Abstract
Background
Public health scientists have used spatial tools such as web-based Geographical Information System (GIS) applications to monitor and forecast the progression of the COVID-19 pandemic and track the impact of their interventions. The ability to track SARS-CoV-2 variants and incorporate the social determinants of health with street-level granularity can facilitate the identification of local outbreaks, highlight variant-specific geospatial epidemiology, and inform effective interventions. We developed a novel dashboard, the University of Massachusetts’ Graphical user interface for Geographic Information (MAGGI) variant tracking system that combines GIS, health-associated sociodemographic data, and viral genomic data to visualize the spatiotemporal incidence of SARS-CoV-2 variants with street-level resolution while safeguarding protected health information. The specificity and richness of the dashboard enhance the local understanding of variant introductions and transmissions so that appropriate public health strategies can be devised and evaluated.
Objective
We developed a web-based dashboard that simultaneously visualizes the geographic distribution of SARS-CoV-2 variants in Central Massachusetts, the social determinants of health, and vaccination data to support public health efforts to locally mitigate the impact of the COVID-19 pandemic.
Methods
MAGGI uses a server-client model–based system, enabling users to access data and visualizations via an encrypted web browser, thus securing patient health information. We integrated data from electronic medical records, SARS-CoV-2 genomic analysis, and public health resources. We developed the following functionalities into MAGGI: spatial and temporal selection capability by zip codes of interest, the detection of variant clusters, and a tool to display variant distribution by the social determinants of health. MAGGI was built on the Environmental Systems Research Institute ecosystem and is readily adaptable to monitor other infectious diseases and their variants in real-time.
Results
We created a geo-referenced database and added sociodemographic and viral genomic data to the ArcGIS dashboard that interactively displays Central Massachusetts’ spatiotemporal variants distribution. Genomic epidemiologists and public health officials use MAGGI to show the occurrence of SARS-CoV-2 genomic variants at high geographic resolution and refine the display by selecting a combination of data features such as variant subtype, subject zip codes, or date of COVID-19–positive sample collection. Furthermore, they use it to scale time and space to visualize association patterns between socioeconomics, social vulnerability based on the Centers for Disease Control and Prevention’s social vulnerability index, and vaccination rates. We launched the system at the University of Massachusetts Chan Medical School to support internal research projects starting in March 2021.
Conclusions
We developed a COVID-19 variant surveillance dashboard to advance our geospatial technologies to study SARS-CoV-2 variants transmission dynamics. This real-time, GIS-based tool exemplifies how spatial informatics can support public health officials, genomics epidemiologists, infectious disease specialists, and other researchers to track and study the spread patterns of SARS-CoV-2 variants in our communities.
Keywords: geographic information science, GIS, COVID-19, SARS-CoV-2, variants, surveillance, dashboard, web mapping, public health, web-based information, digital health, epidemiology
Introduction
COVID-19 has infected 219 million people resulting in over 4.5 million deaths as of October 2021 [1]. The pandemic has highlighted the role of geographic mapping technologies to provide easy-to-understand indicators such as new cases, confirmed tests, and inpatient rates using spatiotemporal visualizations [2]. For example, public agencies [3] and news media [4,5] have used these visualizations to inform the public about the spread of COVID-19 and explain why officials recommend adopting intervention measures such as mask mandate and vaccinations [6].
Health science has long leveraged Geographical Information System (GIS) spatial analysis and applications [7]. GIS offers an interactive and efficient approach to revealing meaningful patterns and associations [8] that would be otherwise difficult to visualize using traditional figures and tables. As a result, researchers are increasingly using spatial analysis to study the impacts of COVID-19 [9,10] or understand the relationship between COVID-19 cases and sociodemographic or economic data and vaccination rates in local environments [11-13].
With the constant technological evolution of GIS platforms, many geospatial online dashboards are available for monitoring COVID-19 worldwide [14,15]. The COVID-19 dashboard developed by John Hopkins University is arguably the most popular and stands out for its effectiveness at capturing new cases around the globe [16]. Meanwhile, the Centers for Disease Control and Prevention’s National Healthcare Safety Network has also developed a geospatial dashboard for COVID-19 infection data analysis and prevention [17]. Most GIS systems, however, track data with coarse geographic resolution (eg, state or county). Further, few GIS systems track the spatiotemporal transition dynamics of SARS-CoV-2 variants [18].
The impact of using genomic epidemiology to monitor the COVID-19 pandemic has been profound, as it provides unprecedented detail into the appearance and global dissemination of SARS-CoV-2 variants [19-24]. In addition, phylogenetic-epidemiological analysis has enabled the reconstruction of super-spreader events [25]. Notably, these analyses have illustrated the power of combining epidemiological analytics with deep viral genome sequencing to gain fundamental insights into mutational dynamics and transmission properties [26]. Meanwhile, researchers have developed visualization tools to analyze phylogenetics spatially [27-29], albeit with a coarse geospatial resolution due to poor detail in data collection.
A detailed analysis of the demographic and social determinants of disease risk provides insights into how social characteristics impact health risks and disparities [30-32]. For many infectious diseases, populations of lower socioeconomic status tend to be associated with higher disease prevalence [33-35]. Typically, GIS dashboards fail to incorporate the socioeconomic correlates of health that could provide a deeper understanding of transmission dynamics when combined with geographic and genomic data.
