Abstract
The impact of the ravages of COVID-19 on people's lives is obvious, and the development of novel potential inhibitors against SARS-CoV-2 main protease (Mpro), which has been validated as a potential target for drug design, is urgently needed. This study developed a model named MproI-GEN, which can be used for the de novo design of potential Mpro inhibitors (MproIs) based on deep learning. The model was mainly composed of long-short term memory modules, and the last layer was re-trained with transfer learning. The validity (0.9248), novelty (0.9668), and uniqueness (0.0652) of the designed potential MproI library (PMproIL) were evaluated, and the results showed that MproI-GEN could be used to design structurally novel and reasonable molecules. Additionally, PMproIL was filtered based on machine learning models and molecular docking. After filtering, the potential MproIs were verified with molecular dynamics simulations to evaluate the binding stability levels of these MproIs and SARS-CoV-2 Mpro, thereby illustrating the inhibitory effects of the potential MproIs against Mpro. Two potential MproIs were proposed in this study. This study provides not only new possibilities for the development of COVID-19 drugs but also a complete pipeline for the discovery of novel lead compounds.
Keywords: de novo drug design, Transfer learning, Virtual screening, Molecular dynamics simulation, Deep learning
List of abbreviations
- Mpro
main protease
- MproIs
Mpro inhibitors
- DL:
deep learning
- PMproIL:
potential MproI library
- ML:
machine learning
- MD
molecular dynamics
- NSPs
non-structural proteins
- AI
artificial intelligence
- LSTM
long short-term memory
- MW
molecular weight
- IC50
half-maximal inhibitory concentration
- CharRNN
char-level recurrent neural network
- SVM
support vector machine
- RF
random forest
- k-NN
k-nearest neighbor
- XGBoost
extreme gradient boosting
- ECFP
extended connectivity fingerprints
- CV
cross-validation
- RMSD
root mean square deviation
- MM/GBSA
molecular mechanics-generalized Born surface area
- ROC:
receiver operating characteristic
- AUC
the area under the ROC curve
- ACC
accuracy
- SEN
sensitivity
- SPC
specificity
- HBAs
number of H-bond acceptors
- HBDs
number of H-bond donors
- QED
quantitative estimate of drug likeness
- TPSA
topological polar surface area
1. Introduction
COVID-19, caused by SARS-CoV-2, has been a pandemic worldwide since 2019, imposing a strong shock on economic and social stability worldwide [1]. According to the ninth version of the living guidelines published by the WHO on January 14, 2022 [2], some drugs, such as Janus kinase inhibitors [3], molnupiravir [4], and sotrovimab [5], are recommended for COVID-19. Pfizer's Paxlovid also received the emergency use authorization in December 2021 and is available in multiple countries [6]. Unfortunately, the existing recommended drugs still exhibit some limitations: (1) considering the urgent need for effective drugs, the development time required for existing drugs is insufficient, so the safety of these drugs has yet to be validated; (2) uncertainty remains regarding the therapeutic effects of existing drugs on patients with different symptoms. Therefore, the development of more potential inhibitors against SARS-COV-2 is urgently needed to provide potential dugs for COVID-19. Empirical trials involving trial-and-error are often costly, which is the major reason why drug development is time- and money-consuming. To meet the urgent need for the development of drugs to defend against COVID-19, it is necessary to rapidly discover potential lead compounds with computational methods.
Two overlapping polyproteins in SARS-CoV-2, pp1a and pp1b, are used to encode the replicates that are essential for viral replication and transcription. The main protease (Mpro) of SARS-CoV-2 operates at pp1a and pp1b for intramolecular cleavage, resulting in several non-structural proteins (NSPs). These NSPs are involved in the synthesis of viral subgene RNA and four structural proteins (the envelope protein, membrane protein, spike protein, and nucleocapsid protein), thereby completing the reproduction and release of progeny viruses [[7], [8], [9]]. Considering that Mpro plays a crucial role in the viral life cycle and that no homologous protein is possessed by humans, Mpro is an ideal target for antiviral drug development. In this study, Mpro was selected as the drug target for the design of an anti-SARS-CoV-2 drug.
Research related to drug design and screening has been conducted around the clock [10,11]. Jin et al. designed a Michael acceptor inhibitor (N3) for SARS-CoV-2 Mpro, which was used as the positive control in this study [7]. Additionally, Ma et al. screened the Selleckchem bioactive compound library with a FRET-based enzymatic assay and identified several potential inhibitors, including boceprevir, GC-376, and calpain inhibitors II and XII [12,13].
Most drug discovery efforts concerning SARS-CoV-2 have focused on repurposing existing drugs [[14], [15], [16]]. For example, corticosteroids, IL-6 receptor blockers (tocilizumab and sarilumab), and Janus kinase inhibitors (baricitinib, ruxolitinib, and tofacitinib) are recommended as drugs for COVID-19. However, uncertainty remains regarding these drugs. For example, the safety of the drugs in different patients, such as children, pregnant individuals, and immunocompromised people, cannot be determined, and it is impossible to determine whether these drugs are suitable for patients with different disease severity levels [2]. These results suggest the need to design better and more potent Mpro inhibitors (MproIs) that specifically target SARS-CoV-2. The de novo design of compounds aims to automatically design compounds with structural diversity, synthetic accessibility, and specific biological activities. In recent years, with developments in the field of artificial intelligence (AI), it has become possible to mine knowledge from unlimited chemical spaces and use this information to develop novel small molecules with the desired biological and physicochemical properties [[17], [18], [19], [20], [21], [22]]. Zhavoronkov et al. developed a deep generative model for the de novo design of small molecules and used this model to discover potent inhibitors of discoidin domain receptor 1 in 21 days [18]. Godinez et al. developed a generative model named JARGER based on the junction tree variational autoencoder for discovering novel molecules with desired bioactivity properties and designed novel small antimalarial molecules. They selected, synthesized, and experimentally validated the inhibitory activities of molecules designed by JARGER against malaria, and the results demonstrated the validity of the developed model [23]. These works have widely applied deep learning (DL) and achieved success [24,25]. Additionally, to solve the problem regarding a lack of inhibitor data on SARS-CoV-2 Mpro, we also used transfer learning. Transfer learning was proposed to solve the problem of data scarcity by exploiting the knowledge contained in related datasets, and it has been widely used to address tasks with low data volumes in many fields, such as computer vision [26], natural language processing [27,28], and drug discovery [29].
In this study, we used DL and transfer learning methods to achieve the de novo design of SARS-CoV-2 Mpro inhibitors. The molecules contained in the ZINC database were used to train the small-molecule de novo design model named General-GEN, which was composed of long short-term memory (LSTM) to generate novel and valid molecules. Then, General-GEN was fine-tuned with SARS-CoV and SARS-CoV-2 MproIs to derive a target-specific generation model named MproI-GEN, which was used to design specific molecules for targeting Mpro. Finally, the molecules designed by MproI-GEN were filtered with machine learning (ML) models and molecular docking to obtain potential SARS-CoV-2 MproIs.
2. Methods and materials
2.1. Data sources and usage
Four datasets were used in this paper: one for training General-GEN, one for fine-tuning General-GEN to obtain MproI-GEN, one for training ML models that could classify Mpro inhibitors and non-inhibitors, and one for evaluating the performance of molecular docking. An overview of these four datasets is shown in Table 1 .
Table 1.
Overview of the datasets used in this study.
| Name | Size | Note | Source |
|---|---|---|---|
| DATASET-1 | 575, 815 | Train the General-GEN | [30] |
| DATASET-2 | 249 | Fine-tune the General-GEN to get the MproI-GEN | [12,[31], [32], [33], [34], [35]] |
| DATASET-3 | 494 | Train the ML models | [12,[31], [32], [33], [34], [35]] |
| DATASET-4 | 38 | Evaluate the performances of different scoring functions in molecular docking | [12,[31], [32], [33], [34], [35]] |
These four datasets were collected from the ZINC database [30], BindingDB database [31], PubChem database [32], and some papers [12,[33], [34], [35]] that were recently published. DATASET-1 consisted of about 5 million compounds randomly sampled from the ZINC database. Considering the distribution of the inhibitors to be designed should be similar to the existing inhibitors in chemical space, the molecular properties of the existing inhibitors were calculated, and the filtering criteria were formulated accordingly. Then, the molecules were filtered according to the following criteria: (1) the molecular weight (MW) need to be in the range from 200 to 800; (2) the molecular Log P needed to not be greater than 6.5; (3) the number of rotatable bonds could not be greater than 8; (4) the molecules whose charges needed to be neutralized were exploded; (5) only molecules without atoms other than C, N, O, S, F, Cl, Br, and H were included; and (6) the first dataset was filtered via medicinal chemistry filters and PAINS filters [36]. The final DATASET-1 contained 575,815 molecules, which were used to train General-GEN. Considering that the sequence identity of Mpro in SARS-CoV and SARS-CoV-2 is as high as 96.1%, the inhibitors of these two enzymes might be similar. The inhibitors and non-inhibitors against SARS-CoV and SARS-CoV-2 were collected from the BindingDB database [31], PubChem AID1890 assay [32], and some papers [12,[33], [34], [35]]. The molecules whose half-maximal inhibitory concentration (IC50) values were less than 10 μM were treated as positive data (with inhibition abilities), and the molecules whose IC50 values were greater than 50 μM were treated as negative data (without inhibition abilities). Under these settings, a total of 645 molecules were obtained. Then, the above filter criteria were applied to filter these molecules. After filtering, DATASET-3 was obtained, which was used to develop the ML models. DATASET-3 contained 495 molecules, among which 253 are positive, and DATASET-2 (used to fine-tune General-GEN) was composed of these 253 molecules. DATASET-4 was composed of 38 experimentally validated inhibitors and non-inhibitors against SARS-CoV-2 Mpro to evaluate the performance of molecular docking with different scoring functions.
