA-DRA+, HLA-DRB1+, HLA-DPB1+, CD3D+, CD82+, ARL3+, HMOX1+, ALOX5AP+, DUSP4+, CARS+, LSP1+, CCND2+, TPI1+, GAPDH+, ITM2A+, HMGN3+, CHST12+, NAP1L4+, IL7R−, TPT1−, RPS12−, RPS16−, S100A10−) (E) CD8 protein and transcriptome joint signature (F) CD4 cell surface protein signature (Signature index = CD39+, PD1+, CD45RA−, CD8A−, CD137−) (G) CD4 transcriptome signature (Signature index = CXCL13+, NR3C1+, ADGRG1+, NMG+, ITM2A+, ETV7+, COTL1+, B2M+, IGFL2+ VIM−, MT-ND1−, MALAT1−, EMP3−) (H) CD4 cell surface protein and transcriptome joint signature are shown. See also related Figure S2, S3, S4 and S5.
Figure 8. Enrichment of CXCL13-expressing cells by FACS-based cell sorting.
Dot plots of cell surface proteins that were used for the virtual FACS sort of CXCL13+ cells. Red dots represent all proven neoantigen-reactive T-cells for each patient. CD8 T cells are shown in (A) and CD4 T cells in (B). In (A), each gating line in the plot represents CD39+, CD45RA−, PD1+, CD103+ and CD4low (below the median value for all CD8+) and in (B), CD39+, PD1+, and below the median value of total CD4+ T cells for CD45RA, CD8, and CD137. (C) and (E) show the percentage of CXCL13+ cells within the selected cells and (D) and (F) the fold enrichment of CXCL13+ cells through the serial gating. (patient 1: orange line with circles, patient 2: brown line with squares, patient 3: pink line with triangles, patient 4: blue line with inverted triangles)
Transcriptome analysis of neoantigen-reactive CD8+ T cells (Figure 7A) revealed that markers of terminally exhausted T cells (Miller et al., 2019; Sade-Feldman et al., 2018; Siddiqui et al., 2019; Thommen et al., 2018) CXCL13, ENTPD1 (encodes CD39), LAYN (Thommen et al., 2018), TIGIT, BATF, GZMB, CD27, PHLDA1 (Thommen et al., 2018) were up-regulated and CD127 was down-regulated in the reactive T cells. Multiple genes from the MHC class II gene family including CD74, HLA-DMA, DRA, DRB1, and DPB1, and genes related TCR signaling such as CD3D, CD82, and ARL3, cell cycle controlling genes (CCND2), and glycolysis genes (TPI1 and GAPDH) that may reflect the metabolic status of terminally differentiated T cells were also up-regulated in neoantigen-reactive CD8+ T cells. Additional genes that included ALOX5AP, HMOX1, LSP1, ITM2A, CARS, DUSP4, HMGN3, CHST12, and AP1L4 of unknown significance were also up-regulated in neoantigen-reactive CD8+ T cells.
Figure 7. In depth analysis of neoantigen-reactive CD4+ and CD8+ T cell transcriptome.
Analysis on the transcriptome of CD8+ and CD4+ neoantigen-reactive T cells. (A) CD8+ and (B) CD4+ neoantigen-reactive clonotypes identified in the study. Blue lines on the right side of heat maps indicate down-regulated genes, and red lines up-regulated genes. For each patient, the left-most column is gene expression in non-reactive T-cells. Differentially expressed genes were identified by comparing reactive and non-reactive T-cells in each patient, and a gene was deemed significant in CD8+ T cells if it demonstrated a log2 fold change between groups of at least 0.25, expression in at least 10% of the cells for at least one group, and an adjusted p-value of at least 0.05; the expression threshold was set to 0.05 for CD4+ T cells.
Analysis on the transcriptome of CD4+ T cells led to the identification of 13 genes that demonstrated differential expression between neoantigen-reactive cells and all other CD4+ T cells (Figure 7B). One out of the 4 genes that were down-regulated in neoantigen-reactive CD4+ cells, MT-ND1(DRP1), was previously found to regulate the positioning of mitochondria at the peripheral supramolecular activation cluster (pSMAC) on TCR stimulation (Baixauli et al., 2011) and a conditional knockout of this gene reduced T cell proliferation in response to anti-CD3 stimulation (Simula et al., 2018). MALAT1 (Metastasis-associated lung adenocarcinoma transcript 1) encodes a long intergenic noncoding RNA (lincRNA) that is highly expressed in CD4+ T cells but is down-regulated following T-cell activation (Hewitson et al., 2020). Nine genes, including CXCL13, were up-regulated. One of the up-regulated genes NR3C1, encodes the glucocorticoid receptor (GR), a nuclear receptor that has been found to transactivate immune-inhibitory genes such as Tim-3, PD-1, Lag-3 and IL-10 (Acharya et al., 2020). ADGRG1 was reported to be up-regulated in human CD4+ TEM and TEMRA and was included in a signature of exhausted CD4+ T cells that also contained KLRB1, KLRG1, and KLRF1 (Truong et al., 2019). Additional genes that were up-regulated in CD4+ neoantigen-reactive T cells do not have known functions in T cells, but NMB, ITM2A, NR3C1, and ETV7 were found to be up-regulated together with CXCL13, ADGRG1, ENTPD1, and PDCD1 in follicular helper T cells from melanoma TIL (Li et al., 2018). These findings suggest the coordinated up-regulation of CXCL13, ENTPD1, ADGRG1, NMB, ITM2A, NR3C1, and ETV7 in CD4+ TIL.
We developed transcriptome-based signatures, and again they showed high AUC scores in both CD8 (Figure 6D) and CD4 (Figure 6G). When protein-based and the transcriptome-based signatures were combined, AUC scores were similar or further improved (Figure 6E and 6H). Although the signatures we developed in this study are still preliminary due to the limited number of samples analyzed, the higher AUC scores by the joint signature showed the potential advantage of using cell surface protein and transcriptome to identify tumor-reactive T cells from fresh tumors.
Enrichment of CXCL13+ cells by the cell-surface protein expression
In this study, we showed the simple signature of CD39+, CXCL13+ clonotypes with high-frequency includes neoantigen-reactive T-cells in both CD4+ and CD8+ T cells. However, the low frequency of CD39+, CXCL13+ cells in tumor fresh digests (Figure 4B, 5C, G, L, Q) makes single-cell analysis an inefficient method. We evaluated the potential of cell-surface protein-based enrichment of CXCL13+ cells by utilizing the CITE-seq data. Gates that are applicable to FACS-based sorting were selected from the CD8 and CD4 cell surface protein signatures (Figure 8A, B) and the enrichment potential of CXCL13+ cells was assessed (Figure 8C-F). The frequency of CD39+ CXCL13+ cells in the fresh digest varied between 2.5 and 11.1% in CD8+ (Figure 4B) and 3.5 and 10.1% in CD4+ (Figure 5C, G, L, Q). After the hypothetical cell sorting by CD39+, CD45RA−, PD1+, CD103+, CD4low for CD8 T-cells and CD39+, PD1+, CD45RAlow, CD8low, CD137low for CD4 T-cells, CXCL13+ cells were between 18.0 and 57.1% in CD8+ and 35.4 and 83.0% in CD4+ T cells (Figure 8C and E). The fold enrichment varied between 3.9 and 6.4 in CD8+ and 5.4 and 9.0 in CD4+ T cells (Figure 8D and F). This indicates that simple sorting for cell surface markers can enrich for CXCL13 expressing TIL which otherwise can only be evaluated by transcriptomic analysis at present.
