Abstract
Bats are reservoirs for zoonotic viruses, which they tolerate without experiencing disease. Research focused on deciphering mechanisms of virus tolerance in bats has rarely considered the influence of their gastrointestinal tract (GIT) microbiome. In mammals, GIT microbiomes influence infections through their effect on host physiology, immunity, nutrition, and behavior. Bat GIT microbiomes more closely resemble the Proteobacteria-dominated GIT microbiomes of birds than those of other mammals. As an adaptation to flight bats have rapid GIT transit times which may reduce the stability of their microbiome, constrain nutrient uptake, and affect pathogen exposure and evolution of tolerance mechanisms. Experimental and longitudinal studies are needed to understand the function of bats’ GIT microbiomes, and their role in modulating viral infection dynamics.
Keywords: Chiroptera, bats, bacteriome, microbiome
With over 1,400 documented species, bats (order Chiroptera) collectively constitute about 20% of mammalian species diversity (https://batnames.org/) and are unique as the only mammals capable of powered flight. Bats are long-lived for their body size and exploit diverse dietary strategies, including frugivory, nectarivory, insectivory, carnivory, and sanguivory [1]. Bats provide crucial ecosystem services like pollination, seed dispersal, and consumption of agricultural pests and disease vectors [1]. However, bats also act as reservoirs for emerging zoonoses, including Hendra and Nipah henipaviruses, and SARS, MERS, and SARS-CoV-2 coronaviruses [2]. Much current research is focused on deciphering the mechanisms by which bats host viruses that are highly virulent to other mammals without themselves experiencing clinical disease. The extent to which gastrointestinal (GIT) microbiomes may modulate bat health and viral hosting capacity is a burgeoning field with many unanswered questions. Here, we postulate how bat GIT physiology may influence their microbiome composition, function, and stability to modulate infection dynamics. Future experimental, longitudinal, and functional research will help elucidate microbial contributions to bat health and viral infection susceptibility.
Gastrointestinal microbiomes play a role in mammalian health and disease
Microbial communities (microbiomes; see Glossary) that colonize the GIT play a vital role in mammalian health and immune function. Mammalian GIT microbiota have been shown to affect the nutritive, physiological, behavioral, and immunological status of their host in ways that may influence the probability of infection with and subsequent transmission of infectious pathogens (Figure 1). Studies performed in vitro and in model animals have found that GIT microbes can competitively exclude potential pathogens, directly inhibit pathogen replicative ability and infectious potential [3], and support the development of host mucosal and systemic immunity [4,5]. Intestinal bacteria can also facilitate coinfection of enteric viruses by promoting recombination [6], or providing structures like lipopolysaccharide (found on the surface of Gram-negative bacteria), which some viruses co-opt to evade host immune responses [7]. Thus, under differing conditions, intestinal bacteria may either exclude potential pathogens from the host, or, facilitate pathogen persistence [7].
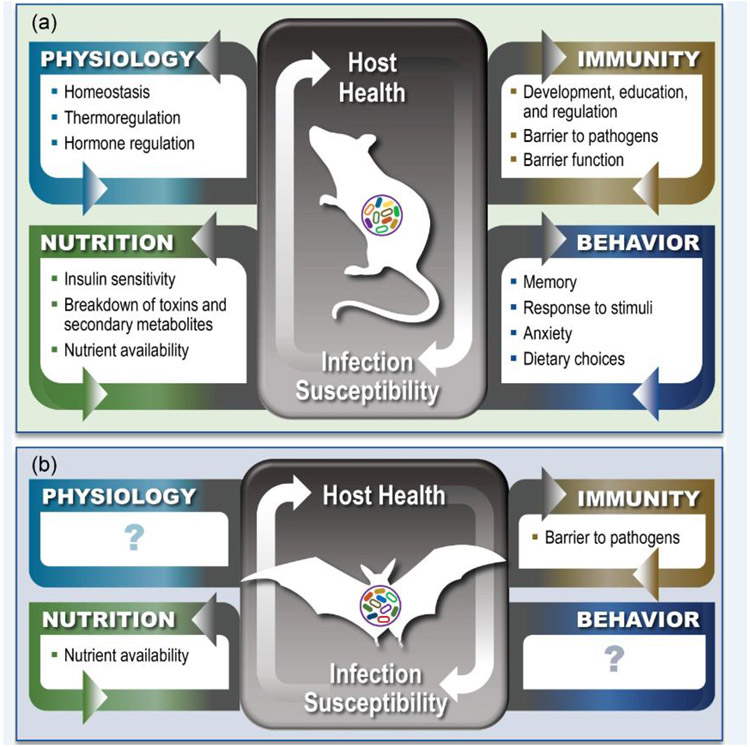
Figure 1. Microbiomes contribute to host health through several mechanisms.
Panel a shows known bidirectional communication between the model and well-studied system hosts (e.g., mice and humans) and their host-associated microbiomes. Host-associated microbiomes are known to contribute to host health by influencing or modulating host physiological processes, immune function, nutritional status, and behavior. Panel b shows that in bats, GIT microbes have been shown to contribute to host nutrition and skin microbes can serve as a barrier to external pathogens (Box 1). However, relative to mice and humans, much less is known about the specific contributions of microbiomes to their chiropteran hosts, highlighting a need for future research in these areas.
Nutrition plays a critical role in host immunity [8] offering an indirect mechanism by which the GIT microbiome can further modulate infection. Host nutrition is affected by GIT microbiota [9], as GIT-dwelling microbes biosynthesize B- and K-vitamins [10], which contribute to mineral homeostasis [11] and help break down dietary fiber to produce short-chain fatty acids (SCFAs) [9]. In particular, SCFAs can directly impede the development of respiratory viral infections [12]. In humans, uptake of B- and K-vitamins improves immune efficiency [13,14], while micronutrient deficiency impairs cellular immune responses [15].
