Highlights
-
•
Multi-site MRI data encode confounding information which may mask case-control differences.
-
•
The impact of the NeuroHarmonize method is evaluated in the ASD-control classification.
-
•
We verified the successful removal of the site effect by the harmonization protocol.
-
•
The increment in the classification performance is quantified after data harmonization.
-
•
We identified the anatomical features that contributed to the two-class separation.
Keywords: Harmonization, ASD, Machine learning, FreeSurfer, Multi-site
Abstract
Machine Learning (ML) techniques have been widely used in Neuroimaging studies of Autism Spectrum Disorders (ASD) both to identify possible brain alterations related to this condition and to evaluate the predictive power of brain imaging modalities. The collection and public sharing of large imaging samples has favored an even greater diffusion of the use of ML-based analyses. However, multi-center data collections may suffer the batch effect, which, especially in case of Magnetic Resonance Imaging (MRI) studies, should be curated to avoid confounding effects for ML classifiers and masking biases. This is particularly important in the study of barely separable populations according to MRI data, such as subjects with ASD compared to controls with typical development (TD). Here, we show how the implementation of a harmo- nization protocol on brain structural features unlocks the case-control ML separation capability in the analysis of a multi-center MRI dataset. This effect is demonstrated on the ABIDE data collection, involving subjects encompassing a wide age range. After data harmonization, the overall ASD vs. TD discrimination capability by a Random Forest (RF) classifier improves from a very low performance (AUC = 0.58 ± 0.04) to a still low, but reasonably significant AUC = 0.67 ± 0.03. The performances of the RF classifier have been evaluated also in the age-specific subgroups of children, adolescents and adults, obtaining AUC = 0.62 ± 0.02, AUC = 0.65 ± 0.03 and AUC = 0.69 ± 0.06, respectively. Specific and consistent patterns of anatomical differences related to the ASD condition have been identified for the three different age subgroups.
1. Introduction
Autism spectrum disorders (ASD) is a diagnostic category of neurodevelopmental disorders defined by persistent social communication and social interaction deficits, as well as restricted, repetitive patterns of behaviour, interests or activities that must be present in the early developmental period and cause clinically significant impairment in social, occupational or other important areas of functioning (American Psychiatric Association et al., 2013). One of the key characteristics of autism is its great heterogeneity across multiple levels, including genetic background (Sullivan et al., 2012), neuroanatomical substrates (Pagnozzi et al., 2018), and phenotypic profile (Georgiades et al., 2013).
Despite the diagnosis of ASD is still made on the basis of direct behavioral evaluation of the child and parent/caregiver interview, neuroimaging has been playing a fundamental role in identifying the neural correlates of this condition since the early 2000s (Courchesne et al., 2001).
In the last decade, machine leaning (ML) techniques have been implemented in the attempt to discover neuroimaging-based biomarker of ASD, intended to either support, facilitate or shorten the diagnostic process (Ecker et al., 2015, Li et al., 2017). After the first encouraging results obtained for adults (Deshpande et al., 2013, Ecker et al., 2010a, Ecker et al., 2010b) and children with ASD (Calderoni et al., 2012, Gori et al., 2015, Ingalhalikar et al., 2011, Jiao et al., 2010, Uddin et al., 2013) on rather limited size datasets, the need of replicating the findings on larger samples emerged. However, inconsistent findings have been reported about the predictive power of ML techniques on neuroimaging data, and also regarding the possible patterns of alteration in the neuro-anatomy and in connectivity measures in ASD (Arbabshirani et al., 2017, Wolfers et al., 2019).
The aggregation of large data samples has been seen as a potential solution to overcome the fragmentation and lack of reproducibility of the previous studies. Large data samples are funda- mental especially to conduct analyses based on ML techniques. Arbabshirani et al. (Arbabshirani et al., 2017) reported that the highest classification performances in case-control discrimination based on neuroimaging data are reached only in studies using small datasets. These performances drop significantly in larger samples, especially in multi-site databases. This observation holds also in the field of ASD research, where several large-scale studies (Abraham et al., 2017, Heinsfeld et al., 2018, Katuwal et al., 2016, Nielsen et al., 2013) reported classification accuracy quite lower than previous studies conducted on smaller samples.
In the field of ASD research, a large and public accessible resource of neuroimaging and phenotypic information has been collected within the Autism Brain Imaging Data Exchange (ABIDE) ini- tiative1. Two world-wide multi-site and large-scale collections have been released so far, ABIDE I (Di Martino et al., 2014) and ABIDE II (Di Martino et al., 2017), jointly consisting in more than a thousand cases and as many controls. In spite of the increased sample sizes, studies based on the ABIDE cohort continued to report highly variable classification performances (Vargason et al., 2020). In the work by Haar et al. (Haar et al., 2016) the modest accuracy in the case-control dis- crimination (<60%) suggested that anatomical measures are of limited diagnostic utility for ASD. It was highlighted afterwards that multi-center MRI data collections suffer from the so-called batch effect (Ferrari et al., 2020a, Ferrari et al., 2020b, Lombardi et al., 2020). In brief, MRI data acquisitions made with different scanners and/or with different acquisition protocols encode confounding information in data which, if not accounted for, may completely mask case-control differences. In the specific case of ASD vs. control comparisons, the possible differences are so tiny that they can be completely obscured by the batch (or site) effect. In this context, Ferrari and colleagues observed that the acquisition site heavily confounds the ML classifiers, which instead need to be trained on a cleaned and controlled data sample. By adopting this method, i.e. limiting the ML training to a cohort of subjects acquired at one single site and controlling for all other confounding variables, the case-control discrimination performance of AUC = 0.79 was obtained on an independent test set (Ferrari, Bosco et al., 2020).
A data harmonization protocol devoted to the elimination of the site effect in neuroimaging studies has been introduced by Fortin et al. (Fortin et al., 2017), as an adaptation of the ComBat method developed by Johnson et al. (Johnson et al., 2007) to remove batch effects in genomics data. Pomponio et al. (Pomponio et al., 2020) have recently presented a modified version of the harmonization protocol, the NeuroHarmonize tool, which is suitable to harmonize pooled dataset in the presence of non-linear age trends. They developed and validated the methodology on a dataset of structural brain scans of more than 10 thousands subjects without known neurological or psychiatric disorders, covering the entire lifespan.
In this study, we evaluated the impact of the implementation of the NeuroHarmonize data har- monization protocol in the ASD vs. control discrimination problem tackled with ML. We implemented a standard Random Forest (RF) classifier to this purpose, and we analyzed the multi-center ABIDE I and ABIDE II data collections. First of all, we verified the successful removal of the site effect by the harmonization protocol. To this purpose, we first observed the confounding effect of the acquisition site on a ML classifier by evaluating non-null performances (AUC = 0.5) in the site vs. site discrimination by a RF trained on non-harmonized data of control subjects. Then, we observed the AUC values return to the expected range (AUC ∼0.5) for the same classification problem after the harmonization process. Secondly, we quantified the increment in the two-class (i.e. ASD vs. control) RF classification performance after data harmonization. We evaluated this increment for both the whole sample and within each of three age-specific subgroups, namely children, adolescents and adults. Finally, for each age-specific subgroup, we identified the neu- roanatomical features that contributed the most to the two-class separation. We thus highlighted specific patterns of brain feature involvement in the ASD condition across the lifespan.
2. Materials and methods
2.1. Participants and data description
We analyzed the structural MRI (T1-weighted) brain scans of the ABIDE I (Di Martino et al., 2014) and ABIDE II (Di Martino et al., 2017) publicly available collections. The total dataset is composed by 2226 subjects (1060 subjects with ASD and 1166 controls with typical development (TD)), collected across 26 international institutions. The MRI scans belong to 39 different samples, which in this paper will be referred to as different sites, each of them containing images acquired with a particular scanner type and specific acquisition parameters. We performed the recon-all Freesurfer preprocessing pipeline, which was unsuccessful for the images of poor quality. We performed manual quality control of these scans by visual inspection and we verified the presence of motion artifacts and of low signal to noise ratio. Hence, we discarded 65 subjects out of the 2226 scans available.
To allow the use of NeuroHarmonize tool, it is necessary that both data from case and control subjects are available within each site; thus, we had to exclude from the analysis two sites (KUL-II and NYU2-II of the ABIDE II cohort) that contributed to the collection exams of control subjects only. Since 97% of the subjects were under the age of 40 years, we limited our study to subjects aged 6 to 40 years only, similarly to other studies in the field (Haar et al., 2016, Katuwal et al., 2016). Moreover, we restricted our analysis to male subjects, due to the limited representation of female subjects in the ABIDE collection (<20% of subjects, spread over different sites and a wide age range). Due to the significant differences in neuroanatomy between males and females both in children (Retico et al., 2016) and in adults (Lai et al., 2013), we preferred to avoid including in this study the additional heterogeneity factor attributable to gender effects.
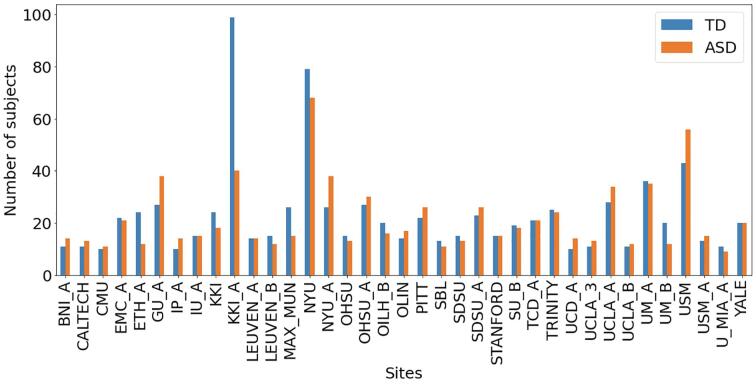
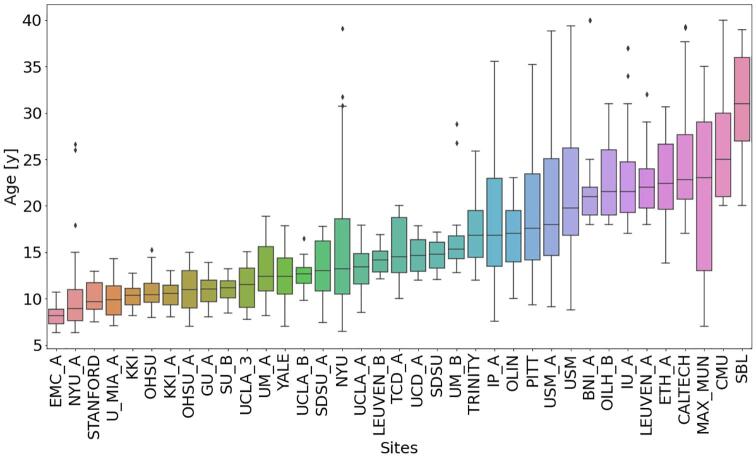
Thus, we obtained a final sample of N = 1638 subjects from 37 sites, including N = 845 typically developing participants with mean age = 15.6 years, standard deviation (STD) of age = 7.0 years and age range = [6.3- 40] years, and N = 793 subjects with ASD with mean age = 15.2 years, STD of age = 6.3 years and age range = [6.4- 40] years. A summary of the sample sizes of the ABIDE I and II cohorts included in this study and of the participants’ average age is reported in Table 1. Fig. 1 shows a bar diagram reporting the number of subjects belonging to each site grouped by diagnosis, whereas Fig. 2 shows the age distribution within each site in terms of box plots. To allow the reproducibility of the analysis, the identification numbers (IDs) of the participants selected in the final sample are reported in Supplementary Materials.
