Abstract
We have previously reported the construction of a food-grade cloning vector for Lactococcus using the ochre suppressor, supB, as the selective marker. This vector, pFG1, causes only a slight growth inhibition in the laboratory strain MG1363 but is unstable in the industrial strains tested. As supB suppresses both amber and ochre stop codons, which are present in 82% of all known lactococcal genes, this undesirable finding may result from the accumulation of elongated mistranslated polypeptides. Here, we report the development of a new food-grade cloning vector, pFG200, which is suitable for overexpressing a variety of genes in industrial strains of Lactococcus lactis. The vector uses an amber suppressor, supD, as selectable marker and consists entirely of Lactococcus DNA, with the exception of a small polylinker region. Using suppressible pyrimidine auxotrophs, selection and maintenance are efficient in any pyrimidine-free medium including milk. Importantly, the presence of this vector in a variety of industrial strains has no significant effect on the growth rate or the rate of acidification in milk, making this an ideal system for food-grade modification of industrially relevant L. lactis strains. The usefulness of this system is demonstrated by overexpressing the pepN gene in a number of industrial backgrounds.
Lactococcus lactis is an important organism for the dairy industry, where it is used for the production of many cheese varieties as well as cultured milk products such as buttermilk. Lactococcus contributes to the successful manufacture of these products through the production of lactic acid and a variety of flavor components. While excellent industrial strains do exist, there is a constant desire in the dairy industry for strains with improved properties. One useful method for strain improvement is genetic engineering, provided, of course, that the genetic modifications are food grade (10). A number of characteristics, such as flavor and acid production, bacteriophage resistance, proteolytic activity, and the autolytic properties of strains, can be improved by overexpressing the relevant genes (3, 4, 5, 10). A simple method for overexpressing a gene is to transfer it to a multicopy cloning vector.
We have previously described the construction of a food-grade cloning vector for Lactococcus (5). This vector, pFG1, uses an ochre suppressor gene, supB, as the selectable marker. It exists in five to nine copies in the cell and has allowed the overexpression of several Lactococcus genes in strains derived from the laboratory strain MG1363. The use of pFG1 in industrial strains of Lactococcus has not been successful, however, due to a reduced acidification rate in strains bearing pFG1 derivatives (unpublished results). Whereas in MG1363 derivatives pFG1 causes a slight growth inhibition, in industrial strains a severe growth inhibition is seen and the vector is very unstable. Since an ochre suppressor suppresses both amber and ochre codons and 82% of Lactococcus genes terminate in either amber or ochre stop codons (2, 5), this growth inhibition is probably caused by the accumulation of elongated, mistranslated polypeptides. Based on these observations, we have developed a new food-grade cloning vector, pFG200, using an amber suppressor, supD, as a selectable marker. supD encodes an altered tRNASer (5) and suppresses only amber codons, which are present at the ends of approximately 10% of Lactococcus genes (2).
A powerful selection system has been constructed in parallel with the vector. By introducing an amber codon into the pyrF gene, we generated pyrimidine auxotrophs to use as host strains for pFG200. This marker allows the selection and maintenance of pFG200 in pyrimidine-free medium. The pyrimidine content of milk is insufficient for growth of these pyrimidine auxotrophs, so the selection also works in milk.
In this paper, we demonstrate the potential of this system by illustrating that pFG200 is stably maintained in the host cells in media including milk, has virtually no impact on growth and acidification rates, and, finally, enables a cloned gene such as pepN (15) to be overexpressed.
MATERIALS AND METHODS
Bacterial strains and media.
Strains of L. lactis and Escherichia coli are listed in Table 1. L. lactis was grown in M17 (16) or DN minimal medium (5). Erythromycin was used at 1 μg/ml when required. E. coli was grown in Luria-Bertani medium, supplemented with ampicillin to 50 μg/ml when required. pyrF(Am) mutants were cultivated in M17 or DN minimal medium, both with 20 μg of uracil per ml and 40 μg of thymidine per ml added. Selection and maintenance of plasmids bearing supD in pyrF(Am) strains were in DN minimal medium without added pyrimidines. Growth was at 30°C and was measured by monitoring the increase in optical density at 600 nm. Experiments to determine the plasmid stability and acidification rate were carried out in reconstituted skim milk (RSM) and pasteurized whole milk, respectively.
