Abstract
It is essential to identify specific food components that inhibit PCR in order to increase the sensitivity of the PCR method for rapid detection of pathogens contaminating a food. We found that collagen, a major component of several foods, inhibited PCR. The inhibitory action of collagen on PCR could be partially reversed by adjusting the concentration of magnesium ion in the reaction mixture and by the use of various DNA extraction methods to remove the collagen from the DNA. Also, the source of thermostable DNA polymerase was affected by the presence of collagen. These results suggest the need to optimize the extraction and assay conditions for rapid detection of enterotoxigenic Clostridium perfringens by PCR with respect to the kind of food being analyzed.
Clostridium perfringens type A food poisoning is among the most common of the human gastrointestinal illnesses (3, 9). Symptoms associated with C. perfringens type A food poisoning are diarrhea and severe abdominal pain. The symptoms are mediated by an enterotoxin, a 35-kDa single polypeptide produced only during sporulation of the organism in the small intestine following ingestion of food contaminated with large numbers of vegetative cells (12, 13). C. perfringens is a typical gram-positive, rod-shaped, spore-forming, anaerobe commonly found in the intestines of humans and mammals. C. perfringens is a part of the microflora of soil, and many reports have revealed the widespread occurrence of this organism in raw and processed foods. However, most of these isolates are nonenterotoxigenic strains. Recent surveys suggest that only about 6% of all C. perfringens isolates carry the gene (cpe) encoding enterotoxin (8, 19). It therefore is necessary to distinguish the enterotoxigenic organisms from the nonenterotoxigenic ones to confirm food poisoning by C. perfringens.
Rapid and highly sensitive techniques based on PCR have been developed recently for the detection of foodborne pathogens (2, 7, 14, 17). A PCR-based detection system is highly sensitive and eliminates the need for enrichment culturing (17). PCR can be used to detect genes involved in the virulence of foodborne bacteria. However, the complex nature of food components offers unique challenges in the application of PCR to rapid detection of pathogens in a food (16). A variety of components—for example, heme and its metabolic products (15), acidic polysaccharides (5, 6), humic substances (18), bean sprout homogenates (7), oysters (7), and soft cheese (20)—have been shown to inhibit PCR amplification.
We have tried to develop a rapid PCR-based method for detection of enterotoxigenic C. perfringens contamination in a food by testing for the presence of the cpe gene directly without preenrichment. Four Korean ethnic foods, man-doo (a bun stuffed with seasoned meat and vegetables), soon-dae (a sausage made of beef and bean curd, stuffed in pig intestine), kim-bab (rice, meat, and vegetables wrapped with seaweed), and steamed pork hock, were tested. Food samples were artificially contaminated with enterotoxigenic C. perfringens type A strain NCTC 8239 (Hobb's serotype 3 [H3]) cells at densities ranging from 101 to 108 CFU/g of food. The food samples were homogenized in a Waring blender for 2 min in distilled water at a ratio of 1 g of food to 5 ml of water, and the cells were collected by filtration through Whatman no. 41 filter paper followed by centrifugation of the filtrate at 15,000 × g for 10 min. The pellet was used for preparation of a DNA template for PCR. Traditionally, DNA templates have been prepared by phenol-chloroform extraction. However, various easy and rapid DNA isolation methods have been developed to replace the time-consuming procedure of phenol-chloroform extraction and ethanol precipitation. We prepared a DNA template for PCR by using two commercially available DNA extraction kits, a QIAamp tissue kit (Qiagen, Hilden, Germany) and a GeneReleaser kit (Bio Ventures, Inc., Murfreesboro, Tenn.), according to the instructions of the manufacturers. Primers specific for the C. perfringens enterotoxin gene (cpe; GenBank accession no. X71844) were designed to include the promoter region and part of the structural cpe gene. These were cpe11 (5′-ACTTAGAGTATCTATAAACTTGATACTC-3′) and cpe12 (5′-TAAATTGTTACTAAGCATATTATAATTAACATC-3′). The size of the PCR product made with these two primers was 599 bp.