2.2. Model training and fine-tuning
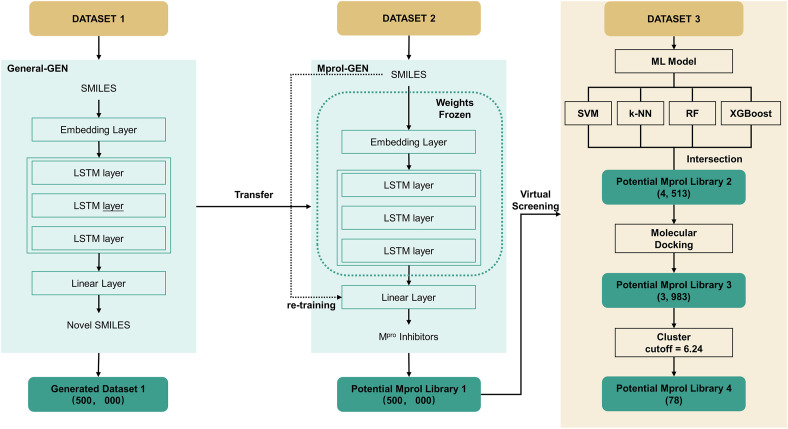
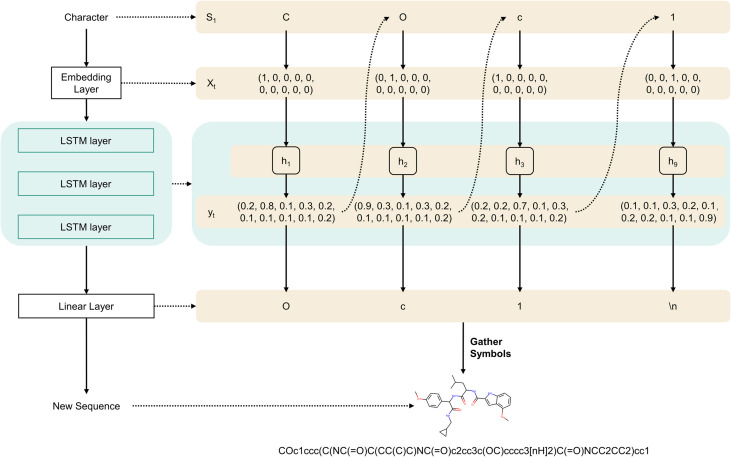
This study developed a generative model specifically for the de novo design of SARS-CoV-2 MproIs. Four steps were involved in this work (Fig. 1 ): (1) General-GEN, which could generate novel and valid compounds, was developed based on a char-level recurrent neural network (CharRNN) [37]; (2) General-GEN was fine-tuned with DATASET-2 to obtain MproI-GEN, which could design novel MproIs for SARS-CoV-2; (3) 500,000 potential MproIs were designed by MproI-GEN to form potential MproI library (PMproIL) 1 and then filtered by ML models and molecular docking to obtain PMproIL 2 and PMproIL 3; (4) PMproIL 3 was clustered to obtain PMproIL 4, and the potential MproIs were validated with molecular dynamics (MD) simulations.
Fig. 1.
The workflow of this study. First, DATASET-1 was used to develop General-GEN to generate novel and reasonable molecules. Afterward, DATASET-2 was used to fine-tune General-GEN to generate potential MproIs. Finally, the generated MproIs (PMproIL 1) needed to be filtered and validated.
General-GEN was implemented with the framework of a CharRNN, which could model the distribution of the next character based on the given character, and the model was used to generate novel compounds [37]. Based on the CharRNN, General-GEN consisted of an embedding layer, a linear layer, and four LSTM layers. General-GEN took molecular SMILES as inputs, and the embedding layers encoded these inputs as vectors. Later, the LSTM layers modeled the distribution of these strings, which enabled the model to predict the next character based on the given character (Fig. S1). General-GEN was trained on DATASET-1 and was implemented with PyTorch [38].
General-GEN, trained on DATASET-1, generated only reasonable molecules with structurally unknown activities against SARS-CoV-2 Mpro. Therefore, General-GEN needed to be transferred to generate active molecules against SARS-CoV-2 Mpro, forming MproI-GEN. In this part, we used a fine-tuning technique to achieve our goal. After obtaining General-GEN, the parameters of all layers except the last linear layer were frozen, and the linear layer was retrained on DATASET-2. This step was also implemented with PyTorch.
2.3. Evaluation metrics for the generated molecules
In this study, the performance of General-GEN and MproI-GEN were evaluated from two perspectives: (1) their generation performance and (2) their active molecule design performance. The evaluation metrics of generation performance mainly refer to the MOSES [39].
To evaluate the generation performance of the generated molecules, the DATASET-1 was split into three non-intersecting parts: train set (477,297 molecules), test set (53,034 molecules), and scaffold test set (45,484 molecules). The molecules in the scaffold test set all have Bemis-Murcko scaffolds [40] mainly containing the ring structures in molecules and the linker fragments connecting with the ring. The scaffold test set was used to assess whether the model could produce novel scaffolds that were not present in the training set. Among them, the test set and the scaffold test set would be used as reference sets.
The validity was defined as the proportion of valid molecules among all generated molecules, and the valid molecules were defined as those for which the valences of the atoms and bonds in their rings were consistent. The atom valency and the consistency of the bonds in the rings of the generated molecules were checked with RDKit 2019.03.2 [41].
The novelty measure was defined as the proportion of the generated molecules that did not appear in DATASET-1 and DATASET-2; this metric was used to evaluate the originality of the generated molecules.
The uniqueness was defined as the proportion of unique compounds among the first 10,000 valid compounds in the generated set; this metric was used to measure the diversity of the generated molecules to ensure that multiple patterns of generated molecules were formed.
BRICS similarity was designed to compare the distributions of BRICS fragments [42] in generated sets and reference sets, which was denoted as SIM (frag). SIM (frag) will be large if there are similar BRICS fragments in both sets, and the limits of this metric are [0, 1].
Bemis-Murcko scaffold similarity, denoted as SIM (scaffold), is similar to BRICS similarity, except that the SIM (scaffold) evaluates the Bemis-Murcko scaffold distributions in generated sets and reference sets.
It is worth noting that both SIM(frag) and SIM(Scaffold) are compared from the substructure of the molecules, so it is possible to have high similarity even if the compared molecular structures are different.
In this study, the activity of the molecules generated by General-GEN and MproI-GEN were evaluated by ML models, and activity was defined as the proportion of the active molecules among all valid generated molecules.
2.4. Screening with ML and molecular docking
In this study, MproI classification models based on ML and molecular docking were used to further screen the PMproIL 1 set generated by MproI-GEN.
2.4.1. MproI classifiers based on ML
Four different ML models were trained on DATASET-3 for MproI prediction: a support vector machine (SVM), a random forest (RF), a k-nearest neighbor (k-NN) classifier, and an extreme gradient boosting (XGBoost) model. These four models were trained with the 2,048 bits extended connectivity fingerprints whose radius was equal to 4 (ECFP4) [43], and the calculation of the ECFP4 was implemented with RDKit [41]. DATASET-3 was split into 80% and 20% subsets. Eighty percent of DATASET-3 was used to train these four ML models, and 20% of DATASET-3 was used as the external validation dataset to validate the predictive abilities of these ML models on new data. The parameters were searched through a grid search during the training process (Table S1), and the search process was evaluated by 5-fold cross-validation (CV), which was implemented with scikit-learn 0.24.2 [44].
2.4.2. Filtering with molecular docking
Current molecular docking-based screening methods typically use the scores given by a scoring function integrated into the docking software to rank the obtained compounds. Therefore, these scores were used as the main basis for the selection of potential inhibitors. In recent years, new scoring functions based on ML methods have been introduced and have been shown to outperform a wide range of classic scoring functions [45]. RF-Score is a scoring function built with the RF algorithm that has outperformed 22 state-of-the-art scoring functions on the PDBbind benchmark [46].
