Abstract
A functional pyc gene was isolated from Lactococcus lactis subsp. lactis C2 and was found to complement a Pyc defect in L. lactis KB4. The deduced lactococcal Pyc protein was highly homologous to Pyc sequences of other bacteria. The pyc gene was also detected in Lactococcus lactis subsp. cremoris and L. lactis subsp. lactis bv. diacetylactis strains.
Lactococci are widely used as starters for milk fermentations and are characterized by their requirements for multiple nutritional factors, especially exogenous organic nitrogen sources for growth. We previously described a Lactococcus lactis C2 derivative, designated L. lactis KB4, which required aspartate or asparagine for growth in milk. This strain lacked pyruvate carboxylase (Pyc) activity (33). Pyc is a biotin-containing enzyme that catalyzes the ATP-dependent carboxylation of pyruvate to form oxaloacetate (OAA) (4). The primary function of Pyc in L. lactis is to form OAA, which serves as a precursor for aspartate synthesis. Aspartate is a precursor for the synthesis of five other amino acids, as well as for pyrimidine synthesis. Aspartate is also a nitrogen donor for purine biosynthesis. These building blocks are further involved in synthesizing molecules such as proteins, DNA, RNA, and ATP.
In the late 1970s, Hillier et al. (13, 14, 15) studied the CO2 requirement of L. lactis C10, which led to the discovery of the aspartate biosynthetic pathway in lactococci. Using [14C]bicarbonate, they found that CO2 was incorporated into pyruvate to form OAA, which was then converted to aspartate. The first reaction was catalyzed by the enzyme pyruvate carboxylase. The pyc gene has been characterized for only a few bacteria (9, 17, 18, 25); thus, genetic characterization of the lactococcal pyc gene would be a significant step in understanding the conservation of this anaplerotic enzyme during evolution. Furthermore, genetic identification of the lactococcal pyc gene is critical for our understanding of the physiology of lactic acid bacteria, especially with regard to their pyruvate metabolism, and may have an application in milk fermentations.
In this study, we report the identification, cloning, and nucleotide sequence of the L. lactis C2 pyc gene and its use in complementing the defect in the aspartate-requiring mutant L. lactis KB4.
DNA manipulations.
Chromosomal DNA was isolated from L. lactis as previously described (26). Plasmid DNA was isolated from Escherichia coli by using a DNA isolation kit (Qiagen Inc., Valencia, Calif.). Plasmids from L. lactis were isolated as described by Anderson and McKay (2). DNA restriction, dephosphorylation, and cloning were performed according to standard procedures (28). DNA fragments were purified from agarose gels by using the Geneclean kit (Bio 101, La Jolla, Calif.).
Lactococcal pyc gene probe development.
The amino acid sequences of reported pyruvate carboxylases (9, 10, 16, 22, 30) and products of prokaryote gene fragments with the same functional domains (6, 19) were aligned. Degenerate oligonucleotide primer pairs were designed based on conserved amino acid sequences from aligned functional domains. The primer pairs and C2 DNA (used as a template) were used in a PCR to obtain a lactococcal pyc-specific gene probe.
Genomic library construction.
To build a genomic library, L. lactis C2 DNA was partially digested with Sau3AI and was ligated to the BamHI-digested lambda pGEM11 vector (Promega Corp., Madison, Wis.). The recombinant DNA was then packaged with lambda phage proteins.
Plaque hybridization and nucleotide sequence determination.
The lactococcal pyc probe was used to identify plaques possessing the pyc sequences from the lambda-C2 library. Phage plaque hybridization, phage DNA isolation (with a kit from Promega Corp.), and nonisotopic digoxigenin labeling and detection (with a kit from Boehringer Mannheim Biochemicals, Indianapolis, Ind.) were performed according to the manufacturers' directions. The filter washing temperature was 65°C. DNA sequencing was performed by the dideoxy method of Sanger et al. (29). Double-stranded sequencing of L. lactis C2 pyc was performed by the Analytical Biotechnology Center, University of Minnesota, St. Paul. Nucleotide and protein sequence homology searches were conducted using the BLAST program (1) via the National Institute for Biotechnology Information server. Multiple protein sequence alignments were made with the Genetics Computer Group (Madison, Wis.) program package, version 8.
Cloning of the pyc gene.
