Abstract
Delta (B.1.617.2), a highly infectious variant of SARS-CoV-2, has been sweeping the world, and threatening the safety of human life seriously. It is urgent to develop a highly selective and sensitive assay to accurately identify the SARS-CoV-2 variant Delta. In this work, we constructed a graphene/CRISPR-dCas9 electrochemical biosensor to accurately identify SARS-CoV-2 variant Delta, where the signal was further amplified by embedded electrochemical probe [Ru(phen)2dppz]BF4. This detection assay could be finished within 47 min totally, with the detection limit of 1.2 pM and good reproducibility with a C·V.% of 2.48% (n = 5). And the biosensor could selectively identify Delta among SARS-CoV-2 and other variants, including Alpha, Beta, Gamma. This assay was further validated by 26 real clinical samples, showing 100% clinical sensitivity and 100% clinical specificity, which provides a new direction for identifying other SARS-CoV-2 variants in the future.
Keywords: SARS-CoV-2, Variant delta, CRISPR-dCas9, Accurate identification, Electrochemical biosensor
Graphical abstract
1. Introduction
The first case of SARS-CoV-2 variant Delta (B.1.617 lineage) was found in October in India, which then triggered a new outbreak of epidemic in United Kingdom and India before spreading to other countries [[1], [2], [3], [4]]. As Delta variants are more infectious than Alpha variants and more resistant to vaccines, highly sensitive and specific assays to distinguish different variants of SARS-CoV-2 are vital for actual epidemic prevention and control [5,6].
The existing gold-standard approach for SARS-CoV-2 is quantitative reverse transcription polymerase chain reaction (RT-PCR) [7,8], next genome sequencing (NGS) [9] and enzyme-linked immunosorbent assay (ELISA) [1,10]. Considering the cost of time and resources, Banada et al. reported a RT-PCR melting temperature assay for N501Y and E484K mutations, which can identify SARS-CoV-2 variant Alpha and Delta from wild type [11]. Compared with RT-PCR, the cost of RT-LAMP assay was reduced, but the detection accuracy was only 89.2% [12]. While conventional nucleic acid amplification methods, such as RT-PCR and RT-LAMP mentioned above, require to operate the sophisticated and precise equipment in a centralized laboratory, electrochemical assay can be applied to reduce the footprint and complexity of devices [8]. Zhang et al. proposed an electrochemical impedance sensor to detect 1000 viral particles/mL with 100% clinical specificity and 80.5% clinical sensitivity [13]. Nevertheless, the existing electrochemical methods are not compatible with the accurate identification of the variant Delta nucleic acid.
Clustered regularly interspaced short palindromic repeats (CRISPR), as a Nobel Prize technique, has been widely used for the edition of target nucleic acids with high specificity and accuracy [14]. This technique also provides a potential method for sequence-specific nucleic acid analysis, which has been widely used in pathogen detection in recent years: (I) nanoparticles combined with CRISPR system, such as Ru complex [15] and Pt nanoparticles [16]; (II) pre-amplification integrated with CRISPR, such as HUDSON-SHERLOCK [17,18] and DETECTR [19]; (III) direct detection [[20], [21], [22], [23]], graphene-CRISPR-Cas9 field-effect transistor [22], CRISPR-Cas13a platform with multiple crRNA for SARS-CoV-2 detection [23], tandem CRISPR nucleases of Cas13 and Csm6 for RNA detection [24]. Hence, CRISPR methodology could offer a promising approach for the accurate identification of SARS-CoV-2, especially for variant Delta.
In this work, we describe a graphene/CRISPR-dCas9 electrochemical biosensor to detect SARS-CoV-2 variant Delta. We believe that this highly selective, sensitive and accurate diagnostic method could provide a new approach to identify Delta or other variants of SARS-CoV-2. The graphene has high carrier mobility, which is advantageous to the adsorption, interaction and signal conversion transmission of biomolecules at its surface [25]. The combination of graphene and CRISPR-dCas9 may be a novel strategy for the accurate identification of Delta virus.
2. Methods
2.1. Synthesis of Complex [Ru(phen)2dppz]BF4
Synthesis of dppz (Dipyrido[3,2-a:2′,3′-c]phenazine) as ligand. 0.5 g o-phenylenediamine (Aladdin Co., Shanghai) and 0.5 g 1,10-phenanthroline-5,6-dione (Aladdin Co., Shanghai) were completely dissolved in ethanol (Aladdin Co., Shanghai) respectively. o-phenylenediamine solution was added slowly into 1,10-phenanthroline-5,6-dione solution. It was heat in water bath, kept boiling for 5 min and cooled to crystallize (brown) at room temperature. Then brown-orange needle-like hemihydrate, dipyrido[3,2-a:2′,3′-c]phenazine, was obtained by recrystallization in ethanol.
Synthesis of Ru(phen)2Cl2. 0.217 g RuCl3·nH2O, 0.494 g 1.10-phenanthroline, 0.298 g Lithium chloride were dissolved in 15 mL N,N-dimethylformamide (DMF, Aladdin Co., Shanghai), heated to 140 °C under the protection of nitrogen and stirred for 4 h, kept away from light. And then it was cooled to room temperature, diluted with 20 mL acetone and placed in the refrigerator overnight. The black precipitate was obtained by vacuum suction filtration and washed with ultra pure H2O and anhydrous ether three times respectively. Finally, Ru(phen)2Cl2 was dried and stored.
Synthesis of Complex [Ru(phen)2dppz]BF4. 0.229 g dppz and 0.468 g Ru(phen)2Cl2 were dissolved in 95 mL methanol and ultrapure H2O (V:V = 2:1), with reflux and stirring at 110 °C for 5 h. Then the obtained solution was concentrated to 10% of the original volume at 180 °C. After adding 35 mL ultrapure H2O, the solution was boiled for 10 min, cooled to room temperature and filtered with a triangular funnel. The filtrate was added with 7.5 mL 10% NaBF4 solution. The precipitate was obtained as [Ru(phen)2dppz]BF4.
2.2. Fabrication of CRISPR electrode
Before CRISPR system modification, the surface of the gold electrode (diameter of 2 mm) was polished with finer-grade aqueous alumina slurries (1.0, 0.3 and 0.05 μm grain sizes) successively on a chamois leather, followed by alternately ultrasound washing with ethanol and double-distilled water.