For molecular docking, the 3D structures of the ligands were generated with Open Babel 3.1.0 [47]. The crystal structure of the receptor (SARS-CoV-2 Mpro) was downloaded from the Protein Data Bank (PDB ID: 7BQY). The water molecules were removed with PyMol (version 2.6), and the hydrogen atoms and Gasteiger charges were added with MGLTools (version 1.5.6). Then, the prepared structure was converted into PDBQT format for subsequent study. The binding conformations of the receptor and ligands were predicted with the molecular docking software AutoDock Vina 1.1.2. The centre and the size of the grid box were determined based on the position of the complex crystal structure of Mpro-N3 (PDB ID: 7BQY). The size of the grid box was set to with a spacing of 1.000 Å, and its centre was located at (X = 5.914, Y = 0.576, and Z = 22.883) to cover all the main key amino acids that were combined with drugs (Fig. S2). The options were set to energy_range = 3, exhaustiveness = 8, and num_modes = 9. In this study, the performances of the RF-Score and AutoDock Vina's default scoring function (Vina-Score) were evaluated on DATASET-3. After the comparison, the RF-Score function was used to measure the binding affinities of the protein-ligand complexes and screen PMproIL 2 based on the yielded scores.
2.5. MD simulations combined with Gibbs free energy calculation
To pick the molecules which could stabilize binding within the pocket, the MD simulation analysis was performed. In this study, an 2 ns MD simulation was performed for the complex structure of the receptor with the molecules in PMproIL 4, and the molecules with binding free energies below −40 kcal/mol were picked for a further 100 ns MD simulation, which was implemented with AMBER 20 [29]. The protein-ligands systems were solvated in a TIP3P water model within an orthorhombic box with buffer dimensions of , and the systems were neutralized by adding Na+ or Cl−. The steepest descent method with 500 steps and the conjugate gradient method with 500 steps were selected during the energy minimization processes. After conducting energy minimization, these systems were heated from 0 K to 300 K over 30,000 steps with a 2 fs step. The solvated complex was balanced by a density equilibrium of 50 ps, the complex was weakly constrained, and then a constant pressure equilibrium of 10 ns was achieved at 300 K [48]. The above steps were performed by the pmemd.cuda module.
The stability of the protease-inhibitor complexes during the MD simulations was evaluated by calculating the ligand-receptor root means square deviation (RMSD), the number of H-bonds, and the molecular mechanics-generalized Born surface area (MM/GBSA) binding free energies. The RMSD and H-bond lifetime analyses were performed with the cpptraj module, and the binding free energies and contribution energies of individual residues were decomposed by the MMPBSA.py script [49]. For generalized Born calculations, igb2 was employed, and MM/GBSA was calculated from the first frame to the 1000th frame with a one-frame step. To calculate the binding free energy, the MM/GBSA method was applied. According to MM/GBSA theory, the binding free energy required between a ligand and a receptor to form a complex is calculated as:
where is the sum of van der Waals and electrostatic interactions and and are the polar and nonpolar solvation energies, respectively [50].
3. Results and discussion
3.1. Generation performance of General-GEN and MproI-GEN
The DATASET-1 collected from ZINC was used to train a general generation model named General-GEN, which could effectively generate compounds. The process of training General-GEN started with pretraining. After pretraining, General-GEN was fine-tuned to obtain MproI-GEN. The fine-tuning process was achieved by freezing the previous weights and retraining the weights of the last layer (linear layer) on DATASET-2. The outputs included 500,000 molecules designed by General-GEN and MproI-GEN, and these molecules were evaluated (Table 2 ).
Table 2.
The generation performance of General-GEN and MproI-GEN.
| General-GEN | MproI-GEN | ||
|---|---|---|---|
| Validity | 0.9973 | 0.9248 | |
| Novelty | 0.9525 | 0.9668 | |
| Uniqueness | 0.9996 | 0.0652 | |
| SIM(Frag) | SIM(Frag)/test | 0.9998 | 0.8584 |
| SIM(Frag)/scaffold | 0.9927 | 0.8310 | |
| SIM(Scaf) | SIM(Scaf)/test | 0.7949 | 0.0000 |
| SIM(Scaf)/scaffold | 0.1234 | 0.0004 | |
For General-GEN, the validity, novelty, and uniqueness of the generated molecular set were 0.9973, 0.9525, and 0.9996, respectively. That is, 95% of the generated molecules were not duplicates of those in DATASET-1. Among the first 10,000 valid generated molecules, 99.96% of the molecules did not repeat each other. Therefore, General-GEN could generate reasonable and novel molecules. Additionally, we also evaluated the generation performance of MproI-GEN. For MproI-GEN, the validity, novelty, and uniqueness were 0.9248, 0.9668, and 0.0652, respectively. In other words, 92.48% of the molecules were valid among the molecules generated by MproI-GEN, and 96.68% of the generated molecules were not duplicates of those in DATASET-2, which meant that MproI-GEN could be used to design structurally valid and novel potential MproIs. The uniqueness ratio was very low, which was to be expected. The fine-tuning technique was performed to make the model generate specific kinds of molecules, so it was reasonable that the uniqueness decreased after the fine-tuning operation; this problem could be eliminated by designing as many molecules as possible. The SIM(Frag) of generated molecules with General-GEN (0.9998 and 0.9927) are higher than generated molecules with MproI-GEN (0.8584 and 0.8310), implying fewer BRICS fragments in the molecules generated with MproI-GEN. BRICS fragments are related to drug-like properties of molecules, and the result indicates that the molecular drug-like properties of the MproI-GEN are lower than those of the General-GEN. Considering the relatively drug-likeness of the molecules in the DATASET-2, it is reasonable for the model to produce such changes after fine-tuning. The SIM(Scaf) could illustrate how similar the scaffolds are in the generated set and the reference set. In the molecular set generated with General-GEN, the SIM(Scaf) is 0.7949 with the test set and 0.1234 with the scaffold test set. In the molecular set generated with MproI-GEN, the SIM(Scaf) is approximately zero in both the test set (0.0000) and the scaffold test set (0.0004). This means that the chemotypes of molecules generated by MproI-Gen are very different from those in the test set and scaffold test set.
3.2. The ability of MproI-GEN to generate PMproILs
As described in section 3.1, both General-GEN and MproI-GEN could generate novel and valid molecules. The difference between the molecules generated by these two models was that the molecules generated by MproI-GEN needed to be active molecules against SARS-CoV-2 Mpro.
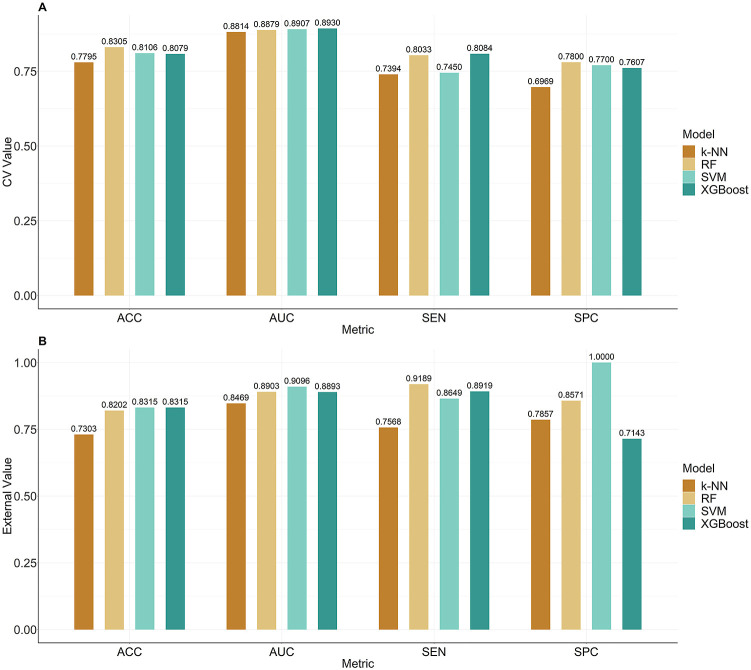
To evaluate whether MproI-GEN could generate active molecules, we trained four different ML models as target prediction models: an RF model, an SVM model, an XGBoost model, and a k-NN model. The ranges of the areas under the ROC curve (AUC), accuracy (ACCs), sensitivity (SEN) and specificity (SPCs) of these four ML models in the CV ranged from 0.8814 (k-NN) to 0.8930 (XGBoost), 0.7795 (k-NN) to 0.8305 (RF), 0.7394 (k-NN) to 0.8084 (XGBoost), and 0.6969 (k-NN) to 0.7800 (RF), respectively. The ranges of the AUC, ACC, SEN and SPC values of these four ML models in the external validation were from 0.8469 (k-NN) to 0.9096 (SVM), 0.7303 (k-NN) to 0.8315 (RF and XGBoost), 0.7568 (k-NN) to 0.9189 (RF), and 0.7143 (XGBoost) to 1.0000 (SVM), respectively (Fig. 2 ). These statistics show that these four ML models could predict whether the molecules had inhibitory activity against Mpro with high accuracy.
Fig. 2.
