Abstract
Application of flow cytometry (FCM) to microbial analysis of milk is hampered by the presence of milk proteins and lipid particles. Here we report on the development of a rapid (≤1-h) FCM assay based on enzymatic clearing of milk to determine total bacteria in milk. When bacteria were added to ultra-heat-treated milk, a good correlation (r ≥ 0.98) between the FCM assay and the more conventional methods of plating and direct microscopic counting was achieved. Raw milk data showed a significant correlation (P < 0.01) and a good agreement (r = 0.91) between FCM and standard plate count methods. The detection limit of the FCM assay was ≤104 bacteria ml of milk−1. This limit is below the level of detection required to satisfy legislation in many countries and states.
The microbiological content of raw milk affects quality, shelf life, and safety of processed milk and other dairy products (3, 21, 24). There are several methods available for detection and enumeration of microorganisms in raw and processed milks. Culture techniques are the most common, but a major disadvantage of these is the time needed to produce results (27). Another significant disadvantage of traditional culture methods is their failure to isolate viable but nonculturable organisms (7). On the other hand, total microscopic count methods are relatively fast, but limitations of these techniques include operator fatigue from prolonged use of microscopes and inability to discriminate between living and dead bacteria. To alleviate problems associated with culture-based detection systems and direct microscopic methods, various other methods have been developed (11, 15, 26). These include colorimetric assay methods based on liberation of dyes from substrates by enzymes, dye reduction tests, and ATP bioluminescence (12, 18, 22, 30). Although biochemically and physiologically based assays are fast, they lack absolute specificity. It is therefore doubtful whether these nonculturing approaches provide complete quality and safety assurance.
Interest in rapid methods and automation in food microbiology has been growing in the past several decades (15). There have been some attempts to introduce direct, automated enumeration methods into dairy testing (6, 23, 26), and BactoScan was developed as an automated instrument for routine testing of the bacteriological quality of raw milk (6, 25). However, the current instrumentation is limited to measuring either total microbial or somatic cell counts, and it has not been developed for more diversified work such as microbial differentiation. By contrast, flow cytometers may suit the broad and specific needs of microbial analysis of milk and dairy products within one type of instrument (15, 23). Flow cytometry (FCM) is extremely sensitive, avoids the need for culturing or enrichment procedures, and can be both qualitative and quantitative (2, 23). Use of fluorescent stains or fluorogenic substrates in combination with FCM allows the detection and discrimination of viable culturable, viable nonculturable, and nonviable organisms (2, 7, 28). Furthermore, there is the possibility that numerous (or even rare) microbial cells could be detected against a background of other bacteria or nonbacterial particles by combining FCM and specific fluorescently-labeled antibodies or oligonucleotide probes (1, 10, 23, 29). As an early step towards demonstrating the potential application of flow cytometers in milk analyses, we have developed a rapid method for detecting total bacteria.
Milk-clearing treatments and detection of bacteria in milk by using FCM.
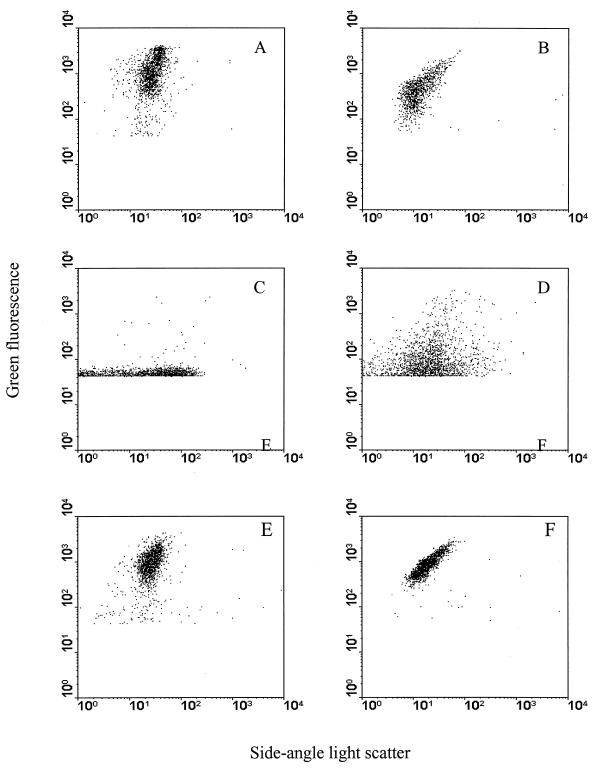
