ABSTRACT
As a self-degradative mechanism, macroautophagy/autophagy has a role in the maintenance of energy homeostasis during critical periods in the development of cells. It also controls cellular damage through the eradication of damaged proteins and organelles. This process is accomplished by tens of ATG (autophagy-related) proteins. Recent studies have shown the involvement of non-coding RNAs in the regulation of autophagy. These transcripts mostly modulate the expression of ATG genes. Both long non-coding RNAs (lncRNAs) and microRNAs (miRNAs) have been shown to modulate the autophagy mechanism. Levels of several lncRNAs and miRNAs are altered in this process. In the present review, we discuss the role of lncRNAs and miRNAs in the regulation of autophagy in diverse contexts such as cancer, deep vein thrombosis, spinal cord injury, diabetes and its complications, acute myocardial infarction, osteoarthritis, pre-eclampsia and epilepsy.
Abbreviations: AMI: acute myocardial infarction; ATG: autophagy-related; lncRNA: long non-coding RNA; miRNA: microRNA.
KEYWORDS: Autophagy, biomarker, cancer, lncRNAs, microRNAs
Introduction
Autophagy is a degradative mechanism that regulates the energy resources at crucial times during development and in periods of nutrient deficiency [1]. This process is also involved in the removal of protein aggregates, elimination of impaired organelles, as well as intracellular pathogens. Autophagy is regarded as a recycling mechanism to enhance energy proficiency through ATP production and governs cellular damage through the eradication of damaged proteins and organelles [1]. Autophagy is accomplished through multiple steps. First, stress-related pathways regulate phagophore formation through modulation of the BECN1/Beclin 1-PIK3C3/VPS34-containing phosphatidylinositol 3-kinase complex at the endoplasmic reticulum. Subsequent multimerization of proteins coded by ATG (autophagy-related) genes and MAP1LC3/LC3 occurs at the phagophore membrane. Then, a number of targets are selected to be degraded and the autophagosome is fused with the lysosome to degrade the trapped molecules through proteolytic reactions [1]. Several ATG proteins participate in autophagy. Notably, many of the corresponding genes are conserved between species [2].
Macroautophagy, microautophagy, and chaperone-mediated autophagy are the principal types of autophagy. All three types lead to proteolytic destruction of cytosolic apparatuses in the cellular lysosomes [3]. Yet, the route of delivery of cytoplasmic elements to the lysosomes differs between these types as in the macroautophagy autophagosome delivers these elements while in the micro-autophagy cytosolic apparatuses are directly delivered to the lysosome. In chaperone-mediated autophagy, targeted proteins are delivered in a complex with chaperone proteins that interact with the lysosomal membrane receptor. This interaction leads to protein unfolding and destruction [3]. Autophagy is regulated by several mechanisms. Among the recently appreciated mechanisms is the involvement of non-coding RNAs (ncRNAs) [4]. It has been revealed that 98% of the genome is transcribed. However, the majority of these transcripts do not encode proteins, thus being described as ncRNAs [5]. Regulatory ncRNAs comprise a significant portion of ncRNAs, with long non-coding RNAs (lncRNAs) and microRNAs (miRNAs) being the most important classes of this group of transcripts. These transcripts can regulate the expression of several genes at the epigenetic, transcriptional, and post-transcriptional levels [5]. Nearly all miRNAs are considered as post-transcriptional suppressors of gene expression. However, lncRNAs can regulate expression of protein-coding genes at both positive and negative directions via different interactions with RNA, protein and chromatin structures [6]. Several lncRNAs have been shown to be evolutionarily conserved [7], albeit to a lesser extent compared with protein-coding genes [8]. It is worth mentioning that the levels of conservation in the promoter areas of lncRNAs are similar to the promoters of several protein-coding genes [6]. Numerous miRNAs have been identified in mammalian genomes, several of them being highly conserved even between remotely related species [9]. While lncRNAs are generated by POLR2 (RNA polymerase II) and POLR3 (RNA polymerase III) [10], miRNAs are transcribed from genomic DNA into primary miRNAs, then being processed into precursor miRNAs and mature miRNAs in a sequential process [10]. Both lncRNAs and miRNAs have been shown to modulate the autophagy mechanism. In the present review, we discuss the role of lncRNAs and miRNAs in the regulation of autophagy in diverse contexts such as cancer, deep vein thrombosis, spinal cord injury, diabetes and its complications, acute myocardial infarction, osteoarthritis, pre-eclampsia and epilepsy. In order to find the relevant literature, we searched PubMed and Google Scholar with the keywords “autophagy” AND “miRNA” or “lncRNA”. Then, we assessed the full texts of the articles to extract data regarding type of disorder, clinical samples, animal models and the molecular pathways being influenced by miRNAs/lncRNAs. Finally, we tabulated the extracted data in order to make the data more comprehensible. It is worth mentioning that the majority of the included studies have assessed the role of miRNA/lncRNAs through functional studies, thus providing enough evidence for contribution of these ncRNAs in the regulation of autophagy.
miRNAs and autophagy
These transcripts have sizes of approximately 22 nucleotides and principally regulate the expression of their target genes at the post-transcriptional level [11]. Several experiments have shown the role of miRNAs in the regulation of autophagy. Dysregulation of miRNAs has been associated with a wide range of disorders, including cancers and nonmalignant disorders.
miRNA and autophagy in cancer
Expression of MIR100 is decreased in renal cell carcinoma cell lines and clinical samples compared with adjacent non-cancerous tissues, while the expression of its target gene, NOX4, is increased in malignant samples. Overexpression of this miRNA or knockdown of its target in the mentioned cell lines has enhanced autophagy while reducing the expression of MTOR (mechanistic target of rapamycin kinase) pathway-associated genes and cancer cell migration and invasion [12]. MIR126 is downregulated in colorectal cancer cells and tissues compared with normal tissues. Forced upregulation of this miRNA compromised viability and growth of these cells and enhanced both autophagy and apoptosis through modulation of expression of the MTOR gene [13]. MIR30A regulates autophagy in hepatocellular carcinoma [14] and gastrointestinal stromal tumor [15]. miRNAs with regulatory roles on the autophagy can also affect epithelial-mesenchymal transition (EMT), thus influencing the metastatic ability of cancer cells [16]. Figure 1 depicts the underlying mechanism of the contribution of two miRNAs in the autophagy and EMT process in the context of gastric cancer.
Figure 1.
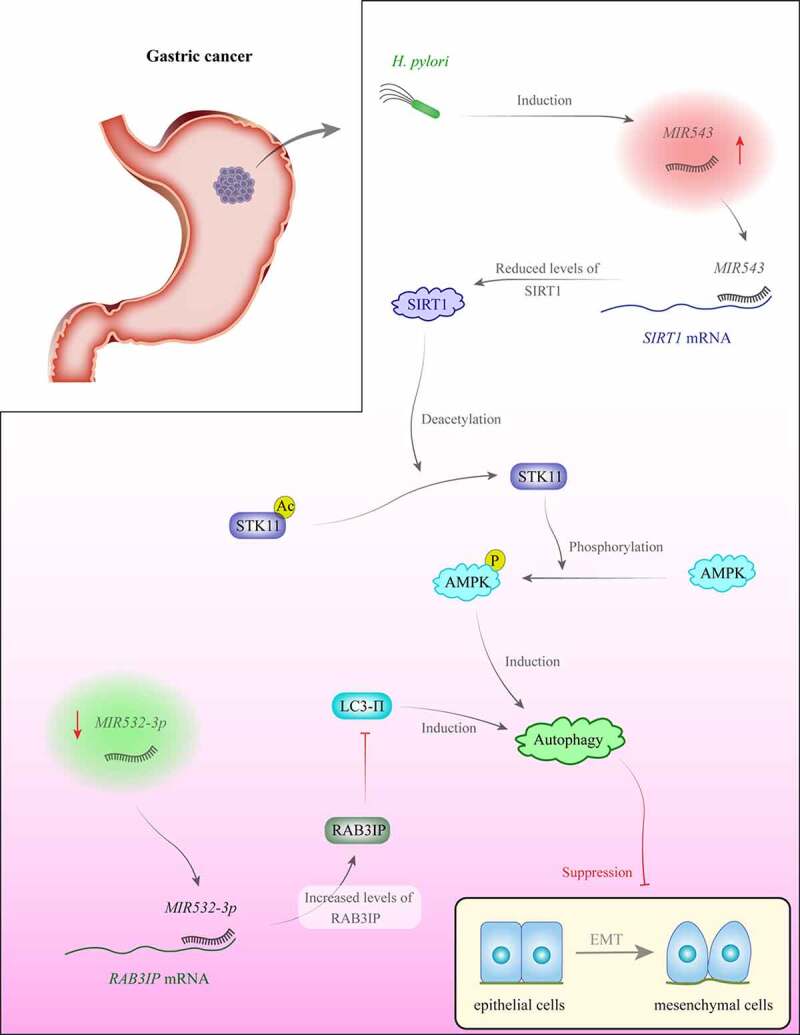
H. pylori increases MIR543 levels in gastric cancer. This miRNA binds with the 3ʹ UTR of SIRT1 to inhibit its expression. Autophagy has a role in the inhibition of epithelial-mesenchymal transition (EMT) in some situations [16]. Conversely, MIR532-3p levels are decreased in gastric cancer. This miRNA inhibits the expression of RAB3IP. Overexpression of RAB3IP is associated with a decrease in autophagy and enhancement of EMT [79].
miRNAs and autophagy in cardiac disorders
Overexpression of MIR26B-5p, MIR204-5p, and MIR497-3p reduces IGF1 (insulin like growth factor 1)-induced cardiomyocyte hypertrophy by inhibiting autophagy [17]. Several miRNAs have been identified that regulate autophagy in the context of acute myocardial infarction (AMI). For instance, overexpression of MIR139-5p could prevent cell autophagy induced by hypoxia-reoxygenation injury [18]. Moreover, MIR638 and MIR384 have functional roles in the reduction of cell autophagy by modulating the expression of ATG5 and activation of the phosphoinositide 3-kinase (PI3K)-AKT/protein kinase B pathway, respectively [19,20]. Conversely, the downregulation of MIR30A can prevent autophagy in myocardial cells [21]. Additionally, MIR30A suppresses BECN1-associated autophagy in diabetic cataract [22]. MIR9-5p has a role in increasing migration, invasion, and angiogenesis of endothelial progenitor cells by lessening TRPM7 transcription through induction of PI3K-AKT-related autophagy. Based on the role of endothelial progenitor cells in resolving thrombi, this miRNA has been suggested as a therapeutic target in deep vein thrombosis [23].
miRNAs and autophagy in osteoarthritis
Several miRNAs have been implicated in the pathogenesis of osteoarthritis via different mechanisms. For instance, MIR27A has a role in the down-regulation of PI3K and subsequent increase in autophagy in IL1B/IL-1β-treated chondrocytes [24]. Conversely, MIR128-1 can suppress chondrocyte autophagy by disturbing ATG12 [25]. MIR4262 also has a role in the development of osteoarthritis by modulating cell autophagy [26]. Expression of MIR375 has been increased in cartilage tissues obtained from osteoarthritis cases, while ATG2B expression has been diminished in these samples. MIR375-mediated suppression of ATG2B in the chondrocytes inhibits autophagy and enhances endoplasmic reticulum stress, thus exacerbating osteoarthritis clinical symptoms [27].
miRNA and autophagy in inflammatory bowel diseases
Several miRNAs have been shown to affect autophagy, thus contributing to the pathogenesis of inflammatory bowel disease. For instance, MIR196A and MIR196B can reduce the expression of IRGM and inhibit autophagy by decreasing the accumulation of LC3-II [28]. Besides, the expression of MIR665 has been increased in the intestinal mucosa of patients with inflammatory bowel disease. This miRNA can decrease the expression of XBP1 and ORMDL3 in the course of endoplasmic reticulum stress, enhancing autophagy sensitivity [29]. Finally, the upregulation of MIR221-5p in colitis tissues has been associated with overexpression of SP, implying its role in inflammatory bowel disease autophagy [30].
Table 1 shows the list of miRNAs that are involved in the process of autophagy.
Table 1.
List of autophagy-associated miRNAs
| microRNA | Disease | Numbers of clinical samples | Gain- or loss-of-function studies/animal models | Targets/Regulators | Signaling Pathways | Function | Ref |
|---|---|---|---|---|---|---|---|
| MIR100 | renal cell carcinoma (RCC) | 113 pairs of RCC and adjacent normal tissues | ± | NOX4, MAP1LC3B | MTOR | Upregulation of MIR100 by targeting NOX4 and inactivating the MTOR could increase the autophagy of RCC cells. | [64] |
| MIR126 | colorectal cancer (CRC) | 30 pairs of RCC and adjacent normal tissues | ± | MAP1LC3B | MTOR, SQSTM1 | MIR126 could regulate the activity of CRC cells via autophagy. | [13] |
| MIR431 | CRC | 24 pairs of CRC tissues and adjacent tumor tissues | ± | ATG3, MAP1LC3B | - | ATG3 upregulation, caused by downregulated MIR431, could promote proliferation and invasion via an autophagy-dependent manner in colon cancer. | [65] |
| MIR221 | CRC | - | -/- | TP53INP1, MAP1LC3B | - | MIR221 could inhibit autophagy and target TP53INP1 in CRC cells. | [31] |
| MIR221 | diabetic cardiac hypertrophy | Mouse | -/+ | MAP1LC3B | MAPK8-JUN, CDKN1, MTOR | MIR221 affects autophagy in diabetic cardiac hypertrophy. | [66] |
| MIR221 | pancreatic cancer (PaC) | - | -/- | HDAC6, MAP1LC3B | - | Downregulation of MIR221 may serve an oncogenic function in the apoptosis and autophagy of PaC cells by inducing the expression of HDAC6. | [67] |
| MIR381 | prostate cancer (PCa) | Mouse | -/+ | RELN, MAP1LC3B | PI3K-AKT-MTOR | Overexpression of MIR381 could suppress PCa cell proliferation while promoted autophagy of PCa cells. | [68] |
| MIR361 | PCa | - | -/- | PKM, SP1, MAP1LC3B | - | MIR361 has affected the progression of PCa and the metabolism and autophagy of PCa cells. | [69] |
| MIR519D | hepatocellular carcinoma (HCC) | Mouse/human; 76 pairs of HCC and adjacent normal tissues | +/+ | RAB10, MAP1LC3B | AMPK | MIR519D could induce autophagy of human HCC cells. | [70] |
| MIR30A | HCC | Mouse/human; 9 pairs of HCC and adjacent normal tissues | +/+ | BECN1, ATG5, MAP1LC3B | - | MIR30A could suppress autophagy-mediated anoikis resistance and metastasis in HCC. | [14] |
| MIR30A | gastrointestinal stromal tumor | Mouse | -/+ | BECN1, ATG5, ATG12, MAP1LC3B | - | MIR30A could target BECN1 to inactivate autophagy gastrointestinal stromal tumor cells. | [15] |
| MIR30A | diabetic cataract | - | -/- | BECN1, MAP1LC3B | ALP | MIR30A could inhibit BECN1-mediated autophagy in diabetic cataract. | [22] |
| MIR30A | AMI | Rat | -/+ | ULK1, BECN1 | - | Downregulation of MIR30A could suppress the myocardial apoptosis in rats by reducing autophagy. | [21] |
| MIR135A | HCC | 103 pairs of RCC and adjacent normal tissues | ± | ATG14, MAP1LC3B | TF-F7/FVII-F2RL1/PAR2 | The upregulation of MIR135A by targeting ATG14 could inhibit autophagy in HCC. | [32] |
| MIR106A | cervical squamous cell carcinoma (CSCC) | 91 CSCC patients and 56 normal cervical squamous epithelium samples | ± | STK11/LKB1, MAP1LC3B | MTOR, AMPK | Upregulation of MIR106A could suppress cell autophagy in CSCC associated with HPV-16. | [71] |
| MIR20A | cervical cancer (CC) | 20 pairs of CC and adjacent normal tissues | ± | THBS2, MAP1LC3B | - | Downregulation of MIR20A by targeting THBS2 could suppress autophagy and induced apoptosis in CC cells. | [72] |
| MIR20A | osteoarthritis (OA) | 30 pairs of OA and adjacent normal tissues | ± | ATG10, MAP1LC3B | PI3K-AKT-MTOR | Inhibition of MIR20A could promote proliferation and autophagy in articular chondrocytes by the PI3K-AKT-MTOR pathway. | [73] |
| MIRG1 | CC | Mouse | -/+ | GRSF1, TMED5, LMNB1, MAP1LC3B | WNT-CTNNB1/CTNNB | MIRG1 could promote serum starvation-induced nuclear macroautophagy/autophagy in CC cells. | [74] |
