Abstract
Objective
The goals of this study were to harmonize data from electronic health records (EHRs) into common units, and impute units that were missing.
Materials and Methods
The National COVID Cohort Collaborative (N3C) table of laboratory measurement data—over 3.1 billion patient records and over 19 000 unique measurement concepts in the Observational Medical Outcomes Partnership (OMOP) common-data-model format from 55 data partners. We grouped ontologically similar OMOP concepts together for 52 variables relevant to COVID-19 research, and developed a unit-harmonization pipeline comprised of (1) selecting a canonical unit for each measurement variable, (2) arriving at a formula for conversion, (3) obtaining clinical review of each formula, (4) applying the formula to convert data values in each unit into the target canonical unit, and (5) removing any harmonized value that fell outside of accepted value ranges for the variable. For data with missing units for all the results within a lab test for a data partner, we compared values with pooled values of all data partners, using the Kolmogorov-Smirnov test.
Results
Of the concepts without missing values, we harmonized 88.1% of the values, and imputed units for 78.2% of records where units were absent (41% of contributors’ records lacked units).
Discussion
The harmonization and inference methods developed herein can serve as a resource for initiatives aiming to extract insight from heterogeneous EHR collections. Unique properties of centralized data are harnessed to enable unit inference.
Conclusion
The pipeline we developed for the pooled N3C data enables use of measurements that would otherwise be unavailable for analysis.
Keywords: reference standards, SARS-CoV-2, electronic health records, data accuracy, data collection
OBJECTIVE
Quantitative data in the National COVID Cohort Collaborative (N3C) originate from data partners who submit electronic health record (EHR) data via different Common Data Models (CDMs), which are then harmonized into the Observational Medical Outcomes Partnership (OMOP) CDM. Our objective was to harmonize measurement units and to reclaim usable data via unit inference for values missing units.
BACKGROUND AND SIGNIFICANCE
N3C has built a repository of EHR data from a growing number of data partners across the United States to facilitate research on coronavirus disease of 2019 (COVID-19).1 Our data partners submit clinical and laboratory data, using one of several CDMs used for distributed EHR-based research, which are then mapped to the OMOP CDM.2,3 While the CDMs specify the structure for storing data, not all fields are required, and what data can be entered in these fields is not tightly controlled.2
One of the key resources of N3C is laboratory measurement data, central to almost all research on COVID-19. Sites submit these data in a variety of units even within the same measurement concept. Most sites map their local lab data to the Logical Observation Identifiers Names and Codes (LOINC) system.4 As part of the LOINC standard, properties being measured, such as mass concentration (mass/volume), number concentration (count/volume), or rate (count/time), is specified as part of the code but the units to be used are not specified. Instead, for many codes, example units are provided, often including units in the Unified Code for Units of Measure (UCUM) format.5 UCUM formatting is designed to remove ambiguity (g vs gm for example) and to integrate with electronic messaging standards. However, UCUM does not dictate which unit is preferred for any particular analyte. Because of this lack of uniformity in reporting, it is almost always necessary to convert units before comparing measurement results from different sites. As an example of OMOP unit concept name diversity in the N3C measurement table, units for body weight can include kilogram, gram, ounce (avoirdupois), oz and pound (US).
Additionally, units of measure are often missing. Dropping these entries from analyses would result in significant data loss and even bias, especially if the units were consistently missing for a given result or from a given data partner. Another option would be to solicit each data partner for their missing units, which would be feasible if working on a small number of data elements and with a small number of data partners, but is not a sustainable solution as the number of data contributors grows over time (55, at as of June 10, 2021). A third option would be to assume a unit based on what is commonly used in clinical practice; while this may work for simple measurements such as heart rate, it is highly error prone for most cases.
In order to preserve as much data as possible without placing an undue burden on the data partners or individual researchers, we determined that an automated method is needed to automatically convert units in a systematic fashion as well as determine, on a site-by-site basis, what the most likely unit is for a given OMOP measurement concept where units are absent. To address this problem of missing units, we propose a method comparing the value distribution of measurements6 which are missing units to those from the same concept set with known units. In general, if the distributions are not found to be significantly different we infer that they use the same unit.
LOINC unit conversion has been addressed by other research groups in a variety of contexts including analysis of aggregated datasets,7–9 Hauser describes creation of a standard set of conversions between compatible LOINC laboratory codes expressed in different units10,11 along with publicly accessible conversions to support conversion of common LOINC codes. We contribute and extend the state of the art by making available reproducible computational pipelines to impute units in cases where units were missing.
In summary, we aimed to develop approaches that incidentally have a broad impact beyond the COVID-19 research of N3C, addressing the need to harmonize units by grouping similar laboratory tests and by converting all data points for each measurement to a single predetermined canonical unit. In addition, we aimed to reclaim as much data as possible missing units of measure to maximize their use. The resulting methods are broadly applicable to contexts of pooled EHR data.
MATERIALS AND METHODS
The design of the N3C data set and a comprehensive characterization of the data available prior to December 2020 have been previously described.1,12 In the current study, we included data ingested as of June 10, 2021. Our analysis did not put any restrictions on the patient population, and included all available measurements. The N3C ingestion pipeline includes comprehensive mapping of measurement concepts and unit concepts to standard concepts in the OMOP CDM. While we had access to information from source vocabularies, all work discussed in this manuscript began with the data after conversion to OMOP.
Working with a community of physicians and informatics experts, we created an initial list of measurements that were of high priority for harmonization, driven primarily by the needs to describe the N3C cohort.12 We gained subject matter expert (SME) consensus on broad sets of semantically similar analytes (and, where relevant, of specimen type) expected by the SMEs to have interoperoberable and convertible measurement units, even though the particulars may differ. For example, differences between venous and arterial measurements are meaningfully different, but their analytes are measured with the same units. Measurement concepts with relative results (eg, ratio or percent) were grouped into different concept sets than absolute measures. All concept sets were reviewed by a clinical SME to ensure that only concepts which would be interpreted similarly in a clinical setting, despite potential differences in sampling time and sample specimen, were included. We also required that the concepts be unique to a single concept set, so that values did not get harmonized in multiple ways. The full list of concepts per concept set can be found in Supplementary Table S1. Notably, the concept sets were used only for unit harmonization optimization and implementation purposes and are not intended to represent clinically synonymous collections of concepts for downstream analysis. In fact, the grouping of concepts for research and clinical use is often necessarily different from the broader grouping that suits unit harmonization and inference purposes. As an extra validation of concept set membership we analyzed measurement concept cumulative distribution functions (CDFs) for each concept set (Supplementary Figure S1), and the few concept sets that appeared to contain an outlier were examined in depth. These outlier concepts in reviewed concept sets included Leukocytes [#/volume] in Blood by Manual count, whose CDF differed from others’, because a manual count is performed under circumstances where automated counts fail due to low or unusual blood cell counts. This distribution was not, however, considered sufficiently different to deserve its own concept set, at least for the purposes of unit harmonization and inference due to (A) being synonymous in terms of the range of plausible units and (B) for reasons detailed in Results.