Escherichia coli (XL1-Blue MRF−) and Staphylococcus aureus (NCTC 4163) were chosen to represent gram-negative rods and gram-positive cocci that are potential contaminants of milk (13, 20). Bacteria were grown overnight in Trypticase soy broth (Oxoid, Sydney, Australia) at 28°C on a rotary shaker (180 rpm) for 16 h. Pure populations of E. coli and S. aureus were easily detected by FCM when they were suspended in phosphate-buffered saline (PBS) (Fig. 1A and B), but when they were inoculated into ultra-heat-treated (UHT) milk, no distinct separation appeared (Fig. 1C). This is due to the presence of proteins and lipid globules that can bind nonspecifically to fluorescent stains and interfere with staining and detection of bacteria. Treatment of milk by centrifugation to remove lipids without also treating samples with proteases was insufficient to allow definition of bacteria (Fig. 1D). Thus, the most critical barrier to FCM analysis of milk is the presence of protein globules. Therefore, we applied enzymatic treatment to remove or modify proteins and thereby enable distinction of bacteria by flow cytometry. We used 0.05 mg of proteinase K (EC 3.4.21.64; Sigma-Aldrich, Sydney, Australia) or 10 μl of savinase (EC 3.4.21.52; Novo Nordisk Bioindustrial Pty. Ltd., Sydney, Australia) to treat 100 μl of UHT milk samples and 50 μl of savinase plus 50 μl of 0.1% Triton X-100 to treat 100 μl of raw milk samples. Treated milk samples were incubated at 37°C for 30 to 45 min, after which 900 μl of 150 mM NaCl was added and mixed by inversion of tubes. Samples were then centrifuged at 14,000 × g for 10 min and lipids (top layer) and digested proteins of the milk were drawn off with a micropipette without disturbing the pelleted material, which contained bacteria. The pellet was resuspended in 100 μl of 150 mM NaCl. The samples were then stained and analyzed using FCM (see below for staining protocol and FCM analyses). After the application of proteases in concert with lipid removal, the bacteria in UHT milk appeared in exactly the same positions on bivariate dot plots as bacteria in PBS (Fig. 1A and E and 1B and F). Sorting of putative bacterial populations coupled with subsequent microscopic analysis and plating on selective agar confirmed that the sorted regions represented bacteria inoculated into the UHT milk samples (data not shown).
FIG. 1.
Detection of bacteria in UHT milk by FCM. (A) E. coli in phosphate-buffered saline; (B) S. aureus in phosphate-buffered saline; (C) untreated milk plus E. coli; (D) milk plus E. coli with fat removed; (E) milk plus E. coli with fat removed and with protease treatment; (F) milk plus S. aureus with fat removed and with protease treatment. Results shown are typical of two repeated experiments assayed in triplicate.
Fluorescent staining and FMC.
Bacterial cells in treated milk samples were stained with SYTO BC (Molecular Probes Inc., Bioscientific Pty. Ltd., Sydney, Australia). SYTO BC (excitation and emission maxima of 480 and 500 nm) is a high-affinity nucleic acid stain that penetrates both gram-positive and gram-negative bacteria, giving a bright green fluorescent signal (Molecular Probes [Eugene, Ore.] handbook). SYTO BC, diluted 1:20 in dimethylsulfoxide, was mixed 1:50 with resuspended bacteria and incubated in the dark at 37°C for 5 to 10 min. The viabilities of bacterial cultures were determined by dual staining of the subsamples with propidium iodide (PI) and SYTO BC. A solution of PI (1 mg ml−1) was prepared in dimethyl sulfoxide, and 1 μl of this was added per ml of bacterial suspension. PI is generally excluded by intact plasma membranes; thus, uptake of PI (orange/red fluorescence) indicates cell death (23; Molecular Probes handbook).
