Abstract
Bacteriophages (phages) are obligate parasites that use host bacterial translation machinery to produce viral proteins. However, some phages have alternative genetic codes with reassigned stop codons that are predicted to be incompatible with bacterial translation systems. We analysed 9422 phage genomes and found that stop-codon recoding has evolved in diverse clades of phages that infect bacteria present in both human and animal gut microbiota. Recoded stop codons are particularly over-represented in phage structural and lysis genes. We propose that recoded stop-codons might function to prevent premature production of late-stage proteins. Stop-codon recoding has evolved several times in closely related lineages, which suggests that adaptive recoding can occur over very short evolutionary timescales.
Introduction
The genetic code is highly conserved and considered to be evolutionarily static1. However, some organisms have alternate genetic codes that reassign one or more codons2. Alternative genetic codes are seen in the nuclear genomes of some ciliates3–5, diplomonads6, green algae7 and yeasts8,9 , as well as genomes of some endosymbionts and mitochondria2. Among bacteria, Mycoplasma10,11 and Spiroplasma12 have reassigned the TGA stop codon to tryptophan (genetic code 4), and members of the Candidate Phyla Radiation (CPR) Gracilibacteria and Absconditabacteria have reassigned the TGA stop codon to glycine13–15 (genetic code 25). Furthermore, a computational screen revealed that some bacterial lineages have also reassigned codons that encode amino acids16.
Alternate genetic codes and genetic code expansion can be beneficial. Programmed incorporation of selenocysteine (the 21st amino acid) into selenoproteins is directed at specific in-frame TGA codons in both prokaryotes17 and eukaryotes18, and pyrrolysine (the 22nd amino acid) is inserted at in-frame TAG codons in some Archaea, where it boosts enzyme activity19,20. In an unusual case, the pathogenic yeast Candida albicans has almost completely reassigned a serine CTG codon to leucine, but still decodes CTG as serine at low levels. This codon-level ambiguity expands the yeast proteome and generates phenotypic diversity, potentially increasing adaptability21.
Some large, uncultivated phages of the gut microbiome - predominantly Lak phages and crAssphages - have recoded the TAG or TGA stop codon22–26 (genetic codes 15 and 4). A 2014 analysis of a single recoded phage genome22 (now classified as a crAssphage25) proposed alternative coding could be a manifestation of phage-host antagonism. In this model, the TAG-recoded crAssphage was hypothesized to infect TGA-recoded bacteria, with both phage and host disrupting translation of the others’ genes. However, recent analyses25 revealed that recoded crAssphages infect standard code hosts (genetic code 11), which has cast doubt on that model. Thus, it remains unclear why some phages have evolved genetic codes that are incompatible with host translation systems. In-frame stop codons should induce phage lethality by preventing translation of full length gene products, motivating our study of phages that employ alternative genetic codes with recoded stop codons.
Here, we carry out an analysis of stop-codon recoding in 9422 phage genomes recovered from human and animal gut metagenomes. We identify diverse lineages of phages with recoded stop codons that are predicted to infect bacteria that use standard code, and use gene and genome level analyses to propose a regulatory role for stop-codon recoding in the phage life cycle.
Results
Genome recoding in gut microbiome phage
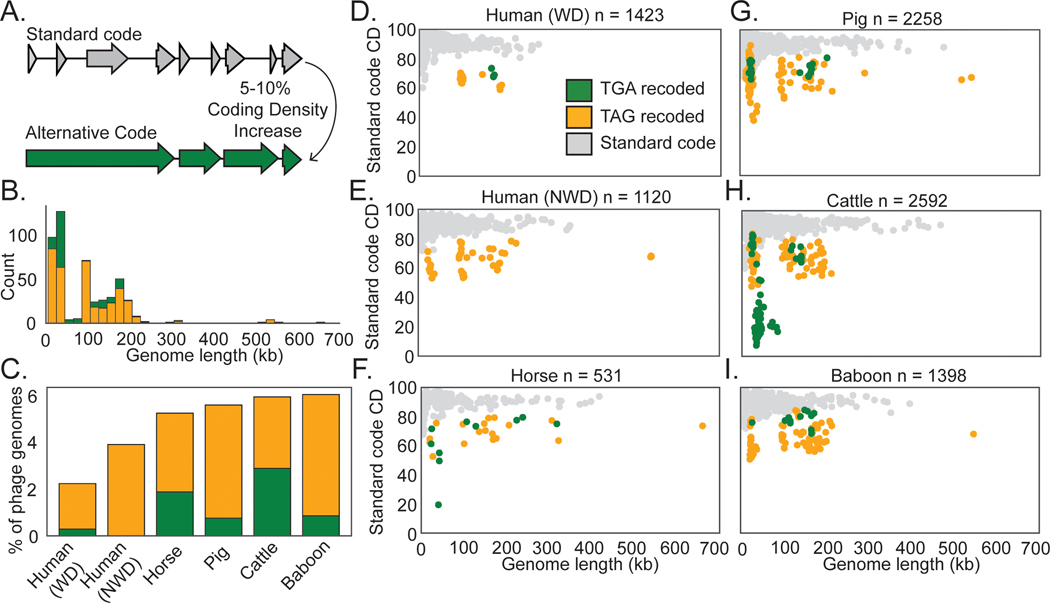
We recovered 9422 complete or nearly complete (≥90% complete) dereplicated phage genomes27 from 726 human and animal gut metagenomes (Supplementary Table 1). To broadly sample phage diversity within the human gut, we analyzed gut microbiomes from individuals inferred to consume westernized28–31 and non-westernized diets23,29,30,32, based on diet and location-related metadata provided in the original studies. To sample phage diversity beyond the human gut, we recovered phages from gut microbiomes of baboons33, pigs34,35, cattle36, horses, and giant tortoises. To identify instances of TAG or TGA stop-codon recoding, we predicted phage genes in standard code (code 11), or alternative genetic codes with TAG or TGA stop codons recoded (code 15 or code 4, respectively) and calculated coding density for each phage genome in each code. As stop codon recoding leads to gene fragmentation in standard code, we identified genomes that underwent a 5–10% coding density increase when genes were predicted with an alternative code (Fig. 1A, Supplementary Information Fig. 1A-C). We then manually verified these putative alternatively coded genomes (Methods), arriving at a final set of 473 recoded double-stranded DNA phage genomes.
Fig. 1: Identification of recoded phage in human and animal microbiomes.
A. A 5–10% coding density increase between standard code and alternative code was used to identify putative recoded phages, followed by manual confirmation of code.
B. Recoded phage genomes spanned a wide size range from 14.7 kilobases (kb) to 660 kb.
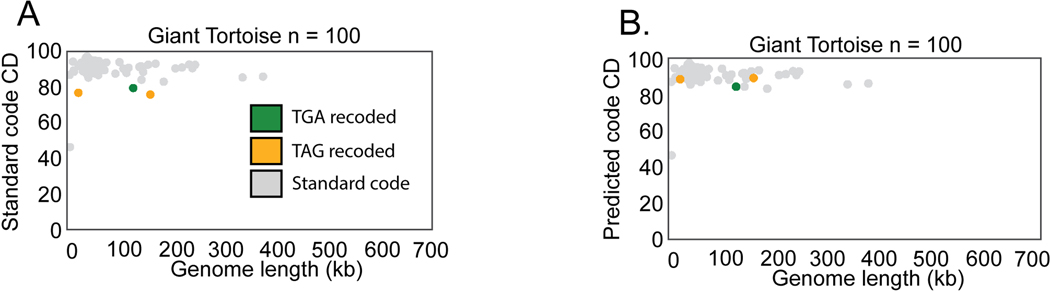
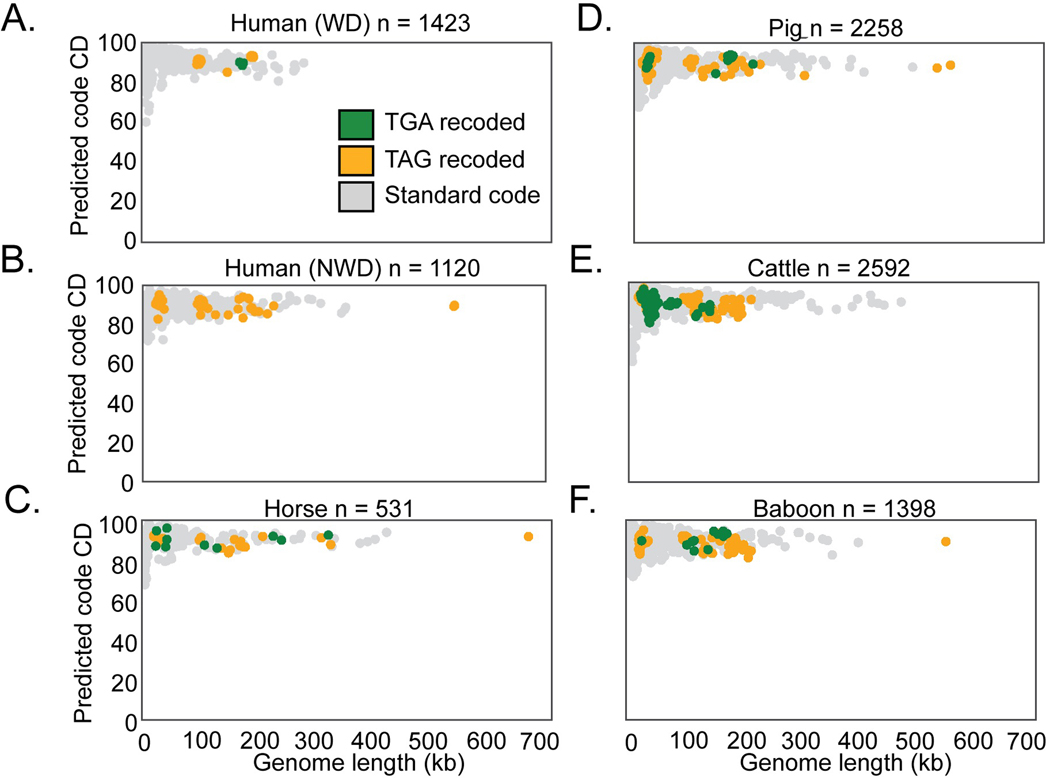
C. Abundance of recoded phages varied from ∼2–6 % of the total phage population in the gut microbiome types surveyed in this study. WD = westernized diet, NWD = non-westernized diet. D-I. Phage genomes recovered from the indicated human or animal microbiome. The number of phage genomes (n) recovered after dereplication from each environment is indicated in the title of each plot. Individual phage genomes are represented by single points and plotted by genome size and coding density (CD) in standard code (code 11). In all plots, phage genomes have been dereplicated and are complete or near complete (>=90%). Symbol color represents genetic code (TGA recoding = green, TAG recoding = orange, standard code = grey). See Extended Data Fig. 1A-F for plots with coding density re-calculated using the predicted genetic code.
