Abstract
Background
The response to neoadjuvant chemotherapy (NAC) differs substantially among individual patients with non‐small cell lung cancer (NSCLC). Major pathological response (MPR) is a histomorphological read‐out used to assess treatment response and prognosis in patients NSCLC after NAC. Although spatial metabolomics is a promising tool for evaluating metabolic phenotypes, it has not yet been utilized to assess therapy responses in patients with NSCLC. We evaluated the potential application of spatial metabolomics in cancer tissues to assess the response to NAC, using a metabolic classifier that utilizes mass spectrometry imaging combined with machine learning.
Methods
Resected NSCLC tissue specimens obtained after NAC (n = 88) were subjected to high‐resolution mass spectrometry, and these data were used to develop an approach for assessing the response to NAC in patients with NSCLC. The specificities of the generated tumor cell and stroma classifiers were validated by applying this approach to a cohort of biologically matched chemotherapy‐naïve patients with NSCLC (n = 85).
Results
The developed tumor cell metabolic classifier stratified patients into different prognostic groups with 81.6% accuracy, whereas the stroma metabolic classifier displayed 78.4% accuracy. By contrast, the accuracies of MPR and TNM staging for stratification were 62.5% and 54.1%, respectively. The combination of metabolic and MPR classifiers showed slightly lower accuracy than either individual metabolic classifier. In multivariate analysis, metabolic classifiers were the only independent prognostic factors identified (tumor: P = 0.001, hazards ratio [HR] = 3.823, 95% confidence interval [CI] = 1.716–8.514; stroma: P = 0.049, HR = 2.180, 95% CI = 1.004–4.737), whereas MPR (P = 0.804; HR = 0.913; 95% CI = 0.445–1.874) and TNM staging (P = 0.078; HR = 1.223; 95% CI = 0.977–1.550) were not independent prognostic factors. Using Kaplan‐Meier survival analyses, both tumor and stroma metabolic classifiers were able to further stratify patients as NAC responders (P < 0.001) and non‐responders (P < 0.001).
Conclusions
Our findings indicate that the metabolic constitutions of both tumor cells and the stroma are valuable additions to the classical histomorphology‐based assessment of tumor response.
Keywords: cancer metabolism, machine learning, mass spectrometry imaging, metabolic classifier, Non‐small cell lung cancer, prognosis, spatial metabolomics, treatment response
Abbreviations
- Chemo‐naïve
chemotherapy‐naïve
- CI
confidence interval
- CPA
cyclic phosphatidic acid
- EGFR
epidermal growth factor receptor
- FFPE
formalin‐fixed paraffin‐embedded
- H&E
hematoxylin and eosin
- HR
hazards ratio
- IQR
interquartile range
- LUAD
lung adenocarcinoma
- LUSC
lung squamous cell carcinoma
- LysoPA
lysophosphatidic acid
- LysoPC
lysophosphatidylcholine
- LysoPE
lysophosphatidylethanolamine
- LysoPI
lysophosphatidylinositol
- MALDI‐FT‐ICR‐MSI
matrix‐assisted laser desorption/ionization fourier‐transform ion cyclotron resonance mass spectrometry imaging
- MC
metabolic classifier
- MPR
major pathological response
- NAC
neoadjuvant therapy
- NSCLC
non‐small cell lung cancer
- OCFAs
odd‐chain fatty acids
- OS
overall survival
- PA
phosphatidic acid
- PC
phosphatidylcholine
- PE
phosphatidylethanolamine
- PGP
phosphatidylglycerophosphate
- PI
phosphoinositol
- RF
random forest
- SM
sphingomyelin
- SPACiAL
spatial correlation image analysis
- TG
triglyceride
- TNM
tumor‐node‐metastasis
1. BACKGROUND
Neoadjuvant chemotherapy (NAC), with or without radiotherapy, followed by surgical resection, improves survival in patients with locally advanced non‐small cell lung cancer (NSCLC) compared with surgery alone, particularly among patients with complete pathological response or major pathological response (MPR), which is classically defined as a residual tumor burden of <10% [1, 2, 3, 4]. NAC has become a vital strategy for reducing tumor size and facilitating surgical resection. NAC also allows for intermediate evaluations of treatment response and prevents the development of micrometastases [5, 6]. Along with the recent successes reported for targeted and immune checkpoint therapies in advanced inoperable NSCLC, recent studies indicate that the adjuvant use of these regimens is also beneficial [7, 8]. However, evidence supporting the therapeutic efficacy of these regimens remains scarce in the neoadjuvant setting, although preliminary outcomes reported for immune checkpoint inhibitors [9] and epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors have been promising [10]. The development of more accurate biomarkers for patient stratification or use as surrogate endpoints would likely result in the efficient identification of patients with resectable or potentially resectable NSCLC.
Accurate patient stratification is becoming increasingly important. The pathological tumor‐node‐metastasis (pTNM) classification is the most important and routinely applied prognosis prediction tool for malignant disease. MPR has been associated with long‐term overall survival (OS) among patients with NSCLC who undergo NAC [4, 11, 12]. MPR represents an estimate of residual tumor cell quantity, used to evaluate tumor regression following NAC. Generally, MPR is used to evaluate the therapeutic response and is often used as a surrogate endpoint in clinical studies of resected NSCLC following preoperative therapies [13, 14, 15, 16]. Until recently, 10% of the baseline tumor has been used as the cutoff value for defining residual tumor, which has been associated with significant prognostic value [4, 17‐19]. However, we [20] and others [16, 21] have shown that although a 10% cutoff is appropriate for lung squamous cell carcinoma (LUSC), the cutoff for lung adenocarcinoma (LUAD) should be greater than 50% [16, 20, 21]. The response to NAC presents substantial variability, ranging from complete to subtotal residual tumor regression, and using a cutoff value of 10%, MPR is only achieved in 17%–33% of patients, depending on the therapeutic strategy [13, 22, 23]. Due to clinicopathologic heterogeneity and different biological behaviors within the tumor, patients with NSCLC at similar pathological stages may have notable prognostic differences [24].
Metabolomics is recognized as a crucial scientific field, offering a promising avenue for identifying diagnostic and prognostic biomarkers for use in clinical practice and may serve as a powerful method for screening potential biomarkers in LUAD [25, 26]. Spatial metabolomics is an emerging domain of omics research. It could be used to investigate tumor heterogeneity [27], evaluate surgical resection margins [28], and contribute to tumor classification [29]. Spatial metabolomics and its enabling technology—matrix‐assisted laser desorption/ionization mass spectrometry imaging (MALDI‐MSI)—localize hundreds to thousands of different metabolites directly from biological tissue sections with cellular spatial resolution [30, 31, 32, 33]. The comprehensive analysis of metabolic heterogeneity and the use of MALDI‐MSI have improved our understanding of tumor metabolism [34]. Previous studies using MALDI‐MSI to analyze human specimens, including tumor tissue and body fluids, identified several biomarkers associated with lung cancer and clinical outcomes [35, 36, 37, 38]. Our previous study found that the high‐mass‐resolution matrix‐assisted laser desorption/ionization Fourier‐transform ion cyclotron resonance mass spectrometry imaging (MALDI‐FT‐ICR‐MSI) was suitable for deciphering therapeutic effects and allowed for the assessment of metabolic changes that occur during the treatment of idiopathic pulmonary fibrosis [39]. The metabolic compositions of both tumor cells and stroma were able to provide rich molecular information and may contribute to estimating prognosis in patients with NSCLC following NAC. Spatial metabolomics enables immunophenotype‐guided in situ metabolomics, facilitating the automated identification of histological and functional features in intact tissue sections and the comprehensive analyses of metabolic constitutions of tumor cells and the stroma [27].
To date, no published studies have examined the ability of spatial metabolomics and metabolite identification to characterize treatment response and differentiate patients into non‐responders and responders in NSCLC. Therefore, the purpose of the present study was to investigate whether spatial metabolomics could be applied for the evaluation of NAC treatment response and prognosis in NSCLC.
2. MATERIALS AND METHODS
2.1. Patient samples and tissue microarrays
This study included two retrospective single‐center patient cohorts (Figure 1), comprising cases diagnosed at the Institute of Pathology of the University of Bern (Bern, Switzerland) between 2000 and 2016. All eligible patients had a pathology‐confirmed diagnosis. The NAC cohort included consecutive patients who received at least one cycle of platinum‐based chemotherapy prior to resection [20]. All applied drug combinations for this cohort are summarized in Table 1. The chemotherapy‐naïve cohort included consecutive patients who underwent primary resection for NSCLC without prior chemotherapy or radiotherapy and were histologically and biologically matched with the NAC cohort, as previously described [40]. Biological matching was accomplished by including only locally advanced NSCLC (at least stage IIIA), ensured by mediastinal lymph node metastasis (pN2), which qualifies for a multi‐disciplinary treatment approach integrating neoadjuvant or adjuvant systemic therapy. We did not statistically compare the stage distribution and pN distribution between the neoadjuvant and chemotherapy‐naïve cohorts due to the downstaging that resulted by NAC and the study design inherent bias towards only locally advanced tumors in the chemotherapy‐naïve cohort, chosen for better biological comparability. Exclusion criteria included patients who died within 30 days after surgery and patients without materials for appropriate evaluations of tumor regression. For the NAC cohort, patients were excluded if chemotherapy was applied without neoadjuvant intent prior to resection.