Table 1.
Number of subjects of the ABIDE I and II cohorts considered in this study. Only male subjects in the age range of [6–40] years (y) are considered. The number of participants are provided per site and per diagnostic group, together with the average age and standard deviation of each group. Abbreviation: STD - standard deviation.
| Centers | N |
Average age (y) |
STD age (y) |
|||
|---|---|---|---|---|---|---|
| ASD | TD | ASD | TD | ASD | TD | |
| BNI_A | 14 | 11 | 22.1 | 22.5 | 5.6 | 6.3 |
| CALTECH | 13 | 11 | 24.9 | 24.6 | 6.8 | 6.8 |
| CMU | 11 | 10 | 26.2 | 27.1 | 5.9 | 6.1 |
| EMC_A | 21 | 22 | 8.2 | 8.3 | 1.2 | 1.0 |
| ETH_A | 12 | 24 | 20.6 | 23.9 | 3.5 | 4.5 |
| GU_A | 38 | 27 | 11.0 | 10.8 | 1.5 | 1.6 |
| IP_A | 14 | 10 | 15.4 | 23.2 | 5.1 | 8.4 |
| IU_A | 15 | 15 | 22.2 | 24.3 | 5.3 | 5.5 |
| KKI | 18 | 24 | 10.1 | 10.3 | 1.4 | 1.3 |
| KKI_A | 40 | 99 | 10.5 | 10.4 | 1.5 | 1.3 |
| LEUVEN_A | 14 | 14 | 21.9 | 23.4 | 4.1 | 3.0 |
| LEUVEN_B | 12 | 15 | 13.9 | 14.6 | 1.4 | 1.6 |
| MAX_MUN | 15 | 26 | 20.5 | 23.3 | 9.5 | 7.8 |
| NYU | 68 | 79 | 14.0 | 16.0 | 6.5 | 6.2 |
| NYU_A | 43 | 28 | 9.7 | 9.1 | 4.6 | 1.9 |
| OHSU | 13 | 15 | 11.7 | 10.1 | 2.2 | 1.1 |
| OHSU_A | 30 | 27 | 12.1 | 10.3 | 2.1 | 1.7 |
| OILH_B | 16 | 20 | 21.4 | 24.2 | 3.9 | 3.9 |
| OLIN | 17 | 14 | 16.3 | 16.9 | 3.1 | 3.8 |
| PITT | 26 | 22 | 19.9 | 19.8 | 7.3 | 6.8 |
| SBL | 11 | 13 | 29.9 | 32.5 | 3.4 | 6.3 |
| SDSU | 13 | 15 | 14.9 | 14.5 | 1.7 | 1.5 |
| SDSU_A | 26 | 23 | 12.6 | 13.4 | 3.3 | 3.1 |
| STANFORD | 15 | 15 | 10.1 | 10.2 | 1.6 | 1.7 |
| SU_B | 18 | 19 | 11.0 | 11.1 | 1.2 | 1.3 |
| TCD_A | 21 | 21 | 14.8 | 15.6 | 3.3 | 3.1 |
| TRINITY | 24 | 25 | 17.3 | 17.1 | 3.6 | 3.8 |
| UCD_A | 14 | 10 | 14.6 | 15.0 | 2.1 | 1.9 |
| UCLA_A | 34 | 28 | 13.3 | 12.3 | 2.4 | 2.2 |
| UCLA_B | 12 | 11 | 12.8 | 12.2 | 1.9 | 1.2 |
| UCLA_3 | 13 | 11 | 12.1 | 9.9 | 2.1 | 2.2 |
| UM_A | 35 | 36 | 12.5 | 13.6 | 2.3 | 3.3 |
| UM_B | 12 | 20 | 14.7 | 16.9 | 1.5 | 4.0 |
| USM | 56 | 43 | 21.8 | 21.4 | 6.3 | 7.6 |
| USM_A | 15 | 13 | 17.5 | 23.9 | 7.0 | 8.6 |
| U_MIA_A | 9 | 11 | 10.5 | 9.5 | 2.0 | 2.0 |
| YALE | 20 | 20 | 12.7 | 12.3 | 3.1 | 2.8 |
| Total | 793 | 845 | 15.2 | 15.6 | 6.3 | 7.0 |
Fig. 1.
Bar diagram showing the number of subjects acquired at each site for each diagnostic group.
Fig. 2.
Box plots showing the distribution of the age of the subjects belonging to each site, sorted by increasing median age.
2.2. Image processing and feature extraction
The MRI scans selected from ABIDE I and ABIDE II cohorts have been processed with Freesurfer (Fischl, 2012) version 6.0 with the recon-all pipeline2. This procedure includes cortical surface mod- elling, spherical coordinate transformation, non-linear curvature registration, automated volumetric segmentation and cortical reconstruction. Among the outputs generated by the Freesurfer processing pipeline, the following brain features have been selected: the global measures and the subcortical features available in the file aseg.stats and the cortical features available in the bilateral files aparc.stats. In this way, a total number of 221 brain morphometric features have been obtained.
These brain descriptive characteristics can be grouped into3:
-
•
9 global quantities: left (L) and right (R) mean thickness, L and R cortex volumes, L and R cerebral white matter volume, cerebrospinal fluid volume, total gray volumes and the volume of segmented brain without ventricles;
-
•
26 volumes of sub-cortical structures and corpus callosum;
-
•
186 measures, including the volume, the mean and standard deviation of the thickness of 62 structures (31 per hemisphere) from the Desikan–Killiany–Tourville Atlas (Klein & Tourville, 2012): 14 in the temporal lobe, 20 in the frontal lobe, 10 in the parietal lobe, 8 in the occipital lobe and 10 in the cingulate cortex.
2.3. Multi-center data harmonization procedure
In this study, we used the publicly available Python package NeuroHarmonize4, the state-of-the- art tool for multi-site neuroimaging analysis developed by Pomponio et al. (Pomponio et al., 2020), to reduce potential biases and non-biological variability induced by site and scanner effects. This approach combines the ComBat harmonization pipeline (Fortin et al., 2018, Fortin et al., 2017, Johnson et al., 2007), which removes unwanted sources of variability, such as site differences, while preserving variations due to other biologically-relevant covariates, with the generalized additive model (GAM) (Hastie and Tibshirani, 1986, Wood, 2017). The latter introduces a penalized nonlinear term to describe age effects in order to capture non-linearities in age-related volume differences in brain anatomy.
The application of the harmonization process to studies focused on case-control comparisons requires the availability of data from an appropriate control population. Indeed, the harmonization model parameters are calculated from the TD population, and then the harmonization transfor- mation is applied to the group of patients. In fact, the assumption behind the NeuroHarmonize approach is that each sample measurement is drawn from the same reference distribution, al- though subjects in each sample may differ in age, sex, and intracranial volume (ICV). Patients with structural brain alterations could violate this assumption and, further, including them in the harmonization process would attenuate disease-related effects (Pomponio et al., 2020). Indeed, for small sample sizes, distinguishing between effects related to the heterogeneity of a disease and site effects might be infeasible. Thus, the use of a relatively more stable TD population to normalize data has been shown to improve the case-control discrimination performances (Fortin et al., 2017, Linn et al., 2016).
The objective of our analysis was to discard from the Freesurfer brain measures the confounding effect attributable to different acquisition sites, while preserving the biological variability of the brain features; thus, following the approach proposed in Pomponio et al. (Pomponio et al., 2020), we estimated the NeuroHarmonize model parameter on the entire cohort of control subjects, by specifying the age as a covariate whose effect is to be preserved during the harmonization process. Finally, we applied the estimated model on the entire sample of subjects with ASD and TD controls.
2.4. Binary classification with Random Forests
We implemented in this study RF binary classifiers to distinguish subjects belonging to two different cohorts. The RF classification method uses bagging of decision trees in order to reduce the variance of single trees, and thus improve the prediction accuracy (Breiman, 2001). The RF training process consists in training a number of decision trees on randomly selected data samples, getting a prediction from each tree, and then selecting the best solution by means of voting. Ran- dom Forests are considered a highly accurate and robust method because of the number of decision trees participating in the process. Moreover, they are less prone to overfitting problems (Breiman, 2001). The main reason is that they take the average of the predictions by all trees, thus reducing the possible biases.
The classification performances can be evaluated in terms of sensitivity (true-positive ratio) and specificity. The trade-off between the sensitivity and the false-positive ratio (which corresponds to one minus the specificity), obtained by varying the decision threshold of the classifier, is known as the receiver operating characteristic (ROC) curve (Metz, 2006). From the ROC curve, the area under curve (AUC) can be estimated. The AUC is a global index to compare the ROC curves of different classifiers and represents the probability of correctly ranking a [case, control] pair (Hanley & McNeil, 1982).
We used the RandomForestClassifier in Scikit-learn (Pedregosa et al., 2011), a Python open-source machine learning library, and we set the number of trees to the default value of 500 and the number of candidate predictors considered at each split to , where nP is the number of predictors (Breiman, 2001). The RF model has been trained according to a stratified 5-fold cross- validation scheme, which accounts for a comparable number of examples of the two classes in each subset to allow a balanced training. We implemented a feature scaling function (the Scikit-learn RobustScaler), that involves the subtraction of the median and the scaling with respect to the interquartile range (IQR). The IQR was computed within each fold of the 5-fold cross validation scheme. The AUC is computed within each fold; then, results across the 5 test folds are used to calculate the mean and the standard deviation of AUC.
2.4.1. Assessment of the effectiveness of feature harmonization
A RF binary classification of control subjects from Sitei vs. Sitej (with i ≠ j) has been carried out to evaluate whether and up to what extent the acquisition site is a confounding information for a ML classifier. A null discrimination ability (AUC ∼ 0.5) is expected to be observed in case Sitei and Sitej are populated by control subjects with similar demographic characteristics, and in the absence of confounding site effects. In case AUC ≠ 0.5 are detected, this could be ascribed either to differences in sample composition (e.g. in terms of age) or to confounding information encoded in the raw data during the acquisition. We expect that the data harmonization protocol is successful in removing the site effects, and this results in a reduced discrimination capability when attempting to predict site. Thus, in order to assess the effectiveness of the harmonization pipeline, we compared the Sitei vs. Sitej (with i ≠ j) RF classification performances obtained before and after harmonization. The RF models have been trained in this case according to a stratified 50% hold-out validation scheme. The classification procedure was repeated 10 times, and the results were averaged to calculate the final value of AUC.