Working with our SMEs, we then chose a single “canonical” unit for each measurement concept set; in most cases the unit where the values are in the most easily interpretable scale, or units where derived variables can subsequently be calculated, for example kilogram for weight was selected due to familiarity, and since it is the most commonly used unit by the clinicians within N3C to calculate the derived variable BMI. Additionally, we identified acceptable value ranges, beyond which a measurement would be discarded, for example percents below 0% or above 100%, and values that are not clinically possible in any patient as determined by our SMEs. These ranges were made as liberal as possible to avoid losing extreme cases yet conservative enough to allow us to filter out poor-quality data that could affect downstream analyses. A list of the measurements and values for the required fields are shown for selected examples in Table 1, and in full, within Supplementary Table S2. Lastly, we manually curated conversion formulae suitable for converting from other units to this canonical unit (Supplementary Table S3).
Table 1.
Example canonical units table
| Measured variable | Enclave codeset ID | Canonical unit concept ID | Canonical unit concept name | Maximum plausible value | Minimum plausible value | Measurement table row count |
|---|---|---|---|---|---|---|
| Respiratory rate | 286601963 | 8483 | Counts per minute | 200 | 0 | 201 976 073 |
| Sodium, mmol/L | 887473517 | 8753 | Millimole per liter | 250 | 50 | 147 177 271 |
| SpO2 | 780678652 | 8554 | Percent | 100 | 0 | 145 403 614 |
| Systolic blood pressure | 186465804 | 8876 | Millimeter mercury column | 400 | 0 | 136 188 546 |
| Temperature | 656562966 | 586323 | Degree celsius | 45 | 25 | 123 986 764 |
| Glucose, mg/dL | 59698832 | 8840 | Milligram per deciliter | 1000 | 0 | 104 743 184 |
| Heart rate | 596956209 | 8483 | Counts per minute | 500 | 0 | 67 530 040 |
| Height | 754731201 | 9546 | Meter | 3 | 0 | 53 998 207 |
| Body weight | 776390058 | 9529 | Kilogram | 500 | 0.1 | 42 113 217 |
| Diastolic blood pressure | 573275931 | 8876 | Millimeter mercury column | 200 | 0 | 42 024 537 |
Note: Chosen canonical units and plausible value range for the top 10 most frequent measured variables in the data out of those selected for unit harmonization and inference.
Unit harmonization
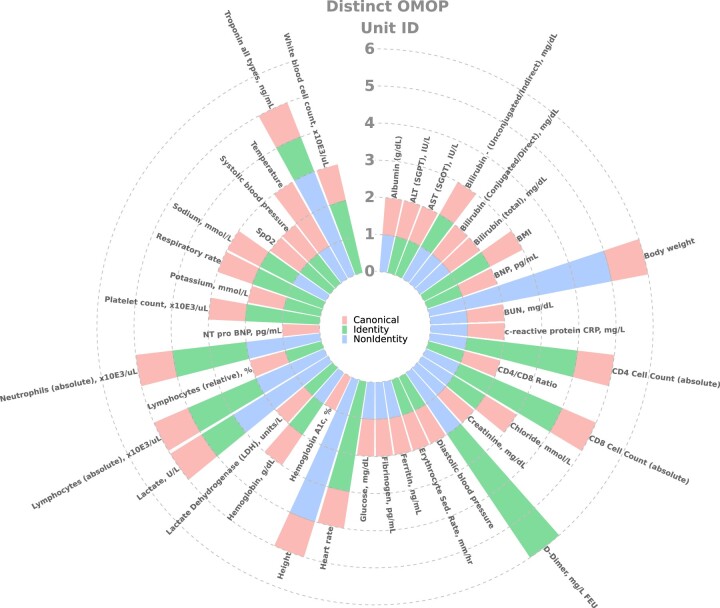
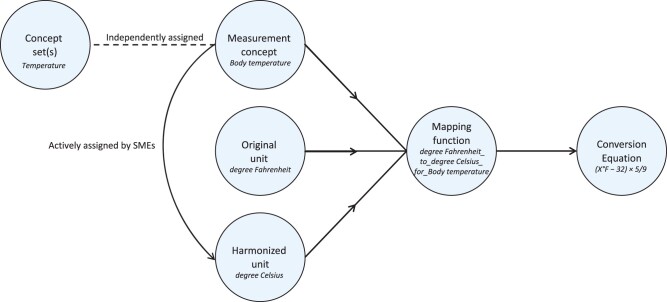
Using the concept sets, canonical units, acceptable ranges and formulae from Supplementary Tables S2 and S3, we assessed the diversity of units present in the data (Figure 1) and implemented a pipeline (Figure 2) that converted value data (ie, value_as_number) to harmonized value data. In our implementation, we created 2 new fields in our measurement table, harmonized_unit_concept_id and harmonized_value_as_number, to preserve the original value data for maximum transparency and flexibility. Conversions were performed using a mapping function, composed of the units to convert and the measurement concept as a lookup for the corresponding conversion equation (Figure 2). After conversion, if the value was outside of the accepted value range, the harmonized_unit_concept_id and harmonized_value_as_number fields were set to null.
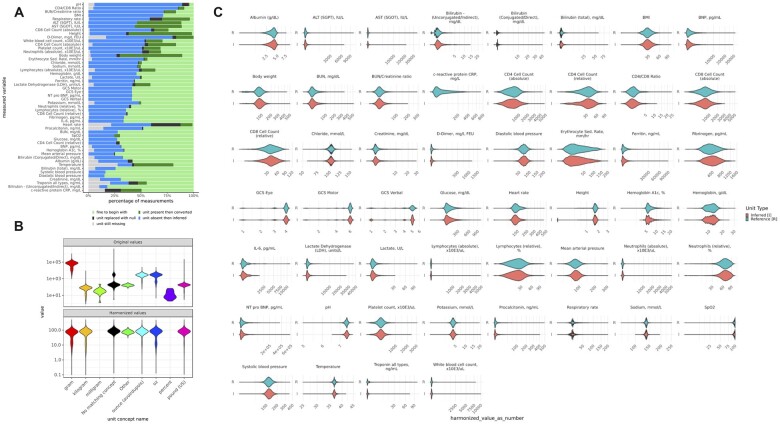
Figure 1.
Diversity of equivalent and nonequivalent units across measured variables: Units present per measurement variable and their equivalency to the selected canonical unit. Equivalent units to the canonical unit are described as “identity” and those with nonequivalent units are referred to as “non-identity.”
Figure 2.
Unit conversion workflow summary. Overview of the process for harmonizing unit in the OMOP measurement table. SME: subject matter expert.