TABLE 1.
Bacterial strains
| Strain | Description | Source (reference) |
|---|---|---|
| L. lactis | ||
| MG1363 | Plasmid-free derivative of NCDO 712 | Gasson (6) |
| FA4-1-1 | Pyrimidine auxotroph of MG1363 (CAA154 → TAG) | This study |
| CHCC4210 | Pyrimidine auxotroph of MG1363 (TCT127 → TAG) | This study |
| CHCC377 | Industrial strain of L. lactis subsp. lactis (major component of a highly successful commercial culture) | CHCCa |
| CHCC4146 | Pyrimidine auxotroph of CHCC377 (TCT127 → TAG) | This study |
| CHCC4223 | CHCC4146(pFG200) | This study |
| CHCC3052 | Industrial strain of L. lactis subsp. lactis | CHCC |
| CHCC4205 | Pyrimidine auxotroph of CHCC4205 (TCT127 → TAG) | This study |
| CHCC270 | Industrial strain of L. lactis subsp. lactis | CHCC |
| CHCC4424 | Pyrimidine auxotroph of CHCC4205 (TCT127 → TAG) | This study |
| E. coli DH5α | deoR endA1 gyrA96 hsdR17 (rK− mK+) relA1 recA1 supE44 thi-1 Δ(lacZYA-argF)U169/φ80dlacΔ(lacZ)M15 | Hanahan (7) |
CHCC, Chr. Hansen Culture Collection.
Competent cells and electroporation.
Preparation of competent L. lactis cells in rich medium containing glycine and electroporation were as described previously (8). For MG1363, CHCC3052, CHCC270, and derivatives, 1% (wt/vol) glycine was used, while for CHCC377 and derivatives, 1.5% (wt/vol) glycine was used. Pyrimidine auxotrophs were grown in medium containing 20 μg of uracil per ml and 20 μg of thymidine per ml to reduce the frequency of revertants. Transformation of E. coli was done essentially as described by Sambrook et al. (14).
DNA purification and manipulation.
Small-scale plasmid preparations from E. coli were by alkaline lysis, and large-scale preparations were with the Qiagen system. Plasmid DNA and chromosomal DNA were isolated from L. lactis as described previously (5) with the modification that 50 μl of 5 M NaCl was added before the phenol extraction when plasmids were purified. DNA sequencing was by cycle sequencing as recommended by Perkin-Elmer. The resulting products were purified on Bio-Rad Micro-Bio-Spin-Chromatography columns before being run on an ABI PRISM 310 Genetic Analyzer. The primers used for sequencing and PCRs are listed in Table 2.
TABLE 2.
Primers
| Primer | Sequence |
|---|---|
| For MG1363 | |
| Fam3 | 5′-AACAGCCTAGGCCGGACTTGATGG-3′ |
| Fam4 | 5′-GTCCGGCCTAGGCTGTTTTTTGTGCG-3′ |
| pyrF1 | 5′-GCAGATCTAAGCTTGATTCAAGAAGTAAAAGAAGGC-3′ |
| pyrF2 | 5′-ATAGATCTACTCGATGCCAAGAATGGACCGC-3′ |
| For CHCC377 | |
| pyrF3 | 5′-AAAGGCCTGTNATNGCNCTNGAYTTYCC-3′ |
| pyrF4 | 5′-TGGACGAATTCCNGGNGT-3′ |
| pyrF5 | 5′-CATAGTAAACGACTTGGGG-3′ |
| pyrF6 | 5′-TACGCACAAAAAACCGCT-3′ |
| pyrF7 | 5′-GGTCGCCTTTACTTGCACC-3′ |
| pyrF8 | 5′-GATTATATTGTTGTCGGCCG-3′ |
| pyrD-degn | 5′-GCTCTAGAGCMWATYGWWATDGGN-3′ |
| llagidB2 | 5′-GGTNGARTGGAAYGARAARATH AAY-3′ |
| Fam5 | 5′-CCTCAACCTAGGAGAAAATTATGC-3′ |
| Fam6 | 5′-TCTCCTAGGTTGAGGTTAATTGTG-3′ |
| pyrD/BglII | 5′-ATAGATCTGCTTAGAAAACTTG-3′ |
| pyrF11/BglII | 5′-ATAGATCTGCATGTAAGCAAAAACC-3′ |
Sequencing of the pyrF gene.