Discussion
The recent awareness of the central role of tumor-specific mutations in generating immune responses to cancer in patients has stimulated interest in defining these tumor rejection antigens and their T-cell responses. This has proven very difficult due to the patient-specific nature of these antigens and the fact that only a small number of mutations are demonstrably antigenic. Tumor-infiltrating lymphocytes (TIL) have been shown to contain highly enriched populations of these tumor-reactive T cells compared to peripheral blood. However, for common epithelial cancers, the frequency of such cells can still be low. The lack of cultured lines of patient-specific tumors as laboratory reagents has also hampered these studies. One comprehensive method for defining true neoantigens and cloning the T cells that recognize them involves WES and the reexpression of the mutations on autologous antigen-presenting cells in the form of transfected minigenes or exogenous synthetic peptides. Yet even this suffers from the fact that APCs do not reflect the sometimes-corrupted biology of tumors, and the TIL must be expanded in vitro for analysis. The phenotypic signature and method presented here uses TIL directly from fresh tumor specimens and dramatically accelerates the process of T-cell identification and TCR sequencing.
An important feature of the CD39+, CXCL13+ phenotypic signature is that it was able to identify both CD8+ and CD4+ neoantigen-reactive T cells from fresh tumor samples. There are data showing that CD4+ neoantigen reactive TIL constitute a greater portion of the reactivity in the common adenocarcinomas than in melanoma where most studies have focused (Parkhurst et al., 2016). The possibility of underestimating the frequency of both CD8+ and CD4+ neoantigen reactive TIL found by this approach is also significant because screening signature-positive TCRs with WES, TMGs, and mutated peptides would miss non-mutated post-translationally modified epitopes, out-of-frame gene products, and products of introns, all of which are known to be immunogenic in tumors (Haen et al., 2020). In addition, the screening method used only looks for mutated epitopes, excluding other antigens such as tumor-germline and tissue differentiation antigens.
CXCL13 (initially described as a B-cell chemotactic factor) is known to be highly expressed in many cancers, including metastatic melanoma (Tirosh et al., 2016), liver (Zheng et al., 2017b), ovarian (Yang et al., 2021), breast (Panse et al., 2008) and NSCLC (Litchfield et al., 2021a; Thommen et al., 2018). The high frequency of CXCL13 expression by our neoantigen reactive TIL contrasts to the overall frequency of CXCL13 expression in all TIL (ranging from 3.6-10.8%). Although some CD39+ CXCL13+ TCR clonotypes did not recognize neoantigens, we cannot rule out that they are recognizing other types of tumor-associated antigens or neoantigens missed by our classical screening method. In one NSCLC study (Thommen et al., 2018), the authors discovered CXCL13 is expressed in T cells highly expressing PD-1 and suggested its role in immune cell recruitment to tertiary lymphoid structures. In another NSCLC study (Litchfield et al., 2021b), the authors showed highly selective expression of CXCL13 in T cells purified by a neoantigen MTFR2 multimer and further identified CXCL13 as the best marker of the T-cell-intrinsic features of immune-checkpoint inhibitor (ICI) sensitivity in the study on over 1,000 patients across seven tumor types. Interestingly, in another study on the ICI sensitivity in NSCLC, CXCL13 was one of the genes most highly correlated with neoantigen-reactive TIL from patients without major pathological responses (Caushi et al., 2021) yet was nevertheless reported to be associated with improved overall survival (Huang et al., 2019). In these studies, the phenotype of reactive cells was studied as a correlate of clinical responses to checkpoint inhibition. In addition, the majority of studies carried out to date have focused on CD8+ T cells and none of these studies validated their signatures by demonstrating that they could prospectively detect neoantigen-reactive T-cell clonotypes or immune responses to mutated epitopes. One difficulty of analyzing CXCL13+ cells is its relatively low frequency in TIL. CITE-seq analysis was helpful in selecting cell surface proteins that are associated with CXCL13 expression. Our analysis suggests the usefulness of FACS sorting by itself for enriching CXCL13+ T cells. Easier ways to screen single cells or better enrich populations for CXCL13 expression are areas of future investigation. Although CD39 was the most consistent marker in the signature, other markers associated with exhaustion were frequently elevated in neoantigen-reactive T cells. Many investigators postulate that chronically stimulated T cells can diverge into a ‘progenitor exhausted’ subset (typically PD1hiTIM3lowTCF1+) with proliferative potential and a ‘terminally exhausted’ subset (typically PD1hiTIM3hiTCF1−) that can no longer proliferate (Blank et al., 2019; Wherry and Kurachi, 2015). Studies of bulk melanoma TIL (where a high mutation rate and a high frequency of neoantigen reactivity may obviate the need to identify and select neoantigen reactive clonotypes) have shown that T cells with less differentiation and more “stemness” are associated with both responses to checkpoint inhibitors and TIL adoptive therapy (Krishna et al., 2020; Miller et al., 2019; Sade-Feldman et al., 2018; Siddiqui et al., 2019). These findings do not necessarily conflict with the results of this study. Again, the goal of other studies was to predict responses to checkpoint inhibition or TIL administration, whereas the goal of this study was to detect and define neoantigen reactivity. The TCRs we identify are verified to be neoantigen reactive, but the epigenetic state of the cells in which those TCRs are found may not have high therapeutic potential as they may represent a terminally differentiation population that possess a poor proliferative potential. On the other hand, stem-like populations of TIL with neoantigen reactivity presumably exist in relatively small numbers in fresh tumors, rendering their isolation more difficult. One solution to this problem is to use the cloned TCRs from exhausted TIL to genetically reconstruct younger T cells from PBL for adoptive therapy, and a clinical protocol pursuing this is in progress (Clinicaltrials.gov ID: NCT03412877).
T cells recognizing tumor-specific mutated epitopes appear to be the primary drivers of the immune response to cancer in patients. The phenotype associated with neoantigen-reactive un-cultured TIL from NSCLC appears to be able to enrich such cells greatly. These findings in TIL from NSCLC need to be expanded to other tumor types, but preliminary data from gastrointestinal cancers support the importance of CD39 and CXCL13. Additional studies also need to be done to determine the impact of current immunotherapies on this signature. Although many attributes of T cells may contribute to their ability to reject tumors, antigen recognition is paramount. The generalizability of signatures designed in this study needs further confirmation due to the limited number of cases analyzed. However, having a rapid method with high sensitivity and specificity for identifying clonotypes with neoantigen reactivity represents a potentially valuable tool for in vitro studies and may lead to the development of effective cancer immunotherapies based upon the adoptive cell transfer of neoantigen-reactive T cells.
STAR Methods
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources should be directed to and will be fulfilled by the lead contact, Ken-ichi Hanada (hanada@nih.gov).