This paper presents the development of the University of Massachusetts’ Graphical user interface for Geographic Information (MAGGI) variant tracking system, a GIS dashboard that integrates genomics and socioeconomic data into a high-resolution geographical dashboard. Our aims for developing MAGGI were to (1) track the transition dynamics of SARS-CoV-2 variants across space and time, (2) identify geographical areas at high risk for transmission using variant cluster risk analysis, and (3) assess socioeconomic risk factors within these high-risk areas.
Methods
Ethics Approval
This study was approved by the University of Massachusetts (UMass) Chan Medical School Institution Review Board (protocol H00021561).
Data Source
Data incorporated into MAGGI originates from multiple sources, including the American Community Survey [36], Massachusetts Department of Public Health (DPH) [37], Centers for Disease Control and Prevention [38], and the UMass Chan Medical School clinical research data warehouse (RDW). The RDW combines data from the UMass Memorial electronic health record system from Epic, Allscripts, and Soarian. Next, we used the 2018 American Community Survey 5-year data to extract zip code–level social demographic data, including race, ethnicity, median family income, housing, language, poverty, and population density. We downloaded the ZIP Code Tabulation Areas (ZCTA) data from the census, and ZCTA are what we used in the MAGGI system. However, we used zip code as a proxy of ZCTA since the latter will be the same as its zip code in most cases and the term zip code is more commonly used and known. Finally, we obtained the COVID-19 vaccination rate data from the Massachusetts DPH and the social vulnerability index from the Centers for Disease Control and Prevention [33].
Remnant positive SARS-CoV-2 test patient samples (swab and saliva) from the UMass Memorial Health Care system were collected and archived by the UMCCTS Biospecimen bank. Sample aliquots were transferred to the UMass Center for Microbiome research for SARS-CoV-2 genomic extraction and sequencing. Sequencing libraries were prepared using NEBNext ARTIC SARS-CoV-2 FS Library Prep Kits and sequenced on the Illumina NextSeq 500 platform as 75nt paired-end reads per manufacturer protocols. Sequence data were analyzed using the Cecret workflow developed at the Utah Public Health Laboratory that provides SARS-CoV-2 genomic sequence and lineage determinations [39]. The genomic results were then linked to patient data.
Geocoding
Geocoding is the technology used to transform physical street addresses into geographic coordinates such as latitude and longitude, enabling us to place markers on the map. After determining the SARS-CoV-2 genomic sequence and lineage, we geocoded the addresses extracted from electronic medical records using the ArcGIS Pro Geocode function on the secure UMass Amazon Web Service private cloud. All geocoding processes used a local geolocator from the Environmental Systems Research Institute (ESRI). Approximately 10%-15% of the extracted addresses contained errors or represented recent addresses that the ESRI geolocator had yet to capture; we manually corrected these errors with GPS coordinates obtained from Google Maps search results. Next, we randomly geocoded Post Office Box addresses to the area defined by the corresponding zip code. Last, we deidentified the geocoded layer to include only longitude and latitude, strain lineage, sample collection date, and variant type to further protect patient information.
Data Integration
The UMass Memorial Health Care System (UMMHC) is the primary health care provider in Central Massachusetts and has used the Epic electronic health record system [40] beginning in 2017. The Epic Clarity database is our primary source for clinical and demographic data. Viral genomic data resides in a separate database. Consequently, we created a Structured Query Language Server database to link genomic data with clinical data using a universal identifier number. This Structured Query Language database remits data to ArcGIS Pro for geocoding. The polygon geometry from zip code and census tracts was integrated into the geodatabase project. Finally, we designed the geodatabase to organize and store spatial databases, tables, and vector data sets. The geodatabase combines geocoding results, social demographics, social vulnerability, and vaccination data, and then passes the data to ArcGIS Online for web-based mapping (Figure 1).
Figure 1.
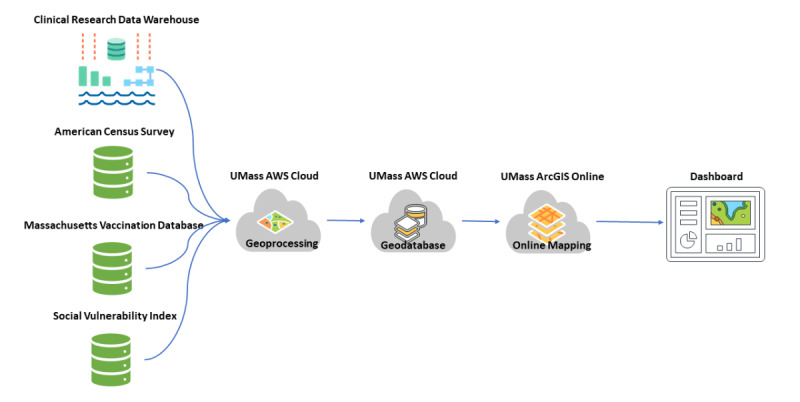
Dashboard workflow and technology stack. UMass: University of Massachusetts; AWS: Amazon Web Service.