The nucleotide sequence of the CHCC377 pyrF gene was obtained by first producing a 550-bp internal pyrF fragment via PCR using two degenerate primers (pyrF3 and pyrF4), which were designed based on amino acid sequence comparison of the pyrF proteins of a number of microorganisms. The fragment was sequenced with the same primers. New primers (pyrF5 and pyrF6) were obtained based on this sequence and used for inverse PCR with Sau3AI-digested DNA, giving additional sequence data. The sequence of the amino-terminal end of pyrF was completed using a degenerate primer designed from the sequence of pyrD (pyrD-dgen), which is immediately upstream of pyrF, and two additional primers (pyrF7 and pyrF8). The carboxy-terminal end of pyrF was sequenced using a degenerate primer (llagidB2) designed from the sequence of gidB, which is immediately downstream of pyrF, and pyrF6.
Plasmid constructions.
The construction of pFG200 is described in detail in Results. The pepN gene of L. lactis strain Wg2 was subcloned from pSTO3 (15) as a 3.5-kbp BamHI-SacI fragment, including the promoter region, into the corresponding sites in the pFG200 polylinker to generate pFG202. The promoterless lacLM genes were subcloned from pAK80 (9) as a HindIII-SalI fragment either into HindIII-SalI-digested pFG200, generating pFG209, or into HindIII-XhoI-digested pFG200, generating pFG208. These two plasmids represent the two possible orientations of lacLM in pFG200.
Plasmid copy number determination.
Total genomic DNAs isolated from fresh saturated cultures of CHCC4210 and CHCC4210/pFG200 grown in DN medium were digested with HindIII. After separation by agarose gel electrophoresis, the DNA was transfered to a GeneScreen Plus nylon filter (E.I. du Pont de Nemours). A 298-bp PCR fragment containing supD, generated using the primers amber-3 and amber-6 with pAK93 DNA as the template, was used as a hybridization probe. The probe was labeled with [α-32P]dATP, using the Megaprime labeling kit (Amersham), for 3 h at 37°C. Hybridization was carried out at 60°C in 6× Denhardt's solution and the filter was washed three times, as described by Sambrook et al. (14). The level of 32P in each band was quantified with an Instant Imager (Packard Instrument Co.). The copy number of the vector was calculated as the ratio of counts per minute in the vector band to counts per minute in the chromosomal band (average of three determinations).
Enzyme assays.
Cultures were grown to saturation in DN medium. After sonication, the cell extracts were used to determine either PepN activity (using l-lysine-p-nitroanilide [15]) or β-galactosidase activity (using o-nitrophenyl-β-d-galactoside [9]). One unit of lysine aminopeptidase will hydrolyze 1 nmol of substrate min−1. β-Galactosidase activity, in Miller units, was calculated as described previously (9). Protein determinations were done with a Protein Assay Kit I (Bio-Rad).
Acidification rate.
Evaluation of acidification rates was by measuring the pH following inoculation of cultures as single strains into pasteurized whole milk. The temperature was controlled by an automatic temperature controller, generating a modified cheddar cheese temperature profile. The milk had been stored overnight at 4 to 7°C in bottles with loose lids, in order to ensure equal oxygen levels in all bottles. The pH was measured semicontinuously throughout 16 h of incubation with an AAC data-logger from Intab A/B. Acidification curves were generated by the Easyview software package, version 3.2.0.4.
Nucleotide sequence accession number.
The DNA sequence of the pyrF gene appears in the EMBL and GenBank nucleotide sequence data library under accession number AF174425.
RESULTS
Sequencing and cloning of pyrF genes from industrial strains.
