Abstract
Immunity is governed by fundamental genetic processes. These processes shape the nature of immune cells and set the rules that dictate the myriad complex cellular interactions that power immune systems. Everything from the generation of T cell receptors and antibodies, control of epitope presentation, and recognition of pathogens by the immunoediting of cancer cells is, in large part, made possible by core genetic mechanisms and the cellular machinery that they encode. In the last decade, next-generation sequencing has been used to dissect the complexities of cancer immunity with potent effect. Sequencing of exomes and genomes has begun to reveal how the immune system recognizes “foreign” entities and distinguishes self from non-self, especially in the setting of cancer. High-throughput analyses of transcriptomes have revealed deep insights into how the tumor microenvironment affects immunotherapy efficacy. In this Review, we discuss how high-throughput sequencing has added to our understanding of how immune systems interact with cancer cells and how cancer immunotherapies work.
Introduction
The role of genetics in determining cancer immunotherapy response is born from two fundamental principles: (a) cancer is at its core a genetic disease, and (b) genetic mechanisms underlie the ability of the immune system to recognize diverse targets. These concepts evolved alongside the growing capabilities of next-generation sequencing (NGS) technologies that were increasingly used to identify targetable oncogenic driver mutations with the intent to expand the utility of genetic profiling in the clinic. The enthusiasm for this approach in immuno-oncology was amplified by the observation that the expression of the immune checkpoint inhibitor targets alone — i.e., PD-1, PD-L1, and CTLA-4 — was really quite insufficient to optimally identify patients with cancer who do or do not respond to immune checkpoint blockade (ICB) (1, 2).
The hypothesis that tumor genetics can influence immunotherapy response in patients with cancer was first observed in the setting of anti–CTLA-4 therapy for metastatic melanoma (2). It was observed that tumor mutational burden (TMB), which reflects the number of immunogenic neoantigens, was predictive of therapeutic response. Following the demonstration of this foundational principle — that alterations of the somatic genetic landscape can directly affect immunogenicity — biomarker development in immuno-oncology expanded rapidly. Investigations examining the effect of many types of genetic alterations — individual gene mutations, mutational signatures, genomic instability, and host genetics — have revealed a constellation of factors that influence the efficacy of ICB (3).
Clinically, immune checkpoint inhibitors (ICIs) have improved outcomes in oncology, including prolonging the survival of patients across various cancers whether alone or in combination with other agents (4). Despite the progress made, the majority of patients do not benefit from ICIs. Our ability to predict therapeutic benefit has improved via development of tumor- and patient-specific biomarkers in recent years; however, much improvement is needed. For instance, PD-L1 IHC is the most widely used biomarker in clinical practice for patients receiving ICB; however, many tumors with high PD-L1 expression do not respond to PD-1/PD-L1 inhibitors. Furthermore, there is an unmet clinical need to develop combinatorial biomarkers that integrate multiple determinants of response to more effectively predict ICI response above and beyond what is achievable with single biomarkers. In this Review, we acknowledge the vast and diverse efforts toward satisfying this unmet need and highlight biomarker development through the lens of genomics, given its integral role in identifying clinically relevant biomarkers as well as its potential for furthering our understanding of immuno-oncology.
Tumor cell–intrinsic determinants of immunotherapy response
TMB.
TMB most commonly refers to the number of nonsynonymous single-nucleotide variants (nsSNVs) in a tumor. It can be assessed by multiple NGS techniques, including whole-exome sequencing (WES), whole-genome sequencing, or NGS panels that sequence predetermined sets of cancer-related genes, such as MSK-Impact, Tempus xT, and the FoundationOne CDx panel. Our group first discovered that the presence of high TMB or mutations in DNA damage repair genes is likely to respond better to ICI treatment (2, 5–8). High TMB (>100 nonsynonymous mutations per exome) and the presence of specific neoepitope signatures correlated with response to and survival with CTLA-4 inhibitors in a discovery set of 25 patients and a validation set of 39 patients with melanoma. Patients with a high versus low TMB had a 50% versus 10% survival at 40 months in the discovery set (9). In June 2020, the US FDA approved pembrolizumab (anti–PD-1) for treatment of patients with malignant solid tumors of any type given 10 or more mutations per megabase (TMB ≥10), based on the KEYNOTE 158 study (10). In this prospective trial, 102 of 790 evaluable patients had tumors with a TMB ≥10. Patients with tumors with TMB ≥10 demonstrated significantly greater response rates (29% vs. 6%) than patients with tumors with TMB less than 10. In an analysis of 1678 anti–PD-1/PD-L1–treated patients with 16 different solid tumors, 25% of patients were found to have a high TMB using the TMB ≥10 cutoff. TMB-high tumors had higher response rates than TMB-low tumors in 11 of the 16 cancers. Using a cancer type–specific cutoff, TMB-high tumors showed numerically higher response rates in 14 of 16 cancers (11). Associations between high TMB and ICI response have been reported across various cancers (12), including urothelial carcinoma (13), small cell lung cancer (14), melanoma (9), and non–small cell lung cancer (NSCLC) (5). Numerous studies have validated our initial findings and, together, unequivocally validated that TMB helps drive ICI benefit.
As larger cohorts of patients are treated with ICIs, it is becoming evident that the prognostic utility of TMB is context dependent; ICI-treated patients have a better outcome with high TMB, whereas non-ICI-treated patients could potentially have worse prognosis with high TMB (8). Interestingly, frameshift insertions/deletions (fs-indels), although less common, are thought to be highly immunogenic mutations (15). These are usually degraded through the nonsense-mediated decay pathway. However, a fraction of fs-indels can escape degradation and have been found to predict response to ICIs even in TMB-low tumors (16). While TMB has provided the ability to select for patients likelier to respond to ICIs, better prediction approaches will necessitate the use of this biomarker in conjunction with others. Furthermore, harmonization of TMB reporting is needed, and substantial efforts are under way to accomplish this (17).
PD-L1 expression.
IHC measurement of tumoral PD-L1 is one of the most commonly used clinical tests in guiding the use of anti–PD-1 and anti–PD-L1 therapy. Several conflicting studies reported both positive correlation and no correlation between PD-L1 expression and ICI response, potentially owing to variable PD-L1 staining (14, 18–25). A meta-analysis by Liu et al. of 24 randomized trials of patients who received PD-1/PD-L1 blockade found an improved overall survival (OS) in both PD-L1–positive and –negative patients. The magnitude of benefit was dependent on the expression of PD-L1 (26). In addition to its role as an immunosuppressive molecule, PD-L1 has several tumor-intrinsic roles in cancer initiation, metabolism, inhibition of proapoptotic signals, tumor growth, epithelial-mesenchymal transition, and metastasis through downstream signaling (27–29). Although the focus of this Review is not on PD-L1 IHC, it is nonetheless important to mention that PD-L1 staining and high TMB are not identifying identical sets of patients. These biomarkers are complementary in their predictive value and can be used together to improve identification of ICI responders (30).
Tumor neoantigens and immunogenicity.
Tumor neoantigens are somatic mutation–generated peptides presented on the surface of tumor cells and are absent from normal tissues (31). These neoantigens can be presented by major histocompatibility complexes (MHCs) and trigger a neoantigen-specific T cell receptor–mediated (TCR-mediated) response (32). Neoantigens are thought to arise as a result of genetic alterations such as nonsynonymous single nucleotide variants (nsSNVs), insertions/deletions (indels), recombination events, gene fusions, or defective mRNA alternative splicing (AS) (31). Defective AS can lead to neoantigens that might change temporally as the tumor splicing machinery evolves (33). WES can be used to identify candidate neoantigens produced from nsSNVs, gene fusions, or indels. However, WES may not capture subtler neoantigenic changes from posttranscriptional or splicing-related alterations (34). Combining NGS, RNA sequencing, and proteomics can improve neoantigen prediction, and several groups have elucidated features of mutated peptides that are likelier to be immunogenic (35, 36). Although neoantigen levels generally track with TMB, it is our opinion that the current generation of prediction algorithms does not have sufficient accuracy for routine use as biomarkers (37).
