Abstract
Phosphatidylserine (PS) in the plasma membrane plays an important role in cell signaling and apoptosis. Cell degeneration is also linked to numerous amyloid diseases, pathologies that are associated with aggregation of misfolded proteins. In this work, we examine the effect of both saturated PS (DMPS) and unsaturated PS (DOPS and POPS) on the aggregation properties of insulin, as well as the structure and toxicity of insulin aggregates formed in the presence of these phospholipids. We found that the degree of unsaturation of fatty acids in PS alters the rate of insulin aggregation. We also found that toxicity of insulin–DMPS aggregates is significantly lower than the toxicity of DOPS– and POPS–insulin fibrils, whereas all these lipid-containing aggregates exert lower cell toxicity than insulin fibrils grown in a lipid-free environment.
Keywords: AFM-IR, amyloid, insulin, phospholipids, toxicity
Amyloid diseases constitute a large group of pathologies that are caused by abrupt accumulation of misfolded proteins in different organs and tissues [1–3]. In such deposits, proteins are assembled into long linear polymers, which are known as amyloid fibrils [4,5]. Cryo-electron microscopy and solid-state nuclear magnetic resonance (ss-NMR) revealed that fibrils have β-sheet secondary structure [6–8]. These aggregates may exhibit a large diversity of topologies ranging from laterally assembled filaments to both right- and left-handed coils.
In vitro experiments confirmed that denatured proteins can spontaneously self-assemble forming aggregates with similar structures and morphologies as those observed in amyloid deposits. These experiments also reveal that a large number of biological molecules can uniquely alter rates of protein aggregation, as well as modify toxicity and secondary structure of fibrils [9–11]. For instance, Cataldi and co-workers showed that 3,4-dihydroxyphenylacetaldehyde, a product of dopamine oxidation, can uniquely change the secondary structure of amyloid β oligomers significantly increasing their toxicity [9].
High-speed AFM (HS-AFM) [12,13] atomic force microscopy infrared (AFM-IR) [14–18] and tip-enhanced Raman spectroscopy [19–23] are scanning probe methods that can be utilized to unravel nanoscale structural organization of amyloid oligomers. Specifically, HS-AFM was able to reveal structural transformations in islet amyloid precursor peptide (IAPP), amyloid β1–42 (β1–42), and α-synuclein (α-Syn) that lead to fibril formation [12,24,25]. These proteins that are directly linked to diabetes type 2, Alzheimer, and Parkinson diseases, respectively. Furthermore, HS-AFM revealed two aggregation mechanisms of Aβ1–42 that led to formation of straight and spiral fibrils [13]. Using AFM-IR, Zhou and Kurouski [18] resolved the structural heterogeneity of α-Syn oligomers formed at different stages of protein aggregation. It has been found that at early stages, oligomers with a mixture of unordered and α-helix secondary structure, as well as parallel and antiparallel β-sheet were observed. However, α-Syn aggregates present at late stages of protein aggregation were dominated by parallel β-sheet secondary structure. Using TERS, Kurouski and co-workers were able to probe the exterior surface of insulin fibrils [21]. The researchers found high abundance of unordered and α-helical protein present on the fibrils surface, whereas fibril core was entirely dominate by the β-sheet secondary structure. Kurouski and co-workers also were able to resolve different fibril polymorphs using TERS [22].
A growing body of evidence shows that lipids can either accelerate or decelerate the rates of protein aggregation [26–28]. For instance, Zhang and co-workers found that low levels of anionic lipids accelerated the aggregation of islet amyloid precursor protein (IAPP), as well as enhanced membrane permeabilization properties of IAPP aggregates. At the same time, zwitterionic lipid did not alter the rate of IAPP aggregation, whereas cholesterol at or below physiological levels significantly decelerated IAPP amyloid formation, as well as lowered the propensity of IAPP aggregates to cause membrane leakage. Lipids can also uniquely modify the secondary structure of amyloid β1–40 (Aβ1–40) [29] and α-Syn aggregates [5].
Our group found that an aggregation of insulin, a small protein hormone that regulates glucose metabolism, in the presence of cardiolipin (CL) and phosphatidylcholine (PC) results in inclusion of these lipids in the structure of insulin oligomers and fibrils [30]. Furthermore, in the equimolar mixture of CL and PC, insulin exclusively binds to PC. In addition to these two phospholipids, plasma membranes of eukaryotic cells contain ~ 10% of phosphatidylserine (PS) [31]. This lipid is primarily localized on the interior part of the membrane via ATP-dependent flippase-mediated transport. In the absence of ATP, which is a sign of cell malfunction, PS gradually appears on the exterior part of the plasma membrane [32]. The presence of PS attracts and activates phagocytes, which trigger cell degradation. Expanding upon this, we hypothesize that the exposure of PS on the exterior part of the plasma membrane upon neurodegeneration may influence aggregation properties of extracellular proteins such as insulin [33]. Abrupt aggregation of this hormone is observed upon diabetes type 2 and injection amyloidosis. In the former case, an overproduction of insulin in pancreas triggers its misfolding and aggregation [34], whereas in the latter case, high local concentration of insulin in the skin dermis triggers the formation of fibrils [35,36]. These insulin aggregates, in turn, can catalyze aggregation of other proteins present in cell media which often results in systemic amyloidosis [37].
Cell membranes contain PS with different degrees of saturation of the phospholipid fatty acids (FA) [31,38]. In addition to the fully saturated FA PS, which is known as 1,2-dimyristoyl-sn-glycero-3-phospho-L-serine (DMPS), plasma membranes contain PS with one [1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-L-serine (POPS)] and two [1,2-dioleoyl-sn-glycero-3-phospho-L-serine (DOPS)] double bonds [33,39,40]. Expanding upon this, we aim to determine the extent to which DMPS, POPS, and DOPS can alter the rate of insulin aggression, as well as modify the structure and toxicity of insulin aggregates that are formed in the presence of these lipids. For this, we aggregated DMPS, POPS, and DOPS with equimolar concertation of insulin in a presence of thioflavin T (ThT), a fluorophore that allows for monitoring fibril formation. Next, we aggregated insulin under the same experimental conditions without ThT and examined the topology and secondary structure of the corresponding aggregates using atomic force microscopy (AFM), infrared (IR), and AFM-IR, we employed an array of biochemical assays to examine the toxicity of Ins:DMPS, Ins:POPS, and Ins:DOPS aggregates.
