Abstract
Escherichia coli is a widely utilized indicator of the sanitary quality of bivalve molluscan shellfish sold for human consumption. However, it is now well documented that shellfish that meet the E. coli standards for human consumption may contain human enteric viruses that cause gastroenteritis and hepatitis. In this study we investigated using F-specific RNA bacteriophage (FRNA bacteriophage) to indicate the likely presence of such viruses in shellfish sold for consumption. FRNA bacteriophage and E. coli levels were determined over a 2-year period for oysters (Crassostrea gigas) harvested from four commercial sites chosen to represent various degrees of sewage pollution. Three sites were classified as category B sites under the relevant European Community (EC) Directive (91/492), which required purification (depuration) of oysters from these sites before sale. One site was classified as a category A site, and oysters from this site could be sold directly without further processing. Samples were tested at the point of sale following commercial processing and packaging. All of the shellfish complied with the mandatory EC E. coli standard (less than 230 per 100 g of shellfish flesh), and the levels of contamination for more than 90% of the shellfish were at or below the level of sensitivity of the assay (20 E. coli MPN per 100 g), which indicated good quality based on this criterion. In contrast, FRNA bacteriophage were frequently detected at levels that exceeded 1,000 PFU per 100 g. High levels of FRNA bacteriophage contamination were strongly associated with harvest area fecal pollution and with shellfish-associated disease outbreaks. Interestingly, FRNA bacteriophage contamination exhibited a marked seasonal trend that was consistent with the trend of oyster-associated gastroenteritis in the United Kingdom. The correlation between FRNA bacteriophage contamination and health risk was investigated further by using a reverse transcription-PCR assay for Norwalk-like virus (NLV). NLV contamination of oysters was detected only at the most polluted site and also exhibited a seasonal trend that was consistent with the trend of FRNA bacteriophage contamination and with the incidence of disease. The results of this study suggest that FRNA bacteriophage could be used as viral indicators for market-ready oysters.
Sewage contamination of filter-feeding bivalve shellfish produces a well-documented human health risk due to microorganisms transmitted by the fecal-oral route, especially when the shellfish are consumed raw or lightly cooked (38, 40). Shellfish-associated infectious disease outbreaks continue to occur both in the developed world and in the developing world and may be large scale (19, 27, 34). The etiological agents most frequently associated with such outbreaks are enteric viruses that cause gastroenteritis, such as the Norwalk-like caliciviruses (NLVs) and hepatitis A virus (11, 18, 31, 32, 42). To deal with these health risks, most countries impose legislative controls on the harvesting and placing on the market of live bivalve shellfish. European Community (EC) Directive 91/492 (2) includes such controls for the EC, while the United States Food and Drug Administration National Shellfish Sanitation Program (13) provides similar requirements for the United States. These controls rely heavily on using Escherichia coli as an indicator of fecal pollution in shellfish. The EC controls require classification of shellfish harvest areas depending on the degree of fecal pollution, as judged by E. coli monitoring. The classification used determines whether shellfish can be sold for direct consumption or must be treated prior to sale. Treatment most commonly involves controlled self-purification (depuration) in tanks of clean seawater (37, 43); less commonly, treatment involves relaying for an extended period in clean seawater (37). All shellfish sold for consumption must comply with a standard of less than 230 E. coli (or less than 300 fecal coliforms) per 100 g of shellfish flesh. U.S. Food and Drug Administration controls similarly rely on E. coli and fecal coliform monitoring of harvest waters in order to determine approved and restricted harvest areas and treatment requirements prior to sale for consumption.
While current legislation appears to be effective for controlling bacterial illness (43), viral infections associated with shellfish consumption continue to be reported (7). Shellfish that are implicated in disease outbreaks with a viral etiology are frequently compliant with the E. coli standard (less than 230 E. coli per 100 g) (7, 24), particularly when the shellfish are purified prior to sale. Therefore, there is a well-recognized need for a more accurate indicator of the viral risk associated with shellfish sold for consumption.
The human enteric viruses responsible for gastroenteritis and hepatitis following shellfish consumption cannot be cultured by conventional techniques. Although molecular techniques for detection of NLVs (4, 17, 41) and hepatitis virus (4, 28) are now available, these methods are currently too expensive and time-consuming for routine screening of shellfish. The F-specific RNA bacteriophages (FRNA bacteriophages) are a group of single-stranded RNA viruses with simple cubic capsids that are 24 to 27 nm in diameter (14). The genomic and physical properties of these phages are similar to the properties of NLVs and hepatitis A virus. This fact, the abundance of these phages in sewage, and the ease with which they can be enumerated make them attractive indicators of viral contamination in the environment (23). We (9, 10), and other workers (6, 8), have used FRNA bacteriophages to model virus removal from shellfish during depuration. These studies demonstrated that during depuration FRNA bacteriophages are removed from the digestive tract of a contaminated shellfish considerably more slowly than E. coli is removed. The slow elimination kinetics of FRNA bacteriophages appears to be representative of the elimination kinetics of human enteric viruses (37). FRNA bacteriophage persistence following depuration or relaying may, therefore, be useful for predicting the presence of enteric viruses. Similarly, FRNA bacteriophages may persist in shellfish following pollution events not readily detectable by routine E. coli monitoring and thus may indicate potential viral risk.