FIGURE 1.
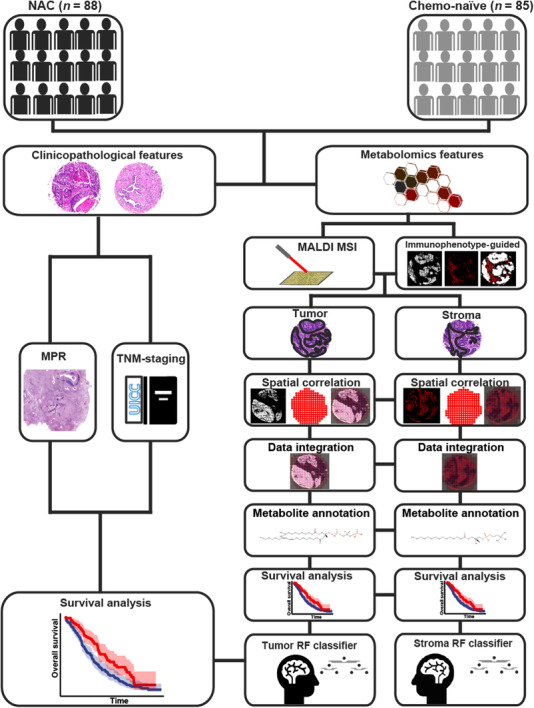
Study design for the development of a metabolic classifier and the assessment of the predictive abilities of clinicopathological features (MPR and TNM staging) and metabolic factors for stratifying patients with NSCLC. Separate metabolic classifiers were established using the metabolic features evaluated in tumors and the stroma, resulting in individual survival risk categories, which were evaluated by Kaplan‐Meier and Cox regression analyses and compared with standard clinicopathological evaluations using MPR and TNM staging. Abbreviations: NAC, neoadjuvant chemotherapy; Chemo, chemotherapy; MALDI MSI, matrix‐assisted laser desorption/ionization mass spectrometry imaging; MPR, major pathological response; TNM, tumor‐node‐metastasis; NSCLC, non‐small cell lung cancer; RF, random forest
TABLE 1.
Baseline clinicopathologic characteristics of the the two cohorts of patients with NSCLC, who either have received neoadjuvant therapy before resection (NAC) or were chemotherapy‐naïve
| Characteristics | NAC (cases [%]) | Chemotherapy‐naïve (cases [%]) | P value |
|---|---|---|---|
| Total | 88 | 85 | |
| Sex | 0.529 | ||
| Male | 59 (63.6) | 53 (62.4) | |
| Female | 29 (36.4) | 32 (37.6) | |
| Smoking status | 0.934 | ||
| Non‐smoker | 12 (13.6) | 9 (10.6) | |
| Ex‐smoker | 29 (33.0) | 26 (30.6) | |
| Active smoker | 37 (42.0) | 33 (38.8) | |
| No record | 10 (11.4) | 17 (20.0) | |
| MPR | NA | ||
| Present | 37 (43.5) | NA | |
| Absent | 51 (56.5) | NA | |
| ypTNM stage * | |||
| 0 | 6 (6.9) | NA | |
| I | 17 (19.5) | NA | |
| II | 20 (23.0) | NA | |
| III | 40 (44.8) | 77 (90.6) | 0.766 |
| IV | 5 (5.8) | 8 (9.4) | |
| ypT stage * | |||
| pT0 | 11 (12.5) | NA | |
| pT1 | 26 (29.5) | 17 (20.0) | 0.259 |
| pT2 | 22 (25.0) | 28 (32.9) | |
| pT3 | 13 (15.9) | 19 (22.4) | |
| pT4 | 16 (18.1) | 21 (24.7) | |
| ypN stage * | |||
| pN0 | 35 (39.8) | NA | |
| pN1 | 20 (22.7) | NA | |
| pN2 | 31 (35.2) | 83 (97.6) | 0.318 |
| pN3 | 2 (2.3) | 2 (2.4) | |
| ypM stage * | 0.352 | ||
| pM0 | 83 (94.3) | 77 (90.6) | |
| pM1 | 5 (5.7) | 8 (9.4) | |
| Subtype | 0.714 | ||
| LUSC | 40 (45.5) | 41 (48.2) | |
| LUAD | 48 (54.5) | 44 (51.8) | |
| Neoadjuvant Chemotherapy | NA | ||
| Cisplatin + docetaxel | 50 (56.8) | NA | |
| Carboplatin + paclitaxel | 3 (3.4) | NA | |
| Cisplatin + pemetrexed | 12 (13.6) | NA | |
| Cisplatin + gemcitabin | 7 (8.0) | NA | |
| Cisplatin + vinorelbin | 9 (10.2) | NA | |
| Others | 7 (8.0) | NA |
pTNM/pT/pN/pM stage for the Chemotherapy‐naïve cohort.
Abbreviations: NSCLC, non‐small cell lung cancer; NAC, neoadjuvant therapy; NA, not applicable; MPR, major pathological response; TNM, tumor‐node‐metastasis; T, component/category coding the extension of the primary tumor; N, component/category coding regional lymph node metastases; M, component/category coding distant metastases; p, pathological TNM staging; yp, pathological TNM staging after neoadjuvant therapy; LUSC, lung squamous cell carcinoma; LUAD, lung adenocarcinoma
Clinicopathological features and follow‐up data were retrieved, as previously described [20], and all data were thoroughly re‐evaluated to update pathological tumor stages according to the Union for International Cancer Control (UICC) and the American Joint Committee on Cancer (AJCC) 8th edition pTNM classification guidelines and harmonize regression grading among other scales [41]. OS was defined as the time elapsed between treatment initiation and death of any cause. Routine clinical follow‐up was performed for all patients, and all available information regarding relapse and disease progression was retrieved from the clinical files.
Response to neoadjuvant therapy was histomorphologically assessed by a pathologist specialized in pulmonary pathology (Sabina Berezowska) and a pulmonary pathology‐experienced student (Philipp Zens) for each case, and the histological data including MPR were previously reported [20]. Residual tumor content was assessed by the histological evaluation of all slides containing the tumor bed, as previously described [4]. MPR was defined as ≤10% residual tumor cells for LUSC or as ≤65% residual tumor cells for LUAD, as previously described [16, 20]. Patients were classified as NAC responders (MPR present) and non‐responders (MPR absent).
Metabolic analysis was performed on tissue microarrays (TMAs) constructed for each cohort, as previously reported [42]. From each patient at least two tumor‐containing tissue cores were collected. Briefly, 0.6‐mm‐diameter tissue cores were annotated on FFPE tissue blocks/slides by a pathologist specialized in lung pathology (Sabina Berezowska) and transposed into an acceptor TMA block. The study was approved by the Cantonal Ethics Commission of the Canton of Bern (KEK 2017‐00830), which waived the requirement for a written informed consent from patients.
2.2. High‐mass‐resolution MALDI‐FT‐ICR‐MSI analysis
MALDI‐MSI was performed as previously described [32, 43]. NSCLC TMA blocks were cut into 4‐μm sections using a microtome (HM 355S, Microm; Thermo Fisher Scientific, Waltham, MA, USA) and mounted onto indium tin oxide‐coated conductive glass slides (Bruker Daltonik GmbH, Bremen, Germany). The slides were coated in 1:1 poly‐L‐lysine (Sigma‐Aldrich, Taufkirchen, Germany) and 0.1% Nonidet P‐40 (Sigma‐Aldrich) before tissue mounting. The tissue sections were incubated at 60°C for 1 h and deparaffinized in xylene (2 × 8 min), followed by drying at room temperature (22°C). Subsequently, the samples were covered with 10 mg/mL 9‐aminoacridine hydrochloride monohydrate matrix (Sigma‐Aldrich) in 70% methanol using a SunCollect sprayer (Sunchrom, Friedrichsdorf, Germany). The matrix application was performed in eight passes using ascending spray rates (flow rates: 10, 20, 30 μL/min for the first three layers, followed by 40 μL/min for the remaining five layers). MALDI‐FT‐ICR‐MSI was performed on a Bruker Solarix 7T FT‐ICR MS (Bruker Daltonik) in the negative ion mode, utilizing 100 laser shots per pixel at a frequency of 1000 Hz. Mass spectra were acquired over a mass range of m/z 75‐1000 Da with a 50‐μm spatial resolution.