Results and Discussion
Kinetics of insulin aggregation
Insulin aggregation in the lipid-free environment shows a well-defined lag-phase (tlag = 16.5 ± 0.4 h) that is followed by a rapid increase in the ThT intensity, which indicates the formation of protein aggregates, Fig. 1. We found that equimolar presence of DMPS (tlag = 8.8 ± 0.56 h) and DOPS (tlag = 14.7 ± 0.5 h) drastically shortened the lag-phase of insulin aggregation, whereas POPS, on the opposite, delayed (tlag = 18.0 ± 0.8 h) fibril formation. Based on these results, we can conclude that the degree of unsaturation of FAs in PS alters the rate of insulin aggregation. We also found that ThT intensity of insulin aggregation in the lipid-free environment has the highest intensity compared to the intensities of ThT signals for Ins:DMPS, Ins:POPS, and Ins:DOPS. This finding suggests that insulin aggregation in the presence of lipids yields fewer ThT active protein aggregates.
Fig. 1.
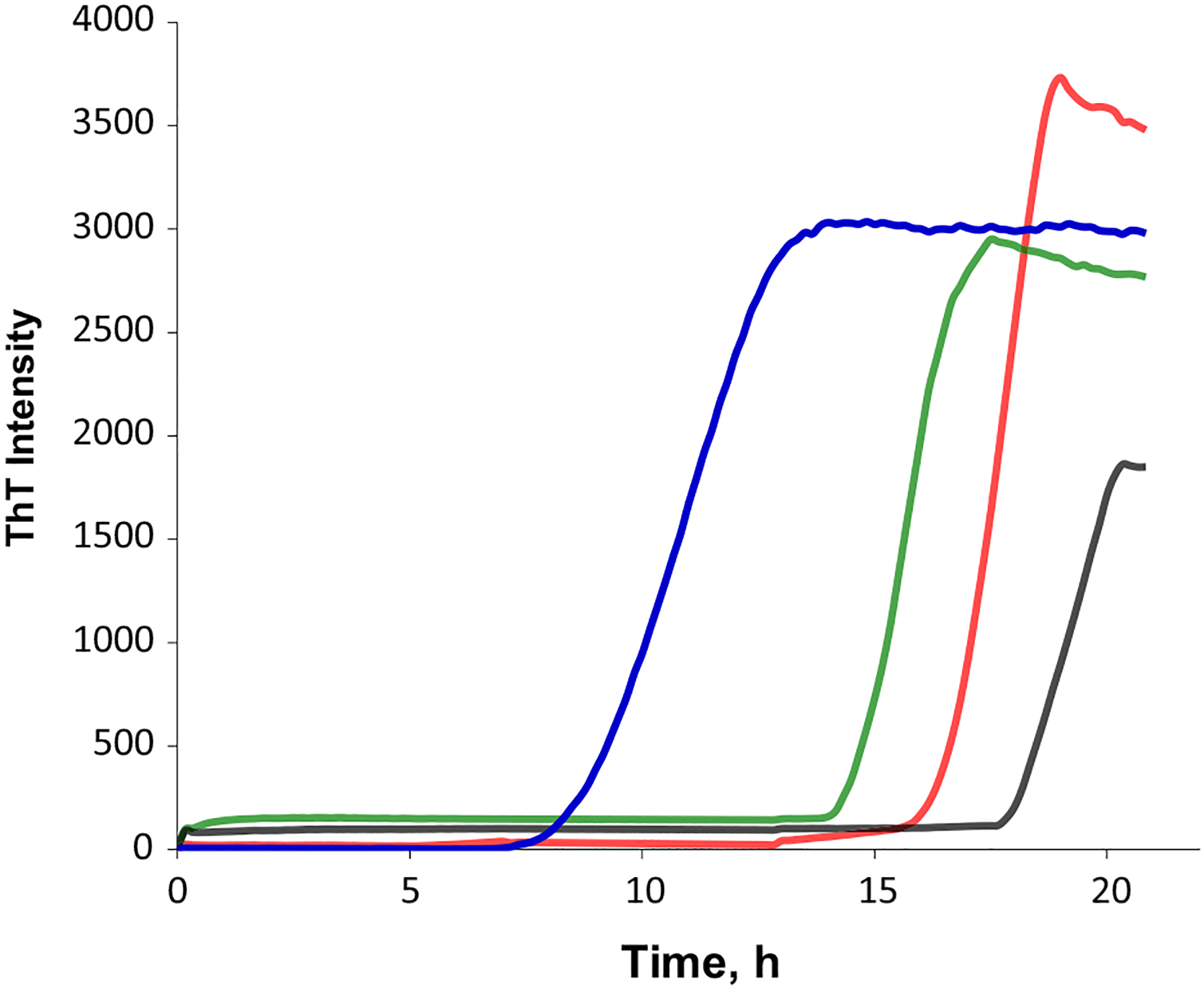
PS with different degrees of unsaturation of FAs alters the rates of insulin aggregation. Averages of triplicates of ThT aggregation kinetics of insulin in the lipid-free environment [(Ins), red] and in the presence of DMPS (blue), POPS (black), and DOPS (green) at 1 : 1 molar ratio. For Ins, 400 μM of protein was dissolved in 1×PBS with 2 mM of ThT; pH adjusted to pH 3.0. For Ins: DMPS, Ins:POPS, and Ins:DOPS, 400 μM of insulin was mixed with an equivalent concentration of the corresponding lipid; pH was adjusted to pH 3.0. All samples were kept at 37 °C under 510 rpm for 24 h.
Morphological analysis of insulin aggregates
Insulin aggregation in the lipid-free environment yielded fibrils with a large distribution of lengths that had on average 12 nm in height, Fig. 2. We found that in the presence of DMPS, shorter but much thicker fibrils were formed. We also observed shot aggregates (6–8 nm in height) and small oligomers (4–6 nm in height) present in Ins:DMPS sample. At the same time, insulin aggregation in the presence of unsaturated phospholipids (POPS and DOPS) yielded aggregates with bead-like appearance, suggesting that numerous oligomeric species self-assembled into fibril-like structures. It should be noted that no significant morphological differences were observed between Ins: DOPS and Ins:POPS aggregates.
Fig. 2.