The most precise way to obtain a pyrF(Am) mutant is to construct the mutation in vitro via PCR (see below) and then to introduce the mutation into the chromosome by homologous recombination (10). Attempts to use MG1363 pyrF(Am) DNA to introduce a pyrF(Am) mutation into some industrial strains were unsuccessful, presumably due to insufficient homology between the pyrF genes of MG1363 and the industrial strains used. We determined the DNA sequence of the CHCC377 pyrF gene and found it to be 86% identical to that of MG1363 (1). Sequencing of pyrF genes from other industrial strains revealed that L. lactis subsp. lactis strains had sequences nearly identical to that of CHCC377 and that L. lactis subsp. cremoris strains had sequences nearly identical to that of MG1363.
Introduction of an amber mutation into a cloned pyrF gene.
The procedure for introduction of the mutation into the pyrF gene is the same for each host strain. The DNA sequence is searched for a serine codon that can be changed to an amber codon and which is flanked by sequences that allow the introduction of a restriction site without affecting the amino acids encoded by the flanking sequences. PCR is then used to introduce the desired alterations.
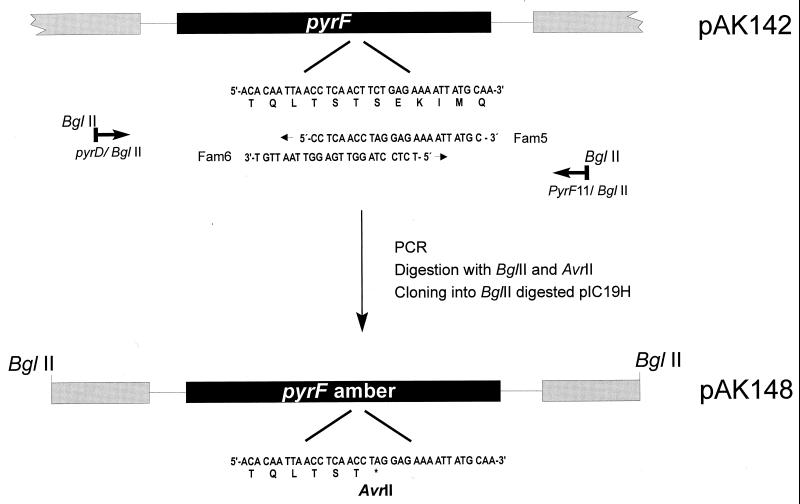
For CHCC377, the entire pyrF gene was cloned as a 1.1-kb PCR fragment in pIC19H (12) to produce pAK142 (Fig. 1). Two overlapping PCR primers, Fam5 and Fam6, contain the substitutions that generate an amber codon and an AvrII restriction site (Fig. 1). When Fam5 is used in combination with a primer upstream of pyrF (PyrD/BglII) and Fam6 is used with a downstream primer (PyrF11/BglII), with pAK142 as template DNA, two PCR products are generated. These were digested with AvrII and BglII, mixed, and cloned into BglII-digested pIC19H. The resulting clone, pAK148, contains the pyrF gene with the desired amber mutation. Suppression of the amber mutation by supD will introduce a serine residue resulting in an enzyme identical to that of the parent strain.
FIG. 1.
Construction of a pyrF(Am) mutation. The given example illustrates the construction made for the CHCC377 pyrF gene. Plasmid pAK142, carrying the entire pyrF gene, is used as template DNA in a PCR. The two overlapping primers, Fam5 and Fam6, containing the substitutions to generate an amber codon within an AvrI site, were combined with a primer upstream of pyrF (PyrD/BglII) and a primer downstream of pyrF (PyrFII/BglII). The two resulting PCR products were digested with AvrII and BglII, mixed, and cloned. The resulting plasmid, pAK148, carries the desired amber mutation in pyrF.
A similar approach was used to construct amber mutations in the pyrF genes of other strains. The host strain, FA4-1-1, was derived from MG1363 but constructed slightly differently to serve as a host for both pFG1 and pFG200 (see below). Using the approach described above and in Fig. 1 with other primers (Table 2), an amber stop codon was introduced at glutamine codon 154 in the pyrF gene. Suppression by supD will insert serine, while suppression by supB, contained on pFG1, causes insertion of glutamine in the synthesized pyrF enzyme.
