Abstract
The pH-neutral cell supernatant of Enterococcus faecalis BFE 1071, isolated from the feces of minipigs in Göttingen, inhibited the growth of Enterococcus spp. and a few other gram-positive bacteria. Ammonium sulfate precipitation and cation-exchange chromatography of the cell supernatant, followed by mass spectrometry analysis, yielded two bacteriocin-like peptides of similar molecular mass: enterocin 1071A (4.285 kDa) and enterocin 1071B (3.899 kDa). Both peptides are always isolated together. The peptides are heat resistant (100°C, 60 min; 50% of activity remained after 15 min at 121°C), remain active after 30 min of incubation at pH 3 to 12, and are sensitive to treatment with proteolytic enzymes. Curing experiments indicated that the genes encoding enterocins 1071A and 1071B are located on a 50-kbp plasmid (pEF1071). Conjugation of plasmid pEF1071 to E. faecalis strains FA2-2 and OGX1 resulted in the expression of two active peptides with sizes identical to those of enterocins 1071A and 1071B. Sequencing of a DNA insert of 9 to 10 kbp revealed two open reading frames, ent1071A and ent1071B, which coded for 39- and 34-amino-acid peptides, respectively. The deduced amino acid sequence of the mature Ent1071A and Ent1071B peptides showed 64 and 61% homology with the α and β peptides of lactococcin G, respectively. This is the first report of two new antimicrobial peptides representative of a fourth type of E. faecalis bacteriocin.
Bacteriocins are ribosomally synthesized bacteriostatic or bactericidal proteins and peptides which are produced by a number of gram-positive and gram-negative bacteria. By definition these proteins exhibit a relatively narrow spectrum of antimicrobial activity and are in general active only against bacteria closely related to the producer strain (18). The bacteriocins of lactic acid bacteria were classified by Klaenhammer (18) into four groups. Most of the bacteriocins isolated so far belong to class I or class II. Class I bacteriocins, named lantibiotics, are small (<5-kDa) membrane-active peptides which contain posttranslationally modified amino acid residues. Nisin is the best-studied lantibiotic (31). Class II bacteriocins are unmodified, heat-stable, low-molecular-mass (<10-kDa), membrane-active peptides, usually characterized by a G-G-Xaa where Xaa is any amino acid, processing site, in the bacteriocin precursor. The class II bacteriocins are divided into three subgroups; IIa comprises peptides that contain a Y-G-N-G-V-Xaa-C motif near their N termini (Listeria-active peptides), e.g., pediocin PA-1 (22) and sakacin A (12); IIb comprises two-peptide bacteriocins, e.g., lactococcin G (25) and brochocin-C (23); and IIc comprises thiol-activated peptides, which require reduced cysteine residues for activity, e.g., lactococcin B (39).
To date, six bacteriocins of Enterococcus faecalis have been described (7, 10, 17, 19, 34, 35, 40), of which only three types have been biochemically and genetically characterized. A hemolysin/bacteriocin, encoded on a 58-kbp conjugative plasmid (pAD1) and originally isolated from Enterococcus faecalis subsp. zymogenes DS16, is classified as type 1 (6, 11, 14, 15). The cyclic peptide antibiotic AS-48, encoded on a 58-kbp plasmid (pMB2) and isolated from Enterococcus faecalis subsp. liquefaciens S-48 (20, 21), and bacteriocin 21, encoded on a 59-kbp plasmid (pPD1) and isolated from E. faecalis 39-5Sa (9, 35), are identical and have been classified as type 2. Bacteriocin 31, an antilisterial peptide encoded on a 57.5-kbp plasmid (pYI17) and isolated from E. faecalis YI17, has been classified as type 3 (34).