The procedures were as followed: (I) 0.2 mg/mL graphene dispersed (Sigma-Aldrich) in 0.1% chitosan was drop-casting on the surface and it was placed in oven (60 °C, 30 min); (II) and 10 μL 1-pyrenebutanoic acid (PBA, dissolved in N,N-dimethylformamide, Aladdin Co., Shanghai) of 5 mM was drop-casted on this surface under 37 °C for 30 min. PBA was stacked on the graphene surface via π-π interaction. And the electrode was washed by ultrapure H2O; (III) accordingly [22], carboxyl group of PBA was activated by EDC (1-(3-dimethylaminopropyl)-3-ethylcarbodiimide hydrochloride, 4 mM): NHS (N-hydroxy succinimide, 11 mM) solution (100 μL:100 μL in 50 μL 50 mM 2-morpholinoethanesulfonic acid buffer, all provided by Aladdin Co., Shanghai) for 30 min under 37 °C; (IV) and the electrode was incubated in 1 μM dCas9 (Tolo Biotechnology) for 30 min under 37 °C. (V) and 1% BSA (bovine serum albumin, Sigma-Aldrich) blocked the non-specific active sites for 30 min under 37 °C. It was washed by 2 mM MgCl2; (VI) the electrode was incubated in 1 μM sgRNA (Sangon Co., synthesized and provided) for 30 min under 37 °C, and washed by 2 mM MgCl2 for 3 min. Thus, a CRISPR electrode was fabricated.
2.3. Sample detection
The CRISPR electrode was incubated in the prepared [Ru(phen)2dppz]BF4 of 30 μM for 3 min at room temperature and then washed with ultrapure H2O for 1 min. Electrochemical scanning was performed on the electrode in PBS buffer (0.01 M, pH 7.4) as background signal, with potential from 1.5 V to 0 V and sensitivity of 1 × 10−5 A/V.
The CRISPR electrode was incubated in the samples under 37 °C for 40 min and washed with 2 mM magnesium chloride solution at room temperature for 3 min. After that, the electrode was incubated in the prepared [Ru(phen)2dppz]BF4 for 3 min at room temperature and then washed with ultrapure H2O for 1 min again. Electrochemical scanning was performed on the electrode in PBS, with potential from 1.5 V to 0 V and sensitivity of 1 × 10−5 A/V.
2.4. Electrochemical measurements
Differential pulse voltammetry (DPV) testing was conducted. The scanning potential range was set from 0 V to 1.2 V at a scan rate of 50 mV/s in a PBS buffer (0.01 M, pH 7.4).
Cyclic voltammetry (CV) measurement was conducted for electrochemical characterization. The scanning potential range was set from −0.2 V to 0.6 V at a scan rate of 50 mV/s in a 0.05 M K3[Fe(CN)6]/K4[Fe(CN)6] solution that contained 0.50 M KCl. Electrochemical impedance spectroscopy (EIS) was performed in the frequency range of 0.1 Hz–100 000 Hz and at an alternating voltage of 5 mV.
2.5. Agarose gel electrophoresis
2% agarose gels were prepared in 1 × TAE buffer and heated in a microwave oven (Galanz, Guangdong). Samples and 6 × Ficoll gel loading buffer III were pre-mixed for 3 min and added to the gel. Finally, standard agarose gel electrophoresis was performed with an EPS 300 electrophoresis apparatus (Tanon, Shanghai) and 4100 digital gel image processing system (Tanon, Shanghai). All the reagents were provided by Sangon Inc. (Shanghai, China).
2.6. sgRNAs and clinical samples pre-treatment
The modeling of sgRNA was specifically designed as followed: (1) the structure as well as principle was confirmed according to the reference (Ran et al., 2013), (II) then its recognition efficiency was evaluated through the provided Zhang's lab website (https://zlab.bio/guide-design-resources). And sgRNAs were provided by Bio-lieesci Co., LTD, Guangzhou and sgRNA sequences were shown in Table S2. The SARS-CoV-2 delta virus samples were collected from COVID-19 infected patients and nucleic acids extraction experiments were conducted in biological safety protection third-level (P3) laboratory by Qingdao customs (Shandong, China). Then all the extracted clinical samples were verified by RT-PCR kit (FP313-01, Tiangen Co., Beijing) and provided by Qingdao Customs (Shandong, China). The RT-PCR details were shown in Supporting Information (Table S2 and S3). Finally, samples of the amplicons were quantified by a nanodrop meter (Merinton SMA4000, Beijing) and diluted to a certain concentration in our laboratory.
2.7. Statistical analysis
All statistical results presented were performed by two-way ANOVA and linear regression analyzed by using origin 9.0 software. These results were performed by mean ± SD. Significance level was implied by *, **, ***, ****, ns for p < 0.05, p < 0.01, p < 0.001, p < 0.0001, no significance respectively.
