Abstract
Neutrophils play a significant role in determining disease severity following SARS-CoV-2 infection. Gene and protein expression defines several neutrophil clusters in COVID-19, including the emergence of low density neutrophils (LDN) that are associated with severe disease. The functional capabilities of these neutrophil clusters and correlation with gene and protein expression are unknown. To define host defense and immunosuppressive functions of normal density neutrophils (NDN) and LDN from COVID-19 patients, we recruited 64 patients with severe COVID-19 and 26 healthy donors (HD). Phagocytosis, respiratory burst activity, degranulation, neutrophil extracellular trap (NET) formation, and T-cell suppression in those neutrophil subsets were measured. NDN from severe/critical COVID-19 patients showed evidence of priming with enhanced phagocytosis, respiratory burst activity, and degranulation of secretory vesicles and gelatinase and specific granules, while NET formation was similar to HD NDN. COVID LDN response was impaired except for enhanced NET formation. A subset of COVID LDN with intermediate CD16 expression (CD16Int LDN) promoted T cell proliferation to a level similar to HD NDN, while COVID NDN and the CD16Hi LDN failed to stimulate T-cell activation. All 3 COVID-19 neutrophil populations suppressed stimulation of IFN-γ production, compared to HD NDN. We conclude that NDN and LDN from COVID-19 patients possess complementary functional capabilities that may act cooperatively to determine disease severity. We predict that global neutrophil responses that induce COVID-19 ARDS will vary depending on the proportion of neutrophil subsets.
Keywords: neutrophil, COVID-19, low density neutrophil, respiratory burst, exocytosis, NET formation, T cell proliferation
Introduction
Acute respiratory distress syndrome (ARDS) is a life-threatening disorder induced by inflammatory injury to the lungs initiated by local (eg. pneumonia) or systemic (eg. sepsis, trauma) disorders (1). Neutrophil recruitment and activation, leading to the release of reactive oxygen species (ROS), degranulation, and neutrophil extracellular trap (NET) formation, are critical components of ARDS pathophysiology (1, 2). Among patients hospitalized with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)-induced coronavirus disease 2019 (COVID-19), approximately 1/3 develop ARDS, making COVID-19 the current leading cause of ARDS (3–6). Despite some potential differences in pathophysiology, accumulating evidence indicates that neutrophils also play a critical role in the development of COVID-19 ARDS. Lung specimens taken at autopsy show a marked infiltration of neutrophils and the presence of NETs. In addition, neutrophils are the predominant immune cells in bronchoalveolar lavage fluid (BALF) from COVID-19 patients with ARDS, and neutrophil serine proteases are present in BALF (7–22).
Patients with severe COVID-19 exhibit marked alterations in innate and adaptive immunity, including changes in neutrophil abundance, phenotype, and function (22–26). Neutrophil characterization by transcriptional and proteomic analysis in COVID-19 indicates significant heterogeneity among mature neutrophils and emergence of a large number of low density neutrophils (LDN) with multiple phenotypes (16, 24, 25, 27–30). The functional capability of these various neutrophil populations has been inferred from gene or protein expression profiles (22–26), however, previous direct functional studies either did not evaluate the various neutrophil populations or disparate results were reported (12–15, 24, 28–37). Thus, the functional heterogeneity among the various neutrophil populations remains to be defined. The emergence of LDN in the circulation and lungs (16, 24, 25, 27, 37), particularly in severe and critical COVID-19, indicates that defining the spectrum of functional alterations of different neutrophil populations is important to understanding the contribution of neutrophils to COVID-19 ARDS pathophysiology. To address this gap in knowledge, the current study compares the functional responses of LDN from COVID-19 patients (COVID LDN) with the responses of normal density neutrophils from COVID-19 patients (COVID NDN) and of NDN from healthy donors (HD NDN). Our results indicate that functional heterogeneity correlates with phenotypic heterogeneity and suggest the possibility that different neutrophil populations cooperate to induce ARDS in COVID-19.
Results
Characteristics of COVID-19 Patients
Between November, 2020 and December, 2021, we recruited 64 patients who tested positive for SARS-CoV-2 by nasopharyngeal swab and were assessed to have severe or critical COVID-19, as defined by the WHO criteria (38). The study participant demographics and summary of clinical information are shown in Table 1. The mean age was 59.8 ± 14.6 years. ICU admission was required in 59 of the 64 patients, 57 required mechanical ventilatory support, and 6 were supported with ECMO. The PaO2:FiO2 was below 100 in 25 patients, between 100 and 200 in 29, between 200 and 300 in 7, and above 300 in 3 who required mechanical ventilation. Comorbidities known to be risk factors for severe COVID-19 were present in 56 of the 64 patients, the most common of which were hypertension (n=39, 61%) and obesity (n=42, 66%). Elevated neutrophil to lymphocyte ratio (NLR), D-dimer, ferritin, and C-reactive protein were common, as previously reported (16, 39–42). The mean/median hospital day on which neutrophils used in this study were obtained was 8.5/6.5. Treatment with corticosteroids, remdesivir, and monoclonal antibodies or convalescent plasma occurred in 59 (92%), 48 (75%), and 29 (45%) patients, respectively. Outcome was poor, as 32 (50%) patients died during hospitalization and 19 (30%) were discharged to a long-term care or rehabilitation facility. A total of 26 healthy donors (HD) were recruited from the students, staff, and faculty at the University of Louisville. Demographic data was not recorded for HD, but the pool of subjects eligible for recruitment was younger than the patient population.
Table 1.
Clinical characteristics and demographics.
| n | 64 |
|---|---|
| Median Age (range) | 59 (24-96) |
| Male (%) | 34 (53%) |
| Race (%) | |
| White or Caucasian | 48 (75%) |
| Black or African American | 13 (20%) |
| Hispanic | 1 (2%) |
| Not recorded | 2 (3%) |
| Weight (Kg) | 99.8 (41-190) |
| BMI median (range) | 35.6 (17.2-66.1) |
| Comorbidity | |
| Obesity, patient number (%) | 42 (66%) |
| Hypertension, patient number (%) | 39 (61%) |
| Diabetes Mellitus, patient number (%) | 26 (41%) |
| Coronary artery disease/arrhymia/CHF, patient number (%) | 20 (31%) |
| Underlying lung disease, patient number (%) | 21 (33%) |
| Chronic kidney disease, patient number (%) | 7 (11%) |
| Hyperlipidemia, patient number (%) | 14 (22%) |
|
Hospital Day Neutrophils Obtained
mean/median (range) |
8.5/6.5 (1-36) |
| Laboratory Values | |
| WBC/Neutrophil (4.1-10.8 x103/μL) mean/median | 13.6/11.7 |
| NLR mean/median | 20.5/12.5 |
| d-dimer (0.19-0.74 μg/ml FEU) | 423, range 0.37-15,107 (n=40) |
| Ferritin (7-350 ng/ml) | 711, range 71-7500 (n=34) |
| C-reactive protein (<3.0 mg/L) | 177, range 2.6-380 (n=30) |
| Hospital Course | |
| APACHE | 21, range 6-38 |
| ICU, patient number (%) | 59 (92%) |
| Ventilator, patient number (%) | 57 (89%) |
| ECMO, patient number (%) | 6 (9%) |
| Dialysis, patient number (%) | 16 (25%) |
| Died, patient number (%) | 32 (50%) |
| Discharge Home, patient number (%) | 9 (14%) |
| Discharge to LTC, patient number (%) | 22 (34%) |
| Treatment | |
| Remdesivir, patient number (%) | 48 (75%) |
| Corticosteroids, patient number (%) | 59 (92%) |
| Monoclonal antibody or Convalescent Plasma, patient number (%) | 29 (45%) |
Phagocytosis and Respiratory Burst
Neutrophil phagocytosis and respiratory burst activity are critical for host defense function (43), and release of ROS is suggested as a contributing mechanism of lung injury leading to ARDS in COVID-19 (44, 45). Previous reports presented contradictory results for both neutrophil phagocytosis and respiratory burst activity (8, 25), and the response of LDN has not been reported. To address this gap in knowledge, we measured bacterial phagocytosis and phagocytic stimulation of H2O2 production in LDN and NDN neutrophils isolated from 11 COVID-19 patients and compared their responses to those of NDN from 6 HD. Figure 1A shows phagocytosis before and after in vitro priming with TNFα. Phagocytosis by unprimed cells was significantly greater in COVID NDN, compared to HD NDN. COVID LDN showed a level of phagocytosis intermediate between the other 2 groups and was not significantly different from either. TNFα priming significantly enhanced phagocytosis of both HD NDN and COVID NDN, with a significantly greater response by COVID NDN. Phagocytosis by COVID LDN did not increase with in vitro priming. Figure 1B shows phagocytosis-stimulated H2O2 production in the three groups of neutrophils. H2O2 production by COVID NDN, but not COVID LDN, was significantly greater than that by HD NDN. TNFα priming significantly enhanced H2O2 production to an equivalent degree in all three groups, with COVID NDN production remaining significantly greater than HD NDN or COVID LDN. We interpret these results to indicate that NDN from severe COVID-19 patients are primed for enhanced phagocytosis and respiratory burst. Despite existing in the same milieu, LDN are not primed for either enhanced phagocytosis or respiratory burst activity. Respiratory burst activity by COVID LDN was enhanced to the same degree as HD NDN in response to in vitro priming, indicating LDN are responsive to some priming agents.