In addition to nutritional and immunological function, GIT microbes also influence host fitness through behavior and physiology. The ‘gut-brain axis’ describes constant cross-talk between GIT microbes and the host brain [16], which occurs through several mechanisms. For example, humans with Inflammatory Bowel Syndrome, characterized by perpetual GIT microbiome dysbiosis, are more likely to experience anxiety [17], and the transplantation of “depression microbiota” into healthy mice can produce depression-like behaviors [18]. GIT microbiota are also important for maintaining homeostasis. Alterations in the GIT microbiome during cold temperature can expand GIT size and absorptive capacity, improve insulin sensitivity, and increase the metabolic activity of white adipose tissue through a process known as “browning” [19]. Thus, through both direct and indirect mechanisms, microbiomes play a large role in maintaining health and preventing infection in mammalian hosts. While most mammalian microbiome research has been focused on human and mouse model systems, exciting new research is beginning to unravel these processes in non-model species, including bats.
Bat microbiome research is a growing field
We systematically reviewed the literature on bats and microbiomes. Most studies on bat microbiomes were published in 2016 or after, likely as a result of increasing global access to sequencing technologies and increasing awareness of the importance of microbiomes more generally [20]. Most bat microbiome literature focuses on the GIT microbiome (78%) with a smaller number of studies investigating external microbiomes (e.g., skin; Box 1) or microbiomes of other organs or fluids (e.g., heart, eyes, urine). Because the first line of mammalian pathogen defenses are often local (e.g., respiratory mucosal surface [21]), the microbes that colonize these other sites are important for understanding host health.
Box 1: Bat microbiomes beyond the gastrointestinal tract.
In humans, the skin microbiota vary by site (e.g., forearm vs armpit) and tend to remain stable over time in healthy adults [77]. These resident bacteria protect the host by preventing the colonization of pathogenic bacteria [77]. In humans, dysbiosis of skin microbiota may result in skin disorders such as acne, eczema, malodor, and chronic wounds [77]. In reptiles and amphibians, changes in the skin microbiota have been associated with sometimes-fatal fungal infections [78].
Bacterial communities on bat skin and fur are dominated by microbes derived from the classes: Actinobacteria, Gammaproteobacteria, and Firmicutes [55,79-84]. Microbial richness associated with bat skin is greater than GIT and oral communities [38]. The external microbiota of bats are host-specific [82,84] and reflects the host environment [79], but can change at the roost-level over time [55]. Bats roosting together tend to have external microbiomes more similar to each other over time rather than themselves from earlier time points [55].
Bats’ skin microbes also protect from pathogens (Figure 1). Bacteria isolated and cultured from bat skin inhibits the growth of Pseudogymnoascus destructans (Pd), the causative agent of white-nose syndrome [85]. The skin microbiome of WNS-positive bats is typically enriched with bacteria with the ability to inhibit Pd-growth [81] but Pd growth can also destabilize skin microbiomes [82]; Grisnik et al. [83] found reduced populations of anti-fungal bacteria on the skin of bats infected with Pd, perhaps evidence of Pd’s successful overpowering of the skin microbiome. In addition to acting as a barrier to pathogens, microbes on bat skin likely aid in reproduction. Sex-related differences in bats’ skin microbiota are associated with specific odors and corresponding sex-selected scent organs [86,87].
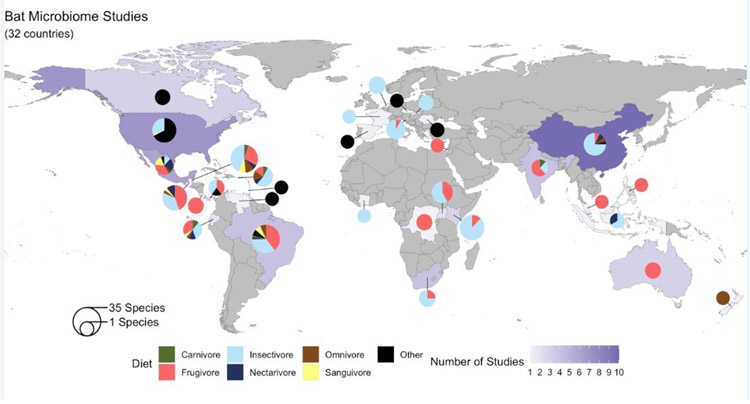
To date, the microbiomes of 205 bat species from 14 bat families have been sampled from 32 countries (Figure 2). Phyllostomidae and Vespertilionidae are highly represented relative to other bat families, respectively accounting for 30% and 32% of all bat species sampled for microbiome analyses. This research skew may be due in part to geographic convenience for North and South American research groups, and could reflect the unique diversity of feeding strategies (nectarivory, frugivory, sanguivory, carnivory, and insectivory) that exist within Phyllostomidae—thus enabling researchers to control for phylogeny while testing associations between diet type and microbial community [22]. Much microbiome research has focused on insectivorous bats, probably because two-thirds of bats globally are insectivorous [1].
Figure 2. Map of the current bat microbiome studies.
Shading of the countries shows the global distribution of bat microbiome studies, where darker shading indicates a greater number of studies from that country. The pie charts show the number of bat species sampled from each country (as indicated by the circumference of the pie) where smaller pies indicate fewer number of species sampled. The corresponding proportions of the pie show the diet types of the bats for which there are GIT microbiome studies; studies focused on non-GIT microbiomes (e.g., skin, urine, or other organs or fluids) are shown in black as “other.”
More longitudinal and experimental studies will elucidate bat GIT microbiome dynamics
Most bat microbiome studies (74%) have been cross-sectional with few longitudinal or experimental studies. Future longitudinal studies will be important in providing insight into the stability of bat GIT microbiomes across seasons and the extent to which bats experience dysbiosis following pathogen infection or dietary shifts. Vouchering specimens in collaboration with natural history museums will further support the establishment of bat microbiome records over longer time horizons [23]. Additionally, experimental studies will be crucial for deciphering the directionality in pathogen-microbiome interactions. For example, adenovirus infection was associated with altered GIT microbial communities in juvenile Artibeus jamaicensis, but not adults and subsequently predicted downstream impacts on nutritional and immunological functions for infected vs. uninfected juveniles [24]. Without experimentation, it is impossible to distinguish whether viral infection drives a change in the GIT microbiome or whether the presence of certain microbial communities render the host more susceptible to infection. Admittedly, experimental approaches in bat microbiome research are challenging as captive bat colonies are rare and difficult to maintain and experimental infections with virulent human pathogens require high biosafety containment [25]. Nonetheless, simple experiments can be undertaken on wild bats held temporarily in field settings [26] and experimental infections can be accomplished using non-zoonotic pathogens [27].