To clone the pyc gene from C2, the DNA fragment containing the upstream and downstream sequences of pyc was obtained by high-fidelity PCR with cloned Pfu polymerase (Stratagene, La Jolla, Calif.). Conditions were as recommended by the manufacturer. The PCR fragment was cloned into the E. coli-lactococcal shuttle vector pCI372 (12).
Growth in SSM and acid production in milk.
Cells were grown in simple synthetic medium (SSM) (33) containing all amino acids required for growth except for aspartate and asparagine in order to study their aspartate-synthesizing capability. To evaluate the rate of acid production, cells were grown at 30°C in 11% (wt/vol) reconstituted skim milk (steamed for 60 min) using a 1% inoculum from an overnight culture, and the pH was monitored.
Immunoblot analysis of Pyc in L. lactis.
L. lactis cells from 10 ml of an overnight culture were collected, washed once with 0.85% NaCl, and suspended in 0.5 ml of Tris-EDTA (28) buffer. The iced cell suspensions were disrupted for 2 min by using a Mini-bead beater-8 cell disrupter (Biospec Products, Bartlesville, Okla.) at maximal power. The cell suspension was centrifuged at 10,000 × g for 2 min to collect the supernatant. The protein content of the supernatant was determined by using a protein assay kit (Bio-Rad, Hercules, Calif.) before the supernatant was mixed with sample buffer. The mixtures were incubated for 10 min in boiling water. About 120 μg of cell proteins was separated on a sodium dodecyl sulfate–10% polyacrylamide gel and electroblotted onto nitrocellulose membranes for 18 h at 30 V as recommended by the manufacturer (Bio-Rad). The lactococcal Pyc protein on the blot was detected by using rabbit anti-Rhodobacter capsulatus Pyc serum (35) and the Bio-Rad amplified alkaline phosphatase goat anti-rabbit immunoblot assay kit as recommended by the manufacturer.
Isolation and cloning of the L. lactis C2 pyc gene.
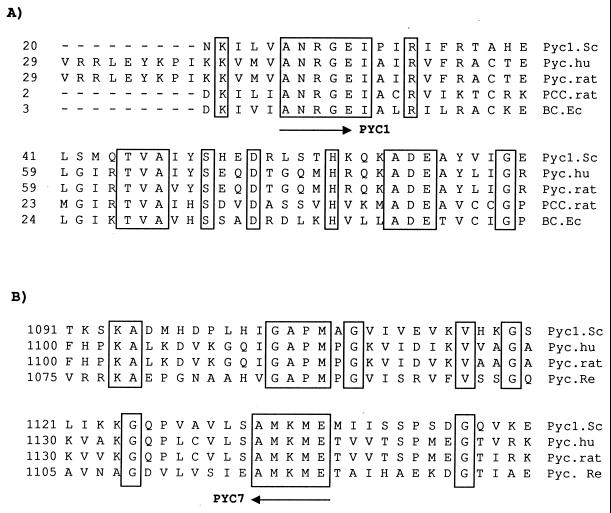
By comparing the amino acid sequences of functional domains of pyruvate carboxylases from various organisms with those of products of prokaryote gene fragments carrying the same functional domains, several conserved regions were selected (Fig. 1). Degenerate oligonucleotides were designed based on these conserved sequences, according to frequent codon usage in lactococci (7). These oligonucleotides were used as primers in PCR. A PCR product of approximately 3 kb was detected when the pair of degenerate oligonucleotides PYC1 (5′-GCNAAMGNGGNGARATH-3′) and PYC7 (5′-YTCCATYTTCATNGC-3′) were used as the primers (N = A+T+G+C, Y = G+T, M = A+C, R = A+G, and H = A+T+C) and C2 chromosomal DNA was used as the template. Partial DNA sequence analysis showed that the PCR fragment encoded a putative protein that was highly homologous to known pyruvate carboxylases. A subfragment (a 600-bp EcoRV digestion fragment) of this PCR product was then used as a lactococcal C2 pyc gene-specific probe. An L. lactis C2 genomic DNA library was constructed, and the lactococcal pyc-specific probe was used to screen phage plaques for DNA insertions containing the pyc gene. One of the positive plaques, S11-1, contained a 20-kb insertion from C2. This 20-kb fragment was cloned into the shuttle vector pTRKH2 (24), and the resulting plasmid was designated pTHPYC1. The DNA sequence of the region containing the pyc gene was determined. In order to study the role of pyc without the interference of other genes on the large fragment, a 3,746-bp fragment that encompassed the putative regulation and termination regions of the lactococcal pyc gene was obtained by high-fidelity PCR with PYC18 (5′-TCC CCC GGG CCA AAT TAT GAA AAC GAT TGA CAA ATG-3′) and PYC20 (5′-TTC TGC AGT TGT AAA ATT CAC TAG GAA TTT GTG-3′). These primers were selected from the end of the open reading frame (ORF) upstream and the beginning of the ORF downstream of pyc (unpublished sequence) coupled with 5′-TCC CCC GGG for SmaI digestion and 5′-TTC TGC AG for PstI digestion, respectively. L. lactis C2 DNA was the template. The PCR product was cloned into the shuttle vector pCI372 (12), resulting in pCPYC1.