In this study we examined whether FRNA bacteriophages could be used as viral indicators in commercially depurated oysters sold for consumption. FRNA bacteriophage and E. coli levels were monitored over a 2-year period in marketed oysters (Crassostrea gigas) harvested from four commercial sites chosen to represent various degrees of risk, as judged by both E. coli levels in the harvest area and shellfish-associated disease outbreaks. Our results were compared with the general trends of oyster-associated food poisoning in England and Wales, with known incidents of food poisoning due to animals from each site during the study period, and with direct NLV monitoring of harvested oysters by a previously described nested reverse transcription (RT)-PCR technique (17). The results were analyzed in relation to the ability of FRNA bacteriophages to indicate potential risks of contamination of oysters with enteric viruses that cause gastroenteritis.
MATERIALS AND METHODS
Site selection and sampling.
Four commercial oyster (C. gigas) production areas (sites 1 to 4) in the United Kingdom were chosen to represent a range of pollution levels. Site 1 was situated in a remote location known, as far as possible, to be free of polluting influences and was classified as a category A site (less than 230 E. coli or 300 fecal coliforms per 100 g of shellfish flesh) under EC Directive 91/492. Although depuration was not required by legislation, shellfish from this site were routinely depurated by the producer. Site 2 was influenced by low-level intermittent sewage pollution. At the beginning of this study site 2 was classified as a category A site, but during the study period it was reclassified as a category B site (less than 4,600 E. coli or 6,000 fecal coliforms per 100 g of shellfish flesh in 90% of the samples). Throughout the study period all shellfish harvested from site 2 were depurated prior to sale. Sites 3 and 4 were known to be affected by sewage contamination and were classified as category B sites, so the shellfish had to be depurated prior to sale. Oysters from sites 2, 3, and 4 were depurated for at least 42 h with conventional depuration systems approved by regulatory authorities and operated in accordance with EC regulations. The water temperature in all depuration systems was kept above 8°C, which was consistent with the legal requirements in the United Kingdom. All study samples were provided by the producers after processing and packaging. Twenty-four market-ready oysters were supplied on a periodic basis by producers at each of the sites over a 2-year period from February 1995 to February 1997. In addition to the marketed oysters, additional samples consisting of 24 oysters were periodically taken directly from the harvest areas and used for E. coli analysis. Oysters were transported by courier at the ambient temperature and were received in the laboratory within 48 h of dispatch.
Sample preparation.
When oysters were received, they were thoroughly washed and scrubbed under running potable water. Dead and open oysters that did not respond to percussion were discarded. Six oysters were aseptically opened with a flame-sterilized shucking knife, and the meat and intravalvular fluid were removed, diluted, and homogenized as described previously (10) and then used for E. coli and FRNA bacteriophage analyses. The remaining oysters were frozen whole at −20°C. Some frozen animals were subsequently processed and used for analysis of NLVs by RT-PCR.
E. coli.
Diluted homogenates were assayed for E. coli by using a standard most-probable-number (MPN) method (33). Briefly, the procedure used was a five-tube three-dilution procedure involving inoculation of tubes containing mineral-modified glutamate broth (catalog no. CM607; Oxoid Ltd., Basingstoke, United Kingdom) with Durham tubes, followed by incubation at 37°C for up to 48 h. We confirmed that tubes in which gas was produced contained E. coli by preparing subcultures in tubes containing brilliant green bile broth (catalog no. CM31; Oxoid Ltd.) with Durham tubes and in tubes containing 1% tryptone water (catalog no. CM87; Oxoid Ltd.) and incubating the preparations at 44°C for 18 h. Tubes containing tryptone water were tested for indole production by using Kovács reagent, and subcultures that produced both indole and gas were considered to be E. coli positive. The limit of assay sensitivity was a MPN of 20 E. coli cells per 100 g of shellfish.
FRNA bacteriophages.
Shellfish were assayed for FRNA bacteriophages by using the host bacterium Salmonella typhimurium WG49 described by Havelaar and Hogeboom (22). This host was genetically modified by inserting a plasmid coding for F-pilus production into it. This produced a bacterial host which is susceptible to FRNA bacteriophages but experiences negligible interference from somatic DNA bacteriophages. Diluted homogenates were prepared as described above and then centrifuged at 1,000 × g for 5 min at room temperature. The supernatant was decanted into a universal bottle, and a 1:10 dilution with peptone water (pH 7.2) (catalog no. L37; Oxoid) was prepared. Ten milliliters of the undiluted supernatant and 4 ml of the 1:10 dilution were then assayed for FRNA bacteriophages by using 1-ml portions, 90-mm petri dishes, and a standard double agar overlay method (3). Briefly, to 2.5-ml portions of molten 1% tryptone–yeast extract agar at 45°C we added replicate 1-ml portions of undiluted or diluted shellfish homogenates and 1-ml portions of a WG49 host culture. The molten agar and sample were mixed by inversion and poured onto previously prepared 2% tryptone–yeast extract–glucose agar base in a 90-mm petri dish. The overlays were inverted and incubated overnight. The quality of the host bacterium was maintained throughout the study by careful adherence to standard procedures (3). Principally, this involved using kanamycin sulfate and nalidixic acid in the culture media when working cultures of the host bacterium were prepared in order to ensure that the plasmid coding for F-pilus production was retained. The sensitivity of the host bacterium during each analysis was determined by using a standardized FRNA bacteriophage culture as a control culture. We confirmed that the bacteriophages that were detected were RNA bacteriophages by assaying samples in parallel with RNase and by obtaining differential counts by the standard procedure (3). The limit of sensitivity of the assay was 30 PFU of FRNA bacteriophages per 100 g of shellfish.