Neoantigenic immunogenicity requires MHC-neoepitope complex presentation and subsequent T cell recognition. Currently, a considerable challenge stems from our inability to distinguish the neoantigens that will trigger a significant T cell immune response from those that bind to MHC without a T cell response. Multiple issues contribute to this challenge. First, immunogenic neoantigens might only be expressed by a minority of tumor cells. Single-cell sequencing techniques might be able to dissect heterogeneous tumor neoantigen expression more precisely (38). This is important because both the degree of clonal frequency and the quality of a neoantigen seem to affect its immunogenic potential (39, 40). Second, surface density of MHC-neoepitope complexes appears to contribute to the extent of T cell activation, and this density is difficult to routinely measure (41). Third, even if these neoantigens are presented, TCR recognition is variable (3). One effort to formalize a model to determine neoantigen immunogenicity has been attempted by the Tumor Neoantigen Selection Alliance (TESLA), which has identified five features that can help differentiate immune recognition of MHC-I–restricted peptides generated by SNVs and indels (36): MHC binding affinity; binding stability; clonality; the differential between the MHC binding affinity of a mutated peptide and that of the wild-type form (i.e., agretopicity); and foreignness. This model does not take some critical factors into consideration, and its construction did not use modern learning approaches, so it is unclear how generalizable it is (42, 43).
In contrast, contemporary approaches to prediction of MHC-binding potentials are increasingly relying on state-of-the-art artificial intelligence algorithms, such as neural networks (NetMHC and ConvMHC), random forest classifiers (ForestMHC), and natural language processing techniques (HLA-CNN) (44–46). MHC binding predictors have markedly improved in the last several years. While these approaches may aid in more precise identification of MHC binding predictors, there remain many challenges to bridging this to routine clinical use, as highlighted by Pearlman et al. (47).
Mutational signatures.
Tumors with mutagen-specific mutational signatures have been shown to respond to ICIs more favorably. For example, in patients with NSCLC treated with pembrolizumab, the smoking-related signature was associated with significantly improved progression-free survival (PFS) (5). Similarly, UV light–generated alterations that characterize melanoma are associated with favorable response to ICIs. In fact, the UV mutational signature has been shown to increase the hydrophobicity of neoantigens, making the neoantigens better presented by the MHC and better recognized by T cells (48). Distinct hypermutable states that result from mutations in mismatch repair (MMR) and DNA polymerase epsilon/delta (POLE/POLD1) genes are associated with improved response to ICIs (49). POLE/POLD1 mutations result in hypermutability due to their role in DNA proofreading and fidelity during replication (50). MMR deficiency and POLE/POLD1 mutations lead to a high TMB, thus conveying a better response to ICIs (51). Because TMB and DNA repair deficiencies such as microsatellite instability (MSI) are causally linked to hypermutation signatures, the quality and quantity of mutation generation and subsequent neoantigen creation cannot be functionally separated. MMR deficiency has been approved by the FDA as a tumor type–agnostic biomarker for pembrolizumab and nivolumab. Furthermore, there are now prospective clinical trials of anti–PD-1 therapy in patients harboring mutations in POLE/POLD1 that are not MSI-high (ClinicalTrials.gov NCT03810339). APOBEC mutational signatures have also been reported to be predictors of ICI response in urothelial cancer, NSCLC, and head and neck cancer (52–53). APOBEC is a family of cytidine deaminases that induce mutations in viral genomes, hindering viral replication. However, off-target activity on the host genome can also lead to a hypermutated status that has been shown in an in silico model to lead to increased hydrophobicity of the neoantigens, leading to more robust immune response (55).
Microsatellite stability.
Mutations in the MMR pathway lead to MSI and a high number of somatic mutations, which generate a high number of tumor neoantigens (56). We first observed that ICIs can result in strong response in tumors with mutations in the MMR pathway (5). This observation was subsequently verified by a clinical trial of anti–PD-1 therapy that enrolled 41 patients with MMR-deficient colorectal cancer (CRC) (cohort A), MMR-proficient CRC (cohort B), and MMR-deficient non-CRC (cohort C). Response rates were 40%, 0%, and 71%, respectively (49). A later study of PD-1 blockade in 12 different MMR-deficient cancers showed 21% complete responses and 33% partial responses. These findings led to the tissue-agnostic FDA approval of anti–PD-1 immunotherapy for solid malignancies with MSI (57). A phase III randomized trial then led to approval of pembrolizumab as first-line therapy for advanced MSI+ CRC (58). Interestingly, 45% to 70% of MSI+ patients do not respond to ICIs (59). Differences in the degree of MSI present in tumors and in indel load (59, 60), disruptions in WNT signaling, and defective antigen presentation as a result of loss of β2-microglobulin or loss of HLA molecules are all likely contributors to differential responses in MSI+ tumors (61).
Aneuploidy.
Aneuploidy or somatic copy number alterations (SCNAs) have been shown to be associated with worse outcomes after ICI treatment (62). More recently, specifically arm- and chromosome-level SCNAs were correlated with immune evasion in 10 of 12 cancers studied based on analysis of The Cancer Genome Atlas. This study also showed that tumor aneuploidy negatively correlated with survival in ICI-treated metastatic patients with melanoma in two clinical trials (63). Interestingly, total SCNA burden was not predictive of response. However, specific losses and amplifications were found to be predictive. For instance, 9p21 amplification was associated with resistance (64), while 9q34 loss was associated with higher response rates (65). Furthermore, SCNAs in IFNG, the gene encoding IFN-γ, were also found to be enriched in ICI-resistant patients with melanoma (66). Using an unbiased machine-learning method, Chowell et al. validated the effect of extent of copy number alteration on ICI response. The degree of effect is observable but smaller than that of TMB and some other factors (67).
Mutations in discrete genes influencing immunotherapy outcomes
BRAF mutations.
Mutant BRAF suppresses intratumoral T cells via overexpression of IL-1α and IL-1β by tumor cells, leading to overexpression of PD-L1 and PD-L2 in tumor-associated fibroblasts (68). Patients with melanoma with BRAF mutations had higher rates of PFS and OS when treated with combination PD-1 and CTLA-4 ICIs compared with patients with wild-type BRAF in the CheckMate 067 trial (68% vs. 53% 3-year OS, respectively; ref. 69). However, it is likely that other factors can modulate the overall influence of BRAF mutation. In a study of 68 patients with melanoma who received nivolumab, pretherapy TMB and clonal mutation load correlated with survival and response in patients who were ipilimumab naive, but not in those who previously progressed on ipilimumab. Context is therefore an important factor influencing the effects of these biomarkers.
In patients with melanoma with BRAF V600 mutation, ICIs and tyrosine kinase inhibitor therapy are both potential options. Which is the better therapy to use in the first-line setting? The DREAMseq trial recently showed that patients with BRAF V600–mutated advanced melanoma who received nivolumab plus ipilimumab followed by dabrafenib plus trametinib experienced greater OS (72%) compared with patients receiving the converse sequence (52%) (70).
Mutations in KRAS and STK11/LKB1.
KRAS is the most frequently mutated oncogene in lung adenocarcinomas. KRAS-mutated lung adenocarcinomas can be subdivided into STK11/LKB1-comutated or TP53-comutated subtypes (71). In the Stand Up To Cancer (SU2C) cohort of 174 patients treated with nivolumab, patients treated with nivolumab with STK11/LKB1 comutation had significantly lower objective response rates (7.4% vs. 28.6%), median PFS (1.8 vs. 2.7 months), and median OS (6.4 vs. 16.1 months) compared with the KRAS-mutated, LKB1 wild-type patients. STK11/LKB1 alterations were also significantly associated with PD-L1–negative status in lung adenocarcinomas with intermediate to high TMB. In addition, patients with positive PD-L1 also had worse outcomes if they had an STK11/LKB1 alteration (71). The interaction of KRAS and STK11 has also been demonstrated to have prognostic importance across cancers as coalteration of these genes is associated with overall worse prognosis (72). Therefore, these alterations may have both prognostic and predictive implications.
PTEN.
PTEN loss of function (LOF) is suggested to decrease T cell infiltration via overexpression of immunosuppressive cytokines in a melanoma preclinical model (73). Analysis of 135 resected melanoma regional metastases found that melanomas with PTEN loss had significantly lower CD8+ T cell tumor infiltration than tumors with PTEN expression (74). PTEN loss causes overactivation of the PI3K/AKT pathway, which promotes immune evasion in various ways, such as sustaining the function of regulatory T cells (75).
SWI/SNF complex, PBAF, and PBRM1.
Several genomic studies have identified various components of the SWI/SNF chromatin remodeling complex, such as polybromo- and BRG1-associated factors (PBAF), which contains PBRM1, ARID2, and BRD7, as frequently mutated genes across cancer types (76). PBAF complexes are ATP-dependent chromatin remodelers that regulate transcription (76).