Introduction of the amber mutation into the chromosome.
The DNA fragments carrying the pyrF amber mutations of MG1363, CHCC377, and CHCC270 were subcloned into pG+Host9 (11) to produce the plasmids pAK133, pAK149, and pRL202, respectively. The resulting pG+Host-derived vectors were then used to integrate the amber mutations into the corresponding chromosomal pyrF genes. Strains with the desired pyrF(Am) allele are found by screening survivors isolated on M17 plates at 30°C for their pyrimidine requirement (10). The pyrF(Am) derivatives of MG1363 were named CHCC4210 (serine to amber) and FA4-1-1 (glutamine to amber). The pyrF(Am) derivatives of CHCC377, CHCC3052, and CHCC270 were named CHCC4146, CHCC4205, and CHCC4424, respectively.
Construction of the food-grade vector pFG200.
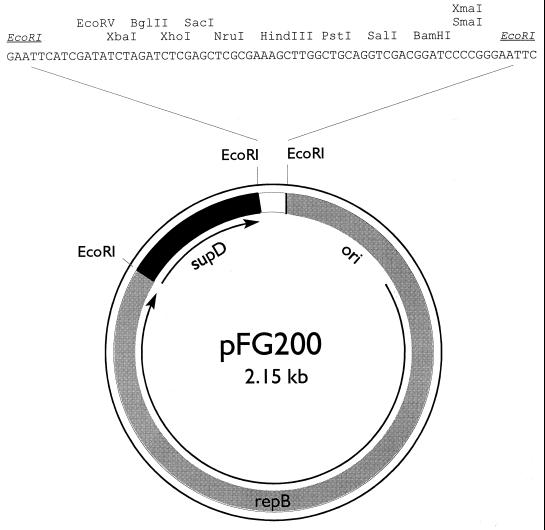
The construction of pFG200 was based on the strategy previously described for pFG1 (5). Plasmid pKR41 contains the L. lactis subsp. lactis biovar diacetylactis citrate plasmid replicon cloned in pIC19H (13). PCR was performed on pAK89.1 using primers amber-2 and amber-3 to produce a fragment with EcoRI ends that contains the supD gene (5). After digestion with EcoRI, the 347-bp PCR product was cloned into the EcoRI site of pIC19H to generate pAK93. Plasmids pKR41 and pAK93 were both digested with EcoRI, mixed, and ligated. The resulting ligation mixture was electroporated into FA4-1-1, selecting Pyr+ transformants. Among the plasmids obtained, approximately half contained the citrate plasmid replicon, supD, and pIC19H. One of these, pMPJ103, was used to construct pFG200. To remove all of pIC19H except the polylinker, pMPJ103 was digested with HindIII, ligated, and transformed into FA4-1-1. The resulting Pyr+ transformants contained a 2.15-kb plasmid, which was named pFG200 (Fig. 2).
FIG. 2.
Map of food-grade vector pFG200. The components used for the construction are described in the text. Arrows indicate the direction of transcription of repB and supD. Except for EcoRI, only those restriction sites that are unique to the polylinker are shown.
Electroporation of pFG200 into FA4-1-1 and CHCC4210 is very efficient and typically results in >106 transformants per μg of DNA. The copy number of pFG200 was found to be 6 to 11. The efficiency of transformation of CHCC4146 and derivatives of other industrial strains is significantly lower due to the difficulty of making good competent cells.
Plasmid stability in milk.
A fresh overnight culture of CHCC4223 (CHCC4146/pFG200) in minimal medium was subcultured 1:100 in RSM and incubated overnight. The resulting culture was plated on M17 plates and also subcultured 1:100 in RSM. This procedure was repeated for five consecutive days. Each outgrowth was taken to be seven generations. After each outgrowth, 100 single colonies were picked and patched onto minimal plates and minimal plates containing uracil to screen for plasmid-harboring cells. Plasmid preparations were used to ensure that the isolated colonies contained pFG200 and had the expected plasmid profiles. No plasmid loss was observed after 35 generations in milk.