NLVs.
Six previously frozen oysters were defrosted at room temperature, and the meat and intravalvular fluid were removed and diluted 1:10 (wt/vol) in 10% tryptose phosphate broth (Lab M, Bury, United Kingdom) containing 0.05 M glycine buffer (pH 9.0 to 9.5). Virus extraction and purification followed by nucleic acid extraction from purified oyster concentrates were then performed by using previously described methods (29). cDNA was synthesized from RNA pellets as previously described (30), and a nested PCR for the NLVs was performed. The strategy used to develop the nested RT-PCR has been described elsewhere (17). Briefly, in the first-round NLV RT-PCR, a broadly reactive primer combination consisting of three primers, G1, G2, and SM31, was used. The sense primers, G1 and G2, were derived from previously described NLV RNA polymerase sequences and were designed to anneal specifically with genogroup I and II strains, respectively. The antisense primer, SM31, has been described previously (35). The internal (nested) primers used were a previously described primer pair (NI and E3) which amplified a 113-bp region of the RNA polymerase gene corresponding to nucleotides 4756 to 4867 of Norwalk virus (16) and preferentially amplified genogroup II strains and a second primer set in the same region that reacted preferentially with genogroup I strains (unpublished data). RT-PCR-positive amplicons of the correct size were cloned and sequenced by using previously described methods (25) to confirm that NLV was present and to determine the genogroups and identities of strains.
RESULTS
The formal harvest area classification for each area under EC Directive 91/492 reflected the perceived pollution status, and the classifications ranged from category A to category A/B to category B (Table 1). Additional E. coli monitoring of the harvest areas was performed in this study in order to more precisely define the degree of harvest area contamination. The results (Table 1) showed that the degrees of fecal pollution in the harvest areas as determined by E. coli monitoring clearly differed; site 1 was the least polluted and site 4 was the most polluted, as determined by both the maximum levels and the geometric mean levels of E. coli in shellfish. The geometric mean levels of E. coli at site 4 were 60-fold higher than the geometric mean levels of E. coli at site 1, and the levels for the other sites were intermediate. These differences in fecal pollution were reflected in the incidence of gastrointestinal illness associated with marketed products from each of the sites reported during the study period. Sites 1 and 2 were not associated with any outbreaks, whereas sites 3 was associated with one outbreak and site 4 was associated with six outbreaks (Table 1). Clearly, therefore, the study sites used represented a spectrum of fecal pollution, and products harvested from the sites differed with respect to the risk of enteric viral contamination.
TABLE 1.
Levels of fecal contamination in oyster harvest areas during the study period as determined by an E. coli analysis of oysters taken directly from each area and reported incidence of gastrointestinal illness associated with products obtained from each site
| Site | Categorya |
E. coli concn (MPN/100 g) in oysters
|
Incidence of illnessb
|
|||
|---|---|---|---|---|---|---|
| Minimum | Maximum | Geometric mean (n) | No. of outbreaks | No. of cases | ||
| 1 | A | <20 | 40 | 6 (20) | 0 | 0 |
| 2 | A/Bc | <20 | 1,300 | 15 (30) | 0 | 0 |
| 3 | B | <20 | 5,000 | 48 (22) | 1 | 10 |
| 4 | B | <20 | 22,000 | 363 (59) | 6 | 97 |
Formal categories for shellfish harvest areas as assigned by the United Kingdom authorities (Ministry of Agriculture) according to EC Directive 91/492.
Officially reported incidents of gastrointestinal disease associated with oysters obtained from each site during the study period. Data were provided by the Public Health Laboratory Service, Communicable Disease Surveillance Centre, Colindale, United Kingdom.
Both category A and category B classifications were assigned during the study period.
