Abstract
White-Sutton syndrome (WHSUS), which is caused by heterozygous pathogenic variants in POGZ, is characterized by a spectrum of intellectual disabilities and global developmental delay with or without features of autism spectrum disorder. Additional features may include hypotonia, behavioral abnormalities, ophthalmic abnormalities, hearing loss, sleep apnea, microcephaly, dysmorphic facial features, and rarely, congenital diaphragmatic hernia (CDH). We present a 6-year-old female with features of WHSUS, including CDH, but with non-diagnostic clinical trio exome sequencing. Exome sequencing reanalysis revealed a heterozygous, de novo, intronic variant in POGZ (NM_015100.3:c.2546–20T>A). RNA sequencing revealed that this intronic variant leads to skipping of exon 18. This exon skipping event results in a frameshift with a predicted premature stop codon in the last exon and escape from nonsense-mediated mRNA decay (NMD). To our knowledge, this case is the first case of WHSUS caused by a de novo, intronic variant that is not near a canonical splice site within POGZ. These findings emphasize the limitations of standard clinical exome filtering algorithms and the importance of research reanalysis of exome data together with RNA sequencing to confirm a suspected diagnosis of WHSUS. As the sixth reported case of CDH with heterozygous pathogenic variants in POGZ and features consistent with WHSUS, this report supports the conclusion that WHSUS should be considered in the differential diagnosis for patients with syndromic CDH.
Keywords: Congenital diaphragmatic hernia, RNA sequencing, genome sequencing, splicing
INTRODUCTION
Heterozygous pathogenic variants in POGZ cause White-Sutton syndrome (WHSUS, OMIM: #616364), a disorder characterized by a wide spectrum of intellectual disabilities and global developmental delay with or without features of autism spectrum disorder (Assia Batzir et al., 2020; Garde et al., 2021; Stessman et al., 2016; White et al., 2016; Ye et al., 2015). Additional reported phenotypic features of WHSUS include hypotonia, behavioral abnormalities, ophthalmic abnormalities, variable hearing loss, sleep apnea, microcephaly, and dysmorphic facial features (Assia Batzir et al., 2020; Stessman et al., 2016). These dysmorphic facial features may include broad forehead, midface hypoplasia, triangular mouth, and a broad and flat nasal bridge (Assia Batzir et al., 2020; Stessman et al., 2016). Other phenotypic features may include abnormal brain imaging, seizures, gait abnormalities, brachydactyly, and gastrointestinal abnormalities (Assia Batzir et al., 2020; Ferretti et al., 2019). Congenital diaphragmatic hernia (CDH) has been described in five individuals with heterozygous pathogenic variants in POGZ and features consistent with WHSUS (Assia Batzir et al., 2020; Garde et al., 2021; Longoni et al., 2017; Murch et al., 2021; White et al., 2016).
Heterozygous missense, nonsense, and frameshift variants in POGZ have been associated with WHSUS (Assia Batzir et al., 2020; Stessman et al., 2016). To date, the only non-coding variants that have been reported in association with this diagnosis affect canonical splice sites or are in close proximity to canonical splice sites (Assia Batzir et al., 2020; Garde et al., 2021; Stessman et al., 2016). The nonsense, frameshift, and splice variants are distributed throughout the gene with a large number of pathogenic variants located in the last exon (Assia Batzir et al., 2020). Although few studies have evaluated these variants at an RNA or protein level (Garde et al., 2021; Tan et al., 2016), some of these variants are predicted to lead to nonsense-mediated mRNA decay (NMD) whereas others are predicted (or in a single case, demonstrated) to escape NMD and to produce a truncated protein (Assia Batzir et al., 2020; Garde et al., 2021; Tan et al., 2016). However, no unequivocal genotype-phenotype correlations have been established (Assia Batzir et al., 2020).
Here we present a 6-year-old female with typical phenotypic features of WHSUS including CDH in the setting of a non-diagnostic evaluation, including clinical trio exome sequencing. Re-analysis of her exome sequencing data within the Undiagnosed Diseases Network (UDN) identified a heterozygous, de novo, intronic variant in POGZ (NM_015100.3:c.2546–20T>A). RNA sequencing in her fibroblasts demonstrated that this intronic variant leads to skipping of exon 18, which causes a frameshift and predicted premature stop codon in the last exon of POGZ. Despite this exon-skipping event, the levels of POGZ expression in her fibroblasts were equivalent to controls suggesting that the mutant transcript escapes NMD as predicted. Overall, our patient represents the first example of WHSUS caused by a de novo, intronic variant that is not near a canonical splice site in POGZ and thus could be missed by standard clinical exome sequencing analysis.