FIG. 1.
Alignment of amino acid sequences of reported pyruvate carboxylases and products of prokaryote gene fragments with the same functional domains. (A) Biotin carboxylase domain; (B) biotin binding domain. Pycl.Sc, S. cerevisiae Pyc1 (33); Pyc.hu, human Pyc (11); Pyc.rat, rat Pyc (19); PCC.rat, rat propionyl coenzyme A carboxylase (6); BC.Ec, E. coli acetyl coenzyme A carboxylase (24); Pyc.Re, R. etli (9). Conserved amino acid sequences (boxed) were used to design degenerate oligonucleotide primers for PCR.
Sequence analysis of the L. lactis pyc gene.
DNA sequence analysis of pTHPYC1 identified an uninterrupted ORF spanning a 3-kb fragment that showed homology to known pyruvate carboxylases at both the DNA and protein levels. The L. lactis C2 pyc gene consisted of 3,411 bp and encoded a 1,137-amino-acid Pyc protein with a predicted molecular mass of 123 kDa. A perfect −10 promoter consensus was identified upstream of the pyc ATG start codon, but no appropriately spaced −35 consensus region was present.
Immediately upstream of the pyc coding region, a canonical lactococcal ribosome binding site (AAGGA; positions 94 to 98) was detected. A stem-loop structure can be formed between positions 3531 and 3547, followed by a string of T's, indicating the possible existence of a rho-independent termination structure at the end of the pyc gene.
Searches for homology with the deduced L. lactis Pyc protein detected a 62% overall protein sequence similarity with Bacillus stearothermophilus Pyc (18) and 60, 50, 49, and 49% similarity with Bacillus subtilis Pyc (GenBank accession no. 2633857), Rhizobium etli Pyc (9), and Saccharomyces cerevisiae Pyc1 (22) and Pyc2 (30), respectively.
The two consensus sequences for carbamoyl-phosphate synthase believed to be involved in ATP binding (11, 22, 27) and found in all pyruvate carboxylases (5, 21, 34) were identified. The conserved Cys residue (nucleotides 786 to 788) suggested to be involved in CO2 fixation was present. It has been proposed that this Cys residue and a conserved Lys eight residues downstream have important roles in the catalytic mechanism of Pyc (4, 32). A putative pyruvate binding region (32) was identified in the L. lactis Pyc sequence that matched very closely with the proposed consensus of RFLXED/CPWXR (32). The only deviation from the consensus was an S in place of D/C. As this is not a conservative change, it indicates that this residue is likely not essential for pyruvate binding. The W residue (nucleotides 1860 to 1862) proposed to be directly involved in pyruvate binding (20) was also conserved. The motif proposed as being involved in metal binding (32) and the biotin binding (9) motif were also identified.
Mutant complementation.