Growth and acidification rates of strains harboring pFG200.
In DN minimal medium, MG1363 and CHCC4210/pFG200 grew with identical growth rates (t1/2 = 63 min), whereas FA4-1-1/pFG200 grew with a lower growth rate (t1/2 = 77 min). The lower growth rate of FA4-1-1/pFG200 is most likely due to a less active PyrF enzyme in that strain resulting from the amino acid substitution at position 154.
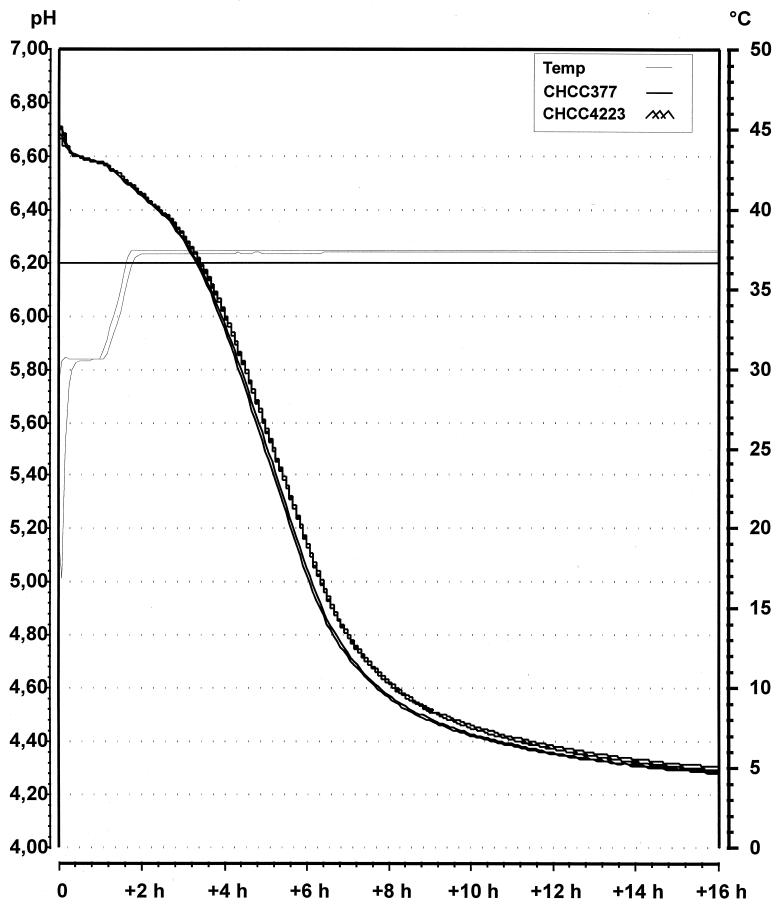
The acidification of milk by CHCC377 and CHCC4223 showed the high rate of acidification characteristic for CHCC377 (Fig. 3). This is a significant improvement over the pFG1 system, where the acidification rates for various CHCC377 derivatives were considerably reduced (unpublished results).
FIG. 3.
Acidification curve showing the change in pH during 16 h in whole milk inoculated with strain CHCC377 or strain CHCC4223 (double determinations). The temperature (Temp) profiles for this modified cheddar cheese model system (water bath and bottle) are also shown.
Measurement of transcriptional readthrough.
To determine if there is any transcriptional readthrough into the polylinker region, promoterless lacLM (9) was cloned in both orientations into the polylinker of pFG200 and the β-galactosidase activity was measured. A very low expression of β-galactosidase, corresponding to 5 to 10 Miller units, was detected, with no significant difference between the orientations, revealing little or no transcription into the polylinker.
Assay of lysine aminopeptidase activity expressed from pFG202.
The pepN gene from L. lactis strain Wg2 was cloned into pFG200 to generate pFG202. After electroporation into CHCC4210, CHCC4146, CHCC4205, and CHCC4424, the levels of lysine aminopeptidase activities were determined (Table 3). Significant increases in pepN activities were seen in all backgrounds.