The presence of various foods had inhibitory effects on PCR, as indicated in Table 1. We could detect as few as 30 cells/ml of C. perfringens culture, but the detection limit was increased in the presence of various foods. The detection limit was increased to 400 cells/g of man-doo, 2.5 × 103 cells/g of soon-dae, and 4.5 × 103 cells/g of kim-bab, respectively (Table 1). Interestingly, the inhibition by pork hock was so strong that even 105 C. perfringens cells per g of pork hock was insufficient for detection by this method. There was virtually no difference in the sensitivities of detection by PCR with GeneReleaser- and QIAamp-prepared DNA templates.
TABLE 1.
Minimum number of C. perfringens cells detectable by PCR when DNA was prepared with a QIAamp kit or by phenol extraction from foods artificially contaminated with C. perfringens
| Detection method | Min. no. of C. perfringens cells detectable in:
|
||||
|---|---|---|---|---|---|
| No food | Man-doo | Soon-dae | Kim-bab | Pork hock | |
| QIAamp kit | 30/ml | 400/g | 2.5 × 103/g | 4.5 × 103/g | NDa |
| Phenol extraction | 10/ml | 104/g | 1 × 103/g | 1 × 103/g | 1 × 104/g |
ND, none detected.
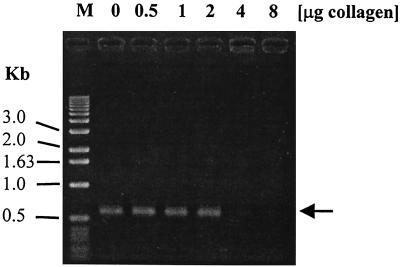
The inhibitory effect of pork hock on PCR was further studied. We tested the effect of collagen, one of the main components of pork hock, and found that it strongly inhibited PCR. Collagen is classified into a number of structurally and genetically distinct types (4), each of which showed a different degree of inhibition of the PCR. Type I collagen (Sigma type III) inhibited PCR if more than 2 μg was added to a reaction mixture and 3.5 mM MgCl2 was used in the PCR buffer (Fig. 1). In contrast, type I collagen from human placenta (Sigma type VIII) did not show inhibition even at 8 μg/reaction. The inhibition of PCR by collagen was specific, since addition of an equivalent amount of bovine serum albumin to the PCR did not affect the reaction. Also, the inhibition of PCR by collagen was not specific to the particular primer set used for the detection of the cpe gene, since the primer set (plc2 [5′-TCCCCTTTCTAGATAACGATTAACAC-3′] and plc4 [5′-GTTAGCATGCTGTTTTCTAAGTTAAAACC-3′]) used for the detection of the plc gene of C. perfringens was also inhibited by collagen.
FIG. 1.
Inhibition of PCR by type I collagen. Type I collagen from calf skin was added to 50 μl of PCR mix in the presence of 3.5 mM MgCl2 at the concentrations indicated above the gel. The positions of molecular size markers (M) are shown on the left. The arrow indicates the position of the 599-bp DNA fragment.
It has been shown that the function of thermostable DNA polymerases from different sources is inhibited differently by known PCR inhibitors (1). Collagen showed different degrees of inhibition of Pwo DNA polymerase. Generally, Pwo DNA polymerase was more sensitive to collagen than Taq DNA polymerase. These results suggest that the appropriate thermostable DNA polymerase should be used for sensitive PCR detection of pathogens contaminating a food.