Previously stop codon recoding had only been found in phages with large genomes: crAssphages22,25 (95–190kb), jumbophages24(200–500 kb), and megaphages23,26 (>500kb to 660 kb). We identified complete recoded phage genomes across a very wide diversity of sizes, ranging down to 14.7 kb (Fig. 1B). We observed that TAG recoding is more common than TGA recoding (75% TAG recoded, 25% TGA recoded, Fig. 1B,C). While each gut microbiome type has recoded phages present, recoding was least common in phages recovered from humans inferred to consume a westernized diet, and was most common in baboon phages (Fig. 1C) . We conclude that alternative coding is a common feature of phage populations in the human and animal gut, and occurs in phages of diverse genome sizes (Fig. 1D-I, Extended Data Fig. 1A).
Diversity and evolution of recoded phages
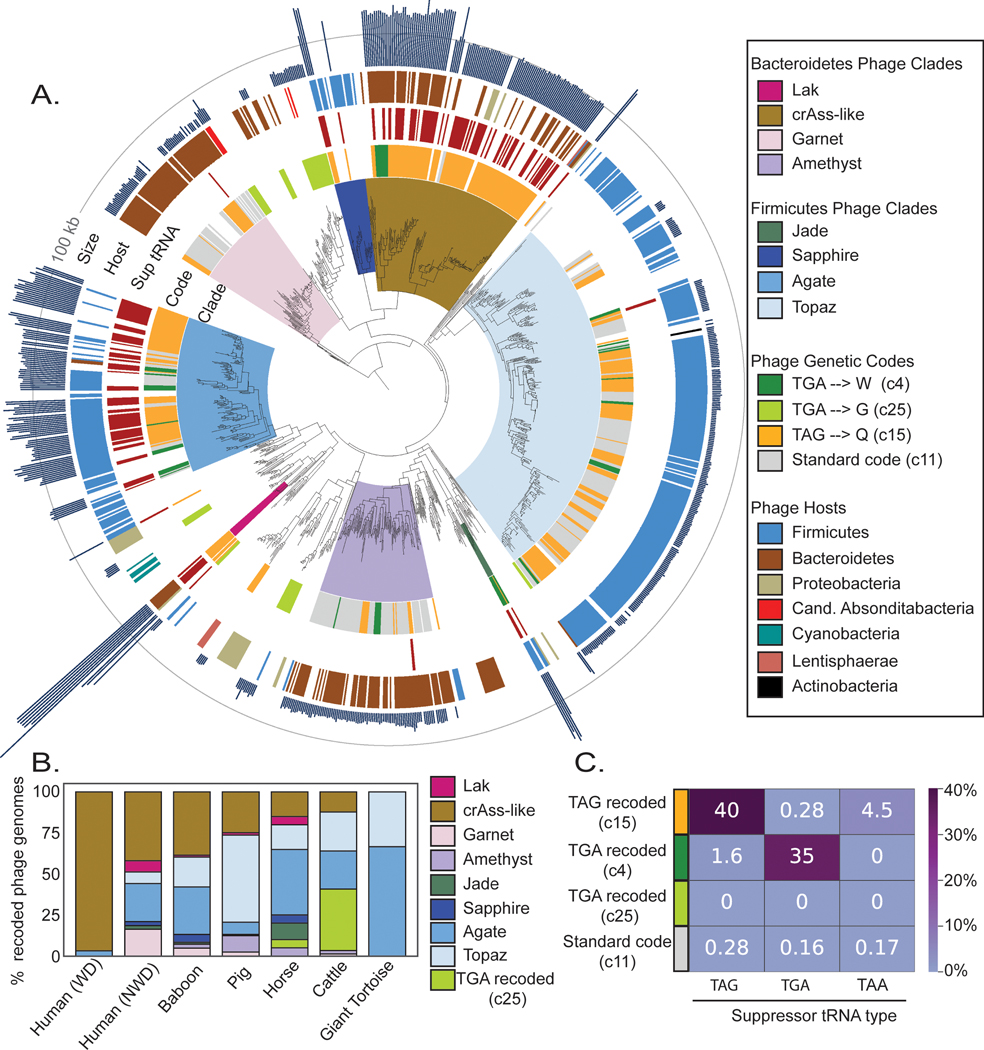
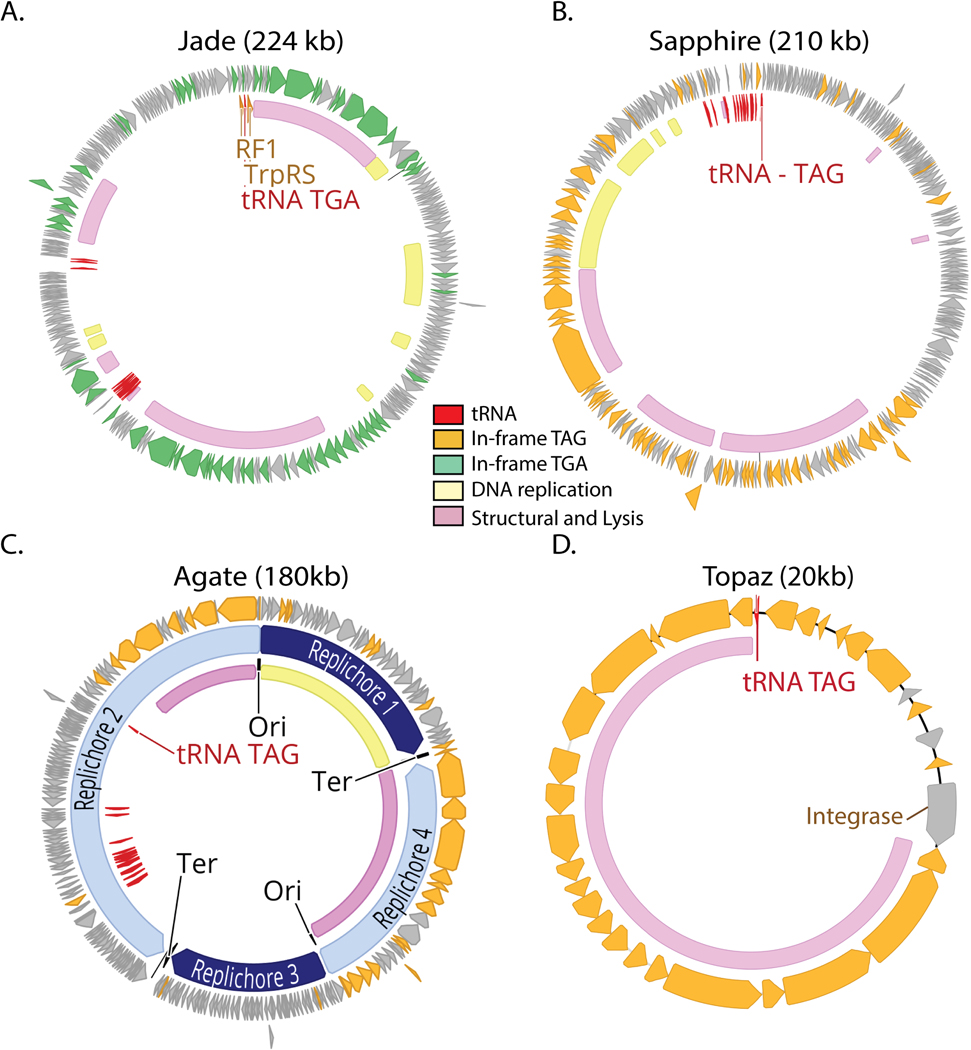
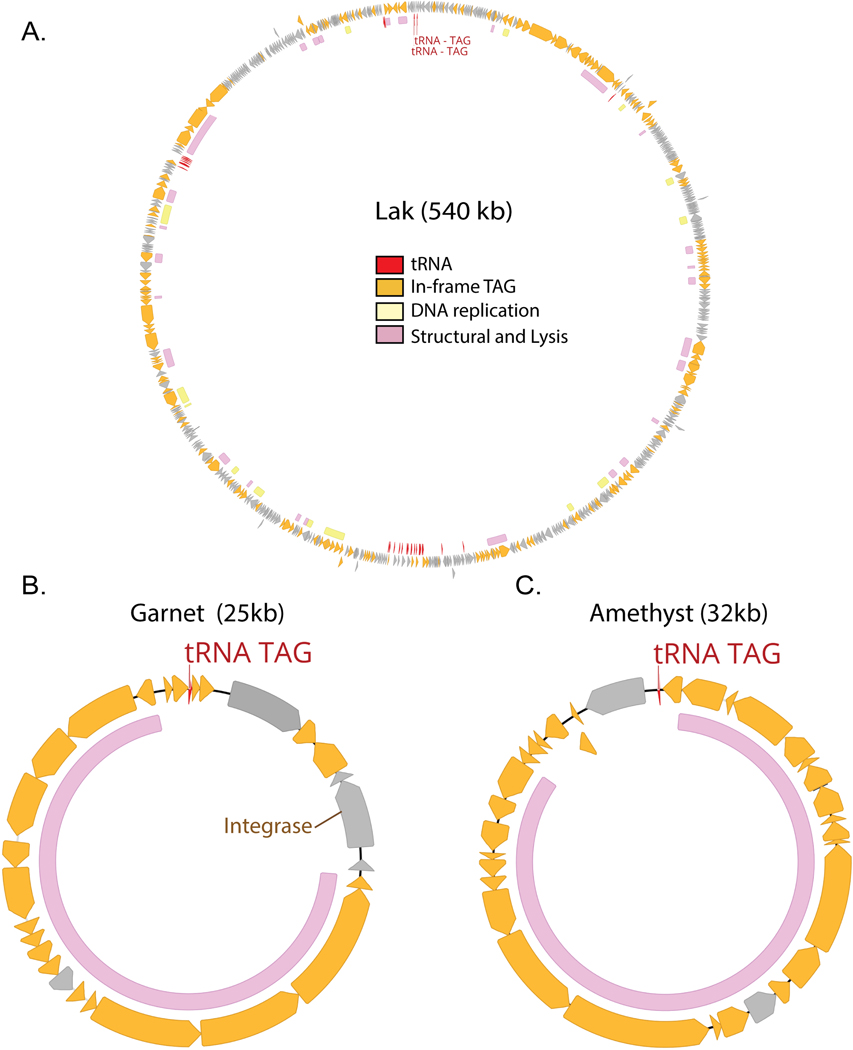
We constructed a phylogenetic tree of large terminase subunits from recoded phages and their standard code relatives, finding many sequences form clades with high bootstrap support (≥95%)(Fig. 2A). Inspired by the historical designation of TAG and TGA as the amber and opal stop codons, we chose to name the six newly discovered clades of TAG and TGA recoded phages after other gemstones (Garnet, Amethyst, Jade, Sapphire, Agate, Topaz). These clade designations are not intended as taxonomic names. Including previously discovered Lak23 and crAss-like families22,25, we describe 8 independent phage clades that use recoded stop codons in human and animal gut microbiomes (see Supplementary Table 2 for clade-level data, Supplementary Table 3 for genome-level data).
Fig. 2: Phylogeny of recoded phages and suppressor tRNA usage.
A. The phylogeny of recoded phages was reconstructed using large terminase sequences from a dereplicated set of complete or near-complete (>=90%) recoded phages (n=444) and their close standard code relatives (n =258), as well as related proteins from Refseq r205 (n=410). Terminase sequences from eukaryotic herpesviruses (n=8) were used to root the tree. The inner to outer ring shows phage clade (>= 95% bootstrap support), genetic code for phages from this study, suppressor tRNA presence, host phylum as predicted by taxonomic profiling and CRISPR spacer matches, and genome size with a grey line at 100 kilobases (kb) for scale. Genetic code, suppressor tRNA presence, and genome size were not included for Refseq proteins since some proteins were derived from prophages and/or incomplete phage genomes.