2.3. Immunophenotype‐guided MSI analysis, data processing, and pathway analysis
The Spatial Correlation Image Analysis (SPACiAL) workflow was used for the immunophenotype‐guided MALDI‐MSI analysis of automatically annotated tumor and stroma regions in NSCLC tissues, as previously described [27]. SPACiAL is a computational multimodal workflow that includes a series of image and MALDI data processing steps to combine molecular imaging data with multiplex immunofluorescence. The SPACiAL workflow includes MALDI and immunofluorescence data integration, multiple image co‐registration, image digitization, and data conversion. After MALDI imaging, the matrix was removed from the section surface by a 5 min incubation in 70% ethanol, and sections were subsequently stained with hematoxylin and eosin (H&E) in a HistoCore SPECTRA ST multistainer (Leica, Wetzlar, Germany). To remove the H&E stain, we incubated the sections in a Coplin jar containing 100% xylene at room temperature (22°C) for 12 h. The slides were then transferred to a second Coplin jar containing 100% xylene for a 2‐min incubation, to a third Coplin jar containing pure propanol for 2 min, to a fourth Coplin jar containing 100% ethanol for 2 min, and to a fifth Coplin jar containing 1% HCl in 100% ethanol for 5 min. The slides were then washed under running tap water for 5 min. The tissue sections were subjected to immunofluorescence after H&E removal and analyzed by double staining using pan‐cytokeratin (monoclonal mouse pan‐cytokeratin plus [AE1/AE3þ8/18] [1:75], cat#CM162, Biocare Medical, Pacheco, CA, USA) and vimentin antibodies (1:500, clone ab92547, Abcam, Berlin, Germany). Signal detection was conducted using fluorescence‐labeled secondary antibodies (anti‐rabbit IgG DyLight 633 [cat# 35563] and anti‐mouse IgG Alexa Fluor 750 antibody [cat# A‐21037], Thermo Fisher Scientific), and Hoechst 33342 was used for nuclear staining.
Automated steps for the analyses and annotation of tumor and stroma regions were applied to mass spectrometry data using SPACiAL as follows: first, the epithelial marker pan‐cytokeratin (white) was used to stain tumor cells, and vimentin was used to stain stroma regions (red); second, single‐channel images of pan‐cytokeratin and vimentin were used to annotate and separate tumor and stroma using fluorescence imaging; third, the digitized and co‐registered fluorescence images were scaled to match the exact MALDI resolution and converted into numerical matrices comprised of values corresponding to the lightness values for each pixel; fourth, objective tissue annotations were assigned based on semantics and function. The entire workflow is applied to the same tissue section, allowing for the automatic integration of morphological and spatial metabolomics data for thousands of molecules. We established this method and have successfully applied it in previous works [27, 44‐46]. Supplementary Figure S1 displays representative immunofluorescence sections used during this process. Fluorescence stains were scanned at 20× magnification using an AxioScan.Z1 digital slide scanner (Mirax Desk, Carl Zeiss MicroImaging GmbH; Jena, Germany) and visualized using the software ZEN 2.3 blue edition (Zeiss; Oberkochen, Germany).
All root‐mean‐square normalized mass spectra were exported from SCiLS Lab v. 2020 (Bruker Daltonics). Peaks in the mass range of m/z 75‐1000 Da were annotated by accurate mass matching with the Human Metabolome Database (http://www.hmdb.ca/) [47] and METASPACE (http://annotate.metaspace2020.eu [48]; ion mode: negative; adduct type: [M−H], [M−H−H2O], [M+Na−2H], [M+Cl], and [M+K−2H]; mass accuracy, ≤4 ppm). Pathway enrichment analysis was performed on tumor and stromal tissue using MetaboAnalyst 5.0 (https://www.metaboanalyst.ca) [49]. Pathway analysis algorithms, the hypergeometric test for overrepresentation analysis, and relative‐betweenness centrality were selected for pathway topology analysis. P value and impact score were calculated for each metabolic pathway, revealing substantial differences in enriched pathways between the tumor and the stroma.
2.4. Random forest classifiers
The random forest classification, a robust machine learning algorithm, was performed for the classifier. Leave‐one‐out cross‐validation (R 4.0.2) was used to predict long‐ or short‐term survival for each patient. The cohort was separated into long‐term (≥35 months) and short‐term survivors (<35 months) according to median OS. For feature selection, molecules included in the random forest (RF) analysis were selected based on their significance level in the log‐rank test (P < 0.05). After performing the RF analysis, the importance of each molecule was calculated as the total reduction of the criterion brought by that feature (Gini importance). The top 100 most important metabolites according to feature selection were re‐selected as the final variables included in the RF classifiers. The analysis for each RF classifier was repeated 100 times, and a majority vote determined the final prediction model. The mean accuracy, sensitivity, and specificity were used to evaluate the performance of each classifier and MPR. Accuracy refers to the percentage of positive predictions made by the classifier that were correct. The correct prediction is determined by classifying patients with an OS longer than median OS in the long‐term group and that shorter than median OS in the short‐term group. Accuracy was expressed as the ratio of true positives and true negatives to the total observations. Sensitivity was calculated by dividing true positives by all observations in the actual class, representing the percentage of actual positive predictions that were correctly classified by the classifier. Specificity refers to the ratio of true negatives to total negatives. The RF classifiers were calculated using the R package randomForest.
2.5. Further statistical analyses
All analyses were performed using R software (version 4.0.2, https://cran.r‐project.org) with suitable packages. Survival analysis was performed using Kaplan‐Meier analysis and Cox proportional hazards regression, with 95% confidence interval (95% CI) estimates (R 4.0.2, survival). Variables in the multivariate Cox regression were included based on their significance in the log‐rank test (P < 0.05) during univariate Cox regression analyses. Comparisons between tumor and adjacent normal lung tissue were performed using the rank‐based Mann‐Whitney U‐test and Spearman's rank‐order correlation for continuous data (R 4.0.2, corrplot). The log‐rank test was used to assess differences. P values < 0.05 were considered significant.
3. RESULTS
3.1. Patients’ characteristics
The cohort of consecutive patients with resected NSCLC following NAC initially included 117 patients who received at least one cycle of platinum‐based chemotherapy before surgery, as previously described [20]. After all inclusion and exclusion criteria were applied, 88 NAC and 85 chemotherapy‐naïve patients were identified with sufficient materials available for metabolic analyses. The median ages were 62 years (interquartile range [IQR], 42‐77 years) and 63 years (IQR, 39‐84 years) for the NAC and chemotherapy‐naïve cohorts, respectively. No significant differences in the distribution of histological subtypes, median age, or sex were observed between the NAC and chemotherapy‐naïve cohorts. Detailed clinicopathological patient characteristics are summarized in Table 1. A total of 59 adjacent normal lung tissue samples from patients with NSCLC in the NAC cohort were included for analyses.
3.2. Metabolic classifiers established for stratifying patients into prognostic risk groups
Metabolites from tumor and stroma regions were automatically extracted using spatial metabolomics analysis, resulting in 5014 distinct molecular features detected in tissues from all patient samples. In the NAC cohort, metabolic classifiers were trained separately on metabolites identified within the tumor and those identified within stroma tissues. The top 100 metabolites for each classifier, ranked in descending order according to feature importance, are shown for tumors in Figure 2A and for the stroma in Figure 2B. Postulated endogenous annotations for the top 100 molecules are listed in Supplementary Table S1.
FIGURE 2.
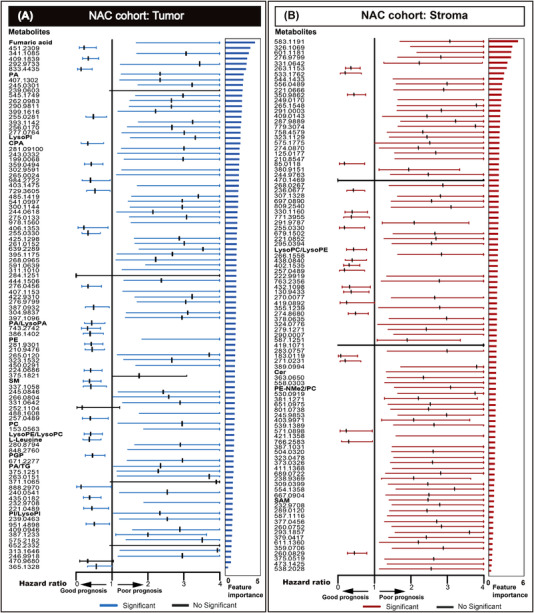
The metabolic classifiers were established for tumor cells (A) and the stroma (B) for stratifying patients with NSCLC who received NAC followed by resection into prognostic risk groups. The forest plot shows the hazard ratio and 95% confidence intervals achieved for best‐performing metabolites for categorizing patients into prognostic risk groups. Metabolites with P < 0.05 are highlighted and ranked in descending order of importance. The feature importance value from 0 to 6, the higher values indicated more impacted on the prediction model. Abbreviations: NSCLC, non‐small cell lung cancer; NAC, neoadjuvant chemotherapy; PA, phosphatidic acid; LysoPI, lysophosphatidylinositol; CPA, cyclic phosphatidic acid; LysoPA, lysophosphatidic acid; PE, phosphatidylethanolamine; SM, sphingomyelin; PC, phosphatidylcholine; LysoPE, lysophosphatidylethanolamine; LysoPC, lysophosphatidylcholine; PGP, phosphatidylglycerophosphate; TG, triglyceride; PI, phosphoinositol; Cer, ceramide; PE‐Nme, dimethylphosphatidylethanolamine; SAM, S‐adenosylmethioninamine
For the metabolic tumor classifier, sphingomyelin (SM, d18:1/15:0 or d16:1/17:0) was shown as an example of a prognosis marker in patients with NSCLC. A high mass intensity for SM was significantly associated with a good prognosis (Figure 3A). Ion distribution maps revealed the specific localization of metabolites in tumor cell regions (Figure 3B). Boxplots displayed the variance in mass intensity values measured for SM (Figure 3C). For the metabolic stroma classifier, a high mass intensity of m/z 480.3091, which can be postulated annotated as either lysophosphatidylcholine (LysoPC, 15:0/0:0) or lysophosphatidylethanolamine (LysoPE, 18:0/0:0), was significantly associated with long survival (Figure 3D) and demonstrated distinct distribution patterns (Figure 3E). Boxplots displayed the variance in mass intensity values measured for LysoPC/LysoPE (Figure 3F). The other important endogenous metabolites used to distinguish between good and poor prognosis in patients with NSCLC following NAC are presented in Supplementary Figure S2.
