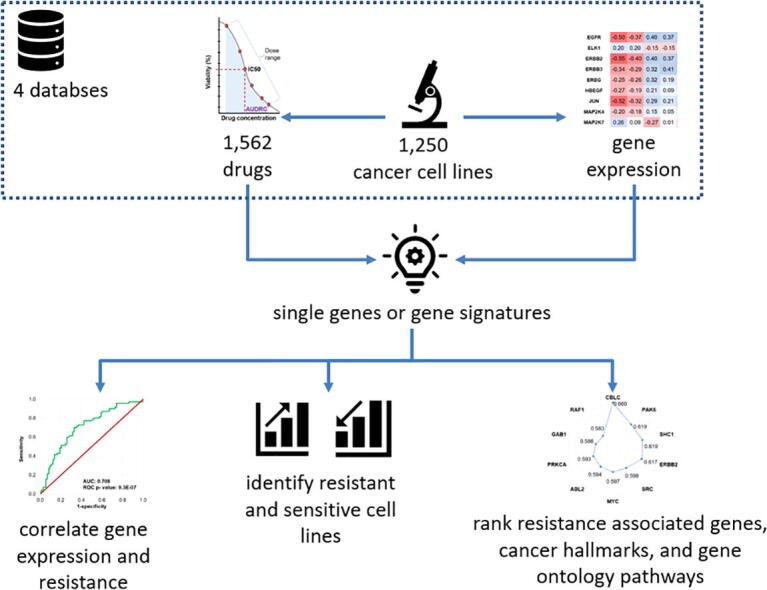
Graphical abstract
Keywords: Chemotherapy, Proliferation, In vitro, Receiver operator characteristics, RNAseq, Machine learning, Random forest
Abstract
Intro
In vitro cell line models provide a valuable resource to investigate compounds useful in the systemic chemotherapy of cancer. However, the due to the dispersal of the data into several different databases, the utilization of these resources is limited. Here, our aim was to establish a platform enabling the validation of chemoresistance-associated genes and the ranking of available cell line models.
Methods
We processed four independent databases, DepMap, GDSC1, GDSC2, and CTRP. The gene expression data was quantile normalized and HUGO gene names were assigned to have unambiguous identification of the genes. Resistance values were exported for all agents. The correlation between gene expression and therapy resistance is computed using ROC test.
Results
We combined four datasets with chemosensitivity data of 1562 agents and transcriptome-level gene expression of 1250 cancer cell lines. We have set up an online tool utilizing this database to correlate available cell line sensitivity data and treatment response in a uniform analysis pipeline (www.rocplot.com/cells). We employed the established pipeline to by rank genes related to resistance against afatinib and lapatinib, two inhibitors of the tyrosine-kinase domain of ERBB2.
Discussion
The computational tool is useful 1) to correlate gene expression with resistance, 2) to identify and rank resistant and sensitive cell lines, and 3) to rank resistance associated genes, cancer hallmarks, and gene ontology pathways. The platform will be an invaluable support to speed up cancer research by validating gene-resistance correlations and by selecting the best cell line models for new experiments.
1. Introduction
Cancer is considered the second most common cause of death worldwide with 10 million cancer-related deaths and 19.3 million new cancer diagnoses according to an estimation in 2021 [1]. Despite improvements of progression-free and overall survival in the past few years in the treatment of certain tumors, we still see a significant rate of tumor types in which no improvement has been made [2]. The majority of anticancer agents are not universally effective, and they have anti-tumor activities only in distinct groups of tumors. Besides, the therapeutic response varies from person to person, which is determined by factors of genetic and environmental variations. A robust resource enabling the application of personalized therapies is the determination of gene expressions levels that are capable of acting as biomarkers to select patients who will most likely benefit from a given therapy as has been demonstrated for breast cancer endocrine therapy [3].
New drug development can be challenging due to high costs and the fact that the mean time from the initial screening to final approval of the drug can take more than ten years. Approval can be based on drug repurposing as well – in this case a therapy already approved for some indication is assessed for another indication [4]. As these drugs already have a regulatory approval, data on their safety profiles and potential interactions with other drugs are readily available, thus the time and the cost needed to introduce the therapy with a new indication can be significantly reduced. There are several drug repurposing candidates with anticancer potential today. For example, the effects of cardiovascular drugs including aspirin, ACE inhibitors, and beta blockers are now under investigation in oncology [5]. Anticancer indications have also been suggested for psychiatric drugs including valproic acid, phenothiazines, selective serotonin reuptake inhibitors, tricyclic antidepressants, and MAO inhibitors [6]. A rational first step in the discovery and validation of such agents is the preclinical analysis of their effect in cancer cell lines.
Cancer cell lines provide clinically useful data by enabling the experimental investigation and modelling of new treatments and therapy resistance related factors [7]. In the past decades, several cell line databases have been established that enable the linking of pharmaceutical agents to tumor growth inhibition. Initial studies had panels of cell lines with sensitivity data for a handful of agents [8]. Large scale anti-tumor drug screening with more than 21,000 agents tested in sixty cell lines was launched by the National Cancer Institute in the 1990′s [9]. The Cancer Cell Line Encyclopedia (CCLE) project, a cooperation between the Broad Institute and Novartis, provides the genetic and pharmacological characteristics of more than 1100 cell lines [10]. The Cancer Therapeutics Response Portal (CTRP) enables access to 860 cell lines [11] while the Genomics of Drug Sensitivity in Cancer (GDSC) project contains data for more than 1000 cell lines and their interactions with more than 500 drugs [12]. The Cancer Dependency Map (Depmap), a multi-institutional project to map genetic dependencies, provides genetic mapping for more than five hundred cell lines and resistance data for more than 4,000 agents [13].
In our work we have set three goals. First, we integrated data from multiple large-scale cell line databases to establish an easy to use online platform that provides swift access and analysis of the data in order to uncover relationships between gene expression and therapeutic response across a large panel of drugs. Second, we established a ranking of cell lines enabling the identification of the most robust preclinical model. Third, we validated our approach by selecting lapatinib and afatinib, two ERBB2 tyrosine kinase domain inhibitors, which were evaluated in each included dataset, and by ranking the significant gene expression-based biomarkers.
2. Methods
2.1. Drug screening and gene expression data
For the setup of the database, we collected data from four publicly available drug screening databases. Drug sensitization data of the Cancer Dependency Map Consortium's DepMap portal (https://depmap.org/) were obtained from the PRISM Repurposing 19Q4 secondary screen dose–response dataset [13]. From the Genomics of Drug Sensitivity in Cancer (GDSC) project [14] both GDSC1 and GDSC2 drug screening datasets were taken, whereas from the Cancer Therapeutics Response Portal (CTRP) the version 2 drug screening dataset was obtained [15].