B. Distribution of recoded phages by clade across the 7 types of gut microbiomes evaluated in this study. WD = westernized diet, NWD = non-westernized diet.
C. Heatmap of the percent of genomes of each genetic code that have tRNAs predicted to suppress translation termination at the TAG, TGA, or TAA stop codons.
To identify the genetic codes used by the recovered recoded genomes, we analyzed the alignments of terminase sequences with in-frame recoded stop codons translated to X, and found that in most cases TGA aligned with tryptophan (genetic code 4) and TAG aligned with glutamine (genetic code 15). Use of genetic code 15 has since been confirmed in crAss-like phages via metaproteomics of human samples37. Many clades encompassed multiple genetic codes (Fig. 2A, Supplementary Table 2). Some recoded phages used code 25 , where TGA is reassigned to glycine. We predict these phages infect Candidatus Absconditabacteria, which also uses code 2514(Supplementary Table 3). In all other cases, the recoded phage clades are predicted to infect bacteria from common standard code gut phyla, Firmicutes and Bacteroidetes (Supplementary Table 2, Supplementary Table 3).
Lak, crAss, Jade, Sapphire and Agate phages have larger than average genome length ( 562 ± 44kb, 210 ± 35 kb, 201 ± 22 kb, 154 ± 27 kb, ± SD) and Garnet, Amethyst, and Topaz phages have smaller than average genomes (34 ± 5kb, 34 ± 6kb, 22 ± 3 kb, ± SD) (Supplementary Table 2). These eight clades have uneven distributions across the environments analyzed here (Fig. 2B). Notably, the recoded phage present in westernized-diet microbiomes mainly comprises crAss-like phages, whereas other gut microbiomes have higher diversity of recoded phages (Fig. 2B).
Mechanisms of phage recoding
In bacteria that use the standard genetic code, the TAG, TGA, and TAA stop codons are recognized by specific release factors (RF1 or RF2), which trigger translation termination. Suppressor tRNAs recognize TAG, TGA, or TAA codons, and have been previously identified in recoded phage genomes22–26, where they presumably mediate code-change. We predicted tRNAs in all phage genomes, and calculated the frequency at which genomes of each code encoded suppressor tRNAs for the TAG, TGA, or TAA stop codons. We found a strong relationship between stop codon recoding and suppressor tRNA usage, detecting TAG suppressor tRNAs in 40% of TAG recoded genomes, and TGA suppressor tRNAs in 35% of TGA recoded (code 4) genomes (Fig. 2C, Supplementary Table 4) Surprisingly, when we analyzed suppressor tRNA occurence across phage phylogeny, we found that suppressor tRNAs were strongly partitioned between phage clades (Fig. 2A). Specifically, almost all the suppressor tRNAs detected were found in the Lak, crAss-like, Sapphire, and Agate phages which have large genomes. In contrast, the small-genome Garnet, Amethyst, and Topaz clades rarely encoded suppressor tRNAs.
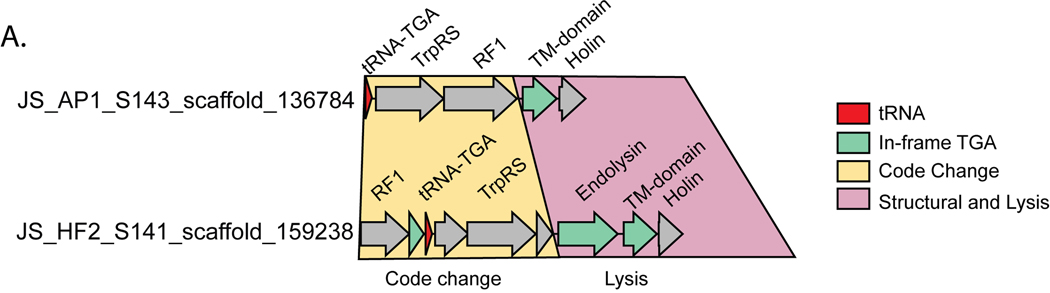
We also searched for phage-encoded release factors (RF), which terminate translation at “true” stop codons and have been previously observed in Lak23 and crAss-like22 phages. We identified RF2 (terminates translation at TAA and TGA) in six TAG recoded Lak phages and two TAG recoded Agate phages (Supplementary Table 5). RF1, which terminates translation at TAG and TAA stop codons was identified in two TGA recoded Jade phages (Supplementary Table 5). We also identified tryptophanyl tRNA synthetases in the same two Jade genomes (Supplementary Table 5), which we predict could ligate the amino acid tryptophan to the TGA suppressor tRNA, thus mediating the TGA → W code change.
Relationships between recoded and standard-code phages
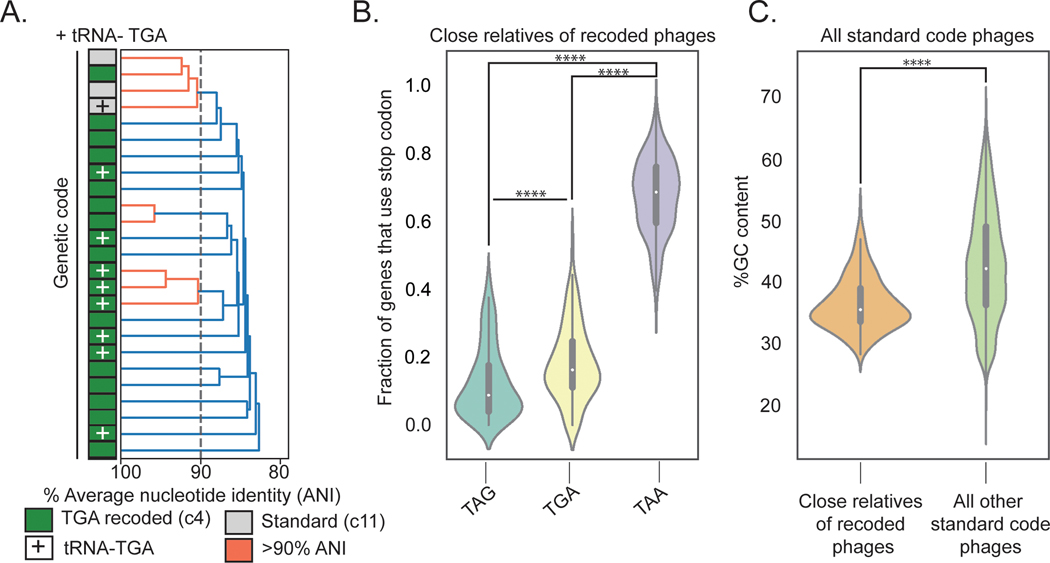
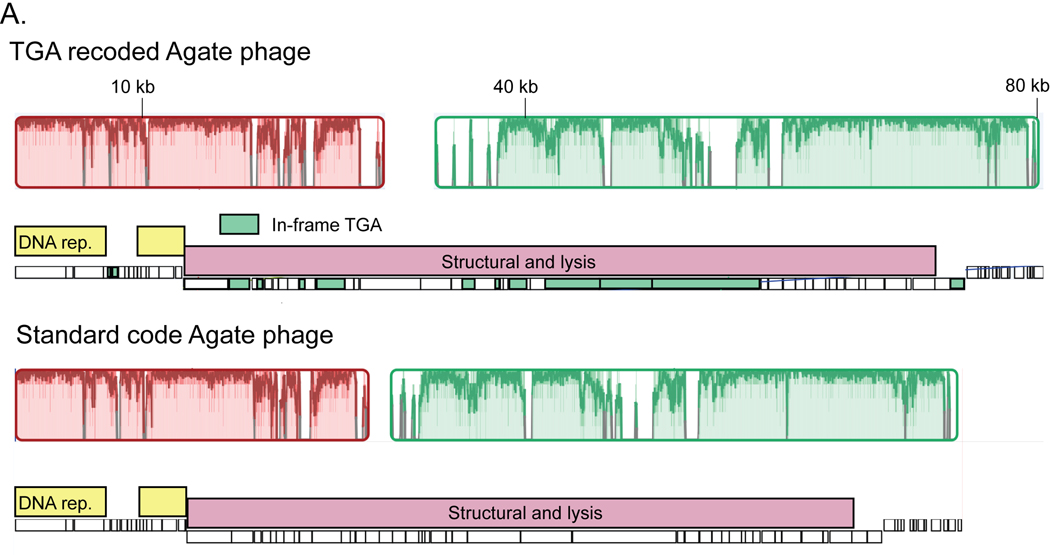
We next calculated the average nucleotide identity (ANI) between all phage in our dataset to identify examples of very closely related genomes that use different genetic codes. We identified a set of Agate clade genomes with greater than 80% ANI that includes TGA-recoded code 4 genomes and standard code genomes (Fig. 3A). One standard code phage had acquired a TGA suppressor tRNA (+), potentially preceding the code change (Fig. 3A). We also found an example where a TGA recoded Agate phage Cattle_ERR2019405_scaffold_1063 and a standard code Agate phage pig_ID_3053_F60_scaffold_12 share greater than 90% ANI (Fig. 3A, Extended Data Fig. 3A). This indicates that genetic code can change over short evolutionary timescales.
Fig. 3: Evolutionary relationships among phages according to genetic code.
A. Dendrogram of average nucleotide identity (ANI) across a set of Agate phage genomes. Standard code (grey) and TGA recoded (green) phages share >80% ANI, and one cluster of >90% ANI genomes (orange) has both standard and TGA recoded genomes, indicating an extremely close evolutionary relationship.
B. Close relatives of recoded phages (n = 260 biologically independent phage genomes) use the TAA stop codon codon at a higher rate than the TAG and TGA stop codons (TAG vs. TAA: Z = −19.71, p = 1.63e-86, TGA vs. TAA: Z = −19.65, p = 5.98e-86, two-sided Wilcoxon Rank-Sum Test). The TAG stop codon is depleted relative to TGA (Z = −6.43, p = 1.24e-10, two-sided Wilcoxon Rank-Sum Test) in these phages. TAG frequencies: Minima = 0.0, Maxima = 0.38, Median = 0.09, IQR = 0.14, Q1 = 0.04 Q3 = 0.18. TGA frequencies: Minima = 0.0, Maxima = 0.44, Median = 0.16, IQR = 0.13, Q1 = 0.11, Q3 = 0.25. TAA frequencies: Minima = 0.40, Maxima = 0.94, Median = 0.68, IQR = 0.16, Q1 = 0.59, Q3 = 0.76.
C. Close relatives of alternatively coded phages have a lower mean GC content relative to all other standard code phages (Z = −12.59, p = 2.33e-36, two-sided Wilcoxon Rank-Sum Test). Close relatives: n = 260 biologically independent phage genomes, Minima = 27.97, Maxima = 45.73, Median = 35.10, IQR = 5.30, Q1 = 33.27, Q3 = 38.57. Unrelated standard code phages: n = 8689 biologically independent phage genomes, Minima = 19.60, Maxima = 67.07, Median = 41.827, IQR = 12.67, Q1 = 35.95, Q3 = 48.62.