FIGURE 3.
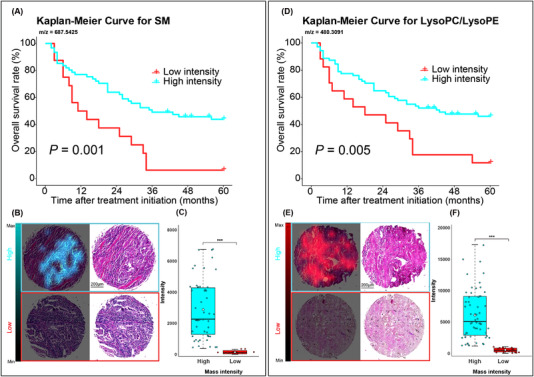
Endogenous metabolites included in the classifiers were used to distinguish between good and poor prognosis groups among NSCLC patients who received neoadjuvant therapy. For tumor classifier, high level of m/z 687.5425 [SM (d18:1/15:0 or d16:1/17:0)] was associated with a good prognosis (A). Ion distribution maps revealed the specific localization of SM in tumor cell regions for high and low mass intensity. The corresponding H&E stains of the same tissue core were shown on the right (B). Boxplot with individual points was shown for variance in different groups for SM (C). For stroma classifier, a high mass intensity of m/z 480.3091 [LysoPC(15:0/0:0)/LysoPE(18:0/0:0)] was associated with a good prognosis (D). The ion map revealed the specific localization of LysoPC/LysoPE in stroma regions (E) and the boxplot was shown for variance in different groups for LysoPC/LysoPE (F). *** P < 0.001. Abbreviations: NSCLC, non‐small cell lung cancer; SM, sphingomyelin; LysoPC, lysophosphatidylcholine; LysoPE, lysophosphatidylethanolamine
We have identified all of the odd‐chain fatty acids (OCFAs) included among the 100 most important molecules used for the metabolic classifier (Supplementary Table S1). The following OCFAs were identified: m/z 687.5425 [SM (d18:1/15:0 or d16:1/17:0)], m/z 480.3091 [LysoPC (15:0/0:0)], m/z 508.3405 [LysoPC (17:0/0:0)], and m/z 852.6496 [PC (15:0/24:0)]. In addition, we performed Spearman's rank‐order correlation analysis between these OCFAs. The results indicated that LysoPC (17:0/0:0) was positively correlated with SM (d18:1/15:0 or d16:1/17:0) and LysoPC (15:0/0:0), whereas PC (15:0/24:0) was negatively correlated with the other OCFAs (Supplementary Figure S3).
3.3. Spatial metabolomics improved the evaluation of therapy response compared with major pathological response
Comparing prognostic efficacy between the two metabolic classifiers and clinicopathological features revealed significant prognostic power for both the tumor (P < 0.001) and stroma (P < 0.001) metabolic classifiers in the Kaplan‐Meier survival analysis, resulting in better prognostic stratification performance than either MPR or TNM staging alone (Figure 4). The tumor metabolic classifier displayed a prediction accuracy of 81.6%, which was similar to that of the stroma metabolic classifier (accuracy = 78.4%) in the NAC cohort (Figure 4A and 4B). The accuracy, sensitivity, and specificity for MPR were all lower than those for either metabolic classifier (Figure 4C). The accuracies achieved when combining the metabolic classifiers with MPR (Supplementary Figure S4) were slightly lower than those achieved by either metabolic classifier alone. In the chemotherapy‐naïve cohort, the accuracies of the tumor (Figure 4D) and stroma metabolic classifier (Figure 4E) were higher than that of TNM staging (Figure 4F).
FIGURE 4.
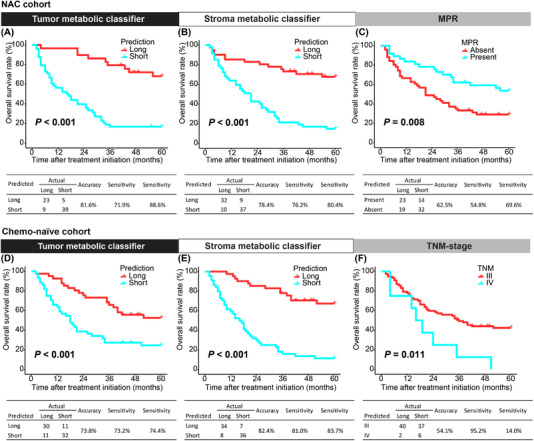
The performance of metabolic classifiers and pathological parameters for stratifying patients with NSCLC into prognostic risk groups. For the NAC cohort, both tumor (A) and stromal (B) metabolic classifiers showed better performance for stratifying prognostic risk groups than MPR (C). For the Chemo‐naïve cohort, the performance of the tumor (D) and stroma (E) metabolic classifiers were superior to TNM staging (F). Kaplan‐Meier survival analyses were used to evaluate differences in patient overall survival. Abbreviations: NSCLC, non‐small cell lung cancer; MPR, major pathological response; NAC, neoadjuvant chemotherapy; Chemo, chemotherapy; TNM, tumor‐node‐metastasis
In univariate analysis, the tumor and stroma metabolic classifiers, MPR, and TNM staging all demonstrated significant predictive efficacy in the NAC cohort (Table 2). However, in the multivariate regression analysis, which included the metabolite‐based classifiers and clinicopathological parameters, the metabolic classifiers were the only independent prognostic factors (tumor metabolic classifier: P = 0.001; stroma metabolic classifier: P = 0.049), whereas MPR and TNM staging were no longer significantly associated with prognosis. In the chemotherapy‐naïve cohort, multivariate analysis confirmed the stroma metabolic classifier as the only independent prognostic factor (P < 0.001), whereas the tumor metabolic classifier and TNM staging were no longer identified as significant factors (Table 2).
TABLE 2.
Univariate and multivariate Cox proportional analyses to identify the OS predictors for patients with NSCLC
| Terms | Univariate Cox analysis | Multivariate Cox analysis | ||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P value | HR | 95% CI | P value | |
| NAC cohort | ||||||
| Tumor metabolic classifier | 5.591 | 2.678‐11.670 | <0.001 | 3.823 | 1.716‐8.514 | 0.001 |
| Stroma metabolic classifier | 4.626 | 2.446‐8.749 | <0.001 | 2.180 | 1.004‐4.737 | 0.049 |
| MPR | 0.465 | 0.260‐0.830 | 0.010 | 0.913 | 0.445‐1.874 | 0.804 |
| TNM staging | 1.304 | 1.082‐1.571 | 0.005 | 1.223 | 0.977‐1.550 | 0.078 |
| Chemo‐naïve cohort | ||||||
| Tumor metabolic classifier | 2.836 | 1.603‐5.016 | <0.001 | 1.767 | 0.971‐3.217 | 0.063 |
| Stroma metabolic classifier | 6.118 | 3.219‐11.630 | <0.001 | 4.953 | 2.517‐9.747 | <0.001 |
| TNM staging | 2.570 | 1.200‐5.502 | 0.015 | 1.417 | 0.653‐3.075 | 0.379 |
Abbreviations: OS, overall survival; NSCLC, non‐small cell lung cancer; HR, hazards ratio; CI, confidence interval; NAC, neoadjuvant chemotherapy; MPR, major pathologic response; TNM, tumor‐node‐metastasis; Chemo, chemotherapy
In addition, we evaluated the abilities of the metabolic classifiers to predict patient survival in relation to the pathological response of MPR. The Kaplan‐Meier survival analyses indicated that the metabolic classifiers were able to further stratify patients according to MPR outcomes. Both tumor and stroma metabolic classifiers predicted significant differences in OS between NAC responders (MPR present; P < 0.001) and non‐responders (MPR absent; P < 0.001) (Figure 5).
FIGURE 5.