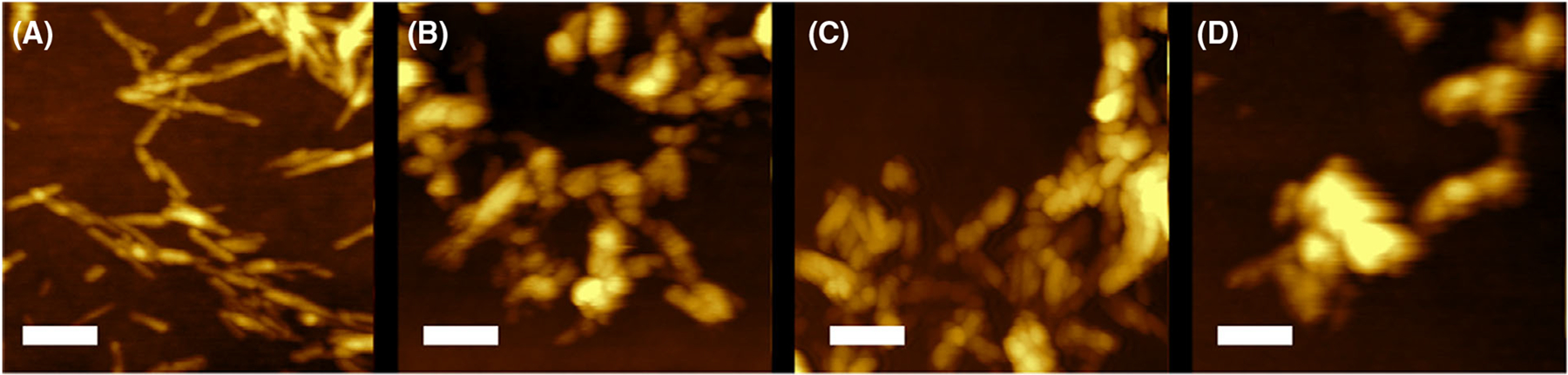
PS with different degrees of unsaturation of FAs alters morphology of protein aggregates. AFM images of insulin aggregates grown in the lipid-free environment (A), as well as in the presence of DMPS (B), POPS (C), and DOPS (D). After 24 h of incubation of insulin (400 μM) with and without lipids at 37 °C under 510 rpm, sample aliquots were diluted with 1×PBS pH 3.0 and deposited onto precleaned silicon wafer. AFM imaging was performed in tapping mode. Scale bars are 200 nm.
These results show that in the lipid-free environment, insulin forms morphologically different fibrils compared to the aggregates that were observed in the presence of phospholipids. These findings also show that the topology of insulin aggregates grown in the presence of phospholipids is determined by the unsaturation of FAs in PS, however, not the degree of unsaturation of FAs in the lipids.
Structural characterization of protein aggregates
We found that ATR-FTIR spectrum of Ins aggregates exhibited amide I at 1628 cm−1. At the same time, amide I band in ATR-FTIR spectra collected from Ins:DMPS, Ins:POPS and Ins:DOPS was centered at ~ 1631 cm−1. This indicates that parallel β-sheet secondary structure dominates in Ins, Ins:DMPS, Ins: POPS and Ins:DOPS, Fig. 3 [41,42]. This finding points on the substantial structural difference between the parallel β-sheet secondary structure in insulin aggregates grown in the lipid-free environment and the aggregates grown in the presence of lipids. We also observed a shoulder at 1657 cm−1 in the amide I region of the spectra collected form the protein aggregates grown in the presence of PS, as well as in the lipid-free environment. This suggests about the presence of unordered protein secondary structure in Ins: DMPS, Ins:POPS and Ins:DOPS [18,43]. Finally, we found that ATR-FTIR spectra collected from Ins: DMPS, Ins:POPS and Ins:DOPS exhibit a small peak at 1735 cm−1, which could be assigned to the carboxyl vibration of the phospholipids [44]. It should be noted that this peak was not observed in the ATR-FTIR spectra collected from Ins, Fig. 3. This spectroscopic evidence suggests about the presence of lipids in the structure of insulin aggregates that were grown in the presence of PS.
Fig. 3.
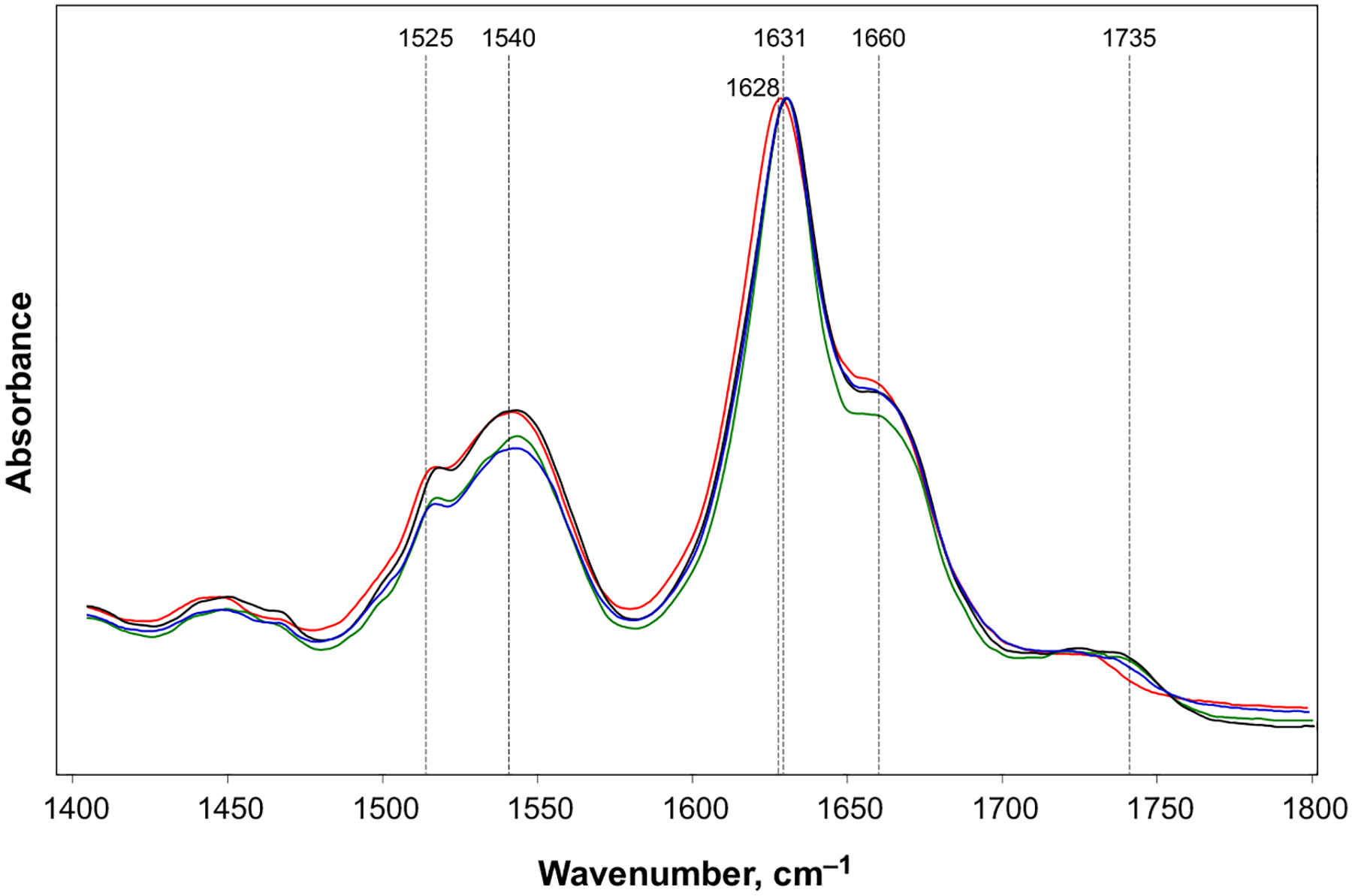
Structural analysis of insulin aggregates. ATR-FTIR spectra of insulin fibrils grown in the lipid-free environment (red), as well as in the presence of DMPS (blue), POPS (black), and DOPS (green). After 24 h of incubation of insulin (400 μM) with and without lipids at 37 °C under 510 rpm, triplicates of samples were diluted with 1×PBS pH 3.0 and directly deposited onto ATR crystal (ATR-FTIR) and dried under room temperature. For each of the presented traces, three independent ATR-FTIR measurements were averaged.