DepMap and CTRP drug screening datasets are based on the CCLE cell lines and for gene expression data the 21Q1 RNAseq data was used as a source [16]. Read count data were normalized with the DESeq algorithm, then a quantile and a scaling normalization method were applied to set the mean gene expression in each cell line to 1000. Genes with a zero-expression value in more than half of the cell lines were excluded from the analysis. For the gene expression of the cell lines in the GDSC drug screening datasets we obtained RMA normalized Affymetrix HGU-219 microarray expression matrix, and applied a second scaling normalization method as above. Pre-processed data were imported into a PostgreSQL database. For the identification of unambiguous gene names, we used the HUGO Gene Nomenclature Committee (HGNC) database (https://www.genenames.org/).
2.2. Treatment response categorization
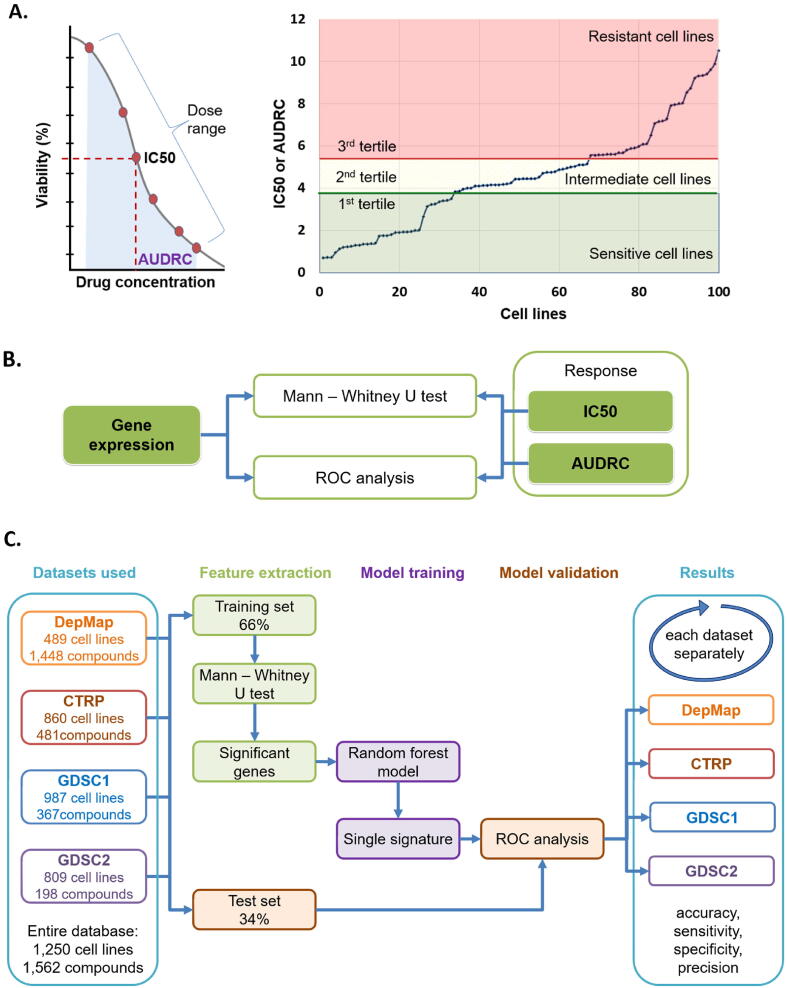
In the analysis of DepMap and GDSC based projects we used the reported half-maximal inhibitory concentration (IC50) and the area under the dose-response curve (AUDRC) values to evaluate therapeutic response. AUDRC is determined using the dose range spanning from the lowest to the highest applied dose for the drug under investigation. Cases where neither IC50 nor AUDRC was determined were excluded from the analysis. For each agent, we defined lower and upper tertile cutoff values based on the IC50 or AUDRC values and cell lines with IC50 or AUDRC values in the lower tertile were considered as sensitive and those in the upper tertile were considered as resistant. Cells belonging to the intermediate tertile were not considered in the analysis. In the analysis of the CTRP project only AUDRC values were reported and we used it to assess therapeutic response with the median and tertile based method as described above. The difference between IC50 and AUDRC is summarized in Fig. 1A.
Fig. 1.
Overview of the analysis pipeline. Summary of response classification using IC50 and AUDRC (area under the dose response curve) values (A). Primary statistical methods for single gene analyses (B), and the setup for the machine learning pipeline for gene signature analysis (C).
2.3. General statistical methods
The analysis was performed in the R statistical software environment (https://www.r-project.org/). Mann-Whitney U test and receiver operating characteristics (ROC) were computed in order to compare single gene expression values between sensitive and resistant samples (Fig. 1B). Spearman rank correlation was applied to compare published AUDRC values with gene expression. The statistical significance cutoff was set at p < 0.05.
2.4. Gene signature analysis
To assess the relation of different pathways and cancer hallmark genes to therapeutic response, we utilized the lists of KEGG pathways (https://www.genome.jp/kegg/) and a previously assembled lists of cancer hallmark genes [17]. In these, 712 genes belonging to seven hallmarks and 4,602 genes belonging to 186 KEGG pathways can be tested per therapeutic agent. Using these genes, the analysis pipeline was extended with a machine learning computation method to analyze the entire signature. As a first step, samples are randomly divided into a training (66%) and a test set (34%). Second, the system selects genes significantly (p < 0.05) correlated to resistance using a Mann-Whitney test. Then, the significant genes are integrated by a random forest classifier into a single signature. Finally, a ROC analysis is used to evaluate the predictive effectiveness of this signature. As a result, the list of genes significant in the signature, the confusion matrix of the test set, and the overall predictive power of the signature including the computed accuracy=(TP + TN)/(TP + FN + FP + FN), sensitivity = TP/(TP + FN), specificity = TN/(TN + FP), and precision = TP/(TP + FP) values are provided (Fig. 1C).
2.5. Online analysis portal
We extended our previously established ROC plotter tool [18] with the cell line database. The portal is set up to require the investigated agent and the biomarker candidate as input. Datasets with available treatment and expression data as well as the most robust response data are automatically selected. Using these input parameters, the ROC AUC plot is generated for each available setting. Furthermore, the sensitivity across all available cell lines is provided as a table. In addition to single genes, simultaneous analysis of multiple genes can be performed by using the mean expression of the included genes as described above. When analyzing a set of genes, false discovery rate is computed and provided in the results page.
2.6. Sample application: Validation of genes related to sensitivity against lapatinib and afatinib
To validate the robustness of the established database, we aimed to analyze two selected pharmaceutical agents acting on an established therapeutic target. For this validation, we selected the drugs afatinib and lapatinib, both targeting the ERBB2 receptor according the DrugBank database [19]. This selection was based on the fact that sensitivity data for these two agents was available in each of the four included databases. The analysis was performed using the tertile- based therapeutic response categorization as described above.