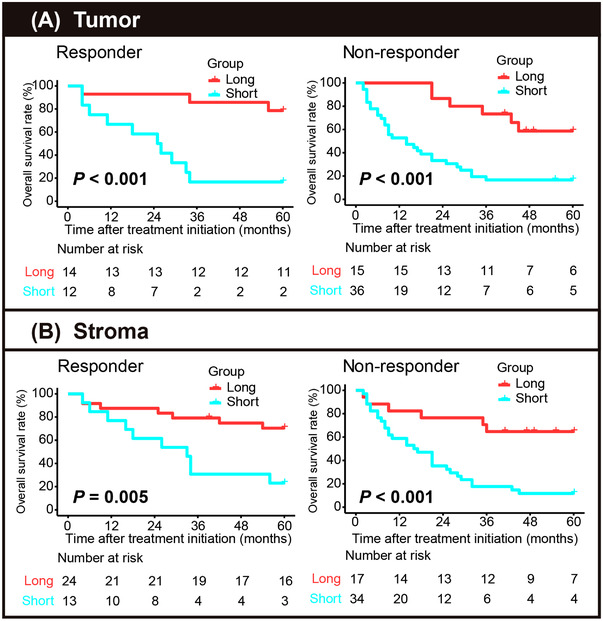
Metabolic classifiers sub‐stratified NSCLC patients with different pathological responses into prognostic risk groups. The tumor (A) and stroma metabolic classifiers (B) could further stratify responder (MPR present) and non‐responder patients (MPR absent) into short‐ and long‐term survivors using the Kaplan‐Meier analysis. Abbreviations: NSCLC, non‐small cell lung cancer; MPR, major pathological response
3.4. Metabolic classifiers were specific to neoadjuvant therapy response
To validate whether the classifiers are specific to NAC response, the metabolite levels measured in the NAC cohort were compared with those measured in the chemotherapy‐naïve cohort (Figure 6). The calculated hazard ratios of the top 100 identified metabolites defined by the NAC classifier were shown for the chemotherapy‐naïve cohort. Significant differences in metabolite levels were identified between the chemotherapy‐naïve and NAC cohorts. Of the 100 included metabolites in the tumor classifier, 93 were significantly associated with OS in the NAC cohort, whereas only 35 were associated with OS in the chemotherapy‐naïve cohort (Figure 6). Of the 100 metabolites included in the stroma classifier, 98 were significantly associated with OS in the NAC cohort, whereas only 32 were significantly associated with OS in the chemotherapy‐naïve cohort (Figure 6). We also compared the 100 most important metabolites between tumor and normal tissues, which showed that 19 metabolites were significantly upregulated and 4 were significantly downregulated in tumor tissues compared with normal tissues. Most of the upregulated metabolites were identified as lipids, including phosphatidylethanolamine (PE), phosphatidic acid (PA), phosphatidylglycerophosphate (PGP), lysophosphatidic acid (LysoPA), lysophosphatidylinositol (LysoPI), phosphoinositol (PI), LysoPC, LysoPE, and SM (Supplementary Figure S5).
FIGURE 6.

Prognostic significance of the metabolites included in the RF classifiers informed by the NAC cohort, compared with the chemotherapy‐naïve cohort. The specificities of the metabolic classifiers were assessed for predicting survival in the NAC and chemotherapy‐naïve cohorts based on the metabolites detected in tumor (A) and stroma tissues (B).The univariate hazard ratios from Cox regression models were calculated for the top 100 metabolites in patients with chemotherapy‐naïve NSCLC, using metabolites in patients treated with NAC as references. Each line in the plot represents the 95% confidence interval, and each row represents the same metabolite. Metabolites with P < 0.05 are highlighted in blue or red. Abbreviations: RF, random forest; NAC, neoadjuvant chemotherapy; Chemo‐naïve, chemotherapy‐naïve; NSCLC, non‐small cell lung cancer
3.5. Pathway enrichment analysis
Metabolic pathway enrichment analysis was performed, and molecules were annotated separately within the tumor and stroma classifiers to investigate metabolic heterogeneity in tumor and stromal tissues. Enriched metabolic pathways in tumor tissues included fructose and mannose metabolism; amino and nucleotide sugar metabolism; starch and sucrose metabolism; galactose metabolism; inositol phosphate metabolism; the pentose phosphate pathway; glycolysis/gluconeogenesis; phosphatidylinositol signaling; and valine, leucine, and isoleucine biosynthesis (Figure 7A). We identified nine enriched pathways in stroma tissues: glycerophospholipid metabolism; purine metabolism; inositol phosphate metabolism; phosphatidylinositol signaling; fructose and mannose metabolism; galactose metabolism; linoleic acid metabolism; starch and sucrose metabolism; and thiamine metabolism (Figure 7B). For the tumor metabolic classifier, the fructose and mannose metabolic pathway showed the highest impact. However, the glycerophospholipid metabolism pathway had the highest impact in the stroma metabolic classifier.
FIGURE 7.
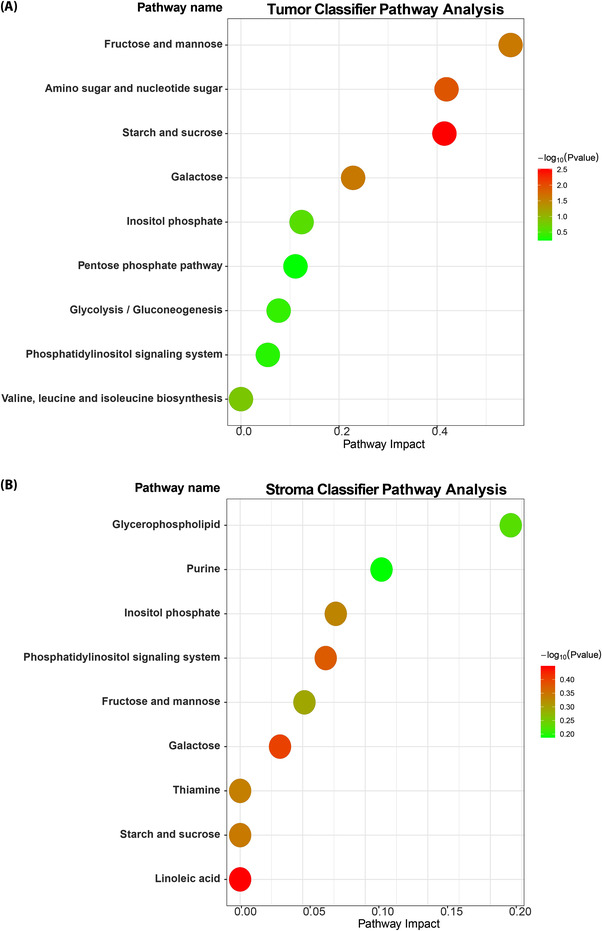
Pathway enrichment analysis shows the distinct metabolic profiles of the tumor and the stroma. Pathway enrichment analysis was performed on the metabolites included in the tumor (A) and stroma classifiers (B). The Y axis indicates the names of the enriched pathways
4. DISCUSSION
The findings of the present study revealed that (1) metabolic response showed superior performance for predicting patient outcomes than the conventional clinicopathological features, such as MPR and TNM stage; (2) our developed metabolic classifiers enabled the stratification of patients within MPR categories; and (3) the metabolic classifiers were specific to NAC‐treated NSCLC compared with chemotherapy‐naïve NSCLC.
The performances of the metabolic classifiers were primarily compared with the performance of MPR, which assesses tumor cell quantity. The metabolic classifiers showed superior performance for the stratification of patient survival than MPR, with higher accuracy, sensitivity, and specificity. Our findings suggest that in addition to the number of residual tumor cells, alterations in the composition of tumor metabolites may determine the treatment response and prognosis of patients with NSCLC following NAC. The metabolites identified in the stroma could also impact the treatment response to NAC among patients with NSCLC. Both metabolic classifiers could provide useful information regarding the treatment response based on the comprehensive metabolite composition. Although the biochemical characteristics of both the tumor and the stroma can be assessed by MALDI‐MSI, MPR assessments have limited ability to evaluate the stroma. However, a classifier trained using both metabolites and MPR showed slightly reduced accuracy compared with the classifier trained on metabolites alone. These comparable results indicate that the inclusion of MPR had minimal impact on classifier performance. Our developed metabolic classifiers also demonstrated the ability to stratify patients according to metabolic response, which is not possible using MPR alone. These results demonstrate that metabolic assessments based on tumor and stromal constitutions represent effective prognostic indicators.
To investigate the specificity of the classifiers relative to treatment status, the metabolites included in the classifiers were compared between the NAC and chemotherapy‐naïve cohorts, which revealed that the majority of the metabolites included in the classifiers were significantly altered by NAC. These results are consistent with previous reports describing altered expression levels of molecular and prognostic biomarkers following neoadjuvant treatment [50, 51]. We observed altered PC and PGP levels between the NAC and chemotherapy‐naïve cohorts. These findings indicated that high levels of PC were associated with poor prognosis in the NAC cohort, which aligns with a previous report that the high levels of PC were associated with the accumulation of energy‐rich molecules, supporting tumor cell proliferation [52]. Our results also indicate that increased PGP corresponded with better patient prognosis. However, another study reported increased PGP in patients with NSCLC than in a healthy control population [53]. LysoPC is a type of lipid primarily derived from PC. We observed that decreased LysoPC was associated with poor prognosis in the NAC cohort but was nonspecific for chemotherapy‐naïve patients. This finding is consistent with plasma LysoPC levels, which are commonly reduced in patients with advanced metastatic cancer [54, 55]. SM is a bioactive molecule that plays key roles in regulating tumor cell signalling to manage cancer suppression or survival [56]. Hydrolysis of SM to generate ceramide and accumulation of ceramide results in tumour suppression and cancer cell death [57]. Our dataset also identified that high levels of SM were related to good prognostic outcomes in patients who receive NAC, which may be due to the regulation of the cell cycle and the inhibition of cancer cell proliferation [58].