| MIR199A | epithelial ovarian cancer (EOC) | 70 EOC samples and 30 normal ovarian samples | ± | circMUC16, BECN1, RUNX1, ATG13, TERF2IP, MAP1LC3B | MAPK, VEGF | CircMUC16 could promote autophagy of EOC via interaction with ATG13 and MIR199A. | [75] |
| MIR199A | parkinson | - | -/- | GSK3B, BECN1, MAP1LC3B | PTEN-AKT-MTOR | Increasing MIR199A expression in PC12 cells could reduce autophagy. | [76] |
| MIR133A | gastric cancer (GC) | - | -/- | FOXP3, MAP1LC3B | - | MIR133A by targeting FOXP3 could promote autophagy in GC. | [77] |
| MIR5100 | GC | Mouse | -/+ | CAAP1, MKL1, MAP1LC3B | - | MIR5100 could promote apoptosis and inhibit autophagy of GC cells. | [78] |
| MIR543 | GC | Mouse/human; 50 pairs of GC and adjacent normal tissues | +/+ | SIRT1, MAP1LC3B | - | MIR543 by targeting SIRT1 could suppress autophagy in GC cells. | [16] |
| MIR532 | GC | Rat/human; 150 pairs of GC and adjacent normal tissues | +/+ | RAB3IP, MAP1LC3B | - | MIR532 directly targets RAB3IP and represses its function in the proliferation of GC cells through autophagy. | [79] |
| MIR375 | GC | Mouse/human; 30 pairs of GC and adjacent normal tissues | +/+ | ATG7, MAP1LC3B | AKT-MTOR | Overexpression of MIR375 could inhibit autophagy through the AKT-MTOR pathway. | [80] |
| MIR375 | osteoarthritis (OA) | Mouse/human; 8 pairs of knee OA patients and normal control group | +/+ | ATG2B, MAP1LC3B | - | MIR375 exacerbates knee osteoarthritis via repressing chondrocyte autophagy by targeting ATG2B. | [27] |
| MIR183 | GC | - | -/- | MAP1LC3B | - | Overexpression of MIR183 by targeting LC3 could reduce starvation-induced autophagy in GC cells. | [81] |
| MIR3657 | GC | Mouse | -/+ | ATG7, MAP1LC3B | circRACGAP1 | MIR3657 could reduce autophagy in GC cells. | [82] |
| MIR150 | non-small cell lung carcinoma (NSCLC) | 54 NSCLC and 30 non-neoplastic lung tissues | ± | EPG5, MYC, MAP1LC3B | - | MIR150 via repressing EPG5 could inhibit the autophagic flux and promote NSCLC tumorigenesis. | [33] |
| MIR16 | NSCLC | 20 pairs of NSCLC and adjacent normal tissues | ± | TGFB1, ATG3, MAP1LC3B | - | MIR16 could inhibit TGFB1-induced EMT via activation of autophagy in NSCLC cell lines. | [83] |
| MIR21 | NSCLC | 46 pairs of NSCLC and adjacent normal tissues | ± | ULK1, PRKAA/AMPKα, MAP1LC3B, p-PRKAA/AMPKα | SQSTM1/p62 | MIR21 could regulate autophagy activities of NSCLC via AMPK/ULK1 pathway. | [84] |
| MIR26 | NSCLC | Mouse/Human; 6 pairs of NSCLC and adjacent normal tissues | +/+ | TGFB1, MAP1LC3B | JNK | MIR26 could inhibit autophagy in human NSCLC cells via the TGFB1-MAPK/JNK pathway. | [85] |
| MIR26A | melanoma | - | -/- | HMGB1, MAP1LC3B | - | MIR26A could reduce autophagy via targeting HMGB1 in melanoma. | [86] |
| MIR26B | cardiac hypertrophy | Rat | -/+ | ULK1, MAP1LC3B, BECN1 | - | Overexpression of MIR26B could attenuate IGF1-induced cardiomyocyte hypertrophy by suppressing autophagy. | [17] |
| MIR26B | breast cancer (BCa) | 3 pairs of BCa and adjacent normal tissues | ± | DRAM1, MAP1LC3B | - | MIR26B could suppress autophagy in BCa cells by targeting DRAM1. | [87] |
| MIR210 | lung cancer (LCa) | 30 pairs of LCa and adjacent normal tissues | ± | ATG7, BECN1, MAP1LC3B | - | MIR210 by targeting ATG7 could reduce autophagy of lung cancer cells. | [88] |
| MIR223 | lung I/R injury | Mouse | -/+ | EPAS1/HIF2A, MAP1LC3B | - | MIR223/HIF2A/CTNNB1 axis could promote autophagy to aggravate H/R-induced injury in mouse PMVECs. | [89] |
| MIR326 | pulmonary fibrosis | Mouse | -/+ | TNFSF14, PTBP1, MAP1LC3B | - | MIR326 could reduce pulmonary inflammation by targeting TNFSF14 and promote autophagy activity of fibroblasts through targeting PTBP1. | [90] |
| MIR192 | asthma | Mouse | -/+ | MMP16, ATG7, MAP1LC3B | - | MIR192 by targeting MMP16 and ATG7 could reduce autophagy in asthma. | [91] |
| MIRLET7A1, MIRLET7D |
glioblastoma (GBM) | Mouse/human; 132 GBM and 20 normal brain tissues | +/+ | MAP1LC3B | STAT3 | Upregulation of cluster MIRLET7A1 ~ MIRLET7D could accelerate cell apoptosis and autophagy in glioma. | [92] |
| MIR449A | GBM | Mouse/human; 72 pairs of GBM and adjacent normal tissues | +/+ | BECN1, CISD2, MAP1LC3B | - | Overexpression of MIR449A affects autophagy by targeting CISD2 in glioma cells. | [34] |
| MIR449A | lymphoma | Mouse | -/+ | ATG4B, MAP1LC3B | - | MIR449A via downregulating ATG4B could reduce the autophagy of T-cell lymphoma cells. | [93] |
| MIR101 | GBM | 32 pairs of GBM and adjacent normal tissues | ± | STMN1, RAB5A, ATG4D, MAP1LC3B | MALAT1 | Downregulation of MIR101 could increase autophagy by targeting MALAT1 in glioma. | [36] |
| MIR181B | gallbladder cancer (GBC) | Mouse/Human; 93 pairs of GBC and adjacent normal tissues | +/+ | CREBRF, CREB3, MAP1LC3B | - | MIR181B could promote autophagy by regulating CREBRF/CREB3 pathway in GBC. | [94] |
| MIR24-1 | melanoma | 77 pairs of melanoma and adjacent normal tissues | ± | UBD, BECN1, BCL2L1/BCLlXL, MAP1LC3B | MAPK/JNK, LC3 | Overexpression of MIR24-1 could promote autophagy in malignant melanoma cells. | [95] |
| MIR24, MIR152 | uterine sarcoma | 101 patients with uterine sarcoma and 54 healthy subjects | ± | SIRT1, MAP1LC3B | - | MIR24 and MIR152 could promote autophagy by activating SIRT1 and deacetylating LC3. | [96] |
| MIR17 | head & neck squamous cell carcinoma (HNSCC) | - | -/- | BECN1 | - | Overexpression of MIR17 could inhibit autophagy under hypoxia in head and neck squamous cell carcinoma cells. | [97] |
| MIR224 | breast cancer (BCa) | 30 metastatic BCa patients, 35 non-metastatic BCa patients, 25 health control patients | ± | SMAD4, MAP1LC3B | - | MIR224 could inhibit autophagy in BCa cells via targeting Smad4. | [98] |
| MIR107 | BCa | Mouse/human; 62 pairs of BCa and adjacent normal tissues | +/+ | HMGB1, BECN1 | SQSTM1/p62 | MIR107 could inhibit cell autophagy of breast cancer cells by targeting HMGB1. | [99] |
| MIR204, MIR497 | cardiac hypertrophy | Rat | -/+ | ULK1, MAP1LC3B, BECN1 | - | Overexpression of MIR204 and MIR497 could attenuate IGF1-induced cardiomyocytes hypertrophy by suppressing autophagy. | [17] |
| MIR128 | cardiac hypertrophy | Rat | -/+ | PPAR, NFKB, MAP1LC3B | AMPK-MTOR | MIR128 has pro-autophagic effects via directly targeting PPAR in cardiac hypertrophy. | [100] |
| MIR128A | osteoarthritis (OA) | Rat/human; 28 OA patient and 17 normal tissues | +/+ | ATG12, MAP1LC3B | - | MIR128A could reduce chondrocyte autophagy by disrupting ATG12. | [25] |
| MIR29B | heart failure (HF) | 35 patients with HF and 35 healthy donors | ± | SPARC, MAP1LC3B | TGFB1,SMAD3 | MIR29B could inhibit autophagy and apoptosis in hypoxia-induced H9c2 cells by targeting SPARC. | [101] |
| MIR9 | deep vein thrombosis (DVT) | Mouse | -/+ | TRPM7, MAP1LC3B | PI3K-AKT | MIR9 could promote EPC angiogenesis via the mediated TRPM7 expression and PI3K-AKT-autophagy pathway. | [23] |
| MIR145 | intimal hyperplasia | Mouse | -/+ | TGFB1, MAP1LC3B | - | Overexpression of MIR145 could inhibit cell autophagy in TGFB1-stimulated VSMCs. | [102] |
| MIR145 | AMI | Rat | -/+ | FGF21, BECN1, ANGPT2, MAP1LC3B | - | MIR145 inhibitor could decrease the inhibitory effect of FGF21 on I/R-induced autophagy. | [103] |
| MIR145 | osteosarcoma (OS) | 30 pairs of OS and adjacent normal tissues | ± | HDAC4, MAP1LC3B | - | Overexpression of MIR145 by targeting HDAC4 could induce the apoptosis and autophagy of OS. | [104] |
| MIR384 | spinal cord injury (SCI) | Rat | -/+ | BECN1, HSPA5/GRP78 | - | MIR384 could promote recovery of rats with SCI by suppressing autophagy via targeting of BECN1. | [105] |
| MIR384 | AMI | Rat | -/+ | BECN1, MAP1LC3B | PI3K-AKT | Overexpression of MIR384 could inhibit I/R-induced autophagy, accompanied by the activation of the PI3K-AKT pathway. | [20] |
| MIR372 | SCI | Rat | -/+ | BECN1, MAP1LC3B | - | MIR372 could reduce nerve cell apoptosis in SCII via increasing autophagy by upregulating BECN1. | [106] |
| MIR202 | intervertebral disc degenerative (IDD) | 65 pairs of nucleus pulposus form patients with IDD and normal intervertebral disc | ± | ATG7, BAX, MAP1LC3B | SQSTM1/p62 | Inhibition of MIR202 could effectively promote autophagy of NP cells. | [107] |
| MIR93 | - | - | -/- | ULK1, MAP1LC3B | - | MIR93 could regulate hypoxia-induced autophagy by targeting ULK1. | [108] |
| MIR376B | chronic kidney disease (CKD) | Mouse | -/+ | ATG5, MAP1LC3B | - | Downregulation of MIR376B could promote macrophage autophagy by negatively regulating ATG5 in mice with CKD. | [109] |
| MIR141 | diabetic kidney disease (DKD) | Rat | -/+ | PTEN, MAP1LC3B | PTEN-AKT-MTOR | Overexpression of MIR141 could decrease autophagy in DKD. | [110] |
| MIR1273G | diabetic retinopathy (DR) | Rat | -/+ | MMP2, MMP9, TNF, LC3-II, CTSB, CTSL, MAP1LC3B | ALP | MIR1273G by modulating the autophagy-lysosome pathway affects the progression of DR. | [111] |
| MIR25 | polycystic kidney disease (PKD) | Mouse | -/+ | ATG14, BECN1, MAP1LC3B | - | Inhibition of MIR25 could enhance autophagy in renal cells. | [112] |
| MIR139 | acute myocardial infarction (AMI) | - | -/- | ATG4D, MAP1LC3B | AMPK-MTOR-ULK1 | Overexpression of MIR139 could inhibit cell autophagy induced by H/R. | [18] |
| MIR638 | AMI | - | -/- | ATG5, MAP1LC3B | - | Overexpression of MIR638 could reduce cell autophagy by regulating the ATG5 in the HCMs. | [19] |
| MIR153 | knee I/R injury | Mouse | -/+ | BCL2, BECN1, MAP1LC3B | - | Overexpression of MIR153 could block the interaction between BCL2 and BECN1 to promote autophagy of chondrocytes. | [113] |
| MIR153 | chronic myeloid leukemia (CML) | 44 CML patients | ± | BCL2, MAP1LC3B | - | Dysregulation of MIR153 may target BCL2 to attenuate apoptosis in CML. | [114] |
| MIR9A | cerebral ischemic stroke (CIS) | - | -/- | ATG5, MAP1LC3B | - | Overexpression of MIR9A could inhibit autophagy in the focal cerebral ischemia model by targeting ATG5. | [115] |
| MIR129 | hypoxia | Rat | -/+ | MAP1LC3B, BECN1 | - | MIR129 overexpression could restore hypoxia-induced autophagy deficiency in H9c2 cardiomyocytes. | [116] |
| MIR129 | ischemic heart disease (IHD) | - | -/- | ATG14, MAP1LC3B | PI3K-AKT-MTOR | MIR129 by targeting ATG14 could inhibit autophagy and apoptosis of H9c2 cells. | [117] |
| MIR129 | osteosarcoma (OS) | Mouse/human; 18 pairs of OS and adjacent normal tissues | +/+ | LHX2, BECN1, ATG3, ATG7, ATG12, LAMP1, MAP1LC3B | MTOR | LHX2 could regulate tumorigenesis and autophagy via MTOR in OS and is negatively regulated by MIR129. | [118] |
| MIR4465 | - | - | -/- | PTEN, MAP1LC3B | AKT-MTOR | MIR4465 could decrease PTEN expression and inhibit autophagy via the AKT-MTOR pathway. | [119] |
| MIR155 | paget disease of bone (PDB) | Mouse | -/+ | Table 2, MAP3K7, MAP1LC3B | - | MIR155 could induce differentiation and autophagy in OC. | [120] |
| MIR155 | atherosclerosis | - | -/- | ox‑LDL, MAP1LC3B | PI3K-AKT-MTOR | MIR155 could promote ox-LDL-induced autophagy in HUVECs by targeting the PI3K-AKT-MTOR pathway. | [121] |
| MIR378 | duchenne muscular dystrophy (DMD) | Mouse | -/+ | PDK1, MAP1LC3B | MTOR/ULK1 | Overexpression of MIR378 was able to enhance autophagy and repress apoptosis in the skeletal muscle of mice. | [122] |
| MIR193B | osteosarcoma (OS) | 53 pairs of OS and adjacent normal tissues | ± | FEN1, MAP1LC3B | - | Overexpression of MIR193B in the OS cells could induce autophagy and apoptosis. | [123] |
| MIR15A | chronic constriction injury (CCI) | Rat | -/+ | AKT3, MAP1LC3B | - | Overexpression of MIR15A could suppress AKT3 and induce autophagy in CCI rats. | [124] |
| MIR27A | osteoarthritis (OA) | 20 OA patients and 10 normal cartilages | ± | PI3K, MAP1LC3B | PI3K-AKT-MTOR | Upregulation of MIR27A via targeting PI3K-AKT-MTOR pathway could lead to apoptosis and autophagy in IL1B-treated chondrocytes. | [24] |
| MIR4262 | OA | Rat | -/+ | SIRT1, MAP1LC3B | PI3K-AKT-MTOR | Upregulation of MIR4262 could promote the occurrence and development of OA in rats by regulating cell autophagy and matrix synthesis. | [26] |
| MIR206 | OA | Rat | -/+ | IGF1, BECN1, ULK1, ATG5, BCL2, CASP3, BAX, MAP1LC3B | PI3K-AKT-MTOR | MIR206 has inhibitory effects on autophagy and apoptosis of articular cartilage in OA via activating the IGF1‐mediated PI3K-AKT-MTOR signaling pathway. | [125] |
| MIR411 | OA | - | -/- | HIF1A, ULK1, BECN1, MAP1LC3B | SQSTM1/p62 | MIR411 could promote chondrocyte autophagy by targeting HIF1A. | [126] |
| MIR590 | OA | - | -/- | TGFB1, MAP1LC3B | - | Suppression of MIR590 could inhibit chondrocytes apoptosis and autophagy in response to mechanical pressure injury. | [127] |
| MIR320 | retinoblastoma (RB) | 30 pairs of RB and adjacent normal tissues | ± | HIF1A, BECN1, MAP1LC3B | PI3K-AKT-MTOR, SQSTM1/p62 | MIR320 could regulate autophagy by targeting HIF-1αand the related mechanism may be associated with the MTOR pathway in RB development. | [128] |
| MIR23A | acute myeloid leukemia (AML) | Mouse/human; 25 primary ALL tissues, 27 AML tissues, 15 APL tissues | +/+ | TLR2, BECN1, ATG12, MAP1LC3B | NFKB1 | Downregulation of MIR23A in leukemic cells could lead to the upregulation of protective autophagy by targeting TLR2 expression. | [129] |
| MIR138 | - | -/- | SIRT1, BECN1, TNF, MAP1LC3B | SQSTM1/p62 | MIR138 could contribute to the TNF-induced insulin resistance, possibly through inducing autophagy in HepG2 cells by regulating SIRT1. | [130] | |
| MIR7 | Mouse | -/+ | CELF1, MBNL1, MAP1LC3B | AKT | MIR7 could affect muscle dysfunction through autophagy in myotonic dystrophy muscle cells. | [131] |
Based on the fundamental roles of autophagy in the development of cancer and its course, expression of autophagy-associated miRNAs can predict cancer patients’ survival. Higher expressions of MIR221, MIR135A1-5p, MIR150, and MIR449A have been associated with unfavorable outcome in patients with colorectal cancer, hepatocellular carcinoma, non-small cell lung carcinoma, and glioma, respectively [31–34]. Table 2 summarizes the results of studies that assessed the association between expression levels of autophagy-related miRNAs and the survival of cancer patients.
Table 2.