**** p ≤ 0.0001, two-sided Wilcoxon Rank-Sum test.
The Agate phage pig_ID_3053_F60_scaffold_12 uses standard code and only 4 out of 146 genes (2.7%) use TGA stop codons. In contrast, 34 genes use TAG and 108 genes use TAA. This suggests divestment in TGA as a stop codon may precede its reassignment in Agate phages, consistent with the codon capture hypothesis of genetic code evolution38. To test for TAG or TGA stop codon loss at a wider scale, we surveyed stop codon usage across all standard code phages that are closely related to recoded phages. These relatives are more likely to use the TAA stop codon than the TAG and TGA stop codons (Fig. 3B, TAG vs. TAA: Z = −19.71, p = 1.63e-86, TGA vs. TAA: Z = −19.65, p = 5.98e-86, two-sided Wilcoxon Rank-Sum Test). We also observed that TAG is rarer than TGA (Fig. 3B, Z = −6.43, p = 1.24e-10, two-sided Wilcoxon Rank-Sum Test). This depletion of TAG and TGA is likely due to the reduced GC content (Fig. 3C, Z = −12.59, p = 2.33e-36, two-sided Wilcoxon Rank-Sum Test) in these phages compared to standard code phages that are not close relatives of recoded phages. Thus, stop codon loss driven by low GC content may be an evolutionary precursor to stop codon recoding.
Recoding may regulate cell lysis
We analyzed functional predictions for genes with in-frame recoded stop codons in the genomes of representatives of each recoded phage clade (Fig. 4A-B, Extended Data Fig. 4A-D, Extended Data Fig. 5A-C). Consistent with previous observations, we saw that both Lak23 and crAss-like22,25 genomes use alternative code for their “late” structural and lysis genes. Furthermore, we observed that Garnet, Amethyst, Jade, Sapphire, Agate, and Topaz phages also use alternative code for structural and lysis genes. In contrast, use of alternative code was variable in the DNA replication machinery. In crAss-like, Garnet, Amethyst, and Topaz phages, all the structural and lysis genes are encoded together a single alternatively-coded genomic unit (Fig. 4A, Extended Data Fig. 4D, Extended Data Fig. 5B-C). In Jade, Sapphire, Agate, and Lak phages, the structural and lysis genes are in multiple alternatively-coded modules that are spread across the genome (Fig. 4B, Extended Data Fig. 4A-C, Extended Data Fig. 5A). As structural and lysis proteins encoded with recoded stop codons cannot be expressed before the code change is manifested, stop codon recoding could effectively regulate the timing of protein expression from related gene modules.
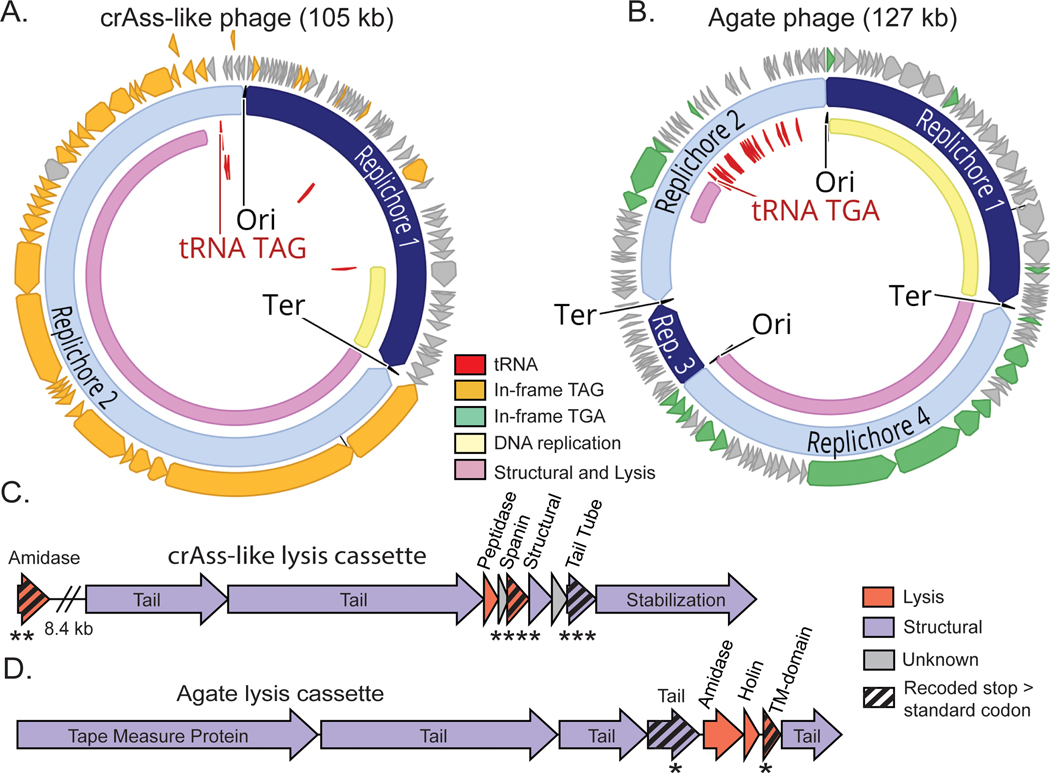
Fig. 4: Preferential recoding of lysis-related genes in recoded phages.
A-B. Genomic maps of manually-curated representatives of crAss-like phages (js4906–23-2_S13_scaffold_20) and Agate phages (GiantTortoise_AD_1_scaffold_344). TAG recoded genomes (A) contain genes with in-frame TAG codons (orange) while TGA recoded genomes (B) have genes with in-frame TGA codons (green). Suppressor tRNAs (red labels) are predicted to suppress translation termination at recoded stop codons. Regions of the genome encoding structural and lysis genes (pink) coincide with high use of alternative code. In these phages, DNA replication machinery (yellow) is encoded in standard code. Origins and termini were identified based on GC skew patterns indicative of bidirectional replication, and unique replichores are marked in alternating shades of blue.
C-D. Genomic maps of highly-recoded lysis cassette neighborhoods from representative TAG-recoded crAss-like phages (C) and TGA-recoded Agate phages (D). Lysis genes (pink) as well as structural genes (purple) that were significantly biased towards use of in-frame recoded stop codons are marked with black striping. In crAss-like phage (C), lytic amidase ( p = 6.82e-3), spanin (p=9.00e-6) and tail tube (p = 7.59e-4) gene families preferentially used TAG to encode glutamine (Q) . In Agate phages, a tail gene family (p = 4.18e-2) and a transmembrane domain protein family (TM-domain, p = 4.64e-2) preferentially use TGA to encode tryptophan (W). **** p ≤ 0.0001,*** p ≤ 0.001, ** p ≤ 0.01, * p ≤ 0.05, Benjamini-Hochberg p-value corrected two-sided Wilcoxon Rank-Sum Test. This statistical test was used to analyze rates of TAG use relative to standard code encoding of glutamine (TAG →Q recoded phage in C) or TGA use relative to the standard code encoding of tryptophan (TGA →W recoded phage in D).
We next identified the gene families most biased towards use of recoded stop codons, as they would be most impacted by this proposed form of gene regulation. We measured the codon preference in two phage types that were represented by a sufficiently large set of related genomes to enable gene-level statistics: ∼105 kb TAG-recoded crAss-like phages and ∼127 kb TGA-recoded Agate phages. While many genes in these phages have at least one in-frame recoded stop codon (Fig. 4A-B), only a few gene families preferentially use recoded stop codons over standard code encodings of glutamine (crAss-like phages, TAG → Q) or tryptophan (Agate phages, TGA → W).
In the crAss-like phage genomes analyzed, only four gene families preferentially use TAG over CAG or CAA to encode Q (two-sided Wilcoxon Rank-Sum Test, corrected for multiple comparisons) (Fig. 4C). Two of these four families are essential components of the lysis cassette: a lysozyme type amidase (Z = 2.91, p = 6.82e-3) and a spanin, which is a critical regulator of lysis of gram-negative bacteria39,40 (Z = 4.82, p=9.00e-6). A tail tube gene family that is encoded two genes downstream (1.1 kb) of the spanin gene is also preferentially recoded (Z = 3.56, p = 7.59e-4). Having multiple strongly alternatively coded genes in the same transcript may amplify the stop codon mediated translation block. The fourth recoded gene family is of unknown function.
In the Agate genomes analyzed, three gene families preferentially use TGA instead of TGG to encode W (two-sided Wilcoxon Rank-Sum Test, corrected for multiple comparisons) (Fig. 4D). One of these is a group I intron endonuclease (Z = 3.88, p = 1.32e-3) inserted in the DNA replication module, which predominately uses standard code. This self-splicing intron is expected to excise itself from the mRNA, but then in-frame recoded stop codons should prevent homing endonuclease production until late in the infection cycle. A tail gene directly upstream of the lysis cassette (Z = 2.69, p = 4.18e-2) is also preferentially recoded, analogous to the tail tube gene in the crAss-like phages. A preferentially recoded transmembrane domain protein (Z = 2.63, p = 4.64e-2) in the lysis cassette belongs to a family of transmembrane proteins that are assigned various lysis and lysis regulation related functions (holin, spanin, lysis regulatory protein, and ATP synthetase B chain precursor). We hypothesize a putative role for this protein in controlling lysis, potentially by depolarizing the cell membrane41–44.
We also identified a “code change module” composed of a suppressor tRNA, a tRNA synthetase, and a release factor directly upstream of the lysis cassette in Jade phages (Extended Data Table 2, Extended Data Fig. 6A-B). These code-change related genes are all encoded in standard code, whereas the lysis genes directly downstream use alternative code. We anticipate that expression of these code change genes would drive expression of the lysis program. Overall, we propose that by changing the genetic code of the infected cell over time, these phages can use stop codon recoding to coordinate protein expression from related late genes and also to suppress misexpression of critical lytic gene products.
Is recoding in prophages a lysogeny switch?
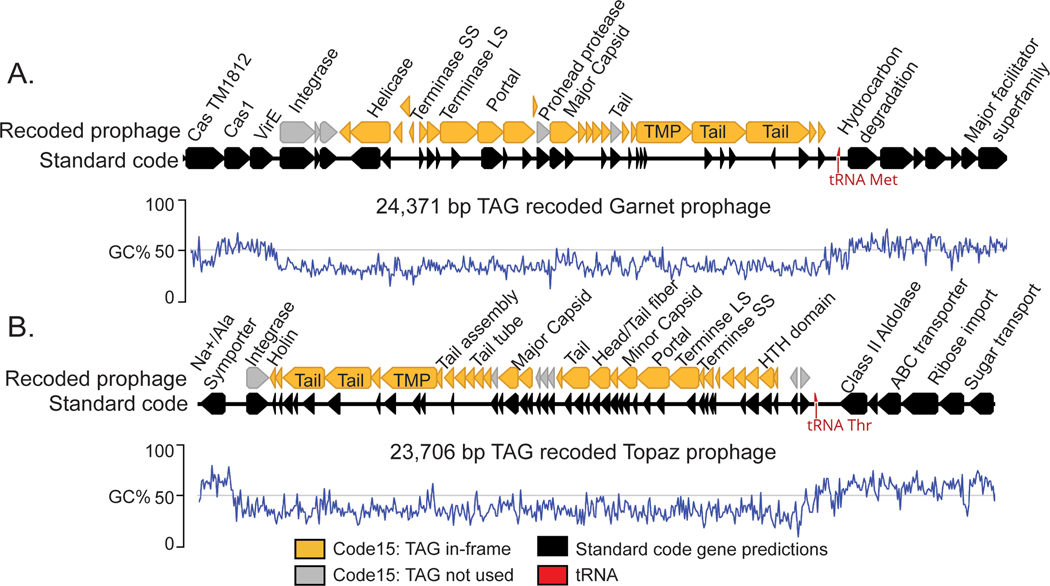
Many phages integrate into their bacterial host chromosome as prophages. Excitingly, we found recoded Garnet and Topaz prophages integrated into standard code bacterial contigs, two of which we analyzed in depth (Fig. 5A-B).