Materials availability
Plasmids generated in this study will be made available upon request and completion of a Material Transfer Agreement.
Data and code availability
scCITE-seq (gene expression and feature barcoding) and scVDJ-seq data have been deposited at dbGaP portal and are publicly available. Accession number is phs002792.v1.p1 (https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs002792.v1.p1)
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Human samples
NSCLC tumor samples and peripheral blood mononuclear cells were derived from patients enrolled on a clinical protocol (NCT02133196) approved by the institutional review board of the National Cancer Institute, National Institutes of Health, Bethesda USA. Informed consent for tissue usage for the research was obtained in the patient enrollment to the clinical protocol.
METHOD DETAILS
Culture of Tumor-Infiltrating Lymphocytes (TIL)
Surgically resected tumors were cut into 2-4 mm fragments and placed individually into 24 wells of a 24-well plate containing 2 ml of complete medium consisted of RPMI1640 with 10% human serum, 2 mM L-glutamine, 25 mM HEPES, and 10 μg/ml gentamicin and human interleukin-2 (6000 IU/ml). 24 fragment cultures were grown separately, and after the initial outgrowth (between 2-4 weeks), the majority of TIL were frozen for the patient treatment purpose. Approximately 1x106 cells left in culture were grown till enough cells (~4e6 cells/fragment) grew for this study.
Alignment, Processing, and Variant calling
Whole exome sequencing (WES) and RNA-seq on tumor tissue and WES on peripheral blood cells were done at the Surgery Branch, NCI. Alignment and variant calling were done as previously described (Parkhurst et al., 2019). Alignments were performed using novoalign MPI from novocraft (http://www.novocraft.com/) to human genome build hg19. Duplicates were marked using Picard’s MarkDuplicates tool. Insertion deletion (Indel) realignment and base recalibration was carried out according to the GATK best practices workflow (https://www.broadinstitute.org/gatk/). Post cleanup of data, samtools mpileup (http://samtools.sourceforge.net) was used to create pileup files and Varscan2, (http://varscan.sourceforge.net), SomaticSniper (http://gmt.genome.wustl.edu/packages/somatic-sniper/), Strelka (https://sites.google.com/site/strelkasomaticvariantcaller/), and Mutect (https://www.broadinstitute.org/gatk/). Following callers VCF files were merged using GATK CombineVariants tools and annotated using Annovar (http://annovar.openbioinformatics.org). Variants were then annotated using Annovar (http://annovar.openbioinformatics.org).
Identification of neoantigen-reactive TCR
Preparation of immature dendritic cells: Immature dendritic cells were prepared by the cell culture flask adherence method. Briefly, the apheresis sample was thawed into 50 ml of AIM-V medium with DNase I (100 Kunitz U/ml) and plated at the cell density of ~1.5 x 106 cells/cm2 (typically 3 x 108 cells in a T175 flask). After 2-hour incubation at 37°C, 5% CO2, non-adherent cells were removed by rinsing with PBS. 25 ml of AIM-V medium with human GM-CSF (1,000 U/ml Peprotech) and human IL-4 (1,000 U/ml, Peprotech) was added. On days 2 and 4, GM-CSF (25,000 U) and IL-4 (25,000 U) were replenished, and DC were frozen on Day 6 in CryoStor CS10 (Biolife Solutions).
Presentation of class I MHC-restricted epitopes
For each non-synonymous mutation identified by WES and RNAseq, a minigene encoding the corresponding amino acid change flanked by 12 amino acids of the wild-type protein sequence (25 amino acids in total) was designed. Multiple (typically less than 10) minigenes were strung together to generate TMG constructs. For insertion/deletions (indels), minigenes encoding the frame-shifted sequences until the next stop codon were prepared. After the codon optimization and the addition of ATG preceded by the Kozak consensus to the 5’ end, double-stranded DNA fragments were synthesized (gBlocks, Integrated DNA Technologies) and cloned into the pCMV-Sport7 vector. The vector pCMV-Sport7 was modified from pCMV-Sport6 to encode the MAGEA3 HLA-DP4 epitope (Yao et al., 2016) in-frame with TMG at its 3’ terminus. Recognition of the MAGEA3 epitope by HLA-DP4-restricted MAGEA3-reactive T cells served as the positive control of the successful RNA synthesis, antigen processing, and presentation for HLA-DP4+ patients. For DP4− patients, allogeneic DP4+ EBVB were used for the quality control. pCMV-Sport7-TMG-MAGEA3 plasmid constructs were linearized by NotI for in vitro transcription by T7 RNA polymerase (mMESSAGE mMACHINE T7 Transcription Kit, ThermoFisher Scientific) followed by poly(A) addition (Poly(A) Tailing Kit, ThermoFisher Scientific). After confirming the size of RNA by TapeStation System (Agilent), mRNA was electroporated to dendritic cells (5x105-2x106 DC, 8μg mRNA in 100μl OPTI-MEM) by a BTX ECM 830 electroporator (150V, 10 ms, and 1 pulse in 2mm gap cuvettes). Electroporated DC were transferred to an ultra-low attachment 24-well plate (Corning, 3473) with 2 ml of AIM-V containing GM-CSF 1,000 IU/ml and IL-4 400 IU/ml and incubated at 37°C, 5% CO2. Co-culture with TIL was set up 4 hours after the electroporation.
Class II MHC-restricted epitope presentation by DC
For single nucleotide variants, 25-mer peptides with the mutated amino acid in the center flanked by 12 wild-type amino acids were synthesized (Genscript or in-house). For frame-shift mutations, series of 5 amino acid-overlapping 25-mer peptides were synthesized to cover the whole open reading frame. Typically, less than 10 peptides were mixed to create peptide pools, and each pool was pulsed onto DC at the concentration of 1μM for each peptide. Two hours after adding peptides, DC were washed three times and used to set up co-culture with TIL.
Co-culture and ELISPOT
Co-culture for IFNγ ELISPOT assay was set up using ~2x104 dendritic cells and 5x104 TIL in the 96-well format. ELISPOT development was performed using reagents from Mabtech following the manufacturer’s protocol. Spots were counted by the ImmunoSpot analyzer (Cellular Technology Limited).
FACS analysis of co-cultured TIL
20~24-hour co-cultured cells were harvested from the 96-well format ELISPOT plates by pipetting, transferred to U-bottom 96-well plates, and spun down at 2,500 RPM for 2 minutes. After removing the supernatant, an antibody cocktail (anti-CD3 APC-H7, clone SK7, BD Bioscience, 0.4μl/well, anti-CD8 PE-Cy7, clone SK1, BD Bioscience, 0.05μl/well, anti-CD4 PE, clone SK3, BD Bioscience, 0.3μl/well, anti-CD137 APC, clone 4B4-1, BD Bioscience, 0.5μl/well, and anti-CD134 FITC, clone ACT35, BD Bioscience, 0.5μl/well) was added. After a 30 minute-incubation at 4°C, cells were washed once and were analyzed on a FacsCanto II (BD Bioscience) flow cytometer.
