Abstract
Corynebacterium glutamicum is able to biotransform demeton-S-methyl, an organophosphorus compound, during cometabolism with more readily metabolizable substrates. Among the cosubstrates used, fructose is the growth substrate that is most favorable for demeton-S-methyl biotransformation. The reaction mechanism of demeton-S-methyl biotransformation involves reductive cleavage of an S-C bond, which leads to accumulation of dimethyl thiophosphate in the culture medium.
Synthetic organophosphorus (OP) compounds are used extensively as agricultural and domestic pesticides and could be used as chemical warfare agents. Most of these xenobiotic compounds have common organic phosphorus-ester bonds. The extremely toxic military OP compounds, such as soman, sarin, and Vx [o-ethyl S-(diisopropylaminoethyl) methyl-phosphonothiolate], are phosphonofluoridates and phosphorothioates which may be quite persistent in nature (4). Since the 1993 International Chemical Warfare Convention, the stockpiles of these chemical warfare agents have not been wanted. Using natural biological systems for OP degradation could result in both environmentally friendly and in situ detoxification (4). Some microorganisms, such as Pseudomonas diminuta MG, Flavobacterium sp. (8), Alteromonas species (1, 2), Bacillus stearothermophilus (4), Nocardia sp. (7), Escherichia coli (10), and Arthrobacter (9), possess OP-hydrolyzing (OPH) activity. Previously, only the pseudomonad OPH enzyme was able to hydrolyze the P-S bond of Vx and demeton-S-ethyl (5, 6), but it hydrolyzed this bond at low rates. Ziegler et al. (11) have suggested that Corynebacterium glutamicum metabolizes a P-S bond-containing OP compound.
Biodegradation of demeton-S-methyl by C. glutamicum.
The ability of C. glutamicum ATCC 13745 to degrade the P-S bond-containing OP compound demeton-S-methyl (O,O-dimethyl-S-2-ethylthiolethyl phosphorothioate), a pesticide and a Vx analogue, was examined in 1.5-liter batch cultures by using the following three growth substrates: acetate, glucose, and fructose. The growth medium used was the medium described previously (3), except that the carbon source was glucose, acetate, or fructose that was added to obtain an initial substrate concentration of 18 g liter−1. Filtered demeton-S-methyl (in 100 mM Tris-HCl [pH 7.0] buffer) was added separately to the bioreactor to obtain final concentrations of 10 to 20 mg liter−1. The temperature was maintained at 27°C, the pH was maintained at 7.0, and both the aeration rate and the stirrer speed were modulated so that the dissolved oxygen concentration did not fall below 50% saturation. The inoculum was grown on the same medium but without demeton-S-methyl. Demeton-S-methyl was extracted from the cell suspensions by using a 50% n-hexane–50% ethylacetate mixture. Malathion (in 100 mM Tris-HCl [pH 7.0]) was used as an internal standard. One microliter of an extracted sample was injected (splitless) into a Hewlett-Packard model 5890A chromatograph equipped with an HP-5 M.S. (cross-linked 5% phenyl methyl silicone) column (30 m by 0.25 mm by 0.25 μm). Helium was the carrier gas (flow rate, 0.9 ml min−1) and the auxiliary gas (flow rate, 20 ml min−1). The hydrogen and air flow rates were 2.9 and 100 ml min−1, respectively. The oven temperature was programmed as follows: 30 s at 80°C, linear increase (at a rate of 30°C min−1) to 200°C, 30 s at 200°C, linear increase to 230°C in 1 min, 30 s at 230°C, linear increase (at a rate of 30°C min−1) to 310°C, and 2 min at 310°C. The injector and nitrogen phosphorus detector (NPD) temperatures were 260 and 300°C, respectively. The demeton-S-methyl quantification threshold was 0.1 mg liter−1.