Most bat microbiome studies describe the microbial community (42%), but few study the functional role of that community. Nonetheless, consensus concerning the state and function of bat GIT microbiomes is beginning to emerge.
Bats break the mammalian mold with bird-like GIT microbiomes
At a broad taxonomic level, bat GIT microbiomes are unique from those of other mammals. Bat GIT microbiomes are dominated by microbes in the phylum Proteobacteria, including families Enterobacteriaceae and Streptococcaceae, [22,28-34], whereas most mammalian GITs are primarily colonized by microbes from the phyla Bacteroidetes and Firmicutes [32]. Bat GIT microbiomes more closely resemble those of flying birds than those of other mammals, though the species-level composition of specific GIT Proteobacteria—and their concomitant functions—may diverge in these two hosts [34]. In humans and mice, increased proportions of Proteobacteria signify dysbiosis and associated diseases (IBD, diabetes, obesity, and some neurological disorders [35], but in bats, Proteobacteria stably dominate apparently healthy GITs, suggesting that model systems cannot be universally extrapolated. Proteobacteria likely do not equate to dysbiosis in bats.
As in other mammal hosts, a combination of intrinsic (e.g., genetics, sex, reproductive status) and extrinsic (e.g., geographic location) factors shape bat GIT microbiomes [28,30,36-41]. Across mammals, GIT microbial diversity decreases as dietary strategies shift from herbivory to carnivory [32,42]. By contrast, bats primarily exhibiting herbivorous (nectarivorous or frugivorous) feeding strategies have lower microbial diversity than bats exhibiting carnivorous (insectivorous, sanguivorous, or omnivorous) feeding strategies [22,28,29,43] regardless of phylogenetic and geographic separation. Early bat microbiome research (primarily within Phyllostomidae) revealed that closely related bats share closely related microbiota (‘phylosymbiosis’) [22,28,36,44]. However, more recent work including a broader range of bat families suggests that bat GIT microbiomes—like those of birds—host communities that vary geographically but do not stably track host phylogeny [34,38] but see Box 2 for discussion of the role that sampling methods may play in these results.
Box 2: Sampling technique matters when defining bat GIT microbiomes.
Sampling the intestinal microbiota of bats requires euthanizing the animal, whereas fecal and rectal samples may be collected from live bats. Fecal samples can be collected non-invasively from underneath roosting bats [49,88]. However, microbial composition differs between bat intestinal and fecal samples (i.e. guano) [22,89]. Shannon richness has shown to be higher in bat fecal samples, but intestinal samples harbor greater microbial phylogenetic diversity (via Faith’s Phylogenetic Diversity index), a measure of diversity within a target (in this case, microbial) class [22]. Importantly, intestinal microbiota is more phylogenetically constrained than fecal microbiota and better reflects the evolutionary history of the bat host (phylosymbiosis) [22]. Feces may not necessarily be a good proxy for understanding the microbes that colonize the bat GIT [22,89] in contrast to taxa, like primates, with longer GIT transit times, for which phylosymbiosis can be deduced from feces [90]. Intestinal samples likely offer a better medium for investigating questions in the context of host evolution, whereas fecal samples may be better suited to addressing questions related to host diet and ecology [22]. The differences in microbiome diversity among feces and intestines relative to rectal swabs, has not been tested. Rectal swabs may offer better insight into the bacteria colonizing bats’ intestines and can be collected sans euthanasia.
Bat GIT microbiome stability—and its effect on bat health—remains unclear
In humans, lack of stability of the GIT microbiome is linked to disease [45]. Human GIT microbiomes exhibit successional changes over the first 3-4 years of age before reaching climactic adult-like microbiomes largely resilient to perturbation [45]. Currently, the literature concerning the stability of bat GIT microbiomes is indeterminant: bat GIT microbiome diversity appears to shift seasonally during during/after hibernation [46,47], and during reproduction in females [39,40]. Some studies have shown age-related changes in bat GIT microbiome diversity [39], though patterns are not consistent. For example, young Rhinolophus hipposideros bats host an over-representation of Klebsiella bacteria relative to adults [33], while Vespertilio sinensis bats show increasing GIT microbial diversity from birth until four weeks of age [48]. Habitat fragmentation and dietary shifts may also impact the variability of bat GIT microbiomes [37] and even within bat species, GIT microbiomes can differ between roosts [49].
By contrast, one study of Myotis myotis demonstrated identical rectal microbiomes in juveniles and adults, indicating a lack of age-related shift in this species [50]. However, all bats used in the study [50] were volant and therefore at least partially weaned. Because early successional dynamics of the GIT microbiome often correspond to dietary transitions [51], it would be interesting to assess whether differences manifest in the GIT microbiomes of nonvolant pups, which consume milk instead of adult foods. These transitions may be particularly important for the development of bat immune systems, as bats inherit maternal immunity via milk [52]. As mentioned previously, adenovirus infection in bats is associated with altered GIT microbiomes in juvenile bats, but not adults [24]. This work highlights the importance of successional changes in the GIT microbiome that could modulate viral shedding events that are associated with the juvenile-adult transition in other bat systems [53,54]. Adult Rousettus aegyptiacus GIT microbiomes more closely reflect microbial communities from previous samples of the same individual than temporally-coincident samples from other bats [55], highlighting the compositional stability of the microbial community in mature bat GITs. Finally, it should be noted that, like the microbiomes of other mammals, bat GIT microbiomes converge in captivity [56,57].