2.4.2. Evaluation of the impact of harmonization on case-control discrimination
Once the capability of the multi-center data harmonization process is demonstrated, the effect of this operation on the case-control discrimination ability of a RF classifier has been assessed. The RF classification of subjects with ASD vs. TDs has been carried out on both non-harmonized and harmonized data, in order to quantify the expected increment in the discrimination performance. To evaluate the significance of the classification performance achieved, we carried out a permutation test with 1000 repetitions. During the permutation testing the labels of the samples are changed randomly at each iteration and the classification task is repeated, thus simulating the null distribution of the performance metric under test, which is the AUC in our case. An empirical p-value is calculated as the percentage of times the score obtained is greater that the one obtained using the data with the original un-permuted class labels (Ojala & Garriga, 2009).
In addition to the analysis of the sample as a whole, we evaluated the RF classification per- formance across the lifespan, by partitioning the dataset into three subgroups by age: children ([6–12] years), adolescents ([12–20] years) and adults ([20–40] years), as summarized in Table 2. In subgrouping subjects by age, the thresholds were chosen because they approximately reflect pre- puberty, adolescence and early adulthood, and also because they allow to generate subgroups with a consistent and comparable number of subjects in each group (Katuwal et al., 2016).
Table 2.
Summary of the number of subjects (N) in each subgroup (Children, Adolescents and Adults). The mean and standard deviation (STD) of the age are reported in years (y) for each diagnostic group.
| Subgroups | N |
Mean age (y) |
STD age (y) |
|||
|---|---|---|---|---|---|---|
| ASD | TD | ASD | TD | ASD | TD | |
| Children | 301 | 345 | 9.8 | 9.9 | 1.5 | 1.3 |
| Adolescents | 352 | 309 | 15.3 | 15.2 | 2.3 | 2.4 |
| Adults | 140 | 191 | 26.2 | 26.5 | 5.2 | 5.1 |
It is worth specifying that the impact of the harmonization protocol in the whole sample and in the three age-specific subgroups is evaluated as follows: the harmonization protocol is applied on the whole sample (the harmonization model parameters are evaluated on the TD population and then the model is applied to the whole sample); then, RF classifiers are trained to distinguish ASD from TD subjects both on the non-harmonized and harmonized data in order to compare the performances. The latter operation is carried out for the whole sample and in the three age-specific subgroups.
2.5. Feature importance
Random Forest classifiers have the relevant advantage of allowing an embedded interpretable feature importance analysis (Chen & Ishwaran, 2012). Indeed, several techniques can be used to identify the particular set of features with relevant role in the classification process. We calculated the importance of each feature by using the permutation importance function implemented in Scikit- learn. The permutation feature importance is defined as the decrease in a model score when a single feature value is randomly shuffled. This procedure breaks the relationship between the feature and the target, thus the drop in the model score is indicative of how much the model depends on that feature. It can be summarized as follows:
-
•
Take as input the fitted predictive model m on training dataset D
-
•
Compute the accuracy score (s) of the model m on data D
-
•
For each feature j:
- For each repetition k in 1, …, K:
∗ Randomly shuffle column j of data-set D to generate a corrupted version of the data named Dk,j.
∗ Compute the score sk,j of model m on corrupted data Dk,j.
- Compute importance ij for feature fj defined as:
| (1) |
For the age-specific subgroups in the case of harmonized image features, we randomly mixed each feature 100 times, thus we obtained a sample of importance scores. Since a feature selection algorithm may be sensitive to changes in the training set, the feature importance was calculated as an average of the importance scores from 10 train folds. As the most important features, we selected the scores above the 90th percentile.
The effect size of ASD vs. TD group difference was quantified using Cohen’s d coefficient. It consists in the standardized difference between two mean values µ defined as (µASD-µTD)/SDpooled, where SDpooled is the weighted average of the standard deviations of the two groups (Cohen, 1988).
3. Results
3.1. Implementation and test of the data harmonization effectiveness
We estimated the harmonization model provided by the NeuroHarmonize package on the control group consisting of 845 subjects, thus obtaining both the model parameters and the harmonized features for the control group. Then, we applied the model on the group of 793 subjects with ASD to obtain the dataset for the case-control comparison entirely harmonized for site effects. In this process, we used the site information as a model covariate and we specified the age parameter as a nonlinear term to be accounted for in the harmonization process in order to preserve the age trend of the brain descriptive features.
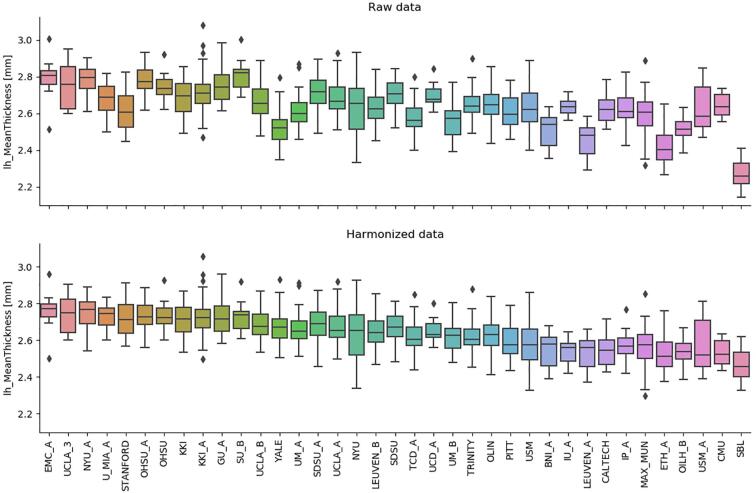
Fig. 3 shows how the harmonization procedure acts on the values of an example feature (the mean cortical thickness of the left hemisphere). It can be noticed that appreciable inter-site biases.
Fig. 3.
The effect of the harmonization process on an example feature, the left hemisphere cortical thickness, is shown. The box plots show of the distributions of the features, grouped by site, the list of which is ordered by increasing median age. The presence of inter-site biases which is visible on raw data (top panel) is canceled by the harmonization process while preserving the expected age trend of the feature (bottom).
in the feature values are removed, whereas the expected age trend of the feature is preserved (in the case of the cortical thickness, a thinning with age occurs in normal neurodevelopment).
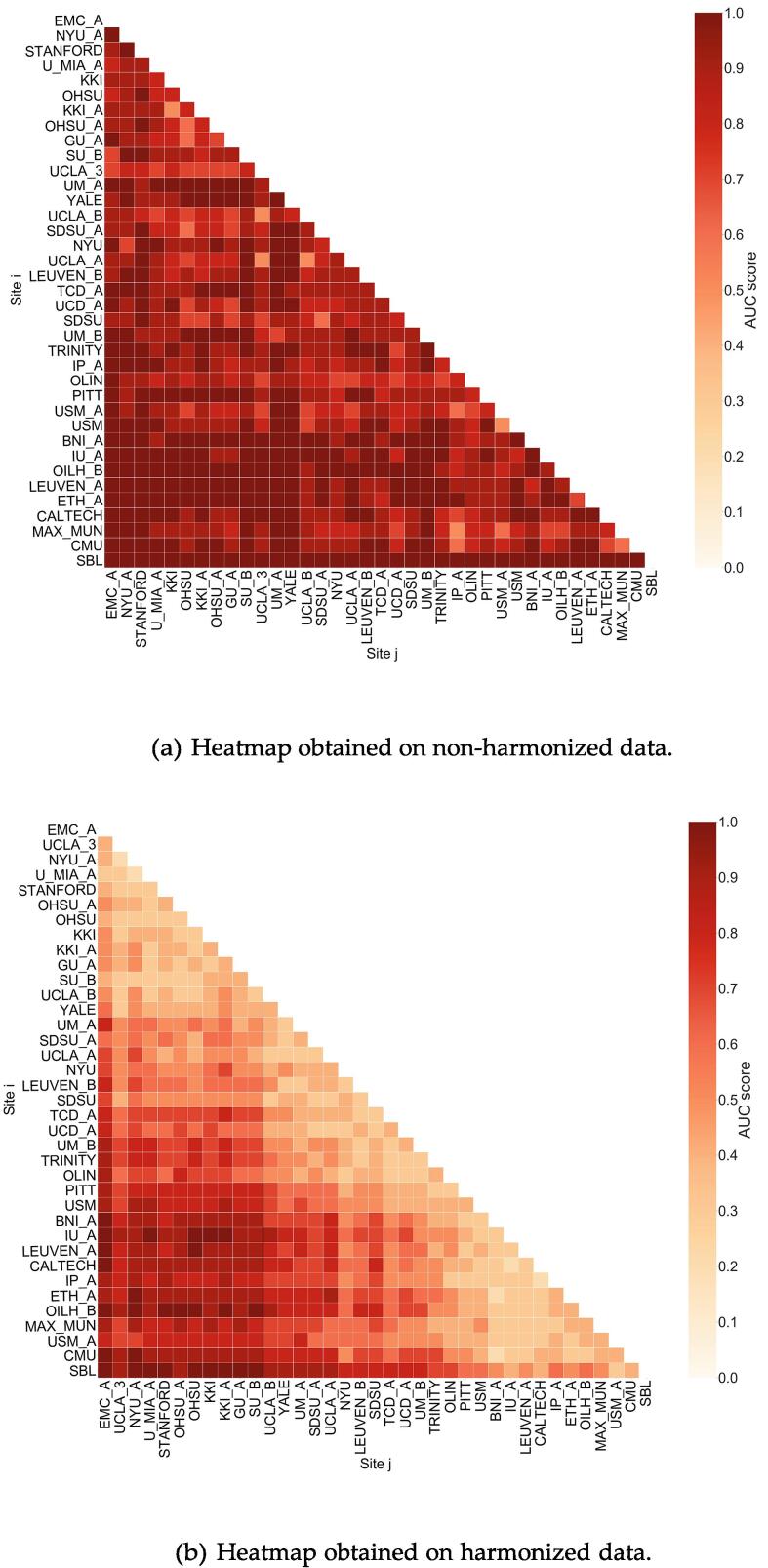
The effectiveness of the harmonization process in removing site-related biases has been quantified in terms of a measurable reduction of the confounding effect that site has on a RF binary classification. Fig. 4 shows the heatmaps of the AUC values obtained on non-harmonized and on harmonized data in the attempt to discriminate TD subjects acquired at Sitei from those acquired at Sitej with a RF classifier, according to ten repetitions of a stratified 50% hold-out validation scheme. As visible in panel (a) of the figure, extremely high AUC values are obtained in the site-by- site discrimination, based on non-harmonized features. Once the features have been harmonized, as shown in panel (b), the Sitei vs. Sitej discrimination capability of a RF classifier decreases to values closer to AUC 0.5, which is the expected null classification performance for indistinguish- able cohorts. As visible in the bottom left corner of the map reported in panel (b), when comparing sites populated by TD subjects in different age ranges, the RF classifier maintains extremely high discrimination ability, as expected. Ultimately, the visual comparison between panel (a) and (b) of Fig. 4 highlights that the data harmonization process allows to recover homogeneity of sample features, thus AUC 0.5, in the Sitei vs. Sitej discrimination for pairs of sites in similar age ranges, i.e. site combinations reported close to the diagonal of the heatmaps.