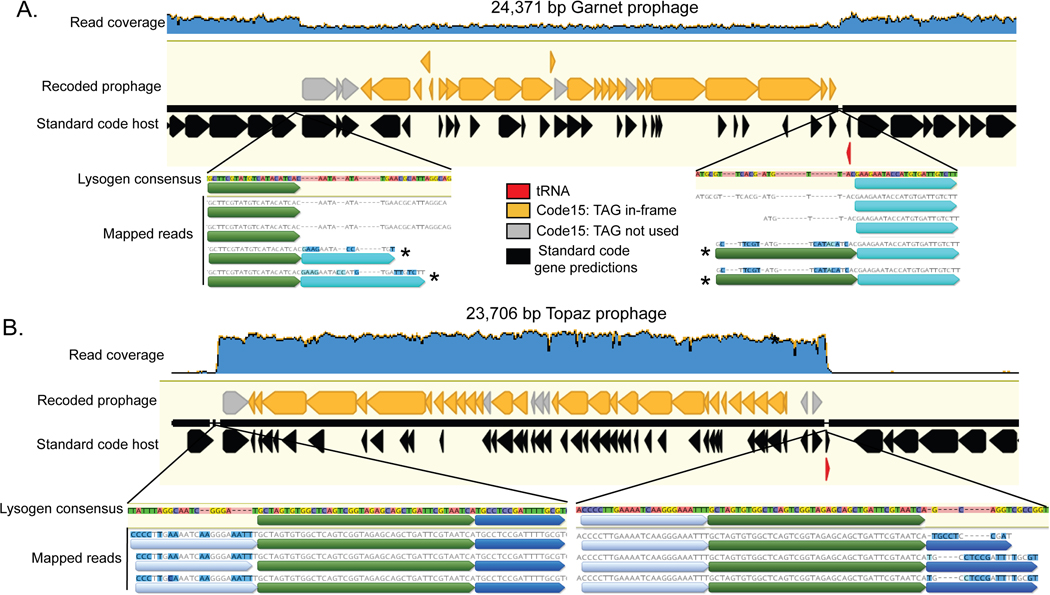
Fig. 5: Recoded prophages integrated into bacterial genomes.
A. A manually curated 24,371 bp TAG-recoded Garnet prophage integrated in a Prevotella sp. genome.
B. A manually curated 23,706 bp TAG-recoded Topaz prophage integrated in a Oscillospiraceae sp. genome. In both A and B, the bacterial hosts use standard code (black gene predictions). Standard code results in highly fragmented gene predictions in the prophages, due to the high number of genes with in-frame TAGs (orange). In both A and B, the integrase is one of the few prophage genes that does not have in-frame TAG codons (grey). An increase in GC content (blue line) and transition from phage to bacterial gene content marks prophage boundaries. LS = Large subunit, SS = Small Subunit, TMP = Tape Measure Protein.
The Garnet prophage is part of a 94 kb Prevotella contig (SRR1747048_scaffold_47) assembled from a baboon metagenome. (Fig. 5A). When we mapped reads to this prophage we observed that the sequencing read depth of the bacterial region was twice that of the integrated prophage (Extended Data Fig. 7A). Some reads spanned the prophage, corresponding to Prevotella genomes that lack the integrated prophage. Thus, the exact prophage 24,371 bp genome could be defined.
The Topaz genome is part of a 36.9 kb Oscillospiraceae contig (SRR1747065_scaffold_956) assembled from a baboon metagenome (Fig. 5B). In this case, sequencing reads coverage over the prophage region is ∼50 times higher than the flanking genome (Extended Data Fig. 7B). We infer the vast majority of phages in the sample were replicating and only a subset remained integrated at the time of sampling. Based on the sequence margins, we determined that the length of this prophage genome is 23,706 bp.
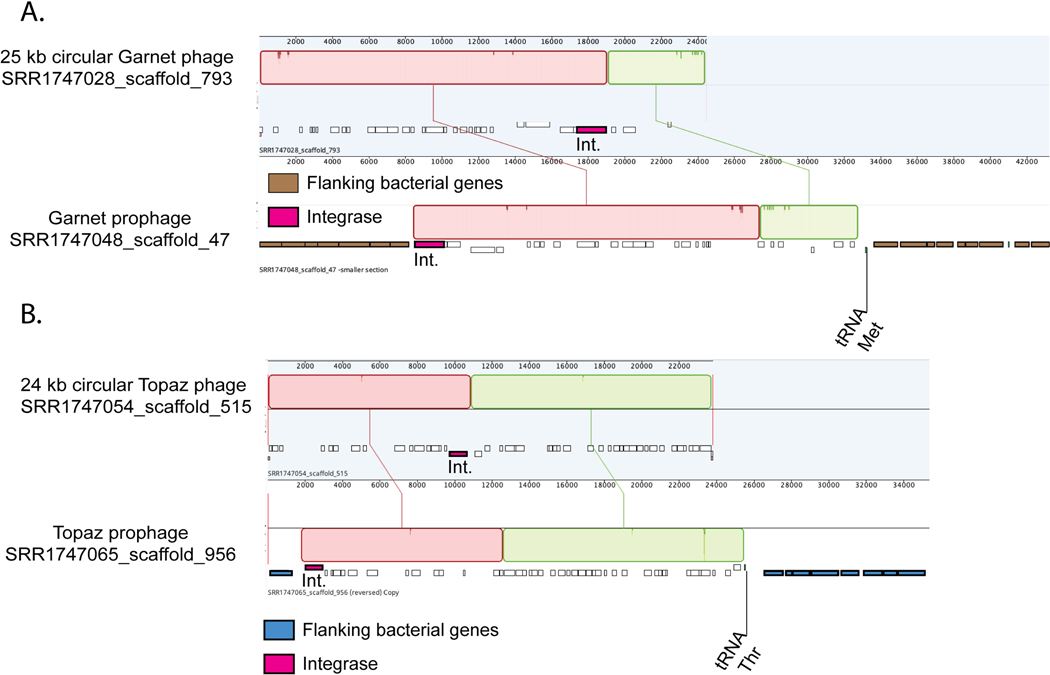
We also identified circular free phage genomes in related baboon samples that were nearly 100% identical to the Garnet and Topaz prophages analyzed here. This supports our conclusion that these prophages represent actively-replicating viable phages (Extended Data Fig. 8A-B) and verifies the lengths determined from the read mapping analysis.
We noticed that while almost all of the prophage genes were extremely fragmented in standard code, the integrase genes did not contain recoded stop codons (Fig. 5A-B, Extended Data Fig. 4D, Extended Data Fig. 5B ). When we measured codon preference across all gene families encoded by alternatively coded Garnet and Topaz phages, we found the integrase gene families strongly avoided use of recoded stop codons. (Garnet: Z = −3.97, p=5.12e-3, Topaz: Z = −8.87, p = 1.23e-16, two-sided Wilcoxon Rank-Sum Test, corrected for multiple comparisons). We hypothesize that these phages are using stop codon recoding as a regulator of the lytic-lysogenic switch. In this scenario, the standard code translation environment of the host promotes expression of the integrase and establishment of lysogeny, with strong suppression of lytic genes. Likewise, a switch to alternative code would promote expression of lysis-related proteins during initial infection or prophage induction. Thus, genetic code may function as a mechanism to partition two distinct arms of the phage life cycle.
Discussion
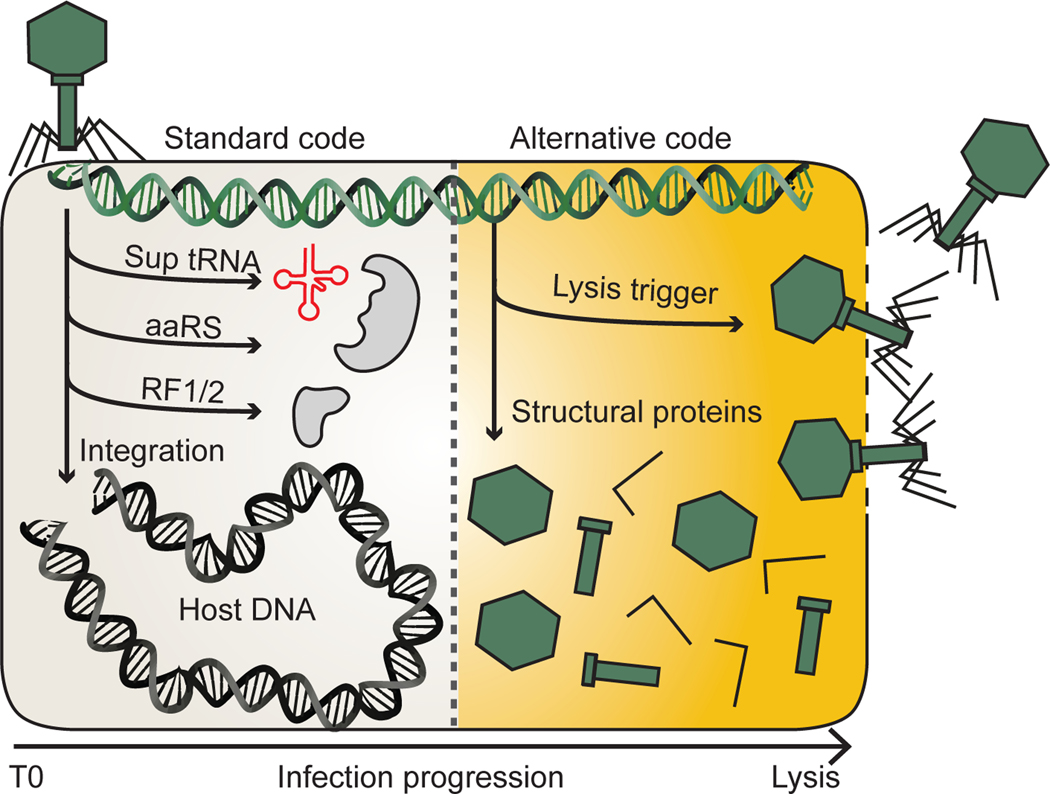
Using a computational analysis, we detected widespread use of recoded stop codons in eight families of phage and prophage present in human and animal gut microbiomes. We hypothesize that evolution of alternate code involves ancestral depletion of TAG and TGA stop codons, and propose a model in which stop codon recoding is a post-transcriptional regulator of protein expression in phages and prophages (Fig. 6).
Fig. 6: A model for recoding in the phage life cycle.
