Abstract
Exosomes are extracellular vesicles that mediate cell-to-cell communication. Bioactive substances such as DNA, RNA, lipids, and proteins are present in it, and they play an essential role in the pathogenesis of colorectal cancer (CRC). The role of RNA and protein in exosomes has been extensively studied. Exosome DNA has recently attracted the attention of a great deal of scientists. According to studies, exosome DNA mainly contains genomic DNA (gDNA) and mitochondrial DNA (mtDNA), of which exosome gDNA is widely used in liquid biopsy of CRC. It includes a variety of clinically relevant tumor-specific mutation genes. In addition to liquid biopsy, researchers find that exosome gDNA regulates immune and metabolic functions in CRC, making it an important research object. However, the primary research on exosome gDNA is still limited. Here, we describe the occurrence and composition of exosomes. Summarize the essential characteristics and mode of action of exosome gDNA. Remarkably, this paper constitutes a comprehensive summary on the role of exosome gDNA on CRC with the intent of providing a theoretical basis and reference for early diagnosis and clinical treatment of cancer.
Keywords: colorectal cancer, exosome, genomic DNA, liquid biopsy, tumor immunity
Introduction
CRC is the third most common malignant tumor in the world. Every year one million people worldwide will develop CRC (Franke et al., 2019; Keum and Giovannucci, 2019). It is a heterogeneous intestinal epithelial disease characterized by the accumulation of mutations and disordered immune responses (Janney et al., 2020; Lichtenstern et al., 2020; Stoffel and Murphy, 2020). Currently, the choice of treatment scheme for CRC is mostly determined by tissue biopsy. Despite tissue biopsy being the gold standard for diagnosis, classification, and treatment decision-making, it is not always available. Tumor tissue also exhibits solid spatial heterogeneity due to the uneven distribution of tumor subclones. Tumor molecular compositions may change dynamically in response to micro-environmental stimuli and therapeutic pressure, making tissue biopsy an unreliable method of diagnosis. Therefore, liquid biopsy is becoming increasingly popular (Dekker and Rex, 2018). A liquid biopsy is a non-invasive procedure in which samples of blood or other body fluids are collected to analyze exosomes, circulating tumor cells, and ctDNA (Marcuello et al., 2019). In the early stages of CRC, there are no obvious symptoms. Most patients with CRC are found in the advanced stage. Liquid biopsy provides a more accurate picture of tumor details in real time, which is particularly important for early detection of cancer and reduction of mortality from it (Marcuello et al., 2019; Kolencik et al., 2020; Martini et al., 2020; Rodriguez-Casanova et al., 2021). Nevertheless, the key to prolonging the survival time of CRC patients is not only to make early diagnosis of the disease, but also to understand the tumor progression (Ogunwobi et al., 2020; Sveen et al., 2020). The identification of a sensitive biomarker is important for observing the immune response and metabolism during the treatment of CRC and assessing the drug resistance and heterogeneity further (Marcuello et al., 2019; Kolencik et al., 2020; Martini et al., 2020; Rodriguez-Casanova et al., 2021).
Exosomes are extracellular nano-sized vesicles containing DNA, RNA, lipids, and protein (Pegtel and Gould, 2019; Jafari et al., 2020; Kalluri and Lebleu, 2020; Ahmadi and Rezaie, 2021; Liu et al., 2021; Yu et al., 2021; Rezaie et al., 2022; Vahabi et al., 2022). CRC exosomes have the ability to transport their contents to recipient cells in the tumor microenvironment, thereby playing an important role in cell-to-cell communication. It plays a vital role in tumor immunity, tumor survival, tumor chemotherapy resistance, and metastasis (Xiao et al., 2020; Liu et al., 2021; Chang et al., 2021). In CRC exosomes, there are a variety of active substances. DNA is one of the most stable substances (Jeppesen et al., 2019). However, there is little research on exosome DNA compared to proteins and RNA. Essentially, exosome DNA is derived from normal DNA metabolism or damage induction, primarily from gDNA from the nucleus and mtDNA from the mitochondria (Kahlert et al., 2014; Thakur et al., 2014; Kalluri and Lebleu, 2016). The gDNA of exosomes contains DNA fragments from multiple chromosomes, including mutant DNA fragments, and is mainly found in body fluids or the genome of some immune cells (Thakur et al., 2014; Waldenstrom et al., 2012) (Table 1). gDNA from exosomes is widely used in liquid biopsy, it also affects tumor immunity and metabolism (Lian et al., 2017; Vanpouille-Box et al., 2017; Zhang et al., 2018; Sharma and Johnson, 2020; Zhao et al., 2021). In this review, we discuss the relevance of exosome gDNA in the early detection, development and prognosis, immune response, and drug resistance of CRC.
TABLE 1.
Exosome gDNA as biomarkers for diagnosis of diseases.
| Disease | Mutantion | Detection mode | Biofluid | Reference |
|---|---|---|---|---|
| Pancreatic Cancer | KRAS TP53 | ddPCR | Plasma | (Kahlert et al., 2014; Yang et al., 2017) |
| Glioma | EGFR | Conventional PCR | Cerebrospinal fluid | Vaidya and Sugaya, (2020) |
| non-small cell lung cancer | EGFR | PNA-PCR | Alveolar lavage fluid and Plasma | Hur et al. (2018) |
| Lung Adenocarcinoma | EGFR | PNA-PCR | Pleural effusion | Lee et al. (2018) |
| Pancreatic cancer | KRAS | ddPCR | Plasma | Allenson et al. (2017) |
| Pulmonary adenocarcinoma | EGFR | ddPCR | Bronchial Washing | Park et al. (2020) |
| Glioma | RET HIF2A VHL SDHB | Conventional PCR | Plasma | Wang et al. (2018) |
| non-small cell lung cancer | EGFR | ddPCR | Plasma | Castellanos-Rizaldos et al. (2018) |
| non-small cell lung cancer | EGFR | ddPCR | plasma and pleural fluid | Kim et al. (2021) |
| Lung Adenocarcinoma | EGFR | Quantitative PCR | Pleural Effusions | Qu et al. (2019) |
| Neuroblastoma | BRAF | Quantitative PCR | Plasma | Degli et al. (2021) |
| Bladder cancer | KRAS | ddPCR | Urine | Zhou et al. (2021) |
| prostate cancer | PTEN TP53 | Quantitative PCR | Plasma | Lazaro-Ibanez et al. (2014) |
| Colorectal cancer | KRAS | ddPCR | Plasma | Lucchetti et al. (2021) |
| non-small cell lung cancer | EGFR | PNA-PCR | bronchoalveolar lavage fluid | Hur et al. (2019) |
| non-small cell lung cancer | EGFR | ddPCR | Plasma | Krug et al. (2018) |
| Pancreatic cancer | KRAS | ddPCR | Plasma | Bernard et al. (2019) |
| Glioma | IDH1 | ddPCR | Plasma | Garcia-Romero et al. (2017) |
Overview of Exosomes
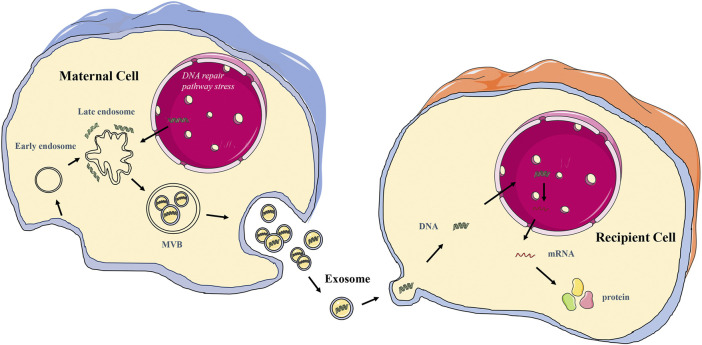
Exosomes are extracellular nano-sized bilayer membrane vesicles with an average diameter of 30–150 nm (Pegtel and Gould, 2019; Kalluri and Lebleu, 2020; Yu et al., 2021). It is produced in the cytoplasm by the classical endosomal sorting complex. It is formed by the inward budding of cell membranes containing ubiquitinated surface receptors, leading to the formation of early endosomes. With the help of the Golgi apparatus, these early endocorpuscles become late endocorpuscles and intracavitary vesicles. Intracavitary vesicles accumulate in endosomes, leading to the formation of multivesicular bodies (MVB). MVB is ultimately transported into lysosomes for degradation or fuses with the cytoplasmic membrane, releasing its contents (including exosomes) into the extracellular space (He et al., 2018; Doyle and Wang, 2019; Zhang and Yu, 2019; Rezaie et al., 2021) (Figure 2). Exosomes are identified by classical molecular markers, such as tetrapeptide (CD63, CD9, CD81), FLOTILIN-1, and heat shock 70 protein (Liang et al., 2021). Numerous literature has been extensively studied to reveal the detailed mechanism of exosome biogenesis.