Bat microbiomes likely benefit their hosts
In many mammals, GIT microbes provide a myriad of essential functions to the host (Figure 1), while receiving habitat and resources in return. Though support for a hypothesis of bat-microbe mutualism is mounting, several studies document evidence of commensal and parasitic interactions between bats and their microbiota, as well. Understanding these relationships is essential for understanding how GIT-dwelling microbes could modulate bat virus infection dynamics.
Bat GIT microbiomes likely contribute to nutrient acquisition
In support of a more mutualistic bat-microbiome relationship, recent work suggests that bat GIT bacteria play a role in host nutrient acquisition. Unique communities of bacteria that inhabit different body sites suggest that site-specific microbiomes have specialized needs, functions, or competitional differences [30]. Numerous studies have shown that bat GIT microbiome composition is associated with host dietary strategy [22,28,36,38,43,58], and Phillips et al. [59] demonstrated that while three sympatric insectivorous bat species had varying GIT bacterial composition, the predicted functions of these communities were consistent across species. Sanguivorous vampire bats (Desmodus rotundus) have GIT microbiomes that are distinct from those of insectivorous bats in both composition and predicted function [44]. Indeed, the GIT microbiomes of vampire bats are more similar to those of sanguivorous birds than they are to those of other bats [60]. Similarly, whole genome sequencing of the common vampire bat and its GIT microbes show that microbial genes are enriched for functions contributing to nutrient acquisition, metabolism, and homeostasis [44]. Specifically, these functions are complementary extensions of endogenously encoded host gene functions suggesting that vampire bats depend on their microbes to function optimally.
Bats with frugivorous or nectarivorous dietary strategies also host bacteria that have predicted functions that may contribute to host nutrient acquisition. Chiefly, Enterobacter identified in the GIT of frugivorous Cynopterus brachyotis are known to metabolize sugars (including xylose, which comprises a large portion of most plant carbohydrates) [61,62] and GIT bacteria isolated from nectarivorous/frugivorous Pteropus giganteus have cellulolytic capabilities which help break down plant cell walls [63]. Nonetheless, studies have yet to categorically demonstrate the digestive functions of these microbes in bat hosts. Additional functional studies are needed to determine whether microbes identified in frugivorous bat GITs truly support nutrient acquisition in their respective hosts.
Bat GIT microbiomes may play a role In host longevity
Further evidence suggests that the functions of bat GIT bacteria may extend beyond nutrient acquisition. Hughes et al. [50] postulated that bats’ GIT microbes may contribute to longevity after identifying stable GIT microbes in long-lived bat GITs with functions including DNA replication and repair, metabolism, and oxidative phosphorylation. However, these processes are ubiquitous across all living organisms, making it difficult to pinpoint unique differences for bats. It should be noted that this study and many other bat microbiome studies summarized here use the program PICRUSt [24,46,48,50,57-60] to predict functions from 16S rRNA sequence data. PICRUSt is a less accurate predictor for non-human samples [64] and does not account for active microbial metabolism further complicated by transcriptional- and post-translational regulatory processes. To our knowledge, no studies have assessed the fecal or intestinal transcriptome or metabolome of bats to determine the specific metabolic pathways encoded in bat GIT microbial genomes. Though more work is needed to accurately assess the direct contributions of the microbiome, evidence to date suggests that GIT microbes support nutrient acquisition in bats which may impact their abilities to host or resist pathogens.
Simple digestive tracts in bats and birds may facilitate pathogen exposure
Both bats and birds have rapid gut transit times, a hypothesized adaptation to facilitate low body mass for flight [66]. Indeed, both bats and birds have simplified digestive tracts which are short in length and have reduced intestinal nominal surface areas (excluding contributions of villi and microvilli) and less intestinal tissue relative to nonvolant mammals [67,68]. Furthermore, most bats lack a cecum or appendix, an organ that houses bacteria beneficial for digestion in several bird species [69]. In general, consumption of difficult-to-digest, highly fibrous foliage is rare in both taxa [66]. Frugivorous bats are known to disgorge most of the fibrous material (pulp) in their diets and concentrate consumption on nectar and juice [1].
Regardless of diet, shorter intestine lengths and reduced gut transit times should pose constraints on both bird and bat nutrient uptake within the gut. Most nonvolant vertebrates acquire nutrients via a process known as transcellular transport, through which nutrients (e.g., glucose) are transported by cells across the gut epithelium. By contrast, paracellular absorption is the process by which nutrients are acquired via passive transport across the gut epithelium’s intracellular spaces [70]. Both birds and bats show evidence of expanded intestinal villi lining the gut epithelium, which could functionally increase the inner surface area of the small intestine and amplify opportunities for both transcellular and paracellular nutrient absorption. Indeed, previous work demonstrates that paracellular absorption is enhanced in both flying bird and bat taxa [67,70], which could reduce the reliance of these taxa on microbiota to enhance these processes. Additionally, heightened use of paracellular absorption may place species at risk of higher toxin or pathogen exposure [71]. Theory predicts that higher pathogen exposure should favor the evolution of tolerance mechanisms in a host [72], therefore, paracellular absorption could have important implications for bats’ evolution as tolerant viral reservoirs [2]. Given bats’ association with several emerging pathogens of importance to human health [2], the particular rapidity of GIT transit exhibited by Chiroptera, with implications for the composition of the GIT microbiome, is of special interest.
Rapid GIT transit times in bats and birds may select for commensal GIT microbiota
The extent to which different hosts rely on GIT microbiota for various functions varies widely across the animal kingdom, with some animals not requiring a microbiome at all [65]. In animals with low microbial biomass and rapid GIT transit times, it can be difficult to tease apart which microbes are transient or resident, and to identify the relative strength of microbial functional contributions to the host [65]. Recent research reporting lack of phylosymbiosis in bat and bat GIT microbiome phylogenies [34,38] may suggest a low reliance of bats on their GIT microbes, but the contributions of the bat GIT microbiome to essential host functions are still being elucidated.