3. Results
3.1. Therapeutic agents in the database
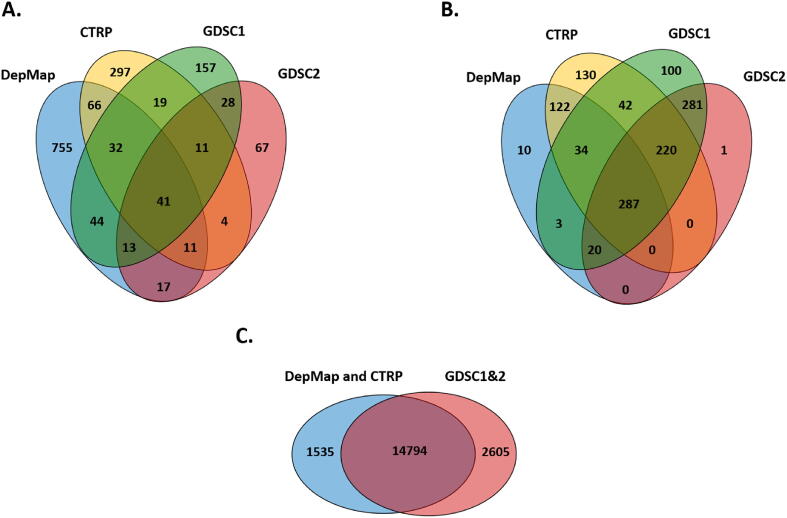
The complete aggregated database contains 1562 compounds tested in at least one drug screening projects in a minimum of 100 cell lines with reported IC50 or AUDRC values. Of these, there are 979 compounds in the DepMap project, 481 compounds in the CTRP, 345 compounds in the GDSC1, and 192 compounds in the GDSC2 projects. A total of 286 (18.3%) compounds were tested in at least two drug screening projects and 41 (2.6%) agents were tested in all four projects (Fig. 2A).
Fig. 2.
Venn diagrams comparing the four included datasets, including The number of investigated agents (A), the number of cell lines (B), and the number of genes (C) used in each included cohort.
The database contains a total of 120 FDA approved anticancer agents. Half of the authorized therapeutic products (n = 61) were categorized as a chemotherapy with antimetabolites being the most common (n = 16). A second major group in medicines authorized for oncology are the targeted therapies (n = 52) and the vast majority (n = 47) of these is involved in the inhibition of a signaling pathway. A complete list of all available oncology licensed compounds is presented in Table 1.
Table 1.
List of all FDA approved oncology drugs with available in vitro resistance data.
| Agent | Mechanism of action | Target/Classification | Category |
|---|---|---|---|
| 5-fluorouracil | antimetabolite | DNA | chemotherapy |
| abemaciclib | CDK inhibitor | CDK inhibitor | targeted |
| abiraterone | antiandrogen | hormonal | hormonal |
| afatinib | EGFR inhibitor | signal transduction inhibitor | targeted |
| alectinib | ALK inhibitor | signal transduction inhibitor | targeted |
| alpelisib | PI3K inhibitor | signal transduction inhibitor | targeted |
| axitinib | anti-angiogenesis | signal transduction inhibitor | targeted |
| azacitidine | antimetabolite, hypomethylating agent | DNA | chemotherapy |
| belinostat | HDAC inhibitor | HDAC inhibitor | chemotherapy |
| bendamustine | alkylating agent | DNA | chemotherapy |
| bexarotene | retinoid receptor agonist | differentiating agent | miscellaneous |
| bicalutamide | antiandrogen | hormonal | hormonal |
| binimetinib | MEK inhibitor | signal transduction inhibitor | targeted |
| bleomycin | antitumor antibiotic | DNA | chemotherapy |
| bortezomib | proteasome inhibitor | proteasome inhibitor | chemotherapy |
| bosutinib | BCR-ABL inhibitor | signal transduction inhibitor | targeted |
| brigatinib | ALK inhibitor | signal transduction inhibitor | targeted |
| busulfan | alkylating agent | DNA | chemotherapy |
| cabazitaxel | antimicrotubular agent | DNA | chemotherapy |
| cabozantinib | multiple receptor tyrosine kinase inhibitor | signal transduction inhibitor | targeted |
| carfilzomib | proteasome inhibitor | proteasome inhibitor | chemotherapy |
| carmustine | alkylating agent | DNA | chemotherapy |
| cetuximab | EGFR inhibitor | signal transduction inhibitor | targeted |
| chlorambucil | alkylating agent | DNA | chemotherapy |
| cisplatin | platinum analog | DNA | chemotherapy |
| cladribine | antimetabolite | DNA | chemotherapy |
| clofarabine | antimetabolite | DNA | chemotherapy |
| cobimetinib | MEK inhibitor | signal transduction inhibitor | targeted |
| crizotinib | multiple receptor tyrosine kinase inhibitor | signal transduction inhibitor | targeted |
| cyclophosphamide | alkylating agent | DNA | chemotherapy |
| cytarabine | antimetabolite | DNA | chemotherapy |
| dabrafenib | BRAF inhibitor | signal transduction inhibitor | targeted |
| dacarbazine | alkylating agent | DNA | chemotherapy |
| dacomitinib | EGFR inhibitor | signal transduction inhibitor | targeted |
| dactinomycin | antitumor antibiotic | DNA | chemotherapy |
| dasatinib | BCR-ABL inhibitor | signal transduction inhibitor | targeted |
| daunorubicin | antitumor antibiotic | DNA | chemotherapy |
| decitabine | antimetabolite | DNA | chemotherapy |
| docetaxel | antimicrotubular agent | DNA | chemotherapy |
| doxorubicin | antitumor antibiotic | DNA | chemotherapy |
| epirubicin | antitumor antibiotic | DNA | chemotherapy |
| erdafitinib | FGFR inhibitor | signal transduction inhibitor | targeted |
| erlotinib | EGFR inhibitor | signal transduction inhibitor | targeted |
| estramustine | antimicrotubular agent | DNA | chemotherapy |
| etoposide | topoisomerase inhibitor | DNA | chemotherapy |
| etoposide-phosphate | topoisomerase inhibitor | DNA | chemotherapy |
| everolimus | mTOR inhibitor | signal transduction inhibitor | targeted |
| fedratinib | JAK inhibitor | signal transduction inhibitor | chemotherapy |
| floxuridine | antimetabolite | DNA | chemotherapy |
| fludarabine | antimetabolite | DNA | chemotherapy |
| fulvestrant | antiestrogen | hormonal | hormonal |
| gefitinib | EGFR inhibitor | signal transduction inhibitor | targeted |
| gemcitabine | antimetabolite | DNA | chemotherapy |
| hydroxyurea | antimetabolite | DNA | chemotherapy |
| ibrutinib | BTK inhibitor | signal transduction inhibitor | targeted |
| idarubicin | antitumor antibiotic | DNA | chemotherapy |
| idelalisib | PI3K inhibitor | signal transduction inhibitor | targeted |
| ifosfamide | alkylating agent | DNA | chemotherapy |
| imatinib | BCR-ABL inhibitor | signal transduction inhibitor | targeted |
| Irinotecan | topoisomerase inhibitor | DNA | chemotherapy |
| ixabepilone | antimicrotubular agent | DNA | chemotherapy |
| ixazomib | proteasome inhibitor | proteasome inhibitor | chemotherapy |
| lapatinib | ERBB inhibitor | signal transduction inhibitor | targeted |
| lenalidomide | immunomodulatory | miscellaneous | miscellaneous |