Association between the survival of cancer patients and miRNAs that functionally affect autophagy (the expression of miRNAs could be associated with the prognosis independently of autophagy regulation)
| Cancer type | miRNA | Number of samples | Prognostic correlation | Ref |
|---|---|---|---|---|
| colorectal cancer | MIR221 | TCGA data | Overexpression predicts short OS rates. | [31] |
| hepatocellular carcinoma | MIR135A | 103 pairs of cancerous and non-cancerous samples | Overexpression predicts short OS rates. | [32] |
| non-small cell lung carcinoma | MIR150 | 54 cancerous and 30 non-cancerous tissues | Overexpression predicts short OS rates. | [33] |
| Glioma | MIR449A | 72 pairs of cancerous and non-cancerous samples | Overexpression predicts short OS rates. | [34] |
| uterine sarcoma |
MIR152 and MIR24 |
101 cancerous and 54 non-cancerous samples | Overexpression predicts better OS rates. | [96] |
| osteosarcoma | MIR129 | 18 pairs of cancerous and non-cancerous samples | Overexpression predicts better OS rates. | [118] |
| ALL, AML, APL | MIR23A | 25 primary ALL, 27 AML, 15 APL samples | Overexpression predicts better OS rates. | [129] |
| hepatocellular carcinoma | MIR30A | 9 pairs of cancerous and non-cancerous samples | Overexpression predicts better OS rates. | [132] |
LncRNAs and autophagy
LncRNAs are transcripts comprising more than 200 nucleotides, devoid of protein-coding capacity, which are expressed in several tissues and exert regulatory roles on the expression of target genes. Several lncRNAs have been identified that influence the process of autophagy. As autophagy is involved in the pathogenic process of several human disorders, these lncRNAs participate in diverse disorders ranging from cancer to age-related pathologies.
lncRNA and autophagy in cancer
HOTAIR can enhance autophagy through the regulation of ATG3 and ATG7 in hepatocellular carcinoma [35]. Also, MALAT1 can activate autophagy in glioblastoma through the MIR101-3p-STMN1-ATG4D and MIR384-GOLM1 axes [36,37]. NEAT1 has a role in conferring resistance to 5-fluorouracil in colorectal cancer cells through modulation of MIR34A [38]. HULC can modulate cisplatin resistance in gastric cancer through the regulation of FOXM1 expression and suppression of autophagy [39]. This lncRNA enhances the malignant progression of hepatocellular carcinoma cells via reducing expression of MIR15A and increasing expression of SQSTM1/p62. Moreover, the overexpression of HULC enhances LC3 levels and subsequently induces LC3 via SIRT1. HULC also promotes the interaction between LC3 and ATG3. Also, HULC enhances the expression of BECN1. Taken together, HULC increases autophagy through SIRT1-mediated overexpression of LC3-II. HULC also suppresses PTEN expression via autophagy-SQSTM1 and ubiquitin–proteasome mechanisms [40]. Figure 2 shows the mechanism of participation of HULC in the carcinogenesis.
Figure 2.
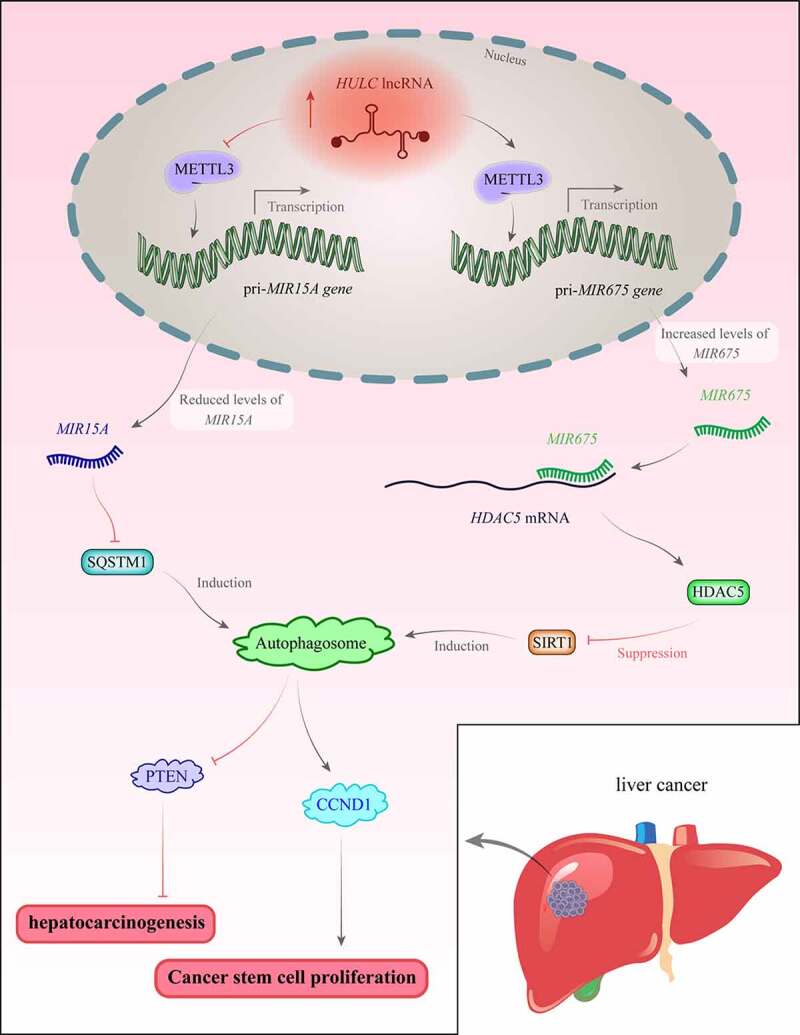
The expression of HULC is increased in hepatocellular carcinoma. This lncRNA inhibits METTL3 binding with pri-MIR15A and decreases methylation of pri-MIR15A. Besides, HULC precludes binding of DGCR8 and DROSHA with this pri-miRNA, leading to a significant reduction in the levels of mature MIR15A. Downregulation of this miRNA results in the upregulation of SQSTM1, which contributes to the formation of autophagosome, suppression of PTEN, and induction of cancer [40]. On the other hand, HULC enhances the binding of METTL3 with pri-MIR675 and increases MIR675 levels. This miRNA binds with 3ʹ UTR of HDAC5 mRNA and decreases its expression. Therefore, SIRT1 levels and the formation of autophagosomes are enhanced. This increases CCND1 synthesis and promotes the proliferation of cancer stem cells [134].
lncRNAs and autophagy in nonmalignant conditions
HOTAIR participates in the pathogenesis of intervertebral disc degeneration through modulation of the AMPK-MTOR-ULK1 pathway and enhancement of autophagy, apoptosis, and senescence in the nucleus pulposus cells [41]. In cerebral ischemic stroke, MALAT1 acts as a molecular sponge for MIR26B and MIR200C-3p to upregulate ULK2 and SIRT1, respectively. Both interactions lead to the enhancement of autophagy and the protection of brain microvascular endothelial cells against oxygen-glucose deprivation [42,43]. NEAT1 has a role in the pathogenesis of diverse disorders, including congenital heart disease and Parkinson disease, through enhancement of autophagy via different pathways [44,45]. A number of lncRNAs such as SPAG5-AS1, Gm5524, Gm15645, and SOX2-OT are involved in the regulation of autophagy in the context of diabetic nephropathy [46–48]. Being upregulated in pre-eclampsia, the lncRNA H19 decreases cell viability but enhances invasion and autophagy in trophoblast cells possibly through induction of the PI3K-AKT-MTOR signaling [49].
Table 3 shows the list and function of autophagy-related lncRNAs.
Table 3.
List of autophagy-associated lncRNAs
| lncRNA | lncRNA Nucleotides | Disease | Animal/human (numbers of clinical samples) | Gain- or loss-of-function studies/animal models | Targets/Regulators | Signaling Pathways | Function | Ref |
|---|---|---|---|---|---|---|---|---|
| HNF1A-AS1 | 2455 | Hepatocellular carcinoma (HCC) | 40 pairs of HCC and adjacent normal tissues | ± | MIR30B-5p, ATG5, MAP1LC3B, BCL2 | SQSTM1/p62 | HNF1A-AS1 via sponging hsa-MIR30B-5p could promote autophagy in HCC. | [133] |
| HULC | 500 | HCC | Mouse/human; 30 pairs of HCC and adjacent normal tissues |
+/+ | PTEN, MIR15A, CTNNB1, PKM2, CDK2, SIRT1, NOTCH1, JUN |
MAPK9/SAPK,MAPK8/JNK | HULC by inhibiting PTEN via autophagy cooperation to MIR15A could accelerate liver cancer. | [40] |
| HULC | 500 | Gastric cancer (GC) | Mouse | -/+ | FOXM1, BECN1, MAP1LC3B | SQSTM1/p62 | METase-HULC-FOXM1 axis by suppressing autophagy could reduce cisplatin resistance in GC. | [39] |
| HULC | 500 | Liver cancer | Mouse | -/+ | CCND1, MIR675, PKM2, SIRT1, RB1 |
CDKN1/WAF1/CIP1 | HULC by upregulating CCND1 through the miR675-PKM2 pathway via autophagy could accelerate the growth of human liver cancer stem cells. | [134] |
| HOTAIR | 2370 | HCC | 54 pairs of HCC and adjacent normal tissues | ± | ATG3, ATG7 | - | HOTAIR by upregulating ATG3 and ATG7 could activate autophagy in HCC. | [35] |
| HOTAIR | 2370 | Intervertebral disc degeneration (IDD) | Rat/human; intervertebral disc tissue samples from DD group (n = 30), idiopathic scoliosis (healthy group, n = 10) | +/+ | ULK1, BECN1, MAP1LC3B | AMPK-MTOR, SQSTM1 | HOTAIR via the AMPK-MTOR-ULK1 pathway could upregulate autophagy to enhance apoptosis and senescence of nucleus pulposus cells. | [41] |
| HOTAIRM1 | 1052 | Acute promyelocytic leukemia (APL) | Mouse/human; 54 APL samples at diagnosis and 25 APL samples after therapy | +/+ | MIR20A, MIR106B, MIR125B, ULK1, E2F1, DRAM2 | - | HOTAIRM1 by enhancing the autophagy pathway could regulate myeloid cell differentiation. | [135] |
| PVT1 | 1957 | HCC | 80 pairs of HCC and adjacent normal tissues | ± | MIR365, ATG3, ATG10, MAP1LC3B | - | PVT1 via the ATG3-MIR365 axis could promote autophagy in HCC. | [136] |
| PVT1 | 1957 | Pancreatic ductal adenocarcinoma (PDA) | Mouse/human, GEO database |
+/+ | ULK1, MIR20A, MAP1LC3B | - | PVT1 via the MIR20A ULK1 axis could trigger cytoprotective autophagy and promote PDA development. | [137] |
| HIF1A-AS1 | 652 | HCC | 50 pairs of HCC and adjacent normal tissues | ± | HIF1A | MTOR | Inhibition of HIF1A-AS1 could promote starvation-induced HCC cell apoptosis. | [52] |
| HAGLROS | 699 | HCC | 68 pairs of HCC and adjacent normal tissues | ± | MIR5095, ATG12, BECN1, BAX, BCL2, CASP3, MAP1LC3B | PI3K-AKT-MTOR, SQSTM1 | HAGLROS could inhibit apoptosis and enhance autophagy in HCC. | [54] |
| HAGLROS | 699 | GC | Mouse/human; 48 pairs of GC and adjacent normal tissues | +/+ | STAT3,MIR100, ATG9A, ATG9B | MTOR | HAGLROS could contribute to the autophagy and malignant progression of GC cells. | [138] |
| DCST1-AS1 | 1202 | HCC | 45 pairs of HCC and adjacent normal tissues | ± | CCNB1, CCND1, BAX, BCL2, CASP3, CDH2, CDH1 |
AKT-MTOR | DCST1-AS1 via the AKT-MTOR pathway could accelerate the proliferation, metastasis, and autophagy of HCC. | [139] |
| LNCRNA-ATB | Unknown | HCC | 72 pairs of HCC and adjacent normal tissues | ± | YAP, ATG5, MAP1LC3B |
- | LNCRNA-ATB by activating YAP and inducing ATG5 could promote autophagy in HCC. | [55] |
| MALAT1 | 8779 | Glioblastoma (GBM) | 32 pairs of GBM and adjacent normal tissues | ± | MIR101, STMN1, RAB5A, ATG4D, SQSTM1, MAP1LC3B | SQSTM1 | MALAT1 by sponging MIR101 and upregulating STMN1, RAB5A, and ATG4D could activate autophagy and promote cell proliferation in GBM. | [140] |
| MALAT1 | 8779 | GBM | 25 pairs of GBM and adjacent normal tissues | ± | MIR384, GOLM1, MAP1LC3B, VIM, CDH1 | SQSTM1 | The knockdown of MALAT1 could inhibit cell migration and invasion by suppressing autophagy in GBM. | [37] |
| MALAT1 | 8779 | Cerebral ischemic stroke (CIS) | Mouse | -/+ |
MIR26B, ULK2 |
SQSTM1 | MALAT1 by sponging MIR26B and upregulating ULK2 could promote autophagy and protect BMECs against OGD/R-induced injury. | [43] |
| MALAT1 | 8779 | CIS | - | -/- | MIR200C, SIRT1, MAP1LC3B | SQSTM1 | MALAT1 by binding to MIR200C and upregulating SIRT1 could induce autophagy and protect BMECs against oxygen-glucose deprivation (OGD). | [42] |
| MALAT1 | 8779 | Multiple myeloma (MM) | Mouse/human; bone marrow samples from 60 untreated MM patients, normal plasma cells as control (n = 60) | +/+ | HMGB1, BECN1, MAP1LC3B | - | MALAT1 by elevating HMGB1 could promote autophagy in multiple myeloma. | [141] |
| MALAT1 | 8779 | B-cell lymphoma | Mouse | -/+ | MAP1LC3B, ATG5 | SQSTM1 | Inhibition of MALAT1 could decrease chemotherapy resistance of diffuse large B-cell lymphoma by enhancing autophagy-related proteins. | [142] |
| MALAT1 | 8779 | Epilepsy | Rat | -/+ |
MIR101, MET |
PI3K-AKT | Downregulation of MALAT1 via activating the PI3K-AKT pathway could protect hippocampal neurons against excessive apoptosis and autophagy. | [143] |
| MALAT1 | 8779 | GC | 57 pairs of GC and adjacent normal tissues | ± | MIR204, MAP1LC3B | SQSTM1 | MALAT1 by downregulating MIR204 could activate autophagy and promote cell proliferation in GC. | [144] |
| MALAT1 | 8779 | Acute myocardial infarction (AMI) | Rat | -/+ | MIR558, ULK1, BECN1, MAP1LC3B | - | MALAT1 via sponging MIR558 to enhance ULK1‐mediated protective autophagy could protect cardiomyocytes from isoproterenol (ISO)‐induced apoptosis. | [145] |
| MALAT1 | 8779 | Atherosclerosis (AS) | Peripheral blood samples from 40 atherosclerotic patients and 40 healthy subjects | ± | MIR216A-5p, BECN1, CASP3, MAP1LC3B | SQSTM1 | Ox-LDL-induced MALAT1 by sponging MIR216A-5p and regulating BECN1 could promote autophagy in HUVECs. | [146] |
| MALAT1 | 8779 | AS | Mouse/human; peripheral blood from 26 atherosclerotic heart disease (CAD) patients and 20 volunteers GEO database |
+/+ | MIR15B, MAPK1, ATG1, MAP1LC3B | MAPK3, MAPK1, MTOR | MALAT1 via MIR15B-MAPK1-MTOR could inhibit EPCs autophagy. | [147] |
| MALAT1 | 8779 | AS | - | -/- | RPS6KB1, MAP1LC3B | PI3K-AKT | MALAT1 by inhibiting the PI3K-AKT pathway could promote ox‐LDL‐induced autophagy in HUVECs. | [148] |
| MALAT1 | 8779 | Vascular endothelial cell injury | - | -/- | MIR19B, HIF1A, MAP1LC3B | SQSTM1 | The knockdown of MALAT1 via the MIR19B-HIF1A axis could reduce the hypoxia-induced HUVECs apoptosis and autophagy. | [149] |
| MALAT1 | 8779 | Retinoblastoma (RB) | - | -/- | MIR124, STX17, BECN1, MAP1LC3B | SQSTM1 | MALAT1 could modulate the autophagy of retinoblastoma cells. | [150] |
| MALAT1 | 8779 | - | - | -/- | MIR23, LAMP1, MAP1LC3B | SQSTM1 | MALAT1-MIR23-LAMP1 axis could be involved in promoting autophagy in macrophages. | [151] |
| MALAT1 | 8779 | - | - | -/- |
MIR142, ATG7 |
- | Downregulation of MALAT1 via targeting ATG7 could attenuate platelet-derived growth factor-BB (PDGF-BB)-induced proliferation and migration in VSMCs. | [152] |
| MEG3 | 1595 | GBM | 79 pairs of GBM and adjacent normal tissues | ± | MAP1LC3B | - | MEG3 could regulate autophagy in GBM. | [153] |
| MEG3 | 1595 | Ovarian cancer (OC) | Mouse/human; normal ovarian tissues (n = 8), benign OC (n = 17), borderline OC (n = 6), OC (n = 95), metastatic momentum (n = 25) | +/+ | ATG3, LAMP1, SQSTM1, MAP1LC3B | SQSTM1 | Overexpression of MEG3 by regulating the activity of ATG3 could induce autophagy to inhibit tumorigenesis of epithelial OC. | [154] |
| MEG3 | 1595 | Ventricular septal defect (VSD) | Rat/human; heart tissues and blood samples from 20 patients with VSD and 24 healthy individuals | +/+ | MIR7, EGFR, AKT3, BECN1, ATG7 | SQSTM1 | Uric acid and sphingomyelin via MEG3-MIR7-EGFR axis could enhance autophagy in iPS cell-originated cardiomyocytes. | [155] |
| PCED1B-AS1 | 2502 | Pulmonary tuberculosis (PTB) | 20 patients with active PTB and 20 healthy controls | ± | MIR155, BAX, BCL2, CASP3, MAP1LC3B | - | PCED1B-AS1 by sponging MIR155 could regulate macrophage apoptosis and autophagy in tuberculosis. | [156] |
| EPS | Unknown | PTB | 120 patients with active PTB and 105 healthy controls | ± | MAP1LC3B | MAPK8 | Lowerexpression of lncRNA EPS via the MAPK8 could regulate autophagy and apoptosis in Bacillus Calmette-Guérin (BCG)-infected RAW264.7 macrophages. | [157] |
| RMRP | 277 | CIS | - | -/- | BCL2, BAX, MAP1LC3B | SQSTM1, PI3K-AKT-MTOR |
Suppression of RMRP by inhibiting autophagy and apoptosis could ameliorate OGD/R-induced neural cell injury. | [158] |
| TCTN2 | Spinal cord injury (SCI) |