Identification of Class I MHC-restricted antigens
Once antigenic TMG was identified, we created plasmids with staggered minigenes such that plasmid 1 contains minigenes 1&2, plasmid 2 minigenes 2&3, and so on. By screening these plasmids by the RNA electroporation method described above, we identified the antigenic minigene. From the minigene, we synthesized predicted epitope peptides (Gartner et al., 2021) and tested their recognition by TIL by pulsing onto the autologous DC. For the peptide titration assay, the autologous dendritic cells were pulsed with serially diluted peptides in the Ultra-Low attachment U-bottom 96-well plates (Corning 7007) for 2 hours at room temperature, washed three times, and co-cultured with cognate T cells. ELISPOT and FACS analysis were performed as described above.
Identification of class II MHC-restricted antigens
Once antigenic peptide pools were identified, these pools were parsed by assaying peptides individually. Autologous DC (2x104 cells) were pulsed with each peptide at 1μM concentration for 2 hours, at 37°C, 5% CO2 in Ultra-low attachment U-bottom 96-well plates (Corning 7007), and after washing three times, DC were transferred to ELISPOT plates, and T cells (5-10x104 cells) were added. After 20-24 hour-co-culture, ELISPOT and FACS analysis were performed as described above.
TCR cloning from FACS-sorted TIL
TIL containing a neoantigen reactivity (≤1x106 cells) were co-cultured with DC (~ 20% cell number of T cells) pulsed with the cognate peptide in a 24-well Ultra-low attachment plate. After 20-24 hour co-culture, cells were stained with anti-CD3 PE, clone SK7, 10 μl, BD Biosciences, anti-CD8 FITC, clone SK1, 10 μl, BD Biosciences, anti-CD4 Alexa Fluor 700, clone SK3, 25 μl, BD Biosciences, and anti-CD137, clone 4B4-1, 5 μl, BD Biosciences. The population that up-regulated CD137 was single-cell sorted on a SONY SH800 or MA900 cell sorter. For the isolation of TCR genes, cell lysis, RT-reaction with template switch, and whole cDNA amplification was done following the SmartSeq2 protocol (Picelli et al., 2014) except a sequence modification to TSO (5’-AAGCAGTGGTATCAACGCAGAGTACATTAATACGACTCACTATAGrGrG+G-3’). Amplified cDNA (2μl each) was used as a template for PCR-amplifying TRA and TRB, respectively. PCR amplification of TRAV was done by the TSO-forward primer 5’-CAACGCAGAGTACATTAATACGAC-3’ and the TRA constant region reverse primer 5’-CTCCAGGCCACAGCACTGTTG-3’. TRBV amplification was done by the TSO-forward primer and the TRB constant region reverse primer 5’-CATTCACCCACCAGCTCAGCTC-3’. TRAV PCR amplicons were sequenced by the TRAC sequence primer 5’-GACTTGTCACTGGATTTAGAGTCTC-3’ and TRBV PCR amplicons by the TRBC sequence primer 5’-GGGAGATCTCTGCTTCTGATGGC-3’. Once the TRAV and TRBV usages were identified, full-length TRA and TRB were PCR amplified using the cDNA created by the SmartSeq2 method as the template. Full-length TRA and TRB were cloned into the MSGV1 vector individually by the InFusion technology (Takara Clontech).
Functional test of TCRs and their information
TCR gene-transduction was conducted as previously described (Tran et al., 2015) except in this study, MSGV1 TRA and MSGV2 TRB retroviral plasmids were individually transfected to 293gp cells with the envelope-encoding plasmid RD114. Two viruses were harvested, mixed and loaded onto Retronectin (Takara Clontech)-coated plates. 5 days after the retroviral TCR transduction, T cells were co-cultured with autologous DC pulsed with HPLC-purified mutated or wild-type peptides (minimal determinant for CD8 and 25-mers for CD4). Peptide pulsing and T-cell co-culture were done in the same way as in TIL described above. The readout was by IFNγ ELISA (R&D Systems) following the manufacturer’s protocol. Structural information of TCR and peptide sequences used for the assay are provided in the supplement table 3 and 10X barcodes are listed in the supplement table 4.
Single-Cell Analysis by CITE-seq
Part of fresh tumor samples were used for TIL culture described above and the rest was cut into 3-4 mm pieces and were mechanically filtered through 2 mm metal mesh. Filtered tumor cells were suspended in RPMI1640 medium with Collagenase (1 mg/ml, Sigma C-5138) and DNase I (100 Kunitz units/ml) and processed by GentleMACS (Miltenyi) to make single-cell suspensions. Cells were processed by Ficoll to remove dead cells and were frozen in the cell freezing medium CryoStor CS10 (Biolife Solutions) for the later analysis. To prepare single-cell analysis libraries, frozen single-cell suspension tumor cells were thawed and cultured overnight in RPMI1640 with 10% human type-AB serum, 1X Antibiotic-Antimycotic (Thermo Fisher Scientific), Gentamicin (10 μg/ml), Deoxyribonuclease I (30 Kunitz units/ml) without any cytokine.
Approximately 1 million cells were incubated with mouse polyclonal immunoglobulin for blocking and were stained with TotalSeq-C FBC antibodies (anti-CD4, CD8, CD62L, CD45RA, CD45RO, CD69, CD103, CD134, CD137, CD39, PD1 and TIM3, 2 μl each) and anti-CD3 APC-H7 antibody (clone SK7, BD Biosciences, 20 μl) at 4°C for 30 minutes. After a centrifuge (1,000g, 5 min, 4°C), cells were suspended in 4°C RPMI with 10% FBS 500μl and CD3+ cells were sorted by a SONY SH800 or MA900 cell sorter. Sorted cells were washed twice and resuspended in 100 μl PBS. Approximately 5x104 CD3+ cells were processed by a 10X Genomics Chromium controller to capture cells for three single-cell libraries targeting 6,000 cell capture per library. (For patient 4, one library was lost due to clogging of the chip.) Gene expression, V(D)J, and Feature Barcoding libraries were prepared following the manufacturer’s protocol. For the CITE-seq analysis, alignment of scRNA sequencing reads to the human genome was conducted with 10X CellRanger using the GRCh38 human genome reference. H5 files of aggregated gene expression (GEX) and feature barcoding (FBC) from Cell Ranger (10X Genomics) were imported to Partek Flow (version 10.0.21.0204). After splitting GEX and FBC data, outliers for each dataset were removed based on total counts, detected feature number and %mitochondrial counts. Data were normalized using the center log-ratio technique with the pseudocount of 1. Dimensionality reduction was performed by 9 and 15 principal components for FBC and GEX, respectively, and projections were created using the tSNE algorithm (Maaten and Hinton, 2008). CDR3A and CDR3B sequences of neoantigen-reactive TIL identified by the classic screening method were linked to 10X barcodes by the Loupe browser with VDJ data to create annotation files. These annotation files were imported to Partek Flow to analyze the properties of neoantigen-reactive T cells. Gene expressions were compared by the Hurdle model and protein expressions by ANOVA. For either case, FDR≤0.01 and (FOLD)2≥4 were used as the selection threshold. TCR frequency analysis was done by starting from the 10X cellranger vdj Filtered_contig_annotations file, considering only cells with productive TCRβ. TCRs were grouped and counted for unique combinations of V region, D region, J region, CDR3, and C region. Frequencies were determined by dividing unique TCRβ by total productive TCRβ.
