Abstract
ICU Cockpit: a secure, fast, and scalable platform for collecting multimodal waveform data, online and historical data visualization, and online validation of algorithms in the intensive care unit. We present a network of software services that continuously stream waveforms from ICU beds to databases and a web-based user interface. Machine learning algorithms process the data streams and send outputs to the user interface. The architecture and capabilities of the platform are described. Since 2016, the platform has processed over 89 billion data points (N = 979 patients) from 200 signals (0.5–500 Hz) and laboratory analyses (once a day). We present an infrastructure-based framework for deploying and validating algorithms for critical care. The ICU Cockpit is a Big Data platform for critical care medicine, especially for multimodal waveform data. Uniquely, it allows algorithms to seamlessly integrate into the live data stream to produce clinical decision support and predictions in clinical practice.
Keywords: clinical research data platform, high resolution waveforms, data dashboards, livestream UI, machine learning integration
INTRODUCTION
Clinical information systems (CIS) are gradually becoming indispensable in intensive care units (ICUs) to record and integrate medical data from patient history, laboratory values, monitoring parameters, and medications. Commercially available CIS, however, do not meet requirements to collect and process high-resolution data streams (full-resolution waveforms). Data logging and storage in many commercial systems is performed only intermittently, capturing information for routine clinical purposes,1,2 thereby limiting potential benefits of clinical decision support systems.
Simultaneously, the advent of deep learning based machine learning (ML) algorithms spurred the development of more accurate but complex models for outcome prediction in critical care.3,4 With more complex models also came the demand for large and detailed datasets. Open source datasets such as Physionet and Physiobank,5 MIMIC III,6 eICU,7 and PIC8 are benchmark datasets used by ML researchers to compare model performance under standardized metrics. Although promising models have been developed, as for early detection and outcome prediction in sepsis,9–12 few have been implemented in ICU practice.13 This phenomenon is called the “AI chasm” and refers to the fact that offline, purely computational validation of models is insufficient for successful clinical deployment.14 The danger of using models developed from ICU context specific, retrospective datasets stems from the inherent problem of “model generalization” to new, as yet unseen patient conditions and hospital environments. Thus, models trained offline still need to be validated online running on real-time data streams, in different data ecosystems, where signals are measured by different devices.14,15
Started in 2014 at the Neurocritical Care Unit, University Hospital Zurich, the aim of the ICU Cockpit project was to develop a secure, fast, and scalable platform for the collection of multimodal waveform data, and deployment of real-time decision support systems integrating live data streams in the ICU. Crucially, ICU Cockpit infrastructure has evolved to cover the entire ML workflow from artificial intelligence (AI) based computational disease modeling, to online, real-time validation of algorithms in clinical trials.
METHODS
Research network
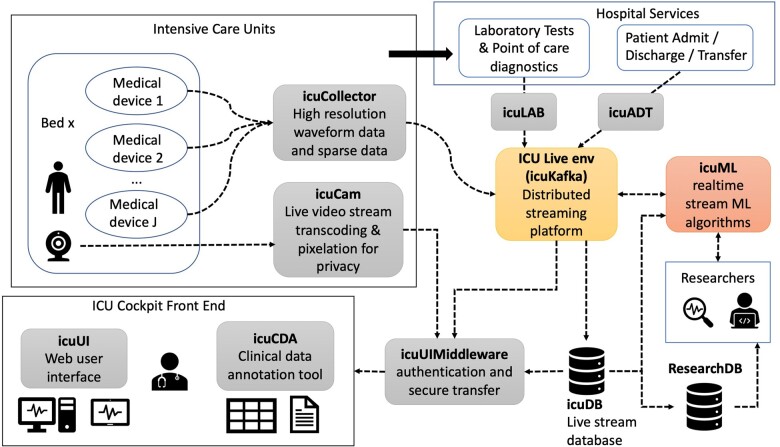
The ICU Cockpit platform is a network of data sources, databases, microservices, and user interfaces (UIs) built around a central data messaging service that allows users to acquire, visualize, compute on, and annotate data from critical care patients. The main components are shown in Figure 1. A detailed description of each component is provided in Supplementary Appendix A1.
Figure 1.