PBRM1 is the second most commonly mutated gene in clear cell renal cell carcinoma (ccRCC) after VHL and is a component of SWI/SNF chromatin remodeling complexes (77). In a cohort of 62 patients with ccRCC, those who had PBRM1 LOF mutations responded better to ICIs (78). Preclinical data suggest that inactivation of components of the PBAF complex help overcome resistance to T cell–mediated cell killing in melanoma (79). However, several studies have shown conflicting implications of PBRM1 mutations in ICI response across different cancers, i.e., no association or worse response through the conferring of a nonimmunogenic phenotype (80–83). These conflicting results are difficult to reconcile but might be explained by the presence of confounding factors in these studies or the weaker effects of PRBM1 mutation on tumor immunity. Indeed, these studies were done before modern systematic efforts to quantify biomarker contribution to ICI response (67). Since it was done at an earlier time, the original study implicating PBRM1 did not account for these factors (78).
CDKN2A.
LOF mutations in CDKN2A were recently found to be associated with decreased response to ICIs in patients with NSCLC despite high PD-L1 expression and high TMB (84). Similarly, genomic alterations in CDKN2A were reported to be associated with a poor response to ICI in urothelial carcinoma in two large cohorts of patients. RNA sequencing data revealed decreased expression of genes involved in immune and inflammatory pathways in patients with CDKN2A alterations, which could provide an explanation for the deleterious effect of these mutations on response to ICIs (85).
BRCA1 and BRCA2.
BRCA1 and BRCA2 mutations are responsible for defects in homologous recombination–based DNA repair (86). Interestingly, BRCA1 and BRCA2 were shown to cause distinct changes in the tumor immune microenvironment, with BRCA2-mutated tumors having higher gene expression of both adaptive immunity– and innate immunity–related pathways. These differential effects on the tumor microenvironment (TME) lead to contrasting outcomes of ICI treatment (6). BRCA2 mutation was associated with better response and survival after ICI treatment in both patients and mice, whereas BRCA1 mutation did not demonstrate these effects. Single-cell sequencing of tumors with these mutations showed differences in myeloid cells that likely regulate the TME. A recent clinical trial (CheckMate 650) treating prostate cancer with nivolumab and ipilimumab validated previous observations on the effects of homologous recombination repair deficiency on ICI response (6, 87). It is important to note that this trial also validated the effects of TMB on ICI response in prostate cancer.
WNT/β-catenin signaling.
Activation of the WNT/β-catenin signaling pathway in melanoma has been correlated with T cell absence in human metastatic melanoma samples (88). The presence of pretreatment T lymphocytes and higher expression of a number of immune-related genes was shown to predict a better outcome to ipilimumab in patients with melanoma (89). Thus, it can be hypothesized that by decreasing T cell infiltration of melanomas, activation of β-catenin signaling can lead to decreased efficacy of ICIs. Large-scale sequencing of patients treated with ICIs also shows that tumors with WNT pathway mutations are associated with worse survival (6).
Tumor cell–extrinsic determinants of response
Along with the ongoing examination of tumor-intrinsic biomarkers, there is a rapidly growing body of evidence supporting the important effects of tumor-extrinsic factors such as host germline variation, HLA genetic variation, the microbiome, and variation in the immune microenvironment. One related observation is the finding that 15% to 20% of the variation in intratumoral immune signaling via interferon and cytotoxic cell infiltration may be explainable by heritable variants in genes including IFIH1, STING1, TMEM108, and RBL1 (90). At the interface of the immune cell–tumor cell interaction is an immune synapse dependent on the generation of an immunogenic antigen and its subsequent presentation via the HLA-I molecules. Just as genetic variations in the cancer genome affect the generation of these tumor peptides, variations in HLA genotypes and diversity shape how these peptides are used via immunopeptidome presentation. This ultimately shapes immunotherapy response (91). HLA-I molecules are encoded by B2M and the HLA-I genes (HLA-A, -B, and -C). The HLA gene family is the most polymorphic set of genes in the human genome (92). Polymorphisms of these genes are concentrated within the peptide-binding domains (93). Diversity in these peptide-binding domains leads to diversity in the presented peptides, which is selected for over evolution (94). HLA-I evolutionary divergence (HED) is a measure of diversity of physiochemical sequence divergence of the HLA molecules. In a cohort of mostly patients with melanoma who received ICIs, HED was a determinant of response and survival (91).
The power in measuring HLA-I variation seems to extend to correlation of benefit with some combination therapies, e.g., combination of tyrosine kinase inhibitors with ICIs in RCC (95). HED predicts aplastic anemia outcomes, bone marrow transplant outcomes, and organ transplant outcomes (96–98). In a meta-analysis of factors that influence ICI efficacy, Litchfield et al. also showed that HLA divergence was a significant factor influencing ICI response in melanoma (64). If HLAs are not actually typed but are estimated by imputation using microarrays, associations may be not apparent (99). Based on more recent modeling data, it appears that HED has a somewhat weaker effect compared with TMB, treatment beforehand with chemotherapy, or neutrophil/lymphocyte ratio. This may explain variation in predictive value between cohorts that are not balanced in all or some of these variables (67).
Tumor immune microenvironment.
While host- and tumor-specific genetic factors form a basic blueprint for immunogenicity, the impact of the immune system is heavily dependent on the local microenvironment, the functional capacity of immune cells, and the balance between immunostimulatory and immunosuppressive stimuli (100–102). Efforts to expand our understanding of these interactions include the development of new computational and high-throughput approaches for immune cell profiling (103–107). These strategies include characterization of the dynamics of the microenvironment in which TCR repertoires may vary, immune-enriched and immune-desert regions that can exert regional immunomodulatory effects, and the influence of the microbiome (1, 3, 108–111). Studies have found that the density of tumor-infiltrating lymphocytes (TILs) can associate with OS regardless of whether patients are treated with immunotherapy (112). An early study found that lack of CD8+ T cells from the vicinity of tumors is associated with poor outcomes in CRC (113). More recently, this finding was refined into a parameter called the Immunoscore, in which T cells are quantified in different vicinities of tumors. This assay was found to be a strong predictor of OS in CRC (112, 114–116).
Tumors can be classified according to the degree of infiltration into various categories: immune-inflamed, immune-excluded, and immune-desert types. Immune-inflamed tumors are characterized by the presence of a dense infiltrate of favorably positioned CD4+ and CD8+ T cells in the proximity of tumor cells. Immune-excluded tumors have CD8+ T cells that are excluded from the tumor parenchyma and instead found in the peritumoral stroma. This phenotype is associated with increased TGF-β signaling in tumor fibroblasts (117). Finally, immune-desert tumors lack the presence of T cells in both the tumor parenchyma and stroma (118, 119). In addition, the exact localization of T cell infiltrate in tumors was found to be associated with response to ICIs. For example, the presence of a dense infiltrate of CD8+ T cells at the invasive margin as opposed to the center of the tumor was associated with response to ICIs (1). Forays into the contribution of different cellular compartments of the microenvironment via spatial transcriptomics have revealed increasing evidence for the role of B cells, unique CD8+ T cells, and tertiary lymphoid structures in the immunotherapy response across various diseases, including melanoma, sarcoma, and RCC (120–122).
Other characteristics, such as the intratumoral and peripheral immune cell receptor repertoire, which can be quantified with metrics such as entropy, richness, and clonality of T cell and B cell populations, were also associated with response to ICIs (123–127). The TCR repertoire was shown to be positively associated with polymorphism of HLA-I loci and negatively associated with CMV positivity and age (128). However, there are conflicting data on the effect of the TCR repertoire on the response to ICIs, likely owing to the complex interactions of the TCR repertoire with ICIs, patient immunologic histories, and environments (110, 129–131). Efforts to understand these conflicting findings have focused on identifying immune cell functional states via differentiation and lineage commitments as well as receptor specificity and clonality in both bulk and single-cell sequencing studies (132–135). Here, conflicting results do not indicate deficiencies in any particular study. Rather, we are likely observing associations affected by population variation and factors we don’t yet fully understand. Furthermore, the aforementioned factors at play in the TME are not yet routinely assessed in a clinical pathology setting and so remain hypothesis-generating given the current lack of standardized validation. It is perhaps most useful to develop combination biomarker suites or nomograms that take into account multiple major biomarkers.
Development of combinatorial approaches to understand immunotherapy efficacy.