Unit inference
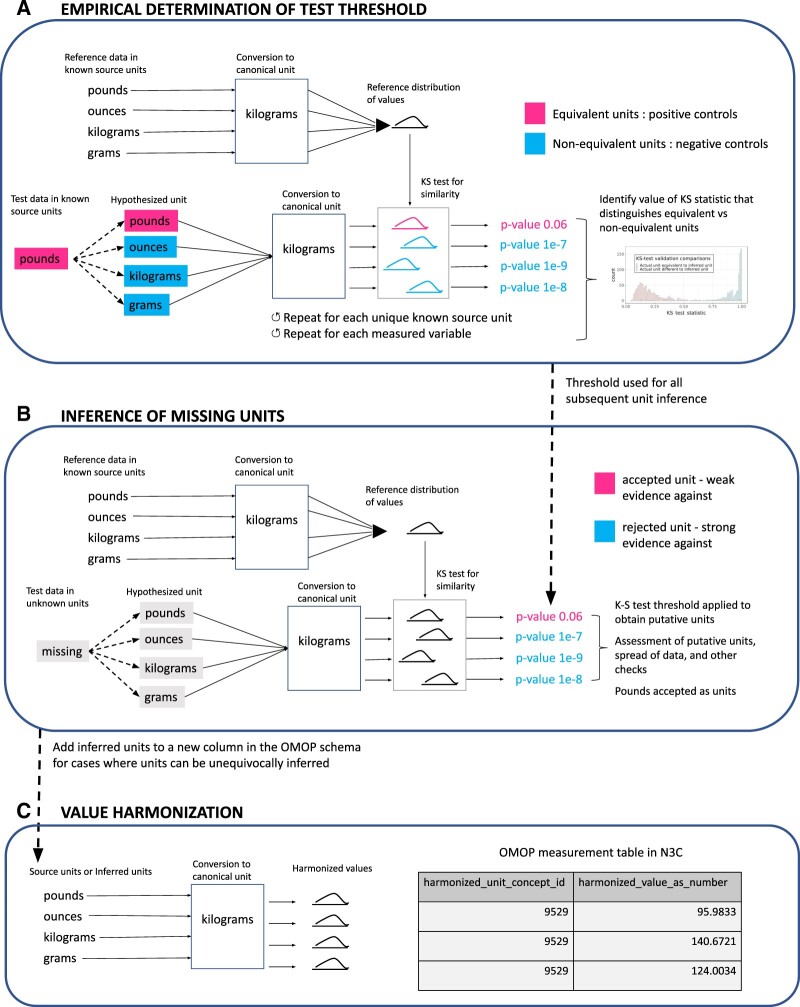
We applied unit inference to measurement records that were missing units in the source data or, missing a valid mapping from the source unit to the OMOP standard unit. The basis of unit inference derives from a previous study6 that developed a method for determining if lab results from 2 different labs represented the same type of measurement, in part, by comparing distributions of the results between the 2 tests using the Kolmogorov-Smirnov test (KS test).13 We adapted this approach, and compared value distributions within each OMOP measurement concept for laboratory tests per data partner, converted to the canonical unit for every plausible unit for that measurement concept (termed “test” value), to a selection of values with known units converted into the canonical unit (termed “reference” value). For each test unit, following the conversion to the canonical unit using the appropriate conversion formula, we assessed whether the distribution of values closely matched the canonical unit reference distribution of values, using the KS test P value, above a threshold, to define “close.” When the match passed our empirically derived threshold and other quality control criteria (described below), the test unit was then assigned as the accepted inferred unit. Figure 3 and Supplementary Figure S2 outline the unit-inference process.
Figure 3.
Unit inference and harmonization workflows. (A) Unit-inference threshold validation workflow. Masking of known units was used as a guide to assess the range of KS test P values that pertain to values in equivalent units across populations. The final threshold selected after plotting all P values together was 1e−5, which was then used for identifying units when they are missing. (B) Unit inference workflow. Process for sampling and performing KS tests on values across data partner and measurement concept combinations, checking for P values above the 1e−5 threshold, and applying thresholds to omit unit inference in cases where units cannot be confidently assigned. (C) Unit harmonization workflow. Conversion of values for each record into the canonical unit. KS test: Kolmogorov-Smirnov test.
Unit-inference threshold and sample-size validation
To determine the correct KS-test threshold, we created a workflow (Figure 3A and Supplementary Figure S2A) that masks known units for each 4-element tuple of measurement variable, data partner, measurement concept, and unit, which were then compared to the reference values (the collection of values of known units where all the appropriate conversions have taken place into the canonical unit). Because of the combinatorial explosion in the number of comparisons, the work was done with samples from each tuple. Within each sample, all possible conversions to the canonical unit take place, each becoming a list of “test” values, simulating the variety of potential originating units and their converted values. The KS test was then performed on each test value list, and the resulting P value (Figure 4) and KS-test statistic (Supplementary Figure S3) were compared for equivalent units (test unit is equivalent to the masked “known” unit) versus nonequivalent units (test unit is not equivalent to the masked “known” unit). This calibration of P values stands instead of the Bonferroni correction.14 The range of KS-test P values or test statistics that uniquely pertain to equivalent unit value distribution comparisons is the potential range to set a threshold in order to avoid false unit assignments; the base of the peak within the left tail of the distribution of P values was chosen as the threshold for inferring units across all variables. The size of the value lists used for value distribution comparisons in the KS test was found empirically to perform similarly at 50 and 100 elements and thus was determined to be robust at those population sizes, and 100 elements was chosen due to being sufficient for stable performance while sufficiently small to avoid long processing times and memory usage. Value-list sizes were held constant as the KS test otherwise has to be corrected for population size to give results that can be compared.
Figure 4.
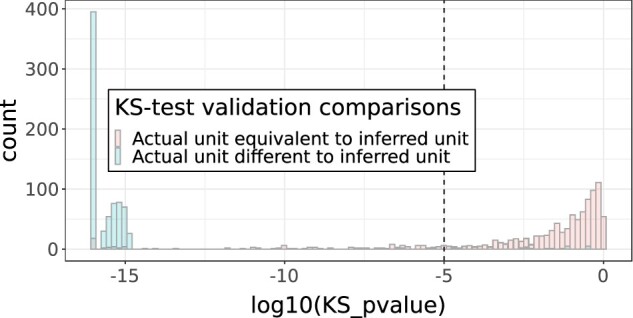
KS test P-value threshold validation. KS P values for equivalent versus nonequivalent units per data partner ID/measurement concept name. CRP was omitted due to having various completely overlapping value distributions in nonequivalent units after visual inspection. CRP: c-reactive protein; KS test: Kolmogorov-Smirnov test.
Unit inference and criteria for unit inference omission
Figure 3B and Supplementary Figure S2B summarize the work done on these sets of 100 values to determine potential units, which included inspection of the test values exceeding the threshold for presence of distinct (multiple) units, in which case unit inference was skipped. Additionally, entire measurement variables for unit inference omission were derived from 2 main filters: (1) We compared relative dispersion of values to the relative unit conversion fold change and excluded measured variables that had a large amount of uncertainty (Supplementary Figure S2B). (2) We used a combination of the number of distinct units over the P value threshold (Supplementary Figure S4), the proportion of fold change differences between the reference and test value lists within a fold change range 1.5–15× indicating nonequivalent units (as supported by Supplementary Figure S5), and the spread of the data, assessed by median absolute deviation from the median within test value lists. The combination of these thresholds is displayed in Supplementary Figure S6.
Incorporating inferred units and implementing the final unit harmonization workflow
Following these quality-control checks, the inferred units were consolidated with the original units, where present, to create a new column in the Measurement OMOP table for unit harmonization (Figure 3C and Supplementary Figure S2C). We retained any variables that were unitless or where only 1 unit was possible and therefore not entered by the sites, for example, BUN/Cr ratio; these variables received a 1:1 mapping from value_as_number to harmonized_value_as_number.