CDR3B deep sequencing
PBL pellets (≤1x106 cells) obtained around the time of tumor resection were sent to Adaptive Biotechnologies (Seattle, USA) for ImmunoSEQ TCRB deep sequencing. The numbers of total productive CDR3B were: patient 1: 125,214, patient 2: 266,713, patient 3: 194,454, and patient 4: 318,838.
Selection of TCRs for analysis
First, T cells were split into two based on the CD39 protein expression. Next, for CD39+ clonotypes, about 10 high-frequency clonotypes were selected after excluding clonotypes with no CXCL13 expression. For CD39- clonotypes, about 10 top frequency clonotypes were selected for analysis. TCR sequence and 10X barcode information is provided in the supplement table 3 and 4.
Evaluation of selected TCRs
DNA sequences of the candidate neoantigen-reactive TCRs were obtained by the Loupe VDJ software (10X Genomics), and TCR sequences were designed in the form of TRBV domain fused to the mouse TRB constant chain, TRAV to the mouse TRA constant domain separated by the furin cleavage site RAKR, linker SGSG and the P2A sequence (final linker sequence: RAKR-SGSG-ATNFSLLKQAGDVEENPGP). Double-stranded DNAs encoding these sequences were synthesized and cloned into the MSGV1 vector. Autologous T cells were retrovirally transduced with TCRs and were co-cultured with DC presenting mutated epitopes by the method described above. T-cell activation was assessed by IFNγ ELISA (R&D Systems).
Signature analysis
Normalized read counts for CD4+ and CD8+ T cells were transferred to R (v3.6.1) for additional analysis with the Seurat package (v3) (Stuart et al., 2019). Based on the cell surface expression pattern, we selected CD39 + PD1 + CD103 − CD4 − CD8A − CD45RA − CD62L − CD134 for CD8, and CD39 + PD1 − CD45RA − CD8 − CD137 for CD4. The counts of surface protein were then combined into a linear signature to generate a single CITESeq-derived score for each cell. Surface proteins that were up-regulated in neoantigen-reactive T cells contributed positively to the signature, while downregulated proteins detracted from the signature score; as such, higher total scores were generally associated with the likelihood of a cell being neoantigen-reactive. Differential expression analysis of genes was performed between neoantigen reactive T cells and non-reactive T cells using the MAST algorithm(Finak et al., 2015). Significantly dysregulated genes were identified in CD8+ T cells as having an adjusted p-value < 0.05, log2 fold change > 0.25, and expression in at least 10% of cells in one cohort; the expression threshold was reduced to 5% of cells for the CD4+ T cell analysis. This analysis was performed for each clonotype of neoantigen-reactive T cells against its respective non-reactive T cells. Genes that were consistently significantly dysregulated for each clonotype comparison were then combined into a linear expression signature to generate an expression score for each cell. To generate a joint transcript- and CITESeq-based signature for the individual patients, the transcript scores and CITESeq scores were independently z-scaled to retain signature rankings while providing similar weight to each of the two scoring systems. The sum of the z-scores for each signature was used to give a joint score for each cell. The strength of the expression signatures in identifying likely neoantigen-reactive T cells was evaluated using the “pROC” package to calculate the area under the curve (AUC) of the receiver operating characteristic (ROC) (Robin et al., 2011) as determined at the per cell level. The similarity of ROC curves was evaluated using DeLong’s test for two correlated ROC curves, as implemented in the pROC package. Additional visualization was performed using the “pheatmap” package in R.
QUANTIFICATION AND STATISTICAL ANALYSIS
GraphPad Prism (version 9.1.2) was used to draw violin plots and bar graphs and statistical analysis in Figure 3, Supplement Figures 4 and 5. For violin plots, non-parametric analyses (Kruskal-Wallis test followed by Dunn’s multiple comparison test or Mann-Whitney test) were performed. For dot plots (cell surface protein expression by FBC), parametric tests (one-way ANOVA followed by Dunnett’s multiple comparison test or Unpaired t-test) were performed. Partek Flow (version 10.0.21.0204) was used for Figure 2a (ANOVA for protein data and Hurdle model for gene expression data).
Supplementary Material
Table S3 T-cell receptors synthesized for screening and peptide sequences used for functional analyses, related to Figures 4 and 5.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-human CD3 APC-H7 (clone SK7) | BD Biosciences | Cat# 560275 |
| Anti-human CD4 PE (clone SK3) | BD Biosciences | Cat# 565999 |
| Anti-human CD8 PE/Cy7 (clone SK1) | BD Biosciences | Cat# 335805 |
| Anti-human CD134 FITC (clone ACT35) | BD Biosciences | Cat# 555837 |
| Anti-human CD137 APC (clone 4B4-1) | BD Biosciences | Cat# 550890 |
| TotalSeq-C0045 anti-human CD4 | Biolegend | Cat# 350006 |
| TotalSeq-C0080 anti-human CD8a | Biolegend | Cat# 301071 |
| TotalSeq-C0063 anti-human CD45RA | Biolegend | Cat# 304163 |
| TotalSeq-C0087 anti-human CD45RO | Biolegend | Cat# 304259 |
| TotalSeq-C0147 anti-human CD62L | Biolegend | Cat# 304851 |
| TotalSeq-C0146 anti-humna CD69 | Biolegend | Cat# 310951 |
| TotalSeq-C0145 anti-human CD103 | Biolegend | Cat# 350233 |
| TotalSeq-C0158 anti-human CD134 | Biolegend | Cat# 350035 |
| TotalSeq-C0137 anti-human CD137 | Biolegend | Cat# 309839 |
| TotalSeq-C0176 anti-human CD39 | Biolegend | Cat# 328237 |
| TotalSeq-C0088 anti-human CD279 | Biolegend | Cat# 329963 |
| TotalSeq-C0169 anti-human CD366 | Biolegend | Cat# 345049 |
| Human IFNγ ELISPOT capture antibody | Mabtech | Cat# 3420-6-1000 |
| Human IFNγ ELISPOT biotinylated antibody | Mabtech | Cat# 3420-3-1000 |
| Anti-human CD3 antibody (OKT3) | Invitrogen | Cat# 13-0037-85 |
| Biological Samples | ||
| Patient tumor and peripheral blood samples | Clinical trial | Trial ID: NCT02133196 |
| Chemicals, peptides, and recombinant proteins | ||
| Custom double stranded DNA for tandem minigenes | IDT DNA | Custom order |
| Custom peptides | Genscript | Custom order |
| Custom peptides | In-house | Custom order |
| Custom retroviral plasmid preparation | Genscript | Custom order |
| Recombinant Human IL-2 (Proleukin) | Prometheus | 65483-116-07 |
| Recombinant Human IL-4 | Peprotech | Cat# 200-04 |
| Recombinant Human GM-CSF | Peprotech | Cat# 300-03 |
| Collagenase | Sigma Aldrich | Cat# C5138 |
| DNase1 | Sigma Aldrich | Cat# D5025 |