| lenvatinib | multiple receptor tyrosine kinase inhibitor | signal transduction inhibitor | targeted |
| mechlorethamine | alkylating agent | DNA | chemotherapy |
| melphalan | alkylating agent | DNA | chemotherapy |
| mercaptopurine | antimetabolite | DNA | chemotherapy |
| methotrexate | antimetabolite | DNA | chemotherapy |
| midostaurin | FLT3 inhibitor | signal transduction inhibitor | targeted |
| mitomycin-c | antitumor antibiotic | DNA | chemotherapy |
| mitoxantrone | antitumor antibiotic | DNA | chemotherapy |
| nelarabine | antimetabolite | DNA | chemotherapy |
| neratinib | ERBB inhibitor | signal transduction inhibitor | targeted |
| nilotinib | BCR-ABL inhibitor | signal transduction inhibitor | targeted |
| niraparib | PARP inhibitor | signal transduction inhibitor | targeted |
| olaparib | PARP inhibitor | signal transduction inhibitor | targeted |
| mepesuccinate | BCR-ABL inhibitor | signal transduction inhibitor | targeted |
| osimertinib | EGFR inhibitor | signal transduction inhibitor | targeted |
| oxaliplatin | platinum analog | DNA | chemotherapy |
| paclitaxel | antimicrotubular agent | DNA | chemotherapy |
| palbociclib | CDK inhibitor | CDK inhibitor | targeted |
| panobinostat | HDAC inhibitor | HDAC inhibitor | chemotherapy |
| pazopanib | multiple receptor tyrosine kinase inhibitor | signal transduction inhibitor | targeted |
| pemetrexed | antimetabolite | DNA | chemotherapy |
| ponatinib | BCR-ABL inhibitor | signal transduction inhibitor | targeted |
| pralatrexate | antimetabolite | DNA | chemotherapy |
| procarbazine | alkylating agent | DNA | chemotherapy |
| regorafenib | multiple receptor tyrosine kinase inhibitor | signal transduction inhibitor | targeted |
| ribociclib | CDK inhibitor | CDK inhibitor | targeted |
| romidepsin | HDAC inhibitor | HDAC inhibitor | chemotherapy |
| rucaparib | PARP inhibitor | signal transduction inhibitor | targeted |
| selinexor | XPO inhibitor | nuclear export inhibitor | targeted |
| selumetinib | MEK inhibitor | signal transduction inhibitor | targeted |
| sirolimus | mTOR inhibitor | signal transduction inhibitor | targeted |
| sonidegib | hedgehog inhibitor | signal transduction inhibitor | targeted |
| sorafenib | multiple receptor tyrosine kinase inhibitor | signal transduction inhibitor | targeted |
| sunitinib | multiple receptor tyrosine kinase inhibitor | signal transduction inhibitor | targeted |
| talazoparib | PARP inhibitor | signal transduction inhibitor | targeted |
| tamoxifen | antiestrogen | hormonal | hormonal |
| tazemetostat | histone lysine methyltransferase inhibitor | methyltransferase inhibitor | targeted |
| temozolomide | alkylating agent | DNA | chemotherapy |
| temsirolimus | mTOR inhibitor | signal transduction inhibitor | targeted |
| teniposide | topoisomerase inhibitor | DNA | chemotherapy |
| thioguanine | antimetabolite | DNA | chemotherapy |
| tipiracil | antimetabolite | DNA | chemotherapy |
| tirbanibulin | microtubule inhibitor | DNA | chemotherapy |
| tivozanib | anti angiogenesis | signal transduction inhibitor | targeted |
| topotecan | topoisomerase inhibitor | DNA | chemotherapy |
| toremifene | antiestrogen | hormonal | hormonal |
| trametinib | MEK inhibitor | signal transduction inhibitor | targeted |
| tucatinib | ERBB inhibitor | signal transduction inhibitor | targeted |
| valrubicin | topoisomerase inhibitor | DNA | chemotherapy |
| vandetanib | multiple receptor tyrosine kinase inhibitor | signal transduction inhibitor | targeted |
| venetoclax | BCL2 inhibitor | signal transduction inhibitor | targeted |
| vinblastine | microtubule inhibitor | DNA | chemotherapy |
| vincristine | microtubule inhibitor | DNA | chemotherapy |
| vinorelbine | microtubule inhibitor | DNA | chemotherapy |
| vismodegib | hedgehog inhibitor | signal transduction inhibitor | targeted |
| vorinostat | HDAC inhibitor | HDAC inhibitor | chemotherapy |
The database also includes therapeutic agents that are licensed for non-oncological indications (n = 233) as well as compounds that are in the experimental and investigational phase (n = 1209).
3.2. Cell lines in the database
Regarding the cell lines in the database, a total of 1250 cell lines were utilized in at least one source dataset. Of these, there are 835 cell lines in the CTRP, 476 in the DepMap, 987 in the GDSC1, and 809 in the GDSC2 projects. A total of 1009 (80.7%) cell lines were tested in at least two drug screening projects and 287 (22.9%) cell lines are available in each source dataset (Fig. 2B). In order to have an adequate sample size for the analyses, certain tumor subtypes were grouped together to create a total of 32 subgroups. A complete list of all 1250 cell lines available in the platform is presented in Supplemental Table S1.
3.3. Gene expression database
The gene expression data table of the CCLE cell lines used by the CTRP and DepMap projects contains 19,148 unique HGNC identifiers, while the gene expression data table used for the GDSC1 and GDSC2 projects contains 17,399 unique HGNC identifiers. Genes whose expression was zero in more than half of the tested cell lines were excluded (n = 2819) from the gene expression table of the CCLE cell lines resulting in a total of 16,329 genes with expression values in the database (Fig. 2C).
3.4. Ranking of genes associated with ERBB2 inhibition resistance
Two drugs in the DrugBank database targeting the ERBB2 tyrosine kinase domain with a known IC50 or AUDRC values were evaluated in all four included datasets, afatinib and lapatinib. The analysis was performed in each included source cohort separately by using the integrated database and platform to uncover gene expression-based markers of resistance in solid tumors.