All codes for unit inference and harmonization were optimized within the N3C pipeline using PySpark v3.0.0 and Spark 3 on the Palantir Foundry platform,1 and deposited to GitHub along with a full package version list (https://github.com/kbradwell/N3C-units).
RESULTS
The June 10, 2021 release set comprised data from 55 data partners, with 2.716 billion rows of quantitative measurement lab data, composed of 12 390 distinct measurement concepts and 361 distinct OMOP unit concepts.
Diversity of measurement units
Unique OMOP units per measurement variable, stratified by equivalency to the selected canonical unit, are shown in Figure 1 (across the 52 concept sets there were 40 canonical, 27 equivalent to canonical, and 23 nonequivalent to canonical units). Height and body weight display the greatest number of nonequivalent units, whereas variables such as diastolic blood pressure and AST had just 2 units, 1 equivalent to the canonical unit and the canonical unit itself. Diversity of units within populations (values from individual data partner and measurement concept combinations) was minimal (Supplementary Figure S7), indicating that generally just 1 unique unit was used per laboratory test at data partner sites.
Unit conversion workflow
A total of 52 measurement variables important to COVID-19 research, including respiratory rate, body weight, and temperature, were selected for unit harmonization (Figure 2), corresponding to a total of 1.608 billion rows of data and 297 measurement concepts. A total of 299 mapping functions (Supplementary Table S3) were manually curated by clinicians and informaticians for use in unit harmonization.
KS test P-value threshold determination
The distributions of validation-step KS test P values were plotted to identify the best threshold (Figure 4). At P values above 0.00001 (1e−5) we saw almost exclusive presence of KS-test results for comparisons between equivalent inferred units versus known units. Supplementary Figure S8A and B highlight the example of body weight, where the only exception to this pattern occurred due to an incorrect unit assignment by the data partner site. Based on this body weight example, Figure 4 distribution profile, and Supplementary Figure S4 that demonstrates lack of false positive outlier variables (aside from CRP, described below), we judged the threshold of .00001 sufficient to accurately distinguish the “true” from “false” units. We additionally checked Leukocytes [#/volume] in Blood by Manual count. As described above, this measurement concept appeared to be an outlier within its concept set, but on inspection of P values for “equivalent” versus “nonequivalent” inferred units versus the masked known units, we obtained good distinction above and below the threshold, thus supporting our decision to avoid separating out these concepts into their own concept set.
Unit inference omission criteria and measurement variables omitted from unit inference
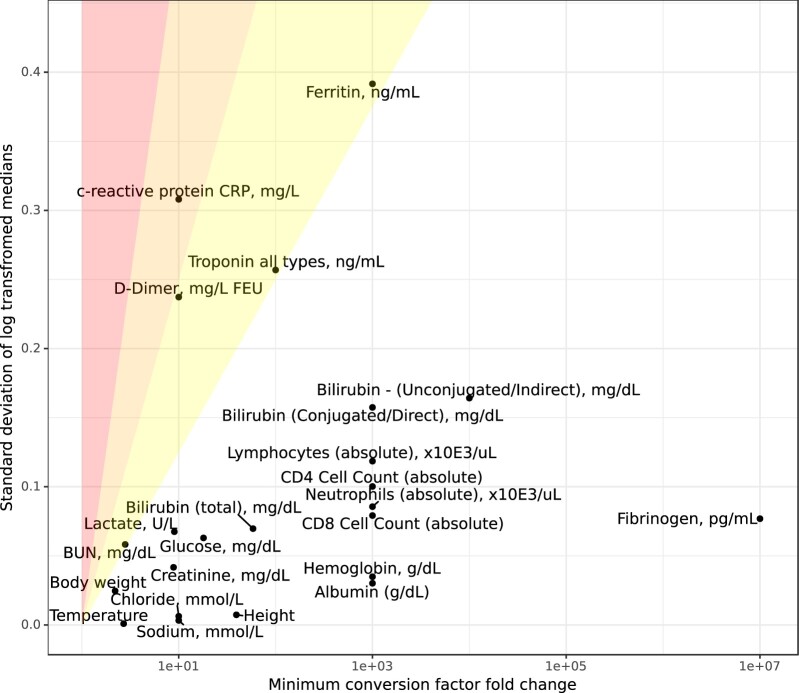
Further analysis of CDFs indicated that, compared to variables such as body weight (Supplementary Figure S9A), c-reactive protein (CRP) has highly overlapping and even interleaved value distributions across its 2 nonequivalent units of milligram per deciliter and milligram per liter, that is distributions for milligram per deciliter can either be found with higher values or lower values than milligram per liter (Supplementary Figure S9B). This overlap indicated that CRP would not be amenable to unit inference. We systematically identified all measurement variables that were refractory to unit inference (Materials and Methods, Figure 3B and Supplementary Figure S2B), including a comparison of transformed measures of dispersion and minimum conversion factor fold change (Figure 5). CRP was the only variable to fall above a threshold of 0.25, indicating substantial overlap across distinct units.
Figure 5.
Omitting variables where units cannot be uniquely assigned; Unit inference omission criteria. The standard deviation of the log median harmonized values (above the KS test P-value threshold) was used as a measure of closeness of different populations of values, and was compared to the log of the minimum conversion factor to determine the level of overlap expected between different units. Ratios: 0.125–0.25 (right-most shaded segment), 0.25–0.5 (middle shaded segment), and >0.5 (left-most shaded segment). KS test: Kolmogorov-Smirnov test.
Proportions of data partner contributions to measured variable reference distributions
The proportion each data partner contributed to the “known units” used for unit inference reference distributions for measured variables is summarized in Supplementary Figure S10, which shows proportion sizes along with their counts. Due to the large number of data partners and general homogeneity in contribution size from each of the data partners, in no case is there a single site that is in the majority for the generation of a reference distribution within 1 unit inference pipeline. The highest proportion found is 0.39, and only 4 concept sets contain any data partner that claims greater than a quarter of the data, 2 being vitals measurements that have only 1:1 or canonical units. The median data partner proportion over all concept sets was 0.017, and mean (SD) of 0.028 ± 0.036, and the vast majority of data partner proportions are of similar size.
Harmonized and inferred units across measurement variables
An overview of the total counts and percentages of harmonized and inferred units can be found in Table 2. There were 1.61 billion input records with values for the harmonization pipeline from the 52 measurement variables of research priority, of which 933 million had valid units. The 675 million that were missing units were processed through the unit inference pipeline, of which 527 million were successfully inferred (78%). In total, 32.8% of the records with values had an inferred unit successfully ascribed. Harmonized values were present for 88% of the data, with 45% coming from nonequivalent units. Figure 6A shows the proportion of inference and harmonization per measurement variable, and identifies large data quality disparities from the source, with temperature particularly problematic for unit missingness.
Table 2.
Counts and percentages of harmonized and inferred units
| Metric | Count | Percentage |
|---|---|---|
| Total measurements with values present | 1 607 758 125 | N/A |
| Total measurements with valid units | 933 030 577 | 58.0 |
| Total measurements without units | 674 727 548 | 42.0 |
| Total nonequivalent units harmonized | 725 051 924 | 45.1 |
| Total harmonized | 1 416 354 459 | 88.1 |
| Total units inferred | 527 400 086 | 78.2a, 32.8b |
Note: Harmonized and inferred unit counts and percentages were calculated across all measured variables out of a total of 1 607 758 125 measurements with values, of which 674 727 548 (42%) had missing units.