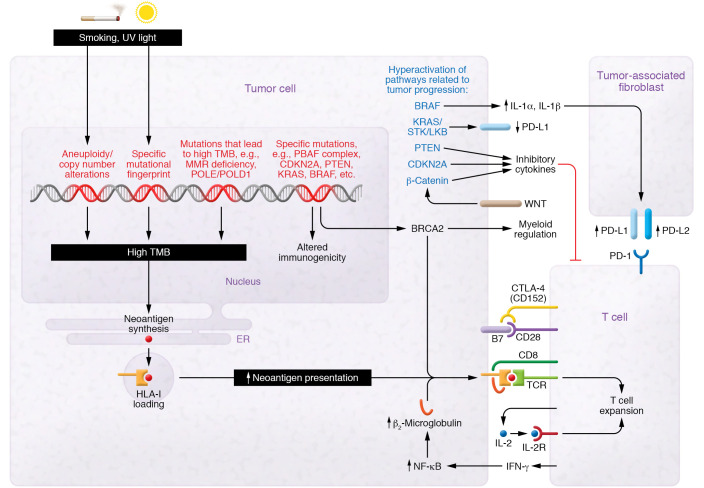
The examples above demonstrate how systematic investigations into immunotherapy biomarkers are enabled by improvement in NGS technologies. With more data available, it became possible to form hypotheses about how multifaceted molecular processes interact with the immune system. However, as more evidence arises, we approach a “long tail” of candidate biomarkers wherein the likelihood of an individual biomarker having substantial predictive capability is diminishing. Not every factor found to influence a biological process is suitable as a biomarker. Biomarkers should (a) be feasible for use in clinical settings, (b) be able to be efficiently measured using cost-efficient means, and (c) provide useful information for decision making in medical practice. Multiple large studies have now clearly shown that, among genomic biomarkers, TMB and a few other factors provide the largest pan-cancer predictive value for ICI response (64, 67). Other genomic alterations, like STK11/LKB1 mutation, can be important for specific cancers or can further modulate immunogenicity. Thus, combinatorial predictive biomarkers are clearly needed, and systems for harmonizing the various validated biomarkers are necessary to improve clinical utility. Figure 1 summarizes some important processes discussed in this Review.
Figure 1. Genetic alterations and immunologic consequences.
Certain mutagens, such as UV light and carcinogens in cigarette smoke, can lead to formation of mutations and aneuploidy, which can cause a high tumor mutational burden (TMB). Other specific alterations can lead to microsatellite instability or POLE/POLD1 mutation, which results in hypermutation. This can result in transcription and translation of tumor neoantigens. These are presented on HLA-I molecules. HLA-I is required to present tumor neoantigens to cytotoxic T cells. Certain mutations, such as KRAS/STK/LKB in lung cancer, have been associated with decreased PD-L1 expression on tumor cells. Other mutations in genes such as BRAF, CDKN2A, and PTEN as well as aberrant activation of the WNT/β-catenin pathway have been implicated in increasing the release of inhibitory cytokines in the tumor microenvironment that act on tumor-infiltrating T lymphocytes or tumor-associated fibroblasts. T cells recognize antigens presented on HLA-I as “non-self” antigens; costimulatory signals are needed for Th cell activation. Costimulatory signals involve the binding of B7 on tumor or antigen-presenting cells to CD28 on T cells. CTLA-4 (CD152) competes with CD28 for the binding of B7, thus inhibiting the necessary costimulatory signal needed. Owing to space constraints, this diagram is not comprehensive.
The promise of combinatorial biomarkers has been highlighted by large-scale efforts from pan-cancer cohorts. For instance, the combination of high TMB and low pretreatment neutrophil/lymphocyte ratio is correlated with greater benefit from ICI (136). These studies demonstrate that select factors can predict response rates across cancer types, although there may be context-specific biomarkers for which the balance and predictive ability of genomic alterations, the TME, TMB, HLA-I diversity, and other markers may vary by tumor type (64, 137). Some have proposed using a combinatorial model based on the triple axis of tumor neoantigens/microenvironment/checkpoints to explain the variance in outcomes of PD-1/PDL-1–directed therapy (137). A meta-analysis of the CPI1000+ cohort demonstrated that at least 80% of the significant biomarkers by tumor type were also significant in the overall pan-cancer cohort and that there were differential predictive potentials of each biomarker by tumor type, such as TMB between melanoma and urothelial carcinoma and loss of 9q34 between RCC and the rest of the cohort (64).
Given the marked complexity of biomarker integration, classic methods of multivariable modeling may need to be supplemented by newer machine-learning methods. The development of artificial intelligence and machine learning in medicine has exploded in recent years, with improvements in modeling and feature selection enabling better prediction of treatment outcome (45). For instance, our recent work in combinatorial biomarker development has enabled superior pan-cancer prediction of immunotherapy response over TMB alone using an exhaustive approach with random forest classifier modeling (67). In the CPI1000+ cohort meta-analysis, a decision tree model was also generated, albeit with gradient boosting–based algorithms (64).
Implications of single-cell profiling technologies for biomarker development.
The use of single-cell profiling technologies has enabled an unprecedented view into the complex dynamics between intratumoral cellular subpopulations (138–140). Recent examples of these studies have identified unique cell types associated with ICI response. These include (a) an abundance of CD8A+ tissue-resident T cells and interferon-stimulated gene–high (ISGhi) tumor-associated macrophages in RCC; (b) tissue-resident macrophages contributing to remodeling of the microenvironment during ICI treatment in lung cancer; (c) distinct neoantigen-specific TILs with specific transcriptional states; and others (103, 105, 106, 108, 138, 141). While these have provided new insights into the composition of the TME, the implications of these findings have yet to translate into clinical use (45, 140, 142). Various cellular atlases have been defined via modern single-cell sequencing, and while they are an enormous research resource, there still remain challenges in the ability to translate these efforts to clinical utility (143, 144). The combination of single-cell sequencing technologies with improved pathologic sampling, single-cell resolution multispectral imaging modalities, and microfluidics will first require new computational tools for interpretation at the biological level and subsequent systematic implementation into clinical trials for robust correlative biomarker identification (138, 142, 145–148).
Dynamic biomarker profiling
While there has been an improved effort for sequential profiling throughout therapy in recent clinical trials, many older studies have been limited to analyses of pretreatment sequencing. We are seeing now that longitudinal sampling paired with molecular profiling can identify peritreatment biomarkers of treatment sensitivity with potential to identify the emergence of adaptive or acquired resistance as well (149–151). Serial profiling has demonstrated that immunoediting is operative in patients treated with ICIs (110). In a study of the pan-cancer INSPIRE cohort, upregulation of PLA2G2D was identified as a marker of resistance to ICIs alongside B2M loss of heterozygosity and copy-number abundance (152). Interestingly, a study showed that enrichment of CX3CR1+CD8+ T cells early during ICI correlates with survival in patients with lung cancer and can be monitored peripherally via blood sampling (135). Longitudinal sampling can also unveil evolutionary dynamics of resistant cells with differential site-specific microenvironments, such as those of NGFRhi versus NGFRlo melanoma cells with differential effect based on PD-L1 expression levels (153). These are only a few recent examples of a vast body of studies that highlight different dynamic elements that change over the course of therapy and are ever-increasingly understood via improvements in single-cell and spatial profiling technologies (138, 142).
The sequencing of circulating tumor DNA (ctDNA) has shown promise for the ability to monitor response to ICI after initiation of treatment. Still, there remain substantial challenges to clinical implementation of dynamic circulating biomarker profiling, including lack of harmonization of reporting metrics, varying kinetics of response from study to study, and varying levels of sensitivity between different assay technologies (154, 155). Improvements in the near future may help make ctDNA sequencing useful for monitoring patients who have already begun treatment and may aid in treatment intensification or even deintensification. Pretreatment ctDNA analysis may also be useful for identifying mutations that can affect ICI sensitivity or calculate blood TMB, which has shown promise in predicting ICI sensitivity (156–158).
Conclusion and future directions
The current state of biomarker development for cancer immunotherapy is excellent, with an ongoing data deluge of considerable potential and ever-improving technologies. Tumor-agnostic FDA approvals for immunotherapy have been achieved, which is a notable achievement in oncology. However, there is still work to do to improve predictive strategies for identifying responders and nonresponders to ICIs, especially in the setting of combination therapies. As discussed above, we are on a path toward understanding the interplay of relevant mechanisms intrinsic to the tumor, the host, and the interaction between tumor and host and how different processes may vary in a context-specific manner. Biomarker models need to be feasible for clinical use and require technological improvements that may be assisted by advances in artificial intelligence and machine learning. Our field’s understanding of immuno-oncology and immunotherapy response has come a long way very quickly, and there is no sign that it will be slowing down anytime soon.
Acknowledgments
We thank the Chan laboratory for helpful discussions. We acknowledge funding from NIH grants R01-CA205426 (to TAC) and R35-CA232097 (to TAC).
Version 1. 06/15/2022
Electronic publication
Footnotes
Conflict of interest: TAC is a cofounder of Gritstone Oncology and owns stock. TAC acknowledges grant funding from Bristol Myers Squibb, AstraZeneca, Illumina, Pfizer, An2H, and Eisai. TAC has served as an advisor for Bristol Myers Squibb, MedImmune, Illumina, Eisai, AstraZeneca, and An2H. TAC is an inventor of intellectual property that uses tumor mutation burden to predict immunotherapy response; the pending patent (11230599) is held by Memorial Sloan Kettering Cancer Center and has been licensed to Personal Genome Diagnostics.