ICU Cockpit platform component diagram. Patient data originate from bedside medical devices installed in ICUs (top left), and via hospital services external to the ICU Cockpit platform (top right). External services such as laboratory analyses and patient metadata from the hospital patient data management system are collected via icuLAB and icuADT. Live bedside webcam video is fed through a separate service (icuCAM) which encodes and optionally blurs the video for patient privacy. The ICU Cockpit Live Environment (ICU Live env) is the central data packaging and distribution module using the icuKafka distributed messaging service. The live stream of any recorded signal and ML algorithm outputs (from icuML) can be visualized in the ICU Cockpit Frontend user interface (UI) (icuUI, bottom left) and stored in the live stream database (icuDB). Patient metadata, admission status, and clinical operations are accessible and edited through the Clinical Data Annotation user interface (icuCDA). Data postprocessing, analysis and algorithm development occur offline by researchers who receive archived and pseudonymized data from the research database (ResearchDB) after authorization. Offline validated algorithms are transferred to the icuML staging environment for integration. Integrated algorithms receive a constant data stream from icuKafka or icuDB, to perform necessary computations and transmit outputs back into the stream to be displayed and/or stored.
RESULTS
Data flow
Patient data originate from patient bedside in one or more ICUs or from hospital services, mainly in 2 forms: streaming and sparse data. Streaming data consist of live patient video and physiological signals such as electrocardiography (ECG), arterial and intracranial pressure, etc., advanced hemodynamic monitoring (eg, continuous cardiac output), parameters from artificial ventilation (fraction of inspired oxygen, positive end expiratory pressure, etc.) and neuromonitoring (eg, electroencephalography (EEG)) acquired at sampling frequencies of once every second to as high as once every 2 ms (=500 Hz, typically for ECG). Sparse data consist of patient metadata (eg, from patient history or clinical events) entered into the clinical data annotation tool (icuCDA, Supplementary Appendix A2) and measurements made infrequently, for example laboratory tests performed once a day, or arterial blood gas analysis performed once every few hours. Various services send streaming and sparse data to a central Apache Kafka message broker (icuKafka) from which it can be accessed by other data processors in the ICU Cockpit Live Environment, Figure 1.
An admission-discharge-transfer service (icuADT) listens for and publishes events that affect patient status, namely admission, discharge, and transfer to another hospital unit. Laboratory results are collected through the icuLAB service.
Streaming data are collected from different medical devices such as Philips IntelliVue (Philips Medical Systems, Boeblingen, Germany), Hamilton-S1, (Hamilton Medical, Bonaduz, Switzerland), Licox Brain Tissue Oxygen Monitor (Integra Princeton, NJ, USA), etc. via Moberg Component Neuromonitoring Systems (Moberg Research Inc, PA, USA). The icuCollector software packs individual data points into sets of formatted messages and sends them via icuKafka to several sinks, such as the icuDBWriter. The icuDBWriter writes data messages to a PostgreSQL database (icuDB). On average, thousands of data points are written into the icuDB every second from each patient. Data stored in the icuDB are accessible for offline inspection. Older data are archived in the ResearchDB for sharing with researchers that receive authorized access clearance.
Data protection
Patients or their representatives must provide informed consent for the use of their data for research purposes. The consent status is recorded in the icuCDA. If consent is not given or withdrawn, any stored data in icuDB or ResearchDB are immediately deleted. Once consent is given, clinicians involved in patient treatment are allowed to access the database via a secure virtual private network (VPN) and visualize data on computers with VPN access enabled. They can view the patient metadata and treatments. For researchers with approved access, data are pseudonymized via the ICUMRNmapper tool and then exported.
Online stream processing and ML-algorithms
Using data collected by the ICU Cockpit platform, algorithms for neurologic prognostication,16,17 detection of epileptic seizures from bedside video,18 detection of clinically relevant EEG patterns,19 statistical analysis of high-resolution data streams20 as well as false alarm reduction21,22 have been already developed, and validated offline. Promising algorithms can be implemented into the ICU Cockpit as stream processors enriching and augmenting the original data streams. Importantly, computed values become part of the original data stream. As an example, we describe the implementation of a heart rate variability computation algorithm23 into the online streams in Supplementary Appendix A3. Its results can be visualized in a historical data dashboard (Supplementary Appendix A4).