Stained samples were analyzed using FACScan (analysis only) or FACSCalibur (cell sorting) flow cytometers (Becton Dickinson, Sydney, Australia). Both were equipped with a 15-mW argon laser emitting light at 488 nm. The sheath fluid was Osmosol (LabAids Pty. Ltd., Sydney, Australia). Both instruments were equipped with forward-angle light scatter (<15°), side-angle light scatter (>15°), and three fluorescence detectors: FL1 (515 to 565 nm), FL2 (565 to 605 nm), and FL3 (>605 nm). The detection threshold was adjusted for FL1 to eliminate particles emitting green fluorescence at a level significantly below that of bacteria suspended in PBS and stained with SYTO BC. Compensation was set so as to remove FL2 fluorescence from the FL1 channel and FL1 from FL2. The settings used routinely for analysis by FACScan FCM of E. coli and S. aureus in UHT milk and for total bacteria in raw milk samples by using the FACSCalibur are given in Table 1. Bacterial counts by FCM were obtained by normalizing the numbers of events occurring in regions on dot plots that defined bacterial populations to the volume of sample analyzed. Data acquired from FCM were converted from Hewlett Packard to PC format by using the computer program HP-Reader (supplied by Becton Dickinson) and analyzed with the computer program Windows Multiple Document Interface FCM application (WinMDI; Joseph Trotter, Salk Institute for Biological Studies, La Jolla, Calif.). For cell sorting, a FACSCalibur flow cytometer was used. Sort regions were defined on bivariate dot plots that delineated distinct populations. Droplets were collected onto 0.02-μm-pore-size filters (supplied by Millipore Pty. Ltd., Sydney, Australia) through a Sort Stage (AusFlow, Sydney, Australia) and examined using epifluorescence microscopy. Sorted samples were also diluted, and 100 μl was spread onto differential agar media to confirm identity and viability (see below).
TABLE 1.
Flow cytometer settings used for defining bacterial populations in milk samplesa
| Sample | Threshold (FL1) | Detector setting
|
% Compensation
|
|||||
|---|---|---|---|---|---|---|---|---|
| FSC | SSC | FL1 | FL2 | FL3 | FL1-FL2 | FL2-FL1 | ||
| E. coli | 416 | E02 | 326 | 770 | 770 | 770 | 88.8 | 24.5 |
| S. aureus | 436 | E02 | 333 | 755 | 763 | 777 | 75.3 | 24.5 |
| Raw milk | 357 | E02 | 505 | 638 | 590 | 646 | 78.1 | 24.7 |
Settings shown for E. coli and S. aureus were used for bacteria in UHT milk or PBS. All gains were logarithmic. FSC, forward-angle light scatter; SSC, side-angle light scatter; FL1, green fluorescence channel; FL2, orange fluorescence channel; FL3, red fluorescence channel (expressed as volts).
Microscopic and plate count methods.
A Carl Zeiss (Sydney, Australia) Axioskop 2 epifluorescence microscope fitted with 10× eyepieces and 40× and 100× (oil immersion) objectives was used to confirm and count cells. Excitation of SYTO BC- or PI-stained cells was by a 100-W Hg vapor arc lamp with an appropriate filter block giving excitation at 450 to 490 nm and examination at 520 nm. Direct microscopic counts of bacterial suspensions were carried out using bright-field microscopy and Thoma counting chamber procedures (5).
Plate count methods were used to determine viable cell numbers. Suspensions of E. coli or S. aureus in milk were serially diluted in PBS, and 100-μl volumes of dilutions were spread plated in triplicate. To differentiate E. coli, Chromocult coliform agar (Merck Pty. Ltd., Sydney, Australia) was used, whereas Baird-Parker agar (Merck Pty. Ltd.) supplemented with egg yolk tellurite emulsion (Oxoid Pty. Ltd., Heidelberg, Victoria, Australia) was used for S. aureus. Colonies were counted after incubation for 24 to 48 h at 37°C. The total viable microbial counts of raw milk were determined by standard plate count using the pour plate method as described in reference 8. Analysis of variance and Student's t test were used to detect significant differences between different methods. Correlations between flow cytometric method and total microscopic counts and plate count methods were calculated with statistical software (SPSS Inc., Chicago, Ill.).
Comparison between FCM, total microscopic count, and plate count methods.