FIGURE 2.
The schematic diagram of exosome gDNA transfer between cells.
Exosomes can be found in almost all cells and body fluids, including blood, sweat, tears, urine, saliva, breast milk, ascites, and cerebrospinal fluid (Liang et al., 2021; Lin et al., 2021). Exosomes carry various macromolecules from different tissues and organs, loaded with nucleic acids (DNA and RNA), structural components of cells (protein and lipids), and cell metabolites. It is a part of the intercellular communication system, which carries and transmits signal molecules that regulate the physiological state of cells, participates in antigen presentation, cell differentiation, growth, and tumor immunity, and is closely related to the occurrence and development of various diseases (Pegtel and Gould, 2019; Liu et al., 2021). After different active substances in exosomes are thrown out of cells, functional protein, RNA, and DNA fragments can be transferred to recipient cells, resulting in cascade changes in the genome and non-genome (Liu et al., 2021; Kalluri and Lebleu, 2020) (Table 2). Various bioactive molecular substances in exosomes are cell signaling effectors and valuable tools for tumor diagnosis. Exosomes can monitor the occurrence and development of tumors and provide new targets and strategies for tumor treatment and diagnosis (Liang et al., 2021; Lin et al., 2021).
TABLE 2.
Exosome gDNA transfer between cells.
| Donor cell | Receptor cell | Exosome gDNA | Integrated into the genome | Reference |
|---|---|---|---|---|
| Cardiomyocyte | Fibroblast | Non specicial | Not sure | Waldenstrom et al. (2012) |
| Colon cancer cell | Fibroblast and epithelial cells | HRAS | No | Lee et al. (2016) |
| Glioma cells | Fibroblast | c-Myc | Yes | Balaj et al. (2011) |
| K562 | HEK293 | AT1 | Yes | Cai et al. (2013) |
| SW480 | In vivo | KRAS RAB | Not sure | Trejo-Becerril et al. (2012) |
| hBMSC | hMSC | A.t.-plasmid | Yes | Fischer et al. (2016) |
| H-ras-driven Intestinal epithelial cells | RAT-1 | HRAS | No | Lee et al. (2014) |
| K562 | Neutrophils | BCR/ABL | Yes | Cai et al. (2014) |
Origin and Characteristics of Exosome Genomic DNA
Exosomes contain many bioactive substances. However, compared with proteins and RNA in exosomes, there is little research on exosome DNA. Essentially, mammalian cells can excrete harmful cytoplasmic DNA through exosomes, thus avoiding cell aging and death (Takahashi et al., 2017). However, tumor cells often accumulate damaged DNA fragments of the cytoplasm because of their aging or abnormal leakage of DNA during division, autophagy, oxidative stress (Riches et al., 2014; Yokoi et al., 2019). For these reasons, tumor cells can secrete more exosomes to avoid the accumulation of nuclear and mitochondrial DNA fragments in the cytoplasm induced by metabolic stress, thus releasing more soluble DNA fragments (Parolini et al., 2009; Whiteside, 2016; Yokoi et al., 2019). There are single-stranded and double-stranded DNA in tumor exosomes (Balaj et al., 2011; Cai et al., 2013; Hur and Lee, 2021), and gDNA, mtDNA, and plasmid DNA all have been found in tumor exosomes (Erdmann et al., 2017; Sansone et al., 2017; Lazaro-Ibanez et al., 2019; Elzanowska et al., 2021).
After analyzing the exosomes in the supernatants of various tumor cell lines and non-tumor-related fibroblasts, scientists found that the exosome DNA in tumor cells are more abundant (Kahlert et al., 2014; Thakur et al., 2014; Kalluri and Lebleu, 2016). Genome-wide sequencing showed that tumor exosomes contained large double-stranded gDNA fragments ranging from 100bp to 20kbp, which could cover the whole chromosome range and reflect the mutation state of tumor parent cells (Kahlert et al., 2014; Thakur et al., 2014; Kalluri and Lebleu, 2016). This gDNA is packed in exosomes in the form of nucleosomes or supercoils. As exosomes contain larger gDNA fragments and originate from living cells, which is more conducive to mutation detection by PCR, they may have advantages over circulating cfDNA (Kahlert et al., 2014; Thakur et al., 2014; Kalluri and Lebleu, 2016). Some of its tumor-related mutations can reflect the progress and prognosis of many kinds of tumors (Table 1), used for molecular map analysis of tumors. As the double-stranded gDNA is very stable. The membrane of the exosome can protect the nucleic acid substances inside it from degradation induced by nuclease. It is also reported that exosome gDNA in serum can remain stable for 1 week at 4°C and 1 day at room temperature, even after repeated freezing and thawing cycles (Jin et al., 2016), so exosome gDNA is more stable. Based on these advantages, scientists can do more in-depth research on it, whether it is screening for early diseases, monitoring drug resistance, or evaluating prognosis, which has its significance.
Extraction, Detection, and Functional Mechanism of Exosome Genomic DNA
As the study of exosome gDNA has attracted extensive attention from scientists, the exosome separation and subsequent exosome gDNA detection still require more specialized studies to ensure optimal performance. When analyzing the gDNA inside exosomes, digestion outside of exosomes by DNase I should be the first procedure. DNase I can reduce the residue of cfDNA in samples and the pollution of DNA outside of exosomes (Spada et al., 2020; Wang et al., 2020) (Figure 1). It has been reported that the number of mutant DNA will be increased if DNase I is not pretreatment. Next, gDNA was extracted from exosomes which were treated with DNase I. The genomic DNA kit for tissues, cells, and blood can be used to extract gDNA from exosomes. Scientists using the Qiagen kit could get a higher concentration of exosome gDNA, but the Qiagen kit may be a bit expensive (Thakur et al., 2014; Kalluri and Lebleu, 2016; Spada et al., 2020; Wang et al., 2020). Our laboratory found that applying the inexpensive Tiangen kit can also successfully obtain a higher concentration of exosome gDNA. Typically, extracting 200 μl exosome according to the instructions will yield at least 20 ng/μl gDNA. After removing gDNA from exosomes, mutation detection of exosome gDNA is carried out. Most detection methods are digital PCR and fluorescence PCR to ensure more accurate mutation detection (Table 1). However, when the mutation to be detected is a point mutation, Sanger sequencing should be adopted as far as possible, and then T-A clones obtained a large number of clones for follow-up detection.