Figure 1.
Phagocytosis and respiratory burst are enhanced in COVID-19 NDN, but not LDN. Results show basal and in vitro TNFα-primed (2 ng/ml) phagocytosis of opsonized, propidium iodide-labeled S. aureus (A) and subsequent H2O2 production (B) in COVID-19 NDN (n=11), LDN from COVID-19 (COVID LDN) (n=11), and NDN from healthy donors (HD NDN) (n=6). Results are expressed as mean ± SEM of mean fluorescence intensity (MFI) of each marker, determined by flow cytometry. *p < 0.05, **p < 0.01, ****p < 0.001.
Degranulation
Previous neutrophil degranulation studies using expression of plasma membrane markers showed enhanced azurophilic granule exocytosis, measured by CD63, in COVID-19 (28, 29, 31). Contradictory results were reported for exocytosis of specific (CD66b) and gelatinase (CD11b) granules (9, 28, 29, 31, 32, 34, 35, 46). To evaluate degranulation of neutrophil populations from COVID-19 patients, we compared expression of plasma membrane markers of the 4 neutrophil granule subsets, CD35 (secretory vesicles), CD11b (gelatinase granules), CD66b (specific granules), and CD63 (azurophilic granules), among HD NDN, COVID NDN, and COVID LDN before and after stimulation (Figure 2A). Basal CD35 expression was significantly greater in COVID NDN and COVID LDN than in HD NDN, indicating that similar secretory vesicle degranulation occurred in vivo in LDN and NDN in COVID-19 patients. Stimulation with TNFα and fMLF increased CD35 expression in HD NDN and COVID NDN, but not COVID LDN, suggesting LDN underwent maximal release of secretory vesicles in vivo. Basal CD11b expression by COVID NDN was significantly greater than expression by HD NDN or COVID LDN. Additionally, TNFα and fMLF stimulation increased CD11b expression in HD NDN and COVID NDN, but not in COVID LDN. Basal CD66b expression was significantly greater in COVID NDN, but not COVID LDN, than in HD NDN. In vitro stimulation resulted in a significant increase in CD66b in all 3 groups of cells. CD63 expression did not differ between HD NDN and COVID NDN. CD63 expression by COVID LDN was significantly lower than either group of NDN. The response of CD63 to in vitro stimulation was not tested, as pharmacologic disruption of the actin cytoskeleton is required (47). Thus, markers for the different neutrophil granule subsets showed distinct patterns of expression, suggesting differences in granule marker expression or in granule mobilization both among granule subsets and between NDN and LDN in COVID-19. We interpret these results to indicate that severe COVID-19 is associated with enhanced granule mobilization in NDN, except for the granule subset requiring the most potent stimulus, azurophilic granules. LDN demonstrate an impaired in vivo response to degranulation, except for the granule subset most easily induced to undergo exocytosis, secretory vesicles.
Figure 2.
Differential degranulation by neutrophil subsets from COVID-19 patients. (A) Basal and stimulated (TNFα 2 ng/ml + fMLF 300 nM) expression of markers for secretory vesicles (CD35), gelatinase granules (CD11b), specific granules (CD66b), and azurophilic granules (CD63) on COVID NDN (n=11-15), COVID LDN (n=11-15), and HD NDN (n=8-9). Results are expressed as mean ± SEM of mean fluorescent intensity (MFI). *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. (B) Comparison of basal expression of CD35, CD11b, CD66b, and CD63 on COVID NDN, CD16Hi LDN, and CD16Int LDN (n=7). Results are expressed as mean ± SEM of MFI. *p < 0.05, **p < 0.01, ***p < 0.001. (C) Plasma levels of MMP9, lactoferrin, and human neutrophil elastase in plasma from HD (n=10) and COVID-19 (n=10-25). Results expressed as mean + SEM in ng/ml. *p < 0.05, **p < 0.01, ***p < 0.001.
We previously reported that an intermediate level of CD16 expression (CD16Int) identifies a distinct subpopulation of LDN that emerges in severe COVID-19 (16). CD16Int LDN have an activated genetic and proteomic phenotype, and their prevalence in the circulation predicts disease severity. CD16Int and CD16Hi LDN obtained by cell sorting and NDN isolated by gradient centrifugation were available from 7 COVID-19 patients to compare expression of neutrophil granule markers. Figure 2B shows CD16Hi LDN express CD35, CD11b, CD66b, and CD63 at the same levels as COVID NDN. On the other hand, CD16Int LDN showed reduced CD35, CD11b, and CD63 expression, while CD66b expression was increased. We interpret these data to indicate that CD16Int LDN represent a unique subpopulation of LDN with impaired mobilization of secretory vesicles, gelatinase granules and azurophilic granules, while specific granule mobilization is enhanced.
Previous studies found elevated levels of granule constituents in the plasma of COVID-19 patients (21, 31, 48–51). To confirm our granule marker results, constituents of gelatinase granules (MMP9), specific granules (lactoferrin), and azurophilic granules (human neutrophil elastase, HNE) were quantified in plasma from HD and severe COVID-19 patients. Figure 2C shows that all three markers were significantly increased in plasma from COVID-19 patients. Thus, plasma concentrations of granule contents and neutrophil plasma membrane expression of granule markers present consistent findings for gelatinase and specific granules, while CD63 expression failed to conform with azurophilic granule mobilization detected by HNE release into plasma.
NET Formation
Enhanced NET formation by circulating neutrophils in COVID-19 is well established, and NETs are observed in the lungs of patients with COVID-19 ARDS and in areas of microthrombosis (10–17). We compared spontaneous and bacterially-stimulated NET formation by isolated HD NDN, COVID NDN and COVID LDN by confocal microscopy. Figure 3A shows a representative merged confocal microscopic image of lactoferrin and DAPI staining of NETs under basal and stimulated conditions, and individual images of each stain are shown in Supplemental Figures 1, 2. Minimal extracellular lactoferrin was observed directly adjacent to unstimulated HD NDN and COVID NDN. On the other hand, NETs were observed emanating from many unstimulated COVID LDN. Incubation with the emerging oral pathogen, Peptoanaerobacter stomatis, for 3 hr resulted in a greater number and larger patches of nets in COVID LDN, compared to either COVID NDN or HD NDN. Enlarged images confirming co-localization of extracellular staining for lactoferrin and DAPI, indicating lactoferrin and dsDNA co-localize outside of cells, are shown in Supplemental Figures 3, 4. To quantify NET release, extracellular dsDNA was measured in the same groups of cells. Figure 3B shows that although NET formation in unstimulated COVID LDN and COVID NDN was numerically greater than in HD LDN, those differences were not statistically significant. All three neutrophil groups showed a significant increase in NET formation following incubation with P. stomatis for 3 hr. The release of dsDNA was significantly greater following bacterial stimulation in COVID LDN, compared to either COVID NDN or HD NDN. Three sets of sorted CD16Hi and CD16Int LDN were available to compare NET formation in LDN subsets. As shown in Figure 3C, NET formation occurred in a greater number of CD16Int LDN for each set, however, those differences were not statistically different in this limited data set. Taken together, our data suggest that COVID LDN possess a higher capacity for stimulated NET formation, and NET formation may be greater in CD16Int LDN than CD16Hi LDN.
Figure 3.

Enhanced NET formation by COVID-19 LDN. Panel (A) shows merged confocal images of lactoferrin staining of NETs (Red) and DAPI staining of DNA (Blue), demonstrating NET formation, in the absence (Basal) or presence of CFSE-labelled P. stomatis, in HD NDN, COVID NDN, and COVID LDN, representative of 4 experiments. Images shown are stacks of the entire collection of acquired z-planes at 20x magnification, scale bar=100 mm. Individual images are shown in Supplemental Figures 1 and 2. Panel (B) shows release of dsDNA into supernatant from basal or P. stomatis stimulated HD NDN, COVID NDN, and COVID LDN. NET formation by COVID LDN was significantly greater following bacterial stimulation, compared to COVID NDN and HD NDN, n=12-15. *p < 0.05, **p < 0.01. Panel (C) shows the results of 3 separate experiments comparing the percent of CD16Hi LDN and CD16Int LDN spontaneously forming NETs, visualized by confocal microscopy.