TABLE 3.
Overexpression of pepN by cloning into pFG200
| Strain | Lysine aminopeptidase activity (U/mg of protein) | Fold induction |
|---|---|---|
| CHCC4210 | 339 | 8 |
| CHCC4210/pFG202 | 2,827 | |
| CHCC4146 | 89 | 20 |
| CHCC4146/pFG202 | 1,822 | |
| CHCC4205 | 104 | 23 |
| CHCC4205/pFG202 | 2,302 | |
| CHCC4424 | 100 | 23 |
| CHCC4424/pFG202 | 2,304 |
DISCUSSION
Existing food-grade cloning vectors for Lactococcus have been optimized for laboratory strains. Reports of their use in industrial strains are absent, possibly because of a lack of success. For example, pFG1 (5) has been successfully used to overexpress a number of genes in MG1363, but introduction of this vector into industrial strains resulted in acidification rates unsuitable for industrial-scale cheese production (unpublished results).
An improved food-grade cloning system for industrial strains of L. lactis has been developed. It is based on a multicopy vector, pFG200 (Fig. 2), constructed without the use of DNA from species other than Lactococcus. It contains the replicon fragment of pCT1138 (13), the amber suppressor supD from Lactococcus strain NJ1 (5) as the selectable marker, and a versatile polylinker. To select for the plasmid, we use the suppression of pyrimidine auxotrophs constructed by introduction of an amber codon in the chromosomal pyrF gene (Fig. 1). This system allows selection and maintenance of the vector in milk, which is essentially pyrimidine free. All the advantages of our previously described food-grade cloning vector, pFG1, are also offered by pFG200. These include (i) easy cloning of genes into a versatile polylinker region; (ii) a small and stable multicopy vector which is electroporated into Lactococcus with high efficiency; and (iii) a selection system allowing selection and maintenance in milk, which allows genetic modification, cloning, and overexpression of Lactococcus DNA in a food-grade manner.
The orientation of a gene inserted in the polylinker of pFG1 affects expression due to a putative promoter downstream of supB (unpublished results). Cloning of lacLM into pFG200 in both orientations and measurement of β-galactosidase activity showed a very low expression, indicating the absence of significant readthrough from either side of the polylinker.
Full tolerance to multiple copies of supD is indicated by several observations. The plasmid is stably maintained in the host cells for more than 35 generations in media including milk. The growth rates of cells with and without pFG200 in both defined media and milk show no differences. Likewise, the presence of pFG200 in important industrial starters has no impact on the acidification rate in milk, an important criterion for the usefulness of these strains in cheesemaking (Fig. 3).
The functionality of the pFG200 system was also tested and demonstrated by cloning the pepN gene from strain Wg2 (15) into pFG200 to generate pFG202. The levels of lysine aminopeptidase in the various host strains either with or without pFG202 were determined (Table 3). The endogenous level of lysine aminopeptidase was found to vary from 89 to 339 U/mg of protein. Introduction of pFG200 did not alter these endogenous levels (data not shown). Thus, the level of lysine aminopeptidase encoded by the chromosomal pepN gene is strain dependent (Søren Herskind, personal communication). In all strains, we found a significant increase in lysine aminopeptidase activity when pFG202 is present, which we presume to result from the increased copy number of pepN.
Promising results have been obtained in cheese trials and taste tests using MG1363-derived strains containing pFG1 derivatives harboring various aminopeptidase genes or the φvML3 lysin gene (unpublished data). We have also cloned the pepA, pepC, pepO, pepT, pepV, and pepX genes, as well as the φvML3 lysin gene, into pFG200 (unpublished results; Per Strøman, personal communication) and will assess the effect of overexpression of these genes in industrial strains.
The improved food-grade cloning system described here thus allows the overexpression of technologically relevant genes in industrially important strains, providing great promise for future food-grade genetic modifications of L. lactis.
ACKNOWLEDGMENTS
We sincerely thank Per Strøman for providing pSTO3, Claus Svane for technical assistance with the acidification studies, Sari Eltong for excellent technical assistance, and the Institute of Microbiology, Technical University of Denmark, for use of their Packard Instant Imager.