To further examine the presence of lipids in Ins: DMPS, Ins:POPS, and Ins:DOPS, we utilized AFM-IR. In this nanoscopy technique, a metalized scanning probe can be positioned above the sample of interest and illuminated by pulsed tunable IR light [45–47]. Next, IR-induced thermal expansions in the sample are recorded by the scanning probe [48,49]. Thus, AFM-IR allows for a direct visualization individual protein aggregates simultaneously enabling their structural characterization [5,16,17,50]. AFM-IR spectra collected from insulin aggregates that were grown in the presence of DMPS, POPS and DOPS exhibited vibrational bands centered ~ 800 and 1000–1270 cm−1, Fig. 4 and Fig. S1. These vibrational bands correspond to C-H and PO2− (L1 and L2) vibrations of lipids, respectively, Fig. S2. [50]. This finding points on the presence of a significant amount of lipids in Ins: DMPS, Ins:POPS and Ins:DOPS [30]. We also observed vibrations around 1300–1460 cm−1, which could be assigned to C-O stretching of lipids or CH2 vibrations of both lipids and proteins [5]. Because these vibrational bands cannot be unambiguously assigned to the certain class of chemical compounds, we omit these vibrations form the future discussion. Finally, AFM-IR spectra show that Ins:DMPS, Ins: POPS and Ins:DOPS possess a different amount of the corresponding lipid in their structures. We found that Ins:DMPS exhibit the highest lipid content, whereas less lipid is observed in Ins:DOPS and Ins: POPS, respectively. These conclusions can be made based on the intensities of ~ 800 and 1000–1200 cm−1 in the corresponding AFM-IR spectra collected from these protein aggregates. It should be noted that we observed no vibrational bands of lipids in the AFM-IR spectra collected from Ins aggregates. We also observed an intense vibration of PS carbonyl (C=O) in the AFM-IR spectrum collected from Ins:DMPS aggregates that was not evident in the spectra acquired from in Ins:DOPS and Ins:POPS. In our previous study, we demonstrated that intensity of this band changes upon lipid-protein interactions [30]. Thus, our current findings show that saturated and unsaturated PS interact differently with insulin. Specifically, DMPS exerts much stronger binding to insulin comparing to DOPS and POPS, which results in its higher content in the corresponding protein: lipid aggregates.
Fig. 4.
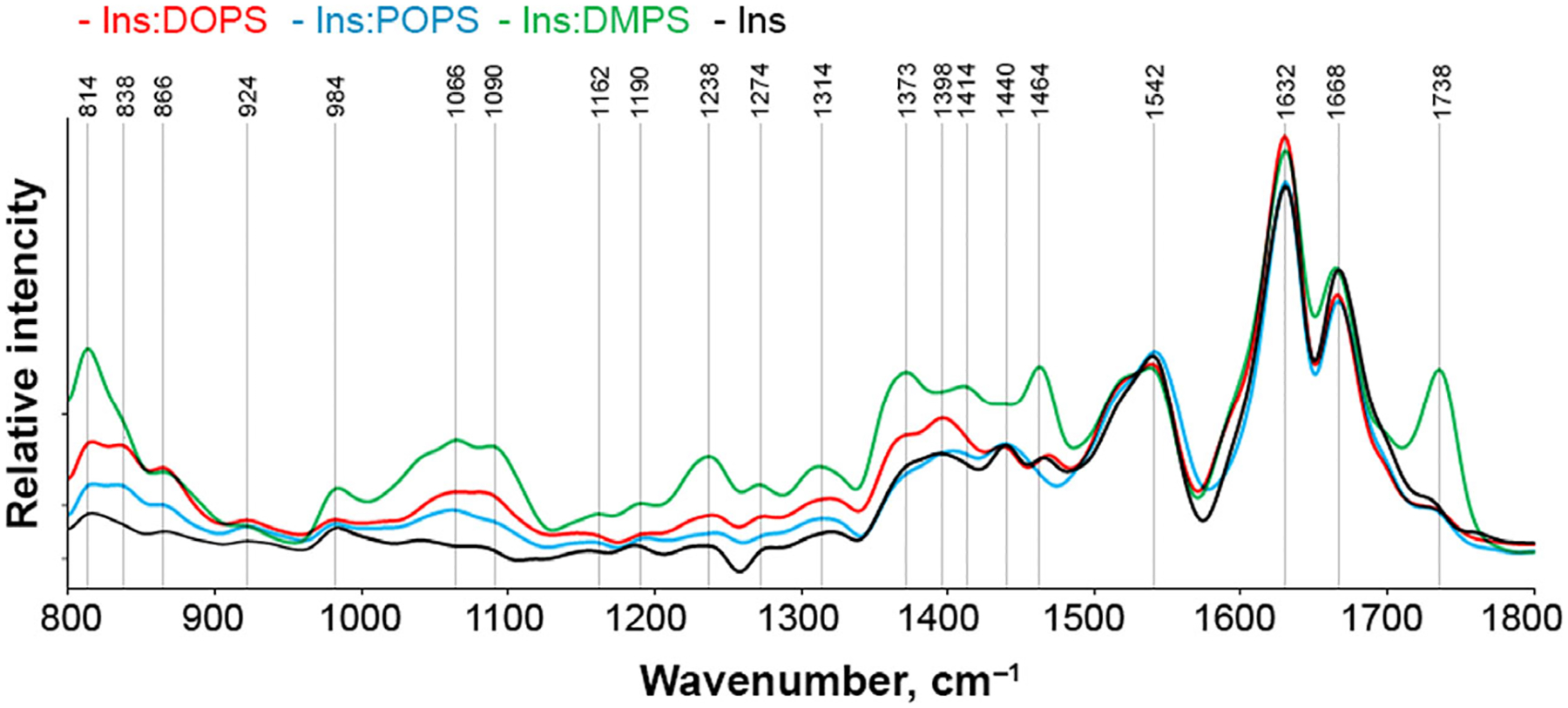
Nanoscale analysis of lipid content of insulin aggregates. Averaged AFM-IR spectra of insulin aggregates (Ins) and Ins:DMPS, Ins: POPS, and Ins:DOPS. Amide I and II bands represent protein secondary structure, whereas L1-L3 bands demonstrate phospholipid vibrations. After 24 h of incubation of insulin (400 μM) with and without lipids at 37 °C under 510 rpm, sample aliquots were diluted with 1×PBS pH 3.0 and deposited onto precleaned silicon wafer. AFM-IR analysis was performed in contact mode. At least 30–40 individual aggregates were analyzed for each sample.