Web user interface for patient monitoring
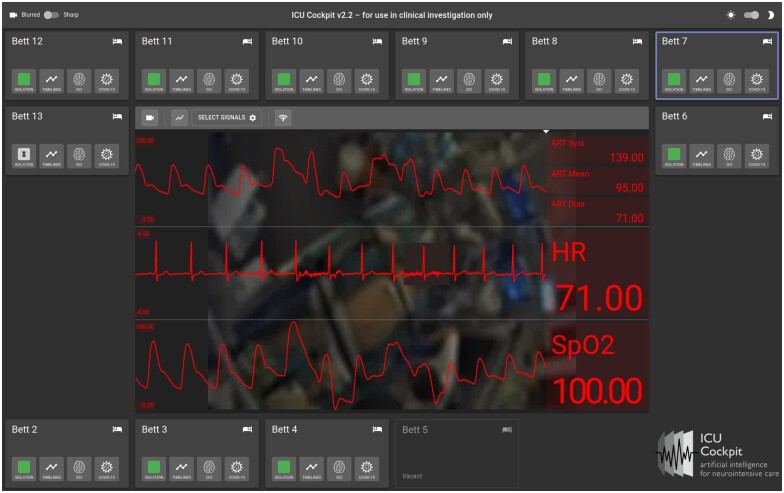
Figure 2 shows a snapshot of the web UI (icuUI) in live use. Each bed of the ICU is represented as a box. In addition to bed number and vacancy, further metadata can be visualized on demand (eg, SARS-CoV-2 status). ML algorithm outputs, such as the predicted probability of delayed cerebral ischemia (DCI) (Figure 2, DCI bed panel button) can be visualized for each bed in a pop-up UI.
Figure 2.
A snapshot of the icuUI feed. The neurocritical care unit has 12 beds. Bed 7 (“Bett 7”) has been selected for display in the central panel. The live webcam video feed from Bed 7 is shown with 3 signals overlaid as charts in the central display panel. The patient face has been masked. Top: ART (arterial blood pressure waveform), Middle: ECG, Bottom: pulse oximetry. Related parametric values (systolic, diastolic and mean arterial blood pressure, heart rate (HR) and Spo2) that are calculated once per second are shown as numbers on the right of each signal. Boxes for each bed additionally contain analytics and metadata such as Covid isolation status (green = not infected), data timelines (‘Timelines’) and algorithm outputs (cf, DCI prediction).
Any bed can be selected for detailed display in the large center panel. A live webcam feed is shown as a backdrop (by default, videos are blurred to protect patient identity). Selected signals are visualized on top of the video feed. Signal charts are backed by the icuKafka layer and are updated as soon as new data are available. Update rates and latencies depend on signal sampling rates as well as network bandwidth. We performed a coarse estimate of signal latency using a stopwatch that used the network time. The ECG sensor on a patient was connected and disconnected several times during routine patient care, and the resulting signal loss was registered within 2 s on the UI.
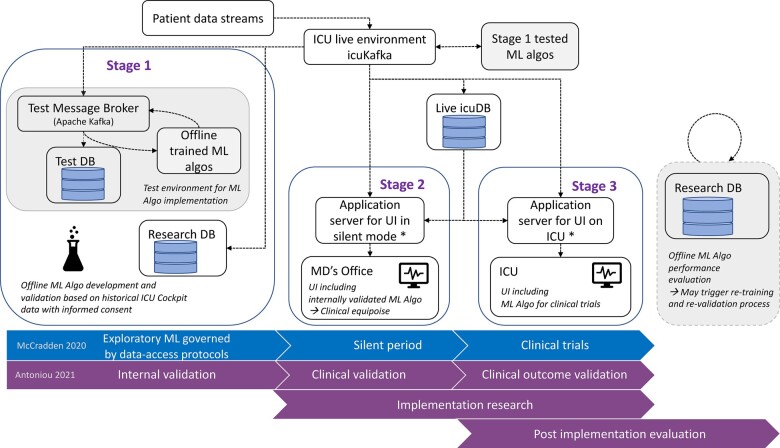
A framework for critical care machine learning model validation
The ICU Cockpit was designed with the aim of implementing, testing, and deploying ML algorithms into the live data stream from ICUs. We have created a framework for model validation that occurs in stages according to the guidelines in references.13,14 First, in Stage 1, models are developed using data stored in ResearchDB. This is also known as internal (offline) validation Figure 3 (bottom). Next, an internally validated model is deployed in a test environment, to ensure that model outputs are produced correctly. Following Stage 1, the model is deployed in the live environment (Stage 2) but with the outputs only visible in the MD’s office where they can be compared to actual patient health status, that is, in “silent” mode. In this stage outputs do not affect the behavior and decision making of clinical staff. Finally, model outputs become visible on the standard icuUI when models are tested in randomized clinical trials (Stage 3) or thereafter. In Stage 3, model outputs are viewed by clinical staff during regular ICU work. Longitudinal monitoring of model performance can be done offline based on model outputs stored in the ResearchDB (Post implementation evaluation). Clinical outcome trials that are only observational studies also fall into this category.