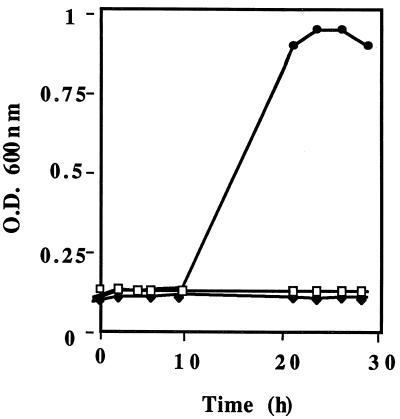
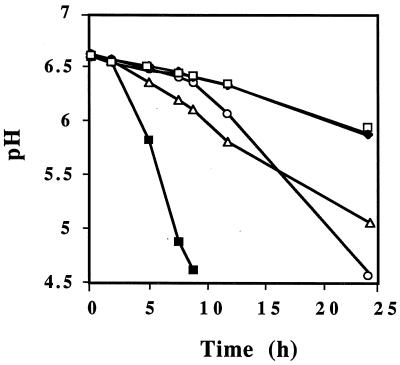
As previously observed, L. lactis KB4 required aspartate for growth and was Pyc− (33). To determine if the cloned pyc gene could complement the aspartate biosynthetic defect in KB4, the vector pCI372, carrying the cloned pyc gene (pCPYC1), was introduced into KB4. The resulting transformant, HWPC001, then grew in a chemically defined medium lacking aspartate (Fig. 2). It was also able to utilize β-casein as a sole nitrogen source (data not shown). However, when the rate of acid production in milk by KB4(pCPYC1) was compared to that of the parental strain, L. lactis C2, KB4(pCPYC1) was found to exhibit a lower rate (Fig. 3). This lower rate was due to the presence of the cloning vector pCI372, as L. lactis C2 carrying pCI372 exhibited a rate of acid production similar to that of KB4 possessing pCI372 with the pyc gene (Fig. 3). Why the vector exhibited this inhibitory effect is unknown. The results do suggest, however, that the pyc gene in KB4 is capable of restoring acid-producing ability similar to that of the wild-type strain, C2, when the latter contains the vector. These results support the view that pyc is capable of reestablishing the Fmc+ phenotype in KB4 by restoring the ability to synthesize aspartic acid. It was previously shown that the addition of aspartic acid to milk restored the fast-acid-producing phenotype to KB4 (33).
FIG. 2.
Growth of L. lactis KB4 and derivatives in SSM with 19 amino acids (Ala, Arg, Cys, Glu, Gln, Gly, His, Pro, Ile, Leu, Lys, Met, Phe, Pro, Ser, Thr, Trp, Tyr, and Val). ⧫, KB4; □, HB372 (transformant of KB4 carrying pCI372); ●, HWPC001 (transformant of KB4 carrying the cloned pyc gene). The results are representative of at least two separate trials. O.D., optical density.
FIG. 3.
Acid production by L. lactis C2 and KB4 derivatives in milk without supplementation of aspartate or asparagine. ▵, HC372 (transformant of C2 carrying pCI372); □, KB4; ⧫, HB372 (transformant of KB4 carrying pCI372); ○, HWPC001 (transformant of KB4 carrying the cloned pyc gene); ■, L. lactis C2. The results are representative of at least two separate trials.
Investigation of Pyc in KB4.
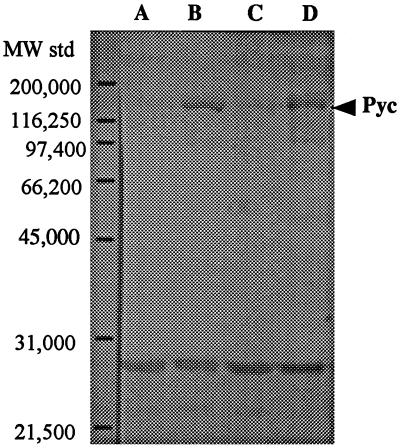
Primers specific to pyc were used to test for the presence of pyc by PCR. A specific PCR product of the expected size for the pyc gene was observed, suggesting the presence of the pyc gene in KB4 (data not shown). As KB4 was missing Pyc activity, we investigated whether a Pyc protein was produced in KB4. On Western blots developed to detect Pyc proteins (using Pyc antibody), a band which migrated with an approximate molecular weight of 127,000 (close to the expected Pyc weight of 123,000) was observed. However, the intensity of the bands from KB4 and the KB4 transformant containing pCI372 (vector only) was less than that of the bands from C2 and the KB4 transformant containing the cloned pyc gene (HWPC001) (Fig. 4). This suggested that KB4 lacked significant Pyc activity because less cellular Pyc protein was present. The absence of Pyc activity in KB4 could be due to changes in the regulation of protein expression, structural mutations in the pyc gene, or polarity effects on gene expression.
FIG. 4.