Rat | -/+ | MIR216B, BECN1, AGO2 | - | Overexpression of TCTN2 by enhancing cell autophagy could protect neurons from apoptosis in SCI. | [159] | |
| GAS8-AS1 | 1000 | Papillary thyroid cancer (PTC) | - | -/- | ATG5, MAP1LC3B | SQSTM1 | GAS8-AS1 via ATG5-mediated autophagy could inhibit cell proliferation in PTC. | [160] |
| HOTTIP | 4665 | Renal cell carcinoma (RCC) | Mouse/human; 42 pairs of RCC and adjacent normal tissues | +/+ | ATG13, LC3B, LAMP2, BECN1 | PI3K-AKT, SQSTM1 |
HOTTIP by regulating autophagy could affect RCC progression. | [161] |
| H19 | 2362 | Severe burn | Mouse | -/+ | MAP1LC3B, BECN1 | EGF | EGF is regulated by H19 in IEC-6 cells after a serious burn. | [114] |
| H19 | 2362 | AMI | Mouse | -/+ | MAP1LC3B, BECN1, ATG7 | - | H19 via activating autophagy could protect acute myocardial infarction in mice. | [162] |
| H19 | 2362 | Diabetic cardiomyopathy (DC) | Rat | -/+ | DIRAS3, EZH2 | MTOR | H19 by epigenetically silencing of DIRAS3 could inhibit autophagy in DC. | [163] |
| H19 | 2362 | Pre-eclampsia (PE) | Placenta tissues of PE patients and healthy pregnant women (n = 20/group) |
± | LC3, RPS6KB1 | PI3K-AKT-MTOR | Overexpression of H19 via the PI3K-AKT-MTOR pathways could promote invasion and autophagy in trophoblast cells. | [49] |
| H19 | 2362 | Breast cancer (BCa) | 23 patients with lymph node (LN)-positive BCa, 20 patients with LN-negative BCa | ± | MIRLET7, LIN28, BECN1, MAP1LC3B | SQSTM1/p62 | H19 via MIRLET7-LIN28 axis could mediate autophagy and inhibit EMT in BCa. | [164] |
| DCRF | 98 | DC | Rat | -/+ | MIR551B, PCDH17, MAP1LC3B | DCRF by upregulating PCDH17 could regulate cardiomyocyte autophagy. | [165] | |
| NEAT1 | 3756 | Congenital heart disease (CHD) | 42 patients with CHD and 32 healthy | ± | MIR181B, BECN1, CASP3, MAP1LC3B | PI3K-AKT-MTOR, JAK1/STAT3, SQSTM1/p62, TP53 | Overexpression of NEAT1 by expediting PI3K-AKT-MTOR and JAK1-STAT3 pathways could ease hypoxia-triggered H9c2 cells apoptosis and autophagy. | [44] |
| NEAT1 | 3756 | Parkinson disease (PD) | Mouse | -/+ | PINK1, MAP1LC3B | - | NEAT1 through stabilizing PINK1 protein could promote autophagy in MPTP-induced Parkinson’s disease. | [45] |
| NEAT1 | 3756 | Colorectal cancer (CRC) |
55 pairs of CRC and adjacent normal tissues | ± | MIR34A, ATG9A, ATG4B, HMGB1, BECN1, CASP3, MAP1LC3B | - | The knockdown of NEAT1 via targeting MIR34A could attenuate autophagy to elevate 5-FU sensitivity in CRC. | [38] |
| BDNF-AS | 2322 | PD | Mouse | -/+ |
MIR125B, BCL2, BAX, CASP3, MAP1LC3B |
SQSTM1 | BDNF-AS via ablating MIR125B could promote autophagy and apoptosis in MPTP-induced Parkinson’s disease. | [166] |
| SNHG1 | 476 | PD | Mouse | -/+ | MIR221, MIR222, CDKN1B, MAP1LC3B | MTOR | Downregulation of SNHG1 could attenuate MPP+-induced cytotoxicity and enhance autophagy in Parkinson disease. | [167] |
| SNHG6 | 727 | Osteosarcoma (OS) | 45 pairs of OS and adjacent normal tissues | ± |
MIR26A, ULK1 |
- | The silencing of SNHG6 by targeting the MIR26A-ULK1 axis could induce cell autophagy in human OS. | [168] |
| SNHG7 | 2176 | Osteoarthritis (OA) | OA cartilage tissues from 15 OA patients, normal cartilage tissues from 10 patients | ± | MIR34A, SYVN1, BECN1, MAP1LC3B | - | Upregulation of SNHG7 by sponging MIR34A could promote cell proliferation and inhibit cell apoptosis and autophagy. | [169] |
| SNHG11 | 1101 | HCC | Mouse/human; 57 pairs of HCC and adjacent normal tissues | +/+ | MIR184, AGO2, BECN1, CASP3, MAP1LC3B | - | SNHG11 by regulating MIR184-AGO2 could promote proliferation, migration, apoptosis, and autophagy in HCC. | [53] |
| SNHG12 | 606 | CIS | Mouse | -/+ | BECN1, MAP1LC3B | SQSTM1 | SNHG12 as a potent autophagy inducer could attenuate cerebral I/R injury. | [170] |
| SNHG14 | 19,263 | CRC | 40 pairs of CRC and adjacent normal tissues | ± | MIR186, ATG14 | - | SNHG14 by regulating the MIR186-ATG14 axis could stimulate cell autophagy to facilitate cisplatin resistance of CRC. | [171] |
| SNHG15 | 860 | OS | 35 pairs of OS and adjacent normal tissues | ± |
MIR141, ATG5, MAP1LC3B |
SQSTM1 | SNHG15 by sponging MIR141 could be contributed to proliferation, invasion, and autophagy in OS cells. | [172] |
| SNHG16 | 860 | Neuroblastoma (NB) | Mouse/human; 45 pairs of NB and adjacent normal tissues | +/+ | MIR542, ATG5, MAP1LC3B | SQSTM1 | SNHG16 via sponging MIR542 and upregulating ATG5 could facilitate proliferation, migration, invasion, and autophagy of NB Cells. | [173] |
| SLCO4A1-AS1 | CRC | Mouse/human; 23 pairs of CRC and adjacent normal tissues | +/+ | MIR508, PARD3 | - | SLCO4A1-AS1 via MIR508-PARD3 axis could promote CRC proliferation by enhancing autophagy. | [174] | |
| CPS1-IT1 | 1440 | CRC | Mouse/human, 24 pairs of CRC and adjacent normal tissues | +/+ | HIF1A, BECN1, MAP1LC3B | EMT | CPS1-IT1 by inhibiting hypoxia-induced autophagy via inactivating HIF1A could suppress EMT and metastasis of CRC. | [175] |
| GAS5 | 501 | CRC | Mouse | -/+ | MIR222, PTEN, BECN1, MAP1LC3B | - | GAS5 via the MIR222-PTEN axis could promote autophagy and inhibit cell migration and invasion in CRC. | [176] |
| GAS5 | 656 | AS | Plasma samples from 30 atherosclerotic patients and 30 healthy subjects | ± | MIR26A, MAP1LC3B | SQSTM1 | Knockdown of GAS5 via upregulating MIR26A could restore ox-LDL-induced impaired autophagy flux in HAECs | [177] |
| GAS5 | 656 | - | - | -/- | ATG3, MIR23A, BECN1 |
MTOR, SQSTM1 |
Knockdown of GAS5 via ATG3-dependent autophagy by regulating MIR23A could attenuate cell viability and inhibit autophagy | [178] |
| UCA1 | 2314 | - | - | -/- |
MIR184, OSGIN1 |
MTOR-RPS6KB/p70S6K | UCA1 via blocking autophagic flux under arsenic stress could attenuate autophagy-dependent cell death. | [179] |
| EGOT | 1529 | Acute kidney injury (AKI) | - | -/- | ATG7, ATG16L1, MAP1LC3B |
- | EGOT by targeting ATG7, and ATG16L1 could regulate autophagy in renal tubular cells. The ELAVL1-EGOT-ATG7-ATG16L1 axis is involved in hypoxia‐induced autophagy in HK‐2. | [15] |
| CCAT1 | 2795 | AKI | - | -/- | MAP1LC3B | PI3K-AKT, SQSTM1 | Exposure to TNFA decreased the expression of CCAT1. CCAT1 via inhibiting autophagy could function as an apoptosis inhibitor in podocytes. | [180] |
| TUG1 | 7598 | AMI | Mouse | -/+ | MIR142, HMGB1, RAC1, BECN1, MAP1LC3B | SQSTM1 | TUG1 via the MIR142-HMGB1-RAC1 axis could play an important role in stimulating autophagic cell apoptosis in myocardial injury induced by I/R. | [181] |
| AK139128 | 1516 | AMI | Rat | -/+ | MIR499, BAX, FOXO4, BCL2, CASP3, MAP1LC3B | SQSTM1 | AK139128 via MIR499-FOXO4 axis could promote cardiomyocyte autophagy and apoptosis in myocardial I/R injury. | [182] |
| AK139328 | 2668 | AMI | Mouse | -/+ | MIR204-3p, ACTA2, ATG7, ATG5, MAP1LC3B | SQSTM1 | Knockdown of AK139328 via modulating MIR204‐3p and inhibiting autophagy could alleviate myocardial I/R injury in diabetic mice. | [183] |
| HRIM | 1470 | AMI | Rat | -/+ | ZDHHC7, PTGIS, KRT23, PHACTR1 | - | Inhibition of HRIM by regulating autophagy levels during hypoxia/reoxygenation could increase cell viability in myocytes. | [184] |
| XIST | 17,918 | AMI | Mouse | -/+ | MIR133A, SOCS2, BECN1, MAP1LC3B | - | Knockdown of XIST via the MIR133A-SOCS2 axis could improve myocardial I/R injury by inhibiting autophagy. | [185] |
| XIST | 17,918 | RB | Mouse/human; 25 RB and 6 matched normal retinal tissues | +/+ |
MIR204, BAX, BCL2, CASP3, CASP9, MAP1LC3B |
SQSTM1 | Silencing of XIST could enhance vincristine sensitivity and also suppress autophagy and proliferation in retinoblastoma cells. | [186] |
| 2810403D21 Rik/Mirf | 1005 | AMI | Mouse | -/+ | MIR26A, SQSTM1, USP15, MAP1LC3B | SQSTM1 | 2810403D21Rik/Mirf via regulating autophagy by targeting MIR26A could promote ischemic myocardial injury. | [187] |
| AK088388 | 3312 | AMI | - | -/- | MIR30A, BECN1, MAP1LC3B | - | AK088388 by targeting MIR30A could regulate autophagy to affect cardiomyocyte injury. | [65] |
| MSTO2P | 2231 | Lung cancer (LCa) | 45 pairs of LCa and adjacent normal tissues | ± | EZH2, ATG5, MAP1LC3B | - | MSTO2P by upregulating EZH2 could promote proliferation and autophagy of LCa cells. | [72] |
| CASC2 | 3284 | Non-small cell lung carcinoma (NSCLC) | 21 pairs of NSCLC and adjacent normal tissues | ± | MIR214, TRIM16, ATG5, MAP1LC3B | SQSTM1 | CASC2 via regulating the MIR214-TRIM16 axis could inhibit autophagy and promote apoptosis in NSCLC cells | [188] |
| SPAG5-AS1 | 1379 | Diabetic nephropathy (DN) | - | -/- | SPAG5, MIR769, PODOCIN, MAP1LC3B |
AKT-MTOR, YY1 | SPAG5-AS1 via the SPAG5-AKT-MTOR pathway could inhibit autophagy and aggravate apoptosis in high-glucose–treated human podocytes. | [46] |
| GM5524 | 1793 | DN | Mouse | -/+ | CASP3, BAX, BCL2, ATG5, ATG7, MAP1LC3B | - | Dysregulation of GM5524 is involved in high-glucose-induced podocyte autophagy and apoptosis in DN. | [47] |
| GM15645 | 2483 | DN | Mouse | -/+ | CASP3, BAX, BCL2, ATG5, ATG7, MAP1LC3B | - | Dysregulation of GM15645 involved in high-glucose-induced podocyte autophagy and apoptosis in DN. | [47] |
| SOX2OT | 2998 | DN | - | -/- | MIR9, SIRT1, BAX, BCL2, CASP3, BECN1, ATG7, MAP1LC3B | SQSTM1 | SOX2OT via autophagy induction by the MIR9/SIRT1 axis could alleviate the high-glucose-induced podocytes injury. | [48] |
| DICER1-AS1 | 830 | OS | Mouse | -/+ | MIR30B, ATG5, MAP1LC3B, BECN1 | - | DICER1-AS1 via MIR30B-ATG5 axis could promote the proliferation, invasion, and autophagy of osteosarcoma cells. | [189] |
| DANCR | 915 | OS | Mouse/human; 45 pairs of OS and adjacent normal tissues | +/+ | MIR216A, SOX5, BECN1, MAP1LC3B | - | DANCR silencing could inhibit SOX5-medicated progression and autophagy in OS. | [190] |
| FEZF1-AS1 | 2653 | Prostate cancer (PCa) | Mouse/human; 47 pairs of PCa and adjacent normal tissues | +/+ |
MIR25, ATG5, ITGB8, BECN1, CDH1, CDH2, VIM, MAP1LC3B |
EMT | FEZF1-AS1 via regulation of MIR2-ITGB8 axis could promote chemoresistance, autophagy, and EMT in PCa. | [191] |
| LINC00337 | 1642 | Esophageal squamous cell carcinoma (ESCC) |
Mouse/human; 74 ESCC and 26 matched mucosal tissues |
+/+ | BECN1, MAP1LC3B, TPX2, E2F4 | - | LINC00337 via upregulating TPX2 by recruiting E2F4 could induce autophagy and chemoresistance to cisplatin in ESCC cells. | [192] |
| FA2H-2 | Unknown | AS | Mouse/human; 20 pairs with atherosclerotic plaque and normal arterial tissues | +/+ | MLKL, LAMP1, VCAM1, IL6, MCP1, IL8, IL18, IL1B, TNFA, RPS6KB1, MAP1LC3B | SQSTM1, MTOR | Silencing FA2H-2 via the MLKL-MTOR axis could activate inflammation and inhibit autophagy flux in atherosclerosis. | [193] |
| DYNLRB2-2 | Unknown | AS | - | -/- | ABCA1, LKB1, AMPK, BECN1, MAP1LC3B | MTOR | DYNLRB2-2 by enhancing autophagy could inhibit THP-1 macrophage foam cell formation. | [194] |
| LINC00460 | 913 | Head and neck squamous cell carcinoma (HNSCC) | 45 pairs of HNSCC and adjacent normal tissues, TCGA database | ± | STC2, MIR206, BECN1, MAP1LC3B | AKT-MAPK | Downregulation of LINC00460 by upregulating MIR206 and downregulating STC2 could promote autophagy of HNSCC. | [195] |
| Lethe | 697 | Sepsis | Mouse | -/+ | IFNG, MAP1LC3B, SQSTM1 | - | Lethe via regulating autophagy of cortical neurons could protect sepsis-induced brain injury. | [196] |
| NKILA | 2615 | Sepsis | Rat | -/+ | MAP1LC3B, BECN1 | PI3K-AKT | NKILA-AKT axis could be involved in promoting autophagy in sepsis-induced kidney injury. | [197] |
| CIR | Unknown | OA | Rat/human; 8 patients undergoing total hip arthroplasty (THA), patients undergoing periacetabular osteotomy (PAO, n = 8) |
+/+ | MMP3, COL2A1, MAP1LC3B, BECN1 | - | CIR by regulating autophagy could promote articular cartilage degeneration in osteoarthritis. | [198] |
| ZNNT1 | 3435 | Uveal melanoma (UM) | Mouse | -/+ | SQSTM1, MAP1LC3B, ATG12 | MTOR, SQSTM1/p62 | ZNNT1 by regulating key autophagy gene expression could inhibit tumorigenesis of UM. | [199] |
| GBCDRlnc1 | Unknown | Gallbladder cancer (GBC) |
45 pairs of GBC and adjacent normal tissues | ± | MAP1LC3B, BECN1, PGK1, ATG3, ATG5, ATG7, ATG12, ULK1 |
SQSTM1 | GBCDRlnc1 by activating autophagy could induce chemoresistance of GBC. | [200] |
| CASC9 | 1471 | Oral squamous cell carcinoma (OSCC) | Mouse/human; 35 pairs of OSCC and adjacent normal tissues | +/+ | MAP1LC3B, BAX, BCL2 | AKT-MTOR | Overexpression of CASC9 by suppressing autophagy-mediated cell apoptosis via the AKT-MTOR pathway could promote tumor progression in OSCC. | [201] |
| NR_003923 | 3025 | Glaucoma | 6 pairs of human fascia and adjacent normal tissues |
± |
MIR760, MIR215, SMA, CDH1, CTNNB1, IL22RA1 |
SQSTM1 | NR_003923 via the MIR760-MIR215-IL22RA1 axis could promote cell fibrosis, proliferation, migration, and autophagy in human tenon’s capsule fibroblast cells (HTFs). | [202] |
| TINCR | 3733 | Cutaneous squamous cell carcinoma (CSCC) | - | -/- | SP3, MAP1LC3B, BECN1, BAX, BCL2 | MAPK1, MAPK3 | TINCR could participate in ALA‐PDT‐induced apoptosis and autophagy in CSCC. | [203] |
| OGFRP1 | 1256 | - | - | -/- | MAP1LC3B, BECN1, BAX, BCL2, CASP3, RPS6KB, CNND1 | AKT-MTOR, SQSTM1 |
Downregulation of OGFRP1 via the AKT-MTOR pathway could induce autophagy and growth inhibition in HCAECs. | [204] |
Vault RNAs (vtRNA) as a group of small ncRNAs being produced by RNA polymerase III can bind with the autophagy receptor SQSTM1 to suppress SQSTM1-dependent autophagy. Mechanistically, vtRNAs binding with SQSTM1 interferes with the oligomerization of SQSTM1 [50]. Notably, the mechanism by which ncRNA may directly regulate protein function in the context of autophagy is implicated in cellular viability [50,51].