SM (d18:1/15:0 or d16:1/17:0) and LysoPC (15:0/0:0) are both OCFAs, and other OCFAs were included among the 100 most important molecules in the metabolic classifiers. We performed correlation analyses among various OCFAs, revealing that LysoPC (17:0/0:0) was positively correlated with both SM (d18:1/15:0 or d16:1/17:0) and LysoPC (15:0/0:0), whereas PC (15:0/24:0) was negatively correlated with other OFCAs. Because LysoPC is primarily derived from PC, this negative correlation makes intuitive sense. However, few studies have adequately addressed the mechanisms involved in OCFA production. Some OCFAs are likely to be expressed in human serum at very low levels [59]. Recently, a study indicated that serum samples from patients with myelodysplastic syndrome displayed reduced levels of SM species containing side‐chain OCFAs than serum samples from healthy control subjects [60], suggesting that the OCFA/even chain fatty acid (ECFA) ratio in cancer patients was decreased. A low OCFA/ECFA ratio reduced cell membrane fluidity due to the lower melting point of OCFAs compared to ECFAs. This suggests that membrane fluidity is lower in patients with malignant hematological diseases. Another study found that some OCFAs, such as heptadecanoic acid and omega‐3 polyunsaturated fatty acids, inhibited the proliferation and migration of lung cancer cells by promoting apoptosis and inhibiting the phosphatidylinositol 3‐kinase (PI3K)/Akt signaling pathway [61]. Heptadecanoic acid was also shown to induce increased apoptosis in lung cancer cells and enhance the cytotoxic effects of gefitinib [62]. However, the oncogenic mechanism that leads to the production of OCFAs remains unknown. Therefore, further studies of OCFA production and anti‐tumor mechanisms in lung cancer remain necessary.
In addition, the comparison of metabolites between tumor and normal tissues in the NAC cohort revealed 19 molecules that were upregulated in tumor cells, most of which were associated with lipids metabolism specific to tumor tissues. For example, LysoPC/LysoPE, SM, and PGP were detected at high levels in tumor tissues compared with normal tissues, suggesting that significant changes in these metabolites may be associated with cancerous tissues. Our results indicate that our developed metabolic classifiers primarily depend on the assessment of lipid metabolites. By contrast, a previous work developing a metabolic classifier to predict the treatment response in patients with gastric cancer depended heavily on molecules associated with DNA metabolism [46]. However, the metabolic classifier designed to assess the treatment response in patients with NAC‐treated esophageal adenocarcinoma relied on amino acids and their analogs [44]. These findings indicate that the important metabolites that can be used to predict response in patients with NSCLC and other cancer types vary substantially across cancer types. The identified metabolites in the present study were able to determine prognosis in response to NAC, indicating that these metabolites are specific to patients with NSCLC who received NAC.
To elucidate metabolic heterogeneity in tumor and stroma tissues, we performed metabolic pathway analysis on tumor cells and stroma. Pathway enrichment analysis demonstrated distinct metabolic pathways profilesbetween tumor cells and stroma. Among the pathways included in the tumor metabolic classifier, the fructose and mannose metabolic pathway had the highest impact score among nine identified pathways. Cancer cells can utilize fructose and mannose as alternative energy sources to fuel growth, and the fructose transporter was reported to be upregulated in LUAD to compensate for low‐glucose conditions [63, 64]. The exposure of less aggressive cancer cells to a fructose‐rich environment enhanced their migration and invasion capacities [65]. These variations can be attributed to fructose‐mediated alterations in glycosylation structures, which have been associated with aggressive and metastatic phenotypes [66]. By contrast, mannose uptake by cancer cells impedes glucose flux into central carbon metabolism pathways, inhibiting cancer cell proliferation [67], and a recent study revealed that mannose supplementation has anticancer effects in NSCLC cells, inhibiting proliferation and increasing cell death [68]. The most important upregulated functional metabolites were related to metabolic pathways involved in energy metabolism and cell proliferation, such as glycerophospholipid metabolism, which had the highest impact score among the pathways associated with the stroma classifier. Alterations in glycerophospholipid metabolism are increasingly recognized as prevalent metabolic hallmarks of cancer associated with tumor progression. The synthesis of glycerophospholipids, which are the main structural and functional components of cell membranes and serve as energy resources for cells, has been associated with tumorigenesis and progression [69, 70]. Hydrolyzed glycerophospholipids are involved in mitochondrial β‐oxidation, which produces ATP as an important energy resource for cell proliferation and metastasis [71]. These results revealed that the tumor and the stroma show metabolic heterogeneity in NSCLC patients.
However, our study has some limitations. Firstly, cases were selected from a single institution. Although the data were of high quality due to thorough clinico‐pathological work‐up, this resulted in a relatively low sample size. Secondly, the identification and characterization of metabolite structures are also limited by the analytical depth of MSI in terms of coverage and identification capabilities [72], and the identification of new, previously uncharacterized metabolites is difficult. Thridly, these identified metabolites would be further validated by tandem mass spectrometry or liquid chromatography–mass spectrometry in larger multicentre studies. Despite these limitations, spatial metabolomics remains a powerful and very useful tool for evaluating the treatment response in NSCLC.
5. CONCLUSIONS
In summary, our findings indicate that spatial metabolomics, combined with machine learning, is a powerful and promising approach and provides valuable information on treatment response and prognosis prediction independent of MPR and TNM stage. Furthermore, we demonstrated that the metabolic response assessed in the tumor and the stroma allows for the stratification of NAC responder and non‐responder patients into different prognosis groups. Therefore, spatial metabolomics might offer an additional method for evaluating the histopathological response in patients with NSCLC.
DECLARATIONS
ETHICS APPROVAL AND PATIENT CONSENT
The present study was conducted according to the Helsinki Declaration of the World Medical Association. The study was approved by the Cantonal Ethics Commission of the Canton of Bern (KEK 2017‐00830), which waived the requirement for a written informed consent.
CONSENT FOR PUBLICATION
Not applicable.
CONFLICT OF INTEREST
Dr. Berezowska received grants from Roche and Basilea and fees to institution from Roche, Eli Lilly and Merck Sharp & Dohme, which are unrelated to the submitted work. The remaining authors declare no conflict of interest.
AUTHOR CONTRIBUTION
AW, SB, NS and JS conceived the study design. JS, NS, AW, AB, SB and PZ wrote the manuscript. JS and NS performed MALDI imaging preparation and measurements. JS, NS and TK performed all bioinformatics analyses. JW, VMP, and QW contributed to metabolomics analyses. AF supported the immunofluorescence experiments. AW, SB, AF, and RH analyzed and interpreted the results. PZ and SB acquired ethical permission, collected the samples and performed the histopathological analyses. AW and SB supervised the project. All authors contributed to the review and approval of the manuscript.
Supporting information
Supporting information
ACKNOWLEDGMENTS
The authors thank Ulrike Buchholz, Claudia‐Mareike Pflüger, Cristina Hübner Freitas, Andreas Voss, and Elenore Samson for excellent technical assistance. The study was supported by the Ministry of Education and Research of the Federal Republic of Germany (BMBF; 01ZX1610B and 01KT1615); the Deutsche Forschungsgmeinschaft (SFB 824 C4, CRC/Transregio 205/1); the Deutsche Krebshilfe (70112617) to A. Walch; Stiftung zur Krebsbekämpfung (SKB425); Cancer Research Switzerland (KFS‐4694‐02‐2019) to S. Berezowska and Cancer Research Switzerland (MD‐PhD‐5088‐06‐2020) to P. Zens.
Shen J, Sun Na, Zens P, Kunzke T, Buck A, Prade VM, et al. Spatial metabolomics for evaluating response to neoadjuvant therapy in non‐small cell lung cancer patients. Cancer Commun. 2022;42:517–535. 10.1002/cac2.12310
Contributor Information
Sabina Berezowska, Email: sabina.berezowska@chuv.ch.
Axel Walch, Email: axel.walch@helmholtz-muenchen.de.
DATA AVAILABILITY STATEMENT
The datasets generated or analyzed during the current study are available on reasonable request from the corresponding authors.