Data Visualization and Dashboard Development
ArcGIS Online, the ESRI web-based mapping software, enables users to build interactive web maps with user interface instead of JavaScript programming [41]. We created the web map to visualize spatial layers with this tool. The layers include the geocoded results of variant type, socioeconomic data, vaccination status, and social vulnerability index data. ArcGIS Dashboard, the essential component to conveying information by presenting location-based analytics using intuitive and interactive data visualization, was used to add interactive charts and functionalities based on the web map. We leveraged the dashboard's versatility to enable users to render a variety of charts and selections, which included the following features (item numbers below correspond to the boxed numbers in Figure 2).
Figure 2.
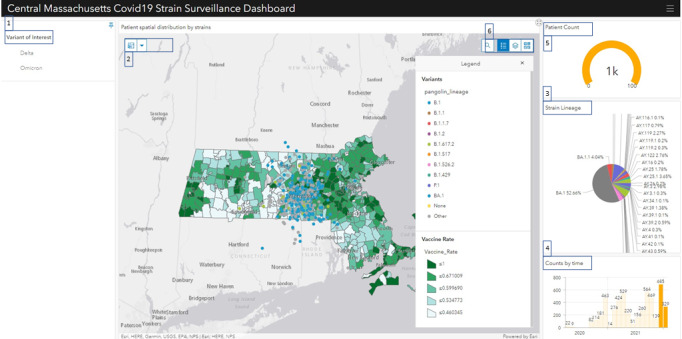
The Massachusetts’ Graphical user interface for Geographic Information (MAGGI) application user interface.
(1) Variants Selection Panel: We built this variant selection panel using the Category Selector function provided by the ArcGIS dashboard. This panel enables researchers to quickly filter the data to the genomic variants of interest (eg, Delta and Omicron).
(2) Spatial Selection Tool: We leveraged the Spatial Selection method in the layer actions section provided by the ArcGIS dashboard to enable the interconnection between the genomic variants and socioeconomic layers. This function allows users to investigate patients in selected geographical units such as zip codes and census units. This tool is useful when genomic epidemiologists study and compare variant population trends through various geolocations.
(3) Strain Lineage Pie Chart function: We created this pie chart using the ArcGIS Pie Chart function to determine the lineage distribution proportion at a given time and zip code. It helps measure the dominated variants and calculate relative risk (RR) in a specific time and space.
(4) Time Serial Chart: We built this chart using the ArcGIS Serial Chart function to render sequenced data by time series. It displays the count of patients by month and works with other selection criteria simultaneously to study the variant transmission dynamic across space and time.
(5) Patient Count: Using the ArcGIS Gauge function, we added the patient count gauge to track patient count based on current selection criteria.
(6) Layer Toggle: We enabled the Basemap Switcher function in the ArcGIS dashboard. This function allows users to change the background map among the socioeconomic layers, social vulnerability index, and vaccination layer. In addition, this function helps researchers easily switch and draw potential correlations between layers.
Cluster Detection: We used the Getis-Ord Gi* hot spot analysis [42] provided by ArcGIS Online to detect the spatial cluster of the variant.
Data Security and Sharing
Users must log in to the ArcGIS Online application via a single sign-on with Microsoft multi-factor authentication technology [43]. Access to the dashboard is based on membership or association with the studies, ensuring that only authorized study team members will see the specific study-related dashboard upon log-in. ArcGIS Online offers several grouping categories to restrict access to appropriate users only: organization, groups within an organization, collaborators with an organization, or everyone (public) [44]. For MAGGI, we used this security protocol to set up private groups for local health department administrators to access only data from their respective jurisdictions. Thus, for example, members of the Massachusetts Fitchburg DPH group can only visualize data from the 11 towns managed by their department. In contrast, administrators from the Massachusetts DPH can access and visualize data from the entire state of Massachusetts.
Results
User Interface
An interactive dashboard provides the user interface that allows researchers to study the spatiotemporal trend of the variants as they work through the investigation process. The user interface depicting data relating to SARS-CoV-2 variants from patient infections between May and October 2001 is presented (Figure 2).
Examples of Use Cases
Use Case 1: Variant Surveillance
MAGGI enables members of public health agencies in Massachusetts and genomic epidemiologists to monitor regional viral spread patterns and the emergence of potential new variants and study the impact of new interventions, including the effectiveness of new policy implementations. For example, genetic epidemiologists use MAGGI to study the relationship between genetic drifts in the genome of SARS-CoV-2 and transmission rate at the local level, thereby revealing unsuspected clusters and evidence for or against suspected transmissions.
Use Case 2: Spatiotemporal Cluster Detection
The Hot Spot Analysis tool calculates the cluster risk using the Getis-Ord Gi* statistic [42]. This calculation’s resulting z scores and P values inform us where to locate areas with high- or low-value clusters spatially. This tool functions by comparing the z scores and P values between any location with its neighbors. Regions labeled as “hot spots” must have a statistically significant score compared to their surrounding neighbors. Based on the cluster risk analysis (Getis-Ord Gi*) results, our first use case objective was to explore the social and vaccination determinants associated with detected clusters. Clusters were identified in the Worcester and Leominster areas served by UMMHC and co-located with areas of lower socioeconomic status and low vaccination rates (Figure 3).