FIGURE 1.
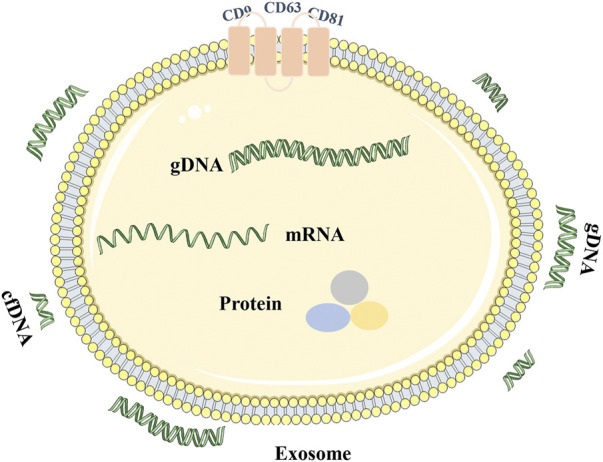
Characterization of exosome gDNA. The short double-stranded DNA is cfDNA and the long double-stranded DNA is gDNA. DNA can be enclosed within exosome, attached to the outer surface of exosome or enclosed and attached to the outer surface of exosome.
Exosomes are an essential form of intercellular communication. Exosomes can deliver both RNA and protein as well as DNA (Kalluri and Lebleu, 2020) (Table 2). The transferred exosome gDNA affects the function of recipient cells by increasing mRNA transcription and protein translation (Cai et al., 2013; Cai et al., 2014; Lee et al., 2014; Fischer et al., 2016). The schematic diagram of the release of exosome gDNA is displayed in Figure 2. In addition to the open reading frame, exosomes should also contain the 5′ promoter region and 3′ untranslated region elements necessary for the transcription mechanism so that the delivered DNA can perform its function. It has been reported that there are DNA fragments in exosomes, including the 5′ promoter region, 3′ untranslated region, and active retrotransposon, which indicates that exosome gDNA may play a role in the genetic instability of receptor cells (Balaj et al., 2011; Cai et al., 2013). It has also been confirmed that exosome gDNA can be located in the recipient nucleus through late endosomal transport related to nuclear membrane invagination (Waldenstrom et al., 2012; Cai et al., 2013). According to these viewpoints, it shows that the function of oncogenes in tumor cells is not only to accumulate in the genome of tumor cells but also to transfer exosome gDNA and spread in tumor and normal cells (Table 2), which may lead to the tumorigenic transformation of normal cells and accelerate the progress of diseases. However, whether the tumor exosome gDNA can be horizontally transmitted to the recipient cells or the changes in the function of the recipient cells caused by the horizontal transmission are still controversial. In terms of the types of recipient cells, it has been reported that not all recipient cells can be horizontally transmitted. When the recipient cells are tumor cells, fibroblasts, or endothelial cells, the exosome gDNA can be easily shared, but it cannot be transmitted when the recipient cells are dormant cells such as epithelial cells (Lee et al., 2016); In the aspect of functional changes after horizontal transmission, some reports suggest that the gDNA transmitted to the recipient cells can be integrated into the genome of the recipient cells for a long time (Balaj et al., 2011; Cai et al., 2013; Cai et al., 2014; Fischer et al., 2016). Some reports suggest that the transmitted gDNA enters the recipient cells only for an instant function and loses its position after 1 month (Trejo-Becerril et al., 2012; Lee et al., 2014; Lee et al., 2016). There is also a report suggests that the transmitted gDNA is a driving factor rather than an initial factor, and only obtaining this driving factor can make the recipient cells deteriorate (Stefanius et al., 2019). The specific mechanism needs further study. From these perspectives, exosome gDNA horizontally may represent a new method of gene transfer and signal transduction between cells, which may be an essential mechanism of tumor occurrence, development, and metastasis, and provide a new direction for discovering new disease mechanisms and developing new treatment strategies.
Colorectal Cancer Microenvironment Plays an Essential Role on Exosome Genomic DNA Packaging and Transmission
Hypoxia and acidification stimulate the tumor microenvironment, further activate the cell stress response mechanism, and increase the production of exosomes (Hoshino et al., 2013; Khaksar et al., 2018; Panigrahi et al., 2018; Soraya et al., 2021). At the same time, it may promote the package of gDNA into exosomes, and transfer it to the recipient cells, thus enhancing the ability of exosomes to induce malignant transformation of the recipient cells (Nemeth et al., 2017).
It has been reported that TP53 mutation in the recipient cells increased after the treatment factors affected the tumor microenvironment, and the transcription in colon epithelial cells and liver cells increased. The difference in mutant TP53 between colon epithelial cells and liver cells was also observed. It seems that liver cells are less likely to absorb mutant genes carried by exosomes than colon epithelial cells. The author also proved that LPS activated TLR4 of colon cancer cell SW480 and promoted the packaging of mutant TP53 in exosomes but could not promote the selective packaging of the KRAS gene. It indicates that the mechanism of gDNA packaging to exosomes is not random (Domenis et al., 2021). More and more experiments proved that exosomes derived from primary tumors could be loaded with specific molecules (Stobiecka et al., 2019; Baris et al., 2021), and the expression or lack of these molecules is helpful to the phenotype transformation of recipient cells. These results indicate that the tumor microenvironment increases the possibility of tumor gene integration by promoting exosome metastasis. Although the integration of mutant genes is still considered a rare event, it may be a related phenomenon in vivo. Further studies are needed to evaluate the effect of tumor-driven genes transferred through the exosome on malignant transformation and their role in tumor progression of microenvironmental stimulation.
Exosome Genomic DNA as a Tool for Colorectal Cancer Liquid Biopsy
Due to the lack of mismatch repair function in CRC, genome instability often leads to the accumulation of frameshift mutation in the microsatellite region. And CRC in an early stage can be prevented and cured, so it is necessary to explore non-invasive tools to help with early detection and treatment monitoring (Janney et al., 2020; Stoffel and Murphy, 2020). A liquid biopsy is a dynamic tool for the non-invasive detection of tumor heterogeneity and mutation over time. Studies have shown that compared with the healthy group, the circulating exosomes in the body fluid of tumor patients are more, and exosome gDNA may provide information about tumor-specific mutations. The relationship between mutated gDNA and types of body fluids is summarized below (Table 1). Therefore, detecting exosome gDNA mutations in tumor progression can help diagnosis early-stage, drug selection, and prognosis analysis.