| Human AB Serum | Valley Biomedical | N/A |
| Propidium Iodide | Sigma Aldrich | Cat# P4170 |
| CryoStor CS10 | BioLife Solutions | Cat# 210102 |
| Streptavidin-ALP for ELISPOT | Mabtech | Cat# 3310-10-1000 |
| Critical commercial assays | ||
| human IFNγ DuoSet ELISA | R&D Systems | Cat#DY285B |
| Chromium Next GEM Single Cell 5’ Kit v2 | 10X Genomics | Cat# 1000265 |
| Chromium Next GEM Chip K Single Cell Kit | 10X genomics | Cat# 1000287 |
| 5’ Feature Barcode Kit v2 | 10X genomics | Cat# 1000256 |
| Chromium Single Cell Human TCR Amplification Kit | 10X Genomics | Cat# 1000252 |
| immunoSEQ Assays, Human TCRB | Adaptive Biotech | Deep Sequencing |
| Deposited data | ||
| Fresh Tumor T-cells Single-cell analysis data | This Paper | dbGaP: phs002792.v1.p1 |
| Oligonucleotides | ||
| TSO-TCR primer: 5’-AAGCAGTGGTATCAACGCAGAGTACATTAATACGACTCACTATAGrGrG+G-3’ | This Paper | N/A |
| TSO-forward primer: 5’-CAACGCAGAGTACATTAATACGAC-3’ | This Paper | N/A |
| TRA constant region reverse primer 5’-CTCCAGGCCACAGCACTGTTG-3’ | This Paper | N/A |
| TRB constant region reverse primer 5’-CATTCACCCACCAGCTCAGCTC-3’ | This Paper | N/A |
| Software and algorithms | ||
| Cellranger | 10x Genomics | N/A |
| Loupe VDJ browser | 10x Genomics | N/A |
| Loupe Genome browser | 10X Genomics | N/A |
| Partek Flow | Partek | Version 10.0.21.0204 |
| FlowJo | Tree Star | Version 10 |
| GraphPad Prism | GraphPad | Version 9.1.2 |
| R | R project | Version 3.6.1 |
| pROC package | R project | Version 1.18.0 |
| pheatmap package | R project | Version 1.0.12 |
| Seurat package | Satija lab | Version 3 |
| Other | ||
| GentleMACS Octo Dissociator | Miltenyi Biotech | |
| GentleMACS C Tubes | Miltenyi Biotech | 130-093-237 |
| Single-Cell Sorter | SONY | MA900 |
| Single-Cell Sorter | SONY | SH800 |
| Flow Cytometer | BD Biosciences | FACSCanto II |
| Electroporation system | BTX | ECM 830 |
| TapeStation System | Agilent | 2200 |
| Immunospot Analyzer (For ELISPOT) | Cellular Technology Limited | |
| Peptide Synthesizer | Intavis | Res Pep SL |
| ELISPOT plates | Millipore | MAIPS4W10 |
| Ultra-low attachment 24-well plates | Corning | 3473 |
| Ultra-low attachment 96-well U-bottom plates | Corning | 7007 |
Highlights.
T-cells in fresh NSCLC tumor was analyzed by CITE-seq with TCR-seq.
Neoantigen-reactive T cells are CD39 protein+, CXCL13+ and high-frequency clonotype
The signature can identify both CD4 and CD8 neoantigen-reactive T cells
This method can expedite the personalized neoantigen-reactive T cell therapy
Acknowledgments
We thank the Surgery Branch TIL Laboratory, and clinical team for generating TIL, and patients enrolled in our clinical protocols. We also would like to thank Samuel Chatmon for his technical help with NGS library construction. Support from CCR Single Cell Analysis Facility and CCR Collaborative Bioinformatics Resource were funded in whole or in part by FNLCR Contract HHSN261200800001E. This work utilized the computational resources of the NIH HPC Biowulf cluster (http://hpc.nih.gov). The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Funding:
This research was supported by the Center for Cancer Research intramural research program of the National Cancer Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
The authors declare no competing interests. A non-provisional international patent has been filed based on the work described in this study.
References
- Acharya N, Madi A, Zhang H, Klapholz M, Escobar G, Dulberg S, Christian E, Ferreira M, Dixon KO, Fell G, et al. (2020). Endogenous Glucocorticoid Signaling Regulates CD8+ T Cell Differentiation and Development of Dysfunction in the Tumor Microenvironment. Immunity 53, 658–671.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baixauli F, Martín - Cófreces NB, Morlino G, Carrasco YR, Calabia - Linares C, Veiga E, Serrador JM, and Sánchez - Madrid F (2011). The mitochondrial fission factor dynamin - related protein 1 modulates T - cell receptor signalling at the immune synapse. Embo J 30, 1238–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blank CU, Haining WN, Held W, Hogan PG, Kallies A, Lugli E, Lynn RC, Philip M, Rao A, Restifo NP, et al. (2019). Defining ‘T cell exhaustion.’ Nat Rev Immunol 19, 665–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Bruggen P, Traversari C, Chomez P, Lurquin C, Plaen ED, den Eynde BV, Knuth A, and Boon T (1991). A gene encoding an antigen recognized by cytolytic T lymphocytes on a human melanoma. Science 254, 1643–1647. [DOI] [PubMed] [Google Scholar]
- Caushi JX, Zhang J, Ji Z, Vaghasia A, Zhang B, Hsiue EH-C, Mog BJ, Hou W, Justesen S, Blosser R, et al. (2021). Transcriptional programs of neoantigen-specific TIL in anti-PD-1-treated lung cancers. Nature 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang TO, Ogunniyi A, Barbee MS, and Drilon A (2015). Pembrolizumab for the treatment of PD-L1 positive advanced or metastatic non-small cell lung cancer. Expert Rev Anticanc 16, 13–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djenidi F, Adam J, Goubar A, Durgeau A, Meurice G, Montpréville V. de, Validire P, Besse B, and Mami-Chouaib F (2015). CD8 + CD103 + Tumor–Infiltrating Lymphocytes Are Tumor-Specific Tissue-Resident Memory T Cells and a Prognostic Factor for Survival in Lung Cancer Patients. J Immunol 194, 3475–3486. [DOI] [PubMed] [Google Scholar]
- Duhen T, Duhen R, Montler R, Moses J, Moudgil T, Miranda N.F. de, Goodall CP, Blair TC, Fox BA, McDermott JE, et al. (2018). Co-expression of CD39 and CD103 identifies tumor-reactive CD8 T cells in human solid tumors. Nat Commun 9, 2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esfahani MS, Lee LJ, Jeon Y-J, Flynn RA, Stehr H, Hui AB, Ishisoko N, Kildebeck E, Newman AM, Bratman SV, et al. (2019). Functional significance of U2AF1 S34F mutations in lung adenocarcinomas. Nat Commun 10, 5712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finak G, McDavid A, Yajima M, Deng J, Gersuk V, Shalek AK, Slichter CK, Miller HW, McElrath MJ, Prlic M, et al. (2015). MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol 16, 278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartner JJ, Parkhurst MR, Gros A, Tran E, Jafferji MS, Copeland A, Hanada K-I, Zacharakis N, Lalani A, Krishna S, et al. (2021). A machine learning model for ranking candidate HLA class I neoantigens based on known neoepitopes from multiple human tumor types. Nat Cancer 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gros A, Robbins PF, Yao X, Li YF, Turcotte S, Tran E, Wunderlich JR, Mixon A, Farid S, Dudley ME, et