METHODS
Editorial Policies and Ethical Considerations.
This study was approved by the Institutional Review Boards (IRBs) at the National Institutes of Health and Baylor College of Medicine (BCM). The patient and both parents were enrolled in the Undiagnosed Diseases Network (UDN) site at BCM. Informed consent was obtained prior to initiating any research procedures, and the family provided consent for the publication of photographs in this report.
Trio Exome Sequencing.
Clinical trio exome sequencing was performed as previously described (Yang et al., 2014). The clinical trio exome sequencing data were transferred to the UDN for research analysis. For the research analysis, all novel de novo variants and biallelic variants with a minor allele frequency of <0.01% were prioritized for review.
RNA Sequencing.
A skin biopsy was performed on the patient and used to establish a fibroblast cell line with standard protocols. RNA was isolated from the fibroblasts, and RNA sequencing was performed as previously described (Murdock et al., 2021). Aberrant gene expression and splicing were identified using a workflow based on the Detection of RNA Outlier Pipeline (DROP) and custom scripts as previously described (Murdock et al., 2021; Yepez et al., 2021). Splice AI (Jaganathan et al., 2019) and Alamut software (Interactive Biosoftware, Rouen, France) were also used to evaluate the potential impact of identified intronic variants.
RESULTS
Clinical Presentation.
The proband was enrolled in the UDN at age 4 years 10 months with a history of microcephaly, global developmental delay, sensorineural hearing loss, CDH, chronic lung disease, skeletal anomalies including mild hip dysplasia, and dysmorphic facial features. She was born at 34 weeks gestation due to severe polyhydramnios and prolonged premature rupture of membranes. Her birth parameters included weight of 1555 grams (Z = −1.44), birth length of 40.7 cm (Z = −1.24), and head circumference of 29 cm (Z = −1.10) on the Fenton growth chart (Chou, Roumiantsev, & Singh, 2020; Fenton & Kim, 2013). A right-sided Bochdalek-type CDH, with liver and bowel and in the right hemithorax, was diagnosed shortly after delivery and was surgically repaired. CDH-associated intestinal malrotation was present. She had a history of failure to thrive and dysphagia as an infant. She ultimately had a gastrostomy tube placed that resulted in improved weight gain and growth. She also had multiple admissions for respiratory illness, and she was eventually diagnosed with recurrent pneumonia and severe obstructive and central sleep apnea.
Global developmental delay was recognized during infancy with gross motor and language skills at the 5 month and 6 month level, respectively, at a corrected age of 13 months. Walking was achieved at 26 months and language skills were limited to single words at age 4 years. At age 3 years, she was noted to have abnormal movements that were diagnosed as stereotypies following neurological evaluation and lack of epileptic features on EEG. A brain MRI showed mild volume loss in the central white matter, midbrain and cerebellar vermis with a simplified gyral pattern in the superior frontal region. Ophthalmic examination was significant for amblyopia, exotropia, myopia, and astigmatism. Audiology examination showed minimal to mild bilateral sensorineural hearing loss.
At the time of the UDN evaluation, she was age 4 years 10 months (Figure 1A). She was continuing to make developmental progress. Her weight was 17 kg (Z = −0.38), her height was at 97 cm (Z = −2.48), and her head circumference was 45.5 cm (Z = −3.95). Her physical examination was notable for microbrachycephaly, sparse hair, and dysmorphic craniofacial features (Figure 1A). Several mild skeletal anomalies were noted on physical examination including a broad anterior chest, kyphotic posture, brachydactyly, sacral dimple, and narrow calcaneous. Her gait was unsteady for age. A skeletal survey performed as part of her UDN evaluation showed brachycephaly, T12 hypoplasia, thoracolumbar kyphosis, chronic bilateral radiocapitellar dislocation, bowing of the radius, hypoplasia of the second and fifth finger middle phalanges, and cone-shaped epiphyses in several distal phalanges of the hands.