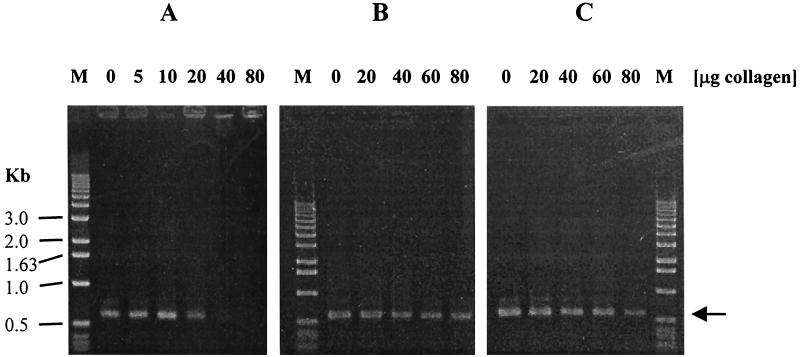
When various types of collagen were added to C. perfringens culture before preparation of DNA by the use of a Gene-Releaser or QIAamp kit, PCR was inhibited by the DNA preparation (data not shown). These results suggest that neither the GeneReleaser nor the QIAamp procedure could remove the collagen from the DNA preparation. However, addition of more MgCl2 to the reaction mixture relieved inhibition of PCR by various types of collagen to some degree, but the PCR product formed a broad band when a high concentration of MgCl2 (more than 5 mM) was used (data not shown). We tested two additional DNA preparation methods (phenol extraction and NaI treatment) as described by Makino et al. (10, 11) in an attempt to remove collagen from the DNA preparation. Even though the presence of collagen inhibited the efficiency of DNA extraction by these methods, the degree of inhibition was much smaller when phenol extraction or NaI treatment was used than when a GeneReleaser or QIAamp kit was employed (Fig. 2). When we tested the phenol extraction and NaI methods for detection of C. perfringens contamination in a food, phenol extraction method worked well but the NaI method did not. We could not detect a PCR product when the C. perfringens DNA template was prepared by NaI treatment in the presence of the above-mentioned foods, which indicates that a PCR inhibitor possibly present in the food could not be removed by NaI treatment. These results indicate the need to evaluate DNA preparation methods and the type of thermal DNA polymerase if PCR-based methods are to be used for the direct detection of microorganisms in foods.
FIG. 2.
Effects of type I collagen on PCR when DNA templates were prepared with a GeneReleaser kit (A), by phenol-chloroform extraction (B), or by NaI treatment (C) in the presence of collagen. One-tenth of the final volume of DNA preparation was used for each PCR, so that the actual amount of collagen present in each PCR was 1/10 the amount indicated at the top of each lane if collagen was not removed by the extraction method. The use of a QIAamp tissue kit produced results similar to those obtained with a GeneReleaser kit. The positions of molecular size markers (M) are indicated on the left. The arrow indicates the position of the 599-bp DNA fragment.
Acknowledgments
This study was supported by a grant (no. HMP-96-F-1-1002) from the 1996 Good Health R&D Project, Ministry of Health & Welfare, Republic of Korea.
REFERENCES
- 1.Abu Al-Soud W, Rådström P. Capacity of nine thermostable DNA polymerases to mediate DNA amplification in the presence of PCR-inhibiting samples. Appl Environ Microbiol. 1998;64:3748–3753. doi: 10.1128/aem.64.10.3748-3753.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baez L A, Juneja V K. Detection of enterotoxigenic Clostridium perfringens in raw beef by polymerase chain reaction. J Food Prot. 1995;58:154–159. doi: 10.4315/0362-028X-58.2.154. [DOI] [PubMed] [Google Scholar]
- 3.Bean N H, Griffin P M. Foodborne disease outbreaks in the United States, 1973–1987: pathogens, vehicles and trends. J Food Prot. 1990;53:804–817. doi: 10.4315/0362-028X-53.9.804. [DOI] [PubMed] [Google Scholar]
- 4.Bornstein P, Traub W. The chemistry and biology of collagen. In: Bornstein P, Traub W, editors. The proteins. IV. New York, N.Y: Academic Press, Inc.; 1979. pp. 412–432. [Google Scholar]
- 5.Demeke T, Adams R P. The effects of plant polysaccharides and buffer additives on PCR. BioTechniques. 1992;12:332–334. [PubMed] [Google Scholar]