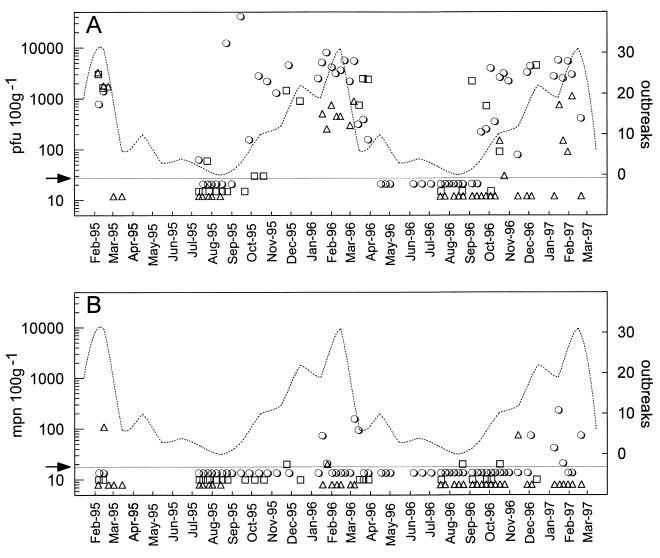
Market-ready oysters from site 1 were consistently negative for all parameters throughout the study period. In contrast, fecal pollution indicators were detected in market-ready oysters from sites 2, 3, and 4. The results obtained for E. coli and FRNA bacteriophages throughout the study period are shown in Fig. 1, and the data are summarized in Table 2. The E. coli contents of market-ready oysters were low, and the values obtained for more than 90% of the samples were at or below the level of sensitivity of the assay (20 E. coli MPN per 100 g). All of the samples from all of the sites complied with the mandatory European E. coli end product standard for human consumption (less than 230 E. coli per 100 g of shellfish flesh). Table 2 shows that there was little or no correlation between the E. coli levels in market-ready products and the levels of fecal pollution in the harvest areas, as judged by both the maximum and geometric mean E. coli levels in marketed products. In contrast, FRNA bacteriophages were frequently detected in market-ready products; the levels often were more than 1,000 PFU per 100 g of shellfish and occasionally were more than 10,000 PFU per 100 g. Furthermore, the extent of contamination with FRNA bacteriophages in market-ready products was closely associated with the degree of fecal pollution in the shellfish harvest area, as judged by the maximum and geometric mean levels of FRNA bacteriophages (Table 2). The highest FRNA bacteriophage levels occurred in products obtained from site 4, the most polluted harvest area, and the lowest levels occurred in products obtained from site 2, the least polluted area. FRNA bacteriophages were not detected in any site 1 sample. Table 1 shows that the FRNA bacteriophage content of market-ready shellfish and the degree of fecal pollution were also correlated with the known incidence of disease associated with products obtained from each harvest area. This suggested that, compared with the E. coli content, the FRNA bacteriophage content of marketed oysters more accurately reflected the consumer health risk due to human enteric viruses. This hypothesis was tested by analyzing the NLV content of a random selection of frozen oyster samples from each site by using the nested RT-PCR. The results are shown in Table 2. Of the samples tested, only the samples from site 4, the most polluted site, gave positive results. We confirmed that RT-PCR-positive amplicons were NLV amplicons by sequence analysis. A total of 37% of 35 samples from site 4 were positive for NLV, and a variety of strains were present (25). These findings are consistent with official disease reports (Table 1) which showed that of the products tested, products from site 4 were most frequently implicated in gastrointestinal illness.
FIG. 1.
Seasonal distribution of FRNA bacteriophages (expressed as number of PFU per 100 g of market-ready oysters), (A), seasonal distribution of E. coli (expressed as E. coli MPN per 100 g of market-ready oysters) (B), and seasonality of infectious disease outbreaks. The fecal indicator test results are shown for site 2 (▵), site 3 (□), and site 4 (○). The arrows and lines indicate the limit of assay sensitivity for FRNA bacteriophages (<30 PFU per 100 g) (A) and E. coli (<20 E. coli MPN per 100 g) (B); the data points below the lines are negative. The dotted lines indicate the distribution by month of outbreaks of infectious disease associated with oyster consumption in the United Kingdom; cumulative data for 1982 to 1997 are shown. Disease data were kindly provided by the Public Health Laboratory Service, Communicable Disease Surveillance Centre, Colindale, United Kingdom.
TABLE 2.
E. coli, FRNA bacteriophage, and NLV data for market-ready oysters obtained from each site during the study period
| Site |
E. coli concn (E. coli MPN/100 g)
|
FRNA bacteriophage concn (PFU/100 g)
|
% Positive for NLVs (n)a | ||||
|---|---|---|---|---|---|---|---|
| Minimum | Maximum | Geometric mean (n) | Minimum | Maximum | Geometric mean (n) | ||
| 1 | <20 | <20 | 0.0 (13) | <30 | <30 | 0.0 (13) | 0 (7) |
| 2 | <20 | 110 | 0.4 (39) | <30 | 3,300 | 11.3 (39) | 0 (15) |
| 3 | <20 | 50 | 0.5 (24) | <30 | 4,500 | 41.4 (24) | 0 (13) |
| 4 | <20 | 220 | 0.9 (58) | <30 | 39,300 | 125.9 (58) | 37 (35) |
Seven-gram samples were used.
Figure 1 also shows that there was a clear seasonal trend in FRNA bacteriophage contamination in market-ready oysters. Levels were frequently elevated during the winter months, but FRNA bacteriophages were largely absent during the summer months. This trend was correlated with the known seasonal incidence of gastroenteric illness due to oyster consumption in the United Kingdom, which is shown in Fig. 1 as the cumulative incidence over a 14-year period. The trends are summarized in Table 3, which shows the seasonal trends for both E. coli and FRNA bacteriophage contents divided into summer months (April to September inclusive) and winter months (October to March inclusive). Although more samples (12 to 28%) were positive for E. coli during winter months than during summer months, the levels were always low; the geometric mean even at the most polluted site (site 4) was only 2.2 E. coli MPN per 100 g of shellfish. In comparison, FRNA bacteriophages were frequently detected in shellfish harvested from polluted sites during the winter months, and the frequencies of detection ranged from 64% (site 2) to 100% (site 4). These high frequencies of detection were coupled with elevated counts in positive samples. The geometric mean levels of FRNA bacteriophages in market-ready shellfish harvested from polluted sites during the winter months ranged from 49 PFU per 100 g at site 2 to 1,865 PFU per 100 g at site 4. Both the frequencies of detection and the geometric mean levels of FRNA bacteriophages in market-ready oysters during the winter months were strongly correlated with the degree of pollution in the harvest area (Table 1). In contrast, during the summer months FRNA bacteriophages were detected in fewer samples (<25%), and the geometric means, even in shellfish harvested at the more polluted sites (sites 3 and 4), were low, only 3.6 PFU per 100 g of shellfish. It is important to note that FRNA bacteriophages were consistently absent, even during the winter months, in shellfish harvested at the pristine site (site 1). These results show that the strong correlation between the FRNA bacteriophage content (but not the E. coli content) of marketed oysters and both the degree of pollution at the harvest area and consumer health risk (Tables 1 and 2) was probably also highly seasonally dependent, with the winter months being the high-risk period. This finding was confirmed by the seasonality of detection of NLVs in marketed shellfish obtained at site 4. NLV contamination was detected only during the winter months, and 62% of the samples were positive during this period (Table 3). These findings were correlated with the known seasonal incidence of gastroenteric illness due to oyster consumption in the United Kingdom (Fig. 1).