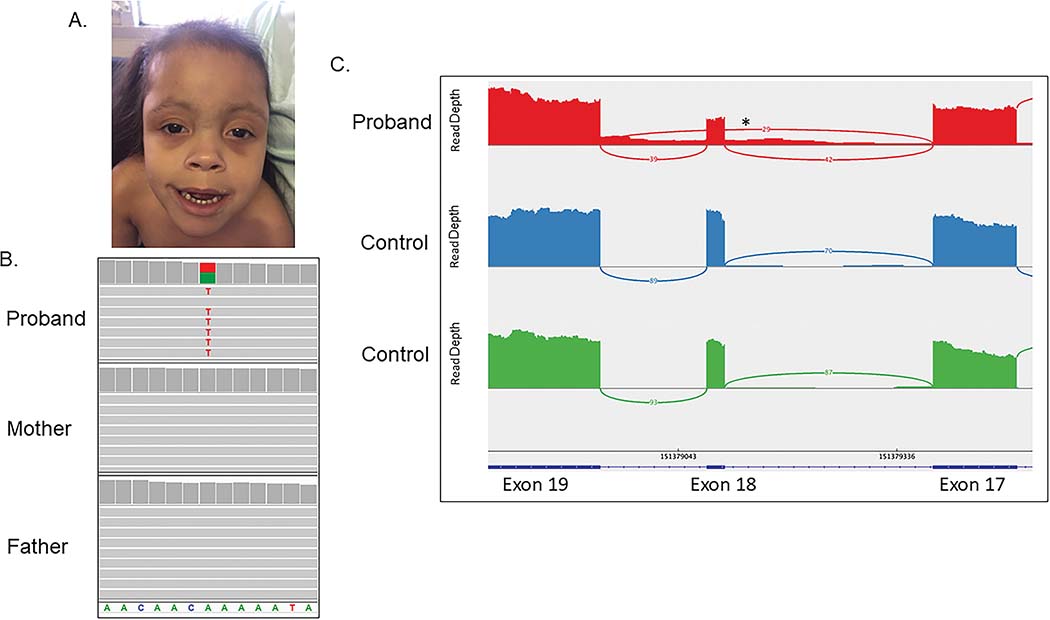
Figure 1. An intronic variant in POGZ impacts splicing and causes WHSUS with CDH.
A) Photograph of patient at 4 years 10 months of age shows a high forehead, bitemporal narrowing, sparse hair, sparse eyebrows with synophrys, a flat nasal root, large anteverted nares, a long philtrum, and a cupid’s bow appearance to her upper lip.
B) Exome sequencing demonstrated that the NM_015100.3:c.2546–20T>A variant in POGZ is present in the patient but not in either parent. C) Sequencing of RNA isolated from her fibroblasts demonstrates exon 18 skipping in 42% of reads and demonstrates low level retention of intron 17. * indicates approximate location of the c.2546–20T>A variant in intron 17.
At the most recent evaluation at age 6 years 9 months she continued to demonstrate delays across all domains. Concerning behaviors emerged with self-injury, aggression, anxiety, and tantrums. She is dependent on gastrostomy tube feedings for nutrition due to oral dysphagia. Due to severe obstructive sleep apnea and BiPAP intolerance, she is prescribed oxygen at night via nasal cannula with overnight oximetry. She is being treated for cyclic vomiting and has frequent gastrointestinal symptoms. Although recurrent infections were frequent as a younger child, these were less frequent with social distancing measures. Immunological evaluation showed nonproductive antibody titers to Diphtheria, Tetanus, and Pneumococcus.
Trio Exome Sequencing.
Clinical trio exome sequencing and a chromosome microarray (CMA, Baylor Genetics, HR+SNP, v10.2) were non-diagnostic. A reanalysis of the clinical trio exome data through the UDN revealed an intronic, de novo variant in POGZ that was 20 base pairs away from the nearest exon-intron boundary (NM_015100.3:c.2546–20T>A, Figure 1B). This variant is located in intron 17, which is the second to last intron of POGZ. The variant is not found in gnomAD v2.1.1 (Karczewski et al., 2020). The CADD score was 14.46 (Rentzsch, Schubach, Shendure, & Kircher, 2021), and Splice AI reported a score of 0.54, which is consistent with a possible effect on splicing (Jaganathan et al., 2019). This variant is predicted to weaken the natural splice acceptor site according to Alamut scores (−14.9% for MaxEnt and −28% for GeneSplicer). Additional variants identified are summarized in Supplemental Table 1.