3. Results and discussion
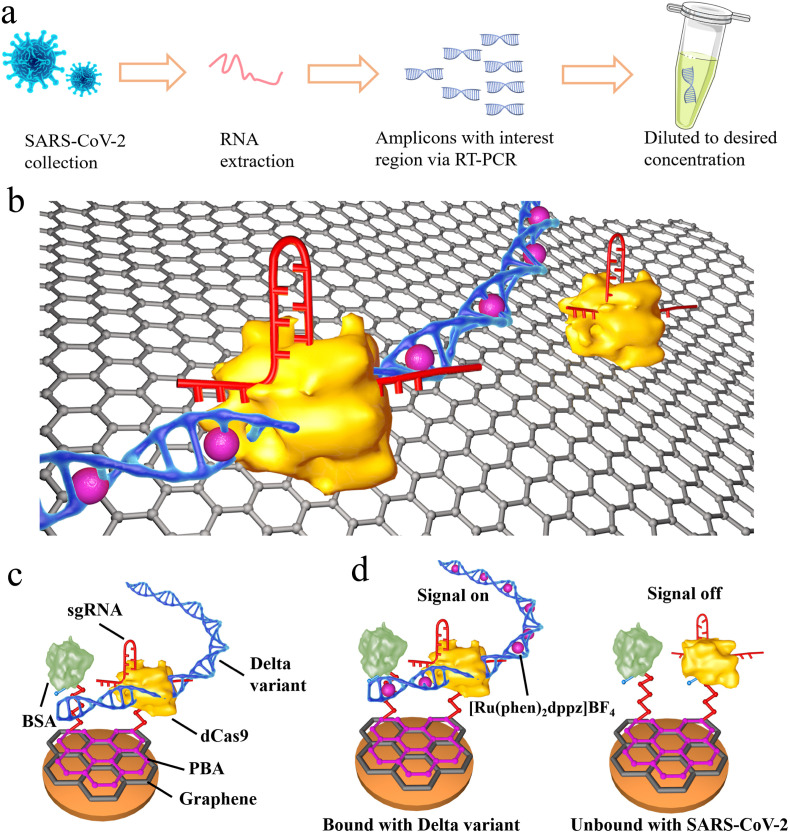
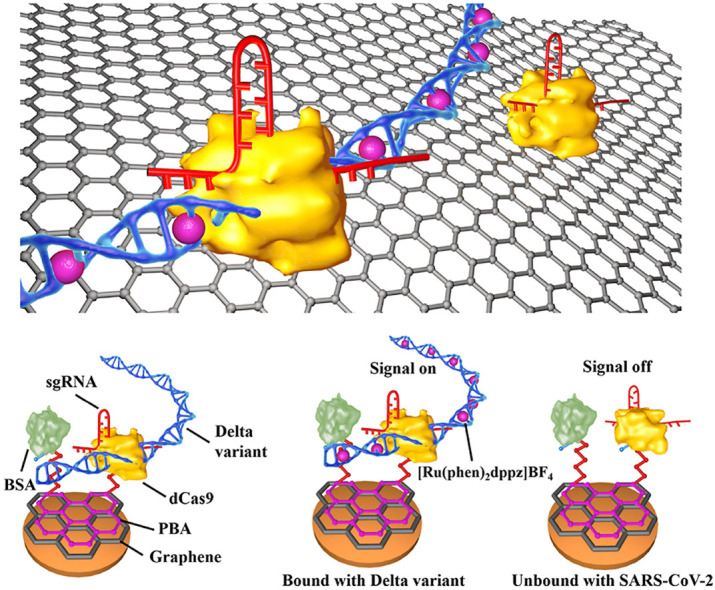
3.1. Principle of the graphene/CRISPR-dCas9 electrochemical biosensor for the identification of delta and wild type SARS-CoV-2
The principle of this biosensor for the identification of SARS-CoV-2 mutation was shown as Fig. 1 a–c. The interest region of target was primarily validated by RT-PCR, then the amplicons were diluted into desired concentration. Thus, the proposed biosensor could directly detect the diluted samples. We first fabricated a biosensor with high specific surface area and high carrier mobility using the graphene/chitosan as the substrate. The sgRNA/dCas9 (denoted as dRNP) immobilized on the sensor substrate could bind the target nucleic acid of SARS-CoV-2 variant Delta (denoted as Delta) specifically, with the assistance of sgRNA and the protospacer adjacent motif (PAM) site. The interaction between dRNP and target DNA results in a change of the electrochemical current signal, which could be further enhanced by the embedded electrochemical probe [Ru(Phen)2dppz]BF4.
Fig. 1.
The schematic and characterizations of the graphene-CRISPR biosensors for the identification of SARS-CoV-2 variant Delta and SARS-CoV-2. (a) The workflow of the sample pretreatment for SARS-CoV-2 variant Delta. (b) The schematic of the graphene-CRISPR biosensor for the identification of SARS-CoV-2 Delta variant (denoted as Delta) by identifying L452R mutation site. (c) The fabrication of the biosensor. (d) The current of “signal-on” type biosensor recorded by the intercalation of [Ru(phen)2dppz]BF4 into the target DNA, producing a current peak at ∼0.5 V.
In this fabrication of graphene/CRISPR electrochemical biosensor, the graphene electrode was first modified with pyrene-1-butyric acid (PBA) via π-π stacking, then modified with dCas9 through 1-(3-dimethylaminopropyl)-3-ethylcarbodiimide hydrochloride (EDC) and N-hydroxy succinimide (NHS). Bovine albumin (BSA) was subsequently used to avoid nonspecific absorption and reduce background signals. Finally, the specific sgRNA was immobilized on dCas9 via tracrRNA domain to search and bind target DNA. When incubated with samples, the CRISPR was primarily triggered by the special binding of Delta target with dRNP. Due to the inactivity of the two domains (HNH and RuvC) of dCas9 [14], the bounded Delta target cannot be gene-edited. Secondly, as shown in Fig. 1d, the electrochemical signal probe of [Ru(phen)2dppz]BF4 could intercalate into the minor-groove of the bounded double-stranded DNA, and could enhance DPV current signal [26].
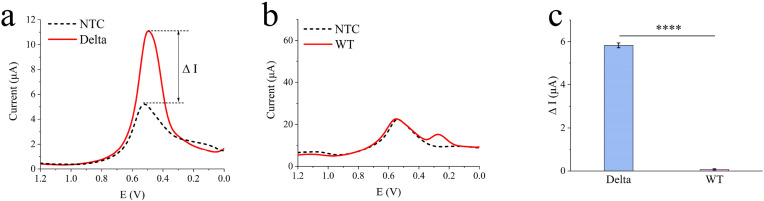
There was no significant DPV signal when wild type SARS-CoV-2 (WT) was incubated with [Ru(phen)2dppz]BF4 (Fig. 2 a and Fig. 2b). As shown in Fig. 2c, the Delta sample had a higher current response (5.83 μA) than that of WT sample (0.07 μA), with significance difference.
Fig. 2.
The investigation of the graphene-CRISPR biosensor. (a) The DPV curve of the biosensor to recognize the Delta DNA from the positive patient. (b) The DPV curve of the biosensor to recognize the SARS-CoV-2 wild type (denoted as WT) from the positive patient. (c) The comparison of the current response between Delta and WT from Fig. 2a and b, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, and ns referring to no significant difference, as analyzed by two-way ANOVA.