Out of total records that were missing units.
Out of total measurements with values.
Figure 6.
Overview and examples of successful harmonized and inferred units. (A) Percentage of values with harmonized and inferred units by measurement variable. Roughly half of the data had correct units and did not require conversion (light green), while half of the data had their units inferred (blue). A minority of values had units that needed conversion (dark green), and the smallest group of data had nonsensical or mislabeled units (black). (B) Original units and their values for body weight and harmonized data for body weight. (C) Inferred versus observed harmonized value distributions.
Sources of unit missingness
Supplementary Figure S11 illustrates the proportion of missing units from the source CDMs, proportion of records with source units that are mapped to standard OMOP units, and proportions of final inferred and noninferred units, showing the high level of data salvage via our unit inference pipeline in cases where units were missing.
Harmonized value distributions indicate successful unit harmonization and inference
An example of successful unit harmonization is shown for body weight in Figure 6B, where grams, kilograms, ounces, and pounds are harmonized to kilograms with similar distributions across the distinct units. Harmonized values for inferred versus reference (known) units across measured variables displayed highly similar distributions (Figure 6C), indicating significant bias or error was not introduced during the unit inference process.
DISCUSSION
We were successful in harmonizing lab data across a very large set of pooled EHR data, and were able to reclaim data that would otherwise have been lost to analysis. With SME consensus, we grouped together sets of semantically similar analytes (and, where relevant, of specimen type) expected by the SMEs to have interoperoberable and convertible measurement units into concept sets for 52 variables important to the understanding of COVID-19. We found, on average (SD), 2.9 ± 1.4 different units used per concept set. Using our pipeline, we harmonized 88.1% of the measurement data that had values present, and inferred units for 78.2% of measurements where units were absent. Missing units pertained to 41.9% of the measurements with values, and our pipeline for inferring missing units shows a false positive rate of 2.7% and a false negative rate of 14.0% (true rates are likely lower due to apparent unit misassignments by sites). We found CRP refractory to unit inference, as previously shown.6
Our approach has several attractions. First, the unit-harmonization pipeline can be integrated with other data-quality review pipelines. The burden of improving lab-data quality is not placed on the sites, except for specific areas of data quality that our harmonization and inference cannot address (eg, Figures 5 and 6A). Misassigned units can be easily identified and shown to sites for correction, which in turn is an example of the potential of this pipeline to improve data partner sites’ own data integrity.
Second, our process reduces or eliminates the need for individual researchers to perform unit conversions.
Third, our work enables consistent research within N3C: all analyses using the harmonized variables can share programming code and the resulting numerical results will be harmonized. For example, calculation of derived variables, such as BMI, was enabled due to the unit harmonization of height and weight.
Fourth, our pipeline enables use of measurements that would otherwise be unavailable for analysis. Reclaiming 78% of data associated with missing data raises the precision of our results (through increased amount of data) and reduces potential bias, due to important factors that may have been associated with the data that were missing units.
Fifth, some of the concept sets we used for unit harmonization are broader than one would use for clinical purposes or analyses and so the distributions are, of necessity, broad. In the case where further distinctions within concept sets may be important (eg, in measurements of venous vs arterial blood), our approach can be applied to assess whether the distinctions matter: If the CDFs of contributing sites lead to nonsignificant KS P values when units are converted then compared to reference values, then no further work on unit harmonization is needed. Researchers should still distinguish the arterial and venous measurements, for example, in their analyses.
Our approach has implications for other researchers beyond N3C. The canonical units, concept sets, conversion formulae, and accepted-value ranges for each measurement variable that we developed can serve as a harmonization resource for the growing number of initiatives aiming to extract insight from patient medical lab records, particularly if present in the widely used OMOP format. The comprehensive and easily interpretable table of unit conversions for labs provided in Supplementary Table S3 can serve as a resource in the context of CDM unit harmonization and any other application that requires unit conversions. The canonical units defined herein have been included as example units for measurement concepts on the LOINC by Regenstrief website.15
Finally, while unit conversions could be applied in a distributed-data environment, our reclaiming of data with missing units relies on the availability of data from all sites. Thus, the ability to reclaim these data is an advantage of a pooled-data architecture. For example, 51.2% of the data partner sites contained body weight records without units (with 7 sites missing units entirely), and 82.4% of data partner sites had only 1 valid source unit to act as a reference. The approach taken of determining P value differences and value distribution differences across units in order to more confidently assign inferred units was thus uniquely enabled by pooling data from all contributing sites, and would be impossible to replicate on a site-by-site basis for the majority of data partner sites. Additionally, even in the case that a site’s unit diversity mirrors centralized unit diversity for a lab measurement, ground truth of the expected value distribution for each unit cannot be accurately obtained on a site-by-site basis due to the potential for misassigned units.
There is further work to determine whether our method for selecting values for the reference distributions per measurement variable for unit inference can be improved. For example, an alternative to random selection over all data partners would be to ensure, where possible, that the same numbers of data points are sampled per data partner, per measured variable, to avoid data partner oversampling and potentially reduce the impact of any one site on the reference distributions. However, the current sampling regime shows good performance for identifying canonical units, rescuing missing data, and identifying outlier sites.
Future work will also involve maintenance of the pipeline over time. As new source units emerge from sites this will result in further standard OMOP unit concepts entering the lab measurement data that require mapping to conversions. Additionally, some sites have included varying amounts of custom units that differ from those expected from the source CDM, and thus are missing from our source to standard OMOP mapping. There is therefore continuous work in N3C to improve the comprehensiveness of source unit mapping.
In cases where a centralized approach is not feasible, or to distribute the resources collected at the centralized level, we also envisage that our centralized approach could be adapted to enable federated use of aggregated resources such as reference distributions to allow for unit inference at the data partner site-level, or the unit harmonization conversions could be shared to the federated sites.
There are some limitations in our approach. The range of units found for each lab measurement may not constitute the full universe of potential units, and thus unit comparisons can only be made for the units we see in the N3C Enclave. In imputing missing units, there is the risk of false positive imputation. Measured variables such as body weight were found to contain nonequivalent reference and inferred units above the assigned KS-test P value threshold (1e−5) during unit inference validation (Supplementary Figure S8A). CDF analysis (Supplementary Figures S8B and S9A) suggested that these were from incorrectly assigned units from the data-contributing sites; for example, one of the CDFs from pounds matched the typical distribution for ounces. Aside from examining false positives at the upper end of the KS-test P value distribution, we also looked for false negatives (units with low P values containing equivalent reference and inferred units), which would lead to data not included in analyses that should be. CDFs of measured variables with P values <1e−5 were plotted (as in Supplementary Figure S8B), and for some variables, such as SpO2, the distributions for certain measurement concepts, for example, oxygen saturation [Pure mass fraction] in Blood, appeared to be different from the average distribution that focuses on arterial blood. While our concept sets largely capture equivalent concepts, this difference in CDFs exemplifies the challenges to inferring units for concepts that may be differentially employed. The same is also true for variables such as body weight in subpopulations such as children, where the correct units are assigned but the overall distribution of values does not match the typical distribution of the general population. These examples highlight the urgent need for more sophisticated unit-inference techniques that take into account specific patient subgroups and samples. Since our unit-inference method assumed just 1 unique unit per measured variable—data partner—measurement concept triple (as supported by Supplementary Figure S7), there also may be poor unit inference in the few cases where there are instead multiple distinct unknown units, although we did not rigorously test how mixtures of units affect unit inference via the KS test. Bayesian inference or machine learning using values from other measurement variables within each patient or patient subgroup as a cross check to infer likely units would be a possible next step for our workflow, for example values from height and BMI could help infer a missing weight measurement within an individual patient.