Fig. 4.
AUC values obtained in the RF classification of non-harmonized (a) and on harmonized (b) features of control subjects of Sitei vs. Sitej, according to a 5-fold cross validation protocol. The site list is sorted according to increasing median age of subjects at each site (see Fig. 2).
3.2. ASD vs. TD discrimination performance
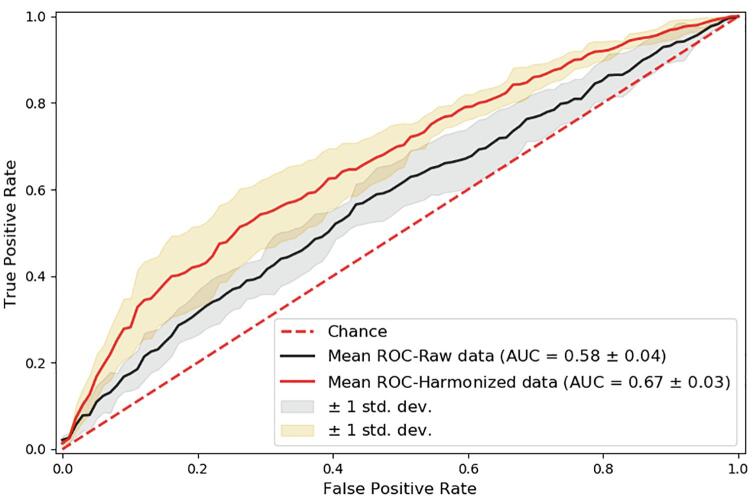
Random Forest classifier have been trained to distinguish subjects with ASD from TD controls on the whole sample and in the three age-specific subgroups, according to a 5-fold cross-validation scheme. The classification was performed both on the non-harmonized and on the harmonized datasets in order to evaluate the impact of the harmonization on the problem of ASD vs. TD categorization. Fig. 5 shows the ROC curves obtained on the whole sample by averaging the ROC curves computed on each of the 5 folds of the cross validation. The mean AUC values and the standard deviations are reported. An AUC of 0.58 ± 0.04 was achieved on non-harmonized data and an AUC of 0.67 ± 0.03 on harmonized data.
Fig. 5.
ROC curves obtained for the ASD vs. TD classification within 5-fold cross-validation scheme on the whole dataset.
The performance in the ASD vs. TD discrimination by RF classifiers trained according to a 5- fold cross validation scheme, are reported in terms of the AUC, accuracy, sensitivity and specificity for the whole sample and in the three age-specific subgroups in Table 3. Both the results obtained on non-harmonized and on harmonized data are provided.
Table 3.
Classification performances (AUC, accuracy, sensitivity and specificity) obtained in the ASD vs. TD discrimination by a RF classifier for the whole sample and in the subgroups of children, adolescents and adults on non-harmonized and on harmonized data. The average value and the standard deviation of each metric are computed according to a 5-fold cross validation scheme.
| Sample | Data | AUC | Accuracy | Sensitivity | Specificity |
|---|---|---|---|---|---|
| Whole | Raw | 0.58 ± 0.04 | 0.56 ± 0.02 | 0.54 ± 0.04 | 0.58 ± 0.02 |
| Harm | 0.67 ± 0.03 | 0.62 ± 0.03 | 0.61 ± 0.06 | 0.63 ± 0.07 | |
| Children | Raw | 0.52 ± 0.09 | 0.53 ± 0.05 | 0.68 ± 0.15 | 0.40 ± 0.21 |
| Harm | 0.62 ± 0.02 | 0.59 ± 0.03 | 0.58 ± 0.05 | 0.60 ± 0.04 | |
| Adolescents | Raw | 0.47 ± 0.07 | 0.49 ± 0.03 | 0.52 ± 0.25 | 0.46 ± 0.24 |
| Harm | 0.65 ± 0.03 | 0.61 ± 0.01 | 0.59 ± 0.04 | 0.62 ± 0.03 | |
| Adults | Raw | 0.62 ± 0.07 | 0.58 ± 0.04 | 0.54 ± 0.10 | 0.60 ± 0.06 |
| Harm | 0.69 ± 0.06 | 0.68 ± 0.06 | 0.65 ± 0.07 | 0.70 ± 0.08 |
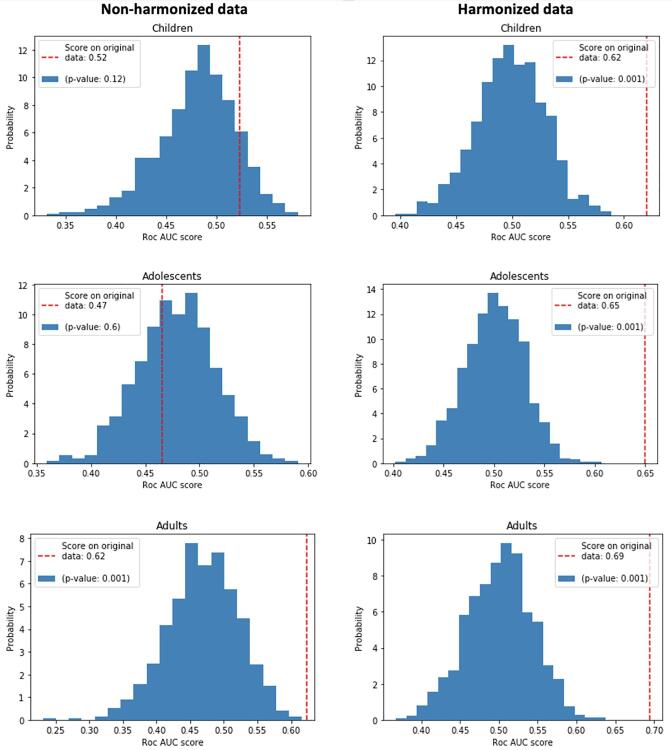
It is apparent from the results reported in the table that for the groups of children and ado- lescents the AUC values obtained on non-harmonized data are consistent with the chance level. To assign a statistical significance to the null hypothesis that the ASD and TD cohorts cannot be distinguished by a RF classifier, we carried out the permutation tests, whose results are shown in Fig. 6. The histograms of the AUC scores obtained at each permutation are reported in the figure. For each histogram, the red line indicates the score obtained by the RF classifier on data with the original un-permuted class labels. The empirical p values are thus computed for each AUC classification performance reported in Table 3, showing that in all cases, except for the classification of non-harmonized data of children and adolescents, as mentioned above, significant p values have been obtained.
Fig. 6.
Histograms reporting the AUC values obtained in the permutation test (with 1000 permutations) for the non-harmonized (left column) and the harmonized data (right column) of subjects belonging to groups of children, adolescents and adults. The vertical dashed red lines indicate the scores obtained by the RF classifiers on data with the original un-permuted class labels. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.3. Relevant brain features in the ASD vs. TD discrimination problem
The most important features in the ASD vs. TD discrimination problem have been identified for the three age-specific subgroups by exploiting the permutation importance function of Scikit-learn. The features whose importance scores exceeded the 90th percentile are reported in Table 4. In addition to the specification of the feature type (e.g. thickness or volume, average or standard deviation), the table reports the sign of the Cohen’s d, thus indicating whether a feature mean is larger/smaller (+/−) in the sample of subjects with ASD with respect to TD controls.
Table 4.
The most important features (importance scores over the 90th percentile) in the RF classi- fication for the three age-specific sub-groups are reported. The reported sign indicates whether the feature mean is larger/smaller (+/−) in the group of subjects with ASD with respect to TD controls.
| Hemisphere | Anatomical region | Measurement |
||
|---|---|---|---|---|
| Children | Adolescents | Adults | ||
| lh | Caudal anterior cingulate | GrayVol(+) | GrayVol(+) | |
| rh | Caudal anterior cingulate | ThickAvg(+) | ||
| lh | Entorhinal | ThickStd(+) | ||
| rh | Entorhinal | ThickAvg(−) | ||
| lh | Fusiform | ThickStd(+) | ThickAvg(+) | |
| rh | Fusiform | ThickStd(+) | ||
| lh | Inferior parietal | GrayVol(−) | ||
| rh | Inferior parietal | GrayVol(−) | ThickStd(+) | |
| lh | Inferior temporal | ThickStd(+) | ThickStd(+) | |
| rh | Inferior temporal | ThickAvg(−) | ||
| lh | Isthmus cingulate | ThickStd(+) | ||
| rh | Isthmus cingulate | GrayVol(+) | ||
| lh | Lateral occipital | ThickAvg(+) | ThickAvg(+) | GrayVol(+) |
| lh | Lateral orbitofrontal | ThickAvg(+) | ||
| rh | Lateral orbitofrontal | GrayVol(−) | ||
| lh | Lingual | ThickStd(+) | ThickStd(+) | |
| rh | Lingual | ThickAvg(+) | ThickAvg(+) | |
| lh | Medial orbitofrontal | ThickAvg(+), GrayVol(+) |
ThickAvg(+) | |
| rh | Medial orbitofrontal | ThickStd(+) | ThickAvg(+) | |
| rh | Middle temporal | ThickAvg(−), ThickStd(+) |
||
| lh | Paracentral | GrayVol(+), ThickStd(+) |
||
| rh | Paracentral | ThickAvg(−) | ||
| rh | Parahippocampal | ThickAvg(−) | ||
| lh | Pars orbitalis | GrayVol(+) | ||
| rh | Pars orbitalis | GrayVol(+) | ||
| rh | Pars triangularis | ThickStd(+) | ||
| lh | Pericalcarine | ThickAvg(+) | ||
| rh | Pericalcarine | GrayVol(−) | ||
| lh | Posterior cingulate | ThickStd(+) | ||
| rh | Posterior cingulate | ThickAvg(+) | ||
| rh | Precentral | ThickAvg(−) | ||
| lh | Precuneus | ThickStd(+) | ||
| rh | Precuneus | ThickStd(+) | ||
| rh | Rostral anterior cingulate | ThickStd(+) | ||
| lh | Rostral anterior cingulate | GrayVol(+) | ||
| lh | Superior frontal | GrayVol(+) | ||
| rh | Superior frontal | GrayVol(+) | ||
| rh | Superior temporal | ThickStd(+) | ThickAvg(+), ThickStd(+) |
|
| lh | Supramarginal | ThickStd(+) | ||
| rh | Supramarginal | GrayVol(+) | ||
| lh | Transverse temporal pole | ThickAvg(+) | GrayVol(+), ThickAvg(+) |
|
| Central corpus callosum | Volume(−) | |||
| Middle anterior corpus callosum | Volume(−) | |||
| lh | Amygdala | Volume(+) | ||
| rh | Caudate | Volume(+) | ||
| rh | Hippocampus | Volume(+) | ||
| lh | Nucleus Accumbens | Volume(+) | ||
| rh | Nucleus Accumbens | Volume(+) | ||
| lh | Pallidum | Volume(+) | ||
| rh | Putamen | Volume(+) | ||
| lh | Thalamus | Volume(+) | ||
| rh | Ventral diencephalon | Volume(+) | ||
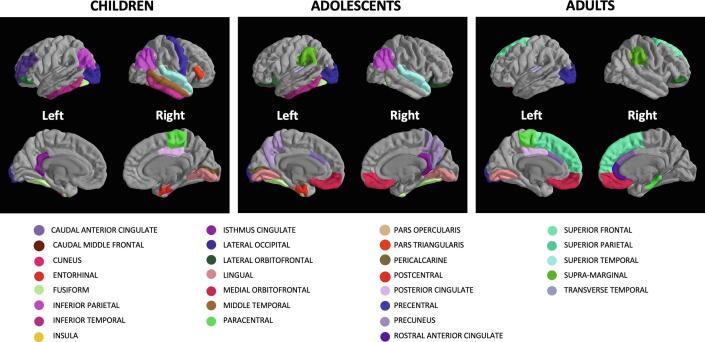
A visual representation of the relevant features is shown in Fig. 7, which allows an immediate identification of the set of features common to the different age-specific subgroups. It can be noticed from the table and the figure that the features identified as important in the ASD vs. TD discrimination problem were mainly from the frontal, parietal and temporal regions.