EGFR mutation was seen in exosomes of malignant pleural effusion (Lee et al., 2018), and T790M mutation was also detected in exosomes of bronchoalveolar lavage fluid of patients with non-small cell lung cancer (Park et al., 2020). The exosome gDNA isolated from ascites of ovarian cancer patients reflects the copy number variation of primary tumors (Crow et al., 2017; Giannopoulou et al., 2019). KRAS mutation is also confirmed in exosomes of pancreatic ductal adenocarcinoma in the early stage and the late phase (Allenson et al., 2017). The serum exosomes of glioma patients also carry the gDNA sequence with the same biological mutation as glioblastoma (Wang et al., 2018; Vaidya and Sugaya, 2020). In addition, the corresponding driving mutation was also found in the exosomes of prostate cancer (Lazaro-Ibanez et al., 2014). The genome-wide sequencing of exosomes of these tumor patients may provide information for diagnosis and prediction results.
In CRC, KRAS and BRAF mutation was detected from the serum exosomes of CRC patients with higher sensitivity and specificity (Hao et al., 2017; Lucchetti et al., 2021). In addition, it has been reported that transforming growth factor receptor 2 (TGFR2) is a part of the key signal pathway in colon epithelial cells. Its double allele frameshift mutation occurs repeatedly in most colorectal tumors, which is thought to drive colorectal cancer. It has been found that the frameshift mutation in the microsatellite region of TGFR2 is wrapped in the exosome derived from CRC cells. Although it can be detected at the DNA level, the author has not observed any frameshift protein in the exosome protein group. And then, acting on the receptor cells can up-regulate the expression of cytokines (Fricke et al., 2017). It has also been reported that the level of exosome gDNA of patients with KRAS mutation in CRC increases significantly in the course of the disease and shows more changes after treatment. Moreover, in CRC patients who meet the surgical conditions, the tumor size is related to the copy number of KRAS mutation. The copy number and abundance score of KRAS mutation in exosomes of patients with liver metastasis is significantly increased. The author also confirmed that KRAS mutation disappeared rapidly after the first chemotherapy cycle. However, it is necessary to closely monitor secondary drug resistance after anti-EGFR treatment to stop this treatment as soon as possible (Lucchetti et al., 2021). Therefore, exosome gDNA can be used as an innovative tool to monitor the diagnosis, drug resistance, and prognosis of CRC patients during treatment.
Exosome Genomic DNA Regulates Immunity and Metabolism of Colorectal Cancer
Tumor immunosuppression is a sign of cancer progression, signaled by immune checkpoints on immune cell subsets (Nikfarjam et al., 2020). In recent years, targeted checkpoint immunotherapy has therapeutic effects on cancer patients (Li et al., 2016; Li et al., 2018). Exosomes play a significant role in tumor immunity by paracrine signal regulators. Among many components of exosomes, exosome gDNA can be used as a potent regulator of the STING pathway, which can regulate tumor immunity. During radiotherapy, gDNA fragments accumulated in the cytoplasm of tumor cells can induce an IFN-1 response and then activate dendritic cells through paracrine to trigger an anti-tumor response. This process is regulated by the feedback loop that DNA exonuclease Trex1 degrades cytosolic DNA fragments to prevent anti-tumor reactions. However, to avoid this phenomenon, tumor cells can export gDNA pieces to the exosome. Once the exosome gDNA is internalized by tumor-infiltrated dendritic cells, it will activate the STING signal path in dendritic cells. Once dendritic cells are activated by exosome gDNA, they will produce IFN-1 and recruit more CD8+T lymphocytes to prevent tumor growth further (Vanpouille-Box et al., 2017). Crohn’s disease is related to the risk of colorectal cancer. Exosome gDNA internalized into macrophages can activate the STING signal path. The research also shows that exosome gDNA can’t work after inhibiting STING, which further confirms the importance of the STING pathway in exosome gDNA. It shows that exosome gDNA plays a vital role in the immunity of CRC and can be used as a potential biomarker and therapeutic target for Crohn’s disease (Zhao et al., 2021). Reports suggest CRC will produce intestinal syndrome after being treated with irinotecan and fluorouracil, and the severity of diarrhea is closely related to the content of exosome gDNA. It is suggested that drug therapy can trigger the release of gDNA by intestinal epithelial cells through exosomes. Then the AIM2 inflammasome in immune cells will be activated, which will promote the body’s inflammatory response. Therefore, exosome gDNA is not only a direct tumor immunomodulatory but also can affect the inflammatory reaction related to chemotherapy (Lian et al., 2017). Exosomes gDNA can indeed destroy the influence of checkpoint inhibitors. Therefore, in drug development, we should pay more attention to the signal of cell autonomic paracrine. In CRC, KRAS mutation will participate in metabolism in vitro and vivo by inducing glucose transporter 1 (Zhang et al., 2018). However, it is not sure if the exosome gDNA works, which needs further study in tumor metabolism.
Further Perspective
The discovery of high-frequency tumor mutant genes in exosome gDNA has attracted the attention of clinicians and basic researchers. Exosome gDNA is long and stable so that it can be used as excellent biomaterials for liquid biopsy (Jin et al., 2016) (Table 1). Many experts predict that in the near future, liquid biopsy based on exosome gDNA may improve the individualized treatment and prognosis of patients, especially for CRC patients, as the essential feature of CRC patients is the accumulation of mutations (Lichtenstern et al., 2020). Exosome gDNA can also be transferred from one cell to another by endocytosis or fusion. The transferred exosome gDNA can increase mRNA and protein expression in recipient cells and affect the function of recipient cells (Table 2). This may explain new pathogenesis of CRC and provide a new method for diagnosis or treatment in the future. However, it is necessary to conduct a larger-scale clinical study on these markers and targeted substances to verify the transformation of exosome gDNA into high-throughput practical solutions in the clinical environment.
Before exosome gDNA is officially applied in the clinic, it needs to meet the repeatable and verifiable conditions. However, due to the small amount of gDNA in exosomes and the different extraction methods, there is no uniform extraction and detection method, so its application is limited. Some hospitals have set up some large-scale analysis platforms to analyze it, which can provide helpful information for doctors’ diagnoses and make it possible to specialize in the analysis of exosome gDNA.
However, if we don’t know the mechanism of exosome gDNA, we can’t get the clinical effect. Therefore, the primary research on the packaging of exosome gDNA is the premise of developing the clinical treatment of exosome gDNA. Finally, further studies on the length, base composition, and stability of exosome gDNA under different physiological conditions will open up a new field for the diagnosis and treatment of colorectal cancer.
Conclusion
This article systematically reviewed the origin, mode of action of exosome gDNA, and its relationship with diagnosis, drug resistance, prognosis, immunity, and metabolism of CRC. The schematic diagram of the function of exosome gDNA is displayed in Figure 3. So far, we may have only seen the tip of the iceberg, and most of it is unknown. Therefore, more research is needed to reveal anonymous information about the application of exosome gDNA in the diagnosis and treatment of CRC.
FIGURE 3.