In this paper we report the isolation and characterization of two new antimicrobial peptides which belong to a fourth type of E. faecalis bacteriocin.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
E. faecalis BFE 1071 was isolated from the feces of minipigs in Göttingen. The indicator strains used in this study are listed in Table 1. E. faecalis FA2-2 and OGX1 were obtained from D. B. Clewell, Department of Microbiology and Immunology, School of Medicine, University of Michigan. Lactococcus lactis subsp. lactis IL1403 was obtained from J. Kok, Department of Genetics, University of Groningen, Groningen, The Netherlands.
TABLE 1.
Spectrum of antimicrobial activity of enterocins 1071A and 1071B
| Organism | Strain(s)a | Sensitivityb |
|---|---|---|
| Bacillus cereus | LMG 13569 | − |
| Bifidobacterium gallicum | LMG 11596 | − |
| Clostridium sporogenes | LMG 13570 | − |
| Clostridium tyrobutyricum | LMG 13571 | − |
| LMG 1285T | + | |
| Enterococcus durans | RC1 | ++ |
| Enterococcus faecalis | LMG 13566, RC2 | +++ |
| BFE 1071c | − | |
| FA2-2, OGX1 | +++ | |
| Enterococcus faecalis subsp. liquefaciens | ATCC 10100 | +++ |
| Enterococcus faecium | RC3 | ++ |
| Escherichia coli XL1-Blue MRF′ | Stratagene | − |
| Lactobacillus acidophilus | LMG 13550 | − |
| Lactobacillus casei | LMG 13552 | − |
| Lactobacillus curvatus | LMG 13553 | − |
| Lactobacillus fermentum | LMG 13554 | − |
| Lactobacillus helveticus | LMG 13555 | − |
| Lactobacillus plantarum | LMG 13556 | − |
| Lactobacillus reuteri | LMG 13557 | − |
| Lactobacillus sake | LMG 13558 | − |
| Lactobacillus salivarius subsp. salivarius | NCIMB 702747T | +++ |
| Lactococcus lactis subsp. lactis | IL 1403 | − |
| Leuconostoc carnosum | NCIMB 702776T | − |
| Leuconostoc cremoris | LMG 13562, LMG 13563 | − |
| Leuconostoc mesenteroides subsp. cremoris | NCIMB 4543 | − |
| Leuconostoc mesenteroides subsp. mesenteroides | NCIMB 4518 | − |
| Listeria innocua | LMG 13568 | +++ |
| Micrococcus sp. | DF 132 | + |
| Pediococcus pentosaceus | LMG 13560, LMG 13561 | − |
| Peptostreptococcus aerogenes | MKB 12 | ++ |
| Propionibacterium acidipropionici | LMG 13572 | − |
| Propionibacterium freudenreichii subsp. shermanii | LMG 16424 | + |
| Propionibacterium spp. | LMG 13573, LMG 13574 | − |
| Staphylococcus aureus | MKB 38 | − |
| Staphylococcus carnosus | LMG 13567 | − |
| Streptococcus agalactiae | ATCC 13813 | + |
| Streptococcus mutans | MKB THSM | − |
| Streptococcus thermophilus | LMG 13564, LMG 13565 | − |
Abbreviations: ATCC, American Type Culture Collection, Rockville, Md.; LMG, Laboratorium voor Microbiologie, University of Ghent, Ghent, Belgium; NCIMB, National Collection of Industrial and Marine Bacteria, Reading, United Kingdom; MKB, Department of Microbiology, University of Stellenbosch, Stellenbosch, South Africa; RC, Red Cross Children Hospital, Cape Town, South Africa; DF, F. Dellaglio, Istituto Policattedra, Universitá degli Studi di Verona, Verona, Italy.
+, sensitive to enterocins 1071A and 1071B (+, ++, and +++ reflect the degree of sensitivity); −, resistant to enterocins 1071A and 1071B.
Enterocin 1071A and 1071B producer strain.