Figure 3.
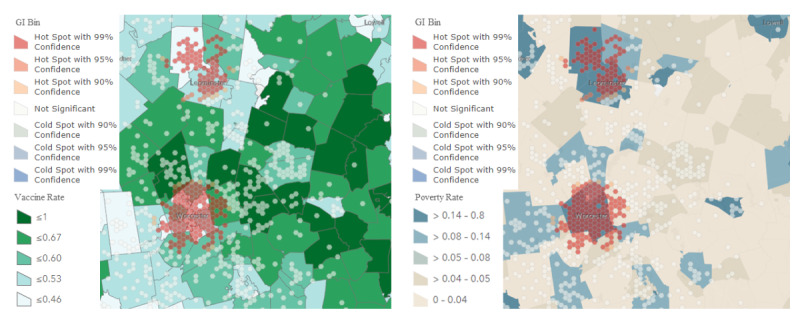
Hot spot analysis (Getis-Ord Gi*) layer overlapping with the social and vaccination determinants layer.
Users can further investigate the dissemination of emerging variants. First, a user specifies the variant of interest and selects the zip codes from the map. The map then updates to visualize the infections meeting the criteria. The user then visualizes the distribution in the time series chart and individually inspects each interval by making the appropriate selections. Finally, the user may browse the filtered results via the map and further probe the distribution of emerging variants or clusters.
Use Case 3: Spatial Distribution of Variant Prevalence and RR
The objective of this use case was to measure the prevalence of variants. Users may use the strain lineage pie chart by configuring the zip code and time interval of interests. They can then observe variant transition dynamics across space and time by opting for a comparison to visualize the progression pattern categorized by demographics and vaccination status. For example, as shown in Figure 4, the Alpha variant progressed from 3% to 48% and from 0% to 69% in Worcester and Marlborough, respectively, between February and April 2021. A comparative visualization as represented in Figure 4 is not available on MAGGI’s user interface. To avoid distraction from an overly complicated pie chart with too many variants, which would be available in MAGGI, we created Figure 4, which only displayed the Alpha variant versus others.
Figure 4.
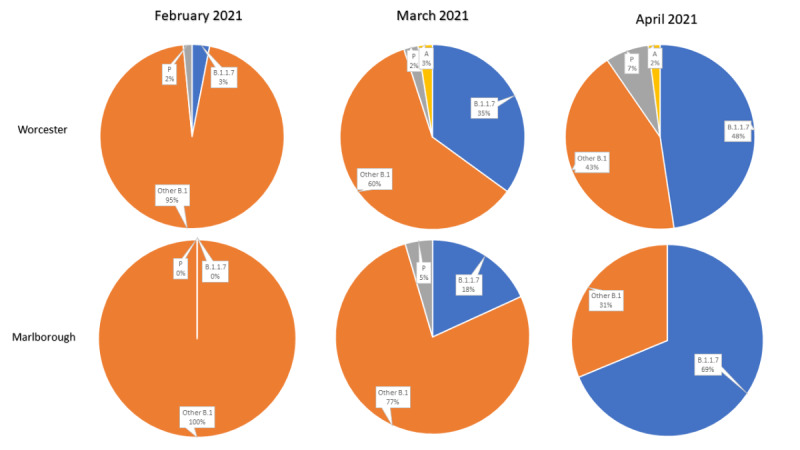
Variant tracking from February to April based on Massachusetts’ Graphical user interface for Geographic Information (MAGGI) data.
We also calculated the RR of variants for selected geographical regions over time. The RR estimates provide insights on how prevalent a variant is in a specific location compared to the other areas. We used the risk from Central Massachusetts to determine our reference group, and then calculated the RR by dividing the risk from Worcester by the risk from the reference group. For example, we compared the RR of the Delta variant in Worcester with those in other Central Massachusetts regions between May and August 2021 (Table 1). We found that people living in Worcester, the largest city in Central Massachusetts, had a 214% increased risk of Delta infection in May 2021 than those living in other regions in Central Massachusetts. The risk of Delta infection increased rapidly through June and July, and individuals in both Worcester and other Central Massachusetts regions were 20 times more likely to contract the Delta variant in July compared to May 2021. We aimed to probe the social determinants of high RR in the early Delta era at the zip code level, but the small sample size after grouping by zip code and month limited the RR calculation at the zip code level.
Table 1.
Relative risk of infection with the Delta variant from May to August 2021.
| Month | Central Massachusetts except Worcester, RRa (95% CI) | Worcester, RR (95% CI) |
| May | Refb | 3.14 (2.63-3.64) |
| June | 5.76 (5.13-6.38) | 12.75 (11.99-13.51) |
| July | 20.29 (19.86-20.72) | 22.56 (21.77-23.35) |
| August | 22.33 (21.90-22.77) | 22.34 (21.82-22.87) |
aRR: relative risk.
bRef: reference group.