The selectivity of the method was extremely high because CRISPR-dCas9 system could highly recognize the Delta by the perfectly matched sgRNA-DNA complex. Moreover, the sensitivity of this method was greatly improved by graphene that had high carrier mobility and [Ru(phen)2dppz]BF4 that was inserted into double-stranded DNA minor-groove. Therefore, this graphene-based CRISPR-dCas9 technology could lead to the development of a promising signal-on biosensor for the accurate identification of SARS-CoV-2 variant Delta.
3.2. Characterization of the graphene-CRISPR biosensor
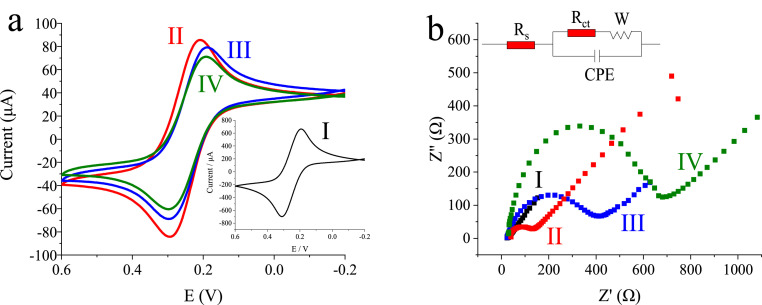
The electrochemical behaviors of the graphene-CRISPR biosensor in the preparation and the detection were investigated by cyclic voltammetry (CV) and EIS in a reversible redox couple of [Fe(CN)6]3–/4–. As shown in Fig. 3 a, the current of redox peaks for graphene electrode (curve II) was lower than that of the bare electrode (curve I), indicating the successful immobilization of graphene/chitosan on the electrode surface by drop-casting process. The molecule recognition of the nucleic acid based on CRISPR system on the surface of the electrode hindered electron transfer between the surface of electrode and the electrolyte solution, which led to the current change of the system. Thus, after modification of the CRISPR-graphene biosensor with dRNP, a decreased current of redox peaks (curve III) was observed, which indicated the dRNP was successfully immobilized on the surface of the graphene modified gold electrode. Then the sensor was incubated with Delta DNA, the obviously decreased current of redox peaks was observed (curve IV), indicating the successful recognition of target DNA through CRISPR-dCas9 system.
Fig. 3.
The electrochemical characterization of the graphene-CRISRP biosensor. (a) Cyclic voltammograms and (b) Nyquist plots of EIS for I) the bare gold electrode, II) the graphene modified gold electrode, III) the graphene-CRISPR biosensor, IV) the graphene-CRISPR biosensor + 2 nM Delta DNA, the electrolyte solution of 0.05 M [Fe(CN)6]3−/4− and 0.5 M KCl. Scan rate: 100 mV/s.
As a sensitive tool to detect the interfacial properties, EIS was also conducted to detect the changes in electron transfer resistance (Ret) of the biosensor in this study. Fig. 3b showed Nyquist plots of the biosensors measured by EIS. All the plots exhibited a semicircle portion at high frequencies, indicating a limited electron transfer process, and a linear portion at low frequencies, indicating a limited diffusion. The semicircles diameters at high frequency region were closely correlative to the Ret. As illustrated in Fig. 3b, a randle model was chosen as the equivalent electrical circuit to fit all the data measured by EIS. Here, Rs, Rct, Zw, CPE represented the solution resistance, the electron-transfer resistance of the redox probes, Warburg impedance and constant phase element, respectively. The increased Rct value of graphene electrode (144 Ω, curve II) than that of GE (6 Ω, curve I) indicated the formation of graphene/chitosan membrane on the electrode surface. When assembled with dRNP, the Rct value of the electrode increased to 387 Ω (curve III), implying the formation of CRISPR-dCas9 system on the surface of the graphene electrode. After incubated with Delta target DNA, the increased Rct value of the graphene-CRISPR biosensor (669 Ω, curve IV) indicated that the binding of the target DNA blocked the electron transfer from the redox probes to the graphene modified gold electrode.
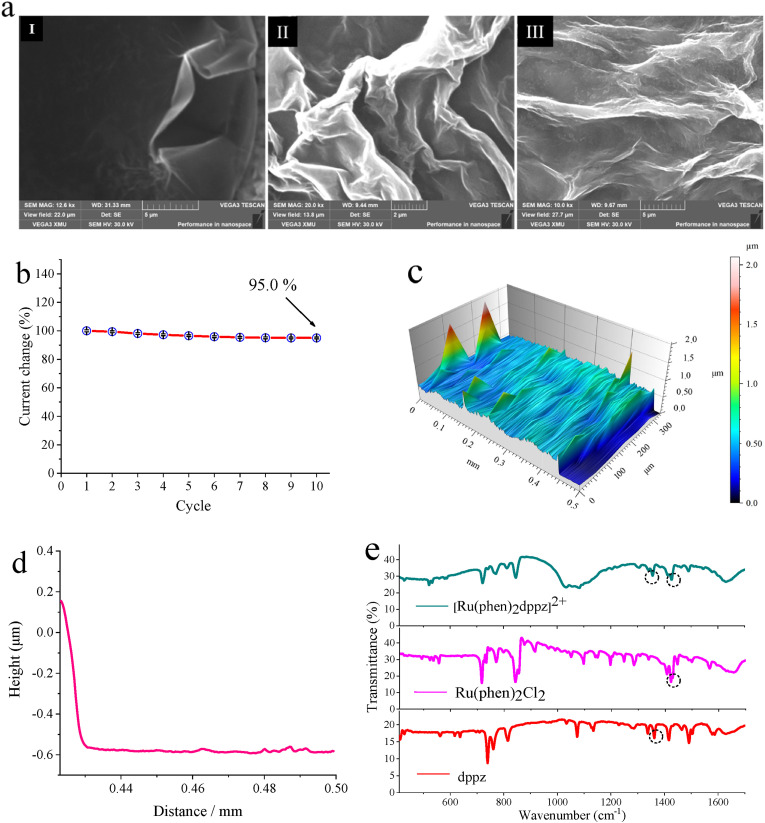
The morphology of graphene may be closely related to the performance of biosensors. In this study, we investigated the morphology of the graphene membrane in different condition by means of scanning electron microscopy (SEM), as shown in Fig. 4 a. We can see that there was a lot of wrinkles for the pristine graphene (Fig. 4a, I). After dispersion in chitosan solution, a significant increase in surface wrinkles of the biosensor could be observed (Fig. 4a, II). Nevertheless, compared with photograph II, there was little change in surface morphology of the biosensor after incubation in PBS (37 °C, pH 7.4, 75 min) (Fig. 4a, III). In order to further investigate the electrochemical performance of the graphene/chitosan membrane, the graphene/chitosan modified biosensors were fabricated in different batches, and applied to electrochemical testing (n = 3). As shown in Fig. 4b, during the 10 cycles testing, the current change maintained at 95.0%, averagely with a C.V. of 1.93%, indicating the high reproducibility of the drop-casting method for the graphene membrane. We also examined the thickness of the graphene membrane using a stylus profiler to further understand the morphology of the drop-casting layer. From Fig. 4c and d, we could see an obvious stage on the surface of graphene membrane with a height of ∼0.55 μm, indicating the well-defined reproducibility, stability, and rigidity of working electrode, based on the drop-casting method. And the as-prepared [Ru(phen)2dppz]BF4 as electrochemical probe was characterized by FTIR spectrum as shown in Fig. 4e, indicating the successful synthesis.