Copyright: © 2022, Halima et al. This is an open access article published under the terms of the Creative Commons Attribution 4.0 International License.
Reference information: J Clin Invest. 2022;132(12):e154945.https://doi.org/10.1172/JCI154945.
Contributor Information
Ahmed Halima, Email: HALIMAA@ccf.org.
Winston Vuong, Email: VUONGW2@ccf.org.
References
- 1.Tumeh PC, et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature. 2014;515(7528):568–571. doi: 10.1038/nature13954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Snyder A, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med. 2014;371(23):2189–2199. doi: 10.1056/NEJMoa1406498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Havel JJ, et al. The evolving landscape of biomarkers for checkpoint inhibitor immunotherapy. Nat Rev Cancer. 2019;19(3):133–150. doi: 10.1038/s41568-019-0116-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Twomey JD, Zhang B. Cancer immunotherapy update: FDA-approved checkpoint inhibitors and companion diagnostics. AAPS J. 2021;23(2):39. doi: 10.1208/s12248-021-00574-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rizvi NA, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348(6230):124–128. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Samstein RM, et al. Mutations in BRCA1 and BRCA2 differentially affect the tumor microenvironment and response to checkpoint blockade immunotherapy. Nat Cancer. 2020;1(12):1188–1203. doi: 10.1038/s43018-020-00139-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Samstein RM, et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat Genet. 2019;51(2):202–206. doi: 10.1038/s41588-018-0312-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Valero C, et al. The association between tumor mutational burden and prognosis is dependent on treatment context. Nat Genet. 2021;53(1):11–15. doi: 10.1038/s41588-020-00752-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Snyder A, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med. 2014;371(23):2189–2199. doi: 10.1056/NEJMoa1406498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marabelle A, et al. Association of tumour mutational burden with outcomes in patients with advanced solid tumours treated with pembrolizumab: prospective biomarker analysis of the multicohort, open-label, phase 2 KEYNOTE-158 study. Lancet Oncol. 2020;21(10):1353–1365. doi: 10.1016/S1470-2045(20)30445-9. [DOI] [PubMed] [Google Scholar]
- 11.Valero C, et al. Response rates to anti–PD-1 immunotherapy in microsatellite-stable solid tumors with 10 or more mutations per megabase. JAMA Oncol. 2021;7(5):739–743. doi: 10.1001/jamaoncol.2020.7684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Legrand FA, et al. Association of high tissue TMB and atezolizumab efficacy across multiple tumor types. J Clin Oncol. 2018;36(15 suppl):12000 [Google Scholar]
- 13.Rosenberg JE, et al. Atezolizumab in patients with locally advanced and metastatic urothelial carcinoma who have progressed following treatment with platinum-based chemotherapy: a single-arm, multicentre, phase 2 trial. Lancet. 2016;387(10031):1909–1920. doi: 10.1016/S0140-6736(16)00561-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hellmann MD, et al. Tumor mutational burden and efficacy of nivolumab monotherapy and in combination with ipilimumab in small-cell lung cancer. Cancer Cell. 2018;33(5):853–861. doi: 10.1016/j.ccell.2018.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Turajlic S, et al. Insertion-and-deletion-derived tumour-specific neoantigens and the immunogenic phenotype: a pan-cancer analysis. Lancet Oncol. 2017;18(8):1009–1021. doi: 10.1016/S1470-2045(17)30516-8. [DOI] [PubMed] [Google Scholar]
- 16.Litchfield K, et al. Escape from nonsense-mediated decay associates with anti-tumor immunogenicity. Nat Commun. 2020;11(1):3800. doi: 10.1038/s41467-020-17526-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vega DM, et al. Aligning tumor mutational burden (TMB) quantification across diagnostic platforms: phase II of the Friends of Cancer Research TMB Harmonization Project. Ann Oncol. 2021;32(12):1626–1636. doi: 10.1016/j.annonc.2021.09.016. [DOI] [PubMed] [Google Scholar]
- 18.Topalian SL, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med. 2012;366(26):2443–2454. doi: 10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reis H, et al. PD-L1 expression in urothelial carcinoma with predominant or pure variant histology: concordance among 3 commonly used and commercially available antibodies. Am J Surg Pathol. 2019;43(7):920–927. doi: 10.1097/PAS.0000000000001264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garon EB, et al. Pembrolizumab for the treatment of non-small-cell lung cancer. N Engl J Med. 2015;372(21):2018–2028. doi: 10.1056/NEJMoa1501824. [DOI] [PubMed] [Google Scholar]
- 21.Brahmer JR, et al. Phase I study of single-agent anti-programmed death-1 (MDX-1106) in refractory solid tumors: safety, clinical activity, pharmacodynamics, and immunologic correlates. J Clin Oncol. 2010;28(19):3167–3175. doi: 10.1200/JCO.2009.26.7609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carbone DP, et al. First-line nivolumab in stage IV or recurrent non–small-cell lung cancer. N Engl J Med. 2017;376(25):2415–2426. doi: 10.1056/NEJMoa1613493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hanna GJ, et al. Frameshift events predict anti-PD-1/L1 response in head and neck cancer. JCI Insight. 2018;3(4):e98811. doi: 10.1172/jci.insight.98811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brahmer J, et al. Nivolumab versus docetaxel in advanced squamous-cell non-small-cell lung cancer. N Engl J Med. 2015;373(2):123–135. doi: 10.1056/NEJMoa1504627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Motzer RJ, et al. Nivolumab versus everolimus in advanced renal-cell carcinoma. N Engl J Med. 2015;373(19):1803–1813. doi: 10.1056/NEJMoa1510665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu X, et al. Association of PD-L1 expression status with the efficacy of PD-1/PD-L1 inhibitors and overall survival in solid tumours: a systematic review and meta-analysis. Int J Cancer. 2020;147(1):116–127. doi: 10.1002/ijc.32744. [DOI] [PubMed] [Google Scholar]
- 27.Escors D, et al. The intracellular signalosome of PD-L1 in cancer cells. Signal Transduct Target Ther. 2018;3(1):26. doi: 10.1038/s41392-018-0022-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gato-Cañas M, et al. PDL1 signals through conserved sequence motifs to overcome interferon-mediated cytotoxicity. Cell Rep. 2017;20(8):1818–1829. doi: 10.1016/j.celrep.2017.07.075. [DOI] [PubMed] [Google Scholar]
- 29.Yadollahi P, et al. Current understanding of cancer-intrinsic PD-L1: regulation of expression and its protumoral activity. BMB Rep. 2021;54(1):12–20. doi: 10.5483/BMBRep.2021.54.1.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yarchoan M, et al. PD-L1 expression and tumor mutational burden are independent biomarkers in most cancers. JCI Insight. 2019;4(6):e126908. doi: 10.1172/jci.insight.126908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gupta RG, et al. Exploiting tumor neoantigens to target cancer evolution: current challenges and promising therapeutic approaches. Cancer Discov. 2021;11(5):1024–1039. doi: 10.1158/2159-8290.CD-20-1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiang T, et al. Tumor neoantigens: from basic research to clinical applications. J Hematol Oncol. 2019;12(1):93. doi: 10.1186/s13045-019-0787-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang L, et al. The evolution of alternative splicing in glioblastoma under therapy. Genome Biol. 2021;22(1):48. doi: 10.1186/s13059-021-02259-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu SX, et al. Pharmacologic modulation of RNA splicing enhances anti-tumor immunity. Cell. 2021;184(15):4032–4047. doi: 10.1016/j.cell.2021.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bulik-Sullivan B, et al. Deep learning using tumor HLA peptide mass spectrometry datasets improves neoantigen identification. Nat Biotechnol. doi: 10.1038/nbt.4313. [published online December 17, 2018]. [DOI] [PubMed] [Google Scholar]