Expression levels of autophagy-related lncRNAs can predict the prognosis of patients with diverse cancer types. Most studies in this regard have been performed in patients with hepatocellular carcinoma. HNF1A-AS1, HOTAIR, HAGLROS, SNHG11, and LNCRNA-ATB are among lncRNAs whose upregulation confer unfavorable outcome in this kind of cancer [35,52–55]. Table 4 summarizes the results of studies that assessed the association between expression levels of autophagy-related lncRNAs and patients’ prognosis. Moreover, few studies have appraised the diagnostic role of these lncRNAs in cancer patients. These studies are also summarized in Table 4.
Table 4.
Prognostic/diagnostic value of autophagy-related lncRNAs in patients with cancers
| Sample number | Area under curve | Kaplan-Meier analysis | Multivariate cox regression | Ref |
|---|---|---|---|---|
| 54 pairs of HCC and adjacent normal tissues | - | - | The overexpression of HOTAIR was associated with tumor size. | [35] |
| 40 pairs of HCC and adjacent normal tissues | - | - | High expression of HNF1A-AS1 was associated with larger tumor size, multiple tumor lesions, poor differentiation, and advanced TNM stage. | [133] |
| GEO database | - | Higher expression of PVT1 was associated with a lower OS rate. | - | [137] |
| 50 pairs of HCC and adjacent normal tissues | - | Higher expression of HIF1A-AS1 was associated with a lower OS rate and a worse DFS. | A high level of HIF1A-AS1 was associated with tumor size, TNM stage, lymph node metastasis. | [52] |
| 68 pairs of HCC and adjacent normal tissues | - | Higher expression of HAGLROS was associated with a lower OS rate. | A high level of HAGLROS was associated with tumor stage or tumor differentiation. | [54] |
| 48 pairs of GC and adjacent normal tissues | - | Higher expression of HAGLROS was associated with a lower OS rate. | - | [138] |
| 72 pairs of HCC and adjacent normal tissues | - | Higher expression of LNCRNA-ATB was associated with a lower OS rate. | A high level of LNCRNA-ATB was associated with the advanced TNM stage. | [55] |
| Normal ovarian tissues (n = 8), ovarian cancer (n = 95) | 0.76 | - | MEG3 expression had a negative correlation with FIGO stages. | [154] |
| 42 pairs of RCC and adjacent normal tissues | - | Lower expression of HOTTIP was associated with a lower OS rate | Higher expression of HOTTIP was associated with TNM stage, histological grade, and lymph node metastasis. | [161] |
| 23 patients with lymph node (LN)-positive BCa, 20 patients with LN-negative BCa | - | Lower expression of H19 was associated with a lower OS rate. | - | [164] |
| 30 human OS tissues and 30 corresponding adjacent normal tissues | - | Higher expression of SNHG6 was associated with a lower OS rate. | Higher expression of SNHG6 was associated with tumor invasion depth and lymph node metastasis. | [168] |
| 57 pairs of HCC and adjacent normal tissues | - | Higher expression of SNHG11 was associated with a lower OS rate. | - | [53] |
| 45 pairs of NB and adjacent normal tissues | - | Higher expression of SNHG16 was associated with a lower OS rate. | A high level of SNHG16 was associated with the INSS stage and MYCN status. | [173] |
| 47 pairs of PCa and adjacent normal tissues | 0.7736 | - | - | [191] |
| 45 pairs of HNSCC and adjacent normal tissues, TCGA database | - | Higher expression of LINC00460 was associated with a lower OS rate. | A high level of LINC00460 was associated with the TNM stage and differentiation degree of HNSCC. | [195] |
| 35 pairs of OSCC and adjacent normal tissues | - | Higher expression of CASC9 was associated with a lower OS rate. | A high level of CASC9 was associated with tumor size, regional lymph node metastasis, and clinical stage of OSCC. | [201] |
| 45 pairs of HCC and adjacent normal tissues | - | Higher expression of DCST1-AS1 was associated with a lower OS rate. | - | [205] |
| 79 pairs of GBM and adjacent normal tissues | - | Lower expression of MEG3 was associated with a lower OS rate. | Lower expression of MEG3 was associated with advanced WHO grade, low KPS, tumor recurrence, IDH wild-type. | [206] |
Discussion
As a conserved process for the elimination of misfolded proteins and damaged organelles, autophagy is involved in the pathogenesis of several disorders. Autophagy is regulated by both lncRNAs and miRNAs. LncRNAs mostly regulate autophagy through modulation of expression of ATG genes. Their function is exerted through their ceRNA role in which they alter the function of autophagy-related miRNAs [56]. Notably, autophagy itself can regulate the expression of several lncRNAs. An example of this type of regulation is represented by the lncRNA PVT1. The expression of this upregulated lncRNA in diabetic patients is downregulated by autophagy suppression [57]. Globally, the role of autophagy-associated ncRNAs has been mostly assessed in cancers. Autophagy-related ncRNAs have remarkable survival in patients with diverse types of cancers.
The role of miRNAs/lncRNAs in the regulation of autophagy is mostly appraised in the context of cancer. Autophagy is regarded as a “dual sword” in the pathogenesis of cancer. Mostly, it preserves the homeostasis of the cancer milieu by affording nutritional supplements in situations of hypoxia and nutrient shortage. Yet, in certain conditions, autophagy can repress carcinogenesis [58]. This note should be considered in the appraisal of the role of autophagy-related ncRNAs in the carcinogenic process. Moreover, autophagy has a fundamental role in the pathogenesis of several age-related conditions such as intervertebral disc degeneration, ischemia-related disorders such as myocardial infarction and cerebral ischemia, and diabetic-related complications. Thus, miRNAs/lncRNAs that regulate this process are putative therapeutic targets for a wide range of disorders. It is worth mentioning that while autophagy has a protective role against cell injury in cerebral ischemic stroke, in many of the mentioned conditions, it aggravates the pathogenic situation. Therefore, the direction of effects of autophagy in human pathologies should be considered in the design of therapeutic strategies. Moreover, it is possible that autophagy-related lncRNAs/miRNAs modulate specific targets or pathways in each tissue. This is particularly important for miRNAs as they can have several targets with variable levels of complementarity.
In addition to the regulatory role of ncRNAs on autophagy, recent studies indicate that autophagy regulates ncRNA biology. For example, autophagy selectively targets key components of the miRNA machinery to regulate miRNAs stability and function [59,60]. DICER1 and the principal miRNA effector, AGO2, is degraded through the selective autophagy receptor CALCOCO2/NDP52 [60]. Moreover, the autophagy machinery has been reported to regulate intracellular and extracellular transport of RNA-binding proteins and ncRNAs. For instance, the LC3-conjugation system regulates the packaging of RNA-binding proteins into extracellular vesicles [61]. Furthermore, ATG5 has been demonstrated to diminish nuclear transport of MIR126-5p [51]. Finally, the MTORC1 pathway and autophagy control the proper assembly of RNA-induced Silencing complexes (RISCs), therefore affecting miRNA-related functions [62].
According to the complexity of the autophagy process and involvement of several ncRNAs in the regulation of this process, integrative system biology-based methods are the preferred strategies for assessment of expression profile and function of miRNAs and lncRNAs and identification of the functional networks in this process. Each module in this network can be applied as a therapeutic target for disorders that are associated with autophagy. It is worth mentioning that with the constant influx of novel researchers in this field, it is necessary to outline standards for this kind of research. Importantly, investigators should apply these guidelines to ensure appropriate study design [63].
Finally, autophagy-associated lncRNAs and miRNAs can predict patients’ outcomes in diverse cancer types. However, the prognostic role of these transcripts has not been assessed in other pathologic conditions. Thus, future studies should focus on this field to unravel the diagnostic/prognostic role of miRNAs and lncRNAs in these conditions to design personalized approaches for these disorders.
Acknowledgments
This study was financially supported by Shahid Beheshti University of Medical Sciences.
Disclosure statement
The authors declare they have no conflict of interest.
References
- [1].Glick D, Barth S, Macleod KF.. Autophagy: cellular and molecular mechanisms. J Pathol. 2010;221(1):3–12. PubMed PMID: 20225336. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Nakatogawa H, Suzuki K, Kamada Y, et al. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat Rev Mol Cell Biol. 2009;10(7):458–467. [DOI] [PubMed] [Google Scholar]
- [3].Saftig P, Beertsen W. Eskelinen E-L. LAMP-2: a control step for phagosome and autophagosome maturation. Autophagy. 2008;4(4):510–512. [DOI] [PubMed] [Google Scholar]
- [4].Yao H, Han B, Zhang Y, Shen L, Huang R. Non-coding RNAs and autophagy. Autophagy: biology and diseases. 2019;199–220. [DOI] [PubMed] [Google Scholar]
- [5].Zhang P, Wu W, Chen Q, et al. Non-coding RNAs and their integrated networks. J Integr Bioinform. 2019;16(3):20190027. PubMed PMID: 31301674. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Johnsson P, Lipovich L, Grandér D, et al. Evolutionary conservation of long non-coding RNAs; sequence, structure, function. Biochim Biophys Acta. 2014;1840(3):1063–1071. PubMed PMID: 24184936. Epub 10/27. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Guttman M, Amit I, Garber M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458(7235):223–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Ponjavic J, Ponting CP, Lunter G. Functionality or transcriptional noise? Evidence for selection within long noncoding RNAs. Genome Res. 2007;17(5):556–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Warnefors M, Liechti A, Halbert J, et al. Conserved microRNA editing in mammalian evolution, development and disease. Genome Biol. 2014;15(6):R83–R. PubMed PMID: 24964909. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Dahariya S, Paddibhatla I, Kumar S, et al. Long non-coding RNA: classification, biogenesis and functions in blood cells. Mol Immunol. 2019. Aug;112:82–92. PubMed PMID: 31079005. Epub 2019/05/13. eng. [DOI] [PubMed] [Google Scholar]
- [11].Bartel DP. Metazoan MicroRNAs. Cell. 2018. Mar 22;173(1):20–51. PubMed PMID: 29570994. Pubmed Central PMCID: PMC6091663. Epub 2018/03/24. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Liu X, Zhong L, Li P, et al. microRNA‐100 enhances autophagy and suppresses migration and invasion of renal cell carcinoma cells via disruption of NOX4‐dependent mTOR pathway. Clin Transl Sci. 2020. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Wei L, Chen Z, Cheng N, et al. MicroRNA-126 inhibit viability of colorectal cancer cell by repressing mTOR induced apoptosis and autophagy. Oncol Targets Ther. PubMed PMID: 32273718. Pubmed Central PMCID: PMC7102882 2020;13: 2459–2468. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [14].Fu X-T, Shi Y-H, Zhou J, et al. MicroRNA-30a suppresses autophagy-mediated anoikis resistance and metastasis in hepatocellular carcinoma. Cancer Lett. 2018;412:108–117. [DOI] [PubMed] [Google Scholar]
- [15].Chen W, Li Z, Liu H, et al., MicroRNA-30a targets BECLIN-1 to inactivate autophagy and sensitizes gastrointestinal stromal tumor cells to imatinib., Cell Death Dis, 2020. Mar 23 11:3 198.PubMed PMID: 32251287. Pubmed Central PMCID: PMC7090062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Shi Y, Yang Z, Zhang T, et al. SIRT1-targeted miR-543 autophagy inhibition and epithelial-mesenchymal transition promotion in Helicobacter pylori CagA-associated gastric cancer. Cell Death Dis. 2019. Aug 19;10(9):625. PubMed PMID: 31423013. Pubmed Central PMCID: PMC6698481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Qi J, Luo X, Ma Z, et al. Downregulation of miR-26b-5p, miR-204-5p, and miR-497-3p expression facilitates exercise-induced physiological cardiac hypertrophy by augmenting autophagy in rats. Front Genet. 2020;11(78). PubMed PMID: 32140172. Pubmed Central PMCID: PMC7042403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Wang Y, Sun H, Song J, et al. MiR-139-5p protect against myocardial ischemia and reperfusion (I/R) injury by targeting autophagy-related 4D and inhibiting AMPK/mTOR/ULK1 pathway. Int J Clin Exp Pathol. 2017;10(9): 10140–10151. PubMed PMID: 31966906. Pubmed Central PMCID: PMC6965934. [PMC free article] [PubMed] [Google Scholar]
- [19].Zhao P, Zhang BL, Liu K, et al. Overexpression of miR-638 attenuated the effects of hypoxia/reoxygenation treatment on cell viability, cell apoptosis and autophagy by targeting ATG5 in the human cardiomyocytes. Eur Rev Med Pharmacol Sci. 22(23)PubMed PMID: 30556888.:8462–8471. 2018. Dec;. [DOI] [PubMed] [Google Scholar]
- [20].Zhang C, Liang R, Gan X, et al. MicroRNA-384-5p/Beclin-1 as potential indicators for epigallocatechin gallate against Cardiomyocytes Ischemia reperfusion injury by inhibiting autophagy via PI3K/Akt pathway. Drug Des Devel Ther. 2019;13:3607–3623. [PubMed PMID: 31802847. Pubmed Central PMCID: PMC6802542]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Xu Y, Xu Y, Wang S. Effect of exosome-carried miR-30a on myocardial apoptosis in myocardial ischemia-reperfusion injury rats through regulating autophagy. Eur Rev Med Pharmacol Sci. 2019;23(16):7066–7072. [DOI] [PubMed] [Google Scholar]
- [22].Zhang L, Cheng R, Huang Y. MiR-30a inhibits BECN1-mediated autophagy in diabetic cataract. Oncotarget. 2017. Sep 29;8(44):77360–77368. PubMed PMID: 29100392. Pubmed Central PMCID: PMC5652784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Zhou DM, Sun LL, Zhu J, et al. MiR-9 promotes angiogenesis of endothelial progenitor cell to facilitate thrombi recanalization via targeting TRPM7 through PI3K/Akt/autophagy pathway. J Cell Mol Med. 24(8)PubMed PMID: 32147957. Pubmed Central PMCID: PMC7176881.:4624–4632. 2020. Apr;. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Cai C, Min S, Yan B, et al. MiR-27a promotes the autophagy and apoptosis of IL-1beta treated-articular chondrocytes in osteoarthritis through PI3K/AKT/mTOR signaling. Aging (Albany NY). 2019. Aug 27;11(16):6371–6384. PubMed PMID: 31460867. Pubmed Central PMCID: PMC6738432. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [25].Lian WS, Ko JY, Wu RW, et al. MicroRNA-128a represses chondrocyte autophagy and exacerbates knee osteoarthritis by disrupting Atg12. Cell Death Dis. 2018. Sep 11;9(9):919. PubMed PMID: 30206206. Pubmed Central PMCID: PMC6134128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Sun W, Li Y, Wei S. miR-4262 regulates chondrocyte viability, apoptosis, autophagy by targeting SIRT1 and activating PI3K/AKT/mTOR signaling pathway in rats with osteoarthritis. Exp Ther Med. 15(1)PubMed PMID: 29434702. Pubmed Central PMCID: PMC5772979:1119–1128. 2018. Jan;. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Li H, Li Z, Pi Y, et al. MicroRNA-375 exacerbates knee osteoarthritis through repressing chondrocyte autophagy by targeting ATG2B. Aging (Albany NY). 12(8). PubMed PMID: 32335541. 7248–7261. 2020. Apr 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Brest P, Lapaquette P, Souidi M, et al. A synonymous variant in IRGM alters a binding site for miR-196 and causes deregulation of IRGM-dependent xenophagy in Crohn’s disease. Nature Genet. 2011;43(3):242–245. [DOI] [PubMed] [Google Scholar]