Infection of a standard code host begins with the production of proteins from standard code compatible genes. In some phage, this is a route to integrase production and establishment of lysogeny. In other phage, this early phase involves the production of molecules involved in switching from standard to alternative code such as suppressor tRNAs (Sup tRNA), amino acyl tRNA synthetases (aaRS) and release factors (RF1/2). As infection proceeds, recoded gene products initially suppressed by in-frame recoded stop codons code can be produced. This allows for expression of phage structural proteins and ultimately triggers lysis.
We propose an evolutionary route to recoding that begins with depletion of TAG or TGA stop codons in standard code phages with low GC content. Via acquisition of a suppressor tRNA, in-frame stop codons can accumulate in positions that would previously have been lethal for the phage. We identified TGA suppressor tRNA acquisition by standard code close relatives of TGA recoded phages, which supports this model. We also found that TAG stop codons are more rare than TGA stop codons in standard code relatives of recoded phages, potentially explaining the higher prevalence of TAG recoding compared with TGA recoding.
After in-frame stop codons are “detoxified” by the acquisition of suppressor mechanisms such as tRNAs, selection enriches or depletes recoded stop codons across specific gene families to create patterns of codon use that can be harnessed as a form of gene regulation. Clades of recoded phages have independently converged upon using recoded stop codons to encode lysis and structural proteins. This is consistent with more limited observations of structural gene recoding seen in Lak23 and crAsslike22,25 phages, and supports a model where the genetic code of the infected cell changes throughout the phage infection cycle. Dynamic codon use throughout the infection cycle has been demonstrated in T4-like phages that encode large tRNA arrays, where late-expressed genes have codon use aligned with the phage tRNA repertoire45,46. This may represent a mechanism to toggle translation efficiency of late genes throughout the phage life cycle.
Stop codons have low to no translation efficiency, so we hypothesize that use of recoded stop codons in late expressed genes is an extreme form of codon based regulation in phages. We found two distinct lineages of phages with preferential recoding of the lysis cassette, for which precisely timed expression is crucial. Premature lysis aborts the phage life cycle and limits phage production, and some anti-phage immune systems even exploit this by forcing early lysis44,47. By encoding lysis regulators with in-frame recoded stop codons, these phages block both accidental or host-forced premature expression of these proteins.
We also identified prophages with recoded stop codons that were integrated into standard code hosts. The decision to enter lytic growth or lysogeny is a crucial point in the temperate phage life cycle, and phages have evolved elaborate regulatory mechanisms to precisely control this decision48–50. We hypothesize that alternate coding may function in the lysis-lysogeny switch in these recoded temperate phages.
Most suppressor tRNAs identified here were encoded by phages with large genomes, consistent with previous reports that tRNAs number increases with phage genome size24,51. However, we predict that all recoded phages that infect standard code hosts would require suppressor tRNAs to decode recoded stop codons. One possibility is that small phages “piggy-back” on large phages of the same code, to use larger-phage suppressor tRNAs during coinfection. Some huge phages have been shown to carry CRISPR-Cas systems that target small phages24, consistent with the hypothesis that small phages may parasitize large phages.
Stop codon recoding could allow phages of any size to sense the presence of co-resident phages that use the same genetic code via activity of translation-related molecules such as suppressor tRNAs. This would be beneficial to prophages that are induced in response to a superinfecting lytic phage, or for coinfecting phages to coordinate the timing of their lytic program.
Conclusion
Stop codon recoding may have an important but previously unappreciated role in the phage life cycle. Further, understanding alternative genetic code use in phage is crucial to our ability to detect and classify phage sequences. Broadening our view of genetic code diversity in phages has the potential to augment our understanding of basic phage biology and bacterial translation, as well as improving synthetic biology strategies to design new genetic codes.
Online Methods
Phage prediction
Phage prediction tools Seeker52 (predict-metagenome) and VIBRANT53 were run on assembled metagenomes (contigs > 5kb) using default settings. CheckV54 (end-to-end) was run on predicted phages and trimmed proviruses to evaluate completeness and quality. Contigs evaluated as low quality by both CheckV and VIBRANT were removed from analysis. Contigs < 100 kb with viral genes > host genes and contigs > 100 kb with < 20% host genes were maintained as high confidence phages. All deposited phage genomes are compliant with MIUVIG standards27.
Phage dereplication
Phage scaffolds for each ecosystem were dereplicated at 99% ANI using the dRep55 dereplicate module ( -sa 0.99 --ignoreGenomeQuality -l 5000 -nc 0.5 --clusterAlg single -N50W 0 -sizeW 1).
Identification of phage genomes with recoded stop codons
Prodigal56 (single mode) was used to predict genes on dereplicated >= 90% complete phage genomes using genetic codes 4, 11, and 15. Coding density was calculated by summing the length of genes for each contig and dividing by the total contig length. Contigs 5–100 kb that had an increase of greater than 10% coding density in code 4 or code 15 relative to code 11 were tentatively assigned that genetic code, as were contigs >= 100 kb with a coding density increase >5%. All code assignments were confirmed by manual analysis of each contig. If the alternative genetic code resulted in more contiguous operon structure, reduced strand switching, correct-length genes (as checked by blastp57 against NCBI database), and did not result in gene fusions (as checked by blastp against NCBI database) the phage was confirmed as alternatively coded.
Structural and functional annotations
Coding sequences predicted by prodigal using genetic code 4 for TGA recoded phages and code 15 for TAG recoded phages. HMMER58 (hmmsearch) was used to annotate the resulting sequences with the PFAM, pVOG, VOG, and TIGRFAM HMM libraries. tRNAs were predicted using tRNAscan-s.e. V.2.0 in general mode59.
Host prediction
A combination of CRISPR spacer analysis and taxonomic classification were used to predict putative host phyla for recoded phages and their standard code relatives. Contigs with a minimum length of 5 kb from the human and animal metagenomes analyzed in this study were searched for CRISPR spacers using minCED60. blastn short was used to identify matches between phage and spacer of >90% identity and >90% spacer coverage. Taxonomic profiling was performed by using DIAMOND61 (fast mode, e = 0.0001) to search all phage proteins against a custom version of the UNIREF100 database that retained NCBI taxonomic identifiers. tRep62 was then used to profile the taxonomy of each phage contig. For each contig, the bacterial phylum with most hits was considered to be the putative host, but only if that phylum had more than 3x hits than the second most common phylum24. In almost every case, the CRISPR spacer analysis and the taxonomic profiling agreed on the phage host phyla. In the rare cases that these analyses were not in agreement, the host phyla was considered unknown.
Phage genome clustering by average nucleotide identity (ANI)
Our total dataset of 9422 non-dereplicated phage scaffolds from all ecosystems was augmented with 1428 phage genomes from other animal/human microbiomes from ggkbase, and the genomes clustered using dRep55 compare module ( -sa 0.8 -pa 0.8 -nc .1 --clusterAlg single). Whole genome alignment was visualized using Mauve63 (progressiveMauve algorithm) implemented in Geneious Prime 2021.0.3 (https://www.geneious.com).
Phage clustering with Vcontact2
Phages scaffold from the dereplicated dataset of >= 90% complete phage scaffolds for each ecosystem were clustered into viral clusters with Refseq viruses using Vcontact264 (--rel-mode ‘Diamond’ --db ProkaryoticViralRefSeq201-Merged --pcs-mode MCL --vcs-mode ClusterONE). Standard code phages that were in the same viral cluster (VC) as at least one alternatively coded phage were considered to be close relatives of alternatively coded phages.
Phylogenetic analysis of large terminase subunit of recoded phages and standard code relatives
Terminases were found using two rounds of HMM-based classification. Proteins were initially annotated using PFAM, pVOG, VOG, and TIGRfam HMMs. This did not result in complete recovery of terminases for all phages of interest. To increase sensitivity, we clustered proteins into subfamilies using MMseqs65 (-s 7.5, -c 0.5, -e 0.001), and used HHblits66 to generate hmms of each subfamily based on alignments generated with the MMseqs result2msa parameter. We used HHSearch 67 (-p 50 -E 0.001) to perform an HMM-HMM comparison with the PFAM database. We then identified subfamilies with a best hit to large terminase HMMs with a >95% probability. Putative terminase subfamilies with a low number of primary terminase annotations were confirmed by blastp against the NCBI database. If subfamily members had hits to terminases in known phages, we considered the subfamily to be a true terminase subfamily. In rare cases, the terminase gene was fragmented due to assembly error or mobile intron insertion. In these cases we chose the larger of the gene fragments for downstream analysis. Terminases from recoded phages and these standard code relatives (from vContact264) were searched against the Refseq protein database using blastp, retaining the top 10 hits per protein. The recovered Refseq proteins were dereplicated at 90% using CD-HIT68. Recoded phage, standard code relative, and deprelicated Refseq terminases were combined and aligned using MAFFT69, and the alignment trimmed with trimAL70 (-gt 0.5). IQ-TREE71 was used to build a tree using the VT+F+R10 model and ultrafast bootstrap with 1000 iterations. Tree was visualized using iTol72.
Codon preference analysis
TAG-recoded crAss and TGA-recoded Agate analysis:
ANI-based genome clustering showed high representation of a lineage of TGA recoded ∼127 kb Agate phages as well as a lineage of TAG recoded ∼105 kb crAss-like phages, which were chosen for further analysis. For each phage lineage, proteins were clustered into families created using a two step protein clustering method. First, proteins were clustered into subfamilies using MMseqs65 (-s 7.5, -c 0.5, -e 0.001), and HHBlits66 was used to generate HMMs of each subfamily based on alignments generated with the MMseqs result2msa parameter. Thes HMMs were then compared to one another using HHBlits (-p 50 -E 0.001). MCLclustering (--coverage 0.70 -I 2.0 --probs 0.95 ) was used to generate families from the HMM-HMM comparisons. Two-sided Wilcoxon rank sum test was used to evaluate protein families that preferred the in-frame recoded stop codon to the standard coding for the recoded amino acid. The Benjamini-Hochberg p-value correction was used to correct for multiple hypothesis testing with a false discovery rate of 5%. For TGA → W recoded phages, TGA occurrence was compared to the occurrence of the standard codon for Trytophan (TGG). For TAG → Q recoded phages, TAG occurrence was compared to the occurrence of the standard codons for Glutamine (CAG, CAA). Proteins were annotated by PFAM, pVOG, VOG, and TIGRFAM as well as BLAST searches against the NCBI database. In some cases, the HHPred webserver73 and the Phyre2 webserver74 were used to augment initial annotations. Gene neighborhoods were visualized using Clinker75.
Garnet and Topaz integrase analysis:
Garnet and Topaz proteins were clustered into families using the two step method detailed above. We identified the integrase families for each phage clade using PFAM, pVOG, VOG, and TIGRfam HMM annotations. We observed that the majority of the integrase genes had zero in-frame recoded stop codons. A few genes had one in-frame stop, and when we examined alignments of the integrase families we found that in all cases the in-frame recoded stop was in a N or C terminal extension of the protein. We believe that this corresponds to incorrect start codon prediction (N terminal extensions) or legitimate use of the codon to terminate the integrase gene (C terminal extensions). We used a two-sided Wilcoxon rank sum test to evaluate all protein families in each phage clade for avoidance of in-frame recoded stop codons relative to the rates at which they use the standard codons for Glutamine (for TAG → Q recoded phages) or Tryptophan (for TGA → W recoded phages). The Benjamini-Hochberg p-value correction was used to correct for multiple hypothesis testing with a false discovery rate of 5%. We found that for both Garnet and Topaz phages, the integrase gene families strongly avoided in-frame recoded stop codons relative to the rate at which they used standard code encodings for glutamine (TAG → Q recoded phages) or tryptophan (TGA → W recoded phages)
Origin and terminus determination via GC Skew
GC skew (G-C/G+C) and cumulative GC skew were calculated across the phage genome using the iRep package (gc_skew.py)76. This allowed us to predict origins of replication, replication termini, and define individual replichores. We observed a variety of replication styles: double origin bi-directional replication, single origin bi-directional replication, and unidirectional replication. We also observed GC skew patterns of unknown significance. See Supplementary Figure 2A-I for cumulative GC skew plots from the representatives of each phage clade.