- 36.Wells DK, et al. Key parameters of tumor epitope immunogenicity revealed through a consortium approach improve neoantigen prediction. Cell. 2020;183(3):818–834. doi: 10.1016/j.cell.2020.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Charoentong P, et al. Pan-cancer immunogenomic analyses reveal genotype-immunophenotype relationships and predictors of response to checkpoint blockade. Cell Rep. 2017;18(1):248–262. doi: 10.1016/j.celrep.2016.12.019. [DOI] [PubMed] [Google Scholar]
- 38.Finotello F, et al. Next-generation computational tools for interrogating cancer immunity. Nat Rev Genet. 2019;20(12):724–746. doi: 10.1038/s41576-019-0166-7. [DOI] [PubMed] [Google Scholar]
- 39.McGranahan N, et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science. 2016;351(6280):1463–1469. doi: 10.1126/science.aaf1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Balachandran VP, et al. Identification of unique neoantigen qualities in long-term survivors of pancreatic cancer. Nature. 2017;551(7681):512–516. doi: 10.1038/nature24462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lang F, et al. Identification of neoantigens for individualized therapeutic cancer vaccines. Nat Rev Drug Discov. 2022;21(4):261–282. doi: 10.1038/s41573-021-00387-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McGranahan N, Swanton C. Neoantigen quality, not quantity. Sci Transl Med. 2019;11(506):eaax7918. doi: 10.1126/scitranslmed.aax7918. [DOI] [PubMed] [Google Scholar]
- 43.Richters MM, et al. Best practices for bioinformatic characterization of neoantigens for clinical utility. Genome Med. 2019;11(1):56. doi: 10.1186/s13073-019-0666-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bhinder B, Elemento O. Computational methods in tumor immunology. Methods Enzymol. 2020;636:209–259. doi: 10.1016/bs.mie.2020.01.001. [DOI] [PubMed] [Google Scholar]
- 45.Bhinder B, et al. Artificial intelligence in cancer research and precision medicine. Cancer Discov. 2021;11(4):900–915. doi: 10.1158/2159-8290.CD-21-0090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Joglekar AV, Li G. T cell antigen discovery. Nat Methods. 2021;18(8):873–880. doi: 10.1038/s41592-020-0867-z. [DOI] [PubMed] [Google Scholar]
- 47.Pearlman AH, et al. Targeting public neoantigens for cancer immunotherapy. Nat Cancer. 2021;2(5):487–497. doi: 10.1038/s43018-021-00210-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pham TV, et al. Role of ultraviolet mutational signature versus tumor mutation burden in predicting response to immunotherapy. Mol Oncol. 2020;14(8):1680–1694. doi: 10.1002/1878-0261.12748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Le DT, et al. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med. 2015;372(26):2509–2520. doi: 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rayner E, et al. A panoply of errors: polymerase proofreading domain mutations in cancer. Nat Rev Cancer. 2016;16(2):71–81. doi: 10.1038/nrc.2015.12. [DOI] [PubMed] [Google Scholar]
- 51.Wang F, et al. Evaluation of POLE and POLD1 mutations as biomarkers for immunotherapy outcomes across multiple cancer types. JAMA Oncol. 2019;5(10):1504–1506. doi: 10.1001/jamaoncol.2019.2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Natesan D, et al. APOBEC mutational signature and tumor mutational burden as predictors of clinical outcomes and treatment response in patients with advanced urothelial cancer. Front Oncol. 2022;12:816706. doi: 10.3389/fonc.2022.816706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Faden DL, et al. APOBEC mutagenesis is tightly linked to the immune landscape and immunotherapy biomarkers in head and neck squamous cell carcinoma. Oral Oncol. 2019;96:140–147. doi: 10.1016/j.oraloncology.2019.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang S, et al. APOBEC3B and APOBEC mutational signature as potential predictive markers for immunotherapy response in non-small cell lung cancer. Oncogene. 2018;37(29):3924–3936. doi: 10.1038/s41388-018-0245-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Boichard A, et al. APOBEC-related mutagenesis and neo-peptide hydrophobicity: implications for response to immunotherapy. Oncoimmunology. 2018;8(3):1550341. doi: 10.1080/2162402X.2018.1550341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Le DT, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science. 2017;357(6349):409–413. doi: 10.1126/science.aan6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Marcus L, et al. FDA approval summary: pembrolizumab for the treatment of microsatellite instability-high solid tumors. Clin Cancer Res. 2019;25(13):3753–3758. doi: 10.1158/1078-0432.CCR-18-4070. [DOI] [PubMed] [Google Scholar]
- 58.Andre T, et al. Pembrolizumab in microsatellite-instability-high advanced colorectal cancer. N Engl J Med. 2020;383(23):2207–2218. doi: 10.1056/NEJMoa2017699. [DOI] [PubMed] [Google Scholar]
- 59.Wang Q-X, et al. The degree of microsatellite instability predicts response to PD-1 blockade immunotherapy in mismatch repair-deficient/microsatellite instability-high colorectal cancers. Exp Hematol Oncol. 2021;10(1):2. doi: 10.1186/s40164-020-00193-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mandal R, et al. Genetic diversity of tumors with mismatch repair deficiency influences anti-PD-1 immunotherapy response. Science. 2019;364(6439):485–491. doi: 10.1126/science.aau0447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sahin IH, et al. Immune checkpoint inhibitors for the treatment of MSI-H/MMR-D colorectal cancer and a perspective on resistance mechanisms. Br J Cancer. 2019;121(10):809–818. doi: 10.1038/s41416-019-0599-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Merkel DE, McGuire WL. Ploidy, proliferative activity and prognosis. DNA flow cytometry of solid tumors. Cancer. 1990;65(5):1194–1205. doi: 10.1002/1097-0142(19900301)65:5<1194::AID-CNCR2820650528>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 63.Davoli T, et al. Tumor aneuploidy correlates with markers of immune evasion and with reduced response to immunotherapy. Science. 2017;355(6322):eaaf8399. doi: 10.1126/science.aaf8399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Litchfield K, et al. Meta-analysis of tumor- and T cell-intrinsic mechanisms of sensitization to checkpoint inhibition. Cell. 2021;184(3):596–614. doi: 10.1016/j.cell.2021.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Han G, et al. 9p21 loss confers a cold tumor immune microenvironment and primary resistance to immune checkpoint therapy. Nat Commun. 2021;12(1):5606. doi: 10.1038/s41467-021-25894-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gao J, et al. Loss of IFN-γ pathway genes in tumor cells as a mechanism of resistance to anti-CTLA-4 therapy. Cell. 2016;167(2):397–404. doi: 10.1016/j.cell.2016.08.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chowell D, et al. Improved prediction of immune checkpoint blockade efficacy across multiple cancer types. Nat Biotechnol. 2021;40(4):499–506. doi: 10.1038/s41587-021-01070-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Frederick DT, et al. BRAF inhibition is associated with enhanced melanoma antigen expression and a more favorable tumor microenvironment in patients with metastatic melanoma. Clin Cancer Res. 2013;19(5):1225–1231. doi: 10.1158/1078-0432.CCR-12-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wolchok JD, et al. Overall survival with combined nivolumab and ipilimumab in advanced melanoma. N Engl J Med. 2017;377(14):1345–1356. doi: 10.1056/NEJMoa1709684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Akins M, et al. DREAMseq (Doublet, Randomized Evaluation in Advanced Melanoma Sequencing): A phase III trial—ECOG-ACRIN EA6134. J Clin Oncol. 2021;39(36):356154–356154. https://ascopubs.org/doi/abs/10.1200/JCO.2021.39.36_suppl.356154 Accessessed June 7, 2022.