T-Cell Suppression
The genetic signature of LDN has been interpreted to indicate at least a portion of this neutrophil population has immunosuppressive activity (23, 25). Evaluation of neutrophil supernatants or neutrophil-T cell interaction showed impairment of T cell proliferation (52, 53). To determine the effect of different neutrophil populations on T cell activation, we determined the effect of NDN and CD16Hi/Int LDN from 6 COVID-19 patients or HD NDN on T cell activation. T cells from a single donor were labeled with CellTracker dye CFSE and then incubated with NDN, CD16Hi LDN, or CD16Int LDN from COVID-19 patients or HD NDN. The distribution of those 3 neutrophil populations in 12 COVID-19 patients is shown in Figure 4A. NDN accounted for 37.4 ± 8.9% of total neutrophils, CD16Hi LDN for 54.7 ± 8.6, and CD16Int LDN for 7.4 ± 2.0%. T cell proliferation was assessed based on CFSE dilution, and cell culture supernatants were harvested to measure IFN-γ levels. While co-culture of CD16Int LDN from COVID-19 patients stimulated proliferation of CD4+ and CD8+ T cells to the same degree as HD NDN, CD16Hi LDN and COVID NDN failed to stimulate CD8+ and CD4+ T cell proliferation (Figure 4B). Co-culture with HD NDN significantly stimulated IFN-γ production. On the other hand, COVID NDN, CD16Int LDN, and CD16Hi LDN all failed to stimulate IFN-γ production (Figure 4C).
Figure 4.
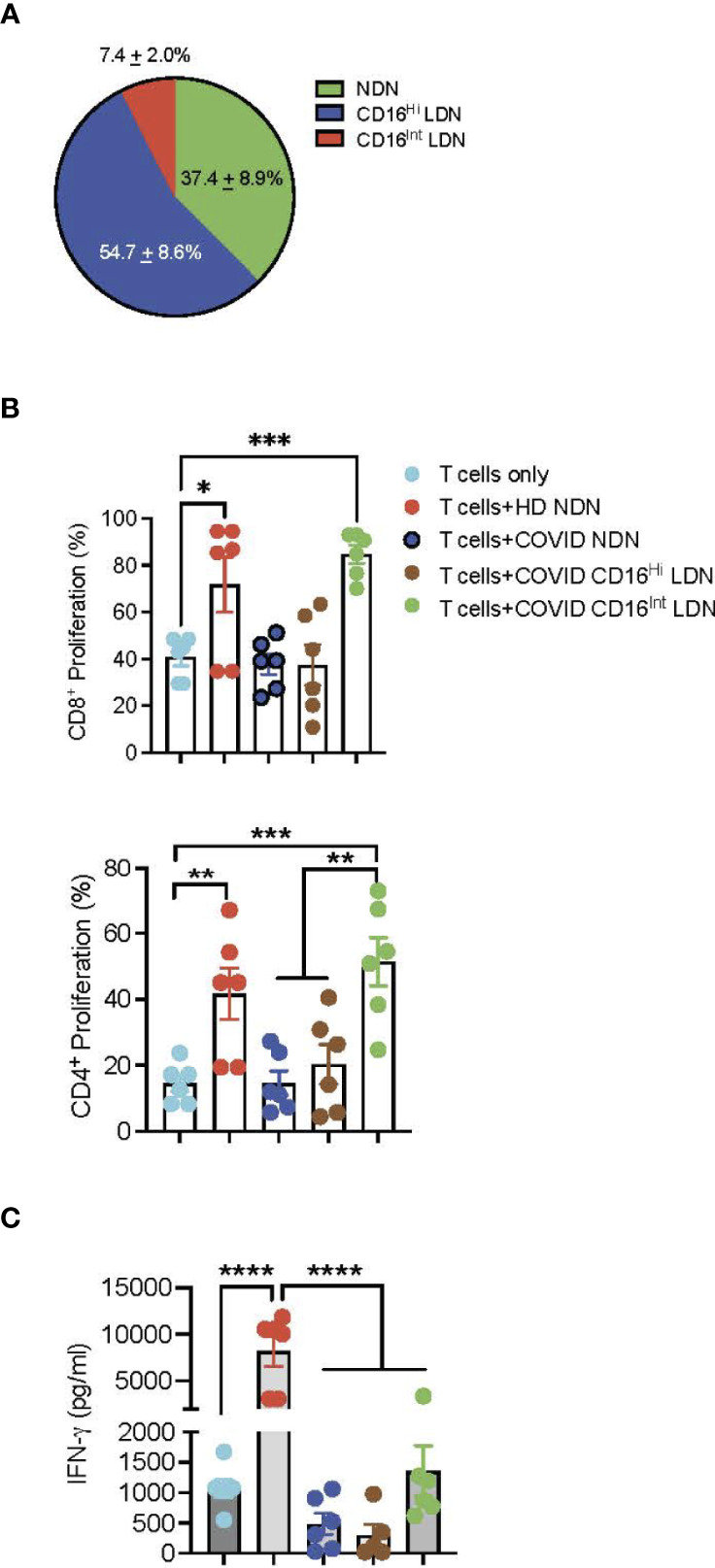
T cell activation assay. (A) Pie chart showing the distribution of NDN, CD16Hi and CD16Int LDN from severe COVID-19 patients (n=12). (B) T cells from the same donor were surface labeled with CellTrace CFSE dye and then co-cultured with ND NDN, COVID NDN, CD16Hi LDN, or CD16Int LDN for 4 days in the presence of plate-bound anti-CD3. Cells were collected and stained with CD4 or CD8. T cell proliferation was determined by flow cytometry and is expressed as mean ± SEM percent proliferation (n=6). (C) Supernatants from those 6 experiments were harvested and the IFN-γ level was determined by ELISA. Results are expressed as mean ± SEM in pg/ml. *p < 0.05, **p < 0.01,***p < 0.001, ****p < 0.0001.
Discussion
Our study provides additional context to the expanding literature implicating neutrophils in the pathogenesis of COVID-19 ARDS. Gene and protein signatures define multiple clusters of mature and immature neutrophils emerging in the PBMC fraction (LDN) from severe COVID-19 patients (16, 22, 23, 25, 27, 28). Those LDN are present in BALF, as well as in the circulation (16, 22). An altered functional status of those neutrophil clusters is inferred from their gene and protein signatures, leading to the hypothesis that dysregulated neutrophil host defense mechanisms and adaptive immune system suppression are major contributors to progression to severe COVID-19 (25, 26, 45, 54). To characterize the dysregulated neutrophil functional responses associated with COVID-19 ARDS, the present study evaluated host defense functions and suppression of T cell activation of NDN and two populations of LDN from COVID-19 patients. The major finding of our study is that neutrophil populations from patients with severe/critical COVID-19 are functionally distinct from each other and from NDN of healthy donors. COVID NDN exist in a primed state, but they failed to support T-cell activation. LDN are generally hyporesponsive, except for more robust NET formation. Separating LDN by their CD16 expression defines subsets with distinct activation and immunosuppressive signatures.
NDN account for an average of 37% (range 1% to 83%) of total neutrophils in our severe/critical COVID-19 patients. They are primed for enhanced phagocytosis, respiratory burst activity, and degranulation. Consistent with our findings, Masso-Silva et al. (8) reported increased basal and stimulated ROS production and increased phagocytosis by neutrophils isolated from COVID-19 patients. On the other hand, Schulte-Schrepping et al. (25) reported that neutrophils from severe COVID-19 patients showed reduced respiratory burst activity, while phagocytosis did not differ from healthy donors. Our findings with granule plasma membrane markers and plasma concentrations of granule cargo suggest robust degranulation of secretory vesicles, gelatinase granules, and specific granules. The presence of elevated levels of HNE in patient plasma, while expression of the plasma membrane marker of azurophilic granules, CD63, did not increase, may be due to the hierarchical nature of neutrophil granule exocytosis, in which fewer than 10% of azurophilic granules are mobilized during an inflammatory response (55, 56). Previous studies of neutrophil degranulation in COVID-19 described conflicting results, depending on the method of analysis and experimental conditions. Using granule marker expression, exocytosis of specific and gelatinase granules was increased, unchanged, or decreased in severe COVID-19, while azurophilic granules were uniformly mobilized (9, 28, 29, 31, 32, 36, 46). On the other hand, reports of elevated concentrations of granule cargo in blood and BALF suggest that azurophilic, specific, and gelatinase granules all undergo exocytosis in vivo in COVID-19 (21, 48–51). Taken together, we suggest that NDN from COVID-19 patients undergo in vivo exocytosis of all granule subsets in the hierarchical manner previously described (55, 56), leading to release of toxic granule cargo and priming of respiratory burst activity (57, 58).