Of the three ERBB receptors, the expression of the ERBB2 and ERBB3 genes had a significant association with the therapeutic response for both drugs regardless of the basis of categorization (IC50 or AUDRC). The EGFR (ERBB1) gene had no statistically significant associations in two datasets for lapatinib and in one dataset for afatinib treatment. Detailed results can be found in Table 2. When generating the list of cell lines with the highest sensitivity and resistance, we used both IC50 and AUDRC based classifications. The lists of top ten cell lines are presented as Table 3.
Table 2.
ROC AUC results and Mann-Whitney test p-values of ERBB receptor tyrosine kinase targeting agents using tertile IC50 and AUDRC based categorization of therapeutic response in each dataset separately.
| Response based on | Dataset | EGFR |
ERBB2 |
ERBB3 |
|||
|---|---|---|---|---|---|---|---|
| Afatinib | Lapatinib | Afatinib | Lapatinib | Afatinib | Lapatinib | ||
| lower vs upper tertile of IC50 | DEPMAP | 0.659 (3.7e-06) | 0.616 (1.8e-03) | 0.735 (8.6e-12) | 0.787 (1.3e-14) | 0.672 (5.7e-07) | 0.679 (1.1e-07) |
| GDSC1 | 0.639 (8.0e-10) | 0.741 (2.5e-08) | 0.658 (2.4e-12) | 0.770 (4.1e-10) | 0.587 (1.1e-04) | 0.609 (5.5e-03) | |
| GDSC2 | n.s. | n.s. | 0.619 (2.1e-05) | 0.577 (7.4e-03) | 0.564 (2.3e-02) | n.s. | |
| CTRP | not applicable | not applicable | not applicable | not applicable | not applicable | not applicable | |
| lower vs upper tertile of AUDRC | DEPMAP | 0.679 (2.0e-07) | 0.655 (3.2e-05) | 0.761 (3.8e-14) | 0.797 (2.0e-15) | 0.652 (9.7e-06) | 0.694 (1.9e-07) |
| GDSC1 | 0.716 (1.6e-16) | 0.728 (3.9e-06) | 0.774 (1.1e-25) | 0.784 (9.2e-09) | 0.683 (2.3e-12) | 0.628 (9.6e-03) | |
| GDSC2 | 0.592 (8.2e-04) | 0.574 (8.5e-03) | 0.657 (1.4e-08) | 0.617 (3.2e-05) | 0.595 (6.2e-04) | 0.575 (8.1e-03) | |
| CTRP | 0.681 (1.9e-10) | 0.614 (7.1e-05) | 0.715 (3.8e-14) | 0.670 (2.8e-09) | 0.659 (2.0e-08) | 0.694 (1.2e-11) | |
n.s.: not significant.
Table 3.
TOP10 lapatinib treated sensitive (upper panel) and resistant (lower panel) cell lines from the CTRP database.
| Cell line | Disease | Standardized AUDRC |
|---|---|---|
| NCIN87 | Gastric Cancer/Adenocarcinoma | 0.178 |
| HCC2218 | Breast Cancer/Breast Ductal Carcinoma | 0.185 |
| LC1F | Non-Small Cell Lung Cancer (NSCLC) | 0.213 |
| ZR7530 | Breast Cancer/Breast Ductal Carcinoma | 0.222 |
| SNU175 | Colon adenocarcinoma | 0.228 |
| YD10B | Head and Neck Cancer/Squamous Cell Carcinoma | 0.236 |
| HCC2935 | Non-Small Cell Lung Cancer (NSCLC) | 0.251 |
| UBLC1 | Bladder carcinoma | 0.251 |
| TE617T | Rhabdomyosarcoma | 0.255 |
| NUGC4 | Gastric adenocarcinoma | 0.257 |
| Cell line | Disease | Standardized AUDRC |
|---|---|---|
| CAL120 | Breast carcinoma | 0.586 |
| MHH-CALL-4 | Acute Lymphoblastic Leukemia (ALL); B-cell | 0.582 |
| BFTC-909 | Renal Carcinoma; transitional cell | 0.576 |
| KMM1 | Multiple myeloma | 0.547 |
| BEN | Non-Small Cell Lung Cancer (NSCLC) | 0.547 |
| HEC265 | Endometrial adenocarcinoma | 0.546 |
| KARPAS620 | Multiple myeloma | 0.545 |
| RERFLCAD1 | Non-Small Cell Lung Cancer (NSCLC) | 0.543 |
| DLD1 | Colon adenocarcinoma | 0.542 |
| TOV21G | Clear cell adenocarcinoma of the ovary | 0.538 |
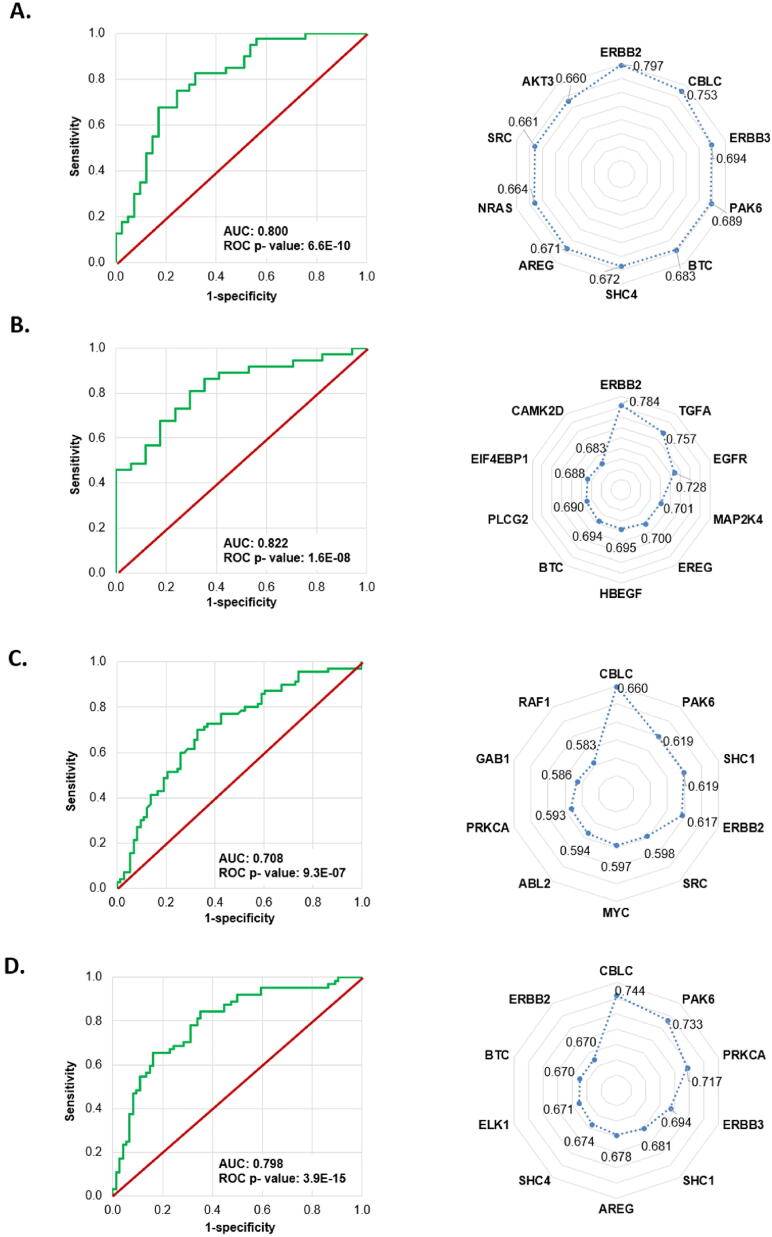
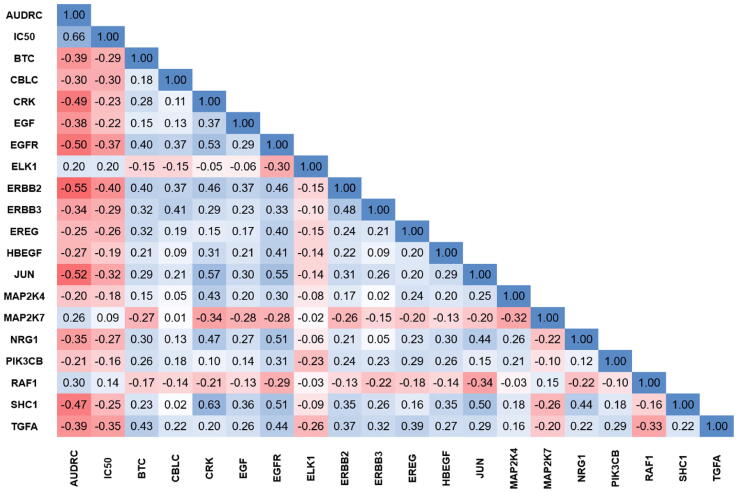
From the 87 genes included in the ERBB pathway we found 25, 30, 24, and 40 genes with significant association with response to lapatinib treatment in DepMap, GDSC1, GDSC2, and CTRP datasets, respectively. The best performing model was observed in the GDSC1 dataset with an overall accuracy of 0.778 and a ROC AUC of 0.822 (p = 1.6E-08). Summary table and radar charts based on the ROC AUC values as well as the ROC plots of the combined models are presented in Fig. 3 and Table 4. Of the significant variables, ERBB2 in the DEPMAP (r = −0.44, p = 7.27E-13) and GDSC1 (r = −0.55, p = 9.99E-22) data set, and CBLC in the GDSC2 (r = −0.27, p = 1.22E-09) and CTRP data sets (r = −0.37, p = 6.89E-18) showed the strongest correlation with the AUDRC values. A correlation matrix between drug screening results (AUDRC and IC50) and gene expression can reveal the influence of individual genes on each other. In Fig. 4 we show a chart depicting only significant genes for which the Spearman correlation coefficient (when compared to AUDRC) was below −0.20 or over ≥ 0.20 in the GDSC1 dataset.