Fig. 7.
The brain regions whose features were identified as relevant (see table 4) in the ASD vs. TD discrimination are highlighted on the MRI scan of the Freesurfer average sample subject. The PySurfer library has been used to produce this figure.
4. Discussion
We showed in this paper that the data harmonization is a necessary preprocessing step in the analysis of brain descriptive features extracted by MRI scans in multi-center studies. Several works demonstrated better performance after harmonization approaches. Qin et al. (Qin et al., 2022) applied ComBat on whole-brain functional networks to identify individuals with major depressive disorder from controls outperforming the accuracy values of other state-of-the-art methods. Wang et al. (Wang et al., 2022) developed a novel deep-learning domain adaptation framework to tackle the confounding effects for both Alzheimer’s disease and Schizophrenia classification tasks by using the whole minimally preprocessed 3D T1-weighted brain MRI scans of the subjects. Monte-Rubio et al. (C. Monte-Rubio et al., 2022) proposed an approach using the predictive probabilities pro- vided by Gaussian processes to harmonize multi-site T1-weighted MRI data for Parkinson’s disease classification. Although the latter two methodologies cannot be applied to data extracted from preprocessed images such as cortical and subcortical features, the authors highlighted that harmo- nization is a crucial preprocessing step to be performed before any clinical classification task. Our work addresses the problem of ASD vs. TD classification task. Our work focuses on the problem of the ASD vs. TD classification task, whose performance significantly improved after the harmoniza- tion procedure. The slightly over the chance-level AUC values obtained without any harmonization indicating the small differences between the two populations were obscured by confounding effects, reached the AUC = 0.67 ± 0.03 after harmonization.
4.1. Comparison with the categorization performances reported in previous studies
Controversial results were reported by other studies regarding the ASD vs. TD discrimination performances of ML classifiers (Arbabshirani et al., 2017, Wolfers et al., 2019). In general, not fully replicated and lower results were reported, thus suggesting that the two groups are not highly separable. Limiting the comparison to previous ML analyses of structural MRI data of large cohorts of subjects such as the ABIDE I and ABIDE II collections, a historical overview is reported below to highlight the evolution of the methodological approaches to the problem:
-
•
the work by Haar et al. (Haar et al., 2016),analyzed a sample restricted to 590 subjects of the ABIDE I collection and then a sample relaxed to 906 subjects, reporting a modest accuracy in the case-control discrimination (<60%). This result is consistent with the almost-chance-level result that we obtained on non-harmonized data;
-
•
in the work by Katuwal et al. (Katuwal et al., 2016) the low accuracy obtained in the study by Haar et al. (Haar et al., 2016) is attributed to the ASD heterogeneity. The authors applied ML classifiers to a group of 734 males (361 ASD vs. 373 TD of the ABIDE I collection), obtaining AUC values in the 61–68% range. Then, they repeated the analysis on more homogeneous subgroups in terms of age, intellectual quotient and autism severity and they obtained very high discrimination performance (AUC greater than 0.8) in specific subgroups. However, they did not take into account possible biases introduced by the site effect;
-
•
the works by Ferrari et al. (Ferrari et al., 2020a, Ferrari et al., 2020b) highlighted that the ABIDE I and ABIDE II multi-center data collections suffer from the so-called batch effect; thus, training the ML model on a subgroup of 86 subjects selected from the most populated site of the ABIDE collection, the NYU1 dataset, an AUC = 0.79 on an independent test set (including subjects under 30 years of age and fully matched on demographic and clinical variables with the subjects of the training set) was obtained in the ASD vs. TD
discrimination problem; despite a trend in the ASD vs. TD discrimination capability of the classification pattern trained on the NYU1 sample was shown in testing it on the whole ABIDE sample, significant classification performances were not achieved in that case, probably due to different demographic composition across centers and to site effects;
-
•
the work by Gao et al. (Gao et al., 2021) implemented a deep convolutional neural network to carry out the ASD vs. TD classification based on the analysis of morphological covariance networks derived by structural MRI scans of a sample of 518 subjects with ASD and 567 TD controls of the ABIDE I collection; no harmonization strategy was implemented in that study, probably since the covariance networks are less sensitive to systematic site effects, and a classification accuracy of 71.8% was achieved. This result, despite provided without an estimate of the variability across the 10-fold cross validation implemented by the authors, slightly outperforms the best accuracy values we obtained. This may be due to the superior classification ability of deep learning classifiers compared to traditional ones.
As a general consideration regarding the modest classification performances achieved in our work, it is worth specifying that the aim of this study was to investigate the impact of a data harmonization strategy in a challenging classification problem. Thus, the systematic search for the best performing classifier was not within the objectives of this work. As an example, deep learning models could certainly lead to superior discrimination performances if a sufficiently large dataset, adequately representing the heterogeneity of the population, is available to properly train them (Avanzo et al., 2021).
4.2. Methodological limitations of this study
A limitation of both the ComBat and NeuroHarmonize tools for multi-site data harmonization consists by the fact that these methods are particularly suitable to harmonize brain descriptive fea- tures (e.g. neuroanatomical (Fortin et al., 2018, Pomponio et al., 2020), connectivity (Ingalhalikar et al., 2021) and diffusivity measures (Fortin et al., 2017)), whereas they are not directly applicable to process original MRI images. An approach devoted to the direct harmonization of the images acquired at different scanners has been proposed by Wrobel et al. (Wrobel et al., 2020). They implemented and evaluated according to a cross-validation scheme a multi-site image harmoniza- tion method based on the alignment of the intensity distributions of images acquired at different sites. Alternatively, the domain adaptation approach could be applied in order to avoid the need of harmonizing multi-center data before ML techniques are applied for data analysis and classifi- cation. This ML approach allows to handle the differences in data distributions between test and train domains and has been successfully applied to analyse several functional connectivity measures derived from the multi-center ABIDE dataset (Bhaumik et al., 2018). Moreover, a deep-learning based implementation of the domain adaptation concept to analyze structural MRI scans has been implemented by Guan et al. (Guan et al., 2021) to eliminate the confounding site effect in a study of the Alzheimer’s Disease.
The relevant features for the discrimination problem have been identified in our study in order to evaluate their consistency between the different age-specific subgroups and across the entire lifespan. An important issue of our work concerns the feature selection strategy: although a RF classifier can handle multicollinearity among features, it may not return all features with the same information content, yielding a minimal set of relevant features to optimize prediction and complicating the biological interpretation of the results. Although we applied the permutation feature importance method to mitigate this problem, further analysis would be required to completely overcome this issue and obtain the set of all regions relevant to discrimination.
4.3. Considerations of age-related class separability and relevance of important features
Despite the discrimination power of a RF classifier found in our study between the two classes of subjects with ASD and TD controls is moderate, the statistical significance of the separation capability has been demonstrated for the whole cohort and for the three age-specific subgroups. When the sample was divided by age group in children, adolescents and adults, we observed the highest discrimination ability of the RF classifier in adults, meaning that the brains of adults with ASD differ from the brains of age-matched controls more than the brains of children/adolescents with ASD differ from those of their peers. Relatively little is known about the factors that shape age-related brain changes in ASD: it is possible that the greater burden of brain alterations in adulthood could be traced back to the frequent association of ASD with other psychiatric disorders, mainly evident with increasing age. Indeed, according to a recent systematic review and meta- analysis (M.-C. Lai et al., 2019), ASD heighten the risk of developing major psychiatric disorders, and some of these (i.e. depressive, bipolar, and schizophrenia spectrum disorders) become more prevalent with increased age. Thus, the brain MRI alterations in adults with ASD could be the result not only of the ASD brain signature, but also of other comorbid psychiatric disorders, which contribute to making the underlying neural alterations greater. Crucially, the cross-sectional design of this investigation did not allow understanding the age-associated trajectory of brain alterations in the same subjects with ASD and longitudinal inferences about development from cross-sectional studies can be seriously misleading (Kraemer et al., 2000): therefore, ad-hoc longitudinal MRI studies in large and well-characterized ASD individuals are needed to answer this research question. Regarding the relevant brain structural features we identified in the ASD vs. TD binary clas- sification with RF, we found out that only one region was consistently found among the most discriminant ones across the whole lifespan: the lateral occipital gyrus of the left (L) hemisphere. Either the volume or the average thickness of this feature were found to be greater in the population of subjects with ASD with respect to controls. This brain region has been previously implicated in the pathogenesis of ASD, although with a decreased value in individuals with ASD compared to TD peers. Indeed, a recent investigation that combined multiple imaging modalities (structural MRI, DTI, and resting state fMRI) to investigate respectively brain anatomy, connectivity and function in a sample of forty boys with ASD detected that individuals with ASD have significantly reduced gray matter surface area, structural connectivity, and resting state brain activity in the lateral occipital cortex (Jung et al., 2019). Additionally, decreases in surface area, structural connectivity, and resting-state brain activity in this region were correlated with increased social symptom severity in subjects with ASD.