LDN are present in a number of disease states, including cancer, sepsis, and autoimmune disorders (59–62), and small numbers of LDN were recently reported in healthy individuals (63). LDN account for an average of 17% of PBMCs in systemic lupus erythematosus (SLE) patients (range 1.2% to 54%) (62), and the number of LDN correlates with disease activity (64). Significant heterogeneity of LDN functional responses appears to be disease dependent (59, 60), and LDN may represent primed, mature neutrophils in healthy individuals (63). Mistry et al. (65) described reduced NET formation, chemotaxis, and phagocytosis in CD10- LDN, compared to CD10+ LDN in SLE, while degranulation was enhanced in both LDN populations. LDN from SLE patients are not immunosuppressive (66), while both immunosuppressive and antitumor LDN phenotypes are described in cancer (59). LDN are reported to account for 20% to 50% of neutrophils in COVID-19 ARDS and 5% to 20% in moderate COVID-19 and non-COVID ARDS (16, 22, 25, 28, 31, 53). Consistent with those reports, LDN account for over 50% of neutrophils in our severe/critical COVID-19 patients, and functional activities differed between LDN and NDN. COVID LDN were not primed for increased phagocytosis, respiratory burst activity, or degranulation. Secretory vesicles showed increased mobilization, which may be explained by their sensitivity to stimuli leading to the earliest and most complete mobilization (55, 56). The responses to in vitro incubation with TNFa indicate an impaired capability of LDN to undergo priming, as only respiratory burst activity and exocytosis of specific granules are enhanced. Previous studies reported increased neutrophil expression of genes encoding proteins involved in neutrophil degranulation in COVID-19, interpreted as indicating enhanced granule mobilization (23–25, 29). Proteins destined to be incorporated into granules are produced during different phases of neutrophil maturation (67). Our results suggest that gene expression of granule constituents in COVID LDN may be due to the presence of immature neutrophil progenitors with incomplete granule formation and does not necessarily predict granule mobilization. A number of studies report increased LDN plasma membrane expression of CD63 and CD66b, compared to NDN (9, 28, 29), while CD11b expression in COVID LDN varied (27, 28, 31, 32, 36, 46). The ‘targeting by timing hypothesis’ predicts granule heterogeneity results from protein synthesis at the time individual granule subsets are formed (67). Thus, granule marker expression is impacted by the proportion of mature and immature neutrophils in various LDN clusters.
Evidence supports NET formation as critical in the development of ARDS and immunothrombosis in COVID-19 patients (10–17). The present study found spontaneous and stimulated NET formation was enhanced in COVID LDN, compared to COVID NDN and HD NDN. NET formation by COVID NDN was numerically, but not statistically, higher than that of HD NDN. Our results are consistent with previous studies showing COVID-19 neutrophils exhibit enhanced spontaneous and stimulated NET formation (12, 13, 15, 16). Our study suggests that LDN are primarily responsible for NET formation in COVID-19, similar to LDN in SLE (61, 65).
We recently reported that expression of intermediate levels of CD16 identifies a unique population of LDN that emerges in severe COVID-19, and that the proportion of CD16Int LDN predicts clinical outcome (16). On average CD16Int LDN accounted for 13% (range 1% to 34%) of LDN in our patients. While sorted cells were not available for phagocytosis and respiratory burst assays in the present study, we reported previously that CD16Int LDN show enhanced phagocytosis, as well as enhanced platelet activation and cytokine production (16). In the present study CD16Int LDN showed reduced secretory vesicle, gelatinase granule, and azurophilic granule exocytosis, while, surprisingly, specific granule exocytosis was enhanced. Although our analysis was limited by the availability of only 3 samples, CD16Int LDN appear more likely to form NETs upon stimulation than CD16Hi LDN. This observation is consistent with our previous study showing spontaneous in vitro NET formation by CD16Int LDN (16). Thus, our results support CD16Int LDN as a unique subset of neutrophils in COVID-19 expressing of both enhanced and impaired pro-inflammatory functional responses.
In a number of diseases LDN contain granulocytic myeloid-derived suppressor cells (G-MDSC) that suppress the adaptive immune response by inhibiting proliferation of T-cells, promoting T-cell anergy, and recruiting regulatory T (Treg) cells (68, 69). Single cell RNA sequencing analysis identified a cluster of circulating LDN with characteristics of G-MDSC that are increased in severe COVID-19 (25–27, 53, 54). Cultivation of activated T cells with COVID-19 LDN or their supernatants inhibit T cell proliferation and impair IFN-γ production (27, 38, 53, 54). The current study found that CD16Hi LDN and COVID NDN failed to support T-cell proliferation, compared to HD NDN. On the other hand, CD16Int LDN promoted T cell proliferation to a level similar to HD NDN. These findings suggest that the proportion of different neutrophil subsets in the lung or circulation determine the level of T-cell proliferation and activation. Thus, changing proportions of neutrophil subsets over the course of COVID-19 may play a role in disease outcome. All 3 neutrophil populations from critically ill COVID-19 patients suppressed stimulation of IFN-γ production, compared to HD NDN. A relationship between neutrophil degranulation and T-cell activation has been established (70–73), providing one potential explanation for the differential T-cell regulation by neutrophil populations in COVID-19. We interpret these results to show differential regulation of the adaptive immune response by different neutrophil populations in COVID-19 patients, while IFN-γ release from effector T-cells is inhibited by all neutrophil populations.
We acknowledge several weaknesses in our study. We could not determine the influence, if any, of corticosteroid therapy on functional changes of neutrophil subsets in COVID-19, as most patients in our cohort received corticosteroids. Recent studies reported disparate results on the effect of steroid treatment on neutrophil functions (28, 74). Many of our patients were recruited when the delta variant was the predominant cause of infection, however, the SARS-CoV-2 variant infecting the patients in our study was not determined. Thus, we cannot conclude that all SARS-CoV-2 variants induce similar changes in neutrophil population functional responses. The time at which blood was obtained after infection with SARS-CoV-2 was not uniform, averaging 8.5 days after hospitalization, and longitudinal sampling was not performed. Due to limitations in the amount of blood obtained from each patient, each functional analysis was only performed on a fraction of patients from the entire cohort. Additionally, the percent of CD16Int LDN varies over time (16), and the distribution of NDN, CD16Int LDN, and CD16Hi LDN in the present study is based on results from a fraction of patients from our cohort. Thus, we could not determine possible relationships between functional responses and neutrophil subset distribution, comorbidities, and patient outcome, nor determine the timing of neutrophil functional changes relative to the onset of ARDS. None of the neutrophil granule plasma membrane markers is restricted to a single granule subset. For example, analysis of granule constituents by Rorvig et al. (75) shows that the distribution of CD63 is 79% in azurophilic granules, 14% in specific granules, and 5% in gelatinase granules, while CD11b distribution is 2% in azurophilic, 37% in specific, 43% in gelatinase granules.
In summary, our study shows that functional capabilities of NDN and LDN differ significantly in COVID-19 patients. NDN are primed for enhanced phagocytosis, respiratory burst, and degranulation, while NET formation and stimulation of T-cell activation are impaired. The presence of both activated and immunosuppressive functions by COVID NDN are consistent with the gene signatures described by Auschenbrenner et al. (23). In contrast, priming and activation of COVID LDN are impaired, with the notable exception of NET formation. We previously reported emergence of a unique subset of LDN, CD16Int LDN, that expresses a pro-inflammatory gene signature (16). CD16Int LDN are functionally unique in that they show enhanced phagocytosis, NET formation, platelet activation, cytokine production, and T-cell activation, while degranulation is impaired. Our data suggest the hypothesis that different neutrophil populations possess complementary pro-inflammatory and immunosuppressive functional signatures, and those neutrophil populations may cooperate to produce ARDS and hypercoagulation in severe COVID-19.
Materials and Methods
Patient Recruitment
The Institutional Review Board at University of Louisville approved the present study and written informed consent was obtained from subjects or their legal authorized representatives (IRB No. 20. 0321). Patients were recruited between November, 2020 and December, 2021. Inclusion criteria were all hospitalized adults (older than 18) who have positive SARS-CoV-2 test results and were consented to this study. Exclusion criteria included age younger than 18 and refusal to participate. COVID-19 patients enrolled in this study were diagnosed with a 2019-CoV detection kit using real-time reverse transcriptase–polymerase chain reaction performed at the University of Louisville Hospital Laboratory from nasal pharyngeal swab samples obtained from patients. The identification of COVID-19 patients with severe/critical disease was based on the requirement for high-flow oxygen support or the need for mechanical ventilation and this group had blood draw along with their standard laboratory work. All COVID-19 patients were followed by the research team daily and the clinical team was blinded to findings of the research analysis to avoid potential bias.
The demographic characteristics (age, sex, height, weight, Body Mass Index (BMI) and clinical data (symptoms, comorbidities, laboratory findings, treatments, complications, and outcomes) were collected prospectively. All data were independently reviewed and entered into a computer database. For hospital laboratory complete blood count (CBC) tests, normal values are the following: white blood cell (4.1-10.8 x103/μL); hemoglobin (13.7-17.5g/dL); platelet (140-370 x103/μL). For hospital laboratory inflammatory and coagulation markers, normal values are the following: D-dimer (0.19-0.74 μg/ml Fibrinogen Equivalent Units); ferritin (7-350 ng/ml); lactate dehydrogenase (LDH) (100-242 Units/Liter); C-reactive protein (<3.0 mg/L).
Neutrophil Isolation
Table 1 shows that blood for neutrophil isolation was collected an average of 8.5 days (median 6.5 days) after hospitalization (range 1 to 36 days). Blood was collected into citrate. Neutrophils were isolated from the blood of healthy donors using plasma-Percoll gradients as previously described (76, 77). Microscopic evaluation of isolated cells showed that >92% of cells were neutrophils. Trypan blue exclusion indicated that >95% of cells were viable. Neutrophils isolated by gradient centrifugation were used in flow cytometric experiments examining phagocytosis, H2O2 generation, and exocytosis, as neutrophils were specifically gated. The Institutional Review Board of the University of Louisville approved the use of human donors who provided informed consent (IRB No. 96.0191).