- [29].Li M, Zhang S, Qiu Y, et al. Upregulation of miR-665 promotes apoptosis and colitis in inflammatory bowel disease by repressing the endoplasmic reticulum stress components XBP1 and ORMDL3. Cell Death Dis. 2017. Mar 23;8(3):e2699. PubMed PMID: 28333149. Pubmed Central PMCID: PMC5386569. Epub 2017/03/24. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Fang K, Sideri A, Law IKM, et al. 196 identification of a novel Substance P (SP)-Neurokinin-1 Receptor (NK-1R) microRNA-221 inflammatory network in human colonic epithelial cells. Gastroenterology. 2014;146(5):S–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Liao D, Li T, Ye C, et al. miR-221 inhibits autophagy and targets TP53INP1 in colorectal cancer cells. Exp Ther Med. 2018. Feb;;15(2):1712–1717. PubMed PMID: 29434757. Pubmed Central PMCID: PMC5774445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Huang KT, Kuo IY, Tsai MC, et al. Factor VII-INDUCED microRNA-135a Inhibits Autophagy and Is Associated with Poor Prognosis in Hepatocellular Carcinoma. Mol Ther Nucleic Acids. 2017. Dec 15 PubMed PMID: 29246306. Pubmed Central PMCID: PMC5675721.;9:274–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Moharamoghli M, Hassan-Zadeh V, Dolatshahi E, et al. The expression of GAS5, THRIL, and RMRP lncRNAs is increased in T cells of patients with rheumatoid arthritis. Clin Rheumatol. 2019;38(11):3073–3080. [DOI] [PubMed] [Google Scholar]
- [34].Sun AG, Meng FG, Wang MG. CISD2 promotes the proliferation of glioma cells via suppressing beclin1mediated autophagy and is targeted by microRNA449a. Mol Med Rep. 2017. Dec;16(6):7939–7948. PubMed PMID: 28983596. Pubmed Central PMCID: PMC5779876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Yang L, Zhang X, Li H, et al. The long noncoding RNA HOTAIR activates autophagy by upregulating ATG3 and ATG7 in hepatocellular carcinoma. Mol Biosyst. 2016;12(8):2605–2612. [DOI] [PubMed] [Google Scholar]
- [36].Fu Z, Luo W, Wang J, et al. Malat1 activates autophagy and promotes cell proliferation by sponging miR-101 and upregulating STMN1, RAB5A and ATG4D expression in glioma. Biochem Biophys Res Commun. 492(3). PubMed PMID: 28834690. 480–486. 2017 Oct 21. [DOI] [PubMed] [Google Scholar]
- [37].Ma R, Zhang B, Zhang Z, et al. MALAT1 knockdown inhibits cell migration and invasion by suppressing autophagy through miR-384/GOLM1 axis in glioma. Eur Rev Med Pharmacol Sci. 2020;24(5):2601–2615. [DOI] [PubMed] [Google Scholar]
- [38].Liu F, Ai FY, Zhang DC, et al. LncRNA NEAT1 knockdown attenuates autophagy to elevate 5‐FU sensitivity in colorectal cancer via targeting miR‐34a. Cancer Med. 2020;9(3):1079–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Xin L, Zhou Q, Yuan Y-W, et al. METase/lncRNA HULC/FoxM1 reduced cisplatin resistance in gastric cancer by suppressing autophagy. J Cancer Res Clin Oncol. 2019;145(10):2507–2517. [DOI] [PubMed] [Google Scholar]
- [40].Xin X, Wu M, Meng Q, et al. Long noncoding RNA HULC accelerates liver cancer by inhibiting PTEN via autophagy cooperation to miR15a. Mol Cancer. 2018;17(1):94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Sheng H, Guo Y, Cao D, et al. MiR-429-5p attenuates the migration and invasion of malignant melanoma by targeting LIMK1. Eur Rev Med Pharmacol Sci. 2020;24(5):2625–2631. [DOI] [PubMed] [Google Scholar]
- [42].Wang S, Han X, Mao Z, et al. MALAT1 lncRNA induces autophagy and protects brain microvascular endothelial cells against oxygen–glucose deprivation by binding to miR-200c-3p and upregulating SIRT1 expression. Neuroscience. 2019;397:116–126. [DOI] [PubMed] [Google Scholar]
- [43].Li Z, Li J, Tang N. Long noncoding RNA Malat1 is a potent autophagy inducer protecting brain microvascular endothelial cells against oxygen-glucose deprivation/reoxygenation-induced injury by sponging miR-26b and upregulating ULK2 expression. Neuroscience. 2017;354:1–10. [DOI] [PubMed] [Google Scholar]
- [44].Lv Y, Liu Z, Huang J, et al. LncRNA nuclear-enriched abundant transcript 1 regulates hypoxia-evoked apoptosis and autophagy via mediation of microRNA-181b. Mol Cell Biochem. 2020;464(1):193–203. [DOI] [PubMed] [Google Scholar]
- [45].Yan W, Chen Z-Y, Chen J-Q, et al. LncRNA NEAT1 promotes autophagy in MPTP-induced Parkinson’s disease through stabilizing PINK1 protein. Biochem Biophys Res Commun. 2018;496(4):1019–1024. [DOI] [PubMed] [Google Scholar]
- [46].Nabipoorashrafi SA, Shomali N, Sadat‐Hatamnezhad L, et al. miR-143 acts as an inhibitor of migration and proliferation as well as an inducer of apoptosis in melanoma cancer cells in vitro. IUBMB Life. 2020;72(9):2034–2044. [DOI] [PubMed] [Google Scholar]
- [47].Feng Y, Chen S, Xu J, et al. Dysregulation of lncRNAs GM5524 and GM15645 involved in high‑glucose‑induced podocyte apoptosis and autophagy in diabetic nephropathy. Mol Med Rep. 2018;18(4):3657–3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Zhang Y, Chang B, Zhang J, et al. LncRNA SOX2OT alleviates the high glucose-induced podocytes injury through autophagy induction by the miR-9/SIRT1 axis. Exp Mol Pathol. 2019;110:104283. [DOI] [PubMed] [Google Scholar]
- [49].Xu J, Xia Y, Zhang H, et al. Overexpression of long non-coding RNA H19 promotes invasion and autophagy via the PI3K/AKT/mTOR pathways in trophoblast cells. Biomed Pharmacother. 2018;101:691–697. [DOI] [PubMed] [Google Scholar]
- [50].Horos R, Büscher M, Kleinendorst R, et al. The small Non-coding vault RNA1-1 acts as a riboregulator of autophagy. Cell. 2019. Feb 21;176(5):1054–67e12. PubMed PMID: 30773316. Epub 2019/02/19. eng. [DOI] [PubMed] [Google Scholar]
- [51].Santovito D, Egea V, Bidzhekov K, et al. Noncanonical inhibition of caspase-3 by a nuclear microRNA confers endothelial protection by autophagy in atherosclerosis. Sci Transl Med. 2020. Jun 3;12(546):eaaz2294. PubMed PMID: 32493793. Epub 2020/06/05. eng. [DOI] [PubMed] [Google Scholar]
- [52].Xu F, Hong F, Li Y, et al. Inhibition of HIF1A-AS1 promoted starvation-induced hepatocellular carcinoma cells apoptosis by reducing HIF-1α/mTOR mediated autophagy. 2020. [DOI] [PMC free article] [PubMed]
- [53].Huang W, Huang F, Lei Z, et al. LncRNA SNHG11 promotes proliferation, migration, apoptosis, and autophagy by regulating hsa-miR-184/AGO2 in HCC. OncoTargets Therapy. 2020;13:413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Wei H, Hu J, Pu J, et al. Long noncoding RNA HAGLROS promotes cell proliferation, inhibits apoptosis and enhances autophagy via regulating miR-5095/ATG12 axis in hepatocellular carcinoma cells. Int Immunopharmacol. 2019;73:72–80. [DOI] [PubMed] [Google Scholar]
- [55].Wang C-Z, Yan G-X, Dong D-S, et al. LncRNA-ATB promotes autophagy by activating Yes-associated protein and inducing autophagy-related protein 5 expression in hepatocellular carcinoma. World J Gastroenterol. 2019;25(35):5310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Yang L, Wang H, Shen Q, et al. Long non-coding RNAs involved in autophagy regulation. Cell Death Dis. 2017;8(10):e3073–e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Li Z, Hao S, Yin H, et al. Autophagy ameliorates cognitive impairment through activation of PVT1 and apoptosis in diabetes mice. Behav Brain Res. 2016;305:265–277. [DOI] [PubMed] [Google Scholar]
- [58].Islam Khan MZ, Tam SY, Law HKW. Autophagy-modulating long non-coding RNAs (LncRNAs) and their molecular events in cancer. Front Genet. 2019;9:750. [PubMed PMID: 30693021. eng.]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Liao CC, Ho MY, Liang SM, et al. Autophagic degradation of SQSTM1 inhibits ovarian cancer motility by decreasing DICER1 and AGO2 to induce MIRLET7A-3P. Autophagy. 2018;14(12):2065–2082. PubMed PMID: 30081720. Pubmed Central PMCID: PMC6984764. Epub 2018/08/08. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Gibbings D, Mostowy S, Jay F, et al. Selective autophagy degrades DICER and AGO2 and regulates miRNA activity. Nat Cell Biol. 14(12)PubMed PMID: 23143396. Pubmed Central PMCID: PMC3771578. Epub 2012/11/13. eng.:1314–1321. 2012. Dec;. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Leidal AM, Huang HH, Marsh T, et al. The LC3-conjugation machinery specifies the loading of RNA-binding proteins into extracellular vesicles. Nat Cell Biol. 2020. Feb;;22(2):187–199. PubMed PMID: 31932738. Pubmed Central PMCID: PMC7007875. Epub 2020/01/15. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].La Rocca G, Olejniczak SH, González AJ, et al. In vivo, Argonaute-bound microRNAs exist predominantly in a reservoir of low molecular weight complexes not associated with mRNA. Proc Natl Acad Sci U S A. 2015. Jan 20;112(3):767–772. PubMed PMID: 25568082. Pubmed Central PMCID: PMC4311832. Epub 2015/01/09. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Klionsky DJ, Abdelmohsen K, Abe A, et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy. 2016;12(1):1–222. PubMed PMID: 26799652. Pubmed Central PMCID: PMC4835977. Epub 2016/01/23. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Liu X, Zhong L, Li P, et al. microRNA-100 enhances autophagy and suppresses migration and invasion of renal cell carcinoma cells via disruption of NOX4-dependent mTOR pathway. Clin Transl Sci. 2020. May 1. PubMed PMID: 32356935. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Huang W, Zeng C, Hu S, et al. ATG3, a Target of miR-431-5p, promotes proliferation and invasion of colon cancer via promoting autophagy. Cancer Manag Res. PubMed PMID: 31849517. Pubmed Central PMCID: PMC6911302. 2019;11(102):75–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Qian LB, Jiang SZ, Tang XQ, et al. Exacerbation of diabetic cardiac hypertrophy in OVE26 mice by angiotensin II is associated with JNK/c-Jun/miR-221-mediated autophagy inhibition. Oncotarget. 2017. Dec 5;8(63):106661–106671. PubMed PMID: 29290979. Pubmed Central PMCID: PMC5739764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Yang Y, Sun Y, Wang H, et al. MicroRNA-221 induces autophagy through suppressing HDAC6 expression and promoting apoptosis in pancreatic cancer. Oncol Lett. 2018. Dec;16(6):7295–7301. PubMed PMID: 30546469. Pubmed Central PMCID: PMC6256293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Liao W, Zhang Y. MicroRNA-381 facilitates autophagy and apoptosis in prostate cancer cells via inhibiting the RELN-mediated PI3K/AKT/mTOR signaling pathway. Life Sci PubMed PMID: 32304760. 2020;254:117672. Apr15. [DOI] [PubMed] [Google Scholar]
- [69].Ling Z, Liu D, Zhang G, et al. miR-361-5p modulates metabolism and autophagy via the Sp1-mediated regulation of PKM2 in prostate cancer. Oncol Rep. 38(3). PubMed PMID: 29094170. 1621–1628. 2017. Sep;. [DOI] [PubMed] [Google Scholar]
- [70].Zhang YJ, Pan Q, Yu Y, et al. microRNA-519d induces autophagy and apoptosis of human hepatocellular carcinoma cells through activation of the AMPK signaling pathway via Rab10. Cancer Manag Res. 2020;12:2589–2602. [PubMed PMID: 32346312. Pubmed Central PMCID: PMC7167370]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Cui X, Wang X, Zhou X, et al. miR-106a regulates cell proliferation and autophagy by targeting LKB1 in HPV-16-associated cervical cancer. Mol Cancer Res. 2020. Apr 28;18(8):1129–1141. PubMed PMID: 32345599. [DOI] [PubMed] [Google Scholar]
- [72].Zhou Q, Dong J, Luo R, et al. MicroRNA-20a regulates cell proliferation, apoptosis and autophagy by targeting thrombospondin 2 in cervical cancer. Eur J Pharmacol. 5(844)PubMed PMID: 30513279.:102–109. 2019 Feb. [DOI] [PubMed] [Google Scholar]
- [73].He W, Cheng Y. Inhibition of miR-20 promotes proliferation and autophagy in articular chondrocytes by PI3K/AKT/mTOR signaling pathway. Biomed Pharmacother. 2018. Jan;97:607–615. PubMed PMID: 29101804. [DOI] [PubMed] [Google Scholar]
- [74].Yang Y, Luo H, Liu S, et al. Platelet microparticles-containing miR-4306 inhibits human monocyte-derived macrophages migration through VEGFA/ERK1/2/NF-κB signaling pathways. Ann Clin Exp Hypertens. 2019;41(5):481–491. [DOI] [PubMed] [Google Scholar]
- [75].Gan X, Zhu H, Jiang X, et al. CircMUC16 promotes autophagy of epithelial ovarian cancer via interaction with ATG13 and miR-199a. Mol Cancer. 2020. Feb 28;19(1):45. PubMed PMID: 32111227. Pubmed Central PMCID: PMC7047414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Ba RQ, Liu J, Fan XJ, et al. Effects of miR-199a on autophagy by targeting glycogen synthase kinase 3beta to activate PTEN/AKT/mTOR signaling in an MPP(+) in vitro model of Parkinson’s disease. Neurol Res. 42(4). PubMed PMID: 32151238. 308–318. 2020. Apr;. [DOI] [PubMed] [Google Scholar]
- [77].Li JP, Zhang HM, Liu MJ, et al. miR-133a-3p/FOXP3 axis regulates cell proliferation and autophagy in gastric cancer. J Cell Biochem. 121(5–6). PubMed PMID: 31904151. 3392–3405. 2020 Jun;. [DOI] [PubMed] [Google Scholar]
- [78].Zhang HM, Li H, Wang GX, et al. MKL1/miR-5100/CAAP1 loop regulates autophagy and apoptosis in gastric cancer cells. Neoplasia. 2020. May;22(5):220–230. PubMed PMID: 32315812. Pubmed Central PMCID: PMC7167518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Guo W, Chen Z, Chen Z, et al. Promotion of cell proliferation through inhibition of cell autophagy signalling pathway by Rab3IP is restrained by MicroRNA-532-3p in Gastric cancer. J Cancer. 2018;9(23):4363–4373. PubMed PMID: 30519341. Pubmed Central PMCID: PMC6277663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Yuan KT, Li BX, Yuan YJ, et al. Deregulation of MicroRNA-375 inhibits proliferation and migration in gastric cancer in association with autophagy-mediated AKT/mTOR signaling pathways. Technol Cancer Res Treat. 2018. Jan 1 1533033818806499. PubMed PMID: 30355273. Pubmed Central PMCID: PMC6202745;17:153303381880649. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [81].Yuan Y, Zhang Y, Han L, et al. miR-183 inhibits autophagy and apoptosis in gastric cancer cells by targeting ultraviolet radiation resistance-associated gene. Int J Mol Med. 42(6)PubMed PMID: 30221685.:3562–3570. 2018. Dec. [DOI] [PubMed] [Google Scholar]
- [82].Ma L, Wang Z, Xie M, et al. Silencing of circRACGAP1 sensitizes gastric cancer cells to apatinib via modulating autophagy by targeting miR-3657 and ATG7. Cell Death Dis. 2020. Mar 5;11(3):169. PubMed PMID: 32139670. Pubmed Central PMCID: PMC7058073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Wang H, Zhang Y, Wu Q, et al. miR-16 mimics inhibit TGF-beta1-induced epithelial-to-mesenchymal transition via activation of autophagy in non-small cell lung carcinoma cells. Oncol Rep. 39(1)PubMed PMID: 29138833.:247–254. 2018. Jan. [DOI] [PubMed] [Google Scholar]