Western immunoblot analysis of Pyc content of L. lactis C2 and KB4 derivatives. Lanes: A, HB372 (transformant of KB4 carrying pCI372); B, HWPC001 (transformant of KB4 carrying the cloned pyc gene); C, KB4; D, C2. The lower band with a molecular weight around 29,000 represents nonspecific binding, which was also detected by A. F. Yakunin (personal communication). MW std, molecular weight standards.
Detection of pyc in other lactococcal strains.
Using the PCR primer pair PYC1 and PYC7, we also tested for the presence of the pyc gene in other lactococcal strains. A 3-kb fragment was detected in all the Lactococcus lactis subsp. cremoris (TR, E8, SK11, and HP) and Lactococcus lactis subsp. lactis bv. diacetylactis (11007, DRC1, and 425A) strains examined, which suggested that the pyc gene also exists in these strains (data not shown). This result suggested that OAA synthesis via pyruvate carboxylase may be an important pathway in lactococci. This result could be of industrial significance, as pyruvate is a central metabolic intermediate and a precursor for various compounds. For instance, the industrially significant flavor compound diacetyl is derived from pyruvate. In order to increase the production of diacetyl, attempts are being made to block pathways that convert pyruvate into other end products. In this way, more pyruvate would be available for diacetyl production. However, the drainage of pyruvate through pyruvate carboxylase has not been considered when designing metabolically engineered strains, possibly due to the lack of knowledge about this pathway. As OAA is an anaplerotic precursor involved in synthesizing amino acids, nucleotides, and other cofactors, the amount of pyruvate utilized could be significant. Thus, an understanding of the role of pyruvate carboxylase in lactococci, especially in L. lactis bv. diacetylactis strains, and of the genetic organization and regulation of this enzyme may open the possibility of bioengineering strains with increased diacetyl production through modification of the pyruvate carboxylase pathway.
Pyruvate carboxylase is a central metabolic enzyme that has attracted extensive research interest since the 1960s. The wide distribution of pyruvate carboxylase among organisms, even among those without a complete tricarboxylic acid cycle, clearly indicates its biological significance (31). The central role of aspartate and OAA in anabolic metabolism might explain why, up to now, there has been no report of an aspartate mutant, or especially of a pyruvate carboxylase mutant, in lactococci.
It is generally believed that in lactic acid bacteria, sugars are metabolized predominantly to generate energy and are used as a carbon source (3), whereas anabolic precursor metabolites are primarily obtained from other components of the medium (8). However, our study on the pyruvate carboxylase-deficient mutant suggests that the primary source of the anabolic precursors OAA and aspartate is biosynthesis. The ability of lactococcal strains to absorb OAA (23, 33) and aspartate (15) may be limited; thus, a functional pyruvate carboxylase is required for fast milk coagulation by lactococci.
Nucleotide sequence accession number.
The sequence of the region containing the pyc gene has been submitted to GenBank under accession no. AF068759.
Acknowledgments
This research was supported, in part, by the Minnesota-South Dakota Dairy Foods Research Center, St. Paul, by Dairy Management Inc., Rosemont, Ill., by a National Institute of General Medical Sciences (NIGMS) Training Grant in Biotechnology, and by the Kraft General Foods Chair in Food Science.
We thank D. Twomey for discussions and suggestions on the manuscript. The Rhodobacter Pyc-specific antibody was kindly provided by Alexander F. Yakunin from Départment de Microbiologie et Immunologie, Université de Montreal, Montreal, Canada.