Fig. 4.
The morphology characterization of the graphene-CRISRP biosensor. (a) SEM for membrane, I to III representing for pristine graphene, the graphene/chitosan modified biosensor, the graphene/chitosan biosensor after incubation in PBS (37 °C, pH 7.4, 75 min). (b) The reproducibility of the graphene modified electrode under different scanning cycles, the graphene/chitosan drop-casted on the surface of the electrode, incubation in 60 °C for 30 min, scanning potential range from −0.2 V to 0.6 V at a scan rate of 50 mV/s in a 0.05 M K3[Fe(CN)6]/K4[Fe(CN)6] solution containing 0.50 M KCl, n = 3. (c) Three-dimensional images of the graphene membrane measured by a stylus profiler, 0.05 mg loading force, 0.1 mm/s scanning speed. (d) Row data of the stylus profiler measurement for graphene membrane. (e) Characterization of multifunctional probe [Ru(phen)2dppz]BF4, FTIR spectrum of synthesized [Ru(phen)2dppz]BF4, –C N- of dppz at 1362 cm−1, –C N- of Ru(phen)2Cl2 at 1422 cm−1, the dppz ligand of [Ru(phen)2dppz]BF4 red-shifted at 1356 cm−1, the phen ligand of [Ru(phen)2dppz]BF4 blue-shifted at 1427 cm−1.
3.3. Specificity of the biosensor for SARS-CoV-2 variant delta
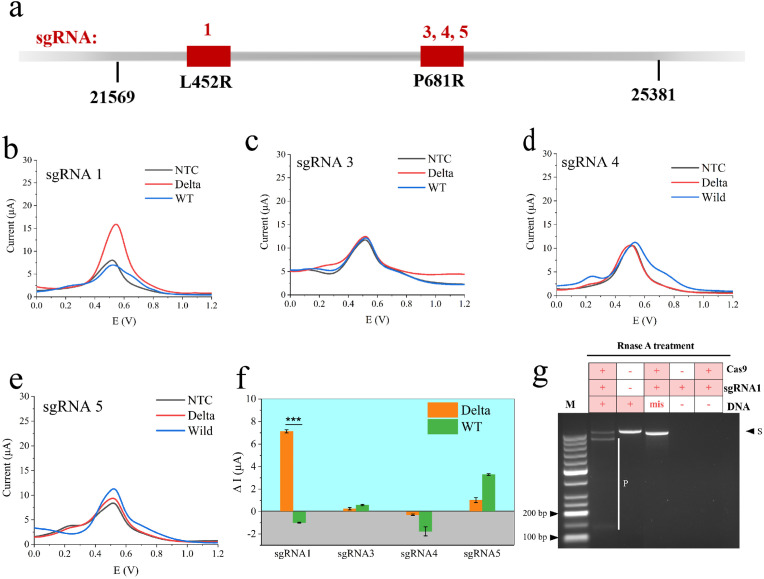
The special sgRNA was most important to the selectivity of the biosensor [23]. To this aim, we investigated the specificity of the graphene-CRISPR biosensor towards Delta and WT gene of SARS-CoV-2. Four sets of sgRNA for two mutations site (sgRNA1 for L452R; sgRNA3, sgRNA4, sgRNA5 for P681R) were designed and screened in vitro by clinical WT and Delta nucleic acids of SARS-CoV-2 (Fig. 5 a). When the [Ru(Phen)2dppz]BF4 electrochemical signal probe was inserted into the DNA minor-groove, the signal increased significantly at the peak of ∼0.5V current, resulting in a synergistic enhancement effect. Thus, we observed an increased curve for the positive samples, whereas little increase or decrease electrochemical signal for the negative samples.
Fig. 5.
Optimization of sgRNA and specificity validation of the graphene-CRISPR biosensor for SARS-CoV-2 variant Delta identification. (a) Schematic of dCas9-sgRNA complex binding Delta DNA. (b) To (e) referring to sgRNA1, sg RNA3, sgRNA4, sgRNA5 for sgRNA primers screening in the identification of Delta and WT of 4 nM, incubation under 37 °C for 40 min, with 30 μM [Ru(phen)2dppz]BF4 electrochemical signal probe. (f) The current response of different sgRNA during the identification of Delta and WT, results from Figure b to Figure e, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, and ns referring to no significant difference, as analyzed by two-way ANOVA. (g) Agarose gel electrophoresis under different conditions, mis referring to WT DNA, 2% agarose, 1 × TAE, 25 °C, 80 min, 100 V.
From the results of Fig. 5b–f, DPV experiments were also performed for the detection of two SARS-CoV-2 virus gene. In Fig. 5b, a significant difference (P = 0.00013) in the current response of Delta was observed for sgRNA1 compared with the template-free control (NTC) and WT, indicating the excellent specificity of sgRNA1. For sgRNA3 (Fig. 5c), there was no significance in current response between Delta (0.26 μA) and WT (0.59 μA). And for sgRNA4 (Fig. 5d), decreased current responses were observed both for Delta and WT, indicating the immobilized dRNP could not bound with target DNA. Although an obvious current increase in the Delta with sgRNA5 (Fig. 5e), the current response in the WT group was higher than Delta, indicating that the dCas9/sgRNA5 system was not selective enough for Delta. From the results of Fig. 5f, sgRNA1 was chosen for the identification of Delta in this study, which had significant difference compared with WT group.