Figure 3.
Clinical validation strategy for medical machine learning models. Data retrieved from the ICU, from the icuCDA and hospital services (eg, icuLab) can be used for model development and validation offline and the learned models can be tested in the data stream in a test environment (Stage 1). All internally validated (Stage 1 tested) models can be run in production and displayed on the UI in the MD’s office (silent period, Stage 2), but only the models validated successfully on prospective data streams (Stage 2) can have model outputs displayed on the UI in the ICU (Stage 3), as in a randomized clinical trial for the model. Purely observational clinical outcome studies and post-implementation evaluation and retraining or fine tuning of algorithms can be performed offline using data from the ResearchDB.
Performance statistics
Since 2016, from a patient cohort of N = 979 admitted to the Neurocritical Care Unit with informed consent, the total number of data points processed and stored by the ICU Cockpit platform was approximately 88.9 billion. These data were collected over 8383 days of admission from 12 ICU beds. Table 1 summarizes statistics across patients.
Table 1.
The following statistics describe the ICU Cockpit platform’s data processing workload
| Statistic | Value (median [min, max]) |
|---|---|
| Number of admitted days | 4 [1, 76] |
| Minimum and maximum number of signals recorded per day | 18 [1, 130], 40 [1, 210] |
| Number of patient hours | 50 [0, 1304] |
| Size of data (giga bytes) | 2.09 [2e−6, 148.41] |
| Sampling rates (min, max in Hz) | 0.5, 500 |
Note: Each statistic is the median [min, max] over 979 patients.
DISCUSSION
Critical care is currently experiencing a big data revolution. Leveraging this data requires IT infrastructure that can (1) support healthcare staff with decision support systems, (2) establish secure access for data to researchers, and (3) allow for the offline and online evaluation of ML algorithms according to the guidelines proposed under CONSORT-AI24 and SPIRIT-AI.25 Recently, the US Food and Drug Administration (FDA) and Health Canada released a 10-point guide for Good Machine Learning Practice for Medical Device Development.26 The ICU Cockpit empowers its users to adhere to these guiding principles, in particular, points which deal with good software engineering practices, model testing in a live environment, the ability to perform statistically sound clinical trials, and continuous monitoring of model performance after deployment.
Few research-oriented data platforms exist that satisfy these requirements. Several private enterprises such as Epic, Nextgen, eClinicalWorks, etc. provide proprietary electronic healthcare record (EHR) management software. These software applications have been successful in facilitating clinical operation, but drawbacks are the typically high cost, low customizability, data security issues arising from patient data stored on external cloud platforms, the absence of online monitoring and interaction with standalone ML algorithms, and most of all the inability to collect high-resolution physiological data. A detailed comparison of the ICU Cockpit platform with other available research-oriented data platforms27–30 is provided in Supplementary Appendix A5.
We have shown that the ICU Cockpit platform processes and stores a large volume of ICU data consisting of signals with variable sampling rates, live webcam video, clinical operations, patient admission status, patient meta data, and lab results. Patient monitoring is complemented with historical data visualization and statistical computations are easily integrated into the processing pipeline, with algorithm outputs appearing as new signals that can be visualized like raw data. Our front end design for the icuUI falls in line with the findings of a user study of clinical data UIs.31
A further advantage of our platform is that it builds on state-of-the-art open-source software such as Apache Kafka, PostgreSQL, ReactJS, Docker, and Grafana, thereby benefiting from the continued development by the respective communities while being easily extendable. Moreover, all UIs are web-based allowing the ICU Cockpit to be deployed in the cloud.