The prediction performances of the ML models. A. The prediction performance of these four models in the CV. B. The prediction performance of these four models in the external validation.
The molecules generated by General-GEN formed the General Molecules Library after the removal of invalid and repeating molecules. The molecules generated by MproI-GEN formed PMproIL 1 after the removal of invalid and repeated molecules. We used these ML models to predict the General Molecules Library and PMproIL 1 (Table 3 ). A small percentage of the molecules were predicted to be active in the General Molecules Library (from 0% to 2.85%), and a larger percentage of the active molecules were predicted to be active in PMproIL 1 (from 73.12% to 83.08%). The increase in the percentage of active molecules meant that MproI-GEN could be used to design potential inhibitors against SARS-CoV-2 Mpro. The intersection among the active molecules predicted by these four ML models was taken as PMproIL 2, which included 4,531 potential MproIs.
Table 3.
The validity and the activity of the ML models were evaluated based on the molecules generated by the General-GEN and MproI-GEN, respectively.
| General-GEN Generation |
MproI-GEN Generation |
|||||
|---|---|---|---|---|---|---|
| Valid | Active | Active Ratio (%) |
Valid | Active | Active Ratio (%) |
|
| SVM | 491, 594 | 3 | 0 | 6, 963 | 5, 091 | 73.12 |
| RF | 491, 594 | 6, 597 | 1.34 | 6, 963 | 5, 785 | 83.08 |
| k-NN | 491, 594 | 13, 990 | 2.85 | 6, 963 | 5, 465 | 78.49 |
| XGB | 491, 594 | 12, 321 | 2.51 | 6, 963 | 5, 762 | 82.75 |
| Inter-section | 491, 594 | 1 | 0 | 6, 963 | 4, 531 | 65.07 |
3.3. Screening based on molecular docking
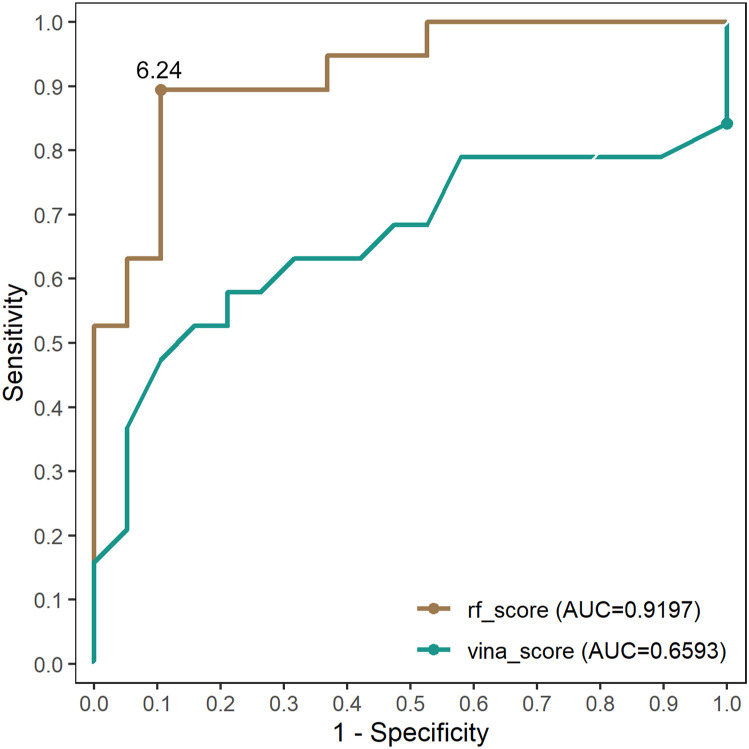
The MproIs in PMproIL were further screened by molecular docking. Before the filtering, the scoring ability of RF-Score and Vina-Score was evaluated, specifically. The data included in DATASET-3 contained experimentally validated inhibitors and non-inhibitors of SARS-CoV-2 Mpro, and the data sources are described in section 2.1. With this dataset, the RF-Score and Vina-Score functions were evaluated regarding their effectiveness in virtual screening for the MproIs of SARS-CoV-2. Briefly, these ligands in DATASET-3 were docked to the crystal of SARS-CoV-2 Mpro using AutoDock Vina. The virtual screening performances achieved by using AutoDock Vina with the two different scoring functions are presented based on their receiver operating characteristic (ROC) curves (Fig. 3 ).
Fig. 3.
The ROC curves of RF-Score (yellow curve) and Vina-Score (green curve).
The AUC value of the RF-Score was 0.920, and that of the Vina-Score was 0.659, which illustrated that the RF-Score could predict the docking of ligands and SARS-CoV-2 Mpro more accurately. In the classification problems, the cut-off values had a direct impact on the confusion matrix, so it is important to determine an appropriate cut-off value. In this study, the best cut-value was determined based on the F1 score. The cut-off of 6.24 was selected at the highest F1 score of 0.8947 (Table S2), which meant that the compounds whose RF-Score values were less than 6.24 were non-inhibitors, and those with values greater than 6.24 were potential MproIs. Under this threshold, the ACC of the RF-Score function was 89.5%, the SEN was 89.5%, and the SPC was 89.5% (Table S3). Considering the effectiveness of the molecular docking approach based on RF-Score for the recognition of MproI, PMproIL 2 was filtered with this method.
The molecules in PMproIL 2 were docked with the crystal structure of SARS-CoV-2 Mpro, and the binding affinity values of these conformations were calculated with the RF-Score functions. The molecules with scores lower than 6.24 were considered non-inhibitors, and the molecules with scores higher than 6.24 were considered potential MproIs against SARS-CoV-2 Mpro. At this point, 3,938 compounds remained in PMproIL 3. After this, the molecules in PMproIL 3 were clustered by their structural similarities via the binning clustering algorithm in ChemMine tools [51], with similarity cut-offs of 0.7, 0.8, and 0.9 (Table S4). When the cut-off value equaled 0.7, the structural similarity differences among the molecules between clusters were the largest. To provide structurally diverse potential MproIs, the clustering results obtained with a cut-off value of 0.7 were selected. After clustering, the molecules with the highest docking scores in each cluster constituted PMproIL 4 (Table S5, Fig. S3), which contained 78 potential MproIs.
3.4. Structural diversity and property analysis of potential inhibitors
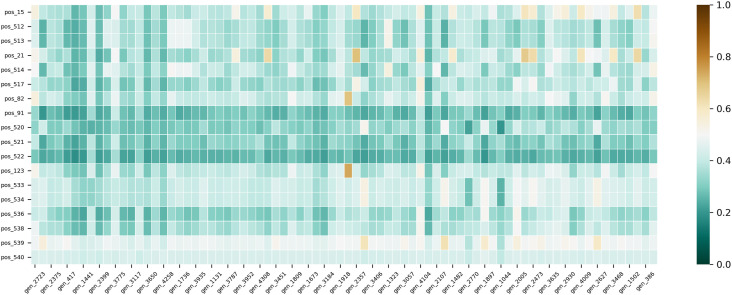
The structural similarity between the inhibitors in DATASET-4 and the molecules in PMproIL 4 (Fig. 4 , Table S6) was determined with RDKit [41] and Seaborn [52]. First, use RDKit to calculate the ECFP of each molecule, and then calculate the Tanimoto similarity scores between ECFPs of different molecules. There are 1,404 () similarity scores in the similarity matrix, of which 1,323 similarity scores were lower than 0.5, and 301 similarity scores were lower than 0.3. The similarity scores between pos_522-gen_2724 and pos_520-gen_1044 were lower than 0.2; these were the lowest similarity scores in the similarity matrix. These results show that most of the molecules in PMproIL 4 were different from the inhibitors in DATASET-4, indicating that the molecules designed with MproI-GEN were structurally novel.
Fig. 4.
A heatmap was used to evaluate the structural similarity among the molecules in PMproIL 4 and the inhibitors in DATASET-4.