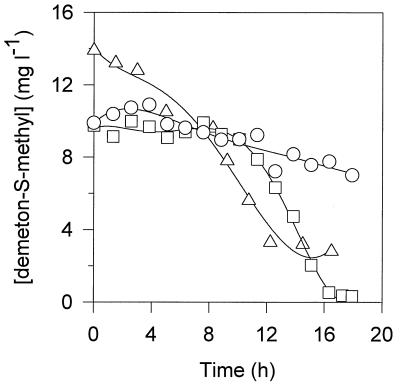
Figure 1 shows the demeton-S-methyl concentrations during batch growth on each growth substrate. No additional peak resulting from demeton-S-methyl cleavage was detected by gas chromatography in the cell suspensions after extraction. Biodegradation was initiated rapidly, indicating that no adaptation period was necessary, but was not complete since some residual demeton-S-methyl remained when growth stopped. The global demeton-S-methyl consumption rates were 0.21, 0.75, and 0.78 mg liter−1 h−1 on glucose, acetate, and fructose, respectively. The abiotic rate of degradation of demeton-S-methyl, as estimated by monitoring concentrations in control cultures lacking C. glutamicum, was less than 4.6 μg liter−1 h−1 over a 200-h period, which indicated that C. glutamicum is able to degrade demeton-S-methyl.
FIG. 1.
Demeton-S-methyl concentrations during C. glutamicum biodegradation under batch conditions when acetate (▵), glucose (○), and fructose (□) were used as growth substrates.
Demeton-S-methyl biodegradation occurs by a cometabolism process.
Demeton-S-methyl (10 mg liter−1) was added to each bioreactor when cells were entering the stationary phase due to substrate exhaustion, and the concentration of demeton-S-methyl was measured for 150 h. The global demeton-S-methyl consumption rates calculated for nonproliferating cells grown on acetate, glucose, and fructose were 0.080, 0.065, and 0.108 mg liter−1 h−1, respectively; these values were considerably lower than the values obtained during the growth phase. Addition of a carbon growth substrate to stationary-phase cultures resulted in an immediate increase in the demeton-S-methyl degradation rate (data not shown), indicating that in nongrowing cells the demeton-S-methyl-degrading enzyme(s) was present but considerably less active. Washed whole cells that were harvested during exponential growth on acetate were incubated at 30°C in a shake flask containing fresh medium supplemented with demeton-S-methyl (12 mg liter−1) in the presence and in the absence of acetate (5 g liter−1). No significant demeton-S-methyl consumption was observed after 65 h of incubation of washed whole cells of C. glutamicum in the absence of growth substrate, while the pesticide was consumed in medium containing acetate, indicating that demeton-S-methyl degradation depends on the functioning of primary metabolism. When fructose was the growth substrate, the rate of demeton-S-methyl consumption was greater than the rate of consumption when either acetate or glucose was the growth substrate (the global rate was 0.78 mg liter−1 h−1, and the maximum instantaneous rate was 1.4 mg liter−1 h−1).
Demeton-S-methyl biotransformation involves reductive cleavage of an S-C bond.
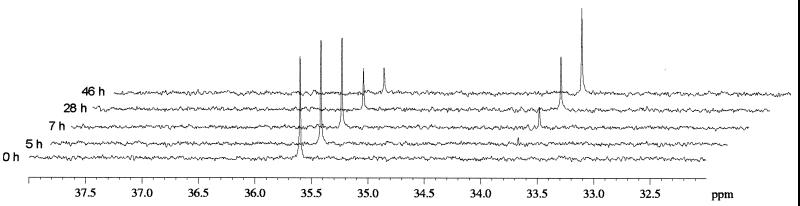
The identities of the degradation products of demeton-S-methyl were investigated by 31P nuclear magnetic resonance (NMR) spectroscopy. 31P NMR was performed at 25°C and 145.7 MHz by using a Brucker Avance model DPX 360 spectrometer equipped with a quadrupole probe (5 mm). A spectrum width of 20,450 Hz was used. Pulses were applied at a 90° flip angle and a 2.6-s repetition rate. The number of scans was set to 16,000, and proton decoupling was accomplished by using the WALTZ16 composite decoupling sequence. Chemical shifts were referenced to external phosphoric acid (δ = 0 ppm). Before analysis 50 μl of D2O was added to each sample (volume, 500 μl). Demeton-S-methyl has a characteristic singlet at 35.7 ppm (Fig. 2); this singlet decreased during incubation with C. glutamicum grown on fructose, and an upfield singlet at 33.8 ppm appeared. This singlet could not be attributed to the spectrum of dimethyl phosphate but was attributed to the spectrum of dimethyl thiophosphate. Abiotic degradation of demeton-S-methyl at pH 14 resulted in a decrease in the 35.7-ppm singlet and the appearance of two upfield singlets at 2.5 and 20.5 ppm (data not shown). The signal at 2.5 ppm corresponded to the spectrum of dimethyl phosphate. These results demonstrated that abiotic degradation at pH 14 resulted from P-S bond hydrolysis but that C. glutamicum did not cleave the P-S bond. Attempts to detect thiol liberation in crude cell extracts were unsuccessful, which confirmed that enzymatic hydrolysis of the P-S bond did not occur. Demeton-S-methyl is biotransformed by breaking the S-C bond, and the resulting dimethyl thiophosphate is not degraded by C. glutamicum. The biotransformation reaction is not a hydrolysis reaction but is reductive cleavage of the S-C bond due to a dehydrogenase-oxidoreductase activity according to the following reaction: OO∥∥CH3-CH2-S-CH2-CH2-S-P-(O-CH3)2 → CH3-CH2-S-CH2-CH3 + HS-P-(O-CH3)2
FIG. 2.