In addition, we observed age-specific cortical abnormalities. In this light, a number of consis- tently altered features between the groups of children and adolescents have been identified. Most of them (if not explicitly differently stated) showed increased values in subjects with ASD with respect to TD controls: the average thickness of the L fusiform gyrus and its standard deviation (SD); the thickness SD of the L inferior temporal gyrus; the volume (decreased in children with ASD) of the right (R) inferior parietal lobule (IPL) and its thickness SD (increased in adolescents with ASD); the average thickness of the R lingual gyrus; the average thickness and its SD of the R superior temporal gyrus. The increased cortical thickness of some brain regions in children and adolescents with ASD is consistent with data reporting an early brain overgrowth in ASD subjects. Both head circumference investigations (Courchesne et al., 2003; Muratori et al., 2012) and structural MRI studies (Courchesne et al., 2001, Redcay and Courchesne, 2005) observed that an increased brain size was typical of children, but not adults with ASD. Specifically, the increased cortical thickness in superior temporal and fusiform gyrus of the temporal lobe we observed is in line with findings of a recent investigation that analyze a subset of the ABIDE I cohort (Khundrakpam et al., 2017),
and could be related to the impairment of face processing, particularly in its dynamic aspects such as gaze, typical of subjects with ASD. In a similar vein, disruption in the lingual gyrus is involved in alterations of object/face recognition and following biological motion cues in ASD (Ecker et al., 2015), since this region (along with lateral occipital cortex, fusiform gyrus and posterior superior temporal sulcus) is part of a network sustaining the aforementioned abilities. On the other hand, the IPL is though to be part of the human Mirron Neuron System (MNS), the set of brain regions which are active both when participants perform an action and when they observe another person performing the same action (Rizzolatti & Craighero, 2004). The human MNS plays a key role in action understanding and imitation (Rizzolatti & Sinigaglia, 2010), and its disruption has been related to impairments in theory of mind and language in subjects with ASD (Gallese, 2007, Gallese, 2008). Finally, we detected increased cortical thickness in the L inferior temporal region (ITG), which is consistent with increased gray matter volume in the same region observed by Cai et al. (Cai et al., 2018), in children with ASD. Since ITG is involved in language acquisition, its abnormal structure could be related to alterations in language development at least in the early stages of ASD.
Finally, consistent findings between the groups of adolescents and adults were detected, consisting in increased values in the population of subjects with ASD regarding: the volume of the L caudal anterior cingulate; the thickness SD of the L lingual gyrus; either the volume, the average and the SD of the thickness of the medial orbital frontal gyrus, bilaterally; either the volume and the average thickness of L transverse temporal gyrus. It is consistently reported that the developmental trajectory of cortical thickness in individuals with ASD deviates from the typical trajectory, even if studies do not agree with each other regarding the direction of the difference. For instance, the longitudinal study by Zielinski and colleagues (Zielinski et al., 2014), detected an overgrowth of the cortical thickness during early childhood, followed by an accelerated decline in mid-childhood, and a phase of normalization during adulthood. Other studies using an age-range similar to ours are in line with current findings, observing that the cerebral cortex thins less in ASD subjects compared to TD peers (Doyle-Thomas et al., 2013, Hardan et al., 2006b, Sussman et al., 2015). As far as relevant findings in our cohort, the increased cortical thickness in the lingual gyrus of L hemisphere is consistent with a recent report on adults with ASD (Arunachalam Chandran et al., 2021), as well as with other studies showing a correlation between structural atypicalities in lingual gyrus of individuals with ASD and visual sensory abnormalities (Habata et al., 2021), or atypical social interaction (Turnbull et al., 2020). Moreover, two structures of the frontal regions seems to mostly differentiate adolescents and adults with ASD from TD peers: i) the medial orbitofrontal cortex is involved in the self-regulation of emotional states in relation to changes in social situa- tions (Bachevalier & Loveland, 2006), and its volumetric alteration has been positively correlated with levels of circumscribed interests –a core feature of ASD (Hardan, Girgis, et al., 2006); ii) the caudal anterior cingulate is involved in processing the value of actions (Amodio & Frith, 2006), and its functional deficits have been linked to altered awareness of emotions and feelings of self and others in ASD (Zhou et al., 2016). The transverse temporal gyri -also known as Heschl’s gyri-, are typically the location of the primary auditory cortex. Abnormal development of auditory cortex has been previously observed in children and adolescents with ASD (Prigge et al., 2013), and may constitute contribution to the core deficits in social communication of ASD subjects.
5. Conclusions
In conclusion, supported by the significant increase in the ASD vs. TD discrimination performance of ML classifiers in case the NeuroHarmonize preprocessing is implemented, we suggest its use in the analyses of multi-center MRI data. This is particularly relevant for studying disorders with very small effects on brain anatomy, which can be easily obscured by the confounding information due to the acquisition site.
CRediT authorship contribution statement
Sara Saponaro: Conceptualization, Methodology, Formal analysis, Software, Investigation, Visualization, Writing – original draft, Writing – review & editing. Alessia Giuliano: Conceptualization, Methodology, Data curation, Validation, Writing – original draft, Writing – review & editing. Roberto Bellotti: Validation, Writing – review & editing. Angela Lombardi: Conceptualization, Methodology, Formal analysis, Validation, Writing – original draft, Writing – review & editing. Sabina Tangaro: Conceptualization, Methodology, Validation, Writing – review & editing, Funding acquisition. Piernicola Oliva: Conceptualization, Methodology, Validation, Writing – review & editing, Funding acquisition. Sara Calderoni: Methodology, Validation, Writing – original draft, Writing – review & editing. Alessandra Retico: Conceptualization, Methodology, Formal analysis, Writing – original draft, Writing – review & editing, Validation, Supervision, Project administration, Funding acquisition.
Acknowledgments
This work has been partially supported by the INFN-CSN5 research project Artificial Intelligence in Medicine (AIM), by the University of Sassari (Italy) (Fondo di Ateneo per la ricerca 2020) and by a grant from the IRCCS Fondazione Stella Maris (Ricerca Corrente, and the 5 1000 voluntary contributions, Italian Ministry of Health, n. 2768566). ALs position is funded by the Program Research for Innovation - REFIN funded by Regione Puglia (Italy) in the framework of the POR Puglia FESR FSE 2014-2020 Asse X - Azione 10.4, project code 928A7C98.
Footnotes
The extensive list of analyzed brain features can be found in the supplementary materials.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.nicl.2022.103082.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Abraham A., Milham M.P., Martino A.D., Craddock R.C., Samaras D., Thirion B., Varo-quaux G. Deriving reproducible biomarkers from multi-site resting-state data: An autism-based example. NeuroImage. 2017;147:736–745. doi: 10.1016/j.neuroimage.2016.10.045. [DOI] [PubMed] [Google Scholar]
- American Psychiatric Association, A. et al. (2013). Diagnostic and statistical manual of mental disorders (5th ed.). American Psychiatric Association Washington, DC.
- Amodio D., Frith C. Amodio, d.m. frith, c.d. meeting of minds: The medial frontal cortex and social cognition. nat. rev. neurosci. 7, 268–277. Nat. Rev. Neurosci. 2006;7:268–277. doi: 10.1038/nrn1884. [DOI] [PubMed] [Google Scholar]
- Arbabshirani M.R., Plis S., Sui J., Calhoun V.D. Single subject prediction of brain disorders in neuroimaging: Promises and pitfalls. NeuroImage. 2017;145:137–165. doi: 10.1016/j.neuroimage.2016.02.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arunachalam Chandran V., Pliatsikas C., Neufeld J., O’Connell G., Haffey A., DeLuca V., Chakrabarti B. Brain structural correlates of autistic traits across the diagnostic divide: A grey matter and white matter microstructure study. NeuroImage: Clinical. 2021;32 doi: 10.1016/j.nicl.2021.102897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avanzo M., Porzio M., Lorenzon L., Milan L., Sghedoni R., Russo G., Massafra R., Fanizzi A., Barucci A., Ardu V., Branchini M., Giannelli M., Gallio E., Cilla S., Tangaro S., Lombardi A., Pirrone G., De Martin E., Giuliano A., Belmonte G., Russo S., Rampado O., Mettivier G. Artificial intelligence applications in medical imaging: A review of the medical physics research in italy. Physica Med. 2021;83:221–241. doi: 10.1016/j.ejmp.2021.04.010. [DOI] [PubMed] [Google Scholar]
- Bachevalier J., Loveland K. The orbitofrontal-amygdala circuit and self-regulation of social-emotional behavior in autism. Neurosci. Biobehav. Rev. 2006;30:97–117. doi: 10.1016/j.neubiorev.2005.07.002. [DOI] [PubMed] [Google Scholar]
- Bhaumik R., Pradhan A., Das S., Bhaumik D.K. Predicting Autism Spectrum Disorder Using Domain-Adaptive Cross-Site Evaluation. Neuroinform. 2018;16(2):197–205. doi: 10.1007/s12021-018-9366-0. [DOI] [PubMed] [Google Scholar]
- Breiman, L., 2001. Random forests.