REFERENCES
- 1.Andersen P, Martinussen J, Hammer K. Sequence analysis and identification of the pyrKDF operon of Lactococcus lactis including a novel gene, pyrK, involved in pyrimidine biosynthesis. J Bacteriol. 1996;178:3975–3982. doi: 10.1128/jb.178.16.5005-5012.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cancilla M R, Hillier A J, Davidson B E. Lactococcus lactis glyceraldehyde-3-phosphate dehydrogenase gene, gap: further evidence for strongly biased codon usage in glycolytic pathway genes. Microbiology. 1995;141:1027–1036. doi: 10.1099/13500872-141-4-1027. [DOI] [PubMed] [Google Scholar]
- 3.Curic M, Stuer-Lauridsen B, Renault P, Nilsson D. A general method for selection of α-acetolactate decarboxylase-deficient Lactococcus lactis mutants to improve diacetyl formation. Appl Environ Microbiol. 1999;65:1202–1206. doi: 10.1128/aem.65.3.1202-1206.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Ruyter P, Kuipers O, Meijer W, de Vos W. Food-grade controlled lysis of Lactococcus lactis for accelerated cheese ripening. Nat Biotechnol. 1997;15:976–979. doi: 10.1038/nbt1097-976. [DOI] [PubMed] [Google Scholar]
- 5.Dickely F, Nilsson D, Hansen E B, Johansen E. Isolation of Lactococcus lactis nonsense suppressors and construction of a food-grade cloning vector. Mol Microbiol. 1995;15:839–847. doi: 10.1111/j.1365-2958.1995.tb02354.x. [DOI] [PubMed] [Google Scholar]
- 6.Gasson M. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J Bacteriol. 1983;154:1–9. doi: 10.1128/jb.154.1.1-9.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 8.Holo H, Nes I F. High-frequency transformation, by electroporation, of Lactococcus lactis subsp. cremoris grown with glycine in osmotically stabilized media. Appl Environ Microbiol. 1989;55:3119–3123. doi: 10.1128/aem.55.12.3119-3123.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Israelsen H, Madsen S M, Vrang A, Hansen E B, Johansen E. Cloning and partial characterization of regulated promoters from Lactococcus lactis Tn917-lacZ integrants with the new promoter probe vector, pAK80. Appl Environ Microbiol. 1995;61:2540–2547. doi: 10.1128/aem.61.7.2540-2547.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johansen E. Genetic engineering. Modification of bacteria. In: Robinson R, Batt C, Patel P, editors. Encyclopedia of food microbiology. London, United Kingdom: Academic Press; 1999. pp. 917–921. [Google Scholar]
- 11.Maguin E, Duwat P, Hege T, Ehrlich S D, Gruss S. New thermosensitive plasmid for gram-positive bacteria. J Bacteriol. 1992;174:5633–5638. doi: 10.1128/jb.174.17.5633-5638.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marsh J L, Erfle M, Wykes E J. The pIC plasmid and phage vectors with versatile cloning sites for recombinant selection by insertional inactivation. Gene. 1984;32:481–485. doi: 10.1016/0378-1119(84)90022-2. [DOI] [PubMed] [Google Scholar]
- 13.Pedersen M L, Arnved K R, Johansen E. Genetic analysis of the minimal replicon of the Lactococcus lactis subsp. lactis biovar diacetylactis citrate plasmid. Mol Gen Genet. 1994;244:374–382. doi: 10.1007/BF00286689. [DOI] [PubMed] [Google Scholar]
- 14.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 15.Strøman P. Sequence of a gene (lap) encoding a 95.3-kDa aminopeptidase from Lactococcus lactis subsp. cremoris Wg2. Gene. 1992;113:107–112. doi: 10.1016/0378-1119(92)90676-g. [DOI] [PubMed] [Google Scholar]
- 16.Terzaghi B E, Sandine W E. Improved medium for lactic streptococci and their bacteriophages. Appl Microbiol. 1975;29:807–813. doi: 10.1128/am.29.6.807-813.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]