- [84].Li S, Zeng X, Ma R, et al. MicroRNA-21 promotes the proliferation, migration and invasion of non-small cell lung cancer A549 cells by regulating autophagy activity via AMPK/ULK1 signaling pathway. Exp Ther Med. 2018 Sep; 16;2038(3):45. PubMed PMID: 30186437. Pubmed Central PMCID: PMC6122184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].He Y, Liu H, Jiang L, et al. miR-26 induces apoptosis and inhibits autophagy in non-small cell lung cancer cells by suppressing TGF-beta1-JNK signaling pathway. Front Pharmacol. 2018;9:1509. PubMed PMID: 30687089. Pubmed Central PMCID: PMC6333751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Yu Y, Xiang N, Lin M, et al. miR- 26a sensitizes melanoma cells to dabrafenib via targeting HMGB1-dependent autophagy pathways. Drug Des Devel Ther. PubMed PMID: 31754297. Pubmed Central PMCID: PMC6825511 2019;13: 3717–3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Meng C, Liu Y, Shen Y, et al. MicroRNA-26b suppresses autophagy in breast cancer cells by targeting DRAM1 mRNA, and is downregulated by irradiation. Oncol Lett. 2018. Feb;;15(2):1435–1440. PubMed PMID: 29399189. Pubmed Central PMCID: PMC5774516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Ju S, Liang Z, Li C, et al. The effect and mechanism of miR-210 in down-regulating the autophagy of lung cancer cells. Pathol Res Pract. 215(3). PubMed PMID: 30573163. 453–458. 2019. Mar;. [DOI] [PubMed] [Google Scholar]
- [89].Ye C, Qi W, Dai S, et al. microRNA-223 promotes autophagy to aggravate lung ischemia-reperfusion injury by inhibiting the expression of transcription factor HIF2alpha. Am J Physiol Lung Cell Mol Physiol. 2020. Apr 8;319(1):L1–L10. PubMed PMID: 32267722. [DOI] [PubMed] [Google Scholar]
- [90].Xu T, Yan W, Wu Q, et al. MiR-326 inhibits inflammation and promotes autophagy in silica-induced pulmonary fibrosis through targeting TNFSF14 and PTBP1. Chem Res Toxicol. 32(11). PubMed PMID: 31642316. 2192–2203. 2019 Nov 18. [DOI] [PubMed] [Google Scholar]
- [91].Lou L, Tian M, Chang J, et al., MiRNA-192-5p attenuates airway remodeling and autophagy in asthma by targeting MMP-16 and ATG7. Biomed Pharmacother, 2020 Feb 122:109692.PubMed PMID: 31918268 . [DOI] [PubMed] [Google Scholar]
- [92].Yang ZY, Wang Y, Liu Q, et al. microRNA cluster MC-let-7a-1~let-7d promotes autophagy and apoptosis of glioma cells by down-regulating STAT3. CNS Neurosci Ther. 2020. Mar;26(3):319–331. PubMed PMID: 31868319. Pubmed Central PMCID: PMC7052808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Zhang N, Qiu L, Li T, et al. MiR-449a attenuates autophagy of T-cell lymphoma cells by downregulating ATG4B expression. BMB Rep. 2020. Mar 16;53(5):254–259. PubMed PMID: 32172731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Wu K, Huang J, Xu T, et al. MicroRNA-181b blocks gensenoside Rg3-mediated tumor suppression of gallbladder carcinoma by promoting autophagy flux via CREBRF/CREB3 pathway. Am J Transl Res. 2019;11(9):5776–5787. PubMed PMID: 31632547. Pubmed Central PMCID: PMC6789245. [PMC free article] [PubMed] [Google Scholar]
- [95].Xiao Y, Diao Q, Liang Y, et al. 2415p promotes malignant melanoma cell autophagy and apoptosis via regulating ubiquitin D. Mol Med Rep. 16(6)PubMed PMID: 28983594.:8448–8454. 2017. Dec;. [DOI] [PubMed] [Google Scholar]
- [96].Tong X, Wang X, Wang C, et al. Elevated levels of serum MiR-152 and miR-24 in uterine sarcoma: potential for inducing autophagy via SIRT1 and deacetylated LC3. BrJ Biomed Sci. 75(1)PubMed PMID: 28929922.:7–12. 2018 Jan;. [DOI] [PubMed] [Google Scholar]
- [97].Pang F, Liu C, Cui Y, et al. miR-17-5p promotes proliferation and migration of CAL-27 human tongue squamous cell carcinoma cells involved in autophagy inhibition under hypoxia. Int J Clin Exp Pathol. 2019;12(6):2084–2091. PubMed PMID: 31934030. Pubmed Central PMCID: PMC6949637. [PMC free article] [PubMed] [Google Scholar]
- [98].Cheng Y, Li Z, Xie J, et al. MiRNA-224-5p inhibits autophagy in breast cancer cells via targeting Smad4. Biochemical and Biophysical Research Communications. 506(4). PubMed PMID: 30389135. 793–798. 2018. Dec 2;. [DOI] [PubMed] [Google Scholar]
- [99].Ai H, Zhou W, Wang Z, et al. microRNAs-107 inhibited autophagy, proliferation, and migration of breast cancer cells by targeting HMGB1. J Cell Biochem. 2018. Dec 2 PubMed PMID: 30506984. doi: 10.1002/jcb.28157 [DOI] [PubMed] [Google Scholar]
- [100].Qi H, Ren J, Ba L, et al. MSTN attenuates cardiac hypertrophy through inhibition of excessive cardiac autophagy by blocking AMPK/mTOR and miR-128/PPARgamma/NF-kappaB. Mol Ther Nucleic Acids. 2020. Mar 6 PubMed PMID: 31923740. Pubmed Central PMCID: PMC6951838;19:507–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Zhou S, Lei D, Bu F, et al. MicroRNA-29b-3p targets sparc gene to protect cardiocytes against autophagy and apoptosis in hypoxic-induced H9c2 cells. J Cardiovasc Transl Res. 12(4)PubMed PMID: 30560317.:358–365. 2019. Aug;. [DOI] [PubMed] [Google Scholar]
- [102].Zhou F, Li Y, Huang Y, et al. Upregulation of CASP9 through NF-κB and its target MiR-1276 Contributed to TNFα-promoted apoptosis of cancer cells induced by doxorubicin. Int J Mol Sci. 2020;21(7):2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Hu S, Cao S, Tong Z, et al. FGF21 protects myocardial ischemia-reperfusion injury through reduction of miR-145-mediated autophagy. Am J Transl Res. 2018;10(11): 3677–3688. PubMed PMID: 30662618. Pubmed Central PMCID: PMC6291727. [PMC free article] [PubMed] [Google Scholar]
- [104].Wu G, Yu W, Zhang M, et al. MicroRNA-145-3p suppresses proliferation and promotes apotosis and autophagy of osteosarcoma cell by targeting HDAC4. Artif Cells Nanomed Biotechnol. 2018;46(sup2):579–586. PubMed PMID: 29893594. [DOI] [PubMed] [Google Scholar]
- [105].Zhou Z, Hu B, Lyu Q, et al. miR-384-5p promotes spinal cord injury recovery in rats through suppressing of autophagy and endoplasmic reticulum stress. Neurosci Lett. 14(727)PubMed PMID: 32243909.:134937. 2020. May. [DOI] [PubMed] [Google Scholar]
- [106].Li X, Lou X, Xu S, et al. Knockdown of miR-372 inhibits nerve cell apoptosis induced by spinal cord Ischemia/Reperfusion injury via enhancing autophagy by up-regulating Beclin-1. J Mol Neurosci. 66(3)PubMed PMID: 30298297.:437–444. 2018. Nov;. [DOI] [PubMed] [Google Scholar]
- [107].Chen J, Sun Q, Liu GZ, et al. Effect of miR-202-5p-mediated ATG7 on autophagy and apoptosis of degenerative nucleus pulposus cells. Eur Rev Med Pharmacol Sci. 24(2). PubMed PMID: 32016953. 517–525. 2020 Jan;. [DOI] [PubMed] [Google Scholar]
- [108].Li W, Yang Y, Ba Z, et al. MicroRNA-93 regulates hypoxia-induced autophagy by targeting ULK1. Oxid Med Cell Longev. 2017;2709053. PubMed PMID: 29109831. Pubmed Central PMCID: PMC5646326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Yang S, Abdulla R, Lu C, et al. Inhibition of microRNA-376b protects against renal interstitial fibrosis via inducing macrophage autophagy by upregulating atg5 in mice with Chronic Kidney disease. Kidney Blood Press Res. 2018;43(6):1749–1764. PubMed PMID: 30472709. [DOI] [PubMed] [Google Scholar]
- [110].Li XY, Wang SS, Han Z, et al. Triptolide restores autophagy to alleviate diabetic renal fibrosis through the miR-141-3p/PTEN/Akt/mTOR pathway. Mol Ther Nucleic Acids. 2017. Dec 15 PubMed PMID: 29246323. Pubmed Central PMCID: PMC5602517;9:48–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Ye Z, Li ZH, He SZ. miRNA-1273g-3p involvement in development of diabetic retinopathy by modulating the autophagy-lysosome pathway. Med Sci Monit. 2017. Dec 3 PubMed PMID: 29197896. Pubmed Central PMCID: PMC5724349;23: 5744–5751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Liu G, Kang X, Guo P, et al. miR-25-3p promotes proliferation and inhibits autophagy of renal cells in polycystic kidney mice by regulating ATG14-Beclin 1. Ren Fail. 42(1). PubMed PMID: 32340512. 333–342. 2020 Nov;. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Qiu S, Liu B, Mo Y, et al. MicroRNA-153-3p increases autophagy in sevoflurane-preconditioned mice to protect against ischaemic/reperfusion injury after knee arthroplasty. J Cell Mol Med. 24(9). PubMed PMID: 32239627. 5330–5340. 2020 May;. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Li YL, Tang JM, Chen XY, et al. MicroRNA-153-3p enhances the sensitivity of chronic myeloid leukemia cells to imatinib by inhibiting B-cell lymphoma-2-mediated autophagy. Hum Cell. 2020. Apr 27;33(3):610–618. PubMed PMID: 32342278. [DOI] [PubMed] [Google Scholar]
- [115].Wang N, Yang L, Zhang H, et al. MicroRNA-9a-5p alleviates ischemia injury after focal cerebral ischemia of the rat by targeting ATG5-mediated autophagy. Cell Physiol Biochem. 2018;45(1):78–87. PubMed PMID: 29316542. [DOI] [PubMed] [Google Scholar]
- [116].Li W, Ren Y, Meng T, et al. miR-129-5p attenuates hypoxia-induced apoptosis in rat H9c2 cardiomyocytes by activating autophagy. J Gene Med. 2020. Apr 16;22(8):e3200. PubMed PMID: 32298509. [DOI] [PubMed] [Google Scholar]
- [117].Zhang H, Zhang X, Zhang J. MiR-129-5p inhibits autophagy and apoptosis of H9c2 cells induced by hydrogen peroxide via the PI3K/AKT/mTOR signaling pathway by targeting ATG14. Biochem Biophys Res Commun. 506(1)PubMed PMID: 30348524.:272–277. 2018 Nov 17. [DOI] [PubMed] [Google Scholar]
- [118].Song H, Liu J, Wu X, et al. LHX2 promotes malignancy and inhibits autophagy via mTOR in osteosarcoma and is negatively regulated by miR-129-5p. Aging (Albany NY). 2019. Nov 13;11(21):9794–9810. PubMed PMID: 31724536. Pubmed Central PMCID: PMC6874432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Tao Z, Feng C, Mao C, et al. MiR-4465 directly targets PTEN to inhibit AKT/mTOR pathway-mediated autophagy. Cell Stress Chaperones. 2019. Jan;;24(1):105–113. PubMed PMID: 30421325. Pubmed Central PMCID: PMC6363616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Sul OJ, Sung YB, Rajasekaran M et al. MicroRNA-155 induces autophagy in osteoclasts by targeting transforming growth factor beta-activated kinase 1-binding protein 2 upon lipopolysaccharide stimulation Bone 2018Nov 116 279–289PubMed PMID: 30144578 [DOI] [PubMed] [Google Scholar]
- [121].Yin S, Yang S, Pan X, et al. MicroRNA155 promotes oxLDLinduced autophagy in human umbilical vein endothelial cells by targeting the PI3K/Akt/mTOR pathway. Mol Med Rep. 18(3). PubMed PMID: 30015881. Pubmed Central PMCID: PMC6102700. 2798–2806. 2018 Sep;. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Li Y, Jiang J, Liu W, et al. microRNA-378 promotes autophagy and inhibits apoptosis in skeletal muscle. Proc Natl Acad Sci U S A. 2018. Nov 13;115(46):E10849–E58. PubMed PMID: 30373812. Pubmed Central PMCID: PMC6243236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Dong S, Xiao Y, Ma X, et al. miR-193b increases the chemosensitivity of osteosarcoma cells by promoting FEN1-mediated autophagy. Onco Targets Ther. PubMed PMID: 31819503. Pubmed Central PMCID: PMC6878930 2019;12: 10089–10098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Cai L, Liu X, Guo Q, et al. MiR-15a attenuates peripheral nerve injury-induced neuropathic pain by targeting AKT3 to regulate autophagy. Genes Genomics. 42(1)PubMed PMID: 31736006.:77–85. 2020. Jan;. [DOI] [PubMed] [Google Scholar]
- [125].Yu Q, Zhao B, He Q, et al. microRNA-206 is required for osteoarthritis development through its effect on apoptosis and autophagy of articular chondrocytes via modulating the phosphoinositide 3-kinase/protein kinase B-mTOR pathway by targeting insulin-like growth factor-1. J Cell Biochem. 120(4)PubMed PMID: 30335903.:5287–5303. 2019 Apr;. [DOI] [PubMed] [Google Scholar]
- [126].Yang F, Huang R, Ma H, et al. miRNA-411 regulates chondrocyte autophagy in osteoarthritis by targeting hypoxia-inducible factor 1 alpha (HIF-1alpha). Med Sci Monit. 2020. Feb 19 PubMed PMID: 32072994. Pubmed Central PMCID: PMC7043337;26: e921155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Mi H, Wang X, Wang F, et al. miR-381 induces sensitivity of breast cancer cells to doxorubicin by inactivation of MAPK signaling via FYN. Eur J Pharmacol. 2018;839:66–75. [DOI] [PubMed] [Google Scholar]
- [128].Liang Y, Chen X, Liang Z. MicroRNA-320 regulates autophagy in retinoblastoma by targeting hypoxia inducible factor-1alpha. Exp Ther Med. 14(3)PubMed PMID: 28962169. Pubmed Central PMCID: PMC5609094.:2367–2372. 2017 Sep;. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Ganesan S, Palani HK, Lakshmanan V, et al. Stromal cells downregulate miR-23a-5p to activate protective autophagy in acute myeloid leukemia. Cell Death Dis. 2019. Sep 30;10(10):736. PubMed PMID: 31570693. Pubmed Central PMCID: PMC6769009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Luan B, Sun C. MiR-138-5p affects insulin resistance to regulate type 2 diabetes progression through inducing autophagy in HepG2 cells by regulating SIRT1. Nutr Res. 2018. Nov;59:90–98. PubMed PMID: 30442237. [DOI] [PubMed] [Google Scholar]
- [131].Sabater-Arcis M, Bargiela A, Furling D, et al. miR-7 restores phenotypes in myotonic dystrophy muscle cells by repressing hyperactivated autophagy. Mol Ther Nucleic Acids. 2020. Mar 6 PubMed PMID: 31855836. Pubmed Central PMCID: PMC6926285.;19: 278–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Fu XT, Shi YH, Zhou J, et al. MicroRNA-30a suppresses autophagy-mediated anoikis resistance and metastasis in hepatocellular carcinoma. Cancer Lett. 1(412). PubMed PMID: 29061507. 108–117. 2018. Jan . [DOI] [PubMed] [Google Scholar]
- [133].Liu Z, Wei X, Zhang A, et al. Long non-coding RNA HNF1A-AS1 functioned as an oncogene and autophagy promoter in hepatocellular carcinoma through sponging hsa-miR-30b-5p. Biochem Biophys Res Commun. 2016;473(4):1268–1275. [DOI] [PubMed] [Google Scholar]
- [134].Wang C, Jiang X, Li X, et al. Long noncoding RNA HULC accelerates the growth of human liver cancer stem cells by upregulating CyclinD1 through miR675-PKM2 pathway via autophagy. Stem Cell Res Ther. 2020;11(1):1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Chen Z-H, Wang W-T, Huang W, et al. The lncRNA HOTAIRM1 regulates the degradation of PML-RARA oncoprotein and myeloid cell differentiation by enhancing the autophagy pathway. Cell Death Differ. 2017;24(2):212–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Yang L, Peng X, Jin H, et al. Long non-coding RNA PVT1 promotes autophagy as ceRNA to target ATG3 by sponging microRNA-365 in hepatocellular carcinoma. Gene. 2019;697:94–102. [DOI] [PubMed] [Google Scholar]
- [137].Huang F, Chen W, Peng J, et al. LncRNA PVT1 triggers Cyto-protective autophagy and promotes pancreatic ductal adenocarcinoma development via the miR-20a-5p/ULK1 Axis. Mol Cancer. 2018;17(1):98. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [138].Chen J-F, Wu P, Xia R, et al. STAT3-induced lncRNA HAGLROS overexpression contributes to the malignant progression of gastric cancer cells via mTOR signal-mediated inhibition of autophagy. Mol Cancer. 2018;17(1):6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Luo D, Li L, Huo H, et al. MicroRNA-29b sensitizes osteosarcoma cells to doxorubicin by targeting matrix metalloproteinase 9 (MMP-9) in osteosarcoma. Eur Rev Med Pharmacol Sci. 2019;23(4):1434–1442. [DOI] [PubMed] [Google Scholar]
- [140].Fu Z, Luo W, Wang J, et al. Malat1 activates autophagy and promotes cell proliferation by sponging miR-101 and upregulating STMN1, RAB5A and ATG4D expression in glioma. Biochem Biophys Res Commun. 2017;492(3):480–486. [DOI] [PubMed] [Google Scholar]
- [141].Gao D, Lv A, Li HP, et al. LncRNA MALAT‐1 elevates HMGB1 to promote autophagy resulting in inhibition of tumor cell apoptosis in multiple myeloma. J Cell Biochem. 2017;118(10):3341–3348. [DOI] [PubMed] [Google Scholar]