TABLE 3.
Detection of E. coli, FRNA bacteriophages, and NLVs in market-ready oysters obtained from each site during summer and winter months
| Season | Site |
E. coli
|
FRNA bacteriophages
|
% Positive for NLVs (n)a | ||
|---|---|---|---|---|---|---|
| % Positive (n) | Geometric mean concn (MPN/100 g) | % Positive (n) | Geometric mean concn (PFU/100 g) | |||
| Summerb | 1 | 0 (6) | 0.0 | 0 (6) | 0.0 | 0 (3) |
| 2 | 0 (14) | 0.0 | 0 (14) | 0.0 | 0 (5) | |
| 3 | 8 (12) | 0.4 | 25 (12) | 3.6 | 0 (6) | |
| 4 | 0 (26) | 0.0 | 23 (26) | 3.6 | 0 (14) | |
| Winterc | 1 | 0 (7) | 0.0 | 0 (7) | 0.0 | 0 (4) |
| 2 | 12 (25) | 0.6 | 64 (25) | 49.1 | 0 (10) | |
| 3 | 17 (12) | 0.7 | 92 (12) | 386.8 | 0 (7) | |
| 4 | 28 (32) | 2.2 | 100 (32) | 1,865.5 | 62 (21) | |
Seven-gram samples were used.
April to September inclusive.
October to March inclusive.
DISCUSSION
The inadequacy of E. coli as an indicator of the viral risk associated with oyster consumption is well documented (7, 15, 24, 31) and has prompted calls for investigations of alternative viral indicators. The inadequacy of the E. coli standards was confirmed in this study; all of our samples met the EC E. coli end product standard (less than 230 per 100 g of shellfish) despite the fact that oysters obtained from two of the sites were implicated in outbreaks of viral gastroenteritis. It is important to note that the shellfish used in this study, like many oysters both in the United Kingdom and in other countries, were purified with commercial depuration systems prior to sale. Depuration has been shown to rapidly and effectively remove bacterial pollution indicators; however, human enteric viruses are known to be more persistent (1, 36, 39; M. D. Sobsey, J. C. Murray, and G. Lovelace, Abstr. 91st Annu. Meet. Am. Soc. Microbiol. 1991, p. 300, 1991). It is not surprising, therefore, that the absence of E. coli in depurated oysters may not guarantee a virus-free product. This is particularly evident during viral infectious disease outbreaks in which shellfish implicated on the basis of epidemiological criteria frequently comply with the bacterial end product standard, less than 230 E. coli per 100 g (4, 7).
In this study we evaluated whether FRNA bacteriophages could be used as alternative viral indicators for shellfish. The FRNA bacteriophage contents of market-ready oysters from four producers were compared with the pollution status of the harvest areas, with reported gastroenteric illness incidents linked to products from each site during the study period, and with NLV contamination, as judged by RT-PCR. FRNA bacteriophages, like E. coli and NLV, were not present in oysters harvested at a pristine site that was free of sewage pollution and was not previously associated with gastroenteric illness. This illustrates that it is possible to obtain shellfish that do not contain FRNA bacteriophages and present a low health risk. In contrast, FRNA bacteriophages were detected, often at high levels, in market-ready oysters harvested at more polluted sites. Furthermore, the frequency and degree of FRNA bacteriophage contamination were closely associated with consumer health risk due to enteric viruses, as judged by the degree of harvest area pollution, the NLV content of shellfish, and the association with reported incidents of gastroenteric illness. These data suggest that FRNA bacteriophages, unlike E. coli, are reliable and effective indicators of the possible presence of human enteric gastroenteritis viruses in depurated market-ready oysters.