Toxicity of insulin aggregates
Amyloid aggregates exert toxicities by enhancing ROS production and inducing mitochondrial dysfunction in cells [4,9]. To determine the extent to which Ins: DMPS, Ins:POPS, and Ins:DOPS engage ROS production and impair mitochondrial activity, we exposed these aggregates, as well as Ins to mice midbrain N27 cell line, Fig. 5.
Fig. 5.
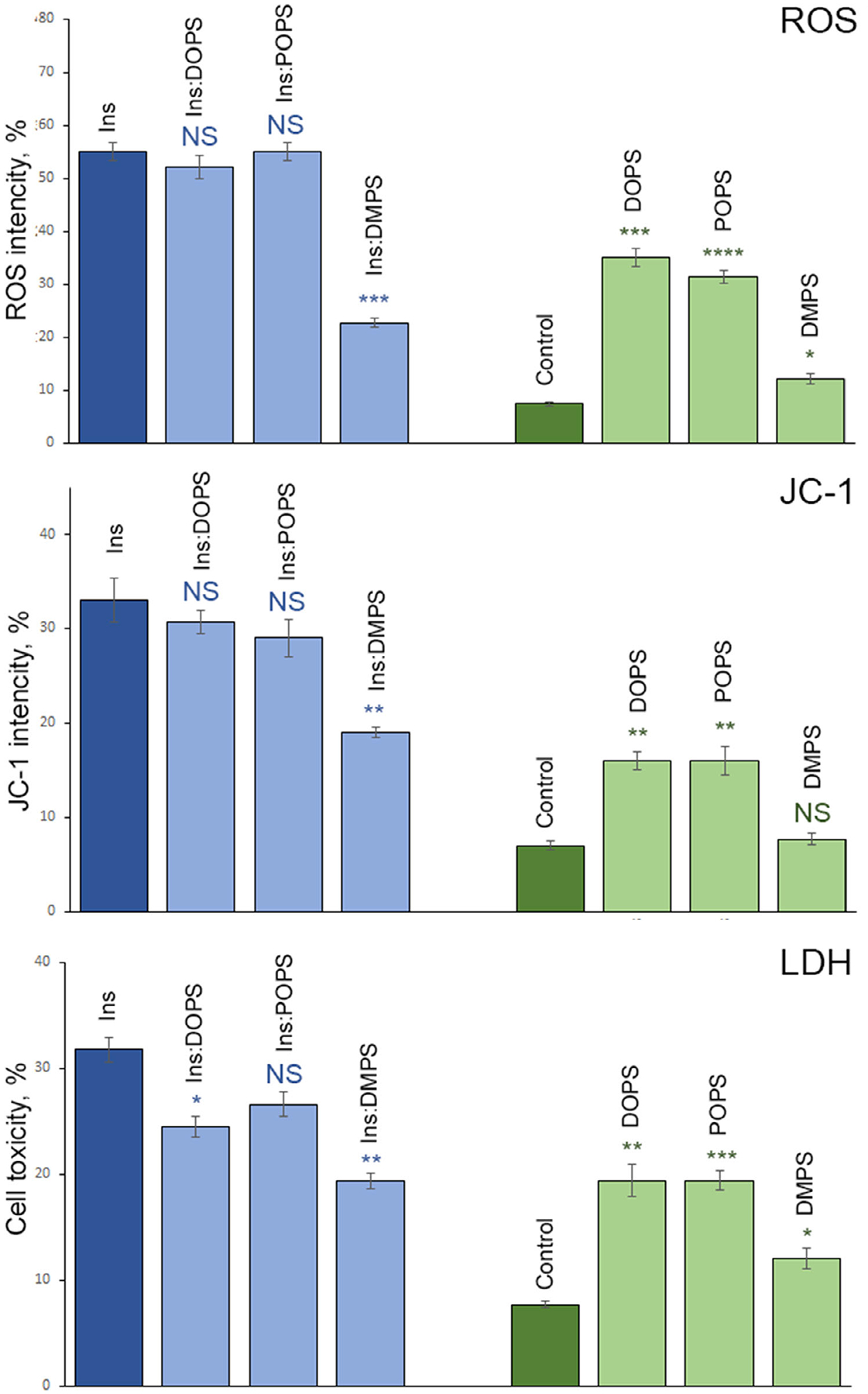
Insulin aggregates grown in the presence of lipids possess lower cell toxicity comparing to the aggregates grown in the lipid-free environment. Histograms of ROS (top), JC-1 (middle), and LDH (bottom) toxicity assays of Ins, Ins:DOPS, Ins:POPS, and Ins:DMPS, as we as DOPS, POPS, and DMPS lipids. After 24 h of incubation of insulin (400 μM) with and without lipids at 37 °C under 510 rpm, sample triplicates were exposed to mice midbrain N27 cells for 48 h. For each of the presented results, three independent measurements were made.
ROS test showed that Ins:POPS and Ins:DOPS engaged similar to Ins levels of ROS in N27 cell line. We also found no significant difference in the ROS levels induced by Ins:POPS and Ins:DOPS. At the same time, ROS levels produced by both Ins:POPS and Ins:DOPS were significantly lower ROS activity of Ins:DMPS. Based on these results, we can conclude that insulin aggregates with saturated and unsaturated FAs exert drastically different ROS activity in cells, whereas the degree of unsaturation of FAs in PS (Ins: POPS vs Ins:DOPS) does not alter ROS levels produced by such lipid:protein aggregates. It should be noted that ROS levels exhibited by such protein:lipid aggregates are significantly greater than the free radical activity exerted by the corresponding lipids. Finally, we found that although DMPS was not engaging any significant ROS activity in N27 cell line, POPS and DOPS lipids induced significantly higher ROS levels than DMPS.