- 71.Skoulidis F, et al. STK11/LKB1 mutations and PD-1 inhibitor resistance in KRAS-mutant lung adenocarcinoma. Cancer Discov. 2018;8(7):822–835. doi: 10.1158/2159-8290.CD-18-0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Krishnamurthy N, et al. STK11 alterations in the pan-cancer setting: prognostic and therapeutic implications. Eur J Cancer. 2021;148:215–229. doi: 10.1016/j.ejca.2021.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Peng W, et al. Loss of PTEN promotes resistance to T cell-mediated immunotherapy. Cancer Discov. 2016;6(2):202–216. doi: 10.1158/2159-8290.CD-15-0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bucheit AD, et al. Complete loss of PTEN protein expression correlates with shorter time to brain metastasis and survival in stage IIIB/C melanoma patients with BRAFV600 mutations. Clin Cancer Res. 2014;20(21):5527–5536. doi: 10.1158/1078-0432.CCR-14-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chapman NM, Chi H. mTOR signaling, Tregs and immune modulation. Immunotherapy. 2014;6(12):1295–1311. doi: 10.2217/imt.14.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hodges C, et al. The many roles of BAF (mSWI/SNF) and PBAF complexes in cancer. Cold Spring Harb Perspect Med. 2016;6(8):a026930. doi: 10.1101/cshperspect.a026930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Carril-Ajuria L, et al. Prognostic and predictive value of PBRM1 in clear cell renal cell carcinoma. Cancers (Basel) 2019;12(1):16. doi: 10.3390/cancers12010016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Miao D, et al. Genomic correlates of response to immune checkpoint therapies in clear cell renal cell carcinoma. Science. 2018;359(6377):801–806. doi: 10.1126/science.aan5951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pan D, et al. A major chromatin regulator determines resistance of tumor cells to T cell-mediated killing. Science. 2018;359(6377):770–775. doi: 10.1126/science.aao1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.McDermott DF, et al. Clinical activity and molecular correlates of response to atezolizumab alone or in combination with bevacizumab versus sunitinib in renal cell carcinoma. Nat Med. 2018;24(6):749–757. doi: 10.1038/s41591-018-0053-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hakimi AA, et al. A pan-cancer analysis of PBAF complex mutations and their association with immunotherapy response. Nat Commun. 2020;11(1):4168. doi: 10.1038/s41467-020-17965-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhou H, et al. PBRM1 mutation and preliminary response to immune checkpoint blockade treatment in non-small cell lung cancer. NPJ Precis Oncol. 2020;4:6. doi: 10.1038/s41698-020-0112-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Liu XD, et al. PBRM1 loss defines a nonimmunogenic tumor phenotype associated with checkpoint inhibitor resistance in renal carcinoma. Nat Commun. 2020;11(1):2135. doi: 10.1038/s41467-020-15959-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gutiontov SI, et al. CDKN2A loss-of-function predicts immunotherapy resistance in non-small cell lung cancer. Sci Rep. 2021;11(1):20059. doi: 10.1038/s41598-021-99524-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Adib E, et al. CDKN2A alterations and response to immunotherapy in solid tumors. Clin Cancer Res. 2021;27(14):4025–4035. doi: 10.1158/1078-0432.CCR-21-0575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Roy R, et al. BRCA1 and BRCA2: different roles in a common pathway of genome protection. Nat Rev Cancer. 2011;12(1):68–78. doi: 10.1038/nrc3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sharma P, et al. Nivolumab plus ipilimumab for metastatic castration-resistant prostate cancer: preliminary analysis of patients in the checkmate 650 trial. Cancer Cell. 2020;38(4):489–499. doi: 10.1016/j.ccell.2020.08.007. [DOI] [PubMed] [Google Scholar]
- 88.Spranger S, et al. Melanoma-intrinsic β-catenin signalling prevents anti-tumour immunity. Nature. 2015;523(7559):231–235. doi: 10.1038/nature14404. [DOI] [PubMed] [Google Scholar]
- 89.Ji RR, et al. An immune-active tumor microenvironment favors clinical response to ipilimumab. Cancer Immunol Immunother. 2012;61(7):1019–1031. doi: 10.1007/s00262-011-1172-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sayaman RW, et al. Germline genetic contribution to the immune landscape of cancer. Immunity. 2021;54(2):367–386. doi: 10.1016/j.immuni.2021.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chowell D, et al. Evolutionary divergence of HLA class I genotype impacts efficacy of cancer immunotherapy. Nat Med. 2019;25(11):1715–1720. doi: 10.1038/s41591-019-0639-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Robinson J, et al. The IMGT/HLA database. Nucleic Acids Res. 2013;41(d1):D1222–1227. doi: 10.1093/nar/gks949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hughes AL, Yeager M. Natural selection at major histocompatibility complex loci of vertebrates. Annu Rev Genet. 1998;32:415–435. doi: 10.1146/annurev.genet.32.1.415. [DOI] [PubMed] [Google Scholar]
- 94.Paul S, et al. HLA class I alleles are associated with peptide-binding repertoires of different size, affinity, and immunogenicity. J Immunol. 2013;191(12):5831–5839. doi: 10.4049/jimmunol.1302101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lee CH, et al. High response rate and durability driven by HLA genetic diversity in patients with kidney cancer treated with lenvatinib and pembrolizumab. Mol Cancer Res. 2021;19(9):1510–1521. doi: 10.1158/1541-7786.MCR-21-0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pagliuca S, et al. The similarity of class II HLA genotypes defines patterns of autoreactivity in idiopathic bone marrow failure disorders. Blood. 2021;138(26):2781–2798. doi: 10.1182/blood.2021012900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Roerden M, et al. HLA evolutionary divergence as a prognostic marker for AML patients undergoing allogeneic stem cell transplantation. Cancers (Basel) 2020;12(7):1835. doi: 10.3390/cancers12071835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Feray C, et al. Donor HLA class 1 evolutionary divergence is a major predictor of liver allograft rejection: a retrospective cohort study. Ann Intern Med. 2021;174(10):1385–1394. doi: 10.7326/M20-7957. [DOI] [PubMed] [Google Scholar]
- 99.Chhibber A, et al. Germline HLA landscape does not predict efficacy of pembrolizumab monotherapy across solid tumor types. Immunity. 2022;55(1):56–64. doi: 10.1016/j.immuni.2021.12.006. [DOI] [PubMed] [Google Scholar]
- 100.Philip M, Schietinger A. CD8+ T cell differentiation and dysfunction in cancer. Nat Rev Immunol. 2021;CD8(4):209–223. doi: 10.1038/s41577-021-00574-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ghorani E, et al. The T cell differentiation landscape is shaped by tumour mutations in lung cancer. Nat Cancer. 2020;1(5):546–561. doi: 10.1038/s43018-020-0066-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Au L, et al. Determinants of anti-PD-1 response and resistance in clear cell renal cell carcinoma. Cancer Cell. 2021;39(11):1497–1518. doi: 10.1016/j.ccell.2021.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bi K, et al. Tumor and immune reprogramming during immunotherapy in advanced renal cell carcinoma. Cancer Cell. 2021;39(5):649–661. doi: 10.1016/j.ccell.2021.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Han J, et al. Resident and circulating memory T cells persist for years in melanoma patients with durable responses to immunotherapy. Nat Cancer. 2021;2(3):300–311. doi: 10.1038/s43018-021-00180-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Caushi JX, et al. Transcriptional programs of neoantigen-specific TIL in anti-PD-1-treated lung cancers. Nature. 2021;596(7870):126–132. doi: 10.1038/s41586-021-03752-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Krishna C, et al. Single-cell sequencing links multiregional immune landscapes and tissue-resident T cells in ccRCC to tumor topology and therapy efficacy. Cancer Cell. 2021;39(5):662–677. doi: 10.1016/j.ccell.2021.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sun D, et al. TISCH: a comprehensive web resource enabling interactive single-cell transcriptome visualization of tumor microenvironment. Nucleic Acids Res. 2021;49(d1):D1420–D1430. doi: 10.1093/nar/gkaa1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Pelka K, et al. Spatially organized multicellular immune hubs in human colorectal cancer. Cell. 2021;184(18):4734–4752. doi: 10.1016/j.cell.2021.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lam KC, et al. Microbiota triggers STING-type I IFN-dependent monocyte reprogramming of the tumor microenvironment. Cell. 2021;184(21):5338–5356. doi: 10.1016/j.cell.2021.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Riaz N, et al. Tumor and microenvironment evolution during immunotherapy with nivolumab. Cell. 2017;171(4):934–949. doi: 10.1016/j.cell.2017.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Routy B, et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science. 2018;359(6371):91–97. doi: 10.1126/science.aan3706. [DOI] [PubMed] [Google Scholar]
- 112.Fridman WH, et al. The immune contexture in human tumours: impact on clinical outcome. Nat Rev Cancer. 2012;12(4):298–306. doi: 10.1038/nrc3245. [DOI] [PubMed] [Google Scholar]
- 113.Naito Y, et al. CD8+ T cells infiltrated within cancer cell nests as a prognostic factor in human colorectal cancer. Cancer Res. 1998;58(16):3491–3494. [PubMed] [Google Scholar]
- 114.Galon J, et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science. 2006;313(5795):1960–1964. doi: 10.1126/science.1129139. [DOI] [PubMed] [Google Scholar]