RNA Sequencing.
RNA sequencing from fibroblasts was performed as previously described (Murdock et al., 2021) and revealed an abnormal splicing pattern with skipping of exon 18 in approximately 42% of the reads. In addition, there was evidence of low-level retention of intron 17, suggesting that multiple splice isoforms may be produced by this de novo variant (Figure 1C). Skipping of exon 18 (c.2546_2570del) alters the reading frame, resulting in a frameshift and early stop codon in the last exon of the gene (p.Leu850Metfs*13) where it would likely escape NMD and produce a truncated protein. Consistent with NMD escape, there was no statistical difference in overall expression of POGZ in this sample compared to controls (p>0.05).
DISCUSSION
An increasing number of reports have demonstrated that the use of RNA sequencing can lead to a 7.5%−36% enhancement in genetic diagnosis (Cummings et al., 2017; Fresard et al., 2019; Gonorazky et al., 2019; Kremer et al., 2017; Lee et al., 2020; Maddirevula et al., 2020; Murdock et al., 2021). This improvement in diagnostic yield is due partly to the identification of intronic variants that affect splicing even though they are not within or near canonical splice sites. These types of intronic variants may not be identified by exome sequencing because sequence coverage decreases rapidly outside of exons. When they are identified by exome sequencing, they may be filtered out during the analysis step given that they are difficult to interpret in the absence of additional data.
In our case, research reanalysis of trio exome data within the UDN revealed a heterozygous, de novo variant in the second to last intron of POGZ (intron 17) that was filtered out of the clinical analysis given its location (−20) within the intron. In addition, RNA sequencing demonstrated skipping of exon 18 of POGZ in this patient’s fibroblasts, which is consistent with the predicted weakening of the natural splice acceptor site from the intronic variant. This exon-skipping event is predicted to cause a frameshift and results in a premature stop codon in the last exon of POGZ. In addition, low-level intronic retention was evident suggesting that multiple splice isoforms result from this variant. These new results combined with the phenotypic features in our patient including developmental delays, microcephaly, facial features, ophthalmologic abnormalities, hearing loss, and sleep apnea allowed us to secure both a clinical and a molecular diagnosis of WHSUS.
WHSUS is associated with wide phenotypic variability, and numerous variants in POGZ have been associated with the disorder (Assia Batzir et al., 2020). POGZ encodes a protein with multiple domains including a zinc finger cluster, an HP1-binding motif, a centromere protein-B-like DNA binding domain, and a transposase-derived DDE domain (Nozawa et al., 2010). This protein plays a role in chromatin remodeling and regulates gene expression networks during brain development and has also been shown to function in the progression of mitosis (Ibaraki et al., 2019; Matsumura et al., 2020; Nozawa et al., 2010). Missense variants, truncating variants, splice site variants, and indels in POGZ have been reported with WHSUS (Assia Batzir et al., 2020). Although there appears to be an increased number of truncating variants in exon 19, truncating variants in other exons have been reported (Assia Batzir et al., 2020). Thus, those variants that lead to premature stop codons are located throughout the gene. Some of these variants are expected to lead to NMD, and others are predicted to escape NMD and result in a truncated protein (White et al., 2016). However, to date, no definitive genotype-phenotype correlations have been identified (Assia Batzir et al., 2020). To our knowledge, this is the first use of RNA sequencing in WHSUS to demonstrate that a premature stop codon in the last exon of this gene leads to an escape from NMD as predicted, and likely results in a truncated protein, at least in fibroblasts. Interestingly, truncated protein has also been detected by western blot in lymphocytes from a patient with a premature stop codon in exon 9 (Tan et al., 2016), and likewise, although abnormalities of splicing were noted, NMD could not be confirmed in a blood sample from an individual with a variant in intron 9 at the −3 position (Garde et al., 2021). It is unclear if the resulting truncated protein produces the WHSUS phenotype as a result of a loss of function or dominant negative effect or a gain of function mechanism.