Rapid GIT transit times [68] may limit the extent to which some bat GIT bacteria are able to provide beneficial metabolites to their hosts, as many of these metabolites (e.g., SCFAs) are produced through food fermentation in the GIT, which is generally a time-consuming process (depending on GIT transit time and amount of fiber ingested [73]). Consequently, it is possible that bat-GIT microbiome relationships are largely commensal, with bats providing habitat for GIT microbes but receiving few essential functions in return. Low bacterial biomass in oral and fecal samples [30] and high inter-individual variation in bat GIT microbiomes [22,36,55] offer evidence of a lack of dependence of bat hosts on their microbiota, but samples with low biomass are likely to be overwhelmed by contaminants during processing and may be more reflective of environmental (i.e., transient) microbes from diet and grooming [65]. Furthermore, some work indicates that microbiome composition and abundance appear relatively consistent throughout various regions of the bat GI tract (excepting some frugivores which demonstrate moderate shifts in relative abundance of different microbial taxa across the length of the GIT [28]). In many other taxa, distinctive GIT microbial communities colonize different regions of the digestive tract and perform specific digestive functions [20]. The homogeneity of bat GIT microbiomes could indicate a more commensal host-microbe relationship or could simply highlight that the low fecal biomass of bat GITs [30] has, to date, hindered our ability to identify important differences in microbial community composition and function.
In humans, microbiota inhabiting the more rapidly transiting small intestine demonstrate faster growth rates and more rapid utilization of simple substrates to avoid expulsion with transit [74]. Rapid transit rates in bird and bat GITs could alter the abiotic conditions in the GIT to select for “weedy” microbial taxa able to quickly utilize available substrates [65]. Support for this hypothesis in bats is seen in the dominance of bat GITs with Proteobacteria, which are also highly represented in the human small intestine, include species with some of the fastest growth rates, and commonly employ phosphoenol-pyruvate transport systems that enable rapid utilization of simple carbohydrate sources [75]. Proteobacteria may offer substantial benefits to chiropteran hosts, but these benefits are not fully characterized in the scientific literature. Without understanding which microbes are transient vs resident, it is difficult to tease apart the contributions of the GIT microbiomes to bat hosts.
More work is needed to characterize the contributions of Proteobacteria to bat health
As mentioned previously, high proportions of Proteobacteria that dominate bat GITs are associated with inflammation and poor health outcomes (e.g., IBD) in humans and mice [76]. Nonetheless, no study identifying the presence of these GIT microbial communities in bats has reported that any of the bat hosts investigated appeared unhealthy. Bats’ unique anti-inflammatory mechanisms evolved to cope with flight may also help them tolerate GIT bacteria associated with inflammation in other mammals. Even so, fitness effects are notoriously difficult to assess in wild animals, and few studies on this topic have been conducted, making it challenging to assess whether high proportions of Proteobacteria are indicative of dysbiosis in bats. Studies interrogating the functional role that these GIT microbes play in bat health and disease thus represent a critical research priority.
Concluding Remarks
The field of bat microbiome research is growing rapidly and exciting new breakthroughs in technique, approach, and understanding are emerging. As evidence of bat-microbe mutualisms continues to grow, future work in this area should progress towards a deeper understanding of the functional contributions offered by bat microbial communities in the GIT and elsewhere. Adoption of more -omics-based approaches (i.e., transcriptomics, metabolomics, proteomics, lipidomics) applied to both bat hosts and their microbiomes will help generate insights concerning microbial contributions to bat health and the dynamics of bat infections. Moreover, inclusion of techniques to assess host viromes and mycobiomes will provide insight into microbial dynamics within hosts. A future emphasis on longitudinal and experimental work will help reveal whether the concept of a “dysbiotic” state applies to bats and, if so, what health implications result from this disturbance (Outstanding Questions). Experimental studies may also enable better assessment of the causal underpinnings of observed associations between bat pathogen infection and changes in the microbial community. Most likely, bat microbiomes play a critical role in bat health, physiology, and immunity. It will be up to future research to demonstrate how.
Outstanding Questions:
How stable are bat GIT microbiomes over time? Are these dynamics sex- or species-specific?
Are rapid gut transit times the causal factor in the dominance of Proteobacteria in the bat GIT microbiome?
Do high proportions of Proteobacteria in the bat GIT microbiome signify a “dysbiotic state”? To what extent do dietary shifts (e.g. from nectar to fruit) perturb the GIT microbiome?
Do Proteobacteria play a role in GIT “leakiness” in bats and enable increased rates of paracellular transport through the epithelium for quicker access to nutrients from food?
Does increased paracellular transport in bat GITs increase their exposure to pathogens and potentially influence tolerance?
Do bat GIT microbes contribute to tolerance or resistance of pathogens?
How heritable are GIT microbiomes from mothers to pups and are there successional changes associated with weaning?
How do bat GIT microbes affect intestinal permeability and barrier function? How much does this matter for bats?
Does the GIT microbiome play a role in educating or modulating bats’ immune systems?
Highlights.
To date, most bat microbiome research is limited to cross-sectional and descriptive studies but recent work is shifting towards longitudinal and experimental investigations aimed to decipher bat microbiome function.
Bats and flighted birds have much shorter gastrointestinal tract (GIT) transit times than non-flying vertebrates, likely influencing the composition of their microbial communities.
Like birds (and distinct from other mammals), bat GIT microbiomes are largely dominated by Proteobacteria, early gut colonizers associated with inflammation in other species.
Bat microbiome studies based on intestinal samples show evidence of host-microbiome phylosymbiosis, while those based on fecal or rectal samples do not.
Consensus is emerging that bat GIT microbiomes likely contribute to nutrient acquisition for their hosts, but the role of the GIT microbiome in bat immune function remains largely unexplored.