Fig. 3.
ROC curves of the random forest models in the test sets and radar chart of the most significant genes correlated with lapatinib resistance in each dataset, including DepMap (A), GDSC1 (B), GDSC2 (C), and CTRP (D). The values presented in the radar chart are the ROC AUC values for the individual genes.
Table 4.
Summary performance of random forest models for lapatinib resistance in the test set in each dataset.
| Dataset | Number of cell lines | Accuracy | Kappa | Sensitivity | Specificity | Precision | ROC AUC | ROC AUC p-value |
|---|---|---|---|---|---|---|---|---|
| DepMap | 240 | 0.741 | 0.482 | 0.683 | 0.800 | 0.778 | 0.800 | 6.60E-10 |
| GDSC1 | 160 | 0.778 | 0.450 | 0.529 | 0.892 | 0.692 | 0.822 | 1.60E-08 |
| GDSC2 | 422 | 0.671 | 0.344 | 0.630 | 0.714 | 0.697 | 0.708 | 9.30E-07 |
| CTRP | 409 | 0.710 | 0.417 | 0.730 | 0.688 | 0.730 | 0.798 | 3.90E-15 |
Fig. 4.
Correlation between genes related to resistance against the ERBB tyrosine kinase inhibitor lapatinib. A correlation matrix between drug screening results (AUDRC and IC50) and gene expressions using the GDSC1 dataset is shown. The chart includes only significant genes of the KEGG ERBB pathway for which the Spearman correlation coefficient (when compared to AUDRC) was ≤ -0.20 or ≥ 0.20.
4. Discussion
Resistance against systemic therapy is a main limitation of current cancer treatment. The utilization of in vitro models can provide two important advantages: one can explore the off-target effects of non-oncology drugs related to their potential anticancer repurposing [20] and one can pinpoint new biomarkers of resistance to established agents. Drug repurposing is a common concept also reflected by the numerous studies included in the oncology drug repurposing database [21].
The aim of our study was to enable straightforward utilization of in vitro results by establishing a tool to link gene expression and drug sensitivity in a cohort of cell lines from four large cohorts. The complete analysis platform is set up in a way that all available databases and all available cell lines will be used regardless of the selected drug. In addition to the analysis of single genes, we also established a pipeline for the ranking and validation of gene signatures. To enable prompt utilization of the platform we extended our online predictive biomarker discovery application, which previously used clinical samples from breast [18], ovarian [22], glioblastoma [23] and colorectal cancer patients to link gene expression and therapeutic response.
To assess the role of different pathways and cancer hallmark genes in therapeutic responses of drugs we have set up a machine learning-based ranking and validation and utilized this feature to evaluate genes related to anti-ERBB2 therapy resistance. In this, genes related to the resistance against two tyrosine kinase inhibitors were investigated, afatinib and lapatinib. Afatinib is an orally administered irreversible inhibitor of ERBB1 (EGFR), ERBB2, and ERBB4 first approved in 2013 [24]. The ERBB1 and ERBB2 inhibitor lapatinib was approved in 2007 after it showed improved outcome in breast cancer patients whose tumors expressed the ERBB2 (HER2) receptor [25]. Although all four ERBB receptors were implicated in cancer, only ERBB1, ERBB2, and ERBB4 have intracellular tyrosine kinase domains [26]. Despite high success rate, a significant proportion of patients develop resistance against these tyrosine kinase inhibitors [27].