All lactic acid bacteria were grown in MRS broth (Biolab Diagnostics, Midrand, South Africa) except L. lactis subsp. lactis IL1403, which was grown in M17 broth (Oxoid, Basingstoke, Hampshire, England). Other bacteria were cultured in brain heart infusion (BHI) broth (Biolab), except Clostridium spp., which were grown in differential reinforced clostridial broth (Merck, Darmstadt, Germany), and Propionibacterium spp., which were cultured in glucose yeast peptone medium, containing the following (per liter): glucose, 5 g; yeast extract, 3 g; peptone, 10 g; meat extract, 10 g; and NaCl, 5 g (pH 6.5). For Escherichia coli XL Blue MRF′ (Stratagene, La Jolla, Calif.), the BHI medium was supplemented with 200 μg of erythromycin (Sigma, St. Louis, Mo.) per ml.
Classification of E. faecalis BFE 1071.
The biochemical identification of E. faecalis BFE 1071 was confirmed by numerical analysis of total soluble cell protein patterns and molecular typing by random amplified polymorphic DNA (RAPD)-PCR. The methods of Pot et al. (28) and Van Reenen and Dicks (37) were used. The primers used for RAPD-PCR were TGGGCGTCAA (OPL-02), ACGCAGGCAC (OPL-05), and ACGATGAGCC (OPL-11) of Operon Kit L (Operon Technologies, Alameda, Calif.).
Inhibitory activity.
The spot-on-lawn method was used to determine the inhibitory activity of enterocin 1071A and enterocin 1071B. The tests were performed as described by Van Reenen et al. (38). A clear inhibition zone at least 2 mm in diameter was recorded as positive. One arbitrary unit (AU) of enterocin activity was defined as the reciprocal of the greatest dilution of the bacteriocin that produced an inhibition zone at least 2 mm in diameter.
Sensitivity to heat, pH, and proteolytic enzymes.
A crude extract containing enterocins 1071A and 1071B (3,200 AU ml−1) was used in these tests. Aliquots of the enterocins (3,200 AU ml−1) were exposed to heat treatments of 40, 60, 80, and 100°C for 10, 30, and 60 min and 121°C for 15 min. The samples were then tested for activity against E. faecalis LMG 13566, as described above. In a separate experiment, samples of the enterocins were adjusted to pH values ranging from 3 to 12, incubated at 37°C for 30 min, neutralized to pH 7, and then tested for antimicrobial activity. Resistance of the enterocins to proteolytic enzymes was determined by incubating samples in the presence of proteinase K (10 U/mg of enterocin), pronase (3,500 U/mg of enterocin), pepsin (1,250 U/mg of enterocin), papain (15 U/mg of enterocin), α-chymotrypsin (45 U/mg of enterocin), and trypsin (55 U/mg of enterocin) at 37°C for 1 h. All enzymes were from Boehringer Mannheim South Africa Ltd. (Howard Place, South Africa). After incubation, the enzymes were heat inactivated for 3 min at 100°C, and the bacteriocins were tested for antimicrobial activity against all the strains listed in Table 1.
Mechanism of activity.
A crude extract containing enterocins 1071A and 1071B (1.5 ml) was added to a 100-ml culture of E. faecalis LMG 13566 at the beginning of the lag and the mid-exponential growth phases. This resembled a final enterocin concentration of 3,072 AU ml−1. Sterile demineralized water (1.5 ml) was added to the control flask. Changes in the turbidity of the cultures were recorded at an optical density at 600 nm, and the number of cells (CFU per milliliter) was determined by plating the samples onto MRS agar.
Production of hemolysin was determined by plating actively growing cells of E. faecalis BFE 1071 onto Columbia blood agar (Oxoid) supplemented with 5% (vol/vol) human blood. Plates were incubated at 37°C in an anaerobic jar (Oxoid) with a gas-generating kit. Results were recorded after 24 and 72 h. A clear zone of hemolysis on blood agar plates was considered a positive result.
Isolation and purification of enterocins 1071A and 1071B.