Discussion
Principal Findings
We developed MAGGI, a secure platform that enables the spatiotemporal surveillance of the dissemination of SARS-CoV-2 variants. The UMass RDW and ArcGIS Ecosystem provided the foundational tool set for developing this platform. MAGGI is a research tool that is proving to be helpful for research into the socioeconomic and environmental determinants of the COVID-19 pandemic in our local region. Compared to other business intelligence tools, such as Tableau [45], Qlik [46], or Microsoft Power BI [47], the ArcGIS Ecosystem provides more powerful spatial capabilities, such as interactive spatial selection and hot spot cluster analysis. However, the interactive spatiotemporal solution we used can alternatively be developed through a traditional web-mapping approach using JavaScript and open-source spatial databases, such as PostGIS. However, that approach would require several months for a full stack developer and a geospatial team to create and deploy with limited change flexibility due to its resource-intensive coding nature. Although traditional web mapping provides better flexibility in user experience/user interface design, the cost to set up, administer, and support compared to the ArcGIS Ecosystem is greater. Furthermore, we presented our user interface designs in 3 separate meetings to epidemiologists and members of public health agencies, allowing them to provide feedback on the user interface. In the end, they were satisfied with the resulting user interface. For these reasons, we opted to take the ArcGIS route. With the recent development of ArcGIS Online Dashboard, the complexity to develop and deploy an interactive spatial dashboard for disease surveillance or public health management has been significantly reduced. The agility of the whole dashboard creation process enables epidemiologists to promptly determine where and when an infection outbreak occurs and which population it impacts the most.
Our development has multiple strengths. We accrued and sequenced over 5000 clinical samples and engaged genomic epidemiologists to generate questions examining variant transition dynamics in the context of social determinants and vaccination. Currently, the data sets used by the application are updated monthly. Our focus on providing high-resolution GIS is unique and relevant to current times given the COVID-19 pandemic. The Massachusetts DPH is interested in adopting MAGGI. With the continual emergence of fast-spreading SARS-CoV-2 variants such as Delta and Omicron, GIS technology around COVID-19 will only become more important as this technology advances and the adoption of the technology increases.
Limitations
MAGGI is limited to UMMHC data. Therefore, we only captured patients who got their COVID-19 tests at UMMHC, and MAGGI is more likely to include patients who live near UMass Memorial Hospitals in Central Massachusetts. COVID-19 testing data becomes less accurate as we get further away from the hospital, thus limiting the generalizability of the findings using our tool. We hope to solve this issue by accessing data at the state level. In that case, sequencing data from all hospitals in Massachusetts will populate MAGGI, allowing researchers to make more informed investigations on variants, leading to breaking down the barrier. Another limitation of our tool is the small cohort size that it tracks, with 5000 specimens sequenced at this time.
Future Direction
In the future, we plan to enhance the application in several ways. First, we will be automating data transfer and geoprocessing from the UMass Center for Microbiome Research into the MAGGI system to provide near real-time data analysis with a daily turnaround time instead of monthly. Second, we plan to aggregate street-level data to census geographical units (block groups, tracts) or zip code for the entire MAGGI system and create a public version of this tool so that it can be accessed by the general public. Third, we plan to create ArcGIS StoryMaps, a tool that enables researchers to create interactive narratives around their ideas with a strong sense of place [48]. We identified the ease and efficiency of making an informative presentation using StoryMaps when presenting to Massachusetts DPH officials. With collective feedback from epidemiologists and public officers, we will further polish and enhance the user experience of our tool. Finally, we will add new layers that could expose spatial clusters and hot spots of variants to enrich the geospatial mining ability to detect abnormal regions. Our goal is to enable deep phenotyping analytics using MAGGI to ultimately help our researchers identify actionable interventions.
Acknowledgments
The research reported in this publication was supported by the National Center for Advancing Translational Sciences of the National Institutes of Health under award number UL1-TR001453. However, the content is solely the authors' responsibility and does not necessarily represent the official views of the National Institutes of Health. This study was also supported by the COVID-19 pandemic research fund and institutional funds from the University of Massachusetts Chan Medical School.
Abbreviations
- ESRI
Environmental Systems Research Institute
- GIS
Geographic Information System
- MAGGI
Massachusetts’ Graphical user interface for Geographic Information
- RDW
Research Data Warehouse
- RR
Relative Risk
- UMass
University of Massachusetts
- UMMHC
UMass Memorial Health Center
- ZCTA
ZIP Code Tabulation Areas
Footnotes
Conflicts of Interest: None declared.
References
- 1.Coronavirus disease (COVID-19) weekly epidemiological update and weekly operational update. World Health Organization. 2020. [2022-06-06]. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports .
- 2.Bernasconi A, Grandi S. A conceptual model for geo-online exploratory data visualization: the case of the COVID-19 pandemic. Information. 2021 Feb 06;12(2):69. doi: 10.3390/info12020069. [DOI] [Google Scholar]
- 3.COVID data tracker. Centers for Disease Control and Prevention. 2022. [2022-06-06]. https://covid.cdc.gov/covid-data-tracker/#datatracker-home .
- 4.Corum J, Zimmer C. Tracking Omicron and other coronavirus variants. New York Times. 2022. May 24, [2022-06-06]. https://www.nytimes.com/interactive/2021/health/coronavirus-variant-tracker.html .