Meanwhile, the specificity of dRNP was further demonstrated by agarose gel electrophoresis. The sgRNA screening was shown in Figure S1. And in Fig. 5g, it could be seen that there were new bands in the line of Cas9/sgRNA1, referring to CRISPR products (denoted as P), compared with the Delta target and WT target (both denoted as S). Moreover, The CRISPR reaction could only be performed in the presence of Cas9, sgRNA1 and target DNA. All these results indicated that Cas9/sgRNA1 could specifically bind to then cut nucleic acids of Delta. And sgRNA as well as [Ru(phen)2dppz]BF4 as the molecular beacon was indispensable to selectively identify Delta for the graphene-CRISPR biosensor.
3.4. Analytical performance of the graphene-CRISPR biosensor for the detection of delta
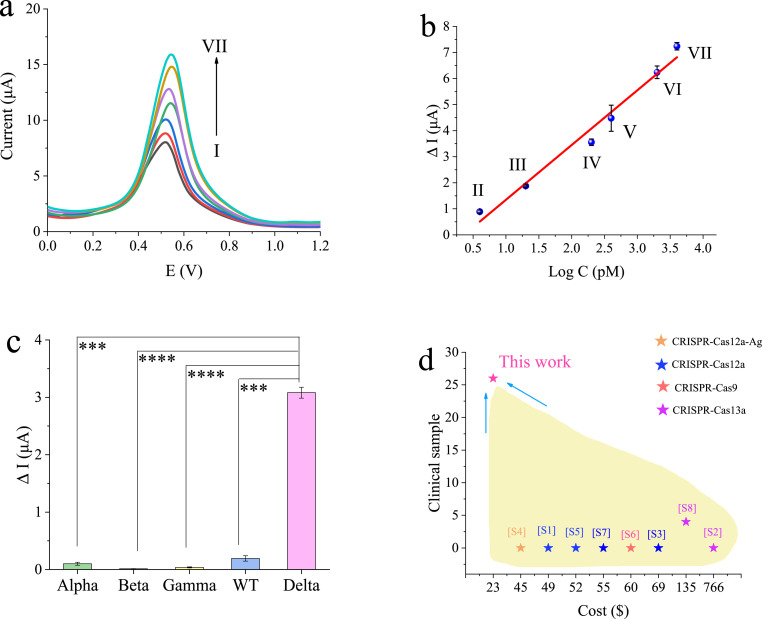
For quantitative detection of Delta (960-bp PCR amplicons provided by Qingdao customs, China), different concentrations of Delta sample (4 pM–4 nM) and the blank sample were incubated with the biosensor and detected. The DPV response of the biosensor was shown in Fig. 6 a. With the synergetic effect of graphene film and CRISPR-dCas9, the current response increased as the concentration of Delta increased, which resulted from the intercalation of [Ru(phen)2dppz]BF4 into the target DNA minor-groove. To explore the sensitivity of the biosensor, the concentration of Delta in the range from 4 pM to 4 nM was recorded, which showed a linear relationship between the changes of current (ΔI) and target DNA concentration. In Fig. 6b, a linear regression equation was ΔI (μA) = 2.1021·lgC (pM)-0.7571 (R = 0.9889). Thus, a limit of detection (LOD) was 1.2 pM (DL = 3δb/K). The high sensitivity of the biosensor may be attributed to the high carrier mobility of graphene and signal enhancement of [Ru(phen)2dppz]BF4 molecular beacon.
Fig. 6.
Analytical performance of the graphene-CRISPR biosensor. (a) The sensitivity of the graphene-CRISPR biosensor for the detection of Delta DNA, including DPV responses incubated with different concentrations of target DNA, from I to VII representing 0, 4, 20, 200, 400, 2000, 4000 pM of Delta DNA. (b) The calibration curve, error bars, mean ± SD (n = 3). (c) Specificity of the graphene-CRISPR biosensor for the identification of 108 copies/μL of SARS-CoV-2 variant Alpha, 108 copies/μL of SARS-CoV-2 variant Beta, 108 copies/μL of SARS-CoV-2 variant Gamma, 108 copies/μL of SARS-CoV-2 (WT), 107 copies/μL of SARS-CoV-2 variant Delta. (d) Performance comparison of the graphene-CRISPR electrochemical biosensor with other reported CRISPR-based electrochemical biosensors for the detection of nucleic acid.
To investigate the specificity of the as-fabricated biosensor, the conserved sequences of SARS-CoV-2 variant Alpha (B.1.1.7 Alpha-N501Y), SARS-CoV-2 variant Beta (B.1.351 Beta-K417 N), SARS-CoV-2 variant Gamma (P.1 Gamma-K417T), SARS-CoV-2 (WT), and SARS-CoV-2 variant Delta (B.1.617.2 Delta-L452R) were cloned into the PUC57 plasmid for the subsequent testing. As shown in Fig. 6c, compared with negative groups (108 copies/μL), the Delta group (107 copies/μL) exhibited an obvious current signal change, which validated the high specificity of the graphene-CRISPR biosensor for the target nucleic acid detection.
The anti-interference ability of the biosensor was detected by DPV in the target sample with the same concentration (4 nM) under the interference conditions of, such as Na+, K+, Cl−, glucose, urea, BSA (2 mM). In Figure S2, compared with the target Delta DNA, the current response generated by the interference is negligible, with a coefficient of variation (C·V.) of <5%.
The reproducibility of the biosensor was tested by recording the current response five times, using one electrode in similar conditions for detection of 4 nM Delta DNA during one batch, with a C.V. of 4.71%, indicating a good reproducibility of each biosensor. And five different biosensors that were prepared in similar conditions for detection of 4 nM Delta DNA, with a C.V. of 2.48%, implying well-defined sensor-to-sensor reproducibility. In Fig. 6d and Table S5, to our best of knowledge, in comparison of previously reported electrochemical biosensors, the graphene-CRISPR biosensor showed advantages in cost and real clinical applications.