One liter of dialyzed casein glucose broth (3) was inoculated with 10 ml of an actively growing culture of E. faecalis BFE 1071 and incubated overnight at 37°C. Cells were removed by centrifugation, and the bacteriocins were precipitated from the supernatant by ammonium sulfate (75% [wt/vol], final concentration). The precipitate was collected by centrifugation at 14,300 × g, resuspended in MilliQ water, and desalted overnight at 8°C by using a 1-kDa-cutoff dialysis bag (Spectrum, Los Angeles, Calif.). The dialyzed sample was stored at −80°C.
Further purification of enterocins 1071A and 1071B was performed by separation on an SP-Sepharose Fast Flow (Amersham Pharmacia Biotech, Uppsala, Sweden) column. Sixty milliliters of a crude extract which contained both bacteriocins (9.6 mg of protein ml−1) was suspended in ammonium acetate buffer (0.05 mol liter−1, pH 5.78) and applied onto a 15-ml SP-Sepharose Fast Flow matrix which had been equilibrated with the same buffer. The column was washed with 40 ml of acetate buffer, and the proteins were eluted with an ammonium acetate step gradient of 0.1 to 0.8 mol liter−1 (pH 5.78). Fractions of 4 ml each were collected, and the protein content was determined by measuring the optical density at 280 nm and tested for activity against E. faecalis LMG 13566, as described above. Fractions containing the bacteriocins were pooled and concentrated by freeze-drying.
Such a concentrated sample containing enterocins 1071A and 1071B was subjected to tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), according to the method of Schägger and von Jagow (33). Protein markers ranging from 2.35 to 46 kDa (Rainbow marker; Amersham Pharmacia Biotech) were used. One half of the gel was stained with Coomassie brilliant blue R250. The position of the active peptide band was determined by overlaying the other half of the gel, prewashed as described by Van Belkum et al. (36), with cells of E. faecalis LMG 13566 (approximately 106 cells ml−1), embedded in MRS agar (0.7% agar, wt/vol).
Molecular mass determination.
Approximately 100 pmol of purified sample containing the two bacteriocins was diluted in 10 μl of 10:90 acetonitrile-water containing 0.01% formic acid and injected via the Rheodyne injection port of a Quattro triple quadropole mass spectrometer (Micromass, Manchester, United Kingdom). The carrier solvent was 10:90 acetonitrile-water at a flow rate of 20 μl min−1, delivered by a Pharmacia-LKB 2249 high-pressure liquid chromatography pump. The capillary voltage and the cone voltage were set at 3.5 kV and 60 V, respectively. Data were collected by scanning from 400 to 1,500 m/z at 2 s/scan. The multiple charged spectra were deconvoluted to obtain the accurate mass of the peptides. The mass spectrometer was calibrated by using the multiply charged spectrum of horse heart myoglobin (Sigma).
Plasmid curing.
Curing experiments were conducted as described by Ruiz-Barba et al. (30). Cells of E. faecalis BFE 1071 were incubated in the presence of novobiocin (1 to 25 μg ml−1) for 72 h at 37°C. The culture which grew at the highest concentration of novobiocin was serially diluted with sterile saline and plated onto MRS agar plates. After overnight incubation at 37°C, the colonies were replica plated, and the original plates were overlaid with cells of E. faecalis LMG 13566. After a further 16 h of incubation at 37°C, the colonies were checked for loss of antimicrobial activity and plasmids, as described below.
Conjugative transfer experiments.