Utilization of JC-1 dye allows for examination of the mitochondrial dysfunction caused by protein and protein:lipid aggregates. We found that Ins:POPS and Ins:DOPS exhibit similar degree of the mitochondrial dysfunction comparing to the insulin aggregates grown in the lipid-free environment. However, Ins:DMPS caused significantly lower mitochondrial dysfunction than Ins:POPS and Ins:DOPS. Our results also show that unsaturated lipids themselves (POPS and DOPS) cause higher mitochondrial dysfunction than the saturated phospholipid (DMPS).
We also investigated the extent to which cell viability is affected by Ins, Ins:DMPS, Ins:POPS, and Ins: DOPS, Fig. 5. Our results show that Ins:DMPS are least toxic to cells, whereas toxicity of Ins:POPS and Ins are similar. Finally, our results revealed significant differences between the toxicities caused by Ins and Ins:DOPS aggregates. Similar to the outcomes of ROS and JC-1 assays, we found that DOPS and POPS are significantly higher to cell than DMPS, whereas the last one also exerted small cell toxicity. These results show that that insulin aggregates with saturated and unsaturated FAs exert drastically different cell toxicity, which in turn is determined by the degree of unsaturation of FAs in the corresponding phospholipid.
Conclusions
To summarize, our results show although DMPS drastically accelerates insulin aggregation, it substantially lowers the toxicity, ROS levels, and the degree of mitochondrial dysfunction exerted by the corresponding protein:lipid aggregates that are formed in the presence of this phospholipid. DOPS and POPS can also alter the rates of protein aggregation, accelerating, and decelerating fibril formation, respectively. Similar to DMPS, these unsaturated phospholipids uniquely modify the toxicity of insulin aggregates that were formed in their presence. We infer that such a striking difference in toxicity of Ins:DMPS, Ins:POPS, and Ins:DMPS is determined by the chemical nature and the amount of corresponding phospholipids in their structure. Finally, we found that DMPS, POPS, and DMPS uniquely alter the topology of insulin aggregates reducing the length of fibrils (DMPS) or yielding oligomer-like structures (POPS and DOPS). These results suggest that PS may play an important role in stability of amyloidogenic proteins present on or in lipid bilayers.
Materials and methods
Materials
Bovine insulin was purchased from Sigma-Aldrich (St. Louis, MO, USA), 1,2-dioleoyl-sn-glycero-3-phospho-L-serine (DOPS), 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-L-serine (POPS), and 1,2-dimyristoyl-sn-glycero-3-phospho-L-serine (DMPS) were purchased from Avanti (Alabaster, AL, USA).
Liposome preparation
DMPS, POPS, and DOPS large unilamellar vesicles (LUVs) were prepared accordingly to the method reported by Galvagnion et al. [51]. Briefly, 0.6 mg of the lipid was dissolved in 2.6 mL of phosphate-buffered saline (PBS) pH 7.4. Lipid solutions were heated in water bath to ~ 50 °C for 30 min and then placed into liquid nitrogen for 3–5 min. This procedure was repeated 10 times. After this, lipid solutions were passed 15 times through a 100-nm membrane that was placed into the extruder (Avanti). LUV sizes were determined by dynamic light scattering.
Insulin aggregation
In the lipid-free environment, 400 μM of insulin was dissolved in PBS; solution pH was adjusted to pH 3.0 using concentrated HCl. For Ins:DMPS, Ins:POPS and Ins:DOPS, 400 μM of insulin was mixed with an equivalent concentration of the corresponding lipid; solution pH was adjusted to pH 3.0 using concentrated HCl. Next, the solutions were placed in the plate reader (Tecan, Männedorf, Switzerland) and incubated at 37 °C under 510 rpm for 24 h.
Kinetic measurements
Insulin aggregation was monitored using thioflavin T (ThT) fluorescence assay. Briefly, protein samples were mixed with 2 mM of ThT solution and placed in the plate reader (Tecan) where samples were incubated at 37 °C under 510 rpm for 30 h. Fluorescence measurements were taken every 10 min.
AFM imaging
Atomic force microscopy imaging was performed using silicon AFM probes with related parameters force constant 2.7 N·m−1 and resonance frequency 50–80 kHz were purchased from Appnano (Mountain View, CA, USA) on AIST-NT-HORIBA system (Edison, NJ, USA). Analysis of collected images was performed using AIST-NT software.
AFM-IR
Atomic force microscopy infrared spectroscopy imaging was conducted using a Nano-IR3 system (Bruker, Santa Barbara, CA, USA). The IR source was a QCL laser. Contact-mode AFM tips (ContGB-G AFM probe, NanoAndMore, Watsonville, CA, USA) were used to obtain all spectra and maps. Treatment and analysis of collected spectra were performed in matlab (The Mathworks, Inc. Natick, MA, USA).
Attenuated total reflectance-Fourier-transform Infrared (ATR-FTIR) spectroscopy
An aliquot of the protein sample was placed onto ATR crystal and dried at room temperature. Spectra were measured using Spectrum 100 FTIR spectrometer (Perkin-Elmer, Waltham, MA, USA). Three spectra were collected from each sample and averaged using Thermo Grams Suite software (Thermo Fisher Scientific, Waltham, MA, USA).
Cell toxicity assays
Mice midbrain N27 cells were grown in RPMI 1640 Medium (Thermo Fisher Scientific) with 10% fetal bovine serum (FBS) (Invitrogen, Waltham, MA, USA) in 96 well-plate (5000 cells per well) at 37 °C under 5% CO2. After 24 h, the cells were found to fully adhere to the wells reaching ~ 70% confluency. Next, 100 μL of the cell culture was replaced with 100 μL RPMI 1640 Medium with 5% FBS containing protein samples. After 48 h of incubation, lactate dehydrogenase (LDH) assay was performed on the cell medium using CytoTox 96 nonradioactive cytotoxicity assay (G1781, Promega, Madison, WI, USA). Absorption measurements were made in plate reader (Tecan) at 490 nm. Every well was measured 25 times in different locations.
In parallel, reactive oxygen species (ROS) assay was performed using the same cell culture. Briefly, ROS reagent (C10422, Invitrogen) was added to reach the final concentration of 5 μM and incubated at 37 °C under 5% CO2 for 30 min. After the supernatant was removed, cells were washed with PBS and resuspended in 200 μL of PBS in the flow cytometry tubes. Sample measurements were made in Accuri C6 Flow Cytometer (BD, San Jose, CA, USA) using red channel (λ = 633 nm). Percentages of ROS cells was determined using Acura software.
For JC-1 staining, 1 μL of JC-1 reagent (M34152A, Invitrogen) was added to cells and incubated at 37 °C under 5% CO2 for 30 min. After the supernatant was removed, cells were washed with PBS and resuspended in 200 μL of PBS in the flow cytometry tubes. Sample measurements were made in Accuri C6 Flow Cytometer (BD, San Jose, CA, USA) using red channel (λ = 633 nm). Percentages of ROS cells were determined using accuri software.