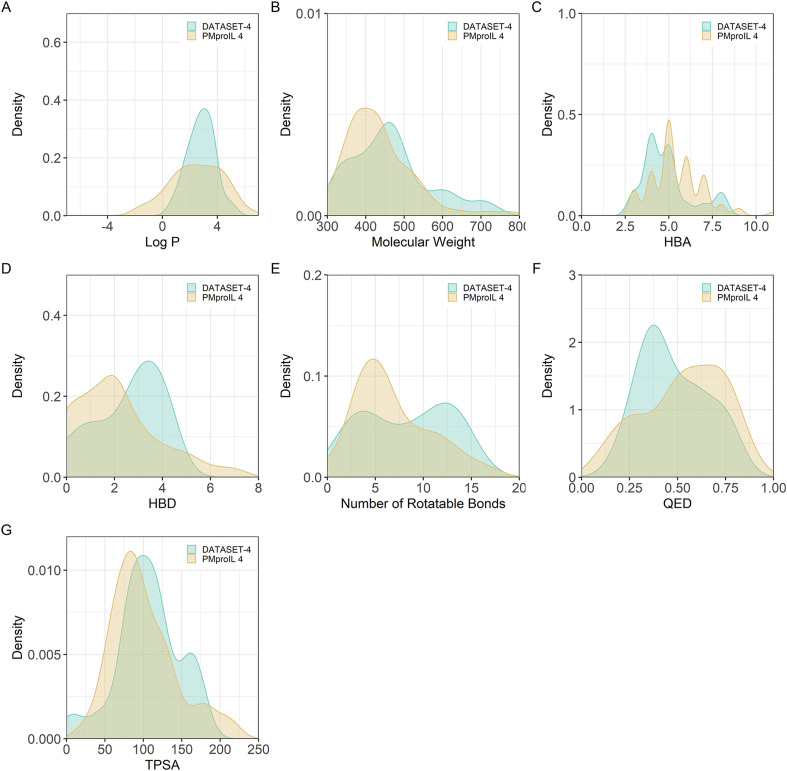
Additionally, we computed the molecular properties of the screened molecules and DATASET-4, such as the Log P, MW, number of H-bond acceptors (HBAs), number of H-bond donors (HBDs), number of rotatable bonds, quantitative estimate of drug-likeness (QED), and topological polar surface area (TPSA) of these potent inhibitors (Table S7 and Table S8). The molecular property distributions of the molecules in PMproIL 4 were consistent with those of the inhibitors in DATASET-4 (Fig. 5 ).
Fig. 5.
Distribution of the molecular properties of PMproIL 4 (yellow) and DATASET-4 (green). Distributions of the calculated molecular properties: A. Log P, B. molecular weight, C. number of HBAs, D. number of HBDs, E. number of rotatable bonds, F. QED, and G. TPSA.
3.5. MD simulation study of potential inhibitors
To obtain better approximations, 2-ns MD simulation snapshots were collected for the MM/GBSA calculation process, and 11 molecules with binding free energies below −40 kcal/mol were selected for a further 100-ns MD simulation (Table 4 , Fig. S4, and Table S9). The final estimated binding energy (ΔG Bind) is influenced by various types of nonbonded interactions, including electrostatic energy (ΔG Bind Coulomb), solvation energy (ΔG Bind Solv GB), and van der Waals contributions (ΔG Bind vdW).
Table 4.
Calculated energy components and MM/GBSA free energy (kcal/mol) values for the SARS-CoV-2 Mpro complexes with potent inhibitors against the N3 snapshots collected from the 100-ns MD simulation trajectories.
| No. | ID | ΔG Bind (kcal/mol) | ΔG Bind Coulomb (kcal/mol) | ΔG Bind Solv GB (kcal/mol) | ΔG Bind vdW (kcal/mol) |
|---|---|---|---|---|---|
| 1 | gen_3854 | −47.59 ± 11.60 | −37.70 ± 12.09 | 48.16 ± 9.96 | −58.05 ± 9.83 |
| 2 | gen_1502 | −46.60 ± 4.43 | −30.94 ± 4.25 | 39.44 ± 3.33 | −55.10 ± 4.44 |
| 3 | gen_2723 | −42.43 ± 5.26 | −28.62 ± 6.81 | 49.55 ± 5.66 | −63.35 ± 4.85 |
| 4 | gen_3946 | −39.89 ± 7.84 | −34.51 ± 7.20 | 38.06 ± 5.57 | −43.44 ± 7.57 |
| 5 | gen_1617 | −39.20 ± 4.51 | −17.26 ± 5.21 | 28.70 ± 4.51 | −50.64 ± 4.45 |
| 6 | gen_3052 | −37.99 ± 3.86 | −14.66 ± 5.51 | 27.57 ± 4.00 | −50.90 ± 3.87 |
| 7 | gen_4104 | −35.40 ± 4.40 | −28.24 ± 5.17 | 37.20 ± 4.52 | −44.37 ± 4.47 |
| 8 | gen_1369 | −34.35 ± 5.43 | −29.31 ± 13.17 | 40.69 ± 11.17 | −45.73 ± 4.58 |
| 9 | gen_976 | −33.73 ± 7.90 | −35.59 ± 14.73 | 47.26 ± 11.04 | −45.40 ± 5.44 |
| 10 | gen_2717 | −32.72 ± 3.88 | −31.88 ± 6.52 | 43.75 ± 5.89 | −44.59 ± 3.95 |
| 11 | gen_1482 | −29.32 ± 8.21 | −12.64 ± 8.30 | 23.23 ± 8.52 | −39.91 ± 8.68 |
| Control | N3 | −49.69 ± 6.66 | −35.75 ± 9.18 | 52.11 ± 8.83 | −66.05 ± 6.80 |
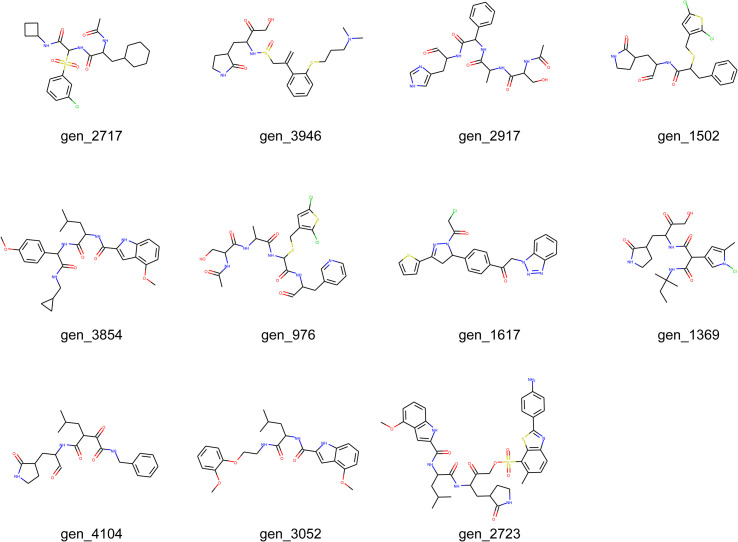
The complexes with binding free energies below −40 kcal/mol were gen_3854-Mpro, gen_1502-Mpro, and gen_2723-Mpro, where gen_3854-Mpro and gen_1502-Mpro had binding energies below −45 kcal/mol. In addition, the recorded binding affinity values for each complex were analyzed; among all the types of interactions, the ΔG Bind Coulomb and ΔG Bind vdW energies contributed most to achieving the average binding energy. Hence, these binding free energy values suggested the potential of gen_3854 and gen_1502 as SARS-CoV-2 Mpro inhibitors against SARS-CoV-2 infection.
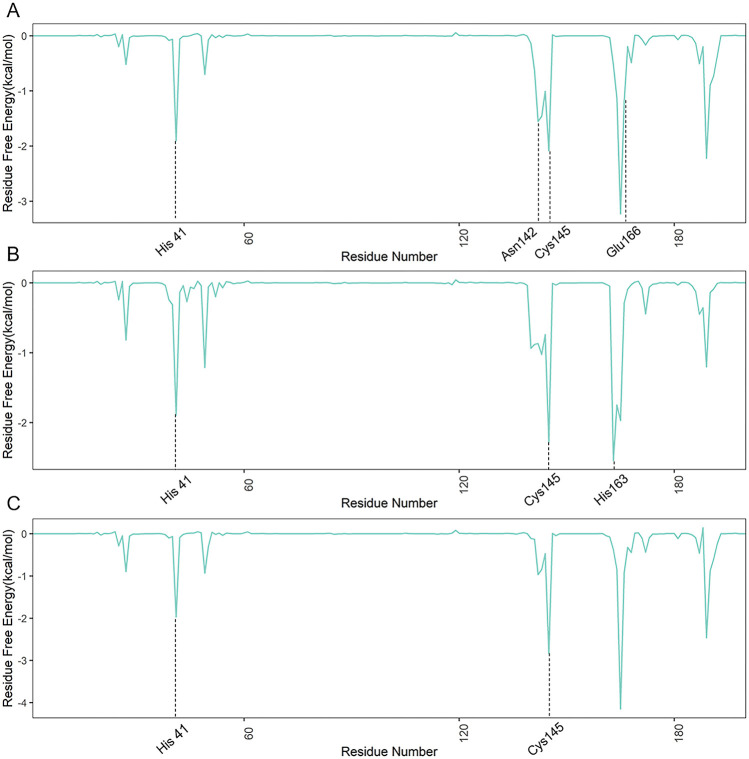
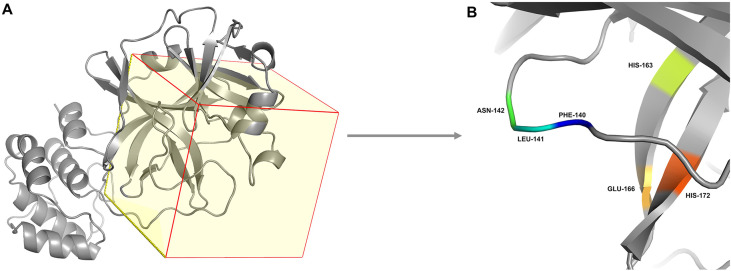
To further analyze the individual residue contribution energies of SARS-CoV-2 Mpro, energy decomposition was performed on the MD-simulated trajectories (Fig. 6 ). Compared with the affinity energy in N3-Mpro, gen_1502-Mpro and gen_3854-Mpro slightly increased the binding free energies on residues MET 49, ASN 142, GLY 143, SER 144, HIE 163, HID 164, and GLU 166. Among them, ASN 142, HIE 163, and GLU 166 were the key residues of SARS-CoV-2 Mpro, and the contribution of these residues was less than −1.0 kcal/mol, which meant that gen_1502 and gen_3854 could bind into the active pocket of SARS-CoV-2 Mpro. Therefore, these two molecules could be potential inhibitors.