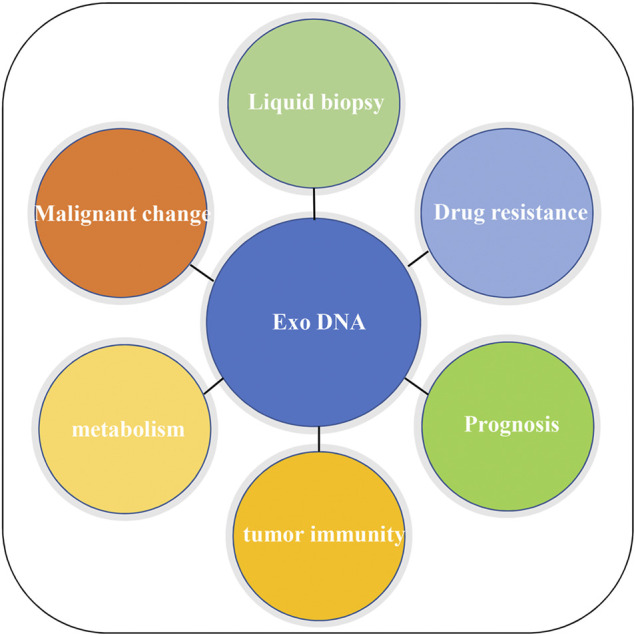
Application of exosome gDNA in CRC.
Author Contributions
RW contributed to the text and tables, QW contributed to revise the review manuscript, and XL contributed to the figures in the review manuscript.
Funding
This work was supported by Double First-class Fund from the Educational Department of Liaoning Province.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- Ahmadi M., Rezaie J. (2021). Ageing and Mesenchymal Stem Cells Derived Exosomes: Molecular Insight and Challenges. Cell biochem. Funct. 39 (1), 60–66. 10.1002/cbf.3602 [DOI] [PubMed] [Google Scholar]
- Allenson K., Castillo J., San Lucas F. A., Scelo G., Kim D. U., Bernard V., et al. (2017). High Prevalence of Mutant KRAS in Circulating Exosome-Derived DNA from Early-Stage Pancreatic Cancer Patients. Ann. Oncol. 28 (4), 741–747. 10.1093/annonc/mdx004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaj L., Lessard R., Dai L., Cho Y. J., Pomeroy S. L., Breakefield X. O., et al. (2011). Tumour Microvesicles Contain Retrotransposon Elements and Amplified Oncogene Sequences. Nat. Commun. 2, 180. 10.1038/ncomms1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barıs I. C., Hacıoglu S., Turk N. S., Cetın G. O., Zencır S., Bagcı G., et al. (2021). Expression and DNA Methylation Profiles of EZH2-Target Genes in Plasma Exosomes and Matched Primary Tumor Tissues of the Patients with Diffuse Large B-Cell Lymphoma. Clin. Transl. Oncol. 23 (6), 1152–1166. 10.1007/s12094-020-02504-6 [DOI] [PubMed] [Google Scholar]
- Bernard V., Kim D. U., San Lucas F. A., Castillo J., Allenson K., Mulu F. C., et al. (2019). Circulating Nucleic Acids Are Associated with Outcomes of Patients with Pancreatic Cancer. Gastroenterology 156 (1), 108–e4. 10.1053/j.gastro.2018.09.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai J., Han Y., Ren H., Chen C., He D., Zhou L., et al. (2013). Extracellular Vesicle-Mediated Transfer of Donor Genomic DNA to Recipient Cells Is a Novel Mechanism for Genetic Influence between Cells. J. Mol. Cell Biol. 5 (4), 227–238. 10.1093/jmcb/mjt011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai J., Wu G., Tan X., Han Y., Chen C., Li C., et al. (2014). Transferred BCR/ABL DNA from K562 Extracellular Vesicles Causes Chronic Myeloid Leukemia in Immunodeficient Mice. PLoS One 9 (8), e105200. 10.1371/journal.pone.0105200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellanos-Rizaldos E., Grimm D. G., Tadigotla V., Hurley J., Healy J., Neal P. L., et al. (2018). Exosome-Based Detection of EGFR T790M in Plasma from Non-small Cell Lung Cancer Patients. Clin. Cancer Res. 24 (12), 2944–2950. 10.1158/1078-0432.CCR-17-3369 [DOI] [PubMed] [Google Scholar]
- Chang Y.-C., Chan M.-H., Li C.-H., Fang C.-Y., Hsiao M., Chen C.-L. (2021). Exosomal Components and Modulators in Colorectal Cancer: Novel Diagnosis and Prognosis Biomarkers. Biomedicines 9 (8), 931. 10.3390/biomedicines9080931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow J., Atay S., Banskota S., Artale B., Schmitt S., Godwin A. K. (2017). Exosomes as Mediators of Platinum Resistance in Ovarian Cancer. Oncotarget 8 (7), 11917–11936. 10.18632/oncotarget.14440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degli Esposti C., Iadarola B., Maestri S., Beltrami C., Lavezzari D., Morini M., et al. (2021). Exosomes from Plasma of Neuroblastoma Patients Contain Doublestranded DNA Reflecting the Mutational Status of Parental Tumor Cells. Ijms 22 (7), 3667. 10.3390/ijms22073667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker E., Rex D. K. (2018). Advances in CRC Prevention: Screening and Surveillance. Gastroenterology 154 (7), 1970–1984. 10.1053/j.gastro.2018.01.069 [DOI] [PubMed] [Google Scholar]
- Domenis R., Cifù A., Mio C., Fabris M., Curcio F. (2021). Pro-Inflammatory Microenvironment Modulates the Transfer of Mutated TP53 Mediated by Tumor Exosomes. Int. J. Mol. Sci. 22 (12). 10.3390/ijms22126258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle L. M., Wang M. Z. (2019). Overview of Extracellular Vesicles, Their Origin, Composition, Purpose, and Methods for Exosome Isolation and Analysis. Cells 8 (7). 10.3390/cells8070727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elzanowska J., Semira C., Costa-Silva B. (2021). DNA in Extracellular Vesicles: Biological and Clinical Aspects. Mol. Oncol. 15 (6), 1701–1714. 10.1002/1878-0261.12777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdmann S., Tschitschko B., Zhong L., Raftery M. J., Cavicchioli R. (2017). A Plasmid from an Antarctic Haloarchaeon Uses Specialized Membrane Vesicles to Disseminate and Infect Plasmid-free Cells. Nat. Microbiol. 2 (10), 1446–1455. 10.1038/s41564-017-0009-2 [DOI] [PubMed] [Google Scholar]
- Fischer S., Cornils K., Speiseder T., Badbaran A., Reimer R., Indenbirken D., et al. (2016). Indication of Horizontal DNA Gene Transfer by Extracellular Vesicles. PLoS One 11 (9), e0163665. 10.1371/journal.pone.0163665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke A. J., Skelton W. P., Starr J. S., Parekh H., Lee J. J., Overman M. J., et al. (2019). Immunotherapy for Colorectal Cancer: A Review of Current and Novel Therapeutic Approaches. J. Natl. Cancer Inst. 111 (11), 1131–1141. 10.1093/jnci/djz093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fricke F., Lee J., Michalak M., Warnken U., Hausser I., Suarez-Carmona M., et al. (2017). TGFBR2-dependent Alterations of Exosomal Cargo and Functions in DNA Mismatch Repair-Deficient HCT116 Colorectal Cancer Cells. Cell Commun. Signal. 15 (1), 14. 10.1186/s12964-017-0169-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Romero N., Carrión-Navarro J., Esteban-Rubio S., Lázaro-Ibáñez E., Peris-Celda M., Alonso M. M., et al. (2017). DNA Sequences within Glioma-Derived Extracellular Vesicles Can Cross the Intact Blood-Brain Barrier and Be Detected in Peripheral Blood of Patients. Oncotarget 8 (1), 1416–1428. 10.18632/oncotarget.13635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannopoulou L., Zavridou M., Kasimir-Bauer S., Lianidou E. S. (2019). Liquid Biopsy in Ovarian Cancer: the Potential of Circulating miRNAs and Exosomes. Transl. Res. 205, 77–91. 10.1016/j.trsl.2018.10.003 [DOI] [PubMed] [Google Scholar]