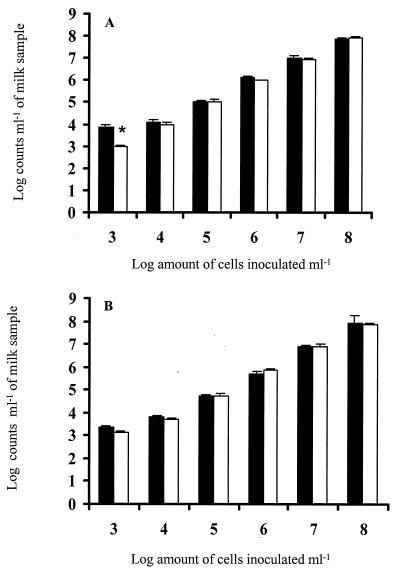
Dual staining of pure bacterial cultures with PI and SYTO BC showed that inocula (16-h bacteria cultures) were >99% viable. UHT milk was inoculated with E. coli and S. aureus cells at different cell concentrations between 103 and 108 ml−1 and analyzed using FCM and traditional techniques such as plate count and direct microscopy. Strong correlations (r = ≥0.98) between FCM and plate counts or direct microscopic counts were obtained for both E. coli and S. aureus (data not shown). E. coli counts measured by FCM in the range of 104 to 108 ml−1 were not significantly (P < 0.05) different from those given by the Chromocult coliform agar plate count method, although FCM estimated a significantly (P < 0.05) greater number of cells at the level of 103 bacteria ml−1 (Fig. 2A). For S. aureus, the numbers measured by FCM were not significantly (P < 0.05) different from those obtained by the Baird-Parker agar plate count methods in the range of 103 to 108 bacteria ml−1 (Fig. 2B).
FIG. 2.
Comparison of bacterial counts given by FCM with plate count methods. Bacteria were inoculated into UHT milk at levels between 103 and 108 ml−1. Data (three replicates) were analyzed using Student's t test (an asterisk denotes significant difference at a P value of <0.05). ■, FCM counts; □, plate counts. (A) E. coli; (B) S. aureus.
Bacterial counts in raw milk.
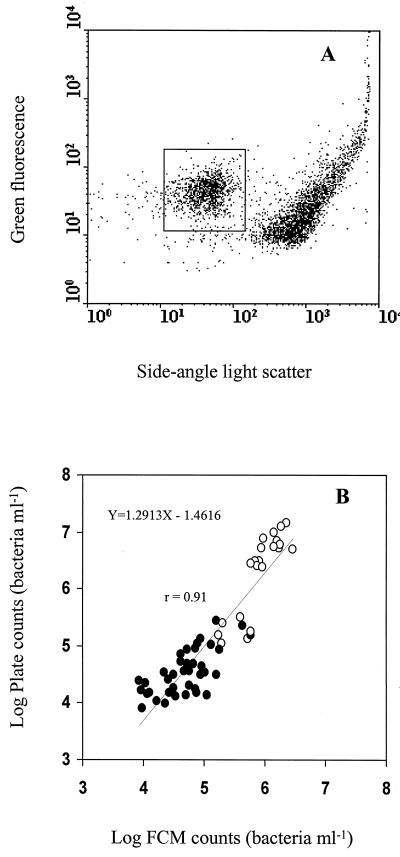
Raw milk was obtained refrigerated fresh from the dairy plant and tested the same day or, where indicated, after 48 h of cold (4°C) storage. Unlike in UHT milk, when proteinase K or savinase was used separately on raw milk, no regions of distinct particle populations could be observed with FCM (data not shown). It is likely that heat-treated proteins that are already partly or fully denatured are more sensitive to proteolysis than native proteins in raw milk. However, following the modified proteolytic treatment (protease and a detergent), a bacterial population could be separated from other milk particles based on light-scattering characteristics (Fig. 3A). Statistical treatment of data showed there was a significant correlation (P < 0.01) and good agreement (r = 0.91) between FCM and the standard plate count method (Fig. 3B).
FIG. 3.
(A) Detection of bacteria in raw milk using FCM. The boxed region defines the bacterial population. (B) Correlation between total bacterial counts for raw milk samples obtained by FCM and standard plate count methods. ●, Bacterial counts of fresh raw milk samples (n = 38); ○, bacterial counts after cold storage (4°C) for 48 h (n = 21). Assays of individual samples were performed in triplicate.
The FCM procedure described here estimates numbers of total bacteria in the processed sample, since SYTO BC binds to live culturable, live nonculturable, and dead cells. On the other hand, plate count numbers represent only culturable cells. Therefore, the plate count method would be expected to produce lower numbers than the FCM method. In both techniques, there is a tendency for bacterial clumps or chains to be enumerated as one unit, resulting in a slight underestimation of total cell counts.
Potential of FCM in milk microbiology.