Acknowledgements:
We would like to thank Dr. Britt Koskella, Dr. Dan Becker, and two anonymous reviewers for helpful comments that improved the manuscript. We would also like to thank Dawn H. Jones, Will Rogers, Manuel Ruiz-Aravena, C. Reed Hranac, and Wyatt Madden for their help creating figures. DNJ was funded by the Montana Academy of Science Student Research Grant and Montana Institute on Ecosystems Graduate Enhancement Fund. DNJ and RKP are funded by the DARPA PREEMPT program Cooperative Agreement #D18AC00031, the U.S. National Science Foundation (DEB-1716698), and the USDA National Institute of Food and Agriculture (Hatch project 1015891). CJY was supported in the preparation of this manuscript by the United States Department of Agriculture’s Agriculture and Food Research Initiative (USDA-AFRI) (2020-67016-31676), the Montana Agricultural Experiment Station (MONB00113), and the Montana INBRE Bioinformatics and Biostatistics core via the National Institute of General Medical Sciences of the National Institutes of Health (NIH-NIGMS; award number P20GM103474). CEB was supported by the Miller Institute for Basic Research and the Branco Weiss Science in Society postdoctoral fellowships; NAFR is supported by a L’Oréal USA For Women in Science fellowship and an NIH grant (1R01AI129822-01) to CEB.
Glossary Box:
- Dysbiosis
Imbalance in the composition and function of the microbiome that may enhance susceptibility to pathogen infection.
- Gut-Brain Axis
The bidirectional communication between the enteric and nervous system whereby emotional and cognitive function and intestinal function are linked.
- Host-associated microbiome
microbial communities that have been demonstrated to have meaningful, mutualistic relationships with the host that harbors the microbes. The host provides a niche for these microbes to colonize. In turn, the microbes provide essential functions such as metabolite anabolism and catabolism.
- Microbiome
Communities of microbial organisms residing on and within humans, plants, animals, or other systems. These microscopic communities include bacteria, viruses, archaea, fungi, and protists.
- Paracellular absorption
The passage of solutes or molecules between epithelial cells. In the GIT, tight junctions (multiprotein complex). Bat (and birds) GITs utilize high rates of paracellular absorptions relative to non-flying mammals. Regarding human health, tight junction function is essential in preventing a “leaky gut” or increased intestinal permeability which is associated with inflammation.
- Phylosymbiosis
a pattern of simultaneous host and microbiome phylogenetic branching align so that more closely related hosts contain more closely related microbes.
- Transcellular transport
The passage of solutes and molecules through a cell either through active transport or passive diffusion.
- White adipose tissue
subcutaneous tissue that stores and releases fatty acids to supply fuel.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kunz TH et al. (2011) Ecosystem services provided by bats. Ann. N.Y. Acad. Sci 1223, 1–38 [DOI] [PubMed] [Google Scholar]
- 2.Irving AT et al. (2021) Lessons from the host defences of bats, a unique viral reservoir. Nature. 589, 363–370 [DOI] [PubMed] [Google Scholar]
- 3.Li D et al. (2016) Anti-viral effect of Bifidobacterium adolescentis against Noroviruses. Front. Microbiol 7, 864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pabst R et al. (1988) Postnatal development and lymphocyte production of jejunal and ileal Peyer’s patches in normal and gnotobiotic pigs. Immunology. 64, 539–544 [PMC free article] [PubMed] [Google Scholar]
- 5.Schachtschneider KM et al. (2013) Modulation of systemic immune responses through commensal gastrointestinal microbiota. PLoS ONE. 8, e53969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Erickson AK et al. (2018) Bacteria facilitate enteric virus co-infection of mammalian cells and promote genetic recombination. Cell. Host. Microbe 23, 77–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilks J et al. (2015) Mammalian lipopolysaccharide receptors incorporated into the retroviral envelope augment virus transmission. Cell Host Microbe 18, 456–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chandra RK (1996) Nutrition, immunity and infection: From basic knowledge of dietary manipulation of immune responses to practical application of ameliorating suffering and improving survival. Proc. Nat. Acad. Sci 93, 14304–14307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flint HJ et al. (2012) The role of the gut microbiota in nutrition and health. Nat Rev Gastroenterol Hepatol 9, 577–589 [DOI] [PubMed] [Google Scholar]
- 10.Hill MJ (1997) Intestinal flora and endogenous vitamin synthesis. Eur. J. Cancer Prev 6 Suppl 1, S43–45 [DOI] [PubMed] [Google Scholar]
- 11.Takahashi K et al. (2020) Effect of gut microflora on nutritional availability of selenium. Food Chem 319, 126537. [DOI] [PubMed] [Google Scholar]
- 12.Antunes KH et al. (2019) Microbiota-derived acetate protects against respiratory syncytial virus infection through a GPR43-type 1 interferon response. Nat. Commun 10, 3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yoshii K et al. (2019) Metabolism of dietary and microbial vitamin B family in the regulation of host immunity. Front. Nutr 6, 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Namazi N et al. (2019) Vitamin K and the immune system. In Nutrition and Immunity (Mahmoudi M and Rezaei N, eds), pp. 75–79, Springer International Publishing. [Google Scholar]
- 15.Hamer DH et al. (2009) Micronutrient deficiencies are associated with impaired immune response and higher burden of respiratory infections in elderly Ecuadorians. J. Nutr 139, 113–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morais LH et al. (2021) The gut microbiota–brain axis in behaviour and brain disorders. Nat. Rev. Microbiol 19, 241–255 [DOI] [PubMed] [Google Scholar]
- 17.Blanchard EB et al. (1990) The role of anxiety and depression in the irritable bowel syndrome. Behav. Res. Ther 28, 401–405 [DOI] [PubMed] [Google Scholar]
- 18.Zheng P et al. (2016) Gut microbiome remodeling induces depressive-like behaviors through a pathway mediated by the host’s metabolism. Mol. Psyc 21, 786–796 [DOI] [PubMed] [Google Scholar]
- 19.Chevalier C et al. (2015) Gut microbiota orchestrates energy homeostasis during cold. Cell. 163, 1360–1374 [DOI] [PubMed] [Google Scholar]
- 20.Savage DC (1977) Microbial ecology of the gastrointestinal tract. Annu. Rev. Microbiol 31, 107–133 [DOI] [PubMed] [Google Scholar]