- Hao Y. X., Li Y. M., Ye M., Guo Y. Y., Li Q. W., Peng X. M., et al. (2017). KRAS and BRAF Mutations in Serum Exosomes from Patients with Colorectal Cancer in a Chinese Population. Oncol. Lett. 13 (5), 3608–3616. 10.3892/ol.2017.5889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C., Zheng S., Luo Y., Wang B. (2018). Exosome Theranostics: Biology and Translational Medicine. Theranostics 8 (1), 237–255. 10.7150/thno.21945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino D., Kirkbride K. C., Costello K., Clark E. S., Sinha S., Grega-Larson N., et al. (2013). Exosome Secretion Is Enhanced by Invadopodia and Drives Invasive Behavior. Cell Rep. 5 (5), 1159–1168. 10.1016/j.celrep.2013.10.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hur J. Y., Kim H. J., Lee J. S., Choi C. M., Lee J. C., Jung M. K., et al. (2018). Extracellular Vesicle-Derived DNA for Performing EGFR Genotyping of NSCLC Patients. Mol. Cancer. 17 (1), 15. 10.1186/s12943-018-0772-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hur J. Y., Lee J. S., Kim I. A., Kim H. J., Kim W. S., Lee K. Y. (2019). Extracellular Vesicle-Based EGFR Genotyping in Bronchoalveolar Lavage Fluid from Treatment-Naive Non-small Cell Lung Cancer Patients. Transl. Lung Cancer Res. 8 (6), 1051–1060. 10.21037/tlcr.2019.12.16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hur J. Y., Lee K. Y. (2021). Characteristics and Clinical Application of Extracellular Vesicle-Derived DNA. Cancers (Basel) 13 (15). 10.3390/cancers13153827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jafari R., Rahbarghazi R., Ahmadi M., Hassanpour M., Rezaie J. (2020). Hypoxic Exosomes Orchestrate Tumorigenesis: Molecular Mechanisms and Therapeutic Implications. J. Transl. Med. 18 (1), 474. 10.1186/s12967-020-02662-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janney A., Powrie F., Mann E. H. (2020). Host-microbiota Maladaptation in Colorectal Cancer. Nature 585 (7826), 509–517. 10.1038/s41586-020-2729-3 [DOI] [PubMed] [Google Scholar]
- Jeppesen D. K., Fenix A. M., Franklin J. L., Higginbotham J. N., Zhang Q., Zimmerman L. J., et al. (2019). Reassessment of Exosome Composition. Cell 177 (2), 428–e18. 10.1016/j.cell.2019.02.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y., Chen K., Wang Z., Wang Y., Liu J., Lin L., et al. (2016). DNA in Serum Extracellular Vesicles Is Stable under Different Storage Conditions. BMC Cancer 16 (1), 753. 10.1186/s12885-016-2783-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahlert C., Melo S. A., Protopopov A., Tang J., Seth S., Koch M., et al. (2014). Identification of Double-Stranded Genomic DNA Spanning All Chromosomes with Mutated KRAS and P53 DNA in the Serum Exosomes of Patients with Pancreatic Cancer. J. Biol. Chem. 289 (7), 3869–3875. 10.1074/jbc.C113.532267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalluri R., Lebleu V. S. (2016). Discovery of Double-Stranded Genomic DNA in Circulating Exosomes. Cold Spring Harb. Symp. Quant. Biol. 81, 275–280. 10.1101/sqb.2016.81.030932 [DOI] [PubMed] [Google Scholar]
- Kalluri R., Lebleu V. S. (2020). The Biology, Function, and Biomedical Applications of Exosomes. Science 367 (6478). 10.1126/science.aau6977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keum N., Giovannucci E. (2019). Global Burden of Colorectal Cancer: Emerging Trends, Risk Factors and Prevention Strategies. Nat. Rev. Gastroenterol. Hepatol. 16 (12), 713–732. 10.1038/s41575-019-0189-8 [DOI] [PubMed] [Google Scholar]
- Khaksar M., Sayyari M., Rezaie J., Pouyafar A., Montazersaheb S., Rahbarghazi R. (2018). High Glucose Condition Limited the Angiogenic/cardiogenic Capacity of Murine Cardiac Progenitor Cells in In Vitro and In Vivo Milieu. Cell biochem. Funct. 36 (7), 346–356. 10.1002/cbf.3354 [DOI] [PubMed] [Google Scholar]
- Kim Y., Shin S., Lee K. A. (2021). Exosome-based Detection of EGFR T790M in Plasma and Pleural Fluid of Prospectively Enrolled Non-small Cell Lung Cancer Patients after First-Line Tyrosine Kinase Inhibitor Therapy. Cancer Cell Int. 21 (1), 50. 10.1186/s12935-021-01761-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolenčík D., Shishido S. N., Pitule P., Mason J., Hicks J., Kuhn P. (2020). Liquid Biopsy in Colorectal Carcinoma: Clinical Applications and Challenges. Cancers (Basel) 12 (6). 10.3390/cancers12061376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krug A. K., Enderle D., Karlovich C., Priewasser T., Bentink S., Spiel A., et al. (2018). Improved EGFR Mutation Detection Using Combined Exosomal RNA and Circulating Tumor DNA in NSCLC Patient Plasma. Ann. Oncol. 29 (3), 700–706. 10.1093/annonc/mdx765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lázaro-Ibáñez E., Lässer C., Shelke G. V., Crescitelli R., Jang S. C., Cvjetkovic A., et al. (2019). DNA Analysis of Low- and High-Density Fractions Defines Heterogeneous Subpopulations of Small Extracellular Vesicles Based on Their DNA Cargo and Topology. J. Extracell. Vesicles 8 (1), 1656993. 10.1080/20013078.2019.1656993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lázaro-Ibáñez E., Sanz-Garcia A., Visakorpi T., Escobedo-Lucea C., Siljander P., Ayuso-Sacido A., et al. (2014). Different gDNA Content in the Subpopulations of Prostate Cancer Extracellular Vesicles: Apoptotic Bodies, Microvesicles, and Exosomes. Prostate 74 (14), 1379–1390. 10.1002/pros.22853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. S., Hur J. Y., Kim I. A., Kim H. J., Choi C. M., Lee J. C., et al. (2018). Liquid Biopsy Using the Supernatant of a Pleural Effusion for EGFR Genotyping in Pulmonary Adenocarcinoma Patients: a Comparison between Cell-free DNA and Extracellular Vesicle-Derived DNA. BMC Cancer 18 (1), 1236. 10.1186/s12885-018-5138-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T. H., Chennakrishnaiah S., Audemard E., Montermini L., Meehan B., Rak J. (2014). Oncogenic Ras-Driven Cancer Cell Vesiculation Leads to Emission of Double-Stranded DNA Capable of Interacting with Target Cells. Biochem. Biophys. Res. Commun. 451 (2), 295–301. 10.1016/j.bbrc.2014.07.109 [DOI] [PubMed] [Google Scholar]