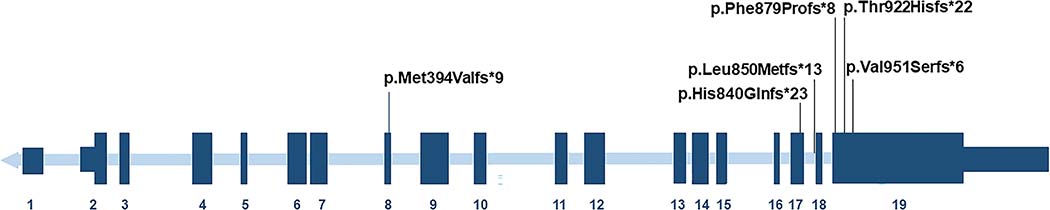
CDH is a rare but increasingly reported feature of WHSUS, our patient being the fifth such reported case of CDH in this disorder (Assia Batzir et al., 2020; Garde et al., 2021; Murch et al., 2021; White et al., 2016). One additional case was reported with a frameshift in POGZ and CDH in the setting of features reminiscent of WHSUS (Supplemental Table 2) (Longoni et al., 2017). All six patients have frameshift variants that result in premature stop codons, four of these stop codons are predicted to be located in exon 19, the last exon of the gene (Figure 2). Likewise, our patient has an intronic variant that leads to exon 18 skipping, a frameshift, and a premature stop codon that is also predicted to occur in the last exon of the gene. In contrast, the sixth patient has a premature stop codon in exon 8. Thus, both nonsense variants predicted to lead to NMD and nonsense variants predicted to lead to an escape from NMD have been reported in the setting of CDH. To date, there does not appear to be an obvious “hot spot” for variants in POGZ associated with CDH. Both Bochdalek and Morgagni-type CDH have been associated with loss-of-function variants in POGZ (Longoni et al., 2017). Two additional individuals with CDH have been reported with heterozygous inherited missense variants in POGZ, but it is unclear whether these two individuals had WHSUS (Longoni et al., 2017). Thus, WHSUS should be considered in the differential diagnosis of individuals who present with syndromic forms of CDH.
Figure 2. Pathogenic variants in POGZ associated with CDH are located throughout the gene.
The six variants in POGZ identified in individuals with WHSUS and CDH are shown. Of note, the variant described in our patient is in intron 17 and results in exon 18 skipping and a premature stop codon in the last exon of the gene.
Overall, this patient is the first example of WHSUS caused by a de novo, intronic variant that is not near a canonical splice site in POGZ and thus, had been missed by clinical exome sequencing and analysis. The use of RNA sequencing combined with rigorous reanalysis of clinical exome sequencing by the UDN revealed the diagnosis of WHSUS for this patient. She also represents the sixth case of CDH in individuals presenting with features of WHSUS. Therefore, our findings together with other previously reported cases demonstrate that WHSUS should be considered in the differential diagnosis for patients with syndromic CDH.
Supplementary Material
Supplemental Table 1. Rare variants identified in the research analysis of trio exome data.
Supplemental Table 2. Individuals with truncating variants in POGZ and congenital diaphragmatic hernia.
ACKNOWLEDGEMENTS.
The authors thank the family for participating in these research studies. Research reported in this manuscript was supported, in part, by the National Institutes of Health (NIH) Common Fund, through the Office of Strategic Coordination/Office of the NIH Director under U01HG007709 and U01HG007530 and the T32GM07526 Medical Genetics Research Fellowship Program. Research reported in this publication was also supported, in part, by the Eunice Kennedy Shriver National Institute of Child Health & Human Development R01HD098458 to D.A.S. and P50HD103555 for use of the Clinical Translational Core facilities at Baylor College of Medicine. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. Lindsay C. Burrage holds a Career Award for Medical Scientists from the Burroughs Wellcome Fund.
Funding sources: NIH 01HG007709, U01HG007530, R01HD098458, T32GM07526, and P50HD10355; Burroughs Wellcome Fund
Footnotes
CONFLICT OF INTEREST STATEMENT. The Department of Molecular and Human Genetics at Baylor College of Medicine receives revenue from clinical genetic testing completed at Baylor Genetics.
See supplementary material for full list of members
DATA AVAILABILITY.