al. (2014). PD-1 identifies the patient-specific CD8+ tumor-reactive repertoire infiltrating human tumors. J Clin Invest 124, 2246–2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gros A, Parkhurst MR, Tran E, Pasetto A, Robbins PF, Ilyas S, Prickett TD, Gartner JJ, Crystal JS, Roberts IM, et al. (2016). Prospective identification of neoantigen-specific lymphocytes in the peripheral blood of melanoma patients. Nat Med 22, 433–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubin MM, Zhang X, Schuster H, Caron E, Ward JP, Noguchi T, Ivanova Y, Hundal J, Arthur CD, Krebber W-J, et al. (2014). Checkpoint blockade cancer immunotherapy targets tumour-specific mutant antigens. Nature 515, 577–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haen SP, Löffler MW, Rammensee H-G, and Brossart P (2020). Towards new horizons: characterization, classification and implications of the tumour antigenic repertoire. Nat Rev Clin Oncol 17, 595–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitson JP, West KA, James KR, Rani GF, Dey N, Romano A, Brown N, Teichmann SA, Kaye PM, and Lagos D (2020). Malat1 Suppresses Immunity to Infection through Promoting Expression of Maf and IL-10 in Th Cells. J Immunol 204, ji1900940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang AC, Orlowski RJ, Xu X, Mick R, George SM, Yan PK, Manne S, Kraya AA, Wubbenhorst B, Dorfman L, et al. (2019). A single dose of neoadjuvant PD-1 blockade predicts clinical outcomes in resectable melanoma. Nat Med 25, 454–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imielinski M, Berger AH, Hammerman PS, Hernandez B, Pugh TJ, Hodis E, Cho J, Suh J, Capelletti M, Sivachenko A, et al. (2012). Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell 150, 1107–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inozume T, Hanada K, Wang QJ, Ahmadzadeh M, Wunderlich JR, Rosenberg SA, and Yang JC (2010). Selection of CD8+PD-1+ Lymphocytes in Fresh Human Melanomas Enriches for Tumor-reactive T Cells. Journal of Immunotherapy 33, 956–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami Y, and Rosenberg SA (1997). Human tumor antigens recognized by T-cells. Immunol Res 16, 313–339. [DOI] [PubMed] [Google Scholar]
- Kortekaas KE, Santegoets SJ, Sturm G, Ehsan I, van Egmond SL, Finotello F, Trajanoski Z, Welters MJP, van Poelgeest MIE, and van der Burg SH (2020). CD39 Identifies the CD4+ Tumor-Specific T-cell Population in Human Cancer. Cancer Immunol Res 8, 1311–1321. [DOI] [PubMed] [Google Scholar]
- Krishna S, Lowery FJ, Copeland AR, Bahadiroglu E, Mukherjee R, Jia L, Anibal JT, Sachs A, Adebola SO, Gurusamy D, et al. (2020). Stem-like CD8 T cells mediate response of adoptive cell immunotherapy against human cancer. Science 370, 1328–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, Skora AD, Luber BS, Azad NS, Laheru D, et al. (2015). PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. The New England Journal of Medicine 372, 2509–2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, van der Leun AM, Yofe I, Lubling Y, Gelbard-Solodkin D, van Akkooi ACJ, van der Braber M, Rozeman EA, Haanen JBAG, Blank CU, et al. (2018). Dysfunctional CD8 T Cells Form a Proliferative, Dynamically Regulated Compartment within Human Melanoma. Cell 176, 775–789.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litchfield K, Reading JL, Puttick C, Thakkar K, Abbosh C, Bentham R, Watkins TBK, Rosenthal R, Biswas D, Rowan A, et al. (2021a). Meta-analysis of tumor- and T cell-intrinsic mechanisms of sensitization to checkpoint inhibition. Cell 184, 596–614.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litchfield K, Reading JL, Puttick C, Thakkar K, Abbosh C, Bentham R, Watkins TBK, Rosenthal R, Biswas D, Rowan A, et al. (2021b). Meta-analysis of tumor- and T cell-intrinsic mechanisms of sensitization to checkpoint inhibition. Cell 184, 596–614.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandapathil M, Lang S, Gorelik E, and Whiteside TL (2009). Isolation of functional human regulatory T cells (Treg) from the peripheral blood based on the CD39 expression. J Immunol Methods 346, 55–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLane LM, Abdel-Hakeem MS, and Wherry EJ (2019). CD8 T Cell Exhaustion During Chronic Viral Infection and Cancer. Annu Rev Immunol 37, 1–39. [DOI] [PubMed] [Google Scholar]
- Miller BC, Sen DR, Abosy RA, Bi K, Virkud YV, LaFleur MW, Yates KB, Lako A, Felt K, Naik GS, et al. (2019). Subsets of exhausted CD8+ T cells differentially mediate tumor control and respond to checkpoint blockade. Nat Immunol 20, 326–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan RA, Dudley ME, Yu YYL, Zheng Z, Robbins PF, Theoret MR, Wunderlich JR, Hughes MS, Restifo NP, and Rosenberg SA (2003). High Efficiency TCR Gene Transfer into Primary Human Lymphocytes Affords Avid Recognition of Melanoma Tumor Antigen Glycoprotein 100 and Does Not Alter the Recognition of Autologous Melanoma Antigens. J Immunol 171, 3287–3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panse J, Friedrichs K, Marx A, Hildebrandt Y, Luetkens T, Bartels K, Horn C, Stahl T, Cao Y, Milde-Langosch K, et al. (2008). Chemokine CXCL13 is overexpressed in the tumour tissue and in the peripheral blood of breast cancer patients. Brit J Cancer 99, 930–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkhurst M, Gros A, Pasetto A, Prickett T, Crystal JS, Robbins P, and Rosenberg SA (2016). Isolation of T-Cell Receptors Specifically Reactive with Mutated Tumor-Associated Antigens from Tumor-Infiltrating Lymphocytes Based on CD137 Expression. Clin Cancer Res 23, 2491–2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkhurst MR, Robbins PF, Tran E, Prickett TD, Gartner JJ, Jia L, Ivey G, Li YF, El-Gamil M, Lalani A, et al. (2019). Unique Neoantigens Arise from Somatic Mutations in Patients with Gastrointestinal Cancers. Cancer Discov 9, 1022–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picelli S, Faridani OR, Björklund AK, Winberg G, Sagasser S, and Sandberg R (2014). Full-length RNA-seq from single cells using Smart-seq2. Nature Protocols 9, 171–181. [DOI] [PubMed] [Google Scholar]