A caveat of our system is the data source bottleneck created by using Moberg CNS monitors as gateways for interfacing medical devices at the bedside. Integrating additional devices can be costly and cumbersome if they are not yet supported, as new partnerships between companies must be established. We recommend greater collaboration between researchers, hospitals, and industry to promote the use of (device) interoperability standards, thereby reducing the cost of data integration (see Supplementary Appendix A6 for details). A second caveat of our system is the lack of an administrative query assistant. For example, doctors often want to filter out patients with certain diseases and attributes for clinical research. We are currently implementing these features into the clinical data annotation tool (icuCDA).
In summary, the ICU Cockpit platform provides clinical staff and researchers with the ability to perform day-to-day clinical monitoring and operations, as well as advance the development and practical use of ML algorithms in patient healthcare through rigorous real-world evaluation.
FUNDING
The development of the ICU Cockpit research platform was supported by the Swiss National Science Foundation grant number 325230_200568, Innosuisse grant number 52441.1 IP-LS, Vontobel-, Helmut Horten-, Gebert-Rüf-, Herzog Egli-, Hasler-, and USZ Foundations.
AUTHOR CONTRIBUTORS
JMB and GN wrote the manuscript with support from EK. CS and EK conceived the general architecture of IT infrastructure. CS carried out the initial implementation and further developed the system with support from DM, JMB, and GN as well as SS and CB. JW has been the medical lead in the development of the CDA (data acquisition and annotation tool) and has contributed to improving the usability of the ICU Cockpit by sharing his experience working with the system in the clinical setting. CM supported the data engineering process for the CDA and designed the corresponding workflows for collecting the medical data via a user interface. JMB and GN conceived and implemented stream processing algorithms. SS and CB implemented the streaming data collection. JA, DB, and RL have developed and implemented the user interface of the ICU Cockpit Dashboard.
SUPPLEMENTARY MATERIAL
Supplementary material is available at Journal of the American Medical Informatics Association online.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dick Moberg, Moberg Research Inc, PA, USA for developing additional interfaces from different medical devices to the Moberg CNS system and inspiring discussions.
CONFLICT OF INTEREST STATEMENT
The authors disclose that the ICU Cockpit is the subject of a pending European patent application (application number EP21213561.0), which relates to the data management, storage, and processing within the ICU Cockpit platform. The patent applicant is the University of Zurich, Switzerland, and the authors JMB, GN, JW, CS, and EK are listed as inventors. The authors also consider commercializing the ICU Cockpit or specific data processors (algorithms) to foster dissemination.
DATA AVAILABILITY STATEMENT
No new data were generated or analyzed in support of this research.
Contributor Information
Jens Michael Boss, Neurocritical Care Unit, Department of Neurosurgery and Institute of Intensive Care Medicine, University Hospital Zurich, Zurich, Switzerland.
Gagan Narula, Neurocritical Care Unit, Department of Neurosurgery and Institute of Intensive Care Medicine, University Hospital Zurich, Zurich, Switzerland.
Christian Straessle, Neurocritical Care Unit, Department of Neurosurgery and Institute of Intensive Care Medicine, University Hospital Zurich, Zurich, Switzerland.
Jan Willms, Neurocritical Care Unit, Department of Neurosurgery and Institute of Intensive Care Medicine, University Hospital Zurich, Zurich, Switzerland.
Jan Azzati, Institute for Medical Engineering and Medical Informatics, School of Life Sciences, University of Applied Sciences and Arts Northwestern Switzerland, Muttenz, Switzerland.
Dominique Brodbeck, Institute for Medical Engineering and Medical Informatics, School of Life Sciences, University of Applied Sciences and Arts Northwestern Switzerland, Muttenz, Switzerland.
Rahel Luethy, Institute for Medical Engineering and Medical Informatics, School of Life Sciences, University of Applied Sciences and Arts Northwestern Switzerland, Muttenz, Switzerland.
Susanne Suter, Life Science and Physics, Supercomputing Systems AG, Zurich, Switzerland.
Christof Buehler, Life Science and Physics, Supercomputing Systems AG, Zurich, Switzerland.
Carl Muroi, Neurocritical Care Unit, Department of Neurosurgery and Institute of Intensive Care Medicine, University Hospital Zurich, Zurich, Switzerland.