Fig. 6.
Individual residue contributions to the binding energies for A. gen_3854-Mpro and B. gen_1502-Mpro. C. The active site and key residues with energy values less than −1.0 kcal/mol are marked.
A hydrogen bond analysis performed on the MD trajectories revealed that SARS-CoV-2 Mpro was responsible for the formation of hydrogen bonds with gen_1502, gen_3854, and N3. The hydrogen bonds with occupancy percentages exceeding 10% were analyzed (Table 5 ).
Table 5.
Hydrogen bond analysis results derived from the 100-ns MD trajectories of the studied systems.
| Complex | H-Bond Acceptor | H-Bond Donor | Percentage Occupancy (%) |
Average Distance |
Average Angle |
|---|---|---|---|---|---|
| gen_3854-Mpro | GLU_166@O | H⋯ N2@gen_3854 | 44.0 | 2.8326 | 156.9342 |
| gen_3854@O1 | H⋯N@GLU_166 | 40.0 | 2.8727 | 161.4542 | |
| gen_3854@O4 | H⋯N@GLY_143 | 28.4 | 2.8488 | 149.0044 | |
| gen_3854@O | HE2 … NE2@HIE_41 | 23.8 | 2.8665 | 152.9741 | |
| GLN_189@OE1 | H18⋯N1@gen_3854 | 14.8 | 2.9015 | 157.3969 | |
| gen_3854@O4 | H⋯N@CYS_145 | 10.5 | 2.9196 | 155.7685 | |
| gen_1502-Mpro | HID_164@O | H9⋯N@gen_1502 | 74.7 | 2.8397 | 158.6999 |
| gen_1502@O2 | HE2 … NE2@HIE_163 | 62.2 | 2.8214 | 151.6997 | |
| gen_1502@O1 | H⋯N@CYS_145 | 38.8 | 2.901 | 159.6612 | |
| PHE_140@O | H8⋯N1@gen_1502 | 15.2 | 2.8945 | 153.0401 | |
| N3-Mpro | N3@O4 | H⋯N@GLU_166 | 46.6 | 2.8790 | 163.6436 |
| N3@O | HE2 … NE2@HIE_41 | 25.8 | 2.8386 | 148.8377 | |
| GLN_189@O | H35⋯N3@N3 | 16.1 | 2.8858 | 157.9821 | |
| GLN_189@OE1 | H26⋯N2@N3 | 12.2 | 2.8865 | 156.3211 |
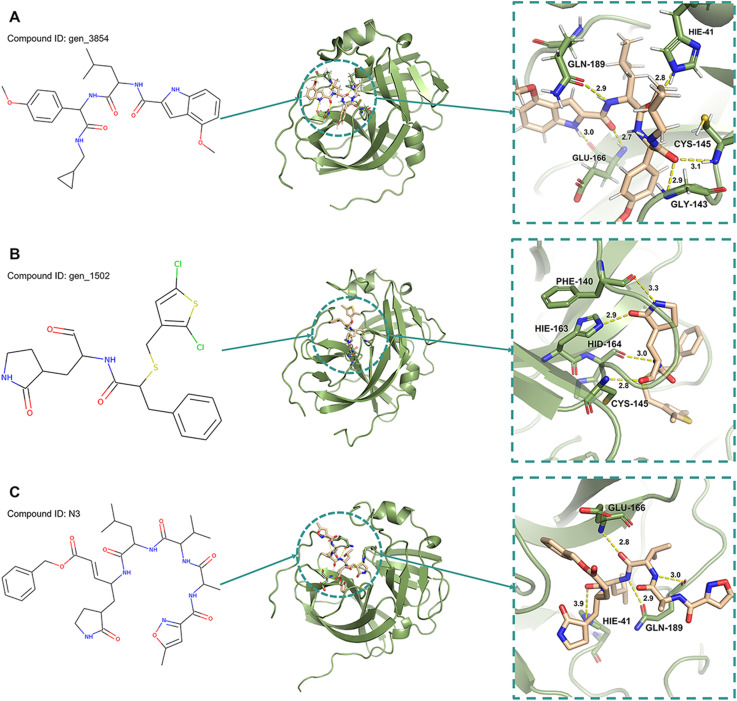
Table 5 shows that the gen_3854 and Mpro of the SARS-CoV-2 structure were stabilized by six hydrogen bonds at residues GLU166 (O–H⋯N, 2.83 Å; O1–H⋯N, 2.87 Å), GLY143 (O4–H⋯N, 2.85 Å), HIE41 (O-HE2 … NE2, 2.87 Å), GLN189 (OE1-H18 … N1, 2.90 Å) and CYS145 (O4–H⋯N, 2.92 Å). However, the N3 and Mpro of the SARS-CoV-2 structure were stabilized by four hydrogen bonds at residues GLU166 (O4–H⋯N, 2.88 Å), HIE41 (O-HE2 … NE2, 2.84 Å), and GLN189 (O–H35⋯N3, 2.89 Å; OE1-H26 … N2, 2.89 Å). The number of hydrogen bonds in gen_3854-Mpro was greater than that in N3-Mpro, which meant that gen_3854 could bind with Mpro effectively. The gen_1502 and Mpro structures were stabilized by four hydrogen bonds at residues HID164 (O–H9⋯N, 2.84 Å), HIE163 (O2-HE2 … NE2, 2.82 Å), CYS145 (O1–H⋯N, 2.90 Å) and PHE140 (O–H8⋯N1, 2.89 Å). The number of hydrogen bonds was the same as that in N3-Mpro, but the occupancy percentage of the hydrogen bonds in gen_1502-Mpro was highest (74.7%, 62.2%, 38.3%, and 15.2%). These results indicated that gen_3854 and gen_1502 could bind well with the Mpro of SARS-CoV-2 (Fig. 7 ). Therefore, it could be suggested that gen_3854 and gen_1502 had a good affinity with the major target (SARS-CoV-2 Mpro).
Fig. 7.
The structures with the lowest complex energies during the MD simulations. A Structure of gen_3854-Mpro at frame 381, B structure of gen_1502-Mpro at frame 847, and C structure of N3-Mpro at frame 371.
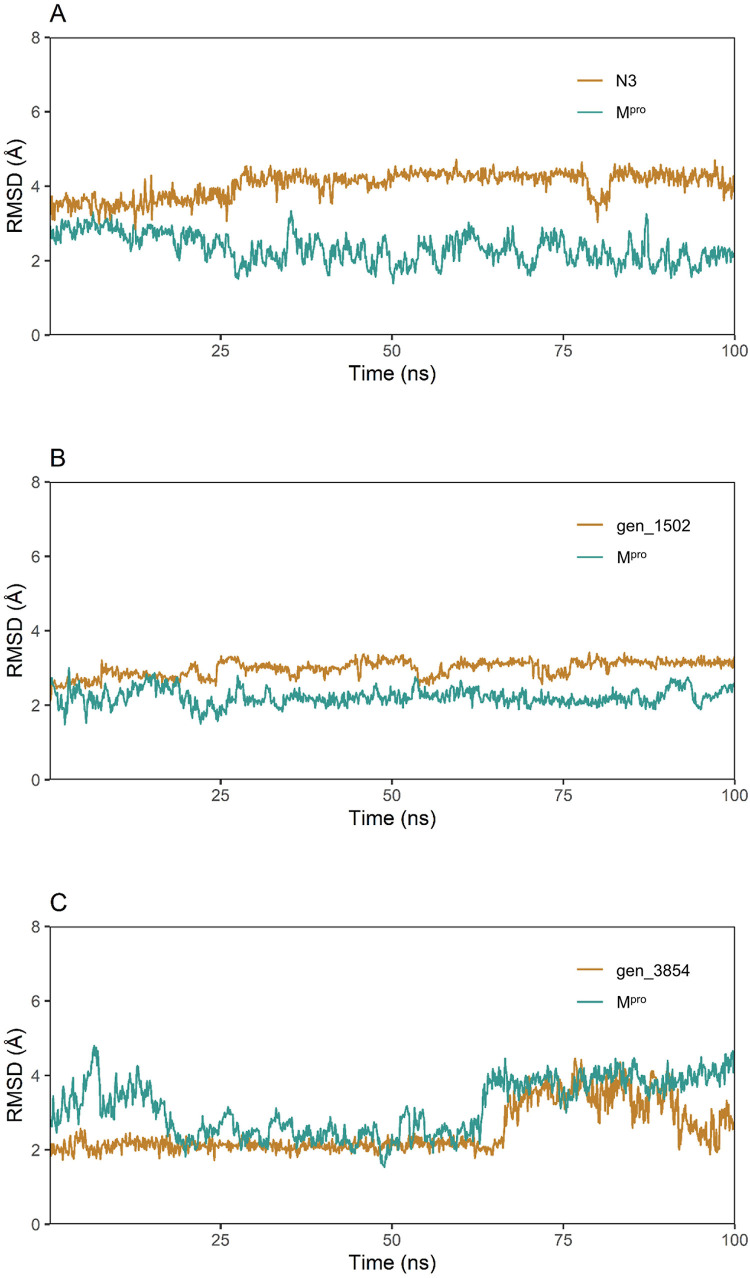
Furthermore, the RMSD was extracted from the MD trajectory for each complex (Fig. 8 ). The Cα atoms of SARS-CoV-2 Mpro produced a constant RMSD in the Mpro-gen_1502 complex, which meant that the viral protease conformation remained stable after the binding of gen_1502. However, the Mpro-gen_3854 complex exhibited a higher variation when the simulation lasted for 75 ns. Additionally, gen_1502 exhibited equilibrium throughout the 100 ns simulation when docked with Mpro, but N3 showed variations at 25 ns and 75 ns. Interestingly, the Mpro-gen_1502 complex did not yield RMSD descriptors above 4 Å, validating the rigid conformation of the drug complexes, and the Mpro-gen_3854 complex had a low RMSD trend between 20 and 60 ns (which thereafter increased slightly), but this complex did not over fluctuate, demonstrating a rigid conformation.