3.5. Clinical sample tests
In this study, real clinical samples (including 16 Delta samples and 10 WT samples) were applied to further evaluate the performance and accuracy of the graphene/CRISPR-dCas9 biosensor. All the real clinical nucleic acid samples provided by Qingdao customs (Shandong, China) have been firstly confirmed by real-time PCR. And then all the clinical samples were conducted by this proposed platform. Regarding these 26 clinical samples, the clinical sensitivity and specificity of the biosensor were 100% and 100%, respectively, as shown in Table 1 and Table S1. And the raw data for clinical samples detection was shown in Figure S3. In Table S4, the recoveries ranged from 93.8% to 98.7% with RSD <5%. Based on the results, we found that this biosensor could highly identify Delta and WT in the clinical applications.
Table 1.
Comparison between the graphene/CRISPR-dCas9 biosensor and hospital disease reports for the detection of SARS-CoV-2 virus among real clinical samples.
| Graphene-CRISPR biosensor | Gold standard |
Total | |
|---|---|---|---|
| Delta | WT | ||
| Delta | 16 | 0 | 16 |
| WT | 0 | 10 | 10 |
| Total | 16 | 10 | 26 |
*:Clinical sensitivity = × 100%; Clinical specificity = × 100%.
4. Conclusion
In summary, we successfully established a signal-on biosensor for accurate identification of SARS-CoV-2 variant Delta, which integrated highly specific CRISPR-dCas9 system and graphene. And graphene with high carrier mobility (>2000 cm2Vs−1) has been widely used in biosensing applications [22,25]. Compared with the gold standard real-time PCR method, this assay had a high clinical sensitivity (100%) and well-defined clinical specificity (100%). Of note, this method had the ability of accurate identification Delta sample, with a detection limit of 1.2 pM, which could avoid the cross-contamination during samples turnaround processes. While some representative advances for SARS-CoV-2 detection have been reported such as field-effect transistor biosensor [27] and multiplexed detection electrochemical platform [28], our proposed electrochemical biosensor was the first with the capability to accurately identify Delta nucleic acid samples from COVID-19 infected subjects. Above all, we believed that this label-free, accurate identification electrochemical platform could offer a highly sensitive, selective, and stable method for Delta or other SARS-CoV-2 variants identification for epidemic disease. And more explorations of quantitative and direct detection will be the following work in the future.
Credit author statement
Bin Yang, Xiaowei Zeng, and Jin Zhang are equally contributed to the paper. Bin Yang conceived the research. Bin Yang and Xiaowei Zeng conducted the experiments. Bin Yang and Xiaowei Zeng prepared the manuscript. Bin Yang, Xiaowei Zeng and Xueen Fang wrote the manuscript. Jin Zhang offered clinical samples and assisted in clinical samples verifications. Xueen Fang and Jilie Kong supervised the research. All the authors read and corrected the manuscript.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We gratefully acknowledge the financial support by the National Natural Science Foundation of China (22174024, 21974028, 22174022, 22127806), Shanghai Scientific and Technological Innovation Action Plan (19441915600) and Shanghai Agricultural Development project (2021-02-08-00-12-F00772); we also thanks China National Postdoctoral Program for Innovative Talents (BX20200090), China Postdoctoral Science Foundation Funded Project (2021M700806) and Shanghai Post-doctoral Excellence Program (2020066).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.talanta.2022.123687.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Planas D., Veyer D., Baidaliuk A., Staropoli I., Guivel-Benhassine F., Rajah M.M., Planchais C., et al. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature. 2021;596:276. doi: 10.1038/s41586-021-03777-9. [DOI] [PubMed] [Google Scholar]
- 2.Hoffmann M., Hofmann-Winkler H., Kruger N., Kempf A., Nehlmeier I., Graichen L., et al. Delta variant (B. 1.617. 2) sublineages do not show increased neutralization resistance. Cell Rep. 2021;36 doi: 10.1038/s41423-021-00772-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Edara V.V., Pinsky B.A., Suthar M.S., Lai L., Davis-Gardner M.E. Infection and vaccine-induced neutralizing antibody responses to the SARS-CoV-2 B. 1.617. 1 variant. N. Engl. J. Med. 2021;385:664–666. doi: 10.1056/NEJMc2107799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wall E.C., Wu M., Harvey R., Kelly G., Warchal S. Neutralising antibody activity against SARS-CoV-2 VOCs B. 1.617. 2 and B. 1.351 by BNT162b2 vaccination. Lancet. 2021;397:2331–2333. doi: 10.1016/S0140-6736(21)01290-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Callaway E. Delta coronavirus variant: scientists brace for impact. Nature. 2021;595:17–18. doi: 10.1038/d41586-021-01696-3. [DOI] [PubMed] [Google Scholar]
- 6.Liu C., Ginn H.M., Dejnirattisai W., Supasa P., Wang B., Tuekprakhon A., et al. Reduced neutralization of SARS-CoV-2 B. 1.617 by vaccine and convalescent serum. Cell. 2021;184:4220–4236. doi: 10.1016/j.cell.2021.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vogels C.B.F., Brito A.F., Wyllie A.L., Fauver J.R., Ott I.M., Kalinich C.C., et al. Analytical sensitivity and efficiency comparisons of SARS-CoV-2 RT–qPCR primer–probe sets. Nat. Microbiol. 2020;5:1299–1305. doi: 10.1038/s41564-020-0761-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yousefi H., Mahmud A., Chang D., Das J., Gomis S., Chen J.B., et al. Detection of SARS-CoV-2 viral particles using direct, reagent-free electrochemical sensing. J. Am. Chem. Soc. 2021;143:1722–1727. doi: 10.1021/jacs.0c10810. [DOI] [PubMed] [Google Scholar]