Supplementary Material
Acknowledgement
We are grateful to the National Institute of Health for the provided financial support (R35GM142869).
Abbreviations
- AFM-IR
atomic force microscopy infrared spectroscopy
- ATR-FTIR
attenuated total reflectance-Fourier transform infrared spectroscopy
- CL
cardiolipin
- DMPS
1,2-dimyristoyl-sn-glycero-3-phospho-L-serine
- DOPS
1,2-dioleoyl-sn-glycero-3-phospho-L-serine
- FA
fatty acid
- HS-AFM
high-speed atomic force microscopy
- IAPP
islet amyloid precursor protein
- LDH
lactate dehydrogenase
- LUV
large unilamellar vesicle
- POPS
1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-L-serine’
- PS
phosphatidylserine
- ROS
reactive oxygen species
- TERS
tip-enhanced Raman spectroscopy
- ThT
thioflavin T
- α-Syn
α-Synuclein
Footnotes
Supporting information
Additional supporting information may be found online in the Supporting Information section at the end of the article.
Data accessibility
The data that support the findings of this study are included in the figures of this article, and any additional data are available from the corresponding author dkurouski@tamu.edu upon request.
References
- 1.Chiti F, Dobson CM. Protein misfolding, amyloid formation, and human disease: a summary of progress over the last decade. Annu Rev Biochem. 2017;86:27–68. [DOI] [PubMed] [Google Scholar]
- 2.Knowles TP, Vendruscolo M, Dobson CM. The amyloid state and its association with protein misfolding diseases. Nat Rev Mol Cell Biol. 2014;15:384–96. [DOI] [PubMed] [Google Scholar]
- 3.Iadanza MG, Jackson MP, Hewitt EW, Ranson NA, Radford SE. A new era for understanding amyloid structures and disease. Nat Rev Mol Cell Biol. 2018;19:755–73. [DOI] [PubMed] [Google Scholar]
- 4.Chen SW, Drakulic S, Deas E, Ouberai M, Aprile FA, Arranz R, et al. Structural characterization of toxic oligomers that are kinetically trapped during alpha-synuclein fibril formation. Proc Natl Acad Sci USA. 2015;112:E1994–2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dou T, Zhou L, Kurouski D. Unravelling the structural organization of individual alpha-synuclein oligomers grown in the presence of phospholipids. J Phys Chem Lett. 2021;12:4407–14. [DOI] [PubMed] [Google Scholar]
- 6.Li B, Ge P, Murray KA, Sheth P, Zhang M, Nair G, et al. Cryo-EM of full-length alpha-synuclein reveals fibril polymorphs with a common structural kernel. Nat Commun. 2018;9:3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guerrero-Ferreira R, Taylor NM, Mona D, Ringler P, Lauer ME, Riek R, et al. Cryo-EM structure of alpha-synuclein fibrils. Elife. 2018;7:e36402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tycko R Solid-state NMR studies of amyloid fibril structure. Annu Rev Phys Chem. 2011;62:279–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cataldi R, Chia S, Pisani K, Ruggeri FS, Xu CK, Šneideris T, et al. A dopamine metabolite stabilizes neurotoxic amyloid-beta oligomers. Commun Biol. 2021;4:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vosough F, Barth A. Characterization of homogeneous and heterogeneous amyloid-beta42 oligomer preparations with biochemical methods and infrared spectroscopy reveals a correlation between infrared spectrum and oligomer size. ACS Chem Neurosci. 2021;12:473–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jinsmaa Y, Sullivan P, Sharabi Y, Goldstein DS. DOPAL is transmissible to and oligomerizes alpha-synuclein in human glial cells. Auton Neurosci. 2016;194:46–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Banerjee S, Sun Z, Hayden EY, Teplow DB, Lyubchenko YL. Nanoscale dynamics of amyloid beta-42 oligomers as revealed by high-speed atomic force microscopy. ACS Nano. 2017;11:12202–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Watanabe-Nakayama T, Ono K, Itami M, Takahashi R, Teplow DB, Yamada M. High-speed atomic force microscopy reveals structural dynamics of amyloid beta1-42 aggregates. Proc Natl Acad Sci USA. 2016;113:5835–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rizevsky S, Kurouski D. Nanoscale structural organization of insulin fibril polymorphs revealed by atomic force microscopy-infrared spectroscopy (AFMIR). ChemBioChem. 2020;21:481–5. [DOI] [PubMed] [Google Scholar]
- 15.Ruggeri FS, Charmet J, Kartanas T, Peter Q, Chia S, Habchi J, et al. Microfluidic deposition for resolving single-molecule protein architecture and heterogeneity. Nat Commun. 2018;9:3890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ruggeri FS, Flagmeier P, Kumita JR, Meisl G, Chirgadze DY, Bongiovanni MN, et al. The influence of pathogenic mutations in alpha-synuclein on biophysical and structural characteristics of amyloid fibrils. ACS Nano. 2020;14:5213–22. [DOI] [PubMed] [Google Scholar]
- 17.Ruggeri FS, Longo G, Faggiano S, Lipiec E, Pastore A, Dietler G. Infrared nanospectroscopy characterization of oligomeric and fibrillar aggregates during amyloid formation. Nat Commun. 2015;6:7831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou L, Kurouski D. Structural characterization of individual alpha-synuclein oligomers formed at different stages of protein aggregation by atomic force microscopy-infrared spectroscopy. Anal Chem. 2020;92:6806–10. [DOI] [PubMed] [Google Scholar]
- 19.Deckert-Gaudig T, Kämmer E, Deckert V. Tracking of nanoscale structural variations on a single amyloid fibril tip-enhanced Raman scattering. J Biophotonics. 2012;5:215–9. [DOI] [PubMed] [Google Scholar]