- [142].Li L-J, Chai Y, Guo X-J, et al. The effects of the long non-coding RNA MALAT-1 regulated autophagy-related signaling pathway on chemotherapy resistance in diffuse large B-cell lymphoma. Biomed Pharmacother. 2017;89:939–948. [DOI] [PubMed] [Google Scholar]
- [143].Wu Q, Yi X. Down-regulation of long noncoding RNA MALAT1 protects hippocampal neurons against excessive autophagy and apoptosis via the PI3K/Akt signaling pathway in rats with epilepsy. J Mol Neurosci. 2018;65(2):234–245. [DOI] [PubMed] [Google Scholar]
- [144].Shao G, Zhao Z, Zhao W, et al. Long non‑coding RNA MALAT1 activates autophagy and promotes cell proliferation by downregulating microRNA‑204 expression in gastric cancer. Oncol Lett. 2020;19(1):805–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Guo X, Wu X, Han Y, et al. MALAT1 protects cardiomyocytes from isoproterenol‐induced apoptosis through sponging miR‐558 to enhance ULK1‐mediated protective autophagy. J Cell Physiol. 2019;234(7):10842–10854. [DOI] [PubMed] [Google Scholar]
- [146].Wang K, Yang C, Shi J, et al. Ox-LDL-induced lncRNA MALAT1 promotes autophagy in human umbilical vein endothelial cells by sponging miR-216a-5p and regulating Beclin-1 expression. Eur J Pharmacol. 2019;858:172338. [DOI] [PubMed] [Google Scholar]
- [147].Zhu Y, Yang T, Duan J, et al. MALAT1/miR-15b-5p/MAPK1 mediates endothelial progenitor cells autophagy and affects coronary atherosclerotic heart disease via mTOR signaling pathway. Aging (Albany NY). 2019;11(4):1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Zheng S-Z, Sun P, Wang J-P, et al. MiR-34a overexpression enhances the inhibitory effect of doxorubicin on HepG2 cells. World J Gastroenterol. 2019;25(22):2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Liu H, Shi C, Deng Y. MALAT1 affects hypoxia-induced vascular endothelial cell injury and autophagy by regulating miR-19b-3p/HIF-1α axis. Mol Cell Biochem. 2020;474(1–2):1–10. [DOI] [PubMed] [Google Scholar]
- [150].Huang J, Yang Y, Fang F, et al. MALAT1 modulates the autophagy of retinoblastoma cell through miR‐124‐mediated stx17 regulation. J Cell Biochem. 2018;119(5):3853–3863. [DOI] [PubMed] [Google Scholar]
- [151].Yuan YL, Yu H, Mu S-M, et al. MiR-26a-5p inhibits cell proliferation and enhances doxorubicin sensitivity in HCC cells via targeting AURKA. Technol Cancer Res Treat. 2019;18:1533033819851833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Song T-F, Huang L-W, Yuan Y, et al. LncRNA MALAT1 regulates smooth muscle cell phenotype switch via activation of autophagy. Oncotarget. 2018;9(4):4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Lang C, Xu M, Zhao Z, et al. MicroRNA96 expression induced by low‑dose cisplatin or doxorubicin regulates chemosensitivity, cell death and proliferation in gastric cancer SGC7901 cells by targeting FOXO1. Oncol Lett. 2018;16(3):4020–4026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Xiu Y-L, Sun K-X, Chen X, et al. Upregulation of the lncRNA Meg3 induces autophagy to inhibit tumorigenesis and progression of epithelial ovarian carcinoma by regulating activity of ATG3. Oncotarget. 2017;8(19):31714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Cao Y, Wen J, Li Y, et al. Uric acid and sphingomyelin enhance autophagy in iPS cell-originated cardiomyocytes through lncRNA MEG3/miR-7-5p/EGFR axis. Artif Cells Nanomed Biotechnol. 2019;47(1):3774–3785. [DOI] [PubMed] [Google Scholar]
- [156].Li M, Cui J, Niu W, et al. Long non-coding PCED1B-AS1 regulates macrophage apoptosis and autophagy by sponging miR-155 in active tuberculosis. Biochem Biophys Res Commun. 2019;509(3):803–809. [DOI] [PubMed] [Google Scholar]
- [157].Ke Z, Lu J, Zhu J, et al. Down-regulation of lincRNA-EPS regulates apoptosis and autophagy in BCG-infected RAW264. 7 macrophages via JNK/MAPK signaling pathway. Infection. Gene Evol. 2020;77:104077. [DOI] [PubMed] [Google Scholar]
- [158].Zhou Z, Xu H, Liu B, et al. Suppression of lncRNA RMRP ameliorates oxygen-glucose deprivation/re-oxygenation-induced neural cells injury by inhibiting autophagy and PI3K/Akt/mTOR-mediated apoptosis. Biosci Rep. 2019;39:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Ren X, Wan C, Niu Y. Overexpression of lnc RNA TCTN 2 protects neurons from apoptosis by enhancing cell autophagy in spinal cord injury. FEBS Open Bio. 2019;9(7):1223–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Qin Y, Sun W, Zhang H, et al. LncRNA GAS8-AS1 inhibits cell proliferation through ATG5-mediated autophagy in papillary thyroid cancer. Endocrine. 2018;59(3):555–564. [DOI] [PubMed] [Google Scholar]
- [161].Su Y, Lu J, Chen X, et al. Long non-coding RNA HOTTIP affects renal cell carcinoma progression by regulating autophagy via the PI3K/Akt/Atg13 signaling pathway. J Cancer Res Clin Oncol. 2019;145(3):573–588. [DOI] [PubMed] [Google Scholar]
- [162].Zhou M, Zou Y, Xue Y, et al. Long non-coding RNA H19 protects acute myocardial infarction through activating autophagy in mice. Eur Rev Med Pharmacol Sci. 2018;22(17):5647–5651. [DOI] [PubMed] [Google Scholar]
- [163].Zhuo C, Jiang R, Lin X, et al. H19 inhibits autophagy by epigenetically silencing of DIRAS3 in diabetic cardiomyopathy. Oncotarget. 2017;8(1):1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Xiong H, Shen J, Chen Z, et al. H19/let‑7/Lin28 ceRNA network mediates autophagy inhibiting epithelial‑mesenchymal transition in breast cancer. Int J Oncol. 2020;56(3):794–806. [DOI] [PubMed] [Google Scholar]
- [165].Feng Y, Xu W, Zhang W, et al. DCRF regulates cardiomyocyte autophagy by targeting miR-551b-5p in diabetic cardiomyopathy. Theranostics. 2019;9(15):4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Fan Y, Zhao X, Lu K, et al. LncRNA BDNF-AS promotes autophagy and apoptosis in MPTP-induced Parkinson’s disease via ablating microRNA-125b-5p. Brain Res Bull. 2020;157:119–127. [DOI] [PubMed] [Google Scholar]
- [167].Qian C, Ye Y, Mao H, et al. Downregulated lncRNA-SNHG1 enhances autophagy and prevents cell death through the miR-221/222/p27/mTOR pathway in Parkinson’s disease. Exp Cell Res. 2019;384(1):111614. [DOI] [PubMed] [Google Scholar]
- [168].Zhu X, Yang G, Xu J, et al. Silencing of SNHG6 induced cell autophagy by targeting miR-26a-5p/ULK1 signaling pathway in human osteosarcoma. Cancer Cell Int. 2019;19(1):82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Tian F, Wang J, Zhang Z, et al. SNHG7/miR-34a-5p/SYVN1 axis plays a vital role in proliferation, apoptosis and autophagy in osteoarthritis. Biol Res. 2020;53(1):1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170].Yao X, Yao R, Huang F, et al. SNHG12 as a potent autophagy inducer exerts neuroprotective effects against cerebral ischemia/reperfusion injury. Biochem Biophys Res Commun. 2019;514(2):490–496. [DOI] [PubMed] [Google Scholar]
- [171].Zeng C, Fan D, Xu Y, et al. Curcumol enhances the sensitivity of doxorubicin in triple-negative breast cancer via regulating the miR-181b-2-3p-ABCC3 axis. Biochem Pharmacol. 2020;174:113795. [DOI] [PubMed] [Google Scholar]
- [172].Liu K, Hou Y, Liu Y, et al. SNHG15 contributes to proliferation, invasion and autophagy in osteosarcoma cells by sponging miR-141. J Biomed Sci. 2017;24(1):46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [173].Wen Y, Gong X, Dong Y, et al. Long non coding RNA SNHG16 Facilitates proliferation, migration, invasion and autophagy of neuroblastoma cells via sponging miR-542-3p and upregulating ATG5 expression. OncoTargets Therapy. 2020;13:263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [174].Wang Z, LncRNA JJ. SLCO4A1-AS1 promotes colorectal cancer cell proliferation by enhancing autophagy via miR-508-3p/PARD3 axis. Aging (Albany NY). 2019;11(14):4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [175].Zhang W, Yuan W, Song J, et al. LncRNA CPS1-IT1 suppresses EMT and metastasis of colorectal cancer by inhibiting hypoxia-induced autophagy through inactivation of HIF-1α. Biochimie. 2018;144:21–27. [DOI] [PubMed] [Google Scholar]
- [176].Liu L, Wang H-J, Meng T, et al. lncRNA GAS5 inhibits cell migration and invasion and promotes autophagy by targeting miR-222-3p via the GAS5/PTEN-signaling pathway in CRC. Mol Ther Nucl Acids. 2019;17:644–656. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [177].Liang W, Fan T, Liu L, et al. Knockdown of growth-arrest specific transcript 5 restores oxidized low-density lipoprotein-induced impaired autophagy flux via upregulating miR-26a in human endothelial cells. Eur J Pharmacol. 2019;843:154–161. [DOI] [PubMed] [Google Scholar]
- [178].Liu E, Sun X, Li J, et al. miR‑30a‑5p inhibits the proliferation, migration and invasion of melanoma cells by targeting SOX4. Mol Med Rep. 2018;18(2):2492–2498. [DOI] [PubMed] [Google Scholar]
- [179].Gao M, Li C, Xu M, et al. LncRNA UCA1 attenuates autophagy-dependent cell death through blocking autophagic flux under arsenic stress. Toxicol Lett. 2018;284:195–204. [DOI] [PubMed] [Google Scholar]
- [180].Su Y, Yao S, Zhao S, et al. CCAT1 functions as apoptosis inhibitor in podocytes via autophagy inhibition. J Cell Biochem. 2020;121(1):621–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [181].Su Q, Liu Y, Lv X-W, et al. Inhibition of lncRNA TUG1 upregulates miR-142-3p to ameliorate myocardial injury during ischemia and reperfusion via targeting HMGB1-and Rac1-induced autophagy. J Mol Cell Cardiol. 2019;133:12–25. [DOI] [PubMed] [Google Scholar]
- [182].Zhu Z, LncRNA ZC. AK139128 promotes cardiomyocyte autophagy and apoptosis in myocardial hypoxia-reoxygenation injury. Life Sci. 2019;116705. DOI: 10.1016/j.lfs.2019.116705 [DOI] [PubMed] [Google Scholar]
- [183].Yu S, Dong B, Fang Z, et al. Knockdown of lnc RNA AK 139328 alleviates myocardial ischaemia/reperfusion injury in diabetic mice via modulating miR‐204‐3p and inhibiting autophagy. J Cell Mol Med. 2018;22(10):4886–4898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [184].Huang Z, Ye B, Wang Z, et al. Inhibition of LncRNA-HRIM increases cell viability by regulating autophagy levels during hypoxia/reoxygenation in myocytes. Cell Physiol Biochem. 2018;46(4):1341–1351. [DOI] [PubMed] [Google Scholar]
- [185].Wu D, Zhang J, Lu Y, et al. miR-140-5p inhibits the proliferation and enhances the efficacy of doxorubicin to breast cancer stem cells by targeting Wnt1. Cancer Gene Ther. 2019;26(3):74–82. [DOI] [PubMed] [Google Scholar]
- [186].Yao L, Yang L, Song H, et al. Silencing of lncRNA XIST suppresses proliferation and autophagy and enhances vincristine sensitivity in retinoblastoma cells by sponging miR-204-5p. Eur Rev Med Pharmacol Sci. 2020;24(7):3526–3537. [DOI] [PubMed] [Google Scholar]
- [187].Liang H, Su X, Wu Q, et al. LncRNA 2810403D21Rik/Mirf promotes ischemic myocardial injury by regulating autophagy through targeting Mir26a. Autophagy. 2019;16(6):1077–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [188].Chen J, Zhou C, Li J, et al. miR‑21‑5p confers doxorubicin resistance in gastric cancer cells by targeting PTEN and TIMP3. Int J Mol Med. 2018;41(4):1855–1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [189].Gu Z, Hou Z, Zheng L, et al. LncRNA DICER1-AS1 promotes the proliferation, invasion and autophagy of osteosarcoma cells via miR-30b/ATG5. Biomed Pharmacother. 2018;104:110–118. [DOI] [PubMed] [Google Scholar]
- [190].Pan Z, Wu C, Li Y, et al. LncRNA DANCR silence inhibits SOX5-medicated progression and autophagy in osteosarcoma via regulating miR-216a-5p. Biomed Pharmacother. 2020;122:109707. [DOI] [PubMed] [Google Scholar]
- [191].Wang Z, Wang J, Wang K, et al. FEZF1-AS1 promoted chemoresistance, autophagy and epithelial-mesenchymal transition (EMT) through regulation of miR-25-3p/ITGB8 axis in prostate cancer. Eur Rev Med Pharmacol Sci. 2020;24(5):2281–2293. [DOI] [PubMed] [Google Scholar]
- [192].Yang C, Shen S, Zheng X, et al. Long non‐coding RNA LINC00337 induces autophagy and chemoresistance to cisplatin in esophageal squamous cell carcinoma cells via upregulation of TPX2 by recruiting E2F4. Faseb J. 2020;34(5):6055–6069. [DOI] [PubMed] [Google Scholar]
- [193].Guo F-X, Wu Q, Li P, et al. The role of the LncRNA-FA2H-2-MLKL pathway in atherosclerosis by regulation of autophagy flux and inflammation through mTOR-dependent signaling. Cell Death Differ. 2019;26(9):1670–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [194].Tu C, Chen W, Wang S, et al. MicroRNA‐383 inhibits doxorubicin resistance in hepatocellular carcinoma by targeting eukaryotic translation initiation factor 5A2. J Cell Mol Med. 2019;23(11):7190–7199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [195].Xue K, Li J, Nan S, et al. Downregulation of LINC00460 decreases STC2 and promotes autophagy of head and neck squamous cell carcinoma by up-regulating microRNA-206. Life Sci. 2019;231:116459. [DOI] [PubMed] [Google Scholar]
- [196].Mai C, Qiu L, Zeng Y, et al. LncRNA Lethe protects sepsis-induced brain injury via regulating autophagy of cortical neurons. Eur Rev Med Pharmacol Sci. 2019;23(11):4858–4864. [DOI] [PubMed] [Google Scholar]
- [197].Yang Y, Li Y, Ding L, et al. Regulatory effect of lncRNA NKILA on autophagy induced by sepsis kidney injury. Eur Rev Med Pharmacol Sci. 2019;23(18):8011–8017. [DOI] [PubMed] [Google Scholar]
- [198].Luan W, Zhou Z, Zhu Y, et al. miR-137 inhibits glutamine catabolism and growth of malignant melanoma by targeting glutaminase. Biochem Biophys Res Commun. 2018;495(1):46–52. [DOI] [PubMed] [Google Scholar]
- [199].Li P, He J, Yang Z, et al. ZNNT1 long noncoding RNA induces autophagy to inhibit tumorigenesis of uveal melanoma by regulating key autophagy gene expression. Autophagy. 2019;16(7):1186–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [200].Cai Q, Wang S, Jin L, et al. Long non-coding RNA GBCDRlnc1 induces chemoresistance of gallbladder cancer cells by activating autophagy. Mol Cancer. 2019;18(1):82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [201].Yang Y, Chen D, Liu H, et al. Increased expression of lncRNA CASC9 promotes tumor progression by suppressing autophagy-mediated cell apoptosis via the AKT/mTOR pathway in oral squamous cell carcinoma. Cell Death Dis. 2019;10(2):1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [202].Zhao Y, Zhang F, Pan Z, et al. NR_003923 promotes cell proliferation, migration, fibrosis, and autophagy via the miR-760/miR-215-3p/IL22RA1 axis in human Tenon’s capsule fibroblasts. Cell Death Dis. 2019;10(8):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [203].Zhou W, Zhang S, Li J, et al. lncRNA TINCR participates in ALA‐PDT‐induced apoptosis and autophagy in cutaneous squamous cell carcinoma. J Cell Biochem. 2019;120(8):13893–13902. [DOI] [PubMed] [Google Scholar]
- [204].Zhang X, Liu J, Gu Y, et al. Down‐regulation of lncRNA OGFRP1 induces autophagy and growth inhibition by AKT/mTOR signaling pathway in HCAECs. Cell Biol Int. 2019;43(2):158–166. [DOI] [PubMed] [Google Scholar]
- [205].Li J, Zhai D, Huang Q, et al. DCST1-AS1 accelerates the proliferation, metastasis and autophagy of hepatocellular carcinoma cell by AKT/mTOR signaling pathways. Eur Rev Med Pharmacol Sci. 2019;23(14):6091–6104. [DOI] [PubMed] [Google Scholar]
- [206].Zhao H, Wang X, Feng X, et al. Long non-coding RNA MEG3 regulates proliferation, apoptosis, and autophagy and is associated with prognosis in glioma. J Neurooncol. 2018;140(2):281–288. [DOI] [PubMed] [Google Scholar]