David Jule Mack, Neurocritical Care Unit, Department of Neurosurgery and Institute of Intensive Care Medicine, University Hospital Zurich, Zurich, Switzerland.
Marko Seric, Neurocritical Care Unit, Department of Neurosurgery and Institute of Intensive Care Medicine, University Hospital Zurich, Zurich, Switzerland.
Daniel Baumann, Neurocritical Care Unit, Department of Neurosurgery and Institute of Intensive Care Medicine, University Hospital Zurich, Zurich, Switzerland.
Emanuela Keller, Neurocritical Care Unit, Department of Neurosurgery and Institute of Intensive Care Medicine, University Hospital Zurich, Zurich, Switzerland.
REFERENCES
- 1. De Georgia M, Loparo KA.. Neurocritical Care Informatics: Translating Raw Data into Bedside Action. New York: Springer; 2020. [Google Scholar]
- 2. De Georgia MA, Kaffashi F, Jacono FJ, Loparo KA.. Information technology in critical care: review of monitoring and data acquisition systems for patient care and research. Sci World J 2015; 2015: 727694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Imhoff M, Fried R, Gather U.. Detecting relationships between physiological variables using graphical models. Proc AMIA Symp 2002: 340–344. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2244539. Accessed June 23, 2021. [PMC free article] [PubMed] [Google Scholar]
- 4. Imhoff M, Fried R, Gather U, Lanius V.. Dimension reduction for physiological variables using graphical modeling. AMIA Annu Symp Proc 2003; 2003: 313–317. [PMC free article] [PubMed] [Google Scholar]
- 5. Goldberger AL, Amaral LA, Glass L, et al. PhysioBank, PhysioToolkit, and PhysioNet: components of a new research resource for complex physiologic signals. Circulation 2000; 101 (23): E215–E220. [DOI] [PubMed] [Google Scholar]
- 6. Johnson AEW, Pollard TJ, Shen L, et al. MIMIC-III, a freely accessible critical care database. Sci Data 2016; 3 (1): 160035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pollard TJ, Johnson AEW, Raffa JD, Celi LA, Mark RG, Badawi O.. The eICU Collaborative Research Database, a freely available multi-center database for critical care research. Sci Data 2018; 5 (1): 180178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zeng X, Yu G, Lu Y, et al. PIC, a paediatric-specific intensive care database. Sci Data 2020; 7 (1): 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shah T, Sterk E, Rech MA.. Emergency department sepsis screening tool decreases time to antibiotics in patients with sepsis. Am J Emerg Med 2018; 36 (10): 1745–1748. [DOI] [PubMed] [Google Scholar]
- 10. Westra BL, Landman S, Yadav P, Steinbach M.. Secondary analysis of an electronic surveillance system combined with multi-focal interventions for early detection of sepsis. Appl Clin Inform 2017; 8 (1): 47–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kidger P, Morrill J, Foster J, Lyons T. Neural controlled differential equations for irregular time series. ArXiv200508926 November 5, 2020. http://arxiv.org/abs/2005.08926. Accessed January 8, 2022.
- 12. Che Z, Purushotham S, Cho K, Sontag D, Liu Y.. Recurrent neural networks for multivariate time series with missing values. Sci Rep 2018; 8 (1): 6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Honeyford K, Cooke GS, Kinderlerer A, et al. ; Sepsis Big Room. Evaluating a digital sepsis alert in a London multisite hospital network: a natural experiment using electronic health record data. J Am Med Inform Assoc 2020; 27 (2): 274–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. McCradden MD, Stephenson EA, Anderson JA.. Clinical research underlies ethical integration of healthcare artificial intelligence. Nat Med 2020;26 (9): 1325–1326. [DOI] [PubMed] [Google Scholar]
- 15. Antoniou T, Mamdani M.. Evaluation of machine learning solutions in medicine. CMAJ 2021; 193 (36): E1425–E1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Natzeder S, Mack DJ, Maissen G, Strässle C, Keller E, Muroi C.. Portable infrared pupillometer in patients with subarachnoid hemorrhage: prognostic value and circadian rhythm of the Neurological Pupil Index (NPi). J Neurosurg Anesthesiol 2019; 31 (4): 428–433. [DOI] [PubMed] [Google Scholar]