Footnotes
Published as paper no. 99-1-18-0023 of the contribution series of the Minnesota Agricultural Experiment Station based on research conducted under project 18-062.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Anderson D G, McKay L L. Simple and rapid method for isolating large plasmid DNA from lactic streptococci. Appl Environ Microbiol. 1983;46:549–552. doi: 10.1128/aem.46.3.549-552.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arnau J, Jørgensen F, Madsen S M, Vrang A, Israelsen H. Cloning, expression, and characterization of the Lactococcus lactis pfl gene, encoding pyruvate formate-lyase. J Bacteriol. 1997;179:5884–5891. doi: 10.1128/jb.179.18.5884-5891.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Attwood P V. The structure and the mechanism of action of pyruvate carboxylase. Int J Biochem Cell Biol. 1995;27:231–249. doi: 10.1016/1357-2725(94)00087-r. [DOI] [PubMed] [Google Scholar]
- 5.Bairoch A, Buchler P, Hofmann K. The PROSITE database, its status in 1997. Nucleic Acids Res. 1997;24:217–221. doi: 10.1093/nar/25.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Browner M F, Taroni F, Sztul E, Rosenberg L E. Sequence analysis, biogenesis, and mitochondrial import of the alpha-subunit of rat liver propionyl-CoA carboxylase. J Biol Chem. 1989;264:12680–12685. [PubMed] [Google Scholar]
- 7.Chopin A. Organization and regulation of genes for amino acid biosynthesis in lactic acid bacteria. FEMS Microbiol Rev. 1993;12:21–37. doi: 10.1111/j.1574-6976.1993.tb00011.x. [DOI] [PubMed] [Google Scholar]
- 8.Cocaign-Bousquet M, Garrigues C, Novak L, Loubiere P, Lindley N D. Physiology of pyruvate metabolism in Lactococcus lactis. In: Venema G, Huis in't Veld J H J, Hugenholts J, editors. Lactic acid bacteria: genetics, metabolism and application. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1996. pp. 157–171. [Google Scholar]
- 9.Dunn M F, Encarnación S, Araíza G, Vargas M C, Dávalos A, Peralta H, Mora Y, Mora J. Pyruvate carboxylase from Rhizobium etli: mutant characterization, nucleotide sequence, and physiological role. J Bacteriol. 1996;178:5960–5970. doi: 10.1128/jb.178.20.5960-5970.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Freytag S O, Collier K J. Molecular cloning of a cDNA for human pyruvate carboxylase. Structural relationship to other biotin-containing carboxylases and regulation of mRNA content in differentiating preadipocytes. J Biol Chem. 1984;259:12831–12837. [PubMed] [Google Scholar]
- 11.Fry D C, Kuby S A, Mildvan A S. ATP-binding site of adenylate kinase: mechanistic implications of its homology with ras-encoded p21, F1-ATPase, and other nucleotide-binding proteins. Proc Natl Acad Sci USA. 1986;83:907–911. doi: 10.1073/pnas.83.4.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hayes F, Daly C, Fitzgerald G F. Identification of the minimal replicon of Lactococcus lactis subsp. lactis UC317 plasmid pCI305. Appl Environ Microbiol. 1990;56:202–209. doi: 10.1128/aem.56.1.202-209.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hillier A J, Jago G R. Metabolism of [14C]bicarbonate by Streptococcus lactis: identification and distribution of labeled compounds. J Dairy Res. 1978;45:231–240. doi: 10.1017/s0022029900016654. [DOI] [PubMed] [Google Scholar]
- 14.Hillier A J, Jago G R. The metabolism of [14C]bicarbonate by Streptococcus lactis: the fixation of [14C]bicarbonate by pyruvate carboxylase. J Dairy Res. 1978;45:433–444. doi: 10.1017/s0022029900016654. [DOI] [PubMed] [Google Scholar]
- 15.Hillier A J, Rice G H, Jago G R. Metabolism of [14C]bicarbonate by Streptococcus lactis: the synthesis, uptake and excretion of aspartate by resting cells. J Dairy Res. 1978;45:241–246. [Google Scholar]
- 16.Jitrapakdee S, Booker G W, Cassady A I, Wallace J C. Cloning, sequencing and expression of rat liver pyruvate carboxylase. Biochem J. 1996;316:631–637. doi: 10.1042/bj3160631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koffas M A G, Famamoorthi R, Pine W A, Sinskey A J, Stephanopoulos G. Sequence of the Corynebacterium glutamicum pyruvate carboxylase gene. Appl Microbiol Biotechnol. 1998;50:346–352. doi: 10.1007/s002530051302. [DOI] [PubMed] [Google Scholar]