The exome sequencing data have been deposited in dbGAP (phs001232.v4.p2), and the novel variant in POGZ described in this manuscript was deposited in Clinvar (SCV001736845.1). Moreover, details regarding the phenotype and genotype were deposited in Phenome Central (P0010291)).
REFERENCES
- Assia Batzir N, Posey JE, Song X, Akdemir ZC, Rosenfeld JA, Brown CW, . . . Sutton VR (2020). Phenotypic expansion of POGZ-related intellectual disability syndrome (White-Sutton syndrome). Am J Med Genet A, 182(1), 38–52. doi: 10.1002/ajmg.a.61380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou JH, Roumiantsev S, & Singh R (2020). PediTools Electronic Growth Chart Calculators: Applications in Clinical Care, Research, and Quality Improvement. J Med Internet Res, 22(1), e16204. doi: 10.2196/16204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings BB, Marshall JL, Tukiainen T, Lek M, Donkervoort S, Foley AR, . . . MacArthur DG (2017). Improving genetic diagnosis in Mendelian disease with transcriptome sequencing. Sci Transl Med, 9(386). doi: 10.1126/scitranslmed.aal5209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenton TR, & Kim JH (2013). A systematic review and meta-analysis to revise the Fenton growth chart for preterm infants. BMC Pediatr, 13, 59. doi: 10.1186/1471-2431-13-59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferretti A, Barresi S, Trivisano M, Ciolfi A, Dentici ML, Radio FC, . . . Specchio N (2019). POGZ-related epilepsy: Case report and review of the literature. Am J Med Genet A, 179(8), 1631–1636. doi: 10.1002/ajmg.a.61206 [DOI] [PubMed] [Google Scholar]
- Fresard L, Smail C, Ferraro NM, Teran NA, Li X, Smith KS, . . . Montgomery SB (2019). Identification of rare-disease genes using blood transcriptome sequencing and large control cohorts. Nat Med, 25(6), 911–919. doi: 10.1038/s41591-019-0457-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garde A, Cornaton J, Sorlin A, Moutton S, Nicolas C, Juif C, . . . Faivre L (2021). Neuropsychological study in 19 French patients with White-Sutton syndrome and POGZ mutations. Clin Genet, 99(3), 407–417. doi: 10.1111/cge.13894 [DOI] [PubMed] [Google Scholar]
- Gonorazky HD, Naumenko S, Ramani AK, Nelakuditi V, Mashouri P, Wang P, . . . Dowling JJ (2019). Expanding the Boundaries of RNA Sequencing as a Diagnostic Tool for Rare Mendelian Disease. Am J Hum Genet, 104(5), 1007. doi: 10.1016/j.ajhg.2019.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibaraki K, Hamada N, Iwamoto I, Ito H, Kawamura N, Morishita R, . . . Nagata KI (2019). Expression Analyses of POGZ, A Responsible Gene for Neurodevelopmental Disorders, during Mouse Brain Development. Dev Neurosci, 41(1–2), 139–148. doi: 10.1159/000502128 [DOI] [PubMed] [Google Scholar]
- Jaganathan K, Kyriazopoulou Panagiotopoulou S, McRae JF, Darbandi SF, Knowles D, Li YI, . . . Farh KK (2019). Predicting Splicing from Primary Sequence with Deep Learning. Cell, 176(3), 535–548 e524. doi: 10.1016/j.cell.2018.12.015 [DOI] [PubMed] [Google Scholar]
- Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alfoldi J, Wang Q, . . . MacArthur DG (2020). The mutational constraint spectrum quantified from variation in 141,456 humans. Nature, 581(7809), 434–443. doi: 10.1038/s41586-020-2308-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer LS, Bader DM, Mertes C, Kopajtich R, Pichler G, Iuso A, . . . Prokisch H (2017). Genetic diagnosis of Mendelian disorders via RNA sequencing. Nat Commun, 8, 15824. doi: 10.1038/ncomms15824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Huang AY, Wang LK, Yoon AJ, Renteria G, Eskin A, . . . Nelson SF (2020). Diagnostic utility of transcriptome sequencing for rare Mendelian diseases. Genet Med, 22(3), 490–499. doi: 10.1038/s41436-019-0672-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longoni M, High FA, Qi H, Joy MP, Hila R, Coletti CM, . . . Donahoe PK (2017). Genome-wide enrichment of damaging de novo variants in patients with isolated and complex