- 17. Hostettler IC, Muroi C, Richter JK, et al. Decision tree analysis in subarachnoid hemorrhage: prediction of outcome parameters during the course of aneurysmal subarachnoid hemorrhage using decision tree analysis. J Neurosurg 2018; 129 (6): 1499–1510. [DOI] [PubMed] [Google Scholar]
- 18. Pediaditis M, Ciubotaru A-N, Brunschwiler T, et al. Machine learning techniques for personalized detection of epileptic events in clinical video recordings. AMIA Annu Symp Proc AMIA Proc 2020; 2020: 1003–1011. [PMC free article] [PubMed] [Google Scholar]
- 19. Narula G, Haeberlin M, Balsiger J, Strässle C, Imbach LL, Keller E.. Detection of EEG burst-suppression in neurocritical care patients using an unsupervised machine learning algorithm. Clin Neurophysiol 2021. https://www.sciencedirect.com/science/article/pii/S1388245721006830. Accessed August 16, 2021. [DOI] [PubMed] [Google Scholar]
- 20. Maissen G, Narula G, Strässle C, Willms J, Muroi C, Keller E.. Functional relationship of arterial blood pressure, central venous pressure and intracranial pressure in the early phase after subarachnoid hemorrhage [published online ahead of print Aug 17, 2021]. Technol Health Care 2021. doi: 10.3233/THC-212956. [DOI] [PubMed] [Google Scholar]
- 21. Muroi C, Meier S, De Luca V, et al. Automated false alarm reduction in a real-life intensive care setting using motion detection. Neurocrit Care 2020; 32 (2): 419–426. [DOI] [PubMed] [Google Scholar]
- 22. Schwab P, Keller E, Muroi C, Mack DJ, Strässle C, Karlen W. Not to cry wolf: distantly supervised multitask learning in critical care. ArXiv180205027 June 7, 2018. http://arxiv.org/abs/1802.05027. Accessed July 29, 2021.
- 23. Hamilton PS, Tompkins WJ.. Quantitative investigation of QRS detection rules using the MIT/BIH arrhythmia database. IEEE Trans Biomed Eng 1986; 33 (12): 1157–65. [DOI] [PubMed] [Google Scholar]
- 24. Liu X, Cruz Rivera S, Moher D, Calvert MJ, Denniston AK, SPIRIT-AI and CONSORT-AI Working Group. Reporting guidelines for clinical trial reports for interventions involving artificial intelligence: the CONSORT-AI extension. Nat Med 2020; 26 (9): 1364–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cruz Rivera S, Liu X, Chan A-W, Denniston AK, Calvert MJ, SPIRIT-AI and CONSORT-AI Consensus Group. Guidelines for clinical trial protocols for interventions involving artificial intelligence: the SPIRIT-AI extension. Nat Med 2020; 26 (9): 1351–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Good Machine Learning Practice for Medical Device Development: Guiding Principles. www.fda.gov/medical-devices/software-medical-device-samd/good-machine-learning-practice-medical-device-development-guiding-principles. Accessed November 12, 2021.
- 27. Smielewski P, Czosnyka Z, Kasprowicz M, Pickard JD, Czosnyka M.. ICM+: a versatile software for assessment of CSF dynamics. In: Schuhmann MU, Czosnyka M, eds. Intracranial Pressure and Brain Monitoring XIV. Vienna: Springer; 2012: 75–79. (Acta Neurochirurgica Supplementum). 10.1007/978-3-7091-0956-4_13. Accessed August 12, 2021. [DOI] [PubMed] [Google Scholar]
- 28. Rehm GB, Kuhn BT, Delplanque J-P, et al. Development of a research-oriented system for collecting mechanical ventilator waveform data. J Am Med Inform Assoc 2018; 25 (3): 295–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kalogeropoulos D, Carson ER, Collinson PO.. Clinical-HINTS: integrated intelligent ICU patient monitoring and information management system. Stud Health Technol Inform 1997; 43 Pt B: 906–910. [PubMed] [Google Scholar]
- 30. Ahmed Z, Kim M, Liang BT.. MAV-clic: management, analysis, and visualization of clinical data. JAMIA Open 2019; 2 (1): 23–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Park J, Rhim S, Han K, Ko J.. Disentangling the clinical data chaos: user-centered interface system design for trauma centers. PLoS One 2021; 16 (5): e0251140. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No new data were generated or analyzed in support of this research.