Lysogen read mapping
Reads from the source metagenome were mapped against lysogenic contigs with Bowtie 277 using default settings. Contigs and mapped reads were visualized in Geneious Prime 2021.0.3 (https://www.geneious.com).
Statistics and Reproducibility
This study was designed to capture a broad range of gut microbiome phage diversity. We recovered phage from 7 gut microbiome ecosystem types that we and others had sampled sufficiently to allow high recovery of near-complete phage genomes. Only high confidence phage genomes were used in this study. Phage-like contigs that were evaluated as low quality by both CheckV and VIBRANT were excluded from this study. Phage-like contigs < 100 kilobases with more host genes than viral genes were excluded from this study. Phage-like contigs > 100 kilobases with > 20% host genes were excluded from this study. We validated these cutoffs by manually inspecting contigs with high host gene content, and found that they often represented plasmids or chromosomal fragments. These cutoffs were employed to ensure we only had phage genomes in our dataset. We also excluded phage genomes that were less than 90% complete from our survey. Since stop codon recoding is often only present in part of the genome, the recoded region of the genome may be greatly reduced or even entirely missing from an incomplete genome. This means that use of genome fragments to determine phage genetic code is unreliable. All phage genomes in our study were dereplicated, to ensure we were measuring independent phage genomes, and were not measuring the “same” phage across multiple different samples. We used a two-sided Wilcoxon Rank-Sum Test to compare differences between groups of genomes (GC content, stop codon use) or gene families (alternative coding bias). When comparing large numbers of gene families, we used Benjamini-Hochberg p-value correction to correct for multiple hypothesis testing with a false discovery rate of 5%. No statistical method was used to predetermine sample size for any analyses. The experiments were not randomized. The Investigators were not blinded to allocation during experiments and outcome assessment.
Extended Data
Extended Data Fig. 1. Identification of recoded phages in the Giant Tortoise microbiome.
A. Dereplicated complete or near complete (>=90%) phage genomes from the Giant Tortoise gut microbiome. Phages are plotted by size and coding density (CD) in standard code (Code11) B. Replotting of phage genomes from panel A, but with coding density of the alternatively coded phage calculated with the predicted genetic code instead of standard code. In all plots, phages that have recoded the TGA stop codon are indicated in green, and phages that have recoded the TAG stop codon are indicated in orange.
Extended Data Fig. 2. True coding density of standard and alternatively coded phages.
A-F. Replotting of phage genomes from Figure 1 in the main text, but this time the coding density of alternatively coded phage was calculated with their predicted genetic code, not standard code. In all plots, symbol color represents genetic code (TGA recoding = green, TAG recoding = orange, standard code = grey).
Extended Data Fig. 3. Evolution of alternative coding.
A. Global alignment of an 80 kilobase (kb) partial TGA recoded Agate genome (Cattle_ERR2019405_scaffold_1063) and a close standard code relative (pig_ID_3053_F60_scaffold_12). Homologous collinear sequences are shown with colored blocks (red and green here), where color corresponds to nucleotide alignment between the two genomes and lack of color represents lack of alignment. Genome structure for each phage is shown under the alignment graph, with DNA replication machinery represented as yellow bars and structural and lysis genes with pink bars. TGA stop codons have predominantly arisen in structural and lysis genes (individual recoded genes below in green).
Extended Data Fig. 4. Genomic maps of Jade, Sapphire, Agate and Topaz phages.
A-D. TGA recoded genomes (A) contain genes with in-frame TGA codons (green) while TAG recoded genomes (B-D) have genes with in-frame TAG codons (orange). Suppressor tRNAs (tRNA TGA or tRNA TAG, red) are predicted to suppress translation termination at TGA and TAG stop codons, respectively. Regions of the genome encoding structural and lysis genes (pink) coincide with high use of alternative code. Contrastingly, genes involved in DNA replication (yellow) are variably encoded in alternative code. Genomes with a GC skew patterns indicative of bidirectional replication and clear origins and termini (C) have unique replichores marked in alternating shades of blue. Genomes with GC skew patterns most consistent with unidirectional replication (A-B,D) have no replication-related annotation. In some cases, unique or interesting genes have been noted with text. Clade representatives: Jade = JS_HF2_S141_scaffold_159238, Sapphire= SRR1747018_scaffold_13, Agate = Cattle_ERR2019359_scaffold_1067472, Topaz = pig_ID_1851_F40_2_B1_scaffold_1589
Extended Data Fig. 5. Genomic maps of Lak, Garnet, and Amethyst phages.
A-C. TAG recoded genomes have genes with in-frame TAG codons (orange). Suppressor tRNAs (tRNA TAG, red) are predicted to suppress translation termination at TAG stop codons. Regions of the genome encoding structural and lysis genes (pink) coincide with high use of alternative code. In Lak phage (A), genes involved in DNA replication (yellow) are mostly encoded in alternative code. Origins and termini are unmarked in these genomes as we were unable to define clear replichores for Lak (A) and Garnet and Amethyst (B-C) appear to utilize unidirectional genome replication based on GC skew patterns. In some cases, unique or interesting genes have been noted with text. Clade representatives: Lak = C1--CH_A02_001D1_final, Garnet = pig_ID_3640_F65_scaffold_1252, Amethyst = pig_EL5596_F5_scaffold_275.
Extended Data Fig. 6. Code change machinery in two TGA-recoded Jade phages.
A. An operon implicated in changing the genetic code from standard code (TGA = Stop) to code 4 (TGA = W) is directly upstream of the lysis cassette. The code change genes themselves are encoded in standard code, while some genes in the lysis cassette have in frame TGA codons (green). TrpRS = Tryptophanyl tRNA synthetase, RF1 = Release Factor 1, TM-domain = Transmembrane domain.
Extended Data Fig. 7. Read mapping to Garnet and Topaz lysogens.
A. Reads were mapped against a manually curated Garnet lysogen. Read coverage for the Prevotella DNA is ∼2x higher than the read coverage of the Garnet prophage, indicating that the bacterial population in this sample is incompletely lysogenized. Supporting this conclusion are paired reads that span the prophage (not shown), as well as some individual reads which show imperfect mapping to the lysogen consensus sequence (marked with asterisk), which represent the contiguous bacterial sequence. Identical sequence blocks are indicated with color. B. Reads were mapped against a manually curated Topaz lysogen. Read coverage for the integrated Topaz phage genome is ∼50x higher than the neighboring Oscillospiraceae sequence. This indicates that the phage is actively replicating in this sample. Supporting this conclusion are paired reads that span the length of the prophage (not shown), as well as individual reads which show imperfect mapping to the lysogen consensus sequence at the 5’ end of the prophage (light blue) and the 3’ end of the prophage (dark blue). The reads correspond to circularized sequences. Identical sequence blocks are indicated with color.
Extended Data Fig. 8. Alignments of free and integrated phage genomes.
A. A 25kb circular TAG-recoded Garnet phage aligned to a prophage integrated in a Prevotella genome (Prevotella genes = brown). The prophage boundaries are marked by the phage integrase (pink) and the host tRNA Met. B. A 24kb circular TAG-recoded Topaz phage aligned to a prophage integrated into a Oscillospiraceae genome (Oscillospiraceae genes = blue). The prophage boundaries are marked by the phage integrase (pink) and the host tRNA Thr.
Supplementary Material
Acknowledgments
We thank Yun Song, Jamie Cate, Kimberly Seed, Grayson Chadwick, Lin-Xing Chen, Jacob West-Roberts, and Spencer Diamond for helpful discussions. We thank Ka Ki Lily Law and Jordan Hoff for technical support. This work was supported by a Miller Basic Research Fellowship to A.L.B, an NSF Graduate Research Fellowship to B.A-S (No. DGE 1752814), and NIH award RAI092531A, a Chan Zuckerberg Biohub award, and Innovative Genome Institute funding to J.F.B.
Footnotes
Code Availability
Python script used to analyze coding density and predict genetic code is available on Github: https://github.com/borgesadair1/AC_phage_analysis/releases/tag/v1.0.0
Competing Interests
J.F.B. is a founder of Metagenomi. The other authors declare no competing interests.
Data Availability
Accessions for MIUVIG-compliant genomes27 and associated reads for alternatively coded phages and relatives are provided in Supplementary Table 3. Genomes and predicted proteins for alternatively coded phages and relatives, the terminase phylogenetic tree file, closely related Agate and crAss-like genomes, and untrimmed lysosgenic contigs are available through Zenodo (10.5281/zenodo.6410225). The UniRef100 database is available through ftp.uniprot.org/pub/databases/uniprot/uniref/uniref100.