- Monte-Rubio C.G., Segura B., Strafella P.A., van Eimeren T., Ibarretxe-Bilbao N., Diez-Cirarda M., Eggers C., Lucas-Jiménez O., Ojeda N., Peña J., Ruppert M.C., Sala- Llonch R., Theis H., Uribe C., Junque C. Parameters from site classification to harmonize mri clinical studies: Application to a multi-site parkinson’s disease dataset. Hum. Brain Mapp. 2022:1–13. doi: 10.1002/hbm.25838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai J., Hu X., Guo K., Yang P., Situ M., Huang Y. Increased left inferior tempo- ral gyrus was found in both low function autism and high function autism. Front. Psychiatry. 2018;9 doi: 10.3389/fpsyt.2018.00542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderoni S., Retico A., Biagi L., Tancredi R., Muratori F., Tosetti M. Female children with autism spectrum disorder: An insight from mass-univariate and pattern classification analyses. NeuroImage. 2012;59(2):1013–1022. doi: 10.1016/j.neuroimage.2011.08.070. [DOI] [PubMed] [Google Scholar]
- Chen X., Ishwaran H. Random forests for genomic data analysis. Genomics. 2012;99(6):323–329. doi: 10.1016/j.ygeno.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen J. (2nd ed.). Routledge; 1988. Statistical power analysis for the behavioral sciences. [Google Scholar]
- Courchesne E., Karns C.M., Davis H.R., Ziccardi R., Carper R.A., Tigue Z.D., Chisum H.J., Moses P., Pierce K., Lord C., Lincoln A.J., Pizzo S., Schreibman L., Haas R.H., Akshoomoff N.A., Courchesne R.Y. Unusual brain growth patterns in early life in patients with autistic disorder: An MRI study. Neurology. 2001;57(2):245–254. doi: 10.1212/wnl.57.2.245. [DOI] [PubMed] [Google Scholar]
- Deshpande G., Libero L.E., Sreenivasan K.R., Deshpande H.D., Kana R.K. Iden- tification of neural connectivity signatures of autism using machine learning. Front. Hum. Neurosci. 2013;7 doi: 10.3389/fnhum.2013.00670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Martino A., Yan C.-G., Li Q., Denio E., Castellanos F.X., Alaerts K., Anderson J.S., Assaf M., Bookheimer S.Y., Dapretto M., Deen B., Delmonte S., Dinstein I., Ertl-Wagner B., Fair D.A., Gallagher L., Kennedy D.P., Keown C.L., Keysers C., Lainhart J.E., Lord C., Luna B., Menon V., Minshew N.J., Monk C.S., Mueller S., Müller R.-A., Nebel M.B., Nigg J.T., O'Hearn K., Pelphrey K.A., Peltier S.J., Rudie J.D., Sunaert S., Thioux M., Tyszka J.M., Uddin L.Q., Verhoeven J.S., Wenderoth N., Wiggins J.L., Mostofsky S.H., Milham M.P. The autism brain imaging data exchange: Towards a large-scale evaluation of the intrinsic brain architecture in autism. Mol. Psychiatry. 2014;19(6):659–667. doi: 10.1038/mp.2013.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Martino A., O’Connor D., Chen B., Alaerts K., Anderson J.S., Assaf M., Balsters J.H., Baxter L., Beggiato A., Bernaerts S., Blanken L.M.E., Bookheimer S.Y., Braden B.B., Byrge L., Castellanos F.X., Dapretto M., Delorme R., Fair D.A., Fishman I., Fitzgerald J., Gallagher L., Keehn R.J.J., Kennedy D.P., Lainhart J.E., Luna B., Mostofsky S.H., Müller R.-A., Nebel M.B., Nigg J.T., O’Hearn K., Solomon M., Toro R., Vaidya C.J., Wenderoth N., White T., Craddock R.C., Lord C., Leventhal B., Milham M.P. Enhancing studies of the connectome in autism using the autism brain imaging data exchange ii. Sci. Data. 2017;4(1) doi: 10.1038/sdata.2017.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle-Thomas K., Duerden E., Taylor M., Lerch J., Soorya L., Wang A.T., Fan J., Hollander E., Anagnostou E. Effects of age and symptomatology on cortical thickness in autism spectrum disorders. Res.. Autism Spectrum Disorders. 2013;7:141–150. doi: 10.1016/j.rasd.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecker C., Marquand A., Mourao-Miranda J., Johnston P., Daly E.M., Brammer M.J., Maltezos S., Murphy C.M., Robertson D., Williams S.C., Murphy D.G.M. Describing the Brain in Autism in Five Dimensions-Magnetic Resonance Imaging-Assisted Diagnosis of Autism Spectrum Disorder Using a Multiparameter Classification Approach. J. Neurosci. 2010;30(32):10612–10623. doi: 10.1523/JNEUROSCI.5413-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecker C., Bookheimer S.Y., Murphy D.G. Neuroimaging in autism spectrum disorder: Brain structure and function across the lifespan. Lancet Neurol.. 2015;14(11):1121–1134. doi: 10.1016/S1474-4422(15)00050-2. [DOI] [PubMed] [Google Scholar]
- Ecker C., Rocha-Rego V., Johnston P., Mourao-Miranda J., Marquand A., Daly E.M., Brammer M.J., Murphy C., Murphy D.G. Investigating the predictive value of whole-brain structural MR scans in autism: A pattern classification approach. NeuroImage. 2010;49(1):44–56. doi: 10.1016/j.neuroimage.2009.08.024. [DOI] [PubMed] [Google Scholar]
- Ferrari, E., Bosco, P., Calderoni, S., Oliva, P., Palumbo, L., Spera, G., Fantacci, M. E., & Retico, A. (2020). Dealing with confounders and outliers in classification medical studies: The autism spectrum disorders case study. Artif. Intell. Med., 108. https://doi.org/10. 1016/j.artmed.2020.101926. [DOI] [PubMed]
- Ferrari E., Retico A., Bacciu D. Measuring the effects of confounders in medical super- vised classification problems: The confounding index (ci) Artif. Intell. Med. 2020;103 doi: 10.1016/j.artmed.2020.101804. [DOI] [PubMed] [Google Scholar]
- Fischl B. FreeSurfer. Freesurfer. 2012;62(2):774–781. doi: 10.1016/j.neuroimage.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortin J.P., Cullen N., Sheline Y.I., Taylor W.D., Aselcioglu I., Cook P.A., Adams P., Cooper C., Fava M., McGrath P.J., McInnis M., Phillips M.L., Trivedi M.H., Weissman M.M., Shinohara R.T. Harmonization of cortical thickness measurements across scanners and sites. NeuroImage. 2018;167:104–120. doi: 10.1016/j.neuroimage.2017.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortin J.P., Parker D., Tunç B., Watanabe T., Elliott M.A., Ruparel K., Roalf D.R., Satterthwaite T.D., Gur R.C., Gur R.E., Schultz R.T., Verma R., Shinohara R.T. Harmonization of multi-site diffusion tensor imaging data. NeuroImage. 2017;161:149–170. doi: 10.1016/j.neuroimage.2017.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallese V. Before and below ’theory of mind’: Embodied simulation and the neural corre- lates of social cognition. Philos. Trans. R. Soc. London Series B, Biol. Sci. 2007;362:659–669. doi: 10.1098/rstb.2006.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallese V. Mirror neurons and the social nature of language: The neural exploitation hypothesis. Soc. Neurosci. 2008;3(3-4):317–333. doi: 10.1080/17470910701563608. [DOI] [PubMed] [Google Scholar]
- Gao J., Chen M., Li Y., Gao Y., Li Y., Cai S., Wang J. Multisite autism spectrum disorder classification using convolutional neural network classifier and individual morpho- logical brain networks. Front. Neurosci. 2021;14:1–10. doi: 10.3389/fnins.2020.629630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiades, S., Szatmari, P., Boyle, M., Hanna, S., Duku, E., Zwaigenbaum, L., Bryson, S., Fom- bonne, E., Volden, J., Mirenda, P., Smith, I., Roberts, W., Vaillancourt, T., Waddell, C., Bennett, T., Thompson, A., & in ASD Study Team, P. (2013). Investigating phenotypic heterogeneity in children with autism spectrum disorder: A factor mixture modeling ap- proach. J. Child Psychol. Psychiatry, 54 (2), 206–215. https://doi.org/https://doi.org/10.1111/j.1469-7610.2012.02588.x. [DOI] [PubMed]
- Gori I., Giuliano A., Muratori F., Saviozzi I., Oliva P., Tancredi R., Cosenza A., Tosetti M., Calderoni S., Retico A. Gray Matter Alterations in Young Children with Autism Spectrum Disorders: Comparing Morphometry at the Voxel and Regional Level. J. Neuroimaging. 2015;25(6):866–874. doi: 10.1111/jon.12280. [DOI] [PubMed] [Google Scholar]
- Guan H., Liu Y., Yang E., Yap P.T., Shen D., Liu M. Multi-site mri harmonization via attention-guided deep domain adaptation for brain disorder identification. Med. Image Anal. 2021;71 doi: 10.1016/j.media.2021.102076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haar S., Berman S., Behrmann M., Dinstein I. Anatomical abnormalities in autism? Cereb. Cortex. 2016;26(4):1440–1452. doi: 10.1093/cercor/bhu242. [DOI] [PubMed] [Google Scholar]
- Habata K., Cheong Y., Kamiya T., Shiotsu D., Omori I.M., Okazawa H., Jung M., Kosaka H. Relationship between sensory characteristics and cortical thickness/volume in autism spectrum disorders. Transl. Psychiatry. 2021;11(1):1–7. doi: 10.1038/s41398-021-01743-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanley J.A., McNeil B.J. The meaning and use of the area under a receiver operating characteristic (roc) curve. Radiology. 1982;143(1):29–36. doi: 10.1148/radiology.143.1.7063747. [DOI] [PubMed] [Google Scholar]
- Hardan A., Girgis R., Lacerda A., Yorbik O., Kilpatrick M., Keshavan M., Minshew N. Magnetic resonance imaging study of the orbitofrontal cortex in autism. J. Child Neurol. 2006;21:866–871. doi: 10.1177/08830738060210100701. [DOI] [PubMed] [Google Scholar]
- Hardan A., Muddasani S., Vemulapalli M., Keshavan M., Minshew N. An mri study of increased cortical thickness in autism. Am. J. Psychiatry. 2006;163:1290–1292. doi: 10.1176/appi.ajp.163.7.1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastie T., Tibshirani R. Generalized Additive Models. Stat. Sci. 1986;1(3):297–310. doi: 10.1214/ss/1177013604. [DOI] [PubMed] [Google Scholar]
- Heinsfeld A.S., Franco A.R., Craddock R.C., Buchweitz A., Meneguzzi F. Identifi- cation of autism spectrum disorder using deep learning and the abide dataset. NeuroImage: Clinical. 2018;17:16–23. doi: 10.1016/j.nicl.2017.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingalhalikar M., Parker D., Bloy L., Roberts T.P., Verma R. Diffusion based abnor- mality markers of pathology: Toward learned diagnostic prediction of ASD. NeuroImage. 2011;57(3):918–927. doi: 10.1016/j.neuroimage.2011.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingalhalikar M., Shinde S., Karmarkar A., Rajan A., Rangaprakash D., Deshpande G. Functional Connectivity-Based Prediction of Autism on Site Harmonized ABIDE Dataset. IEEE Trans. Biomed. Eng. 2021;68(12):3628–3637. doi: 10.1109/TBME.2021.3080259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y., Chen R., Ke X., Chu K., Lu Z., Herskovits E.H. Predictive models of autism spectrum disorder based on brain regional cortical thickness. NeuroImage. 2010;50(2):589–599. doi: 10.1016/j.neuroimage.2009.12.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson W.E., Li C., Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8(1):118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- Jung, M., Tu, Y., Lang, C. A., Ortiz, A., Park, J., Jorgenson, K., Kong, X.-J., & Kong, J. (2019). Decreased structural connectivity and resting-state brain activity in the lateral occipital cortex is associated with social communication deficits in boys with autism spectrum dis- order [Mapping diseased brains]. NeuroImage, 190, 205–212. https://doi.org/https://doi. org/10.1016/j.neuroimage.2017.09.031. [DOI] [PubMed]