Fig. 8.
RMSD values were extracted for the alpha carbon atoms of SARS-CoV-2 Mpro (green curve) and the ligand compounds (yellow curve) from the docked complexes. A. gen_3854, B. gen_1502, and C. N3.
4. Conclusion
Despite the large-scale outbreak of SARS-CoV-2, no effective drug is available. This study used DL and transfer learning to develop a de novo drug design model that could design potential SARS-CoV-2 Mpro inhibitors. PMproIL 1 was filtered by the constructed ML models and the molecular docking method, and MD simulations were used to validate the potential inhibitors.
First, the ZINC dataset was used to train the General-GEN system consisting of an LSTM module for designing novel and valid small molecule compounds. Afterward, General-GEN was fine-tuned to obtain MproI-GEN, which could design specific molecules targeting Mpro. After PMproIL 1 was designed by MproI-GEN, it was filtered with the ML models and molecular docking. Finally, MD simulations were used to validate the effectiveness of the inhibitors.
In this study, PMproIL 1 was designed with MproI-GEN and consisted of 6,963 molecules. After that, four ML models (an RF, an SVM, a k-NN classifier, and an XGBoost model) were used to filter PMproIL 1 to obtain PMproIL 2, which contained 4,513 molecules that were active against both SARS-CoV-2 Mpro and SARS-CoV Mpro. Then, PMproIL 2 was filtered with molecular docking, which was implemented via AutoDock Vina. In the molecular docking process, we used the SARS-CoV-2 Mpro-N3 complex as the control. During molecular docking, SARS-CoV-2 Mpro was used as the receptor to screen out compounds with docking scores lower than the cut-off value (6.24). After performing filtering based on molecular docking, 3,938 molecules remained and made up PMproIL 3. PMproIL 3 was clustered according to structural similarity, and 78 clusters were obtained. The molecules with the highest docking scores were selected from each cluster for a 2-ns MD simulation and a binding free energy calculation. Twelve molecules had binding energies less than −40 kcal/mol, and these molecules were selected for a further 100-ns MD simulation. Among these 12 molecules, the binding free energies of gen_3854 and gen_1502 were less than −45 kcal/mol. Further individual residue contributions, hydrogen bonds, and RMSD analyses involving these two molecules revealed that gen_3854 and gen_1502 could bind to the active pocket of SARS-CoV-2 Mpro. Hence, gen_3854 and gen_1502 could be considered SARS-CoV-2 Mpro inhibitors for further evaluation.
However, this study still has some limitations. First, the molecular structures designed by the MproI-GEN are not very diverse. This is because the molecular structures used for fine-tuning in DATASET-2 are not very different. Second, the ML models used to filter potential MproIs were trained on the inhibitors data of SARS-CoV and SARS-CoV-2. This is because the number of existing SARS-CoV-2 MproIs is too small to train reliable predictive models. It is believed that with the increase of experimentally validated SARS-CoV-2 MproI, MproI-GEN could provide a more novel molecular scaffold for the development of drugs against SARS-CoV-2.
Data availability
The data used in this study and the source codes for General-GEN and MproI-GEN can be found at https://github.com/Shimeng-Li/MproI-GEN.
Declaration of competing interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgements
This study was supported by the National Natural Science Foundation of China (grant number No. 82003655); Liaoning Province Rejuvenating Talents Plan (grant number No. XLYC2002045); the Key R&D Program of Liaoning Province (grant number No. 2019JH2/10300041); Scientific Research Project from Department of Education of Liaoning Province (grant number No. LJKZ0088; No. LQN201906; LJKZ0280); Shenyang Science and Technology Talent Project (grant number No. RC210216).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.compbiomed.2022.105728.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
figs1.
figs2.
figs3.
figs4.
References
- 1.Hu B., Guo H., Zhou P., et al. Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 2021;19:141–154. doi: 10.1038/s41579-020-00459-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Organization W.H. World Health Organization; 2022. Therapeutics and COVID-19: Living Guideline, 14 January 2022. [PubMed] [Google Scholar]
- 3.Cheng L., Zhu Z., Wang C., et al. COVID-19 induces lower levels of IL-8, IL-10, and MCP-1 than other acute CRS-inducing diseases. Proc. Natl. Acad. Sci. USA. 2021;118(21) doi: 10.1073/pnas.2102960118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jayk Bernal A., Gomes da Silva M.M., Musungaie D.B., et al. Molnupiravir for oral treatment of Covid-19 in nonhospitalized patients. N. Engl. J. Med. 2022;386:509–520. doi: 10.1056/NEJMoa2116044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Self W.H., Sandkovsky U., Reilly C.S., et al. Efficacy and safety of two neutralising monoclonal antibody therapies, sotrovimab and BRII-196 plus BRII-198, for adults hospitalised with COVID-19 (TICO): a randomised controlled trial. Lancet Infect. Dis. 2021;22:622–635. doi: 10.1016/S1473-3099(21)00751-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Extance A. Covid-19: what is the evidence for the antiviral Paxlovid? Br. Med. J. 2022;377:o1037. doi: 10.1136/bmj.o1037. [DOI] [PubMed] [Google Scholar]
- 7.Jin Z., Du X., Xu Y., et al. Structure of M(pro) from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582:289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 8.Zhang S., Amahong K., Sun X., et al. The miRNA: a small but powerful RNA for COVID-19. Briefings Bioinf. 2021;22:1137–1149. doi: 10.1093/bib/bbab062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang S., Amahong K., Zhang C., et al. RNA–RNA interactions between SARS-CoV-2 and host benefit viral development and evolution during COVID-19 infection. Briefings Bioinf. 2022;23:bbab397. doi: 10.1093/bib/bbab397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qi C., Wang C., Zhao L., et al. SCovid: single-cell atlases for exposing molecular characteristics of COVID-19 across 10 human tissues. Nucleic Acids Res. 2022;50:D867–D874. doi: 10.1093/nar/gkab881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu Z., Zhang S., Wang P., et al. A comprehensive review of the analysis and integration of omics data for SARS-CoV-2 and COVID-19. Briefings Bioinf. 2022;23:bbab446. doi: 10.1093/bib/bbab446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ma C., Sacco M.D., Hurst B., et al. Boceprevir, GC-376, and calpain inhibitors II, XII inhibit SARS-CoV-2 viral replication by targeting the viral main protease. Cell Res. 2020;30:678–692. doi: 10.1038/s41422-020-0356-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xue W., Fu T., Deng S., et al. Molecular mechanism for the allosteric inhibition of the human serotonin transporter by antidepressant escitalopram. ACS Chem. Neurosci. 2022;13:340–351. doi: 10.1021/acschemneuro.1c00694. [DOI] [PubMed] [Google Scholar]
- 14.Lin S., Wang Y., Zhang L., et al. MDF-SA-DDI: predicting drug-drug interaction events based on multi-source drug fusion, multi-source feature fusion and transformer self-attention mechanism. Briefings Bioinf. 2022;23:bbab421. doi: 10.1093/bib/bbab421. [DOI] [PubMed] [Google Scholar]
- 15.Chu Y., Zhang Y., Wang Q., et al. A transformer-based model to predict peptide–HLA class I binding and optimize mutated peptides for vaccine design. Nat. Mach. Intell. 2022;4:300–311. [Google Scholar]
- 16.Chu Y., Kaushik A.C., Wang X., et al. DTI-CDF: a cascade deep forest model towards the prediction of drug-target interactions based on hybrid features. Briefings Bioinf. 2021;22:451–462. doi: 10.1093/bib/bbz152. [DOI] [PubMed] [Google Scholar]