- 21.Iwasaki A et al. (2017) Early local immune defences in the respiratory tract. Nat. Rev. Immunol 17, 7–2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ingala MR et al. (2018) Comparing microbiome sampling methods in a wild mammal: Fecal and intestinal samples record different signals of host ecology, evolution. Front. Microbiol 9, 803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thompson CW et al. (2021) Preserve a voucher specimen! The critical need for integrating natural history collections in infectious disease studies. mBio 12,12(1):e02698–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wasimuddin et al. (2018) Astrovirus infections induce age-dependent dysbiosis in gut microbiomes of bats. ISME J 12, 2883–2893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paweska JT et al. (2017) South African Ebola diagnostic response in Sierra Leone: A modular high biosafety field laboratory. PLOS Neglect. Trop.l D 11, e0005665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stockmaier S et al. (2015) No fever and leucocytosis in response to a lipopolysaccharide challenge in an insectivorous bat. Biol. Lett 11, 20150576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suu-Ire R et al. (2018) Pathogenesis of bat rabies in a natural reservoir: Comparative susceptibility of the straw-colored fruit bat (Eidolon helvum) to three strains of Lagos bat virus. PLOS Neglect. Trop. D 12, e0006311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carrillo-Araujo M et al. (2015) Phyllostomid bat microbiome composition is associated to host phylogeny and feeding strategies. Front. Microbiol 6, doi: 10.3389/fmicb.2015.00447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Banskar S et al. (2016) Bacterial diversity indicates dietary overlap among bats of different feeding habits. Microbiol. Res 182, 99–108 [DOI] [PubMed] [Google Scholar]
- 30.Dietrich M et al. (2017) The excreted microbiota of bats: evidence of niche specialisation based on multiple body habitats. FEMS Microbiol. Lett 364, fnw284. [DOI] [PubMed] [Google Scholar]
- 31.Fofanov VY et al. (2018) Guano exposed: Impact of aerobic conditions on bat fecal microbiota. Ecol. Evol 8, 5563–5574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nishida AH and Ochman H (2018) Rates of gut microbiome divergence in mammals. Mol. Ecol 27, 1884–1897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vengust M et al. (2018) The fecal bacterial microbiota of bats; Slovenia. PLoS ONE. 13, e0196728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Song SJ et al. (2020) Comparative analyses of vertebrate gut microbiomes Reveal Convergence between Birds and Bats. mBio 11, e02901–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shin N-R et al. (2015) Proteobacteria: microbial signature of dysbiosis in gut microbiota. Trends Biotechnol 33, 496–503 [DOI] [PubMed] [Google Scholar]
- 36.Phillips CD et al. (2012) Microbiome analysis among bats describes influences of host phylogeny, life history, physiology and geography. Molec. Ecol 21,2617–2627 [DOI] [PubMed] [Google Scholar]
- 37.Ingala MR et al. (2019) Habitat fragmentation is associated with dietary shifts and microbiota variability in common vampire bats. Ecol. Evol doi: 10.1002/ece3.5228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lutz HL et al. (2019) Ecology and host identity outweigh evolutionary history in shaping the bat microbiome. mSystems 4, e00511–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dietrich M et al. (2018) Synchronized shift of oral, faecal and urinary microbiotas in bats and natural infection dynamics during seasonal reproduction. R. Soc. open sci 5, 180041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gaona O et al. (2019) Fecal microbiota of different reproductive stages of the central population of the lesser-long nosed bat, Leptonycteris yerbabuenae. PLoS ONE. 14, e0219982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gaona O et al. (2020) Geographical separation and physiology drive differentiation of microbial communities of two discrete populations of the bat Leptonycteris yerbabuenae. MicrobiologyOpen 9, 1113–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ley RE et al. (2008) Worlds within worlds: Evolution of the vertebrate gut microbiota. Nat. Rev. Microbiol 6, 776–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li J et al. (2018) Fecal bacteriome and mycobiome in bats with diverse diets in South China. Curr. Microbiol 75, 1352–1361 [DOI] [PubMed] [Google Scholar]
- 44.Zepeda Mendoza ML et al. (2018) Hologenomic adaptations underlying the evolution of sanguivory in the common vampire bat. Nat. Ecol. Evol 2, 659–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wilmanski T et al. (2021) Gut microbiome pattern reflects healthy ageing and predicts survival in humans. Nat. Metab 3, 274–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xiao G et al. (2019) Seasonal changes in gut microbiota diversity and composition in the Greater Horseshoe Bat. Front. Microbiol 10, 2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maliničová L et al. (2017) The dynamics of Mediterranean Horseshoe Bat (Rhinolophus euryale, Chiroptera) gut microflora during hibernation. Acta Chiropt 19, 211–218 [Google Scholar]
- 48.Yin Z et al. (2020) Changes in the gut microbiota during Asian particolored bat (Vespertilio sinensis) development. PeerJ 8, e9003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Henry R et al. (2018) What’s the risk? Identifying potential human pathogens within grey-headed flying foxes faeces. PLoS ONE. 13, e0191301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hughes GM et al. (2018) Is there a link between aging and microbiome diversity in exceptional mammalian longevity? PeerJ 6, e4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Koenig JE et al. (2011) Succession of microbial consortia in the developing infant gut microbiome. Proc. Nat. Acad. Sci 108, 4578–4585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Epstein JH et al. (2013) Duration of maternal antibodies against canine distemper virus and Hendra virus in Pteropid bats. PLOS ONE. 8, e67584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wacharapluesadee S et al. (2018) Longitudinal study of age-specific pattern of coronavirus infection in Lyle’s flying fox (Pteropus lylei) in Thailand. Virol. J 15, 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Amman BR et al. (2012) Seasonal pulses of Marburg virus circulation in juvenile Rousettus aegyptiacus bats coincide with periods of increased risk of human infection. PLoS Pathog 8, e1002877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kolodny O et al. (2019) Coordinated change at the colony level in fruit bat fur microbiomes through time. Nat. Ecol. Evol 3, 116–124 [DOI] [PubMed] [Google Scholar]