Alternatively, neutrophils were isolated from whole blood of healthy donors and COVID-19 patients using an EasySep Direct Human Neutrophil Isolation Kit (STEMCELL Technologies, Cat# 19666) accordingly to the manufacturer’s instructions and >95% purity was obtained. The purified PMNs were resuspended in colorless RPMI and experiments were performed at 37°C in 5% CO2. Neutrophils isolated by this method were used in NET assays.
Isolation of CD16 LDN by Cell Sorting
PBMCs were isolated from peripheral blood using a Ficoll gradient centrifugation (Lympholyte®-H solution, CEDARLANE) followed by isolation of the buffy-coat layer. The cells were then washed using complete RPMI-1640 (cRPMI) (Sigma). The cell pellet was washed with sterile, cold PBS and added with Fc block (Human TruStain FcX™, Biolegend) to incubate at 4°C for 10 minutes followed by staining with anti-human monoclonal antibodies (mAb) for CD66b-PE (Clone G10F5, Biolegend) and CD16-APC (Clone 3G8, Biolegend) and with the viability dye eFluor 780 (eBioscience), then incubated at 4°C for 20 minutes in dark. The cells were washed with 3-5 ml of MACs buffer (Miltenyi Biotech) by centrifugation at 500 g for 5 minutes. The pellet was then suspended in MACs buffer, filtered, and the cells sorted with a BD FACsAria III sorter to collect the CD16hi (gated as viable, CD66b+CD16hi) and CD16int LDNs (gated as viable, CD66b+CD16int) using separate collection tubes containing a mixture of 50% Fetal Bovine Serum (Atlanta Biologicals), 40% PBS (Sigma) and 10% HEPES buffer (Corning). After collection, the collection tubes were centrifuged at 500 g at 4°C for 8 minutes, washed once with cRPMI, counted and resuspended in required volumes of cRPMI.
Phagocytosis and H2O2 Generation
To measure H2O2 production, isolated neutrophils or PBMCs (4 x 106 cells/ml) were incubated with 2’,7’-dichlorofluorescin diacetate (final concentration 0.5 mM; Molecular Probes/Invitrogen, Carlsbad, CA, USA) for 10 min at 37°C. Fifty microliters of cell suspension was sampled before, and 10 min after, the addition of 50 ml of opsonized, propidium iodide-labeled Staphylococcus aureus (final concentration ~108 bacteria/ml). Samples were analyzed for uptake of labeled bacteria and oxidation of 2’,7’-dichlorofluorescin diacetate to fluorescent 2’,7’-dichlorofluorescein by flow cytometry as previously described (57, 76). The LDN population in the PBMC fraction was defined for analysis by gating on CD66b+ cells.
Degranulation
Exocytosis of secretory vesicles, gelatinase granules, specific granules, and azurophilic granules in HD NDN, COVID NDN, and COVID LDN was determined by measuring the increase in plasma membrane binding of FITC-conjugated monoclonal anti-CD35 (clone E11; Pharmingen, San Diego, CA, USA), ALEXA-Fluor 488-conjugated monoclonal anti-CD11b (BD Biosciences, San Jose, CA, USA), FITC-conjugated monoclonal anti-CD66b (clone CLB-B13.9; Accurate Chemical, Westbury, NY, USA), and FITC-conjugated anti-CD63 (clone AHN16.1/46-4-5; Ancell Corporation, Bayport, MN, USA), respectively, on 4 x 106/ml neutrophils using a FACSCalibur flow cytometer (Becton Dickenson, Franklin Lakes, NJ, USA) as previously described (47, 57). The LDN population in the PBMC fraction was defined for analysis by gating on CD66b+ cells.
For low density neutrophil subsets, Single cell suspensions of PBMCs/Neutrophils were washed with 1 ml PBS (SIGMA) by centrifugation at 500 g for 5 minutes at 4°C. The pellets were then stained with anti-human CD66b-PE (Clone G10F5, Biolegend), anti-human CD16-APC (Clone 3G8, Biolegend), anti-human CD35-FITC (Clone 9H3, Biolegend), anti-human CD11b-PE-Cy7 (Clone CBRM1/5, Biolegend), anti-human CD63-APC-Cy7 (Clone H5C6, Biolegend) and anti-human CD10-PerCP-Cy5.5 (Clone HI10a, Biolegend) monoclonal antibodies, briefly vortexed and incubated for 30 minutes at 4°C in dark. After incubation, cells were washed with ice-cold PBS by centrifugation at 500 g for 5 minutes at 4°C. The pellets were finally suspended in 250-300 µl of cold PBS, briefly vortexed and acquired by BD FACsCanto for analysis.
Plasma concentrations of constituents of gelatinase granules (MMP9, R&D Systems, Minneapolis, MN, USA), specific granules (lactoferrin, Abcam, Cambridge, MA, USA), and azurophilic granules (HNE, Abcam, Cambridge, MA, USA) were determined by commercial ELISA kits, according to the manufacturers’ instructions.
Bacterial Preparation
Peptoanaerobacter stomatis strain CM2 (provided by Dr. Slava Epstein, Northeastern University) was cultivated in tryptic soy broth (TSB) supplemented with 20 g/L yeast extract, 1.0% hemin and 1.0% reducing agent (37.5 g/L NH4Cl, 25 g/L MgCl2 x 6H2O, 5 g/L CaCl2 x 2H2O, 50 g/L L-cysteine HCl, 5 g/L FeCl2 x 4H2O) overnight at 37°C under anaerobic conditions as previously described (78). Overnight culture of P. stomatis was diluted in fresh growth media and incubated under anaerobic conditions for 4-5 hours until reaching an OD600 between 0.4 to 0.6. For confocal assays, P. stomatis was labeled with carboxyfluorescein succinimidyl ester (CFSE, 0.25mg/ml; Invitrogen, USA) for 30 min at room temperature in the dark and washed 3 times with PBS prior to its use.
NET Formation
For confocal imaging, neutrophils 1 × 106 cells/experimental condition in RPMI media without phenol red supplemented with 10mM HEPES were seeded onto poly-L-Lysine-coated 12 mm glass coverslips that were placed in 24-well dishes. Cells were allowed to attach for 30 minutes at 37°C, after which, CFSE-labeled P. stomatis (10 MOI) was added to appropriate wells and the plate centrifuged for 10 min at 600 rcf to synchronize phagocytosis. Untreated and treated cells were incubated for an additional 3h at 37°C. After fixation with paraformaldehyde (2% final concentration) for 30 min at room temperature, cells were blocked in 1% BSA/PBS for 1h at room temperature, followed by incubation with anti-lactoferrin antibody (MP Biomedicals #55040; 1:500), in 1% BSA/PBS, overnight at 4°C. Primary antibody was washed off with three 5 min PBS washes, followed by incubation with donkey anti-rabbit Alexa-Fluor 546 (Thermo Fisher #A10040; 1:1000) in 1% BSA/PBS for 1h at room temperature. Nuclei were stained with DAPI (4′,6-diamidino-2-phenylindole), and coverslips were mounted onto glass slides using ProLong Gold Antifade reagent (Thermo Fisher). Images were acquired using an Olympus Fluoview FV-1000 confocal coupled to an Olympus 1X81 inverted microscope, 20× objective, and FV-10 ASW 2.1 software. A multi-channel scanning configuration with sequential line scanning was setup for acquisition of DAPI, FITC (P.stomatis staining), and AF546 (lactoferrin immunostaining). Optimal brightness setting for each channel was configured to yield maximal intensity without saturation and exclusion of non-specific emission. Images from all planes of cells were acquired and assembled z-stacks of the planes are presented.
For double-stranded DNA (dsDNA) quantification, neutrophils (4×106) were resuspended in 1 ml of colorless RPMI-1640 media. The neutrophils were unstimulated or stimulated with P. stomatis at multiplicity of infection (MOI) of 50. Aliquots of 250 µl of cells (1×106) were plated in triplicate in 96 well plate followed by centrifugation at 600xg at 14°C for 4 mins. The plate was then incubated for 3 h at 37°C, 5% CO2. After incubation the plate was centrifuged at 800 g at 4°C for 2 min and supernatants were transferred to tubes and stored at 4°C until use. The supernatants were quantified using Quant-iT PicoGreen dsDNA assay kit (Invitrogen, USA). The samples were incubated with equal volume of 1X PicoGreen 5 min at room temperature and protected from light. The plates were excited at 485 nm, and the fluorescence emission intensity was measured at 520 nm using a spectrofluorometer (SpectraMax M2; Molecular Devices).