- 20.Krasnoslobodtsev AV, Deckert-Gaudig T, Zhang Y, Deckert V, Lyubchenko YL. Polymorphism of amyloid fibrils formed by a peptide from the yeast prion protein Sup35: AFM and Tip-Enhanced Raman Scattering studies. Ultramicroscopy. 2016;165:26–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kurouski D, Deckert-Gaudig T, Deckert V, Lednev IK. Structure and composition of insulin fibril surfaces probed by TERS. J Am Chem Soc. 2012;134:13323–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kurouski D, Deckert-Gaudig T, Deckert V, Lednev IK. Surface characterization of insulin protofilaments and fibril polymorphs using tip-enhanced Raman spectroscopy (TERS). Biophys J. 2014;106:263–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lipiec E, Perez-Guaita D, Kaderli J, Wood BR, Zenobi R. Direct nanospectroscopic verification of the amyloid aggregation pathway. Angew Chem Int Ed Engl. 2018;57:8519–24. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Y, Hashemi M, Lv Z, Williams B, Popov KI, Dokholyan NV, et al. High-speed atomic force microscopy reveals structural dynamics of alpha-synuclein monomers and dimers. J Chem Phys. 2018;148:123322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kakinen A, Xing Y, Hegoda Arachchi N, Javed I, Feng L, Faridi A, et al. Single-molecular heteroamyloidosis of human islet amyloid polypeptide. Nano Lett. 2019;19:6535–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alza NP, Iglesias Gonzalez PA, Conde MA, Uranga RM, Salvador GA. Lipids at the crossroad of alpha-synuclein function and dysfunction: biological and pathological implications. Front Cell Neurosci. 2019;13:175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Galvagnion C The role of lipids interacting with α-synuclein in the pathogenesis of Parkinson’s disease. J Parkins Dis. 2017;7:433–50. [DOI] [PubMed] [Google Scholar]
- 28.Galvagnion C, Brown JW, Ouberai MM, Flagmeier P, Vendruscolo M, Buell AK, et al. Chemical properties of lipids strongly affect the kinetics of the membrane-induced aggregation of alpha-synuclein. Proc Natl Acad Sci USA. 2016;113:7065–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Avdulov NA, Chochina SV, Igbavboa U, Warden CS, Vassiliev AV, Wood WG. Lipid binding to amyloid beta-peptide aggregates: preferential binding of cholesterol as compared with phosphatidylcholine and fatty acids. J Neurochem. 1997;69:1746–52. [DOI] [PubMed] [Google Scholar]
- 30.Rizevsky S, Matveyenka M, Kurouski D. Nanoscale structural analysis of a lipid-driven aggregation of insulin. J Phys Chem Lett. 2022;13:2467–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fitzner D, Bader JM, Penkert H, Bergner CG, Su M, Weil MT, et al. Cell-type- and brain-region-resolved mouse brain lipidome. Cell Rep. 2020;32:108132. [DOI] [PubMed] [Google Scholar]
- 32.Alecu I, Bennett SAL. Dysregulated lipid metabolism and its role in alpha-synucleinopathy in Parkinson’s disease. Front Neurosci. 2019;13:328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Levental I, Levental KR, Heberle FA. Lipid rafts: controversies resolved, mysteries remain. Trends Cell Biol. 2020;30:341–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.D’Souza A, Theis JD, Vrana JA, Buadi F, Dispenzieri A, Dogan A. Localized insulin-derived amyloidosis: a potential pitfall in the diagnosis of systemic amyloidosis by fat aspirate. Am J Hematol. 2012;87:E131–2. [DOI] [PubMed] [Google Scholar]
- 35.Gupta Y, Singla G, Singla R. Insulin-derived amyloidosis. Indian J Endocrinol Metab. 2015;19:174–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shikama Y, Kitazawa J, Yagihashi N, Uehara O, Murata Y, Yajima N, et al. Localized amyloidosis at the site of repeated insulin injection in a diabetic patient. Intern Med. 2010;49:397–401. [DOI] [PubMed] [Google Scholar]
- 37.Iwaya K, Zako T, Fukunaga J, Sӧrgjerd KM, Ogata K, Kogure K, et al. Toxicity of insulin-derived amyloidosis: a case report. BMC Endocr Dis. 1991;19:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kiechle M, Grozdanov V, Danzer KM. The role of lipids in the initiation of alpha-synuclein misfolding. Front Cell Dev Biol. 2020;8:562241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Michaelson DM, Barkai G, Barenholz Y. Asymmetry of lipid organization in cholinergic synaptic vesicle membranes. Biochem J. 1983;211:155–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Meer G, Voelker DR, Feigenson GW. Membrane lipids: where they are and how they behave. Nat Rev Mol Cell Biol. 2008;9:112–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kurouski D, Lombardi RA, Dukor RK, Lednev IK, Nafie LA. Direct observation and pH control of reversed supramolecular chirality in insulin fibrils by vibrational circular dichroism. Chem Commun. 2010;46:7154–6. [DOI] [PubMed] [Google Scholar]
- 42.Sarroukh R, Goormaghtigh E, Ruysschaert JM, Raussens V. ATR-FTIR: a “rejuvenated” tool to investigate amyloid proteins. Biochim Biophys Acta. 2013;1828:2328–38. [DOI] [PubMed] [Google Scholar]
- 43.Colthup NB, Daly LH, Wiberley SE. Introduction to infrared and raman spectroscopy. Waltham, MA: Academic Press; 1990. [Google Scholar]
- 44.Farber C, Wang R, Chemelewski R, Mullet J, Kurouski D. Nanoscale structural organization of plant epicuticular wax probed by atomic force microscope infrared spectroscopy. Anal Chem. 2019;91:2472–9. [DOI] [PubMed] [Google Scholar]
- 45.Dazzi A, Glotin F, Carminati R. Theory of infrared nanospectroscopy by photothermal induced resonance. J Appl Phys. 2010;107:124519. [Google Scholar]
- 46.Dazzi A, Prater CB. AFM-IR: technology and applications in nanoscale infrared spectroscopy and chemical imaging. Chem Rev. 2017;117:5146–73. [DOI] [PubMed] [Google Scholar]
- 47.Kurouski D, Dazzi A, Zenobi R, Centrone A. Infrared and Raman chemical imaging and spectroscopy at the nanoscale. Chem Soc Rev. 2020;49:3315–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Katzenmeyer AM, Aksyuk V, Centrone A. Nanoscale infrared spectroscopy: improving the spectral range of the photothermal induced resonance technique. Anal Chem. 2013;85:1972–9. [DOI] [PubMed] [Google Scholar]
- 49.Katzenmeyer AM, Holland G, Kjoller K, Centrone A. Absorption spectroscopy and imaging from the visible through mid-infrared with 20 nm resolution. Anal Chem. 2015;87:3154–9. [DOI] [PubMed] [Google Scholar]
- 50.Dou T, Li Z, Zhang J, Evilevitch A, Kurouski D. Nanoscale structural characterization of individual viral particles using Atomic Force Microscopy Infrared Spectroscopy (AFM-IR) and Tip-Enhanced Raman Spectroscopy (TERS). Anal Chem. 2020;92:11297–304. [DOI] [PubMed] [Google Scholar]
- 51.Galvagnion C, Buell AK, Meisl G, Michaels TCT, Vendruscolo M, Knowles TPJ, et al. Lipid vesicles trigger α-synuclein aggregation by stimulating primary nucleation. Nat Chem Biol. 2015;11:229–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are included in the figures of this article, and any additional data are available from the corresponding author dkurouski@tamu.edu upon request.