REFERENCES
- 1. Bozcuk H, Abali H, Coskun S. Lung Cancer Committee of Turkish Oncology G. The correlates of benefit from neoadjuvant chemotherapy before surgery in non‐small‐cell lung cancer: a metaregression analysis. World J Surg Oncol. 2012;10(1):161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Burdett S, Stewart LA, Rydzewska L. A systematic review and meta‐analysis of the literature: chemotherapy and surgery versus surgery alone in non‐small cell lung cancer. J Thorac Oncol. 2006;1(7):611–21. [PubMed] [Google Scholar]
- 3. Group NM‐aC. Preoperative chemotherapy for non‐small‐cell lung cancer: a systematic review and meta‐analysis of individual participant data. Lancet. 2014;383(9928):1561–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Travis WD, Dacic S, Wistuba I, Sholl L, Adusumilli P, Bubendorf L, et al. IASLC Multidisciplinary Recommendations for Pathologic Assessment of Lung Cancer Resection Specimens After Neoadjuvant Therapy. J Thorac Oncol. 2020;15(5):709–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rice JD, Heidel J, Trivedi JR, van Berkel VH. Optimal Surgical Timing After Neoadjuvant Therapy for Stage IIIa Non‐Small Cell Lung Cancer. Ann Thorac Surg. 2020;109(3):842–7. [DOI] [PubMed] [Google Scholar]
- 6. Penault‐Llorca F, Radosevic‐Robin N. Biomarkers of residual disease after neoadjuvant therapy for breast cancer. Nat Rev Clin Oncol. 2016;13(8):487–503. [DOI] [PubMed] [Google Scholar]
- 7. Wu YL, Tsuboi M, He J, John T, Grohe C, Majem M, et al. Osimertinib in Resected EGFR‐Mutated Non‐Small‐Cell Lung Cancer. N Engl J Med. 2020;383(18):1711‐1723. [DOI] [PubMed] [Google Scholar]
- 8. Wakelee HA, Altorki NK, Zhou C, Csőszi T, Vynnychenko IO, Goloborodko O, et al. IMpower010: Primary results of a phase III global study of atezolizumab versus best supportive care after adjuvant chemotherapy in resected stage IB‐IIIA non‐small cell lung cancer (NSCLC). J Clin Oncol. 2021;39(15):8500–8500. [Google Scholar]
- 9. Spicer J, Wang C, Tanaka F, Saylors GB, Chen K‐N, Liberman M, et al. Surgical outcomes from the phase 3 CheckMate 816 trial: Nivolumab (NIVO) + platinum‐doublet chemotherapy (chemo) vs chemo alone as neoadjuvant treatment for patients with resectable non‐small cell lung cancer (NSCLC). J Clin Oncol. 2021;39(15):8503–8503. [Google Scholar]
- 10. Sun L, Guo YJ, Song J, Wang YR, Zhang SL, Huang LT, et al. Neoadjuvant EGFR‐TKI Therapy for EGFR‐Mutant NSCLC: A Systematic Review and Pooled Analysis of Five Prospective Clinical Trials. Front Oncol. 2020;10:586596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Roth JA, Atkinson EN, Fossella F, Komaki R, Bernadette Ryan M, Putnam JB,Jr. , et al. Long‐term follow‐up of patients enrolled in a randomized trial comparing perioperative chemotherapy and surgery with surgery alone in resectable stage IIIA non‐small‐cell lung cancer. Lung Cancer. 1998;21(1):1–6. [DOI] [PubMed] [Google Scholar]
- 12. Rosell R, Gomez‐Codina J, Camps C, Javier Sanchez J, Maestre J, Padilla J, et al. Preresectional chemotherapy in stage IIIA non‐small‐cell lung cancer: a 7‐year assessment of a randomized controlled trial. Lung Cancer. 1999;26(1):7–14. [DOI] [PubMed] [Google Scholar]
- 13. Pataer A, Kalhor N, Correa AM, Raso MG, Erasmus JJ, Kim ES, et al. Histopathologic response criteria predict survival of patients with resected lung cancer after neoadjuvant chemotherapy. J Thorac Oncol. 2012;7(5):825–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chaft JE, Rusch V, Ginsberg MS, Paik PK, Finley DJ, Kris MG, et al. Phase II trial of neoadjuvant bevacizumab plus chemotherapy and adjuvant bevacizumab in patients with resectable nonsquamous non‐small‐cell lung cancers. J Thorac Oncol. 2013;8(8):1084–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hellmann MD, Chaft JE, William WN, Jr. , Rusch V, Pisters KM, Kalhor N, et al. Pathological response after neoadjuvant chemotherapy in resectable non‐small‐cell lung cancers: proposal for the use of major pathological response as a surrogate endpoint. Lancet Oncol. 2014;15(1):e42–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Qu Y, Emoto K, Eguchi T, Aly RG, Zheng H, Chaft JE, et al. Pathologic Assessment After Neoadjuvant Chemotherapy for NSCLC: Importance and Implications of Distinguishing Adenocarcinoma From Squamous Cell Carcinoma. J Thorac Oncol. 2019;14(3):482–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yamane Y, Ishii G, Goto K, Kojima M, Nakao M, Shimada Y, et al. A novel histopathological evaluation method predicting the outcome of non‐small cell lung cancer treated by neoadjuvant therapy: the prognostic importance of the area of residual tumor. J Thorac Oncol. 2010;5(1):49–55. [DOI] [PubMed] [Google Scholar]
- 18. Junker K, Thomas M, Schulmann K, Klinke F, Bosse U, Muller KM. Tumour regression in non‐small‐cell lung cancer following neoadjuvant therapy. Histological assessment. J Cancer Res Clin Oncol. 1997;123(9):469–77. [DOI] [PubMed] [Google Scholar]
- 19. Liu‐Jarin X, Stoopler MB, Raftopoulos H, Ginsburg M, Gorenstein L, Borczuk AC. Histologic assessment of non‐small cell lung carcinoma after neoadjuvant therapy. Mod Pathol. 2003;16(11):1102–8. [DOI] [PubMed] [Google Scholar]
- 20. Zens P, Bello C, Scherz A, Koenigsdorf J, Pollinger A, Schmid RA, et al. A prognostic score for non‐small cell lung cancer resected after neoadjuvant therapy in comparison with the tumor‐node‐metastases classification and major pathological response. Mod Pathol. 2021;34:1333–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liu X, Sun W, Wu J, Feng Y, Mao L, Chen M, et al. Major pathologic response assessment and clinical significance of metastatic lymph nodes after neoadjuvant therapy for non‐small cell lung cancer. Mod Pathol. 2021;34:1990–8. [DOI] [PubMed] [Google Scholar]
- 22. Cascone T, William WN, Weissferdt A, Lin HY, Leung CH, Carter BW, et al. Neoadjuvant nivolumab (N) or nivolumab plus ipilimumab (NI) for resectable non‐small cell lung cancer (NSCLC): Clinical and correlative results from the NEOSTAR study. J Clin Oncol. 2019;37(15):8504–8504. [Google Scholar]
- 23. Weissferdt A, Sepesi B, Cascone T. Pathologic assessment following neoadjuvant immunotherapy or chemotherapy demonstrates similar patterns in non‐small cell lung cancer (NSCLC). Abstracts Tumour Biology and Pathology. 2018;29:1928P. [Google Scholar]
- 24. Yu KH, Zhang C, Berry GJ, Altman RB, Re C, Rubin DL, et al. Predicting non‐small cell lung cancer prognosis by fully automated microscopic pathology image features. Nat Commun. 2016;7:12474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Huang L, Wang L, Hu X, Chen S, Tao Y, Su H, et al. Machine learning of serum metabolic patterns encodes early‐stage lung adenocarcinoma. Nat Commun. 2020;11(1):3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Muranishi Y, Sato T, Ito S, Satoh J, Yoshizawa A, Tamari S, et al. The Ratios of monounsaturated to saturated phosphatidylcholines in lung adenocarcinoma microenvironment analyzed by Liquid Chromatography‐Mass spectrometry and imaging Mass spectrometry. Sci Rep. 2019;9(1):8916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Prade VM, Kunzke T, Feuchtinger A, Rohm M, Luber B, Lordick F, et al. De novo discovery of metabolic heterogeneity with immunophenotype‐guided imaging mass spectrometry. Mol Metab. 2020;36:100953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Abbassi‐Ghadi N, Antonowicz SS, McKenzie JS, Kumar S, Huang J, Jones EA, et al. De Novo Lipogenesis Alters the Phospholipidome of Esophageal Adenocarcinoma. Cancer Res. 2020;80(13):2764–74. [DOI] [PubMed] [Google Scholar]
- 29. Neumann JM, Freitag H, Hartmann JS, Niehaus K, Galanis M, Griesshammer M, et al. Subtyping non‐small cell lung cancer by histology‐guided spatial metabolomics. J Cancer Res Clin Oncol. 2022;148(2):351–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Walch A, Rauser S, Deininger SO, Hofler H. MALDI imaging mass spectrometry for direct tissue analysis: a new frontier for molecular histology. Histochem Cell Biol. 2008;130(3):421–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Miura D, Fujimura Y, Wariishi H. In situ metabolomic mass spectrometry imaging: recent advances and difficulties. J Proteomics. 2012;75(16):5052–60. [DOI] [PubMed] [Google Scholar]
- 32. Ly A, Buck A, Balluff B, Sun N, Gorzolka K, Feuchtinger A, et al. High‐mass‐resolution MALDI mass spectrometry imaging of metabolites from formalin‐fixed paraffin‐embedded tissue. Nat Protoc. 2016;11(8):1428–43. [DOI] [PubMed] [Google Scholar]
- 33. Sun N, Kunzke T, Sbiera S, Kircher S, Feuchtinger A, Aichler M, et al. Prognostic Relevance of Steroid Sulfation in Adrenocortical Carcinoma Revealed by Molecular Phenotyping Using High‐Resolution Mass Spectrometry Imaging. Clin Chem. 2019;65(10):1276–86. [DOI] [PubMed] [Google Scholar]
- 34. Mas S, Torro A, Fernandez L, Bec N, Gongora C, Larroque C, et al. MALDI imaging mass spectrometry and chemometric tools to discriminate highly similar colorectal cancer tissues. Talanta. 2020;208:120455. [DOI] [PubMed] [Google Scholar]