T Cell Suppression of Proliferation and IFN-γ Release Assay
T cells were isolated from healthy human PBMCs, Fc blocked, and surface stained with viability dye eFluor 780 (eBioscience) and anti-human CD3-PerCp-Cy5.5mAb (Clone HIT3a, Biolegend) as mentioned earlier. The cells were then washed with 5 ml of sterile MACs running buffer (Miltenyi Biotech), filtered, and resuspended in appropriate volume of MACs buffer for acquisition by BD FACsAria III sorter. The T cells were gated as viable and CD3+ cells. The isolated T cells were washed (500 g for 5 minutes using a room temperature centrifuge) with pre-warmed 0.1% Bovine Serum Albumin (BSA, Equitech-Bio, Inc). The pellet was then suspended in appropriate volume of pre-warmed 0.1% BSA (1-10 x 106 cells/ml) and CFSE (1µM/ml) (Cell Trace™ CFSE Cell Proliferation Kit, eBioscience) was added to the cell suspension, gently mixed and incubated at 37°C for 10 minutes with gentle mixing 2-3 times during the incubation. Upon the completion of incubation, the CFSE labeling was stopped by addition of 5 ml of ice-cold cRPMI and incubation for 5 minutes on ice and centrifugation at 500 g for 5 minutes at 4°C. The CFSE-labelled T cells were then suspended in appropriate volume of cRPMI and added to U-bottomed 96 well plates (100 µl) pre-coated with anti-human CD3 antibody (Clone OKT3,1µg/ml in PBS at 4°C overnight followed by gentle washing with sterile PBS) to which 100 µl of neutrophils (a ratio of 1:1) was added, gently mixed and incubated at 37°C with 5% CO2 for 4 days. After 4 days, the culture supernatants were collected and stored at -20°C until analyzed. The cells were harvested, washed, Fc-blocked and stained for surface markers. The cells were stained for viability dye eFluor 780 (eBioscience), anti-human CD4-PE-Dazzle™ 594 (Clone A161A1, Biolegend) and anti-human CD8-APC (Clone SK1, Biolegend)mAb and acquired using FACsCanto for analysis of the T cell suppression. The supernatants were used to perform ELISA using a human IFN-γ ELISA Kit (Biolegend). ELISA was performed as per the instructions provided in the Kit protocol.
Statistical Analysis
Results are shown as mean and standard error of the mean (SEM), unless otherwise stated. Analysis of groups was by analysis of variance using SPSS V27 (IBM, Chicago). The dependent variables were phagocytosis, H2O2, CD35, CD11b, CD66b, or CD63 and the independent factors were basal and stimulated. In a separate analysis we performed a repeated measures analysis on basal and P. stomatis stimulated NET formation for the groups HD NDN, COVID LDN, and COVID NDN. A post hoc analysis was performed using a one way analysis of variance comparing NET formation between groups. Difference was determined at p=0.05 using Student-Newman-Keuls. Comparison of 2 data sets was performed by Student t-test using GraphPad Prism v7.0 (La Jolla, California) with significance defined as p < 0.05.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics Statement
The Institutional Review Board at University of Louisville approved the present study and written informed consent was obtained from subjects or their legal authorized representatives (IRB No. 20. 0321 and IRB No. 96.0191). The patients/participants provided their written informed consent to participate in this study.
Author Contributions
Study design and supervision: KM, JY, JH, SU, and MR. Sample collection, data acquisition, experimental work: RS, AV, MB, ML, XH, OC, CW, TS, SB, JI, CH, ST and TM. Data analysis, interpretation, and visualization: MB, KM, JY, SU, MR, and MB. Manuscript writing, review and editing: KM, RS, AV, MR, MB, SU, JH, and JY. All authors contributed to the article and approved the submitted version.
Funding
This work was supported in part by a grant from the Jewish Heritage Fund for Excellence Research Enhancement Grant Program and the Endowment in Translational Research at the University of Louisville School of Medicine (JY). JH is supported by National Center For Advancing Translational Sciences [U18TR003787]; the National Institute of Environmental Health Sciences [P30 (P30ES030283)]; and Gilead Sciences COMMIT COVID-19 RFP Program [IN-US-983-6063].
Author Disclaimer
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or Gilead Sciences.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2022.879686/full#supplementary-material
References
- 1. Matthay MA, Zemans RL, Zimmerman GA, Arabi YM, Beitler JR, Mercat A, et al. Acute Respiratory Distress Syndrome. Nat Rev Dis Primers (2019) 5(1):18. doi: 10.1038/s41572-019-0069-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Grommes J, Soehnlein O. Contribution of Neutrophils to Acute Lung Injury. Mol Med (2011) 17(3-4):293–307. doi: 10.2119/molmed.2010.00138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. Clinical Features of Patients Infected With 2019 Novel Coronavirus in Wuhan, China. Lancet (2020) 395:497–506. doi: 10.1016/S0140-6736(20)30183-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mo P, Xing Y, Xiao Y, Deng L, Zhao Q, Wang H, et al. Clinical Characteristics of Refractory Coronavirus Disease 2019 in Wuhan, China. Clin Infect Dis (2021) 73(11):e4208–13. doi: 10.1093/cid/ciaa270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Matthay MA, Aldrich JM, Gotts JE. Treatment for Severe Acute Respiratory Distress Syndrome From COVID-19. Lancet Respir Med (2020) 8:433–4. doi: 10.1016/S2213-2600(20)30127-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ginestra JC, Mitchell OJL, Anesi GL, Christie JD. COVID-19 Critical Illness: A Data-Driven Review. Annu Rev Med (2022) 73:95–111. doi: 10.1146/annurev-med-042420-110629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Veras FP, Pontelli MC, Silva CM, Toller-Kawahisa JE, de Lima M, Nascimento DC, et al. SARS-CoV-2-Triggered Neutrophil Extracellular Traps Mediate COVID-19 Pathology. J Exp Med (2020) 217(12):e20201129. doi: 10.1084/jem.20201129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Masso-Silva JA, Moshensky A, Lam MTY, Odish M, Patel A, Xu L, et al. Increased Peripheral Blood Neutrophil Activation Phenotypes and NETosis in Critically Ill COVID-19 Patients: A Case Series and Review of the Literature. Clin Infect Dis (2021) 74(3):479–89. doi: 10.1101/2021.01.14.21249831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Leppkes M, Knopf J, Naschberger E, Lindemann A, Singh J, Herrmann I, et al. Vascular Occlusion by Neutrophil Extracellular Traps in COVID-19. EBioMedicine (2020) 58:102925. doi: 10.1016/j.ebiom.2020.102925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Barnes BJ, Adrover JM, Baxter-Stoltzfus A, Borczuk A, Cools-Lartigue J, Crawford JM, et al. Targeting Potential Drivers of COVID-19: Neutrophil Extracellular Traps. J Exp Med (2020) 217(6):e20200652. doi: 10.1084/jem.20200652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Radermecker C, Detrembleur N, Guiot J, Cavalier E, Henket M, d'Emal C, et al. Neutrophil Extracellular Traps Infiltrate the Lung Airway, Interstitial, and Vascular Compartments in Severe COVID-19. J Exp Med (2020) 217(12):e20201012. doi: 10.1084/jem.20201012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Middleton EA, He XY, Denorme F, Campbell RA, Ng D, Salvatore SP, et al. Neutrophil Extracellular Traps Contribute to Immunothrombosis in COVID-19 Acute Respiratory Distress Syndrome. Blood (2020) 136(10):1169–79. doi: 10.1182/blood.2020007008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang J, Li Q, Yin Y, Zhang Y, Cao Y, Lin X, et al. Excessive Neutrophils and Neutrophil Extracellular Traps in COVID-19. Front Immunol (2020) 11:2063. doi: 10.3389/fimmu.2020.02063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ouwendijk WJD, Raadsen MP, van Kampen JJA, Verdijk RM, von der Thusen JH, Guo L, et al. High Levels of Neutrophil Extracellular Traps Persist in the Lower Respiratory Tract of Critically Ill Patients With Coronavirus Disease 2019. J Infect Dis (2021) 223(9):1512–21. doi: 10.1093/infdis/jiab050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nicolai L, Leunig A, Brambs S, Kaiser R, Weinberger T, Weigand M, et al. Immunothrombotic Dysregulation in COVID-19 Pneumonia Is Associated With Respiratory Failure and Coagulopathy. Circulation (2020) 142(12):1176–89. doi: 10.1161/CIRCULATIONAHA.120.048488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Morrissey SM, Geller AE, Hu X, Tieri D, Ding C, Klaes CK, et al. A Specific Low-Density Neutrophil Population Correlates With Hypercoagulation and Disease Severity in Hospitalized COVID-19 Patients. JCI Insight (2021) 6(9):e148435. doi: 10.1172/jci.insight.148435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhou Z, Ren L, Zhang L, Zhong J, Xiao Y, Jia Z, et al. Heightened Innate Immune Responses in the Respiratory Tract of COVID-19 Patients. Cell Host Microbe (2020) 27(6):883–890.e2. doi: 10.1016/j.chom.2020.