Although more sophisticated machine learning approaches to the problem of unit inference can potentially be developed, and maintenance of the pipeline over time will be necessary, our pipeline is easily interpretable, and runs in <2.5 h on the billions of rows of measurement data processed weekly within the N3C Enclave.
CONCLUSION
As collaborative research projects continue to grow and to incorporate larger and more diverse sources of data, we need to minimize time spent preparing data and to maximize its usability. In this work we have developed and implemented a pipeline to harmonize measurements to a canonical unit and to infer missing units of measurement. This pipeline allowed our team to salvage otherwise unusable data and to remove the need for duplicative work converting units for each N3C project. While this work was driven by the specific needs of the N3C, such a pipeline could be incorporated into the analysis of any large dataset of pooled EHR data.
FUNDING
The analyses described in this publication were conducted with data or tools accessed through the NCATS N3C DataEnclave https://covid.cd2h.org and N3C Attribution & Publication Policy v1.2-2020-08-25b, and supported by NCATS U24 TR002306, and NIGMS National Institute of General Medical Sciences, 5U54GM104942-04. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. This work was supported by the National Institutes of Health, National Center for Advancing Translational Sciences Institute grant number U24TR002306.
AUTHOR CONTRIBUTIONS
KRB, BA, AM, EN, ATG, and RAM prepared code utilized in the pipeline. KRB, JW, and HL created the conversion formulas. KRB, RAM, CB, and JY prepared tables and figures. KRB, JTW, HL, and RAM prepared the manuscript. All authors reviewed the manuscript and provided feedback.
SUPPLEMENTARY MATERIAL
Supplementary material is available at Journal of the American Medical Informatics Association online.
Supplementary Material
ACKNOWLEDGMENTS
We would like to thank Swapna Abhyankar from the Regenstrief Institute for her collaboration in the submission of example UCUM units for the LOINC table. This research was possible because of the patients whose information is included within the data from participating organizations (covid.cd2h.org/dtas) and the organizations and scientists (covid.cd2h.org/duas) who have contributed to the ongoing development of this community resource (cite this https://doi.org/10.1093/jamia/ocaa196). The N3C data transfer to NCATS is performed under a Johns Hopkins University Reliance Protocol # IRB00249128 or individual site agreements with NIH. The N3C Data Enclave is managed under the authority of the NIH; information can be found at https://ncats.nih.gov/n3c/resources. We gratefully acknowledge contributions from the following N3C core teams. Principal Investigators: Melissa A. Haendel*, Christopher G. Chute*, Kenneth R. Gersing, Anita Walden; Workstream, subgroup and administrative leaders: Melissa A. Haendel*, Tellen D. Bennett, Christopher G. Chute, David A. Eichmann, Justin Guinney, Warren A. Kibbe, Hongfang Liu, Philip R.O. Payne, Emily R. Pfaff, Peter N. Robinson, Joel H. Saltz, Heidi Spratt, Justin Starren, Christine Suver, Adam B. Wilcox, Andrew E. Williams, Chunlei Wu; Key liaisons at data partner sites: Regulatory staff at data partner sites; Individuals at the sites who are responsible for creating the datasets and submitting data to N3C; Data Ingest and Harmonization Team: Christopher G. Chute*, Emily R. Pfaff*, Davera Gabriel, Stephanie S. Hong, Kristin Kostka, Harold P. Lehmann, Richard A. Moffitt, Michele Morris, Matvey B. Palchuk, Xiaohan Tanner Zhang, Richard L. Zhu; Phenotype Team (Individuals who create the scripts that the sites use to submit their data, based on the COVID and Long COVID definitions): Emily R. Pfaff*, Benjamin Amor, Mark M. Bissell, Marshall Clark, Andrew T. Girvin, Stephanie S. Hong, Kristin Kostka, Adam M. Lee, Robert T. Miller, Michele Morris, Matvey B. Palchuk, Kellie M. Walters. Project Management and Operations Team: Anita Walden*, Yooree Chae, Connor Cook, Alexandra Dest, Racquel R. Dietz, Thomas Dillon, Patricia A. Francis, Rafael Fuentes, Alexis Graves, Julie A. McMurry, Andrew J. Neumann, Shawn T. O'Neil, Usman Sheikh, Andréa M. Volz, Elizabeth Zampino. Partners from NIH and other federal agencies: Christopher P. Austin*, Kenneth R. Gersing*, Samuel Bozzette, Mariam Deacy, Nicole Garbarini, Michael G. Kurilla, Sam G. Michael, Joni L. Rutter, Meredith Temple-O'Connor. Analytics Team (Individuals who build the Enclave infrastructure, help create codesets, variables, and help Domain Teams and project teams with their datasets): Benjamin Amor*, Mark M. Bissell, Katie Rebecca Bradwell, Andrew T. Girvin, Amin Manna, Nabeel Qureshi. Publication Committee Management Team: Mary Morrison Saltz*, Christine Suver*, Christopher G. Chute, Melissa A. Haendel, Julie A. McMurry, Andréa M. Volz, Anita Walden. Publication Committee Review Team: Carolyn Bramante, Jeremy Richard Harper, Wenndy Hernandez, Farrukh M Koraishy, Federico Mariona, Amit Saha, Satyanarayana Vedula. Stony Brook University—U24TR002306 • University of Oklahoma Health Sciences Center—U54GM104938: Oklahoma Clinical and Translational Science Institute (OCTSI) • West Virginia University—U54GM104942: West Virginia Clinical and Translational Science Institute (WVCTSI) • University of Mississippi Medical Center—U54GM115428: Mississippi Center for Clinical and Translational Research (CCTR) • University of Nebraska Medical Center—U54GM115458: Great Plains IDeA-Clinical & Translational Research • Maine Medical Center—U54GM115516: Northern New England Clinical & Translational Research (NNE-CTR) Network • Wake Forest University Health Sciences—UL1TR001420: Wake Forest Clinical and Translational Science Institute • Northwestern University at Chicago—UL1TR001422: Northwestern University Clinical and Translational Science Institute (NUCATS) • University of Cincinnati—UL1TR001425: Center for Clinical and Translational Science and Training • The University of Texas Medical Branch at Galveston—UL1TR001439: The Institute for Translational Sciences • Medical University of South Carolina—UL1TR001450: South Carolina Clinical & Translational Research Institute (SCTR) • University of Massachusetts Medical School Worcester—UL1TR001453: The UMass Center for Clinical and Translational Science (UMCCTS) • University of Southern California—UL1TR001855: The Southern California Clinical and Translational Science Institute (SC CTSI) • Columbia University Irving Medical Center—UL1TR001873: Irving Institute for Clinical and Translational Research • George Washington Children's Research Institute—UL1TR001876: Clinical and Translational Science Institute at Children's National (CTSA-CN) • University of Kentucky—UL1TR001998: UK Center for Clinical and Translational Science • University of Rochester—UL1TR002001: UR Clinical & Translational Science Institute • University of Illinois at Chicago—UL1TR002003: UIC Center for Clinical and Translational Science • Penn State Health Milton S. Hershey Medical Center—UL1TR002014: Penn State Clinical and Translational Science Institute • The University of Michigan at Ann Arbor—UL1TR002240: Michigan Institute for Clinical and Health Research • Vanderbilt University Medical Center—UL1TR002243: Vanderbilt Institute for Clinical and Translational Research • University of Washington—UL1TR002319: Institute of Translational Health Sciences • Washington University in St. Louis—UL1TR002345: Institute of Clinical and Translational Sciences • Oregon Health & Science University—UL1TR002369: Oregon Clinical and Translational Research Institute • University of Wisconsin-Madison—UL1TR002373: UW Institute for Clinical and Translational Research • Rush University Medical Center—UL1TR002389: The Institute for Translational Medicine (ITM) • The University of Chicago—UL1TR002389: The Institute for Translational Medicine (ITM) • University of North Carolina at Chapel Hill—UL1TR002489: North Carolina Translational and Clinical Science Institute • University of Minnesota—UL1TR002494: Clinical and Translational Science Institute • Children's Hospital Colorado—UL1TR002535: Colorado Clinical and Translational Sciences Institute • The University of Iowa—UL1TR002537: Institute for Clinical and Translational Science • The University of Utah—UL1TR002538: Uhealth Center for Clinical and Translational Science • Tufts Medical Center—UL1TR002544: Tufts Clinical and Translational Science Institute • Duke University—UL1TR002553: Duke Clinical and Translational Science Institute • Virginia Commonwealth University—UL1TR002649: C. Kenneth and Dianne Wright Center for Clinical and Translational Research • The Ohio State University—UL1TR002733: Center for Clinical and Translational Science • The University of Miami Leonard M. Miller School of Medicine—UL1TR002736: University of Miami Clinical and Translational Science Institute • University of Virginia—UL1TR003015: iTHRIVL Integrated Translational health Research Institute of Virginia • Carilion Clinic—UL1TR003015: iTHRIVL Integrated Translational health Research Institute of Virginia • University of Alabama at Birmingham—UL1TR003096: Center for Clinical and Translational Science • Johns Hopkins University—UL1TR003098: Johns Hopkins Institute for Clinical and Translational Research • University of Arkansas for Medical Sciences—UL1TR003107: UAMS Translational Research Institute • Nemours—U54GM104941: Delaware CTR ACCEL Program • University Medical Center New Orleans—U54GM104940: Louisiana Clinical and Translational Science (LA CaTS) Center • University of Colorado Denver, Anschutz Medical Campus—UL1TR002535: Colorado Clinical and Translational Sciences Institute • Mayo Clinic Rochester—UL1TR002377: Mayo Clinic Center for Clinical and Translational Science (CCaTS) • Tulane University—UL1TR003096: Center for Clinical and Translational Science • Loyola University Medical Center—UL1TR002389: The Institute for Translational Medicine (ITM) • Advocate Health Care Network—UL1TR002389: The Institute for Translational Medicine (ITM) • OCHIN—INV-018455: Bill and Melinda Gates Foundation grant to Sage Bionetworks • The Rockefeller University—UL1TR001866: Center for Clinical and Translational Science • The Scripps Research Institute—UL1TR002550: Scripps Research Translational Institute • University of Texas Health Science Center at San Antonio—UL1TR002645: Institute for Integration of Medicine and Science • The University of Texas Health Science Center at Houston—UL1TR003167: Center for Clinical and Translational Sciences (CCTS) • NorthShore University HealthSystem—UL1TR002389: The Institute for Translational Medicine (ITM) • Yale New Haven Hospital—UL1TR001863: Yale Center for Clinical Investigation • Emory University—UL1TR002378: Georgia Clinical and Translational Science Alliance • Weill Medical College of Cornell University—UL1TR002384: Weill Cornell Medicine Clinical and Translational Science Center • Montefiore Medical Center—UL1TR002556: Institute for Clinical and Translational Research at Einstein and Montefiore • Medical College of Wisconsin—UL1TR001436: Clinical and Translational Science Institute of Southeast Wisconsin • University of New Mexico Health Sciences Center—UL1TR001449: University of New Mexico Clinical and Translational Science Center • George Washington University—UL1TR001876: Clinical and Translational Science Institute at Children's National (CTSA-CN) • Stanford University—UL1TR003142: Spectrum: The Stanford Center for Clinical and Translational Research and Education • Regenstrief Institute—UL1TR002529: Indiana Clinical and Translational Science Institute • Cincinnati Children's Hospital Medical Center—UL1TR001425: Center for Clinical and Translational Science and Training • Boston University Medical Campus—UL1TR001430: Boston University Clinical and Translational Science Institute • The State University of New York at Buffalo—UL1TR001412: Clinical and Translational Science Institute • Aurora Health Care—UL1TR002373: Wisconsin Network For Health Research • Brown University—U54GM115677: Advance Clinical Translational Research (Advance-CTR) • Rutgers, The State University of New Jersey—UL1TR003017: New Jersey Alliance for Clinical and Translational Science • Loyola University Chicago—UL1TR002389: The Institute for Translational Medicine (ITM) • UL1TR001445: Langone Health's Clinical and Translational Science Institute • Children's Hospital of Philadelphia—UL1TR001878: Institute for Translational Medicine and Therapeutics • University of Kansas Medical Center—UL1TR002366: Frontiers: University of Kansas Clinical and Translational Science Institute • Massachusetts General Brigham—UL1TR002541: Harvard Catalyst • Icahn School of Medicine at Mount Sinai—UL1TR001433: ConduITS Institute for Translational Sciences • Ochsner Medical Center—U54GM104940: Louisiana Clinical and Translational Science (LA CaTS) Center • HonorHealth—None (Voluntary) • University of California, Irvine—UL1TR001414: The UC Irvine Institute for Clinical and Translational Science (ICTS) • University of California, San Diego—UL1TR001442: Altman Clinical and Translational Research Institute • University of California, Davis—UL1TR001860: UCDavis Health Clinical and Translational Science Center • University of California, San Francisco—UL1TR001872: UCSF Clinical and Translational Science Institute • University of California, Los Angeles—UL1TR001881: UCLA Clinical Translational Science Institute • University of Vermont—U54GM115516: Northern New England Clinical & Translational Research (NNE-CTR) Network • Arkansas Children's Hospital—UL1TR003107: UAMS Translational Research Institute.
CONFLICT OF INTEREST STATEMENT
KRB, BA, AM, EN, MB, and ATG are employees of Palantir Technologies. MH is a founder of Pryzm Health.