This study demonstrates the ability of FCM to determine total bacterial numbers after clearing of milk and staining of bacteria with a readily available fluorescent stain. The sensitivity of the FCM procedure was ≤104 total bacteria ml of milk−1. A limit of ≤104 total bacteria ml of raw milk−1 is below the level of detection required to satisfy legislation in many countries and states (14). The assay takes 45 to 60 min depending on whether processed or raw milk is being analyzed. The total FCM analysis time is between 30 s and 2 min, depending on numbers of bacteria in milk. This time frame compares favorably with current culturing methods, which take 72 h (8).
The work reported herein is a first step in developing FCM for use in rapid monitoring of microbiological quality of milk. There have already been some instances in which pathogenic bacteria in milks have been detected by FCM. For example, immunofluorescent labeling techniques have been used to detect Listeria monocytogenes (9) in raw milk and specific Salmonella spp. in raw and processed milks (4, 16, 17, 19). In additional studies, we have found that FCM is suitable for monitoring the growth of bacteria in refrigerated raw, pasteurized, homogenized milk and flavored milk drinks and enumerating viable bacteria (2; T. Gunasekera et al., unpublished results). These combined works indicate the potential breadth of FCM for applications in dairy microbiology. These applications include testing of raw milk for conformation with standards, specifications, and regulatory compliance, monitoring efficiency of manufacturing, cleaning, and sanitation practices, and predicting the shelf life of milk products. The ability to use a single instrument for numerous rapid microbiological assay procedures has obvious advantages for the dairy industry over the current situation, where culturing, microscopy, or several dedicated instruments are needed.
Acknowledgments
We thank Robert Chandler of Dairy Industry Quality Centre, Werribee, Victoria, Australia, for technical advice. We also thank Belinda Chapman and Dan Deere for their input during early stages of this project. We are grateful to Peter Ellis of Perfection Dairies Pty. Ltd., Sydney, Australia, for supplying raw milk samples.
This work was supported by the Australian Dairy Research and Development Corporation in collaboration with Becton Dickinson.
REFERENCES
- 1.Amann R I, Ludwig W, Schleifer K H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Attfield P, Gunasekera T, Boyd A, Deere D, Veal D. Application of flow cytometry to microbiology of food and beverage industries. Australas Biotechnol. 1999;9:159–166. [Google Scholar]
- 3.Celestino E L, Iyer M, Roginski H. The effects of refrigerated storage on the quality of raw milk. Aust J Dairy Technol. 1996;51:59–63. [Google Scholar]
- 4.Clarke R G, Pinder A C. Improved detection of bacteria by flow cytometry using a combination of antibody and viability markers. J Appl Microbiol. 1998;84:577–584. doi: 10.1046/j.1365-2672.1998.00384.x. [DOI] [PubMed] [Google Scholar]
- 5.Cruickshank R, Duguid J P, Marmion B P, Swain R H A. Medical microbiology. 12th ed. Vol. 2. 1975. : the practice of medical microbiology. Churchill Livingstone, Edinburgh, United Kingdom. [Google Scholar]
- 6.Cunningham J D, Saunders C L. Collaborative study of the Bactoscan—an automated method for determinations of total bacteria in raw milk. Can Inst Food Sci Technol J. 1988;21:464–466. [Google Scholar]
- 7.Davey H M, Kell D B. Flow cytometry and cell sorting of heterogeneous microbial populations: the importance of single-cell analyses. Microbiol Rev. 1996;60:641–696. doi: 10.1128/mr.60.4.641-696.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Desmasures N, Gueguen M. Monitoring the microbiology of high quality milk by monthly sampling over 2 years. J Dairy Res. 1997;64:271–280. doi: 10.1017/s0022029996002130. [DOI] [PubMed] [Google Scholar]