- Lee T. H., Chennakrishnaiah S., Meehan B., Montermini L., Garnier D., D'Asti E., et al. (2016). Barriers to Horizontal Cell Transformation by Extracellular Vesicles Containing Oncogenic H-Ras. Oncotarget 7 (32), 51991–52002. 10.18632/oncotarget.10627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Shao C., Shi Y., Han W. (2018). Lessons Learned from the Blockade of Immune Checkpoints in Cancer Immunotherapy. J. Hematol. Oncol. 11 (1), 31. 10.1186/s13045-018-0578-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Li F., Jiang F., Lv X., Zhang R., Lu A., et al. (2016). A Mini-Review for Cancer Immunotherapy: Molecular Understanding of PD-1/PD-L1 Pathway & Translational Blockade of Immune Checkpoints. Int. J. Mol. Sci. 17 (7). 10.3390/ijms17071151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian Q., Xu J., Yan S., Huang M., Ding H., Sun X., et al. (2017). Chemotherapy-induced Intestinal Inflammatory Responses Are Mediated by Exosome Secretion of Double-Strand DNA via AIM2 Inflammasome Activation. Cell Res. 27 (6), 784–800. 10.1038/cr.2017.54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y., Lehrich B. M., Zheng S., Lu M. (2021). Emerging Methods in Biomarker Identification for Extracellular Vesicle-Based Liquid Biopsy. J. Extracell. Vesicles 10 (7), e12090. 10.1002/jev2.12090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenstern C. R., Ngu R. K., Shalapour S., Karin M. (2020). Immunotherapy, Inflammation and Colorectal Cancer. Cells 9 (3). 10.3390/cells9030618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin B., Lei Y., Wang J., Zhu L., Wu Y., Zhang H., et al. (2021). Microfluidic-Based Exosome Analysis for Liquid Biopsy. Small Methods 5 (3), e2001131. 10.1002/smtd.202001131 [DOI] [PubMed] [Google Scholar]
- Liu J., Ren L., Li S., Li W., Zheng X., Yang Y., et al. (2021). The Biology, Function, and Applications of Exosomes in Cancer. Acta Pharm. Sin. B 11 (9), 2783–2797. 10.1016/j.apsb.2021.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucchetti D., Zurlo I. V., Colella F., Ricciardi-Tenore C., Di Salvatore M., Tortora G., et al. (2021). Mutational Status of Plasma Exosomal KRAS Predicts Outcome in Patients with Metastatic Colorectal Cancer. Sci. Rep. 11 (1), 22686. 10.1038/s41598-021-01668-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcuello M., Neves V., Duran-Sanchon R. P. L., Vedeld S., Tham H. M., van Dalum E., et al. (2019). Circulating Biomarkers for Early Detection and Clinical Management of Colorectal Cancer. Mol. Asp. Med. 69, 107–122. 10.1016/j.mam.2019.06.002 [DOI] [PubMed] [Google Scholar]
- Martini G., Dienstmann R., Ros J., Baraibar I., Cuadra-Urteaga J. L., Salva F., et al. (2020). Molecular Subtypes and the Evolution of Treatment Management in Metastatic Colorectal Cancer. Ther. Adv. Med. Oncol. 12, 1758835920936089. 10.1177/1758835920936089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Németh A., Orgovan N., Sódar B. W., Osteikoetxea X., Pálóczi K., Szabó-Taylor K. É., et al. (2017). Antibiotic-induced Release of Small Extracellular Vesicles (Exosomes) with Surface-Associated DNA. Sci. Rep. 7 (1), 8202. 10.1038/s41598-017-08392-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikfarjam S., Rezaie J., Kashanchi F., Jafari R. (2020). Dexosomes as a Cell-free Vaccine for Cancer Immunotherapy. J. Exp. Clin. Cancer Res. 39 (1), 258. 10.1186/s13046-020-01781-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogunwobi O. O., Mahmood F., Akingboye A. (2020). Biomarkers in Colorectal Cancer: Current Research and Future Prospects. Int. J. Mol. Sci. 21 (15). 10.3390/ijms21155311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panigrahi G. K., Praharaj P. P., Peak T. C., Long J., Singh R., Rhim J. S., et al. (2018). Hypoxia-induced Exosome Secretion Promotes Survival of African-American and Caucasian Prostate Cancer Cells. Sci. Rep. 8 (1), 3853. 10.1038/s41598-018-22068-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J., Lee C., Eom J. S., Kim M. H., Cho Y. K. (2020). Detection of EGFR Mutations Using Bronchial Washing-Derived Extracellular Vesicles in Patients with Non-small-cell Lung Carcinoma. Cancers (Basel) 12 (10). 10.3390/cancers12102822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parolini I., Federici C., Raggi C., Lugini L., Palleschi S., De Milito A., et al. (2009). Microenvironmental pH Is a Key Factor for Exosome Traffic in Tumor Cells. J. Biol. Chem. 284 (49), 34211–34222. 10.1074/jbc.M109.041152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegtel D. M., Gould S. J. (2019). Exosomes. Annu. Rev. Biochem. 88, 487–514. 10.1146/annurev-biochem-013118-111902 [DOI] [PubMed] [Google Scholar]
- Qu X., Li Q., Yang J., Zhao H., Wang F., Zhang F., et al. (2019). Double-Stranded DNA in Exosomes of Malignant Pleural Effusions as a Novel DNA Source for EGFR Mutation Detection in Lung Adenocarcinoma. Front. Oncol. 9, 931. 10.3389/fonc.2019.00931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezaie J., Ahmadi M., Ravanbakhsh R., Mojarad B., Mahbubfam S., Shaban S. A., et al. (2022). Tumor-derived Extracellular Vesicles: The Metastatic Organotropism Drivers. Life Sci. 289, 120216. 10.1016/j.lfs.2021.120216 [DOI] [PubMed] [Google Scholar]
- Rezaie J., Aslan C., Ahmadi M., Zolbanin N. M., Kashanchi F., Jafari R. (2021). The Versatile Role of Exosomes in Human Retroviral Infections: from Immunopathogenesis to Clinical Application. Cell Biosci. 11 (1), 19. 10.1186/s13578-021-00537-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riches A., Campbell E., Borger E., Powis S. (2014). Regulation of Exosome Release from Mammary Epithelial and Breast Cancer Cells - a New Regulatory Pathway. Eur. J. Cancer. 50 (5), 1025–1034. 10.1016/j.ejca.2013.12.019 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Casanova A., Costa-Fraga N., Bao-Caamano A., López-López R., Muinelo-Romay L., Diaz-Lagares A. (2021). Epigenetic Landscape of Liquid Biopsy in Colorectal Cancer. Front. Cell Dev. Biol. 9, 622459. 10.3389/fcell.2021.622459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansone P., Savini C., Kurelac I., Chang Q., Amato L. B., Strillacci A., et al. (2017). Packaging and Transfer of Mitochondrial DNA via Exosomes Regulate Escape from Dormancy in Hormonal Therapy-Resistant Breast Cancer. Proc. Natl. Acad. Sci. U. S. A. 114 (43), E9066–E9075. 10.1073/pnas.1704862114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A., Johnson A. (2020). Exosome DNA: Critical Regulator of Tumor Immunity and a Diagnostic Biomarker. J. Cell. Physiol. 235 (3), 1921–1932. 10.1002/jcp.29153 [DOI] [PubMed] [Google Scholar]