- 6.Do N, Adams R P. A simple technique for removing plant polysaccharide contaminants from DNA. BioTechniques. 1991;10:162–166. [PubMed] [Google Scholar]
- 7.Hill W E. The polymerase chain reaction: applications for the detection of foodborne pathogens. Crit Rev Food Sci Nutr. 1996;36:123–173. doi: 10.1080/10408399609527721. [DOI] [PubMed] [Google Scholar]
- 8.Kokai-Kun J F, Songer J G, Czeczulin J R, Chen F, McClane B A. Comparison of Western immunoblots and gene detection assays for identification of potentially enterotoxigenic isolates of Clostridium perfringens. J Clin Microbiol. 1994;32:2533–2539. doi: 10.1128/jcm.32.10.2533-2539.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Labbe R G. Clostridium perfringens. In: Doyle M, editor. Foodborne bacterial pathogens. New York, N.Y: Marcel Dekker, Inc.; 1989. pp. 191–234. [Google Scholar]
- 10.Makino S-I, Iinuma-Okada Y, Maruyama T, Ezaki T, Sasakawa C, Yoshikawa M. Direct detection of Bacillus anthracis DNA in animals by polymerase chain reaction. J Clin Microbiol. 1993;31:547–551. doi: 10.1128/jcm.31.3.547-551.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Makino S-I, Okada Y, Maruyama T. A new method for direct detection of Listeria monocytogenes from foods by PCR. Appl Environ Microbiol. 1995;61:3745–3747. doi: 10.1128/aem.61.10.3745-3747.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McClane B C. Clostridium perfringens. In: Doyle M P, Beuchat L R, Montville T J, editors. Food microbiology: fundamentals and frontiers. Washington, D.C.: ASM Press; 1997. pp. 305–326. [Google Scholar]
- 13.McDonel J L. Clostridium perfringens toxins (type A, B, C, D, E) Pharmacol Ther. 1979;10:617–655. doi: 10.1016/0163-7258(80)90031-5. [DOI] [PubMed] [Google Scholar]
- 14.Olsen J E, Aabo S, Hill W, Notermans S, Wernars K, Granum P E, Popovic T, Rasmussen H N, Olsvik O. Probes and polymerase chain reaction for detection of food-borne bacterial pathogens. Int J Food Microbiol. 1995;28:1–78. doi: 10.1016/0168-1605(94)00159-4. [DOI] [PubMed] [Google Scholar]
- 15.Panaccio M, Lew A. PCR based diagnosis in the presence of 8% (v/v) blood. Nucleic Acids Res. 1991;19:291–292. doi: 10.1093/nar/19.5.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rossen L, Norskov P, Holmstrom K, Rasmussen O F. Inhibition of PCR by components of food samples, microbial diagnostic assays, and DNA-extraction solutions. Int J Food Microbiol. 1992;17:37–45. doi: 10.1016/0168-1605(92)90017-w. [DOI] [PubMed] [Google Scholar]
- 17.Swaminathan B, Feng P. Rapid detection of food-borne pathogenic bacteria. Annu Rev Microbiol. 1994;48:401–426. doi: 10.1146/annurev.mi.48.100194.002153. [DOI] [PubMed] [Google Scholar]
- 18.Tsai Y-L, Olson B H. Rapid method for separation of bacterial DNA from humic substances in sediments for polymerase chain reaction. Appl Environ Microbiol. 1992;58:2292–2295. doi: 10.1128/aem.58.7.2292-2295.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Damme-Jongsten M, Wernars K, Notermans S. Cloning and sequencing of the Clostridium perfringens enterotoxin gene. Antonie Leuwenhoek. 1989;56:181–190. doi: 10.1007/BF00399981. [DOI] [PubMed] [Google Scholar]
- 20.Wernars K, Hevvelman C J, Chakraborty T, Notermans S H W. Use of the polymerase chain reaction for direct detection of Listeria monocytogenes in soft cheese. J Appl Bacteriol. 1991;70:121–126. doi: 10.1111/j.1365-2672.1991.tb04437.x. [DOI] [PubMed] [Google Scholar]