- Katuwal G.J., Baum S.A., Cahill N.D., Michael A.M. Divide and conquer: Sub- grouping of asd improves asd detection based on brain morphometry. PLoS ONE. 2016;11(4):1–24. doi: 10.1371/journal.pone.0153331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khundrakpam B., Lewis J., Kostopoulos P., Carbonell F., Evans A. Cortical thick- ness abnormalities in autism spectrum disorders through late childhood, adolescence, and adulthood: A large-scale mri study. Cereb. Cortex. 2017;27:1–11. doi: 10.1093/cercor/bhx038. [DOI] [PubMed] [Google Scholar]
- Klein A., Tourville J. 101 labeled brain images and a consistent human cortical labeling protocol. Front. Neurosci. 2012 doi: 10.3389/fnins.2012.00171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraemer H.C., Yesavage J.A., Taylor J.L., Kupfer D. How can we learn about devel- opmental processes from cross-sectional studies, or can we? Am. J. Psychiatry. 2000;157(2):163–171. doi: 10.1176/appi.ajp.157.2.163. [DOI] [PubMed] [Google Scholar]
- Lai M.C., Lombardo M.V., Suckling J., Ruigrok A.N., Chakrabarti B., Ecker C., Deoni S.C., Craig M.C., Murphy D.G., Bullmore E.T., Baron-Cohen S. Biological sex affects the neurobiology of autism. Brain. 2013;136:2799–2815. doi: 10.1093/brain/awt216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai M.-C., Kassee C., Besney R., Bonato S., Hull L., Mandy W., Szatmari P., Ameis S.H. Prevalence of co-occurring mental health diagnoses in the autism population: a systematic review and meta-analysis. Lancet Psychiatry. 2019;6(10):819–829. doi: 10.1016/S2215-0366(19)30289-5. [DOI] [PubMed] [Google Scholar]
- Li D., Karnath H.-O., Xu X. Candidate Biomarkers in Children with Autism Spectrum Disorder: A Review of MRI Studies. Neurosci. Bull. 2017;33(2):219–237. doi: 10.1007/s12264-017-0118-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linn K.A., Gaonkar B., Satterthwaite T.D., Doshi J., Davatzikos C., Shinohara R.T. Control-group feature normalization for multivariate pattern analysis of structural MRI data using the support vector machine. NeuroImage. 2016;132:157–166. doi: 10.1016/j.neuroimage.2016.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardi A., Amoroso N., Diacono D., Monaco A., Tangaro S., Bellotti R. Exten- sive evaluation of morphological statistical harmonization for brain age prediction. Brain Sci. 2020;10(6):364. doi: 10.3390/brainsci10060364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metz C.E. Receiver operating characteristic analysis: A tool for the quantitative evaluation of observer performance and imaging systems. J. Am. College Radiol. 2006;3:413–422. doi: 10.1016/j.jacr.2006.02.021. [DOI] [PubMed] [Google Scholar]
- Muratori F., Calderoni S., Apicella F., Filippi T., Santocchi E., Calugi S., Cosenza A., Tancredi R., Narzisi A. Tracing back to the onset of abnormal head circumference growth in italian children with autism spectrum disorder. Res.. Autism Spectrum Disorders. 2012;6:442–449. doi: 10.1016/j.rasd.2011.07.004. [DOI] [Google Scholar]
- Nielsen J., Zielinski B.A., Fletcher P.T., Alexander A.L., Lange N.T., Bigler E.D., Lainhart J.E., Anderson J.S. Multisite functional connectivity mri classification of autism: Abide results. Front. Hum. Neurosci. 2013;7 doi: 10.3389/fnhum.2013.00599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojala M., Garriga G.C. Permutation tests for studying classifier performance. Ninth IEEE International Conference on Data Mining. 2009;2009:908–913. doi: 10.1109/ICDM.2009.108. [DOI] [Google Scholar]
- Pagnozzi A.M., Conti E., Calderoni S., Fripp J., Rose S.E. A systematic review of structural mri biomarkers in autism spectrum disorder: A machine learning perspective. Int. J. Dev. Neurosci. 2018;71(1):68–82. doi: 10.1016/j.ijdevneu.2018.08.010. [DOI] [PubMed] [Google Scholar]
- Pedregosa F., Varoquaux G., Gramfort A., Michel V., Thirion B., Grisel O., Blondel M., Pret-tenhofer P., Weiss R., Dubourg V., Vanderplas J., Passos A., Cournapeau D., Brucher M., Perrot M., Duchesnay E. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 2011;12:2825–2830. [Google Scholar]
- Pomponio R., Erus G., Habes M., Doshi J., Srinivasan D., Mamourian E., Bashyam V., Nasrallah I.M., Satterthwaite T.D., Fan Y., Launer L.J., Masters C.L., Maruff P., Zhuo C., Völzke H., Johnson S.C., Fripp J., Koutsouleris N., Wolf D.H., Gur R., Gur R., Morris J., Albert M.S., Grabe H.J., Resnick S.M., Bryan R.N., Wolk D.A., Shinohara R.T., Shou H., Davatzikos C. Harmonization of large mri datasets for the analysis of brain imaging patterns throughout the lifespan. NeuroImage. 2020;208:116450. doi: 10.1016/j.neuroimage.2019.116450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigge M.D., Bigler E.D., Fletcher P.T., Zielinski B.A., Ravichandran C., Anderson J., Froehlich A., Abildskov T., Papadopolous E., Maasberg K., Nielsen J.A., Alexander A.L., Lange N., Lainhart J. Longitudinal heschl’s gyrus growth during childhood and adolescence in typical development and autism. Autism Res. 2013;6(2):78–90. doi: 10.1002/aur.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin, K., Lei, D., Pinaya, W. H., Pan, N., Li, W., Zhu, Z., Sweeney, J. A., Mechelli, A., & Gong, Q. (2022). Using graph convolutional network to characterize individuals with major depressive disorder across multiple imaging sites. eBioMedicine, 78, 103977. https://doi.org/https://doi.org/10.1016/j.ebiom.2022.103977. [DOI] [PMC free article] [PubMed]
- Redcay E., Courchesne E. When is the brain enlarged in autism? a meta-analysis of all brain size reports. Biol. Psychiatry. 2005;58:1–9. doi: 10.1016/j.biopsych.2005.03.026. [DOI] [PubMed] [Google Scholar]
- Retico A., Giuliano A., Tancredi R., Cosenza A., Apicella F., Narzisi A., Biagi L., Tosetti M., Muratori F., Calderoni S. The effect of gender on the neuroanatomy of children with autism spectrum disorders: A support vector machine case-control study. Molecular Autism. 2016;7 doi: 10.1186/s13229-015-0067-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzolatti G., Craighero L. The mirror-neuron system. Annu. Rev. Neurosci. 2004;27:169–192. doi: 10.1146/annurev.neuro.27.070203.144230. [DOI] [PubMed] [Google Scholar]
- Rizzolatti G., Sinigaglia C. The functional role of the parieto-frontal mirror circuit: Interpretations and misinterpretations. Nat. Rev. Neurosci. 2010;11:264–274. doi: 10.1038/nrn2805. [DOI] [PubMed] [Google Scholar]
- Sullivan P., Daly M., O’Donovan M. Disease mechanisms genetic architectures of psy- chiatric disorders: The emerging picture and its implications. Nat. Rev. Genet. 2012;13:537–551. doi: 10.1038/nrg3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussman D., Leung R.C., Vogan V.M., Lee W., Trelle S., Lin S., Cassel D.B., Chakravarty M.M., Lerch J.P., Anagnostou E., Taylor M.J. The autism puzzle: Diffuse but not pervasive neuroanatomical abnormalities in children with ASD. NeuroImage: Clinical. 2015;8:170–179. doi: 10.1016/j.nicl.2015.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbull A., Garfinkel S., Ho N., Critchley H., Bernhardt B., Jefferies E., Smallwood J. Word up - experiential and neurocognitive evidence for associations between autistic symptomology and a preference for thinking in the form of words. Cortex. 2020;128 doi: 10.1016/j.cortex.2020.02.019. [DOI] [PubMed] [Google Scholar]
- Uddin, L.Q., Supekar, K., Lynch, C.J., Khouzam, A., Phillips, J., Feinstein, C., Ryali, S., & Menon, V. (2013). Salience Network–Based Classification and Prediction of Symptom Sever- ity in Children With Autism. JAMA Psychiatry, 70 (8), 869. https :// doi .org/10.1001/ jamapsychiatry.2013.104. [DOI] [PMC free article] [PubMed]
- Vargason T., Grivas G., Hollowood-Jones K.L., Hahn J. Towards a Multivariate Biomarker-Based Diagnosis of Autism Spectrum Disorder: Review and Discussion of Recent Advancements. Seminars Pediatric Neurol. 2020;34 doi: 10.1016/j.spen.2020.100803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Chaudhari P., Davatzikos C. Embracing the disharmony in medical imaging: A simple and effective framework for domain adaptation. Med. Image Anal. 2022;76 doi: 10.1016/j.media.2021.102309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfers T., Floris D.L., Dinga R., van Rooij D., Isakoglou C., Kia S.M., Zabihi M., Llera A., Chowdanayaka R., Kumar V.J., Peng H., Laidi C., Batalle D., Dimitrova R., Charman T., Loth E., Lai M.-C., Jones E., Baumeister S., Moessnang C., Banaschewski T., Ecker C., Dumas G., O’Muircheartaigh J., Murphy D., Buitelaar J.K., Marquand A.F., Beckmann C.F. From pattern classification to stratification: towards conceptualizing the heterogeneity of Autism Spectrum Disorder. Neurosci. Biobehav. Rev. 2019;104:240–254. doi: 10.1016/j.neubiorev.2019.07.010. [DOI] [PubMed] [Google Scholar]
- Wood, S. N. (2017). Generalized additive models: An introduction with r, second edition.
- Wrobel J., Martin M.L., Bakshi R., Calabresi P.A., Elliot M., Roalf D., Gur R.C., Gur R.E., Henry R.G., Nair G., Oh J., Papinutto N., Pelletier D., Reich D.S., Rooney W.D., Satterthwaite T.D., Stern W., Prabhakaran K., Sicotte N.L., Shinohara R.T., Goldsmith J. Intensity warping for multisite mri harmonization. NeuroImage. 2020;223:117242. doi: 10.1016/j.neuroimage.2020.117242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Y., Shi, L., Cui, X., Wang, S., & Luo, X. (2016). Functional connectivity of the caudal anterior cingulate cortex is decreased in autism. PloS one, 11, e0151879. https://doi.org/ 10.1371/journal.pone.0151879. [DOI] [PMC free article] [PubMed]
- Zielinski, B., Prigge, M., Nielsen, J., Froehlich, A., Abildskov, T., Anderson, J., Fletcher, P., Camp- bell, K., Travers, B., Lange, N., Alexander, A., Bigler, E., & Lainhart, J. (2014). Longitudinal changes in cortical thickness in autism and typical development. Brain: J. Neurol., 137. https://doi.org/10.1093/brain/awu083. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.