The presence of FRNA bacteriophages in depurated oysters indicated that the oysters were subject to fecal contamination in their harvest areas and that any viruses present may not have been eliminated during the depuration process. During this study this was demonstrated for oysters from site 4, the most polluted site, by detection of NLVs in market-ready oysters. NLVs were not detected in oysters from the other sites; however, the numbers of samples examined were low. Contamination of shellfish harvest areas by NLVs is probably sporadic and depends on viral circulation in the community. Like other indicator systems, the presence of FRNA bacteriophages in oysters, therefore, indicates the potential for viral contamination rather than a definitive hazard in a sample being studied. However, it is reasonable to assume that the titer of FRNA bacteriophages found in depurated oysters indicates the relative risk of viral contamination. This was confirmed in this study, in which the average levels of FRNA bacteriophages in depurated oysters were correlated with the frequency of NLV contamination determined by RT-PCR, the number of reported health incidents associated with products from each site, and the degree of harvest area pollution. Conversely, unlike E. coli, the absence of FRNA bacteriophages appears to be a reliable indicator that enteric viruses, such as NLVs, are probably absent.
Outbreaks of gastroenteritis associated with the consumption of oysters in the United Kingdom exhibit a clear seasonal trend, with outbreaks occurring predominantly during the winter months and only rarely in the summer months (Fig. 1). The NLV data obtained for oysters in this study were entirely consistent with this trend. Interestingly, FRNA bacteriophage contamination in depurated oysters also exhibited a marked seasonal trend that was consistent with the high-risk period for contamination by enteric viruses. Traditionally, NLV gastroenteritis has been considered a seasonal disease; it was described in early studies as “winter vomiting disease.” However, in recent years the seasonal nature of the disease has been less consistent, and during our study period peak community infection levels occurred in late spring 1995 and early summer 1996 (12). These peak community levels were not consistent with the period of NLV contamination of oysters observed in this study, suggesting that other factors may also be significant. Likewise, there is little evidence that FRNA bacteriophage levels in sewage effluents are different in different seasons, which makes this an unlikely explanation for the seasonal differences seen in oysters. It seems more likely that NLV and FRNA bacteriophage contents of oysters are influenced either by different winter and summer rates of virus inactivation in the environment or by seasonally dependent uptake and depuration of viral contaminants by molluscs. Since FRNA bacteriophage are resistant to UV irradiation (20), it is unlikely that inactivation in the environment completely accounts for the dramatically decreased levels found in oysters during the summer months. Data on NLV decay in the environment is not available. A stronger possibility is that viruses are eliminated more efficiently during the mollusc depuration process in the summer. This could be associated with factors that affect mollusc metabolism, such as higher summer water temperatures or food availability. Clearly, these possibilities should be investigated further as they may have important implications for improving commercial depuration procedures used for virus removal during the winter months.
One possible criticism of FRNA bacteriophages as indicators of viral risk in oysters is that, like E. coli, these bacteriophages are not human specific. Animal feces originating from land runoff could also cause FRNA bacteriophage contamination (21) but may not pose a health risk due to NLVs. During this study there was little evidence for this as FRNA bacteriophage contamination could be accounted for by known sewage discharges and correlated well with health risk and the presence of NLVs. Oligonucleotide probe hybridization methods for geneotyping FRNA bacteriophages have recently been proposed for differentiating animal-associated and human-associated bacteriophage groups (5, 26). The application of such techniques to shellfish would facilitate differentiation of contamination from human sources and contamination from animal sources. Such techniques could be useful in investigations of sites where sewage pollution sources cannot be identified and contamination from animal feces is suspected.
In conclusion, data obtained in this study suggest that the FRNA bacteriophage content of depurated oysters sold for consumption may reflect the public health risk due to human enteric viruses more accurately than the E. coli content reflects this risk. Data obtained in this study also indicate that currently used commercial depuration practices cannot guarantee removal of human enteric viruses from oysters during the winter months.
ACKNOWLEDGMENT
This work was funded by the Food Hygiene Division of the Ministry of Agriculture, Fisheries and Food, United Kingdom.
REFERENCES
- 1.Abad F X, Pinto R M, Gajardo R, Bosch A. Viruses in mussels—public health implications and depuration. J Food Prot. 1997;60:677–681. doi: 10.4315/0362-028X-60.6.677. [DOI] [PubMed] [Google Scholar]