- 5.Coronavirus in the U.S.: latest map and case count. New York Times. 2022. Jun 05, [2022-06-06]. https://www.nytimes.com/interactive/2021/us/covid-cases.html .
- 6.Bauer C, Zhang K, Lee M, Jones M, Rodriguez A, de la Cerda I, Reininger B, Fisher-Hoch SP, McCormick JB. Real-time geospatial analysis identifies gaps in COVID-19 vaccination in a minority population. Sci Rep. 2021 Sep 13;11(1):18117. doi: 10.1038/s41598-021-97416-y. doi: 10.1038/s41598-021-97416-y.10.1038/s41598-021-97416-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lyseen AK, Nøhr C, Sørensen E M, Gudes O, Geraghty EM, Shaw NT, Bivona-Tellez C, IMIA Health GIS Working Group A review and framework for categorizing current research and development in health related Geographical Information Systems (GIS) studies. Yearb Med Inform. 2014 Aug 15;9:110–24. doi: 10.15265/IY-2014-0008. http://www.thieme-connect.com/DOI/DOI?10.15265/IY-2014-0008 .me2014-0008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shneiderman B, Plaisant C, Hesse BW. Improving healthcare with interactive visualization. Computer. 2013 May;46(5):58–66. doi: 10.1109/MC.2013.38. [DOI] [Google Scholar]
- 9.Franch-Pardo Ivan, Desjardins MR, Barea-Navarro Isabel, Cerdà Artemi. A review of GIS methodologies to analyze the dynamics of COVID-19 in the second half of 2020. Trans GIS. 2021 Jul 11;25(5):2191–2239. doi: 10.1111/tgis.12792. http://europepmc.org/abstract/MED/34512103 .TGIS12792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Franch-Pardo I, Napoletano BM, Rosete-Verges F, Billa L. Spatial analysis and GIS in the study of COVID-19. a review. Sci Total Environ. 2020 Oct 15;739:140033. doi: 10.1016/j.scitotenv.2020.140033. https://linkinghub.elsevier.com/retrieve/pii/S0048-9697(20)33553-1 .S0048-9697(20)33553-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iyanda AE, Adeleke R, Lu Y, Osayomi T, Adaralegbe A, Lasode M, Chima-Adaralegbe NJ, Osundina AM. A retrospective cross-national examination of COVID-19 outbreak in 175 countries: a multiscale geographically weighted regression analysis (January 11-June 28, 2020) J Infect Public Health. 2020 Oct;13(10):1438–1445. doi: 10.1016/j.jiph.2020.07.006. https://linkinghub.elsevier.com/retrieve/pii/S1876-0341(20)30572-4 .S1876-0341(20)30572-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun F, Matthews SA, Yang TC, Hu MH. A spatial analysis of the COVID-19 period prevalence in U.S. counties through June 28, 2020: where geography matters? Ann Epidemiol. 2020 Dec;52:54–59.e1. doi: 10.1016/j.annepidem.2020.07.014. http://europepmc.org/abstract/MED/32736059 .S1047-2797(20)30274-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sannigrahi S, Pilla F, Basu B, Basu AS, Molter A. Examining the association between socio-demographic composition and COVID-19 fatalities in the European region using spatial regression approach. Sustain Cities Soc. 2020 Nov;62:102418. doi: 10.1016/j.scs.2020.102418. http://europepmc.org/abstract/MED/32834939 .S2210-6707(20)30639-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.COVID-19 GIS Hub. Environmental Systems Research Institute. 2020. [2022-06-06]. https://coronavirus-resources.esri.com/
- 15.WHO Coronavirus (COVID-19) Dashboard. World Health Organization. 2022. Jun 06, [2022-06-06]. https://covid19.who.int/
- 16.Dong E, Du H, Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect Dis. 2020 May;20(5):533–534. doi: 10.1016/S1473-3099(20)30120-1. http://europepmc.org/abstract/MED/32087114 .S1473-3099(20)30120-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zheng S, Edwards JR, Dudeck MA, Patel PR, Wattenmaker L, Mirza M, Tejedor SC, Lemoine K, Benin AL, Pollock DA. Building an interactive geospatial visualization application for national health care-associated infection surveillance: development study. JMIR Public Health Surveill. 2021 Jul 30;7(7):e23528. doi: 10.2196/23528. https://publichealth.jmir.org/2021/7/e23528/ v7i7e23528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cahyadi MN, Handayani HH, Warmadewanthi I, Rokhmana CA, Sulistiawan SS, Waloedjo CS, Raharjo AB, Endroyono. Atok M, Navisa SC, Wulansari M, Jin S. Spatiotemporal analysis for COVID-19 delta variant using GIS-based air parameter and spatial modeling. Int J Environ Res Public Health. 2022 Jan 30;19(3):1614. doi: 10.3390/ijerph19031614. https://www.mdpi.com/resolver?pii=ijerph19031614 .ijerph19031614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang P, Croxen MA, Hasan MR, Hsiao WW, Hoang LM. Infection control in the new age of genomic epidemiology. Am J Infect Control. 2017 Feb 01;45(2):170–179. doi: 10.1016/j.ajic.2016.05.015. https://linkinghub.elsevier.com/retrieve/pii/S0196-6553(16)30580-6 .S0196-6553(16)30580-6 [DOI] [PubMed] [Google Scholar]