Here, by analyzing in vitro data, we pinpoint the genes with the highest correlation to resistance against lapatinib and afatinib. In particular, the strongest genes were ERBB2 itself and Cbl Proto-Oncogene C (CBLC). Cbl proteins ubiquitinate and downregulate other tyrosine kinases and regulate ERBB signal transduction [28]. The expression of CBLC is higher in different tumor types including lung, pancreatic, breast, and colorectal cancer cells and has been suggested as a therapeutic target in lung adenocarcinoma [29]. Our results are strongly supported by a previously described link between CBLC and resistance against lapatinib [30]. A third gene among the most significant hits across multiple datasets was PAK6, a gene encoding a serine/threonine-protein kinase. A previous study utilizing HER2 positive cell lines identified the Akt-signaling pathway in cell lines resistant against ERBB2 inhibitors and suggested PAK6 as a biomarker of resistance [31]. Without further elaboration on individual gens we have to emphasize the high proportion of overlapping hits among the different analyses. These results suggest that the resistance mechanisms converge on a few genes and thus provide a support for the utilization of predictive biomarkers for anti-ERBB tyrosine kinase domain inhibitor therapy.
Another important observation is the superior performance of the random-forest derived single signature when compared to individual genes. The signature had higher AUC value than any gene with the exception of ERBB2 itself in the DepMap cohort. The online analysis portal enables the setup of such resistance-associated signatures for each available drug in an automated manner – see, for example, a previous signature of resistance against EGFR inhibitors manually identified in lung cancer [32].
Notable, some analysis for the investigated datasets are already available at the original repositories. In addition, some previous tools enable the analysis of multiple cell line cohorts as well. The GEMiCCL – Gene Expression and Mutations in Cancer Cell Lines portal was set up to mine and visualize gene expression and mutation data of cell lines [33]. The CellminerCDB is a pharmacogenomic data portal primarily based on the NCI-60 cell lines which integrates multiple layers of data [34]. The web portal we present here has a novel unique place among these resources because we have incorporated more recent datasets, we provide a straightforward automated selection for the investigated genes and we also provide a machine learning algorithm for the data analysis.
There are a few limitations to our study. Firstly, only the reported IC50 and AUDRC valued were used to determine the sensitivity or resistance of a cell line to a particular therapy. Using a fixed cutoff of tertiles for determining sensitivity/resistance might be looked upon as artificial. A second limitation is that not all agents are measured in each cell line, thus, depending on the applied filtering, some drugs cannot be investigated by the proposed pipeline. A third limitation is the utilization of transcriptomic data only – some of the results may be affected by mutations in various genes in individual cell lines. For example, we chose afatinib and lapatinib for our analyses. Afatinib was approved for metastatic NSCLC tumors with L858R variants or exon 19 deletion. The cross-tumor analysis of the data for this inhibitor presented in Table 2 could be affected by the proportion of the cell lines carrying EGFR variants affecting sensitivity to afatinib and by the proportion of NSCLC cell lines in each dataset. Multiple ERBB2 mutations may also affect sensitivity or resistance to lapatinib.
Overall, we have collected and created a unified analysis interface enabling simultaneous mining of four cancer cell line database. The registration-free web application is available at http://www.rocplot.com/cells. We utilized this platform to rank genes correlated to resistance against ERBB2 tyrosine kinase domain inhibitors.
CRediT authorship contribution statement
János Tibor Fekete: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. Balázs Győrffy: Conceptualization, Formal analysis, Funding acquisition, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The research was financed by the FIEK_16-1-2016-0005 and 2020-1.1.6-JÖVŐ-2021-00013 grants and by the Higher Education Institutional Excellence Program (2020-4.1.1.-TKP2020) of the Ministry for Innovation and Technology in Hungary, within the framework of the Bionic thematic program of the Semmelweis University. The authors acknowledge the support of ELIXIR Hungary (www.elixir-hungary.org).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2022.06.007.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021 doi: 10.3322/caac.21660. május;71(3):209–49. [DOI] [PubMed] [Google Scholar]
- 2.Falzone L., Salomone S., Libra M. Evolution of Cancer Pharmacological Treatments at the Turn of the Third Millennium. Front Pharmacol. 2018;november 13:9:1300. doi: 10.3389/fphar.2018.01300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mihály Z., Kormos M., Lánczky A., Dank M., Budczies J., Szász M.A., et al. A meta-analysis of gene expression-based biomarkers predicting outcome after tamoxifen treatment in breast cancer. Breast Cancer Res Treat. 2013 doi: 10.1007/s10549-013-2622-y. július;140(2):219–32. [DOI] [PubMed] [Google Scholar]
- 4.Sleire L, Førde HE, Netland IA, Leiss L, Skeie BS, Enger PØ. Drug repurposing in cancer. Pharmacol Res. 2017. Október. 124. 74–91. [DOI] [PubMed]
- 5.Ishida J., Konishi M., Ebner N., Springer J. Repurposing of approved cardiovascular drugs. J Transl Med. 2016 doi: 10.1186/s12967-016-1031-5. december;14(1):269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang J, Zhao D, Liu Z, Liu F. Repurposing psychiatric drugs as anti-cancer agents. Cancer Lett. 2018. Április. 419. 257–65. [DOI] [PubMed]
- 7.Gillet J.P., Varma S., Gottesman M.M. The Clinical Relevance of Cancer Cell Lines. JNCI. J Natl Cancer Inst. 2013:452–458. doi: 10.1093/jnci/djt007. április 3.; 105(7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Györffy B., Surowiak P., Kiesslich O., Denkert C., Schäfer R., Dietel M., et al. Gene expression profiling of 30 cancer cell lines predicts resistance towards 11 anticancer drugs at clinically achieved concentrations. Int J Cancer. 2006:1699–1712. doi: 10.1002/ijc.21570. április 1.; 118(7) [DOI] [PubMed] [Google Scholar]
- 9.Shoemaker R.H. The NCI60 human tumour cell line anticancer drug screen. Nat Rev Cancer. 2006 doi: 10.1038/nrc1951. október;6(10):813–23. [DOI] [PubMed] [Google Scholar]