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shaath H, Vishnubalaji R, Elkord E, Alajez NM. Single-Cell Transcriptome Analysis Highlights a Role for Neutrophils and Inflammatory Macrophages in the Pathogenesis of Severe COVID-19. Cells (2020) 9(11):2374. doi: 10.3390/cells9112374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ronit A, Berg RMG, Bay JT, Haugaard AK, Ahlström MG, Burgdorf KS, et al. Compartmental Immunophenotyping in COVID-19 ARDS: A Case Series. J Allergy Clin Immunol (2021) 147(1):81–91. doi: 10.1016/j.jaci.2020.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cambier S, Metzemaekers M, de Carvalho AC, Nooyens A, Jacobs C, Vanderbeke L, et al. Atypical Response to Bacterial Coinfection and Persistent Neutrophilic Bronchoalveolar Inflammation Distinguish Critical COVID-19 From Influenza. JCI Insight (2022) 7(1):e155055. doi: 10.1172/jci.insight.155055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Seren S, Derian L, Keleş I, Guillon A, Lesner A, Gonzalez L, et al. Proteinase Release From Activated Neutrophils in Mechanically Ventilated Patients With non-COVID-19 and COVID-19 Pneumonia. Eur Respir J (2021) 57:2003755. doi: 10.1183/13993003.03755-2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Silvin A, Chapuis N, Dunsmore G, Goubet AG, Dubuisson A, Derosa L, et al. Elevated Calprotectin and Abnormal Myeloid Cell Subsets Discriminate Severe From Mild COVID-19. Cell (2020) 182(6):1401–18. doi: 10.1016/j.cell.2020.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Aschenbrenner AC, Mouktaroudi M, Krämer B, Oestreich M, Antonakos N, Nuesch-Germano M, et al. German COVID-19 Omics Initiative (DeCOI). Disease Severity-Specific Neutrophil Signatures in Blood Transcriptomes Stratify COVID-19 Patients. Genome Med (2021) 13(1):7. doi: 10.1186/s13073-020-00823-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liao M, Liu Y, Yuan J, Wen Y, Xu G, Zhao J, et al. Single-Cell Landscape of Bronchoalveolar Immune Cells in Patients With COVID-19. Nat Med (2020) 26(6):842–4. doi: 10.1038/s41591-020-0901-9 [DOI] [PubMed] [Google Scholar]
- 25. Schulte-Schrepping J, Reusch N, Paclik D, Baßler K, Schlickeiser S, Zhang B, et al. Deutsche COVID-19 OMICS Initiative (DeCOI). Severe COVID-19 Is Marked by a Dysregulated Myeloid Cell Compartment. Cell (2020) 182(6):1419–1440.e23. doi: 10.1016/j.cell.2020.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mukund K, Nayak P, Ashokkumar C, Rao S, Almeda J, Betancourt-Garcia MM, et al. Immune Response in Severe and Non-Severe Coronavirus Disease 2019 (COVID-19) Infection: A Mechanistic Landscape. Front Immunol (2021) 12:738073. doi: 10.3389/fimmu.2021.738073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cabrera LE, Pekkarinen PT, Alander M, Nowlan KHA, Nguyen NA, Jokiranta S, et al. Characterization of Low-Density Granulocytes in COVID-19. PloS Pathog (2021) 17(7):e1009721. doi: 10.1371/journal.ppat.1009721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Reyes L, Sanchez-Garcia M A, Morrison T, Howden AJM, Watts ER, Arienti S, et al. A Type I IFN, Prothrombotic Hyperinflammatory Neutrophil Signature is Distinct for COVID-19 ARDS. Wellcome Open Res (2021) 6:38. doi: 10.12688/wellcomeopenres.16584.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lourda M, Dzidic M, Hertwig L, Bergsten H, Palma Medina LM, Sinha I, et al. Karolinska Ki/K COVID-19 Study Group. High-Dimensional Profiling Reveals Phenotypic Heterogeneity and Disease-Specific Alterations of Granulocytes in COVID-19. Proc Natl Acad Sci USA (2021) 118(40):e2109123118. doi: 10.1073/pnas.2109123118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wilk AJ, Lee MJ, Wei B, Parks B, Pi R, Martínez-Colón GJ, et al. Multi-Omic Profiling Reveals Widespread Dysregulation of Innate Immunity and Hematopoiesis in COVID-19. J Exp Med (2021) 218(8):e20210582. doi: 10.1084/jem.20210582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Parackova Z, Zentsova I, Bloomfield M, Vrabcova P, Smetanova J, Klocperk A, et al. Disharmonic Inflammatory Signatures in COVID-19: Augmented Neutrophils' But Impaired Monocytes' and Dendritic Cells' Responsiveness. Cells (2020) 9(10):2206. doi: 10.3390/cells9102206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Peruzzi B, Bencini S, Capone M, Mazzoni A, Maggi L, Salvati L, et al. Quantitative and Qualitative Alterations of Circulating Myeloid Cells and Plasmacytoid DC in SARS-CoV-2 Infection. Immunology (2020) 161(4):345–53. doi: 10.1111/imm.13254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mazzitelli I, Bleichmar L, Ludueña MG, Pisarevsky A, Labato M, Chiaradia V, et al. Immunoglobulin G Immune Complexes May Contribute to Neutrophil Activation in the Course of Severe Coronavirus Disease 2019. J Infect Dis (2021) 224(4):575–85. doi: 10.1093/infdis/jiab174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Seery V, Raiden SC, Algieri SC, Grisolía NA, Filippo D, De Carli N, et al. Blood Neutrophils From Children With COVID-19 Exhibit Both Inflammatory and Anti-Inflammatory Markers. EBioMedicine (2021) 67:103357. doi: 10.1016/j.ebiom.2021.103357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Spijkerman R, Bongers SH, Bindels BJJ, Tinnevelt GH, Giustarini G, Jorritsma NKN, et al. Flow Cytometric Evaluation of the Neutrophil Compartment in COVID-19 at Hospital Presentation: A Normal Response to an Abnormal Situation. J Leukoc Biol (2021) 109(1):99–114. doi: 10.1002/JLB.5COVA0820-520RRR [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Vitte J, Diallo AB, Boumaza A, Lopez A, Michel M, Allardet-Servent J, et al. A Granulocytic Signature Identifies COVID-19 and Its Severity. J Infect Dis (2020) 222(12):1985–96. doi: 10.1093/infdis/jiaa591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bost P, De Sanctis F, Canè S, Ugel S, Donadello K, Castellucci M, et al. Deciphering the State of Immune Silence in Fatal COVID-19 Patients. Nat Commun (2021) 12(1):1428. doi: 10.1038/s41467-021-21702-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. World Health Organization (WHO) . Living Guidance for Clinical Management of COVID-19. In: Living Guidance. Geneva: World Health Organization; (2021). [Google Scholar]
- 39. Liu Y, Du X, Chen J, Jin Y, Peng L, Wang HHX, et al. Neutrophil-To-Lymphocyte Ratio as an Independent Risk Factor for Mortality in Hospitalized Patients With COVID-19. J Infect (2020) 81(1):e6–e12. doi: 10.1016/j.jinf.2020.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lei R, Mohan C. Immunological Biomarkers of COVID-19. Crit Rev Immunol (2020) 40(6):497–512. doi: 10.1615/CritRevImmunol.2020035652 [DOI] [PubMed] [Google Scholar]
- 41. Liu J, Liu Y, Xiang P, Pu L, Xiong H, Li C, et al. Neutrophil-To-Lymphocyte Ratio Predicts Critical Illness Patients With 2019 Coronavirus Disease in the Early Stage. J Transl Med (2020) 18(1):206. doi: 10.1186/s12967-020-02374-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kuri-Cervantes L, Pampena MB, Meng W, Rosenfeld AM, Ittner CAG, Weisman AR, et al. Comprehensive Mapping of Immune Perturbations Associated With Severe COVID-19. Sci Immunol (2020) 5(49):eabd7114. doi: 10.1126/sciimmunol.abd7114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nauseef WM. How Human Neutrophils Kill and Degrade Microbes: An Integrated View. Immunol Rev (2007) 219:88–102. doi: 10.1111/j.1600-065X.2007.00550.x [DOI] [PubMed] [Google Scholar]
- 44. Laforge M, Elbim C, Frère C, Hémadi M, Massaad C, Nuss P, et al. Tissue Damage From Neutrophil-Induced Oxidative Stress in COVID-19. Nat Rev Immunol (2020) 20(9):515–6. doi: 10.1038/s41577-020-0407-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Vardhana SA, Wolchok JD. The Many Faces of the Anti-COVID Immune Response. J Exp Med (2020) 217(6):e20200678. doi: 10.1084/jem.20200678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Karawajczyk M, Douhan Håkansson L, Lipcsey M, Hultström M, Pauksens K, Frithiof R, et al. High Expression of Neutrophil and Monocyte CD64 With Simultaneous Lack of Upregulation of Adhesion Receptors CD11b, CD162, CD15, CD65 on Neutrophils in Severe COVID-19. Ther Adv Infect Dis (2021) 8:20499361211034065. doi: 10.1177/20499361211034065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Jog NR, Rane MJ, Lominadze G, Luerman GC, Ward RA, McLeish KR. The Actin Cytoskeleton Regulates Exocytosis of All Neutrophil Granule Subsets. Am J Physiol Cell Physiol (2007) 292:1690–700. doi: 10.1152/ajpcell.00384.2006 [DOI] [PubMed] [Google Scholar]