- 20.Grubaugh ND, Ladner JT, Lemey P, Pybus OG, Rambaut A, Holmes EC, Andersen KG. Tracking virus outbreaks in the twenty-first century. Nat Microbiol. 2019 Jan 13;4(1):10–19. doi: 10.1038/s41564-018-0296-2. http://europepmc.org/abstract/MED/30546099 .10.1038/s41564-018-0296-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chiara M, Horner D, Gissi C, Pesole G. Comparative genomics reveals early emergence and biased spatiotemporal distribution of SARS-CoV-2. Mol Biol Evol. 2021 May 19;38(6):2547–2565. doi: 10.1093/molbev/msab049. http://europepmc.org/abstract/MED/33605421 .6144924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bernasconi A, Mari L, Casagrandi R, Ceri S. Data-driven analysis of amino acid change dynamics timely reveals SARS-CoV-2 variant emergence. Sci Rep. 2021 Oct 26;11(1):21068. doi: 10.1038/s41598-021-00496-z. doi: 10.1038/s41598-021-00496-z.10.1038/s41598-021-00496-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang HC, Chen CH, Wang JH, Liao HC, Yang CT, Chen CW, Lin YC, Kao CH, Lu MYJ, Liao JC. Analysis of genomic distributions of SARS-CoV-2 reveals a dominant strain type with strong allelic associations. Proc Natl Acad Sci U S A. 2020 Dec 01;117(48):30679–30686. doi: 10.1073/pnas.2007840117. https://www.pnas.org/doi/abs/10.1073/pnas.2007840117?url_ver=Z39.88-2003&rfr_id=ori:rid:crossref.org&rfr_dat=cr_pub%3dpubmed .2007840117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang Q, Zhang Q, Bible PW, Liang Q, Zheng F, Wang Y, Hao Y, Liu Y. A new way to trace SARS-CoV-2 variants through weighted network analysis of frequency trajectories of mutations. Front Microbiol. 2022 Mar 16;13:859241. doi: 10.3389/fmicb.2022.859241. doi: 10.3389/fmicb.2022.859241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lemieux JE, Siddle KJ, Shaw BM, Loreth C, Schaffner SF, Gladden-Young A, Adams G, Fink T, Tomkins-Tinch CH, Krasilnikova LA, DeRuff KC, Rudy M, Bauer MR, Lagerborg KA, Normandin E, Chapman SB, Reilly SK, Anahtar MN, Lin AE, Carter A, Myhrvold C, Kemball ME, Chaluvadi S, Cusick C, Flowers K, Neumann A, Cerrato F, Farhat M, Slater D, Harris JB, Branda JA, Hooper D, Gaeta JM, Baggett TP, O'Connell James, Gnirke A, Lieberman TD, Philippakis A, Burns M, Brown CM, Luban J, Ryan ET, Turbett SE, LaRocque RC, Hanage WP, Gallagher GR, Madoff LC, Smole S, Pierce VM, Rosenberg E, Sabeti PC, Park DJ, MacInnis BL. Phylogenetic analysis of SARS-CoV-2 in Boston highlights the impact of superspreading events. Science. 2021 Feb 05;371(6529):eabe3261. doi: 10.1126/science.abe3261. http://europepmc.org/abstract/MED/33303686 .science.abe3261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Popa Alexandra, Genger Jakob-Wendelin, Nicholson Michael D, Penz Thomas, Schmid Daniela, Aberle Stephan W, Agerer Benedikt, Lercher Alexander, Endler Lukas, Colaço Henrique, Smyth Mark, Schuster Michael, Grau Miguel L, Martínez-Jiménez Francisco, Pich Oriol, Borena Wegene, Pawelka Erich, Keszei Zsofia, Senekowitsch Martin, Laine Jan, Aberle Judith H, Redlberger-Fritz Monika, Karolyi Mario, Zoufaly Alexander, Maritschnik Sabine, Borkovec Martin, Hufnagl Peter, Nairz Manfred, Weiss Günter, Wolfinger Michael T, von Laer Dorothee, Superti-Furga Giulio, Lopez-Bigas Nuria, Puchhammer-Stöckl Elisabeth, Allerberger Franz, Michor Franziska, Bock Christoph, Bergthaler Andreas. Genomic epidemiology of superspreading events in Austria reveals mutational dynamics and transmission properties of SARS-CoV-2. Sci Transl Med. 2020 Dec 09;12(573):eabj3222. doi: 10.1126/scitranslmed.abe2555. http://europepmc.org/abstract/MED/33229462 .scitranslmed.abe2555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Argimón Silvia, Abudahab K, Goater R, Fedosejev Artemij, Bhai Jyothish, Glasner Corinna, Feil Edward J, Holden Matthew T G, Yeats Corin A, Grundmann Hajo, Spratt Brian G, Aanensen David M. Microreact: visualizing and sharing data for genomic epidemiology and phylogeography. Microb Genom. 2016 Nov;2(11):e000093. doi: 10.1099/mgen.0.000093. http://mgen.microbiologyresearch.org/pubmed/content/journal/mgen/10.1099/mgen.0.000093 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Theys K, Lemey P, Vandamme AM, Baele G. Advances in visualization tools for phylogenomic and phylodynamic studies of viral diseases. Front Public Health. 