- Quigley M, Pereyra F, Nilsson B, Porichis F, Fonseca C, Eichbaum Q, Julg B, Jesneck JL, Brosnahan K, Imam S, et al. (2010). Transcriptional analysis of HIV-specific CD8+ T cells shows that PD-1 inhibits T cell function by upregulating BATF. Nat Med 16, 1147–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, Lee W, Yuan J, Wong P, Ho TS, et al. (2015). Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science (New York, N.Y.) 348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins PF, Lu Y-C, El-Gamil M, Li YF, Gross C, Gartner J, Lin JC, Teer JK, Cliften P, Tycksen E, et al. (2013). Mining exomic sequencing data to identify mutated antigens recognized by adoptively transferred tumor-reactive T cells. Nat Med 19, 747–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez J-C, and Müller M (2011). pROC: an open-source package for R and S+ to analyze and compare ROC curves. Bmc Bioinformatics 12, 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rooij N, van Buuren MM, Philips D, Velds A, Toebes M, Heemskerk B, van Dijk LJ, Behjati S, Hilkmann H, Atmioui D, et al. (2013). Tumor exome analysis reveals neoantigen-specific T-cell reactivity in an ipilimumab-responsive melanoma. Journal of Clinical Oncology 31, e439–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sade-Feldman M, Yizhak K, Bjorgaard SL, Ray JP, Boer C.G. de, Jenkins RW, Lieb DJ, Chen JH, Frederick DT, Barzily-Rokni M, et al. (2018). Defining T Cell States Associated with Response to Checkpoint Immunotherapy in Melanoma. Cell 175, 998–1013.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma SV, Bell DW, Settleman J, and Haber DA (2007). Epidermal growth factor receptor mutations in lung cancer. Nature Reviews Cancer 7, 169–181. [DOI] [PubMed] [Google Scholar]
- Siddiqui I, Schaeuble K, Chennupati V, Marraco SAF, Calderon-Copete S, Ferreira DP, Carmona SJ, Scarpellino L, Gfeller D, Pradervand S, et al. (2019). Intratumoral Tcf1+PD-1+CD8+ T Cells with Stem-like Properties Promote Tumor Control in Response to Vaccination and Checkpoint Blockade Immunotherapy. Immunity 50, 195–211.e10. [DOI] [PubMed] [Google Scholar]
- Simoni Y, Becht E, Fehlings M, Loh C, Koo S-L, Teng K, Yeong J, Nahar R, Zhang T, and Kared H (2018). Bystander CD8+ T cells are abundant and phenotypically distinct in human tumour infiltrates. Nature 557, 575. [DOI] [PubMed] [Google Scholar]
- Simula L, Pacella I, Colamatteo A, Procaccini C, Cancila V, Bordi M, Tregnago C, Corrado M, Pigazzi M, Barnaba V, et al. (2018). Drp1 Controls Effective T Cell Immune-Surveillance by Regulating T Cell Migration, Proliferation, and cMyc-Dependent Metabolic Reprogramming. Cell Reports 25, 3059–3073.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder A, Makarov V, Merghoub T, Yuan J, Zaretsky JM, Desrichard A, Walsh LA, Postow MA, Wong P, Ho TS, et al. (2014). Genetic Basis for Clinical Response to CTLA-4 Blockade in Melanoma. The New England Journal of Medicine 371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelekati E, Chen Z, Manne S, Kurachi M, Ali M-A, Lewy K, Cai Z, Nzingha K, McLane LM, Hope JL, et al. (2018). Long-Term Persistence of Exhausted CD8 T Cells in Chronic Infection Is Regulated by MicroRNA-155. Cell Reports 23, 2142–2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM, Hao Y, Stoeckius M, Smibert P, and Satija R (2019). Comprehensive Integration of Single-Cell Data. Cell 177, 1888–1902.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thommen DS, Koelzer VH, Herzig P, Roller A, Trefny M, Dimeloe S, Kiialainen A, Hanhart J, Schill C, Hess C, et al. (2018). A transcriptionally and functionally distinct PD-1+ CD8+ T cell pool with predictive potential in non-small-cell lung cancer treated with PD-1 blockade. Nat Med 24, 994–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I, Izar B, Prakadan SM, Wadsworth MH, Treacy D, Trombetta JJ, Rotem A, Rodman C, Lian C, Murphy G, et al. (2016). Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 352, 189–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran E, Ahmadzadeh M, Lu Y-C, Gros A, Turcotte S, Robbins PF, Gartner JJ, Zheng Z, Li YF, Ray S, et al. (2015). Immunogenicity of somatic mutations in human gastrointestinal cancers. Science 350, 1387–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truong K-L, Schlickeiser S, Vogt K, Boës D, Stanko K, Appelt C, Streitz M, Grütz G, Stobutzki N, Meisel C, et al. (2019). Killer-like receptors and GPR56 progressive expression defines cytokine production of human CD4+ memory T cells. Nat Commun 10, 2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Maaten L, and Hinton GE (2008). Visualizing Data using t-SNE. Journal of Machine Learning Research 9, 2579–2605. [Google Scholar]
- Wang H, Guo Y, Dong Z, Li T, Xie X, Wan D, Jiang Z, Yu J, and Guo R (2020). Differential U2AF1 mutation sites, burden and co-mutation genes can predict prognosis in patients with myelodysplastic syndrome. Sci Rep-Uk 10, 18622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wherry EJ, and Kurachi M (2015). Molecular and cellular insights into T cell exhaustion. Nat Rev Immunol 15, 486–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Lu J, Zhang G, Wang Y, He M, Xu Q, Xu C, and Liu H (2021). CXCL13 shapes immunoactive tumor microenvironment and enhances the efficacy of PD-1 checkpoint blockade in high-grade serous ovarian cancer. J Immunother Cancer 9(1), e001136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao X, Lu Y-C, Parker LL, Li YF, El-Gamil M, Black MA, Xu H, Feldman SA, van der Bruggen P, Rosenberg SA, et al. (2016). Isolation and Characterization of an HLA-DPB1*04. J Immunother 39, 191–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacharakis N, Chinnasamy H, Black M, Xu H, Lu Y-C, Zheng Z, Pasetto A, Langhan M, Shelton T, Prickett T, et al. (2018). Immune recognition of somatic mutations leading to complete durable regression in metastatic breast cancer. Nat Med 24, 724–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng C, Zheng L, Yoo J-K, Guo H, Zhang Y, Guo X, Kang B, Hu R, Huang JY, Zhang Q, et al. (2017a). Landscape of Infiltrating T Cells in Liver Cancer Revealed by Single-Cell Sequencing. Cell 169, 1342–1356.e16. [DOI] [PubMed] [Google Scholar]
- Zheng C, Zheng L, Yoo J-K, Guo H, Zhang Y, Guo X, Kang B, Hu R, Huang JY, Zhang Q, et al. (2017b). Landscape of Infiltrating T Cells in Liver Cancer Revealed by Single-Cell Sequencing. Cell 169, 1342–1356.e16. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S3 T-cell receptors synthesized for screening and peptide sequences used for functional analyses, related to Figures 4 and 5.
Data Availability Statement
scCITE-seq (gene expression and feature barcoding) and scVDJ-seq data have been deposited at dbGaP portal and are publicly available. Accession number is phs002792.v1.p1 (https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs002792.v1.p1)
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.