- 2.Anonymous. Council directive laying down the health conditions for the production and placing on the market of live bivalve molluscs (91/492/EEC) Off J Eur Communities. 1991;268:13. [Google Scholar]
- 3.Anonymous. Water quality—detection and enumeration of bacteriophage, part 1: enumeration of F-specific RNA bacteriophages. ISO 10705-1. Geneva, Switzerland: International Organisation for Standardisation; 1996. [Google Scholar]
- 4.Atmar R L, Neill F H, Romalde J L, Leguyader F, Woodley C M, Metcalf T G, Estes M K. Detection of Norwalk virus and hepatitis A virus in shellfish tissues with the PCR. Appl Environ Microbiol. 1995;61:3014–3018. doi: 10.1128/aem.61.8.3014-3018.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beekwilder J, Nieuwenhuizen R, Havelaar A H, van Duin J. An oligonucleotide hybridization assay for the identification and enumeration of F-specific RNA bacteriophages in surface water. J Appl Bacteriol. 1995;80:179–186. doi: 10.1111/j.1365-2672.1996.tb03207.x. [DOI] [PubMed] [Google Scholar]
- 6.Burkhardt W I, Rippey S R, Watkins W D. Depuration rates of northern quahogs Mercenaria mercenaria Linnaeus 1758 and eastern oysters Crassostrea virginica Gmelin 1791 in ozone and ultraviolet light-disinfected seawater systems. J Shellfish Res. 1992;11:105–109. [Google Scholar]
- 7.Chalmers J W T, McMillan J H. An outbreak of viral gastroenteritis associated with adequately prepared oysters. Epidemiol Infect. 1995;115:163–167. doi: 10.1017/s0950268800058222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Mesquita M M F, Evison L M, West P A. Removal of fecal indicator bacteria and bacteriophages from the common mussel (Mytilus edulis) under artificial depuration conditions. J Appl Bacteriol. 1991;70:495–501. doi: 10.1111/j.1365-2672.1991.tb02746.x. [DOI] [PubMed] [Google Scholar]
- 9.Dore W J, Henshilwood K, Lees D N. The development of management strategies for control of virological quality in oysters. Water Sci Technol. 1998;38:29–35. [Google Scholar]
- 10.Dore W J, Lees D N. Behavior of Escherichia coli and male-specific bacteriophage in environmentally contaminated bivalve molluscs before and after depuration. Appl Environ Microbiol. 1995;61:2830–2834. doi: 10.1128/aem.61.8.2830-2834.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dowell S F, Groves C, Kirkland K B, Cicirello H G, Ando T, Jin Q, Gentsch J R, Monroe S S, Humphrey C D, Slemp C, Dwyer D M, Meriwether R A, Glass R I. A multistate outbreak of oyster-associated gastroenteritis—implications for interstate tracing of contaminated shellfish. J Infect Dis. 1995;171:1497–1503. doi: 10.1093/infdis/171.6.1497. [DOI] [PubMed] [Google Scholar]
- 12.Evans H S, Madden P, Douglas C, Adak G K, O'Brien S J, Djuretic T, Wall P, Stanwell-Smith R. General outbreaks of infectious intestinal disease in England and Wales: 1995 and 1996. Commun Dis Public Health. 1998;1:165–171. [PubMed] [Google Scholar]
- 13.Food and Drug Administration. National Shellfish Sanitation Program manual of operations. 1. Sanitation of shellfish growing areas. Washington, D.C.: Shellfish Sanitation Branch, U.S. Public Health Service; 1989. [Google Scholar]
- 14.Furuse K. Distribution of coliphages in the environment: general considerations. In: Goyal S M, Gerba C P, Bitton G, editors. Phage ecology. New York, N.Y: Wiley; 1987. pp. 87–124. [Google Scholar]
- 15.Gill O N, Cubitt W D, Mcswiggan D A, Watney B M, Bartlett C L R. Epidemic of gastroenteritis caused by oysters contaminated with small round structured viruses. Br Med J. 1983;287:1534. doi: 10.1136/bmj.287.6404.1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Green J, Gallimore C I, Norcott J P, Lewis D, Brown D W G. Broadly reactive reverse transcriptase polymerase chain reaction for the diagnosis of SRSV-associated gastroenteritis. J Med Virol. 1995;47:392–398. doi: 10.1002/jmv.1890470416. [DOI] [PubMed] [Google Scholar]
- 17.Green J, Henshilwood K, Gallimore C I, Brown D W G, Lees D N. A nested reverse transcriptase PCR assay for detection of small round-structured viruses in environmentally contaminated molluscan shellfish. Appl Environ Microbiol. 1998;64:858–863. doi: 10.1128/aem.64.3.858-863.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grohmann G S, Greenberg H B, Welch B M, Murphy A M. Oyster associated gastroenteritis in Australia: the detection of Norwalk virus and its antibody by immune electron microscopy and radioimmunoassay. J Med Virol. 1980;6:11–20. doi: 10.1002/jmv.1890060103. [DOI] [PubMed] [Google Scholar]
- 19.Halliday M L, Kang L Y, Zhou T-K, Hu M-D, Pan Q C, Fu T Y, Huang Y S, Hu S L. An epidemic of hepatitis A attributable to the ingestion of raw clams in Shanghai, China. J Infect Dis. 1991;164:852–859. doi: 10.1093/infdis/164.5.852. [DOI] [PubMed] [Google Scholar]
- 20.Havelaar A H. Bacteriophages as model organisms in water treatment. Microbiol Sci. 1999;4:362–364. [PubMed] [Google Scholar]
- 21.Havelaar A H, Furuse K, Hogeboom W M. Bacteriophages and indicator bacteria in human and animal feces. J Appl Bacteriol. 1986;60:255–262. doi: 10.1111/j.1365-2672.1986.tb01081.x. [DOI] [PubMed] [Google Scholar]
- 22.Havelaar A H, Hogeboom W M. A method for the enumeration of male-specific bacteriophages in sewage. J Appl Bacteriol. 1984;56:429–447. doi: 10.1111/j.1365-2672.1984.tb01372.x. [DOI] [PubMed] [Google Scholar]