congenital diaphragmatic hernia. Hum Genet, 136(6), 679–691. doi: 10.1007/s00439-017-1774-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddirevula S, Kuwahara H, Ewida N, Shamseldin HE, Patel N, Alzahrani F, . . . Alkuraya FS (2020). Analysis of transcript-deleterious variants in Mendelian disorders: implications for RNA-based diagnostics. Genome Biol, 21(1), 145. doi: 10.1186/s13059-020-02053-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumura K, Seiriki K, Okada S, Nagase M, Ayabe S, Yamada I, . . . Nakazawa T (2020). Pathogenic POGZ mutation causes impaired cortical development and reversible autism-like phenotypes. Nat Commun, 11(1), 859. doi: 10.1038/s41467-020-14697-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murch O, Jain V, Benneche A, Metcalfe K, Hobson E, Prescott K, . . . Fry AE (2021). Further delineation of the clinical spectrum of White-Sutton syndrome: 12 new individuals and a review of the literature. Eur J Hum Genet. doi: 10.1038/s41431-021-00961-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murdock DR, Dai H, Burrage LC, Rosenfeld JA, Ketkar S, Muller MF, . . . Lee B (2021). Transcriptome-directed analysis for Mendelian disease diagnosis overcomes limitations of conventional genomic testing. J Clin Invest, 131(1). doi: 10.1172/JCI141500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozawa RS, Nagao K, Masuda HT, Iwasaki O, Hirota T, Nozaki N, . . . Obuse C (2010). Human POGZ modulates dissociation of HP1alpha from mitotic chromosome arms through Aurora B activation. Nat Cell Biol, 12(7), 719–727. doi: 10.1038/ncb2075 [DOI] [PubMed] [Google Scholar]
- Rentzsch P, Schubach M, Shendure J, & Kircher M (2021). CADD-Splice-improving genome-wide variant effect prediction using deep learning-derived splice scores. Genome Med, 13(1), 31. doi: 10.1186/s13073-021-00835-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stessman HAF, Willemsen MH, Fenckova M, Penn O, Hoischen A, Xiong B, . . . Kleefstra T (2016). Disruption of POGZ Is Associated with Intellectual Disability and Autism Spectrum Disorders. Am J Hum Genet, 98(3), 541–552. doi: 10.1016/j.ajhg.2016.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan B, Zou Y, Zhang Y, Zhang R, Ou J, Shen Y, . . . Wu L (2016). A novel de novo POGZ mutation in a patient with intellectual disability. J Hum Genet, 61(4), 357–359. doi: 10.1038/jhg.2015.156 [DOI] [PubMed] [Google Scholar]
- White J, Beck CR, Harel T, Posey JE, Jhangiani SN, Tang S, . . . Sutton VR (2016). POGZ truncating alleles cause syndromic intellectual disability. Genome Med, 8(1), 3. doi: 10.1186/s13073-015-0253-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Muzny DM, Xia F, Niu Z, Person R, Ding Y, . . . Eng CM (2014). Molecular findings among patients referred for clinical whole-exome sequencing. JAMA, 312(18), 1870–1879. doi: 10.1001/jama.2014.14601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y, Cho MT, Retterer K, Alexander N, Ben-Omran T, Al-Mureikhi M, . . . Chung WK (2015). De novo POGZ mutations are associated with neurodevelopmental disorders and microcephaly. Cold Spring Harb Mol Case Stud, 1(1), a000455. doi: 10.1101/mcs.a000455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yepez VA, Mertes C, Muller MF, Klaproth-Andrade D, Wachutka L, Fresard L, . . . Gagneur J (2021). Detection of aberrant gene expression events in RNA sequencing data. Nat Protoc, 16(2), 1276–1296. doi: 10.1038/s41596-020-00462-5 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table 1. Rare variants identified in the research analysis of trio exome data.
Supplemental Table 2. Individuals with truncating variants in POGZ and congenital diaphragmatic hernia.
Data Availability Statement
The exome sequencing data have been deposited in dbGAP (phs001232.v4.p2), and the novel variant in POGZ described in this manuscript was deposited in Clinvar (SCV001736845.1). Moreover, details regarding the phenotype and genotype were deposited in Phenome Central (P0010291)).