- 9.Tillett R.L., Sevinsky J.R., Hartley P.D., Kerwin H., Crawford N., Gorzalski A., et al. Genomic evidence for reinfection with SARS-CoV-2: a case study. Lancet Infect. Dis. 2021;21:52–58. doi: 10.1016/S1473-3099(20)30764-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tan C.W., Chia W.N., Qin X., Liu P., Chen M.I., Tiu C., et al. A SARS-CoV-2 surrogate virus neutralization test based on antibody-mediated blockage of ACE2-spike protein-protein interaction. Nat. Biotechnol. 2021;38:1073–1078. doi: 10.1038/s41587-020-0631-z. [DOI] [PubMed] [Google Scholar]
- 11.Banada P., Green R., Banik S., Chopoorian A., Streck D., Jones R., Chakravorty S., Alland D.J. A simple reverse transcriptase PCR melting-temperature assay to rapidly screen for widely circulating SARS-CoV-2 variants. Clin. Microbiol. 2021;59 doi: 10.1128/JCM.00845-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jamwal V.L., Kumar N., Bhat R., Jamwal P.S., Singh K., Dogra S., et al. Optimization and validation of RT-LAMP assay for diagnosis of SARS-CoV2 including the globally dominant Delta variant. Virol. J. 2021;18:178. doi: 10.1186/s12985-021-01642-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Z., Pandey R., Li J., Gu J., White D., Stacey H.D., Ang J.C., Steinberg C.J., et al. High‐affinity Dimeric Aptamers enable the rapid electrochemical detection of wild‐type and B. 1.1. 7 SARS‐CoV‐2 in unprocessed Saliva. Angew. Chem. In. Eng. 2021;60:24266–24274. doi: 10.1002/anie.202110819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anzalone A.V., Koblan L.W., Liu D.R. Genome editing with CRISPR-Cas nucleases, base editors, transposases and prime editors. Nat. Biotechnol. 2020;38:824–844. doi: 10.1038/s41587-020-0561-9. [DOI] [PubMed] [Google Scholar]
- 15.Liu W., Yu H., Zhou X., Xing D. In vitro evaluation of CRISPR/Cas9 function by an electrochemiluminescent assay. Anal. Chem. 2016;88:8369–8374. doi: 10.1021/acs.analchem.6b02338. [DOI] [PubMed] [Google Scholar]
- 16.Shao N., Han X., Song Y., Zhang P., Qin L. CRISPR-Cas12a coupled with platinum nanoreporter for visual quantification of SNVs on a volumetric bar-chart chip. Anal. Chem. 2019;91:12384–12391. doi: 10.1021/acs.analchem.9b02925. [DOI] [PubMed] [Google Scholar]
- 17.Gootenberg J.S., Abudayyeh O.O., Kellner M.J., Joung J., Collins J.J., Zhang F. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science. 2018;360:439–444. doi: 10.1126/science.aaq0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Myhrvold C., Freije C.A., Gootenberg J.S., Abudayyeh O.O., Metsky H.C., Durbin A.F., Kellner M.J., Tan A.L., Paul L.M., Parham L.A., Garcia K.F., et al. Field-deployable viral diagnostics using CRISPR-Cas13. Science. 2018;360:444–448. doi: 10.1126/science.aas8836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen J.S., Ma E.B., Harrington L.B., Da Costa M., Tian X., Palefsky J.M., Doudna J.A. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science. 2018;360:436–439. doi: 10.1126/science.aar6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bruch R., Baaske J., Chatelle C., Meirich M., Madlener S., Weber W., Dincer C., Urban G.A. CRISPR/Cas13a‐powered electrochemical microfluidic biosensor for nucleic acid amplification‐free miRNA diagnostics. Adv. Mater. 2019;31 doi: 10.1002/adma.201905311. [DOI] [PubMed] [Google Scholar]
- 21.Dai Y., Somoza R.A., Wang L., Welter J.F., Li Y., Caplan A.I., Liu C.C. Exploring the trans‐cleavage activity of CRISPR‐cas12a (cpf1) for the development of a universal electrochemical biosensor. Angew. Chem. Int. Ed. 2019;58:17399–17405. doi: 10.1002/anie.201910772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hajian R., Balderston S., Tran T., DeBoer T., Etienne J., Sandhu M., Wauford N.A., et al. Detection of unamplified target genes via CRISPR–Cas9 immobilized on a graphene field-effect transistor. Nat. Biomed. Eng. 2019;3:427–437. doi: 10.1038/s41551-019-0371-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fozouni P., Son S., Derby M.D.D.L., Knott G.J., Gray C.N., Ambrosio M.V., et al. Amplification-free detection of SARS-CoV-2 with CRISPR-Cas13a and mobile phone microscopy. Cell. 2021;184:323–333. doi: 10.1016/j.cell.2020.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu T.Y., Knott G.J., Smock D.C.J., Desmarais J.J., Son S., Bhuiya A., Jakhanwal S., et al. Accelerated RNA detection using tandem CRISPR nucleases. Nat. Chem. Biol. 2021;17:982–988. doi: 10.1038/s41589-021-00842-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Georgakilas V., Tiwari J.N., Kemp K.C., Perman J.A., Bourlinos A.B., Kim K.S., Zboril R. Noncovalent functionalization of graphene and graphene oxide for energy materials, biosensing, catalytic, and biomedical applications. Chem. Rev. 2016;116:5464–5519. doi: 10.1021/acs.chemrev.5b00620. [DOI] [PubMed] [Google Scholar]
- 26.Yang B., Fang X., Kong J. Engineered microneedles for interstitial fluid cell‐free DNA capture and sensing using iontophoretic dual‐extraction wearable patch. Adv. Funct. Mater. 2020;30 [Google Scholar]
- 27.Seo G., Lee G., Kim M.J., Baek S., Choi M., Ku K.B., et al. Rapid detection of COVID-19 causative virus (SARS-CoV-2) in human nasopharyngeal swab specimens using field-effect transistor-based biosensor. ACS Nano. 2020;14:5135–5142. doi: 10.1021/acsnano.0c02823. [DOI] [PubMed] [Google Scholar]
- 28.Torrente-Rodríguez R.M., Lukas H., Tu J., Min J., Yang Y., Xu C., Rossiter H.B., Gao W. SARS-CoV-2 RapidPlex: a graphene-based multiplexed telemedicine platform for rapid and low-cost COVID-19 diagnosis and monitoring. Matter. 2020;3:1981–1998. doi: 10.1016/j.matt.2020.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.