- 35. Rezola A, Pey J, Rubio A, Planes FJ. In‐silico prediction of key metabolic differences between two non‐small cell lung cancer subtypes. PLoS One. 2014;9(8):e103998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Oshita F, Morita A, Ito H, Kameda Y, Tsuchiya E, Asai S, et al. Proteomic screening of completely resected tumors in relation to survival in patients with stage I non‐small cell lung cancer. Oncol Rep. 2010;24(3):637–45. [DOI] [PubMed] [Google Scholar]
- 37. Kriegsmann M, Zgorzelski C, Casadonte R, Schwamborn K, Muley T, Winter H, et al. Mass Spectrometry Imaging for Reliable and Fast Classification of Non‐Small Cell Lung Cancer Subtypes. Cancers (Basel). 2020;12(9):2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Voortman J, Pham TV, Knol JC, Giaccone G, Jimenez CR. Prediction of outcome of non‐small cell lung cancer patients treated with chemotherapy and bortezomib by time‐course MALDI‐TOF‐MS serum peptide profiling. Proteome Sci. 2009;7:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sun N, Fernandez IE, Wei M, Witting M, Aichler M, Feuchtinger A, et al. Pharmacometabolic response to pirfenidone in pulmonary fibrosis detected by MALDI‐FTICR‐MSI. Eur Respir J. 2018;52(3):1702314. [DOI] [PubMed] [Google Scholar]
- 40. Zens P, Bello C, Scherz A, Mv Gunten, Ochsenbein A, Schmid RA, et al. The effect of neoadjuvant therapy on PD‐L1 expression and CD8 lymphocyte density in non‐small cell lung cancer. medRxiv. 2022:2022.04.11.22273684. [DOI] [PMC free article] [PubMed]
- 41. Brierley JD, Gospodarowicz MK, Wittekind C. TNM Classification of Malignant Tumours. 8th ed. Chapter 04: Lung. 8th ed. Wiley‐Blackwell; 2017. p. 105–112: chap 4. [Google Scholar]
- 42. Zlobec I, Suter G, Perren A, Lugli A. A next‐generation tissue microarray (ngTMA) protocol for biomarker studies. J Vis Exp. 2014;23(91):51893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Buck A, Ly A, Balluff B, Sun N, Gorzolka K, Feuchtinger A, et al. High‐resolution MALDI‐FT‐ICR MS imaging for the analysis of metabolites from formalin‐fixed, paraffin‐embedded clinical tissue samples. J Pathol. 2015;237(1):123–32. [DOI] [PubMed] [Google Scholar]
- 44. Buck A, Prade VM, Kunzke T, Feuchtinger A, Kroll D, Feith M, et al. Metabolic tumor constitution is superior to tumor regression grading for evaluating response to neoadjuvant therapy of esophageal adenocarcinoma patients. J Pathol. 2022;256(2):202–13. [DOI] [PubMed] [Google Scholar]
- 45. Kunzke T, Prade VM, Buck A, Sun N, Feuchtinger A, Matzka M, et al. Patterns of Carbon‐Bound Exogenous Compounds in Patients with Lung Cancer and Association with Disease Pathophysiology. Cancer Res. 2021;81(23):5862–75. [DOI] [PubMed] [Google Scholar]
- 46. Kunzke T, Holzl FT, Prade VM, Buck A, Huber K, Feuchtinger A, et al. Metabolomic therapy response prediction in pretherapeutic tissue biopsies for trastuzumab in patients with HER2‐positive advanced gastric cancer. Clin Transl Med. 2021;11(9):e547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wishart DS, Feunang YD, Marcu A, Guo AC, Liang K, Vazquez‐Fresno R, et al. HMDB 4.0: the human metabolome database for 2018. Nucleic Acids Res. 2018;46(D1):D608–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Palmer A, Phapale P, Chernyavsky I, Lavigne R, Fay D, Tarasov A, et al. FDR‐controlled metabolite annotation for high‐resolution imaging mass spectrometry. Nat Methods. 2017;14(1):57–60. [DOI] [PubMed] [Google Scholar]
- 49. Xia J, Wishart DS. Using MetaboAnalyst 3.0 for Comprehensive Metabolomics Data Analysis. Curr Protoc Bioinformatics. 2016;55:14 10 1–14 10 91. [DOI] [PubMed] [Google Scholar]
- 50. Lee HC, Ko H, Seol H, Noh DY, Han W, Kim TY, et al. Expression of Immunohistochemical Markers before and after Neoadjuvant Chemotherapy in Breast Carcinoma, and Their Use as Predictors of Response. J Breast Cancer. 2013;16(4):395–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Krikken E, van der Kemp WJM, van Diest PJ, van Dalen T, van Laarhoven HWM, Luijten PR, et al. Early detection of changes in phospholipid metabolism during neoadjuvant chemotherapy in breast cancer patients using phosphorus magnetic resonance spectroscopy at 7T. NMR Biomed. 2019;32(6):e4086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Podo F, Paris L, Cecchetti S, Spadaro F, Abalsamo L, Ramoni C, et al. Activation of Phosphatidylcholine‐Specific Phospholipase C in Breast and Ovarian Cancer: Impact on MRS‐Detected Choline Metabolic Profile and Perspectives for Targeted Therapy. Front Oncol. 2016;6:171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhang J, Xu J, Lu H, Ding J, Yu D, Li P, et al. Altered phosphatidylcholines expression in sputum for diagnosis of non‐small cell lung cancer. Oncotarget. 2016;7(39):63158–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kuhn T, Floegel A, Sookthai D, Johnson T, Rolle‐Kampczyk U, Otto W, et al. Higher plasma levels of lysophosphatidylcholine 18:0 are related to a lower risk of common cancers in a prospective metabolomics study. BMC Med. 2016;14:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Qiu Y, Zhou B, Su M, Baxter S, Zheng X, Zhao X, et al. Mass spectrometry‐based quantitative metabolomics revealed a distinct lipid profile in breast cancer patients. Int J Mol Sci. 2013;14(4):8047–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Hannun YA, Bell RM. Lysosphingolipids inhibit protein kinase C: implications for the sphingolipidoses. Science. 1987;235(4789):670–4. [DOI] [PubMed] [Google Scholar]
- 57. Jiang W, Ogretmen B. Autophagy paradox and ceramide. Biochim Biophys Acta. 2014;1841(5):783‐92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ogretmen B. Sphingolipid metabolism in cancer signalling and therapy. Nat Rev Cancer. 2018;18(1):33–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Jenkins B, West JA, Koulman A. A review of odd‐chain fatty acid metabolism and the role of pentadecanoic Acid (c15:0) and heptadecanoic Acid (c17:0) in health and disease. Molecules. 2015;20(2):2425–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hori A, Ishida F, Nakazawa H, Yamaura M, Morita S, Uehara T, et al. Serum sphingomyelin species profile is altered in hematologic malignancies. Clin Chim Acta. 2021;514:29‐33. [DOI] [PubMed] [Google Scholar]
- 61. Yin Y, Sui C, Meng F, Ma P, Jiang Y. The omega‐3 polyunsaturated fatty acid docosahexaenoic acid inhibits proliferation and progression of non‐small cell lung cancer cells through the reactive oxygen species‐mediated inactivation of the PI3K /Akt pathway. Lipids Health Dis. 2017;16(1):87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Xu C, Wu P, Gao J, Zhang L, Ma T, Ma B, et al. Heptadecanoic acid inhibits cell proliferation in PC9 nonsmallcell lung cancer cells with acquired gefitinib resistance. Oncol Rep. 2019;41(6):3499–3507. [DOI] [PubMed] [Google Scholar]
- 63. Chen WL, Wang YY, Zhao A, Xia L, Xie G, Su M, et al. Enhanced Fructose Utilization Mediated by SLC2A5 Is a Unique Metabolic Feature of Acute Myeloid Leukemia with Therapeutic Potential. Cancer Cell. 2016;30(5):779–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Weng Y, Fan X, Bai Y, Wang S, Huang H, Yang H, et al. SLC2A5 promotes lung adenocarcinoma cell growth and metastasis by enhancing fructose utilization. Cell Death Discov. 2018;4:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Monzavi‐Karbassi B, Hine RJ, Stanley JS, Ramani VP, Carcel‐Trullols J, Whitehead TL, et al. Fructose as a carbon source induces an aggressive phenotype in MDA‐MB‐468 breast tumor cells. Int J Oncol. 2010;37(3):615–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Dennis JW, Granovsky M, Warren CE. Protein glycosylation in development and disease. Bioessays. 1999;21(5):412–21. [DOI] [PubMed] [Google Scholar]
- 67. Gonzalez PS, O'Prey J, Cardaci S, Barthet VJA, Sakamaki JI, Beaumatin F, et al. Mannose impairs tumour growth and enhances chemotherapy. Nature. 2018;563(7733):719–23. [DOI] [PubMed] [Google Scholar]
- 68. Wang Y, Xie S, He B. Mannose shows antitumour properties against lung cancer via inhibiting proliferation, promoting cisplatinmediated apoptosis and reducing metastasis. Mol Med Rep. 2020;22(4):2957–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Castro‐Gomez P, Garcia‐Serrano A, Visioli F, Fontecha J. Relevance of dietary glycerophospholipids and sphingolipids to human health. Prostaglandins Leukot Essent Fatty Acids. 2015;101:41–51. [DOI] [PubMed] [Google Scholar]
- 70. Yan G, Li L, Zhu B, Li Y. Lipidome in colorectal cancer. Oncotarget. 2016;7(22):33429–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Chen M, Huang J. The expanded role of fatty acid metabolism in cancer: new aspects and targets. Precis Clin Med. 2019;2(3):183‐191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Vaysse PM, Heeren RMA, Porta T, Balluff B. Mass spectrometry imaging for clinical research ‐ latest developments, applications, and current limitations. Analyst. 2017;142(15):2690–2712. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information
Data Availability Statement
The datasets generated or analyzed during the current study are available on reasonable request from the corresponding authors.