- 18.Kondo H, Kazyta Y, Saito A, Fuji K. Cloning and nucleotide sequence of Bacillus stearothermophilus pyruvate carboxylase. Gene. 1997;191:47–50. doi: 10.1016/s0378-1119(97)00027-9. [DOI] [PubMed] [Google Scholar]
- 19.Kondo H, Shiratsuchi K, Yoshimoto T, Masuda T, Kitazono A, Tsuru D, Anai M, Sekiguchi M, Tanabe T. Acetyl-CoA carboxylase from Escherichia coli: gene organization and nucleotide sequence of the biotin carboxylase subunit. Proc Natl Acad Sci USA. 1991;88:9730–9733. doi: 10.1073/pnas.88.21.9730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kumar G K, Haase F C, Phillips N F, Wood H G. Involvement and identification of a tryptophanyl residue at the pyruvate binding site of transcarboxylase. Biochemistry. 1988;27:5978–5983. doi: 10.1021/bi00416a022. [DOI] [PubMed] [Google Scholar]
- 21.Li S, Cronan J E. The gene encoding the biotin carboxylase subunit of Escherichia coli acetyl-CoA carboxylase. J Biol Chem. 1992;267:855–863. [PubMed] [Google Scholar]
- 22.Lim F, Morris C P, Occhiodoro F, Wallace J C. Sequence and domain structure of yeast pyruvate carboxylase. J Biol Chem. 1988;263:11493–11497. [PubMed] [Google Scholar]
- 23.McFeeters R F, Chen K. Utilization of electron acceptors for anaerobic mannitol metabolism by Lactobacillus plantarum. Compounds which serve as electron acceptors. Food Microbiol. 1986;3:73–81. [Google Scholar]
- 24.O'Sullivan D J, Klaenhammer T R. High- and low-copy-number Lactococcus shuttle cloning vectors with features for clone screening. Gene. 1993;137:227–231. doi: 10.1016/0378-1119(93)90011-q. [DOI] [PubMed] [Google Scholar]
- 25.Peters-Wendisch P G, Kreutzer C, Kalinowski J, Patek M, Sahm H, Eikmanns B J. Pyruvate carboxylase from Corynebacterium glutamicum—characterization, expression, and inactivation of the pyc gene. Microbiology. 1998;144:915–927. doi: 10.1099/00221287-144-4-915. [DOI] [PubMed] [Google Scholar]
- 26.Pitcher D G, Saunders N A, Owen R J. Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Lett Appl Microbiol. 1989;8:151–156. [Google Scholar]
- 27.Post L E, Post D J, Raushel F M. Dissection of the functional domains of Escherichia coli carbamoyl phosphate synthase by site-directed mutagenesis. J Biol Chem. 1990;265:7742–7747. [PubMed] [Google Scholar]
- 28.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 29.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stucka R, Dequin S, Salmon J M, Gancedo C. DNA sequences in chromosomes II and III code for pyruvate carboxylase isoenzymes in Saccharomyces cerevisiae: analysis of pyruvate carboxylase-deficient strains. Mol Gen Genet. 1991;229:307–315. doi: 10.1007/BF00272171. [DOI] [PubMed] [Google Scholar]
- 31.Wallace J C. Distribution and biological functions of pyruvate carboxylase in nature. In: Keech D B, Wallace J C, editors. Pyruvate carboxylase. Boca Raton, Fla: CRC Press; 1985. pp. 5–63. [Google Scholar]
- 32.Wallace J C, Jitrapakdee S, Chapmansmith A. Pyruvate carboxylase. Int J Biochem Cell Biol. 1998;30:1–5. doi: 10.1016/s1357-2725(97)00147-7. [DOI] [PubMed] [Google Scholar]
- 33.Wang H, Yu W, Coolbear T, O'Sullivan D, McKay L L. A deficiency in aspartate biosynthesis in Lactococcus lactis subsp. lactis C2 causes slow milk coagulation. Appl Environ Microbiol. 1998;64:1673–1679. doi: 10.1128/aem.64.5.1673-1679.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wexler I D, Du Y, Lisgaris M V, Mandal S K, Freytag S O, Yang B S, Liu T C, Kwon M, Patel M S, Kerr D S. Primary amino acid sequence and structure of human pyruvate carboxylase. Biochim Biophys Acta. 1994;1227:46–52. doi: 10.1016/0925-4439(94)90105-8. [DOI] [PubMed] [Google Scholar]
- 35.Yakunin A F, Hallenbeck P C. Regulation of synthesis of pyruvate carboxylase in the photosynthetic bacterium Rhodobacter capsulatus. J Bacteriol. 1997;179:1460–1468. doi: 10.1128/jb.179.5.1460-1468.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]