References
- 1.Crick FH The origin of the genetic code. J. Mol. Biol. 38, 367–379 (1968). [DOI] [PubMed] [Google Scholar]
- 2.Knight RD, Freeland SJ & Landweber LF Rewiring the keyboard: evolvability of the genetic code. Nat. Rev. Genet. 2, 49–58 (2001). [DOI] [PubMed] [Google Scholar]
- 3.Horowitz S & Gorovsky MA An unusual genetic code in nuclear genes of Tetrahymena. Proc. Natl. Acad. Sci. U. S. A. 82, 2452–2455 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caron F & Meyer E Does Paramecium primaurelia use a different genetic code in its macronucleus? Nature 314, 185–188 (1985). [DOI] [PubMed] [Google Scholar]
- 5.Preer JR Jr, Preer LB., Rudman BM & Barnett AJ Deviation from the universal code shown by the gene for surface protein 51A in Paramecium. Nature 314, 188–190 (1985). [DOI] [PubMed] [Google Scholar]
- 6.Keeling PJ & Doolittle WF A non-canonical genetic code in an early diverging eukaryotic lineage. EMBO J. 15, 2285–2290 (1996). [PMC free article] [PubMed] [Google Scholar]
- 7.Schneider SU, Leible MB & Yang XP Strong homology between the small subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase of two species of Acetabularia and the occurrence of unusual codon usage. Mol. Gen. Genet. 218, 445–452 (1989). [DOI] [PubMed] [Google Scholar]
- 8.Santos MA, Keith G & Tuite MF Non-standard translational events in Candida albicans mediated by an unusual seryl-tRNA with a 5’-CAG-3’ (leucine) anticodon. EMBO J. 12, 607–616 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ohama T et al. Non-universal decoding of the leucine codon CUG in several Candida species. Nucleic Acids Res. 21, 4039–4045 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Inamine JM, Ho KC, Loechel S & Hu PC Evidence that UGA is read as a tryptophan codon rather than as a stop codon by Mycoplasma pneumoniae, Mycoplasma genitalium, and Mycoplasma gallisepticum. J. Bacteriol. 172, 504–506 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamao F et al. UGA is read as tryptophan in Mycoplasma capricolum. Proc. Natl. Acad. Sci. U. S. A. 82, 2306–2309 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stamburski C, Renaudin J & Bové JM Mutagenesis of a tryptophan codon from TGG to TGA in the cat gene does not prevent its expression in the helical mollicute Spiroplasma citri. Gene 110, 133–134 (1992). [DOI] [PubMed] [Google Scholar]
- 13.Wrighton KC et al. Fermentation, hydrogen, and sulfur metabolism in multiple uncultivated bacterial phyla. Science 337, 1661–1665 (2012). [DOI] [PubMed] [Google Scholar]
- 14.Campbell JH et al. UGA is an additional glycine codon in uncultured SR1 bacteria from the human microbiota. Proc. Natl. Acad. Sci. U. S. A. 110, 5540–5545 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hanke A et al. Recoding of the stop codon UGA to glycine by a BD1–5/SN-2 bacterium and niche partitioning between Alpha- and Gammaproteobacteria in a tidal sediment microbial community naturally selected in a laboratory chemostat. Front. Microbiol. 5, 231 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shulgina Y & Eddy SR A computational screen for alternative genetic codes in over 250,000 genomes. Elife 10, e71402 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zinoni F, Birkmann A, Leinfelder W & Böck A Cotranslational insertion of selenocysteine into formate dehydrogenase from Escherichia coli directed by a UGA codon. Proc. Natl. Acad. Sci. U. S. A. 84, 3156–3160 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berry MJ et al. Recognition of UGA as a selenocysteine codon in type I deiodinase requires sequences in the 3’ untranslated region. Nature 353, 273–276 (1991). [DOI] [PubMed] [Google Scholar]
- 19.Hao B et al. A new UAG-encoded residue in the structure of a methanogen methyltransferase. Science 296, 1462–1466 (2002). [DOI] [PubMed] [Google Scholar]
- 20.Sun J et al. Recoding of stop codons expands the metabolic potential of two novel Asgardarchaeota lineages. ISME Communications 1, 1–14 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gomes AC et al. A genetic code alteration generates a proteome of high diversity in the human pathogen Candida albicans. Genome Biol. 8, R206 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ivanova NN et al. Stop codon reassignments in the wild. Science 344, 909–913 (2014). [DOI] [PubMed] [Google Scholar]
- 23.Devoto AE et al. Megaphages infect Prevotella and variants are widespread in gut microbiomes. Nat Microbiol 4, 693–700 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Al-Shayeb B et al. Clades of huge phages from across Earth’s ecosystems. Nature 578, 425–431 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yutin N et al. Analysis of metagenome-assembled viral genomes from the human gut reveals diverse putative CrAss-like phages with unique genomic features. Nat. Commun. 12, 1–11 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Crisci MA et al. Closely related Lak megaphages replicate in the microbiomes of diverse animals. iScience 24, 102875 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roux S et al. Minimum Information about an Uncultivated Virus Genome (MIUViG). Nat. Biotechnol. 37, 29–37 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goltsman DSA et al. Metagenomic analysis with strain-level resolution reveals fine-scale variation in the human pregnancy microbiome. Genome Res. 28, 1467–1480 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Obregon-Tito AJ et al. Subsistence strategies in traditional societies distinguish gut microbiomes. Nat. Commun. 6, 6505 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rampelli S et al. Metagenome Sequencing of the Hadza Hunter-Gatherer Gut Microbiota. Curr. Biol. 25, 1682–1693 (2015). [DOI] [PubMed] [Google Scholar]
- 31.Lou YC et al. Infant gut strain persistence is associated with maternal origin, phylogeny, and traits including surface adhesion and iron acquisition. Cell Rep Med 2, 100393 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.David LA et al. Gut microbial succession follows acute secretory diarrhea in humans. MBio 6, e00381–15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tung J et al. Social networks predict gut microbiome composition in wild baboons. Elife 4, e05224 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Munk P et al. A sampling and metagenomic sequencing-based methodology for monitoring antimicrobial resistance in swine herds. J. Antimicrob. Chemother. 72, 385–392 (2017). [DOI] [PubMed] [Google Scholar]
- 35.Andersen VD et al. Predicting effects of changed antimicrobial usage on the abundance of antimicrobial resistance genes in finisher’ gut microbiomes. Prev. Vet. Med. 174, 104853 (2020). [DOI] [PubMed] [Google Scholar]
- 36.Wallace RJ et al. A heritable subset of the core rumen microbiome dictates dairy cow productivity and emissions. Sci Adv 5, eaav8391 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peters SL et al. Validation that human microbiome phages use alternative genetic coding with TAG stop read as Q. bioRxiv 2022.01.06.475225 (2022) doi: 10.1101/2022.01.06.475225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Osawa S & Jukes TH Codon reassignment (codon capture) in evolution. J. Mol. Evol. 28, 271–278 (1989). [DOI] [PubMed] [Google Scholar]
- 39.Berry J, Rajaure M, Pang T & Young R The spanin complex is essential for lambda lysis. J. Bacteriol. 194, 5667–5674 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Young R Phage lysis: three steps, three choices, one outcome. J. Microbiol. 52, 243–258 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Doermann AH The intracellular growth of bacteriophages. I. Liberation of intracellular bacteriophage T4 by premature lysis with another phage or with cyanide. J. Gen. Physiol. 35, 645–656 (1952). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Heagy FC The effect of 2,4-dinitrophenol and phage T2 on Escherichia coli B. J. Bacteriol. 59, 367–373 (1950). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Park T, Struck DK, Dankenbring CA & Young R The pinholin of lambdoid phage 21: control of lysis by membrane depolarization. J. Bacteriol. 189, 9135–9139 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hays SG & Seed KD Dominant Vibrio cholerae phage exhibits lysis inhibition sensitive to disruption by a defensive phage satellite. Elife 9, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cowe E & Sharp PM Molecular evolution of bacteriophages: Discrete patterns of codon usage in T4 genes are related to the time of gene expression. J. Mol. Evol. 33, 13–22 (1991). [Google Scholar]
- 46.Yang JY et al. Degradation of host translational machinery drives tRNA acquisition in viruses. Cell Syst (2021) doi: 10.1016/j.cels.2021.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Durmaz E & Klaenhammer TR Abortive phage resistance mechanism AbiZ speeds the lysis clock to cause premature lysis of phage-infected Lactococcus lactis. J. Bacteriol. 189, 1417–1425 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zeng L et al. Decision making at a subcellular level determines the outcome of bacteriophage infection. Cell 141, 682–691 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Erez Z et al. Communication between viruses guides lysis-lysogeny decisions. Nature 541, 488–493 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Silpe JE & Bassler BL A Host-Produced Quorum-Sensing Autoinducer Controls a Phage Lysis-Lysogeny Decision. Cell 176, 268–280.e13 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bailly-Bechet M, Vergassola M & Rocha E Causes for the intriguing presence of tRNAs in phages. Genome Res. 17, 1486–1495 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Auslander N, Gussow AB, Benler S, Wolf YI & Koonin EV Seeker: alignment-free identification of bacteriophage genomes by deep learning. Nucleic Acids Res. 48, e121–e121 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kieft K, Zhou Z & Anantharaman K VIBRANT: automated recovery, annotation and curation of microbial viruses, and evaluation of viral community function from genomic sequences. Microbiome 8, 90 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nayfach S et al. CheckV assesses the quality and completeness of metagenome-assembled viral genomes. Nat. Biotechnol. 39, 578–585 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Olm MR, Brown CT, Brooks B & Banfield JF dRep: a tool for fast and accurate genomic comparisons that enables improved genome recovery from metagenomes through de-replication. ISME J. 11, 2864–2868 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hyatt D et al. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11, 119 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Altschul SF, Gish W, Miller W, Myers EW & Lipman DJ Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990). [DOI] [PubMed] [Google Scholar]
- 58.Eddy SR Accelerated Profile HMM Searches. PLoS Comput. Biol. 7, e1002195 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chan PP & Lowe TM tRNAscan-SE: Searching for tRNA Genes in Genomic Sequences. Methods Mol. Biol. 1962, 1–14 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Skennerton CT minced: Mining CRISPRs in Environmental Datasets. (Github). [Google Scholar]
- 61.Buchfink B, Reuter K & Drost H-G Sensitive protein alignments at tree-of-life scale using DIAMOND. Nat. Methods 18, 366–368 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Olm M tRep: Quick get the taxonomy of a genome. (Github). [Google Scholar]
- 63.Darling ACE, Mau B, Blattner FR & Perna NT Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14, 1394–1403 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bin Jang H et al. Taxonomic assignment of uncultivated prokaryotic virus genomes is enabled by gene-sharing networks. Nat. Biotechnol. 37, 632–639 (2019). [DOI] [PubMed] [Google Scholar]
- 65.Hauser M, Steinegger M & Söding J MMseqs software suite for fast and deep clustering and searching of large protein sequence sets. Bioinformatics 32, 1323–1330 (2016). [DOI] [PubMed] [Google Scholar]
- 66.Remmert M, Biegert A, Hauser A & Söding J HHblits: lightning-fast iterative protein sequence searching by HMM-HMM alignment. Nat. Methods 9, 173–175 (2011). [DOI] [PubMed] [Google Scholar]
- 67.Söding J Protein homology detection by HMM-HMM comparison. Bioinformatics 21, 951–960 (2005). [DOI] [PubMed] [Google Scholar]
- 68.Li W & Godzik A Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22, 1658–1659 (2006). [DOI] [PubMed] [Google Scholar]
- 69.Katoh K, Misawa K, Kuma K-I & Miyata T MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30, 3059–3066 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Capella-Gutiérrez S, Silla-Martínez JM & Gabaldón T trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nguyen L-T, Schmidt HA, von Haeseler A & Minh BQ IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Letunic I & Bork P Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49, W293–W296 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zimmermann L et al. A Completely Reimplemented MPI Bioinformatics Toolkit with a New HHpred Server at its Core. J. Mol. Biol. 430, 2237–2243 (2018). [DOI] [PubMed] [Google Scholar]
- 74.Kelley LA, Mezulis S, Yates CM, Wass MN & Sternberg MJE The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gilchrist CLM & Chooi Y-H clinker & clustermap.js: Automatic generation of gene cluster comparison figures. bioRxiv 2020.11.08.370650 (2020) doi: 10.1101/2020.11.08.370650. [DOI] [PubMed] [Google Scholar]
- 76.Brown CT, Olm MR, Thomas BC & Banfield JF Measurement of bacterial replication rates in microbial communities. Nat. Biotechnol. 34, 1256–1263 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Langmead B & Salzberg SL Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Accessions for MIUVIG-compliant genomes27 and associated reads for alternatively coded phages and relatives are provided in Supplementary Table 3. Genomes and predicted proteins for alternatively coded phages and relatives, the terminase phylogenetic tree file, closely related Agate and crAss-like genomes, and untrimmed lysosgenic contigs are available through Zenodo (10.5281/zenodo.6410225). The UniRef100 database is available through ftp.uniprot.org/pub/databases/uniprot/uniref/uniref100.