2019 Aug 2;7:208. doi: 10.3389/fpubh.2019.00208. doi: 10.3389/fpubh.2019.00208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neher RA, Bedford T. Real-time analysis and visualization of pathogen sequence data. J Clin Microbiol. 2018 Nov 25;56(11):e00480–18. doi: 10.1128/JCM.00480-18. http://europepmc.org/abstract/MED/30135232 .JCM.00480-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Norman B, Pedoia V, Majumdar S. Use of 2D U-Net convolutional neural networks for automated cartilage and meniscus segmentation of knee MR imaging data to determine relaxometry and morphometry. Radiology. 2018 Jul;288(1):177–185. doi: 10.1148/radiol.2018172322. http://europepmc.org/abstract/MED/29584598 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walker RJ, Strom Williams J, Egede LE. Influence of race, ethnicity and social determinants of health on diabetes outcomes. Am J Med Sci. 2016 Apr;351(4):366–73. doi: 10.1016/j.amjms.2016.01.008. http://europepmc.org/abstract/MED/27079342 .S0002-9629(15)37995-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Menza T, Hixson L, Lipira L, Drach L. Social determinants of health and care outcomes among people with HIV in the United States. Open Forum Infect Dis. 2021 Jul;8(7):ofab330. doi: 10.1093/ofid/ofab330. http://europepmc.org/abstract/MED/34307729 .ofab330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Du P, Lemkin A, Kluhsman B, Chen J, Roth RE, MacEachren A, Meyers C, Zurlo JJ, Lengerich EJ. The roles of social domains, behavioral risk, health care resources, and chlamydia in spatial clusters of US cervical cancer mortality: not all the clusters are the same. Cancer Causes Control. 2010 Oct 8;21(10):1669–83. doi: 10.1007/s10552-010-9596-4. [DOI] [PubMed] [Google Scholar]
- 34.Kauhl B, Heil J, Hoebe CJPA, Schweikart J, Krafft T, Dukers-Muijrers NHTM. The spatial distribution of hepatitis C virus infections and associated determinants--an application of a geographically weighted Poisson regression for evidence-based screening interventions in hotspots. PLoS One. 2015 Sep 9;10(9):e0135656. doi: 10.1371/journal.pone.0135656. https://dx.plos.org/10.1371/journal.pone.0135656 .PONE-D-15-16540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vermeiren APA, Dukers-Muijrers NHTM, van Loo IHM, Stals F, van Dam DW, Ambergen T, Hoebe CJPA. Identification of hidden key hepatitis C populations: an evaluation of screening practices using mixed epidemiological methods. PLoS One. 2012 Dec 7;7(12):e51194. doi: 10.1371/journal.pone.0051194. https://dx.plos.org/10.1371/journal.pone.0051194 .PONE-D-12-21971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.2014-2018 American Community Survey 5-year estimates. United States Census Bureau. [2022-06-06]. https://www.census.gov/programs-surveys/acs/technical-documentation/table-and-geography-changes/2018/5-year.html .
- 37.Massachusetts COVID-19 vaccination data and updates. Massachusetts Department of Public Health. [2022-06-06]. https://www.mass.gov/info-details/massachusetts-covid-19-vaccination-data-and-updates .
- 38.CDC/ATSDR Social Vulnerability Index. Agency for Toxic Substances and Disease Registry. 2022. Mar 15, [2022-06-06]. https://www.atsdr.cdc.gov/placeandhealth/svi/index.html .
- 39.Cecret. Github. [2022-06-06]. https://github.com/UPHL-BioNGS/Cecret .
- 40.Epic. [2022-06-06]. https://www.epic.com/
- 41.ArcGIS Online. Environmental Systems Research Institute. [2022-06-06]. https://www.esri.com/en-us/arcgis/products/arcgis-online/overview .
- 42.Getis A, Ord JK. The analysis of spatial association by use of distance statistics. In: Anselin L, Rey S, editors. Perspectives on Spatial Data Analysis. Berlin, Heidelberg: Springer; 2010. pp. 127–45. [Google Scholar]
- 43.Multi-factor authentication (MFA) - Microsoft Security. Microsoft. 2022. [2022-06-06]. https://www.microsoft.com/en-us/security/business/identity-access-management/mfa-multi-factor-authentication .
- 44.ArcGIS Online: share items. Environmental Systems Research Institute. 2022. [2022-06-06]. https://doc.arcgis.com/en/arcgis-online/share-maps/share-items.htm .
- 45.Business intelligence and analytics software. Tableau. [2022-06-06]. https://www.tableau.com/
- 46.Qlik. [2022-06-06]. https://www.qlik.com/us/
- 47.Data Visualization: Microsoft Power BI. Microsoft. [2022-06-06]. https://powerbi.microsoft.com/en-us/
- 48.Health care story maps. Environmental Systems Research Institute. [2022-06-06]. https://storymaps-classic.arcgis.com/en/gallery/#s=30&md=storymaps-industry:health-services .