- 115.Mlecnik B, et al. Integrative analyses of colorectal cancer show immunoscore is a stronger predictor of patient survival than microsatellite instability. Immunity. 2016;44(3):698–711. doi: 10.1016/j.immuni.2016.02.025. [DOI] [PubMed] [Google Scholar]
- 116.Pagès F, et al. International validation of the consensus Immunoscore for the classification of colon cancer: a prognostic and accuracy study. Lancet. 2018;391(10135):2128–2139. doi: 10.1016/S0140-6736(18)30789-X. [DOI] [PubMed] [Google Scholar]
- 117.Tauriello DVF, et al. TGFβ drives immune evasion in genetically reconstituted colon cancer metastasis. Nature. 2018;554(7693):538–543. doi: 10.1038/nature25492. [DOI] [PubMed] [Google Scholar]
- 118.Herbst RS, et al. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature. 2014;515(7528):563–567. doi: 10.1038/nature14011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Chen DS, Mellman I. Elements of cancer immunity and the cancer-immune set point. Nature. 2017;541(7637):321–330. doi: 10.1038/nature21349. [DOI] [PubMed] [Google Scholar]
- 120.Helmink BA, et al. B cells and tertiary lymphoid structures promote immunotherapy response. Nature. 2020;577(7791):549–555. doi: 10.1038/s41586-019-1922-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Petitprez F, et al. B cells are associated with survival and immunotherapy response in sarcoma. Nature. 2020;577(7791):556–560. doi: 10.1038/s41586-019-1906-8. [DOI] [PubMed] [Google Scholar]
- 122.Cabrita R, et al. Tertiary lymphoid structures improve immunotherapy and survival in melanoma. Nature. 2020;577(7791):561–565. doi: 10.1038/s41586-019-1914-8. [DOI] [PubMed] [Google Scholar]
- 123.Hu X, et al. Landscape of B cell immunity and related immune evasion in human cancers. Nat Genet. 2019;51(3):560–567. doi: 10.1038/s41588-018-0339-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Pai JA, Satpathy AT. High-throughput and single-cell T cell receptor sequencing technologies. Nat Methods. 2021;18(8):881–892. doi: 10.1038/s41592-021-01201-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wu TD, et al. Peripheral T cell expansion predicts tumour infiltration and clinical response. Nature. 2020;579(7798):274–278. doi: 10.1038/s41586-020-2056-8. [DOI] [PubMed] [Google Scholar]
- 126.Li H, et al. Dysfunctional CD8 T cells form a proliferative, dynamically regulated compartment within human melanoma. Cell. 2019;176(4):775–789. doi: 10.1016/j.cell.2018.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Fairfax BP, et al. Peripheral CD8+ T cell characteristics associated with durable responses to immune checkpoint blockade in patients with metastatic melanoma. Nat Med. 2020;26(2):193–199. doi: 10.1038/s41591-019-0734-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Krishna C, et al. Genetic and environmental determinants of human TCR repertoire diversity. Immun Ageing. 2020;17(1):26. doi: 10.1186/s12979-020-00195-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Reuben A, et al. Genomic and immune heterogeneity are associated with differential responses to therapy in melanoma. NPJ Genom Med. 2017;2:10. doi: 10.1038/s41525-017-0013-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Snyder A, et al. Contribution of systemic and somatic factors to clinical response and resistance to PD-L1 blockade in urothelial cancer: an exploratory multi-omic analysis. PLoS Med. 2017;14(5):e1002309. doi: 10.1371/journal.pmed.1002309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Hopkins AC, et al. T cell receptor repertoire features associated with survival in immunotherapy-treated pancreatic ductal adenocarcinoma. JCI Insight. 2018;3(13):e122092. doi: 10.1172/jci.insight.122092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ehrlich R, et al. SwarmTCR: a computational approach to predict the specificity of T cell receptors. BMC Bioinformatics. 2021;22(1):422. doi: 10.1186/s12859-021-04335-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Lu YC, et al. Direct identification of neoantigen-specific TCRs from tumor specimens by high-throughput single-cell sequencing. J Immunother Cancer. 2021;9(7):e002595. doi: 10.1136/jitc-2021-002595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Sidhom JW, et al. DeepTCR is a deep learning framework for revealing sequence concepts within T-cell repertoires. Nat Commun. 2021;12(1):1605. doi: 10.1038/s41467-021-21879-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Yamauchi T, et al. T-cell CX3CR1 expression as a dynamic blood-based biomarker of response to immune checkpoint inhibitors. Nat Commun. 2021;12(1):1402. doi: 10.1038/s41467-021-21619-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Valero C, et al. Pretreatment neutrophil-to-lymphocyte ratio and mutational burden as biomarkers of tumor response to immune checkpoint inhibitors. Nat Commun. 2021;12(1):729. doi: 10.1038/s41467-021-20935-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Lee JS, Ruppin E. Multiomics prediction of response rates to therapies to inhibit programmed cell death 1 and programmed cell death 1 ligand 1. JAMA Oncol. 2019;5(11):1614–1618. doi: 10.1001/jamaoncol.2019.2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Gohil SH, et al. Applying high-dimensional single-cell technologies to the analysis of cancer immunotherapy. Nat Rev Clin Oncol. 2021;18(4):244–256. doi: 10.1038/s41571-020-00449-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Rozenblatt-Rosen O, et al. The human tumor atlas network: charting tumor transitions across space and time at single-cell resolution. Cell. 2020;181(2):236–249. doi: 10.1016/j.cell.2020.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Davis-Marcisak EF, et al. From bench to bedside: single-cell analysis for cancer immunotherapy. Cancer Cell. 2021;39(8):1062–1080. doi: 10.1016/j.ccell.2021.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Casanova-Acebes M, et al. Tissue-resident macrophages provide a pro-tumorigenic niche to early NSCLC cells. Nature. 2021;595(7868):578–584. doi: 10.1038/s41586-021-03651-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Ding J, et al. Temporal modelling using single-cell transcriptomics. Nat Rev Genet. doi: 10.1038/s41576-021-00444-7. [published online January 31, 2022]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Nieto P, et al. A single-cell tumor immune atlas for precision oncology. Genome Res. 2021;31(10):1913–1926. doi: 10.1101/gr.273300.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Zheng L, et al. Pan-cancer single-cell landscape of tumor-infiltrating T cells. Science. 2021;374(6574):abe6474. doi: 10.1126/science.abe6474. [DOI] [PubMed] [Google Scholar]
- 145.Berry S, et al. Analysis of multispectral imaging with the AstroPath platform informs efficacy of PD-1 blockade. Science. 2021;372(6547):eaba2609. doi: 10.1126/science.aba2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Cleary B, et al. Compressed sensing for highly efficient imaging transcriptomics. Nat Biotechnol. 2021;39(8):936–942. doi: 10.1038/s41587-021-00883-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Hwang B, et al. SCITO-seq: single-cell combinatorial indexed cytometry sequencing. Nat Methods. 2021;18(8):903–911. doi: 10.1038/s41592-021-01222-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Stuart T, Satija R. Integrative single-cell analysis. Nat Rev Genet. 2019;20(5):257–272. doi: 10.1038/s41576-019-0093-7. [DOI] [PubMed] [Google Scholar]
- 149.Sharma P, et al. Primary, adaptive, and acquired resistance to cancer immunotherapy. Cell. 2017;168(4):707–723. doi: 10.1016/j.cell.2017.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Zaretsky JM, et al. Mutations associated with acquired resistance to PD-1 blockade in melanoma. N Engl J Med. 2016;375(9):819–829. doi: 10.1056/NEJMoa1604958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Jacquelot N, et al. Predictors of responses to immune checkpoint blockade in advanced melanoma. Nat Commun. 2017;8(1):592. doi: 10.1038/s41467-017-00608-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Yang SYC, et al. Pan-cancer analysis of longitudinal metastatic tumors reveals genomic alterations and immune landscape dynamics associated with pembrolizumab sensitivity. Nat Commun. 2021;12(1):5137. doi: 10.1038/s41467-021-25432-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Liu D, et al. Evolution of delayed resistance to immunotherapy in a melanoma responder. Nat Med. 2021;27(6):985–992. doi: 10.1038/s41591-021-01331-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Ignatiadis M, et al. Liquid biopsy enters the clinic — implementation issues and future challenges. Nat Rev Clin Oncol. 2021;18(5):297–312. doi: 10.1038/s41571-020-00457-x. [DOI] [PubMed] [Google Scholar]
- 155.Liu MC, et al. Sensitive and specific multi-cancer detection and localization using methylation signatures in cell-free DNA. Ann Oncol. 2020;31(6):745–759. doi: 10.1016/j.annonc.2020.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Si H, et al. A blood-based assay for assessment of tumor mutational burden in first-line metastatic NSCLC treatment: results from the MYSTIC Study. Clin Cancer Res. 2021;27(6):1631–1640. doi: 10.1158/1078-0432.CCR-20-3771. [DOI] [PubMed] [Google Scholar]
- 157.Van Campenhout C, et al. Blood tumor mutational burden: are we ready for clinical implementation? J Thorac Dis. 2019;11(suppl 15):S1906–S1908. doi: 10.21037/jtd.2019.07.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Gandara DR, et al. Blood-based tumor mutational burden as a predictor of clinical benefit in non-small-cell lung cancer patients treated with atezolizumab. Nat Med. 2018;24(9):1441–1448. doi: 10.1038/s41591-018-0134-3. [DOI] [PubMed] [Google Scholar]