- Soraya H., Sani N. A., Jabbari N., Rezaie J. (2021). Metformin Increases Exosome Biogenesis and Secretion in U87 MG Human Glioblastoma Cells: A Possible Mechanism of Therapeutic Resistance. Arch. Med. Res. 52 (2), 151–162. 10.1016/j.arcmed.2020.10.007 [DOI] [PubMed] [Google Scholar]
- Spada S., Rudqvist N. P., Wennerberg E. (2020). Isolation of DNA from Exosomes. Methods Enzymol. 636, 173–183. 10.1016/bs.mie.2020.01.012 [DOI] [PubMed] [Google Scholar]
- Stefanius K., Servage K., de Souza Santos M., Gray H. F., Toombs J. E., Chimalapati S., et al. (2019). Human Pancreatic Cancer Cell Exosomes, but Not Human Normal Cell Exosomes, Act as an Initiator in Cell Transformation. eLife 8. 10.7554/eLife.40226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stobiecka M., Ratajczak K., Jakiela S. (2019). Toward Early Cancer Detection: Focus on Biosensing Systems and Biosensors for an Anti-apoptotic Protein Survivin and Survivin mRNA. Biosens. Bioelectron. 137, 58–71. 10.1016/j.bios.2019.04.060 [DOI] [PubMed] [Google Scholar]
- Stoffel E. M., Murphy C. C. (2020). Epidemiology and Mechanisms of the Increasing Incidence of Colon and Rectal Cancers in Young Adults. Gastroenterology 158 (2), 341–353. 10.1053/j.gastro.2019.07.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sveen A., Kopetz S., Lothe R. A. (2020). Biomarker-guided Therapy for Colorectal Cancer: Strength in Complexity. Nat. Rev. Clin. Oncol. 17 (1), 11–32. 10.1038/s41571-019-0241-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi A., Okada R., Nagao K., Kawamata Y., Hanyu A., Yoshimoto S., et al. (2017). Exosomes Maintain Cellular Homeostasis by Excreting Harmful DNA from Cells. Nat. Commun. 8, 15287. 10.1038/ncomms15287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur B. K., Zhang H., Becker A., Matei I., Huang Y., Costa-Silva B., et al. (2014). Double-stranded DNA in Exosomes: a Novel Biomarker in Cancer Detection. Cell Res. 24 (6), 766–769. 10.1038/cr.2014.44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trejo-Becerril C., Pérez-Cárdenas E., Taja-Chayeb L., Anker P., Herrera-Goepfert R., Medina-Velázquez L. A., et al. (2012). Cancer Progression Mediated by Horizontal Gene Transfer in an In Vivo Model. PLoS One 7 (12), e52754. 10.1371/journal.pone.0052754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vahabi A., Rezaie J., Hassanpour M., Panahi Y., Nemati M., Rasmi Y., et al. (2022). Tumor Cells-Derived Exosomal CircRNAs: Novel Cancer Drivers, Molecular Mechanisms, and Clinical Opportunities. Biochem. Pharmacol. 200, 115038. 10.1016/j.bcp.2022.115038 [DOI] [PubMed] [Google Scholar]
- Vaidya M., Sugaya K. (2020). DNA Associated with Circulating Exosomes as a Biomarker for Glioma. Genes (Basel) 11 (11). 10.3390/genes11111276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanpouille-Box C., Alard A., Aryankalayil M. J., Sarfraz Y., Diamond J. M., Schneider R. J., et al. (2017). DNA Exonuclease Trex1 Regulates Radiotherapy-Induced Tumour Immunogenicity. Nat. Commun. 8, 15618. 10.1038/ncomms15618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldenström A., Gennebäck N., Hellman U., Ronquist G. (2012). Cardiomyocyte Microvesicles Contain DNA/RNA and Convey Biological Messages to Target Cells. PLoS One 7 (4), e34653. 10.1371/journal.pone.0034653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Li Y., Guan X., Zhao J., Shen L., Liu J. (2018). Exosomal Double-Stranded DNA as a Biomarker for the Diagnosis and Preoperative Assessment of Pheochromocytoma and Paraganglioma. Mol. Cancer. 17 (1), 128. 10.1186/s12943-018-0876-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z. Y., Wang R. X., Ding X. Q., Zhang X., Pan X. R., Tong J. H. (2020). A Protocol for Cancer-Related Mutation Detection on Exosomal DNA in Clinical Application. Front. Oncol. 10, 558106. 10.3389/fonc.2020.558106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteside T. L. (2016). Tumor-Derived Exosomes and Their Role in Cancer Progression. Adv. Clin. Chem. 74, 103–141. 10.1016/bs.acc.2015.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y., Zhong J., Zhong B., Huang J., Jiang L., Jiang Y., et al. (2020). Exosomes as Potential Sources of Biomarkers in Colorectal Cancer. Cancer Lett. 476, 13–22. 10.1016/j.canlet.2020.01.033 [DOI] [PubMed] [Google Scholar]
- Yang S., Che S. P., Kurywchak P., Tavormina J. L., Gansmo L. B., Correa de Sampaio P., et al. (2017). Detection of Mutant KRAS and TP53 DNA in Circulating Exosomes from Healthy Individuals and Patients with Pancreatic Cancer. Cancer Biol. Ther. 18 (3), 158–165. 10.1080/15384047.2017.1281499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoi A., Villar-Prados A., Oliphint P. A., Zhang J., Song X., De Hoff P., et al. (2019). Mechanisms of Nuclear Content Loading to Exosomes. Sci. Adv. 5 (11), eaax8849. 10.1126/sciadv.aax8849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W., Hurley J., Roberts D., Chakrabortty S. K., Enderle D., Noerholm M., et al. (2021). Exosome-based Liquid Biopsies in Cancer: Opportunities and Challenges. Ann. Oncol. 32 (4), 466–477. 10.1016/j.annonc.2021.01.074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Yu D. (2019). Exosomes in Cancer Development, Metastasis, and Immunity. Biochim. Biophys. Acta Rev. Cancer 1871 (2), 455–468. 10.1016/j.bbcan.2019.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Jeppesen D. K., Higginbotham J. N., Demory Beckler M., Poulin E. J., Walsh A. J., et al. (2018). Mutant KRAS Exosomes Alter the Metabolic State of Recipient Colonic Epithelial Cells. Cell Mol. Gastroenterol. Hepatol. 5 (4), 627–e6. 10.1016/j.jcmgh.2018.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao F., Zheng T., Gong W., Wu J., Xie H., Li W., et al. (2021). Extracellular Vesicles Package dsDNA to Aggravate Crohn's Disease by Activating the STING Pathway. Cell Death Dis. 12 (9), 815. 10.1038/s41419-021-04101-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Kurywchak P., Wolf-Dennen K., Che S. P. Y., Sulakhe D., D'Souza M., et al. (2021). Unique Somatic Variants in DNA from Urine Exosomes of Individuals with Bladder Cancer. Mol. Ther. Methods Clin. Dev. 22, 360–376. 10.1016/j.omtm.2021.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]