- 56.Edenborough KM et al. (2020) Microbiomes in the insectivorous bat species Mops condylurus rapidly converge in captivity. PLoS ONE 15, e0223629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xiao Y et al. (2019) Captivity causes taxonomic and functional convergence of gut microbial communities in bats. PeerJ 7, e6844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ingala MR et al. (2021) You are more than what you eat: potentially adaptive enrichment of microbiome functions across bat dietary niches. anim microbiome 3, 82. 10.1186/s42523-021-00139-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Phillips CD et al. (2017) Microbiome structural and functional interactions across host dietary niche space. Integr. Comp. Biol 57, 743–755 [DOI] [PubMed] [Google Scholar]
- 60.Song SJ et al. (2019) Is there convergence of gut microbes in blood-feeding vertebrates? Phil. Trans. R. Soc. B 374, 20180249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Daniel DS et al. (2013) Isolation and identification of gastrointestinal microbiota from the short-nosed fruit bat Cynopterus brachyotis brachyotis. Microbiol. Res 168, 485–496 [DOI] [PubMed] [Google Scholar]
- 62.Dhivahar J et al. (2019) Isolation and characterization of hyper-xylanase producing Bacillus spp. from faeces of the Indian Flying Fox (Pteropus giganteus). Acta Chiropt 21, 229–236 [Google Scholar]
- 63.Prem Anand AA and Sripathi K (2004) Digestion of cellulose and xylan by symbiotic bacteria in the intestine of the Indian flying fox (Pteropus giganteus). Comp Biochem. Phys. A 139, 65–69 [DOI] [PubMed] [Google Scholar]
- 64.Sun S et al. (2020) Inference-based accuracy of metagenome prediction tools varies across sample types and functional categories. Microbiome. 8, 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hammer TJ et al. (2019) Not all animals need a microbiome. FEMS Microbiol. Lett 366, fnz117. [DOI] [PubMed] [Google Scholar]
- 66.Price ER et al. (2015) Digestive Adaptations of Aerial Lifestyles. Physiology. 30, 69–78 [DOI] [PubMed] [Google Scholar]
- 67.Caviedes-Vidal E et al. (2007) The digestive adaptation of flying vertebrates: High intestinal paracellular absorption compensates for smaller guts. Proc. Nat. Acad. Sci 104, 19132–19137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Klite PD (1965) Intestinal bacterial flora and transit time of three Neotropical bat species. J. Bacteriol 90, 375–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yeoman CJ et al. (2012) The microbiome of the chicken gastrointestinal tract. Anim. Health Res. Rev 13, 89–99 [DOI] [PubMed] [Google Scholar]
- 70.Caviedes-Vidal E et al. (2008) Paracellular absorption: A bat breaks the mammal paradigm. PLoS ONE. 3, e1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Karasov WH (2011) Digestive physiology: a view from molecules to ecosystem. Am. J. Physiol- Reg. I 301, R276–R284 [DOI] [PubMed] [Google Scholar]
- 72.Boots M and Bowers RG (1999) Three mechanisms of host resistance to microparasites—avoidance, recovery and rolerance—show different evolutionary dynamics. J. Theor. Biol 201, 13–23 [DOI] [PubMed] [Google Scholar]
- 73.Macfarlane S and Macfarlane GT (2003) Regulation of short-chain fatty acid production. Proc. Nutr. Soc 62, 67–72 [DOI] [PubMed] [Google Scholar]
- 74.Zoetendal EG et al. (2012) The human small intestinal microbiota is driven by rapid uptake and conversion of simple carbohydrates. ISME J 6, 1415–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Le Bouguénec C and Schouler C (2011) Sugar metabolism, an additional virulence factor in enterobacteria. Int. J. Med. Microbiol 301, 1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fassarella M et al. (2021) Gut microbiome stability and resilience: elucidating the response to perturbations in order to modulate gut health. Gut 70, 595–605 [DOI] [PubMed] [Google Scholar]
- 77.Byrd AL et al. (2018) The human skin microbiome. Nat. Rev. Microbiol 16, 143–155 [DOI] [PubMed] [Google Scholar]
- 78.Allender MC et al. (2018) Snake fungal disease alters skin bacterial and fungal diversity in an endangered rattlesnake. Sci. Rep 8, 12147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Winter AS et al. (2017) Skin and fur bacterial diversity and community structure on American southwestern bats: effects of habitat, geography and bat traits. PeerJ 5, e3944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lemieux-Labonté V et al. (2016) Environment and host species shape the skin microbiome of captive neotropical bats. PeerJ 4, e2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lemieux-Labonté V et al. (2017) Enrichment of beneficial bacteria in the skin microbiota of bats persisting with white-nose syndrome. Microbiome. 5, 115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lemieux-Labonté V et al. (2020) Antifungal potential of the skin microbiota of hibernating Big Brown Bats (Eptesicus fuscus) infected with the causal agent of white-nose syndrome. Front. Microbiol 11, 1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Grisnik M et al. (2020) The cutaneous microbiota of bats has in vitro antifungal activity against the white nose pathogen. FEMS Microbiol. Ecol 96, fiz193. [DOI] [PubMed] [Google Scholar]
- 84.Avena CV et al. (2016) Deconstructing the bat skin microbiome: Influences of the host and the environment. Front. Microbiol 7, 1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hoyt JR et al. (2015) Bacteria isolated from bats inhibit the growth of Pseudogymnoascus destructans, the causative agent of white-nose syndrome. PLoS ONE. 10, e01213299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Voigt CC et al. (2005) Bats, bacteria, and bat smell: Sex-specific diversity of microbes in a sexually selected scent organ. J. Mamm 86, 745–749 [Google Scholar]
- 87.Gaona O et al. (2019) Microbiota composition of the dorsal patch of reproductive male Leptonycteris yerbabuenae. PLoS ONE 14, e0226239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dietrich M and Markotter W (2019) Studying the microbiota of bats: Accuracy of direct and indirect samplings. Ecol. Evol 9, 1730–1735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wu H et al. (2019) Gut microbial diversity in two insectivorous bats: Insights into the effect of different sampling sources. Microbiology Open 8, e00670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Moeller AH et al. (2016) Cospeciation of gut microbiota with hominids. Science. 353, 380–382 [DOI] [PMC free article] [PubMed] [Google Scholar]