- 48. Meizlish ML, Pine AB, Bishai JD, Goshua G, Nadelmann ER, Simonov M, et al. A Neutrophil Activation Signature Predicts Critical Illness and Mortality in COVID-19. Blood Adv (2021) 5(5):1164–77. doi: 10.1182/bloodadvances.2020003568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ueland T, Heggelund L, Lind A, Holten AR, Tonby K, Michelsen AE, et al. Elevated Plasma sTIM-3 Levels in Patients With Severe COVID-19. J Allergy Clin Immunol (2021) 147(1):92–8. doi: 10.1016/j.jaci.2020.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ueland T, Holter JC, Holten AR, Müller KE, Lind A, Bekken GK, et al. Distinct and Early Increase in Circulating MMP-9 in COVID-19 Patients With Respiratory Failure. J Infect (2020) 81(3):e41–3. doi: 10.1016/j.jinf.2020.06.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Guéant JL, Guéant-Rodriguez RM, Fromonot J, Oussalah A, Louis H, Chery C, et al. Elastase and Exacerbation of Neutrophil Innate Immunity are Involved in Multi-Visceral Manifestations of COVID-19. Allergy (2021) 76(6):1846–58. doi: 10.1111/all.14746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Agrati C, Sacchi A, Bordoni V, Cimini E, Notari S, Grassi G, et al. Expansion of Myeloid-Derived Suppressor Cells in Patients With Severe Coronavirus Disease (COVID-19). Cell Death Differ (2020) 27(11):3196–207. doi: 10.1038/s41418-020-0572-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sacchi A, Grassi G, Bordoni V, Lorenzini P, Cimini E, Casetti R, et al. Early Expansion of Myeloid-Derived Suppressor Cells Inhibits SARS-CoV-2 Specific T-Cell Response and may Predict Fatal COVID-19 Outcome. Cell Death Dis (2020) 11(10):921. doi: 10.1038/s41419-020-03125-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hazeldine J, Lord JM. Neutrophils and COVID-19: Active Participants and Rational Therapeutic Targets. Front Immunol (2021) 12:680134. doi: 10.3389/fimmu.2021.680134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sengeløv H, Kjeldsen L, Borregaard N. Control of Exocytosis in Early Neutrophil Activation. J Immunol (1993) 150(4):1535–43. [PubMed] [Google Scholar]
- 56. Sengeløv H, Follin P, Kjeldsen L, Lollike K, Dahlgren C, Borregaard N. Mobilization of Granules and Secretory Vesicles During In Vivo Exudation of Human Neutrophils. J Immunol (1995) 154(8):4157–65. [PubMed] [Google Scholar]
- 57. Uriarte SM, Rane MJ, Luerman GC, Barati MT, Ward RA, Nauseef WM, et al. Granule Exocytosis Contributes to Priming and Activation of the Human Neutrophil Respiratory Burst. J Immunol (2011) 187(1):391–400. doi: 10.4049/jimmunol.1003112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. McLeish KR, Uriarte SM, Tandon S, Creed TM, Le J, Ward RA. Exocytosis of Neutrophil Granule Subsets and Activation of Prolyl Isomerase 1 are Required for Respiratory Burst Priming. J Innate Immun (2013) 5(3):277–89. doi: 10.1159/000345992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Silvestre-Roig C, Fridlender ZG, Glogauer M, Scapini P. Neutrophil Diversity in Health and Disease. Trend Immunol (2019) 40:565–83. doi: 10.1016/j.it.2019.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Seman BG, Robinson CM. The Enigma of Low-Density Granulocytes in Humans: Complexities in the Characterization and Function of LDGs During Disease. Pathogens (2021) 10:109. doi: 10.3390/pathogens10091091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Tay SH, Celhar T, Fairhurst AM. Low-Density Neutrophils in Systemic Lupus Erythematosus. Arthritis Rheumatol (2020) 72(10):1587–95. doi: 10.1002/art.41395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Carmona-Rivera C, Kaplan MJ. Low Density Granulocytes: A Distinct Class of Neutrophils in Systemic Autoimmunity. Semin Immunopathol (2013) 35:455–63. doi: 10.1007/s00281-013-0375-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Blanco-Camarillo C, Aleman OR, Rosales C. Low-Density Neutrophils in Healthy Individuals Display a Mature Primed Phenotype. Front Immunol (2021) 12:672520. doi: 10.3389/fimmu.2021.672520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Denny MF, Yalavarthi S, Zhao W, Thacker SG, Anderson M, Sandy AR, et al. A Distinct Subset of Proinflammatory Neutrophils Isolated From Patients With Systemic Lupus Erythematosus Induces Vascular Damage and Synthesizes Type I IFNs. J Immunol (2010) 184(6):3284–97. doi: 10.4049/jimmunol.0902199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Mistry P, Nakabo S, O'Neil L, Goel RR, Jiang K, Carmona-Rivera C, et al. Transcriptomic, Epigenetic, and Functional Analyses Implicate Neutrophil Diversity in the Pathogenesis of Systemic Lupus Erythematosus. Proc Natl Acad Sci USA (2019) 116(50):25222–8. doi: 10.1073/pnas.1908576116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Rahman S, Sagar D, Hanna RN, Lightfoot YL, Mistry P, Smith CK, et al. Low-Density Granulocytes Activate T Cells and Demonstrate a non-Suppressive Role in Systemic Lupus Erythematosus. Ann Rheum Dis (2019) 78(7):957–66. doi: 10.1136/annrheumdis-2018-214620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Cowland JB, Borregaard N. Granulopoiesis and Granules of Human Neutrophils. Immunol Rev (2016) 273(1):11–28. doi: 10.1111/imr.12440 [DOI] [PubMed] [Google Scholar]
- 68. Sanchez-Pino MD, Dean MJ, Ochoa AC. Myeloid-Derived Suppressor Cells (MDSC): When Good Intentions Go Awry. Cell Immunol (2021) 362:104302. doi: 10.1016/j.cellimm.2021.104302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Koushki K, Salemi M, Miri SM, Arjeini Y, Keshavarz M, Ghaemi A. Role of Myeloid-Derived Suppressor Cells in Viral Respiratory Infections; Hints for Discovering Therapeutic Targets for COVID-19. BioMed Pharmacother (2021) 144:112346. doi: 10.1016/j.biopha.2021.112346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Minns D, Smith KJ, Hardisty G, Rossi AG, Gwyer Findlay E. The Outcome of Neutrophil-T Cell Contact Differs Depending on Activation Status of Both Cell Types. Front Immunol (2021) 12:633486. doi: 10.3389/fimmu.2021.633486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Minns D, Smith KJ, Findlay EG. Orchestration of Adaptive T Cell Responses by Neutrophil Granule Contents. Mediators Inflamm (2019) 2019:8968943. doi: 10.1155/2019/8968943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Minns D, Smith KJ, Alessandrini V, Hardisty G, Melrose L, Jackson-Jones L, et al. The Neutrophil Antimicrobial Peptide Cathelicidin Promotes Th17 Differentiation. Nat Commun (2021) 12(1):1285. doi: 10.1038/s41467-021-21533-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Singel KL, Emmons TR, Khan ANH, Mayor PC, Shen S, Wong JT, et al. Mature Neutrophils Suppress T Cell Immunity in Ovarian Cancer Microenvironment. JCI Insight (2019) 4(5):e122311. doi: 10.1172/jci.insight.122311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Sinha S, Rosin NL, Arora R, Labit E, Jaffer A, Cao L, et al. Dexamethasone Modulates Immature Neutrophils and Interferon Programming in Severe COVID-19. Nat Med (2022) 28(1):201–11. doi: 10.1038/s41591-021-01576-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Rørvig S, Østergaard O, Heegaard NH, Borregaard N. Proteome Profiling of Human Neutrophil Granule Subsets, Secretory Vesicles, and Cell Membrane: Correlation With Transcriptome Profiling of Neutrophil Precursors. J Leukoc Biol (2013) 94(4):711–21. doi: 10.1189/jlb.1212619 [DOI] [PubMed] [Google Scholar]
- 76. McLeish KR, Knall C, Ward RA, Gerwins P, Coxon PY, Klein JB, et al. Activation of Mitogen-Activated Protein Kinase Cascades During Priming of Human Neutrophils by TNF-α and GM-CSF. J Leukoc Biol (1998) 64:537–45. doi: 10.1002/jlb.64.4.537 [DOI] [PubMed] [Google Scholar]
- 77. Haslett C, Guthrie LA, Kopaniak MG, Johnston RB, Henson PM. Modulation of Multiple Neutrophil Functions by Preparative Methods or Trace Concentrations of Bacterial Lipopolysaccharide. Am J Pathol (1985) 119:101–10. [PMC free article] [PubMed] [Google Scholar]
- 78. Jimenez Flores E, Tian S, Sizova M, Epstein SS, Lamont RJ, Uriarte SM. Peptoanaerobacter Stomatis Primes Human Neutrophils and Induces Granule Exocytosis. Infect Immun (2017) 85(7):e01043–16. doi: 10.1128/IAI.01043-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.