DATA AVAILABILITY
The N3C data transfer to NCATS is performed under a Johns Hopkins University Reliance Protocol # IRB00249128 or individual site agreements with NIH. The N3C Data Enclave is managed under the authority of the NIH; information can be found at ncats.nih.gov/n3c/resources. Enclave data are protected, and can be accessed for COVID-related research with an NIH-approved approved (1) IRB protocol and (2) institutional Data Use Request (DUR). A detailed accounting of data protections and access tiers is found at https://ncats.nih.gov/n3c/resources/data-access. Enclave and data access instructions can be found at https://covid.cd2h.org/for-researchers; all code used to produce the analyses in this manuscript is available within the N3C Enclave to users with valid login credentials to support reproducibility.
Contributor Information
Katie R Bradwell, Palantir Technologies, Denver, Colorado, USA.
Jacob T Wooldridge, Department of Biomedical Informatics, Stony Brook University, Stony Brook, New York, USA.
Benjamin Amor, Palantir Technologies, Denver, Colorado, USA.
Tellen D Bennett, Section of Informatics and Data Science, Department of Pediatrics, University of Colorado School of Medicine, University of Colorado, Aurora, Colorado, USA.
Adit Anand, Department of Biomedical Informatics, Stony Brook University, Stony Brook, New York, USA.
Carolyn Bremer, Department of Biomedical Informatics, Stony Brook University, Stony Brook, New York, USA.
Yun Jae Yoo, Department of Biomedical Informatics, Stony Brook University, Stony Brook, New York, USA.
Zhenglong Qian, Department of Biomedical Informatics, Stony Brook University, Stony Brook, New York, USA.
Steven G Johnson, Institute for Health Informatics, University of Minnesota, Minneapolis, Minnesota, USA.
Emily R Pfaff, Department of Medicine, North Carolina Translational and Clinical Sciences Institute, University of North Carolina at Chapel Hill, Chapel Hill, North Carolina, USA.
Andrew T Girvin, Palantir Technologies, Denver, Colorado, USA.
Amin Manna, Palantir Technologies, Denver, Colorado, USA.
Emily A Niehaus, Palantir Technologies, Denver, Colorado, USA.
Stephanie S Hong, School of Medicine, Section of Biomedical Informatics and Data Science, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
Xiaohan Tanner Zhang, Department of Medicine, Johns Hopkins, Baltimore, Maryland, USA.
Richard L Zhu, Department of Medicine, Johns Hopkins, Baltimore, Maryland, USA.
Mark Bissell, Palantir Technologies, Denver, Colorado, USA.
Nabeel Qureshi, Palantir Technologies, Denver, Colorado, USA.
Joel Saltz, Department of Biomedical Informatics, Stony Brook University, Stony Brook, New York, USA.
Melissa A Haendel, Center for Health AI, University of Colorado, Aurora, Colorado, USA.
Christopher G Chute, Schools of Medicine, Public Health, and Nursing, Johns Hopkins University, Baltimore, Maryland, USA.
Harold P Lehmann, Department of Medicine, Johns Hopkins, Baltimore, Maryland, USA.
Richard A Moffitt, Department of Biomedical Informatics, Stony Brook University, Stony Brook, New York, USA.
REFERENCES
- 1. Haendel MA, Chute CG, Bennett TD, et al. ; N3C Consortium. The National COVID Cohort Collaborative (N3C): rationale, design, infrastructure, and deployment. J Am Med Inform Assoc 2021; 28 (3): 427–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Observational Health Data Sciences and Informatics (OHDSI). Definition and DDLs for the OMOP Common Data Model (CDM) l Version 5.3. Github; 2018. https://github.com/OHDSI/CommonDataModel Accessed April 13, 2020.
- 3. Hripcsak G, Duke JD, Shah NH, et al. Observational Health Data Sciences and Informatics (OHDSI): opportunities for observational researchers. Stud Health Technol Inform 2015; 216: 574–8. [PMC free article] [PubMed] [Google Scholar]
- 4. Huff SM, Rocha RA, McDonald CJ, et al. Development of the Logical Observation Identifier Names and Codes (LOINC) vocabulary. J Am Med Inform Assoc 1998; 5 (3): 276–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schadow G, McDonald CJ. The Unified Code for Units of Measure [Internet]. Indianapolis, IN: Regenstrief Institute, Inc.; c1999–2013 [cited 2021 Sept 01]. http://unitsofmeasure.org/trac/. Jointly published with the UCUM Organization.
- 6. Ficheur G, Chazard E, Schaffar A, Genty M, Beuscart R. Interoperability of medical databases: construction of mapping between hospitals laboratory results assisted by automated comparison of their distributions. AMIA Annu Symp Proc 2011; 2011: 392–401. [PMC free article] [PubMed] [Google Scholar]
- 7. Rajput AM, Ballout S, Drenkhahn C. Standardizing the unit of measurements in LOINC-coded laboratory tests can significantly improve semantic interoperability. Stud Health Technol Inform 2020; 275: 234–5. [DOI] [PubMed] [Google Scholar]
- 8. Drenkhahn C, Ingenerf J. The LOINC content model and its limitations of usage in the laboratory domain. Stud Health Technol Inform 2020; 270: 437–42. [DOI] [PubMed] [Google Scholar]
- 9. Drenkhahn C, Duhm-Harbeck P, Ingenerf J. Aggregation and visualization of laboratory data by using ontological tools based on LOINC and SNOMED CT. Stud Health Technol Inform 2019; 264: 108–12. [DOI] [PubMed] [Google Scholar]
- 10. Hauser RG, Quine DB, Ryder A, Campbell S. Unit conversions between LOINC codes. J Am Med Inform Assoc 2018; 25 (2): 192–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Vreeman DJ, Abhyankar S, McDonald CJ. Response to unit conversions between LOINC codes. J Am Med Inform Assoc 2018; 25 (5): 614–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bennett TD, Moffitt RA, Hajagos JG, et al. , National COVID Cohort Collaborative (N3C) Consortium. Clinical characterization and prediction of clinical severity of SARS-CoV-2 infection among US adults using data from the US National COVID Cohort Collaborative. JAMA Netw Open 2021; 4 (7): e2116901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chakravarti IM, Laha RG, Roy J. Handbook of Methods of Applied Statistics. Vol. 1. New York, NY: Wiley; 1967. [Google Scholar]
- 14. Schuemie MJ, Ryan PB, Hripcsak G, Madigan D, Suchard MA. Improving reproducibility by using high-throughput observational studies with empirical calibration. Philos Trans A Math Phys Eng Sci 2018; 376 (2128): 20170356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.LOINC release notes. LOINC by Regenstrief; 2021. https://loinc.org/kb/loinc-release-notes/. Accessed February 15, 2021.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The N3C data transfer to NCATS is performed under a Johns Hopkins University Reliance Protocol # IRB00249128 or individual site agreements with NIH. The N3C Data Enclave is managed under the authority of the NIH; information can be found at ncats.nih.gov/n3c/resources. Enclave data are protected, and can be accessed for COVID-related research with an NIH-approved approved (1) IRB protocol and (2) institutional Data Use Request (DUR). A detailed accounting of data protections and access tiers is found at https://ncats.nih.gov/n3c/resources/data-access. Enclave and data access instructions can be found at https://covid.cd2h.org/for-researchers; all code used to produce the analyses in this manuscript is available within the N3C Enclave to users with valid login credentials to support reproducibility.