- 9.Donnelly C W, Baigent G J. Method for flow cytometric detection of Listeria monocytogenes in milk. Appl Environ Microbiol. 1986;52:689–695. doi: 10.1128/aem.52.4.689-695.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fouchet P, Jayat C, Héchard Y, Ratinaud M-H, Frelat G. Recent advances of flow cytometry in fundamental and applied microbiology. Biol Cell. 1993;78:95–109. doi: 10.1016/0248-4900(93)90120-4. [DOI] [PubMed] [Google Scholar]
- 11.Fung D Y C. Rapid methods and automation in food microbiology: a review. Food Rev Int. 1994;10:357–375. [Google Scholar]
- 12.Griffiths M W. Applications of bioluminescence in the dairy industry. J Dairy Sci. 1993;76:3118–3125. doi: 10.3168/jds.S0022-0302(93)77651-1. [DOI] [PubMed] [Google Scholar]
- 13.Heeschen W H, Reichmuth J. Mastitis—the disease under aspects of milk quality and hygiene. Kiel Milchwirtsch Forschungsber. 1995;47:221–237. [Google Scholar]
- 14.Hubble I B. Testing and reporting of raw milk quality. Austr J Dairy Technol. 1997;52:102–108. [Google Scholar]
- 15.Karwoski M. Automated direct and indirect methods in food microbiology: a literature review. Food Rev Int. 1996;12:155–174. [Google Scholar]
- 16.McClelland R G, Pinder A C. Detection of low levels of specific Salmonella species by fluorescent antibodies and flow cytometry. J Appl Bacteriol. 1994;77:440–447. doi: 10.1111/j.1365-2672.1994.tb03447.x. [DOI] [PubMed] [Google Scholar]
- 17.McClelland R G, Pinder A C. Detection of Salmonella typhimurium in dairy products with flow cytometry and monoclonal antibodies. Appl Environ Microbiol. 1994;60:4255–4262. doi: 10.1128/aem.60.12.4255-4262.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muir D D. The shelf-life of dairy products. 2. Raw milk and fresh products. J Soc Dairy Technol. 1996;49:44–48. [Google Scholar]
- 19.Pinder A C, McClelland R G. Rapid assay for pathogenic Salmonella organisms by immunofluorescence flow cytometry. J Micros. 1994;176:17–22. doi: 10.1111/j.1365-2818.1994.tb03495.x. [DOI] [PubMed] [Google Scholar]
- 20.Porter J, Mobbs K, Hart C A, Saunders J R, Pickup R W, Edwards C. Detection, distribution and probable fate of Escherichia coli 0157 from asymptomatic cattle on a dairy farm. J Appl Microbiol. 1997;83:297–306. doi: 10.1046/j.1365-2672.1997.00230.x. [DOI] [PubMed] [Google Scholar]
- 21.Ravanis S, Lewis M J. Observation on the effect of raw milk quality on the keeping quality of pasteurized milk. Lett Appl Microbiol. 1995;20:164–167. doi: 10.1111/j.1472-765x.1995.tb00417.x. [DOI] [PubMed] [Google Scholar]
- 22.Richter E R. Biosensors: applications for dairy food industry. J Dairy Sci. 1993;76:3114–3117. doi: 10.3168/jds.S0022-0302(93)77650-X. [DOI] [PubMed] [Google Scholar]
- 23.Shapiro H M. Practical flow cytometry. 3rd ed. New York, N.Y: Wiley-Liss; 1995. [Google Scholar]
- 24.Sørhaug T, Stepaniak L. Psychrotrophs and their enzymes in milk and dairy products: quality aspects. Trends Food Sci Technol. 1997;8:35–41. [Google Scholar]
- 25.Suhren G, Walte H-G. First experiences with automatic flow cytometric determination of total bacterial count in raw milk. Kieler Milchwirtschaftliche Forschungsberichte. 1998;50:249–275. [Google Scholar]
- 26.Vasavada P C. Rapid methods and automation in dairy microbiology. J Dairy Sci. 1993;76:3101–3113. doi: 10.3168/jds.S0022-0302(93)77649-3. [DOI] [PubMed] [Google Scholar]
- 27.Vasavada P C, White C H. Symposium: developing methodology for microbial evaluation of milk and dairy products. J Dairy Sci. 1993;76:3099–3100. doi: 10.3168/jds.S0022-0302(93)77648-1. [DOI] [PubMed] [Google Scholar]
- 28.Vesey G, Narai J, Ashbolt N, Williams K, Veal D. Detection of specific microorganisms in environmental samples using flow cytometry. Methods Cell Biol. 1994;42:489–522. doi: 10.1016/s0091-679x(08)61092-4. [DOI] [PubMed] [Google Scholar]
- 29.Wallner G, Steinmetz I, Bitter-Suermann D, Amann R. Combination of rRNA-targeted hybridisation probes and immuno-probes for the identification of bacteria by flow cytometry. Syst Appl Microbiol. 1996;19:569–576. [Google Scholar]
- 30.White C H. Rapid methods for estimation and prediction of shelf-life of milk and dairy products. J Dairy Sci. 1993;76:3126–3132. [Google Scholar]