- 10.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, és mtsai. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012. március 29.;483(7391):603–7. [DOI] [PMC free article] [PubMed]
- 11.Basu A., Bodycombe N.E., Cheah J.H., Price E.V., Liu K., Schaefer G.I., et al. An Interactive Resource to Identify Cancer Genetic and Lineage Dependencies Targeted by Small Molecules. Cell. 2013 doi: 10.1016/j.cell.2013.08.003. augusztus;154(5):1151–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang W, Soares J, Greninger P, Edelman EJ, Lightfoot H, Forbes S, és mtsai. Genomics of Drug Sensitivity in Cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res. 2012. november 22. 41(D1). D955–61. [DOI] [PMC free article] [PubMed]
- 13.Corsello S.M., Nagari R.T., Spangler R.D., Rossen J., Kocak M., Bryan J.G., et al. Discovering the anticancer potential of non-oncology drugs by systematic viability profiling. Nat. Cancer. 2020 doi: 10.1038/s43018-019-0018-6. február;1(2):235–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iorio F., Knijnenburg T.A., Vis D.J., Bignell G.R., Menden M.P., Schubert M., et al. A Landscape of Pharmacogenomic Interactions in Cancer. Cell. 2016 doi: 10.1016/j.cell.2016.06.017. július;166(3):740–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rees M.G., Seashore-Ludlow B., Cheah J.H., Adams D.J., Price E.V., Gill S., et al. Correlating chemical sensitivity and basal gene expression reveals mechanism of action. Nat Chem Biol. 2016 doi: 10.1038/nchembio.1986. február;12(2):109–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meyers R.M., Bryan J.G., McFarland J.M., Weir B.A., Sizemore A.E., Xu H., et al. Computational correction of copy number effect improves specificity of CRISPR–Cas9 essentiality screens in cancer cells. Nat Genet. 2017 doi: 10.1038/ng.3984. december;49(12):1779–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Menyhárt O., Harami-Papp H., Sukumar S., Schäfer R., Magnani L., de Barrios O., et al. Guidelines for the selection of functional assays to evaluate the hallmarks of cancer. Biochim Biophys Acta BBA – Rev. Cancer. 2016 doi: 10.1016/j.bbcan.2016.10.002. december;1866(2):300–19. [DOI] [PubMed] [Google Scholar]
- 18.Fekete J.T., Győrffy B. ROCplot.org: Validating predictive biomarkers of chemotherapy/hormonal therapy/anti-HER2 therapy using transcriptomic data of 3,104 breast cancer patients. Int J Cancer. 2019 doi: 10.1002/ijc.32369. december;145(11):3140–51. [DOI] [PubMed] [Google Scholar]
- 19.Wishart DS, Feunang YD, Guo AC, Lo EJ, Marcu A, Grant JR, és mtsai. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018. január 4. 46(D1). D1074–82. [DOI] [PMC free article] [PubMed]
- 20.Zhang Z., Zhou L., Xie N., Nice E.C., Zhang T., Cui Y., et al. Overcoming cancer therapeutic bottleneck by drug repurposing. Signal Transduct Target Ther. 2020 doi: 10.1038/s41392-020-00213-8. december;5(1):113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pantziarka P, Verbaanderd C, Sukhatme V, Capistrano R, Crispino S, Gyawali B, és mtsai. ReDO_DB: the repurposing drugs in oncology database. ecancermedicalscience [Internet]. 2018. december 6. [idézi 2022. április 10.];12. Elérhető: https://ecancer.org/journal/12/full/886-redo_db-the-repurposing-drugs-in-oncology-database.php. [DOI] [PMC free article] [PubMed]
- 22.Fekete J.T., Ősz Á., Pete I., Nagy G.R., Vereczkey I., Győrffy B. Predictive biomarkers of platinum and taxane resistance using the transcriptomic data of 1816 ovarian cancer patients. Gynecol Oncol. 2020 doi: 10.1016/j.ygyno.2020.01.006. március;156(3):654–61. [DOI] [PubMed] [Google Scholar]
- 23.Menyhárt O, Fekete JT, Győrffy B. Gene expression-based biomarkers designating glioblastomas resistant to multiple treatment strategies. Carcinogenesis. 2021. június 21. 42(6). 804–13. [DOI] [PMC free article] [PubMed]
- 24.Dungo R.T., Afatinib K.GM. First Global Approval. Drugs. 2013 doi: 10.1007/s40265-013-0111-6. szeptember;73(13):1503–15. [DOI] [PubMed] [Google Scholar]
- 25.Geyer C.E., Forster J., Lindquist D., Chan S., Romieu C.G., Pienkowski T., et al. Lapatinib plus Capecitabine for HER2-Positive Advanced Breast Cancer. N Engl J Med. 2006;355(26):2733–2743. doi: 10.1056/NEJMoa064320. december 28. [DOI] [PubMed] [Google Scholar]
- 26.Roskoski R. Small molecule inhibitors targeting the EGFR/ErbB family of protein-tyrosine kinases in human cancers. Pharmacol Res. 2019. január;139. 395–411. [DOI] [PubMed]
- 27.Zhao Y, Bilal M, Raza A, Khan MI, Mehmood S, Hayat U, és mtsai. Tyrosine kinase inhibitors and their unique therapeutic potentialities to combat cancer. Int J Biol Macromol. 2021. január;168. 22–37. [DOI] [PubMed]
- 28.Kim M., Tezuka T., Suziki Y., Sugano S., Hirai M., Yamamoto T. Molecular cloning and characterization of a novel cbl-family gene, cbl-c. Gene. 1999 doi: 10.1016/s0378-1119(99)00356-x. október;239(1):145–54. [DOI] [PubMed] [Google Scholar]
- 29.Hong S.Y., Lu Y.C., Hsiao S.H., Kao Y.R., Lee M.H., Lin Y.P., et al. Stabilization of AURKA by the E3 ubiquitin ligase CBLC in lung adenocarcinoma. Oncogene. 2022:1907–1917. doi: 10.1038/s41388-022-02180-6. március 25.; 41(13) [DOI] [PubMed] [Google Scholar]
- 30.Masica D.L., Karchin R. Collections of Simultaneously Altered Genes as Biomarkers of Cancer Cell Drug Response. Cancer Res. 2013:1699–1708. doi: 10.1158/0008-5472.CAN-12-3122. március 15.; 73(6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jernström S., Hongisto V., Leivonen S.K., Due E.U., Tadele D.S., Edgren H., et al. Drug-screening and genomic analyses of HER2-positive breast cancer cell lines reveal predictors for treatment response. Breast Cancer Targets Ther. 2017:185–198. doi: 10.2147/BCTT.S115600. március;Volume 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Byers L.A., Diao L., Wang J., Saintigny P., Girard L., Peyton M., et al. An Epithelial-Mesenchymal Transition Gene Signature Predicts Resistance to EGFR and PI3K Inhibitors and Identifies Axl as a Therapeutic Target for Overcoming EGFR Inhibitor Resistance. Clin Cancer Res. 2013:279–290. doi: 10.1158/1078-0432.CCR-12-1558. január 1.; 19(1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jeong I, Yu N, Jang I, Jun Y, Kim MS, Choi J, és mtsai. GEMiCCL: mining genotype and expression data of cancer cell lines with elaborate visualization. Database [Internet]. 2018. január 1. [idézi 2022. május 18.];2018. Elérhető: https://academic.oup.com/database/article/doi/10.1093/database/bay041/4991663. [DOI] [PMC free article] [PubMed]
- 34.Luna A, Elloumi F, Varma S, Wang Y, Rajapakse VN, Aladjem MI, és mtsai. CellMiner Cross-Database (CellMinerCDB) version 1.2: Exploration of patient-derived cancer cell line pharmacogenomics. Nucleic Acids Res. 2021. január 8. 49(D1). D1083–93. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.