- 23.Havelaar A H, Van Olphen M, Drost Y C. F-specific RNA bacteriophages are adequate model organisms for enteric viruses in fresh water. Appl Environ Microbiol. 1993;59:2956–2962. doi: 10.1128/aem.59.9.2956-2962.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heller D, Gill O N, Raynham E, Kirkland T, Zadick P M, Stanwell-Smith R. An outbreak of gastrointestinal illness associated with consumption of raw depurated oysters. Br Med J. 1986;292:1726–1727. doi: 10.1136/bmj.292.6537.1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Henshilwood K, Green J, Lees D N. Monitoring the marine environment for small round structured viruses (SRSVs): a new approach to combating the transmission of these viruses by molluscan shellfish. Water Sci Technol. 1998;38:51–56. [Google Scholar]
- 26.Hsu F C, Shieh Y S C, Vanduin J, Beekwilder M J, Sobsey M D. Genotyping male-specific RNA coliphages by hybridization with oligonucleotide probes. Appl Environ Microbiol. 1995;61:3960–3966. doi: 10.1128/aem.61.11.3960-3966.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kohn M A, Farley T A, Ando T, Curtis M, Wilson S A, Jin Q, Monroe S S, Baron R C, Mcfarland L M, Glass R I. An outbreak of Norwalk virus gastroenteritis associated with eating raw oysters—implications for maintaining safe oyster beds. JAMA. 1995;273:466–471. doi: 10.1001/jama.1995.03520300040034. [DOI] [PubMed] [Google Scholar]
- 28.Lees D N, Henshilwood K, Butcher S. Development of a PCR-based method for the detection of enteroviruses and hepatitis A virus in molluscan shellfish and its application to polluted field samples. Water Sci Technol. 1995;31:457–464. [Google Scholar]
- 29.Lees D N, Henshilwood K, Dore W J. Development of a method for detection of enteroviruses in shellfish by PCR with poliovirus as a model. Appl Environ Microbiol. 1994;60:2999–3005. doi: 10.1128/aem.60.8.2999-3005.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lees D N, Henshilwood K, Green J, Gallimore C I, Brown D W G. Detection of small round structured viruses in shellfish by reverse transcription-PCR. Appl Environ Microbiol. 1995;61:4418–4424. doi: 10.1128/aem.61.12.4418-4424.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mcdonnell S, Kirkland K B, Hlady W G, Aristeguieta C, Hopkins R S, Monroe S S, Glass R I. Failure of cooking to prevent shellfish-associated viral gastroenteritis. Arch Intern Med. 1997;157:111–116. [PubMed] [Google Scholar]
- 32.Mele A, Rastelli M G, Gill O N, Di Bisceglie D, Rosmini F, Pardelli G, Valtriani C, Patriarchi P. Recurrent epidemic hepatitis A associated with consumption of raw shellfish probably controlled through public health measures. Am J Epidemiol. 1989;130:540–546. doi: 10.1093/oxfordjournals.aje.a115368. [DOI] [PubMed] [Google Scholar]
- 33.Ministry of Agriculture, Fisheries and Food, Department of Health and Public Health Laboratory Service working group. Bacteriological examination of shellfish. PHLS Microbiol Digest. 1992;9:76–82. [Google Scholar]
- 34.Morse D L, Guzewich J J, Hanrahan J P, Stricof R, Shayegani M, Deibel R, Grabau J C, Nowak N A, Herrmann J E, et al. Widespread outbreaks of clam- and oyster-associated gastroenteritis: role of Norwalk virus. N Engl J Med. 1986;314:678–681. doi: 10.1056/NEJM198603133141103. [DOI] [PubMed] [Google Scholar]
- 35.Norcott J P, Green J, Lewis D, Estes M K, Barlow K I, Brown D W G. Genomic diversity of small round structured viruses in the United Kingdom. J Med Virol. 1994;44:280–286. doi: 10.1002/jmv.1890440312. [DOI] [PubMed] [Google Scholar]
- 36.Power U F, Collins J K. Differential depuration of poliovirus, Escherichia coli, and a coliphage by the common mussel, Mytilus edulis. Appl Environ Microbiol. 1989;55:1386–1390. doi: 10.1128/aem.55.6.1386-1390.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Richards G P. Microbial purification of shellfish: a review of depuration and relaying. J Food Prot. 1988;51:218–251. doi: 10.4315/0362-028X-51.3.218. [DOI] [PubMed] [Google Scholar]
- 38.Rippey S R. Infectious diseases associated with molluscan shellfish consumption. Clin Microbiol Rev. 1994;7:419–423. doi: 10.1128/cmr.7.4.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schwab K J, Neill F H, Estes M K, Metcalf T G, Atmar R L. Distribution of Norwalk virus within shellfish following bioaccumulation and subsequent depuration by detection using RT-PCR. J Food Prot. 1998;61:1674–1680. doi: 10.4315/0362-028x-61.12.1674. [DOI] [PubMed] [Google Scholar]
- 40.Sockett P N, West P A, Jacob M. Shellfish and public health. PHLS Microbiol Digest. 1985;2:29–35. [Google Scholar]
- 41.Sugieda M, Nakajima K, Nakajima S. Outbreaks of Norwalk-like virus-associated gastroenteritis traced to shellfish—coexistence of two genotypes in one specimen. Epidemiol Infect. 1996;116:339–346. doi: 10.1017/s0950268800052663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tang Y W, Wang J X, Xu Z Y, Guo Y F, Qian W H, Xu J X. A serologically confirmed case-control study of a large outbreak of hepatitis A in China associated with consumption of clams. Epidemiol Infect. 1991;107:651–658. doi: 10.1017/s0950268800049347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.West P A, Wood P C. Control of food poisoning risks associated with shellfish. J R Soc Health. 1985;1:15–21. doi: 10.1177/146642408510500104. [DOI] [PubMed] [Google Scholar]