- 17.Popova M., Isayev O., Tropsha A. Deep reinforcement learning for de novo drug design. Sci. Adv. 2018;4 doi: 10.1126/sciadv.aap7885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhavoronkov A., Ivanenkov Y.A., Aliper A., et al. Deep learning enables rapid identification of potent DDR1 kinase inhibitors. Nat. Biotechnol. 2019;37:1038–1040. doi: 10.1038/s41587-019-0224-x. [DOI] [PubMed] [Google Scholar]
- 19.Stokes J.M., Yang K., Swanson K., et al. A deep learning approach to antibiotic discovery. Cell. 2020;180:688–702. doi: 10.1016/j.cell.2020.01.021. e613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang L., Liu T., Chen H., et al. Predicting lncRNA-miRNA interactions based on interactome network and graphlet interaction. Genomics. 2021;113:874–880. doi: 10.1016/j.ygeno.2021.02.002. [DOI] [PubMed] [Google Scholar]
- 21.Zhang L., Yang P., Feng H., et al. Using network distance analysis to predict lncRNA-miRNA interactions. Interdiscipl. Sci. Comput. Life Sci. 2021;13:535–545. doi: 10.1007/s12539-021-00458-z. [DOI] [PubMed] [Google Scholar]
- 22.Liu W., Jiang Y., Peng L., et al. Inferring gene regulatory networks using the improved Markov blanket discovery algorithm. Interdiscipl. Sci. Comput. Life Sci. 2022;14:168–181. doi: 10.1007/s12539-021-00478-9. [DOI] [PubMed] [Google Scholar]
- 23.Godinez W.J., Ma E.J., Chao A.T., et al. Design of potent antimalarials with generative chemistry. Nat. Mach. Intell. 2022;4:180–186. [Google Scholar]
- 24.Fu J., Zhang Y., Wang Y., et al. Optimization of metabolomic data processing using NOREVA. Nat. Protoc. 2021;17:129–151. doi: 10.1038/s41596-021-00636-9. [DOI] [PubMed] [Google Scholar]
- 25.Xia W., Zheng L., Fang J., et al. PFmulDL: a novel strategy enabling multi-class and multi-label protein function annotation by integrating diverse deep learning methods. Comput. Biol. Med. 2022;145 doi: 10.1016/j.compbiomed.2022.105465. [DOI] [PubMed] [Google Scholar]
- 26.Kan M., Wu J., Shan S., et al. Domain adaptation for face recognition: targetize source domain bridged by common subspace. Int. J. Comput. Vis. 2014;109:94–109. [Google Scholar]
- 27.Dai W., Xue G.-R., Yang Q., et al. Proceedings of the 13th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining. 2007. Co-clustering based classification for out-of-domain documents; pp. 210–219. [Google Scholar]
- 28.Li T., Zhang Y., Sindhwani V. Proceedings of the Joint Conference of the 47th Annual Meeting of the ACL and the 4th International Joint Conference on Natural Language Processing of the AFNLP. 2009. A non-negative matrix tri-factorization approach to sentiment classification with lexical prior knowledge; pp. 244–252. [Google Scholar]
- 29.Case D.A., Cheatham T.E., III, Darden T., et al. The Amber biomolecular simulation programs. J. Comput. Chem. 2005;26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shaw D.E., Grossman J., Bank J.A., et al. SC'14: Proceedings of the International Conference for High Performance Computing, Networking, Storage and Analysis. IEEE; 2014. Anton 2: raising the bar for performance and programmability in a special-purpose molecular dynamics supercomputer; pp. 41–53. [Google Scholar]
- 31.Gilson M.K., Liu T., Baitaluk M., et al. BindingDB in 2015: a public database for medicinal chemistry, computational chemistry and systems pharmacology. Nucleic Acids Res. 2016;44:D1045–D1053. doi: 10.1093/nar/gkv1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim S., Chen J., Cheng T., et al. PubChem in 2021: new data content and improved web interfaces. Nucleic Acids Res. 2020;49:D1388–D1395. doi: 10.1093/nar/gkaa971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu P., Liu H., Sun Q., et al. Potent inhibitors of SARS-CoV-2 3C-like protease derived from N-substituted isatin compounds. Eur. J. Med. Chem. 2020;206 doi: 10.1016/j.ejmech.2020.112702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dai W., Zhang B., Jiang X.-M., et al. Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science. 2020;368:1331–1335. doi: 10.1126/science.abb4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang L., Lin D., Sun X., et al. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020;368:409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baell J.B., Holloway G.A. New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. J. Med. Chem. 2010;53:2719–2740. doi: 10.1021/jm901137j. [DOI] [PubMed] [Google Scholar]
- 37.Segler M.H., Kogej T., Tyrchan C., et al. Generating focused molecule libraries for drug discovery with recurrent neural networks. ACS Cent. Sci. 2018;4:120–131. doi: 10.1021/acscentsci.7b00512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paszke A., Gross S., Massa F., et al. Pytorch: an imperative style, high-performance deep learning library. Adv. Neural Inf. Process. Syst. 2019 [Google Scholar]
- 39.Polykovskiy D., Zhebrak A., Sanchez-Lengeling B., et al. Molecular sets (MOSES): a benchmarking platform for molecular generation models. Front. Pharmacol. 2020;11:1931. doi: 10.3389/fphar.2020.565644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bemis G.W., Murcko M.A. The properties of known drugs. 1. Molecular frameworks. J. Med. Chem. 1996;39:2887–2893. doi: 10.1021/jm9602928. [DOI] [PubMed] [Google Scholar]
- 41.Landrum G. RDKit: Open-source cheminformatics. https://www.rdkit.org
- 42.Degen, Wegscheid-Gerlach C., Zaliani A., et al. On the art of compiling and using 'drug-like' chemical fragment spaces. ChemMedChem: Chem. Enabling Drug Discov. 2008;3:1503–1507. doi: 10.1002/cmdc.200800178. [DOI] [PubMed] [Google Scholar]
- 43.Rogers D., Hahn M. Extended-connectivity fingerprints. J. Chem. Inf. Model. 2010;50:742–754. doi: 10.1021/ci100050t. [DOI] [PubMed] [Google Scholar]
- 44.Pedregosa F., Varoquaux G., Gramfort A., et al. Scikit-learn: machine learning in Python. J. Mach. Learn. Res. 2011;12:2825–2830. [Google Scholar]
- 45.Meng J., Zhang L., Wang L., et al. TSSF-hERG: a machine-learning-based hERG potassium channel-specific scoring function for chemical cardiotoxicity prediction. Toxicology. 2021;464 doi: 10.1016/j.tox.2021.153018. [DOI] [PubMed] [Google Scholar]
- 46.Li H., Leung K.-S., Wong M.-H., et al. Improving AutoDock Vina using random forest: the growing accuracy of binding affinity prediction by the effective exploitation of larger data sets. Mol. Info. 2015;34:115–126. doi: 10.1002/minf.201400132. [DOI] [PubMed] [Google Scholar]
- 47.Hummell N.A., Revtovich A.V., Kirienko N.V. Novel immune modulators enhance Caenorhabditis elegans resistance to multiple pathogens. mSphere. 2021;6 doi: 10.1128/mSphere.00950-20. e00950-00920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hasan M., Parvez M.S.A., Azim K.F., et al. Main protease inhibitors and drug surface hotspots for the treatment of COVID-19: a drug repurposing and molecular docking approach. Biomed. Pharmacother. 2021;140 doi: 10.1016/j.biopha.2021.111742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Miller B.R., III, McGee T.D., Jr., Swails J.M., et al. MMPBSA.py: an efficient Program for end-state free energy calculations. J. Chem. Theor. Comput. 2012;8:3314–3321. doi: 10.1021/ct300418h. [DOI] [PubMed] [Google Scholar]
- 50.Razzaghi-Asl N., Mirzayi S., Mahnam K., et al. Identification of COX-2 inhibitors via structure-based virtual screening and molecular dynamics simulation. J. Mol. Graph. Model. 2018;83:138–152. doi: 10.1016/j.jmgm.2018.05.010. [DOI] [PubMed] [Google Scholar]
- 51.Backman T.W., Cao Y., Girke T. ChemMine tools: an online service for analyzing and clustering small molecules. Nucleic Acids Res. 2011;39:W486–W491. doi: 10.1093/nar/gkr320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hunter J.D. Matplotlib: a 2D graphics environment. Comput. Sci. Eng. 2007;9:90–95. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data used in this study and the source codes for General-GEN and MproI-GEN can be found at https://github.com/Shimeng-Li/MproI-GEN.