31P NMR spectra of demeton-S-methyl and the products of degradation of demeton-S-methyl by C. glutamicum cells grown on fructose.
Acknowledgments
We thank M. Albaret (Centre d'Etudes du Bouchet) for assistance with 31P NMR spectroscopy.
REFERENCES
- 1.Cheng T-C, Harvey S P, Stroup A N. Purification and properties of a highly active organophosphorus acid anhydrolase from Alteromonas undina. Appl Environ Microbiol. 1993;59:3138–3140. doi: 10.1128/aem.59.9.3138-3140.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.DeFrank J J, Cheng T-C. Purification and properties of an organophosphorus acid anhydrase from a halophilic bacterial isolate. J Bacteriol. 1991;173:1938–1943. doi: 10.1128/jb.173.6.1938-1943.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dominguez H, Nezondet C, Lindley N D, Cocaign M. Modified carbon flux during oxygen limited growth of Corynebacterium glutamicum and the consequences for amino acid overproduction. Biotechnol Lett. 1993;15:449–454. [Google Scholar]
- 4.Grimsley J K, Rastogi V K, Wild J R. Biological detoxification of organophosphorus neurotoxins. In: Sikdar S K, Irvine R L, editors. Bioremediation: principles and practice. II. Lancaster, Pa: Technomic Publishing Co.; 1998. pp. 577–613. [Google Scholar]
- 5.Hoskin F C G, Walker J E, Dettbarn W-D, Wild J R. Hydrolysis of tetriso by an enzyme derived from Pseudomonas diminuta as a model for the detoxication of O-ethyl S-(2-diisopropylaminoethyl) methylphosphonothiolate (Vx) Biochem Pharmacol. 1995;49:711–715. doi: 10.1016/0006-2952(94)00496-9. [DOI] [PubMed] [Google Scholar]
- 6.Lai K, Stolowich N J, Wild J R. Characterization of P-S bond hydrolysis in organophosphorothioate pesticides by organophosphorus hydrolase. Arch Biochem Biophys. 1995;318:59–64. doi: 10.1006/abbi.1995.1204. [DOI] [PubMed] [Google Scholar]
- 7.Mulbry W W. The aryldialkylphosphatase-encoding gene adpB from Nocardia sp. strain B-1: cloning, sequencing and expression in Escherichia coli. Gene. 1992;121:149–153. doi: 10.1016/0378-1119(92)90174-n. [DOI] [PubMed] [Google Scholar]
- 8.Mulbry W W, Karns J S, Kearney P C, Nelson J O, McDaniel C S, Wild J R. Identification of a plasmid-borne parathion hydrolase gene from Flavobacterium sp. by Southern hybridization with opd from Pseudomonas diminuta. Appl Environ Microbiol. 1986;51:926–930. doi: 10.1128/aem.51.5.926-930.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ohshiro K, Ono T, Hoshino T, Uchiyama T. Characterization of isofenphos hydrolases from Arthrobacter sp. strain B-5. J Ferment Bioeng. 1997;83:238–245. [Google Scholar]
- 10.Zech R, Wigand K D. Organophosphate-detoxicating enzymes in E. coli. Gel filtration and isoelectric focusing of DFPase, paraoxonase, and unspecific phosphohydrolases. Experientia. 1975;31:157–158. doi: 10.1007/BF01990678. [DOI] [PubMed] [Google Scholar]
- 11.Ziegler W, Engelhardt G, Wallnöfer P R, Oehlmann L, Wagner K. Degradation of demeton S-methyl sulfoxide (metasystox R) by soil microorganisms. J Agric Food Chem. 1980;28:1102–1106. [Google Scholar]