Filter mating experiments were done as described by Reichelt et al. (29). An overnight culture (0.25 ml) of E. faecalis BFE 1071 and FA2-2 was added to 4.5 ml of MRS, mixed, and filtered through a 0.45-μm-pore-size sterile membrane filter (HAWP; Millipore). The membrane was placed onto an MRS agar plate and incubated overnight at 37°C. The cells were washed from the filter into 1 ml of MRS, serially diluted, and plated onto MRS agar plates containing 25 μg of fusidic acid, 25 μg of rifampin, and 2,000 AU of crude enterocin (1071A and 1071B) per ml. The experiment was repeated with E. faecalis OGX1 as the recipient. In this case the selection was done on plates containing 1 mg of streptomycin and 2,000 AU of crude enterocin (1071A and 1071B) per ml. Colonies were selected at random, checked for production of enterocins, and screened for plasmid content, as described above. A colony of conjugated cells of E. faecalis OGX1 which contained pEF1071 (OGX1/pEF1071) was cultured, and the cell supernatant was subjected to enterocin purification and mass spectrometry analysis, as described above. The spectrum of antimicrobial activity of these peptides was tested on plates which had been seeded with sensitive cells, as described above.
Isolation and manipulation of plasmid DNA.
Plasmid DNA of E. faecalis BFE 1071, FA2-2, and OGX1 was isolated by the method of Burger and Dicks (4), followed by CsCl density gradient centrifugation (32). The plasmid DNA was digested with different restriction enzymes and subjected to agarose gel electrophoresis (1% [wt/vol] agarose) (32). EcoRV-digested plasmid DNA was ligated into plasmid pTRKH2, a Lactococcus/E. coli shuttle vector (27) that had been predigested with EcoRV and dephosphorylated. This construct was introduced into E. coli XL1-Blue MRF′ cells by electroporation as described in the GenePulser manual (Bio-Rad, Hercules, Calif.). The restriction and DNA modification enzymes were obtained from Boehringer Mannheim South Africa and used as specified by the supplier. The transformants were selected on BHI agar containing 200 μg of erythromycin (Sigma) per ml, 20 μg of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) (Sigma) per ml, and 1 mmol of 1-isopropyl-β-d-1-thiogalactopyranoside (IPTG) (Sigma) per liter. White colonies were isolated and further analyzed.
Southern blot hybridization.
Southern blot hybridizations were performed as described by Sambrook et al. (32). The plasmid DNA of E. faecalis BFE 1071 was hybridized with a probe made from the cloned EcoRV fragment of plasmid pEF1071, which contains the genes encoding enterocin 1071A and enterocin 1071B. Detection was performed by using the digoxigenin High Prime labeling and detection kit of Boehringer Mannheim South Africa.
DNA sequencing and analysis.
Plasmid DNA for sequencing was purified by CsCl density gradient centrifugation (32). Sequencing was performed with an ABI Prism BigDye Terminator Cycle Sequencing Ready Reaction kit on a ABI Prism 377 DNA sequencer (PE Applied Biosystem, Foster City, Calif.) and initiated with pUC/M13 forward and reverse sequencing primer (17-mer) (Promega, Madison, Wis.). A database search was performed by using the BLASTN and BLASTX programs (2) of the National Center for Biotechnology Information, Bethesda, Md. (http://www.ncbi.nlm.nih.gov).
Nucleotide sequence accession number.
The nucleotide sequences reported here have been submitted to GenBank with accession numbers AF164559 for ent1071A and AF164560 for ent1071B.
RESULTS
Classification of E. faecalis BFE 1071.
Numerical analysis of total soluble cell protein patterns and RAPD-PCR clearly indicated that strain BFE 1071 is a member of E. faecalis and not Enterococcus faecium (data not shown).
Inhibitory activity.
Enterocins 1071A and 1071B were found to be active against all strains of Enterococcus spp. used in this study and against six other gram-positive organisms (Table 1). No enterocin activity was recorded after treatment with proteolytic enzymes.
Sensitivity to heat, pH, and proteolytic enzymes.
Enterocins 1071A and 1071B are resistant to heat treatments of up to 100°C for 60 min. Approximately 50% of the antibacterial activity was retained after 15 min at 121°C. The enterocins are not drastically affected by incubation at pH values ranging from 3 to 12, but they are sensitive to α-chymotrypsin, papain, pepsin, pronase, proteinase K, and trypsin.
Mechanism of activity.
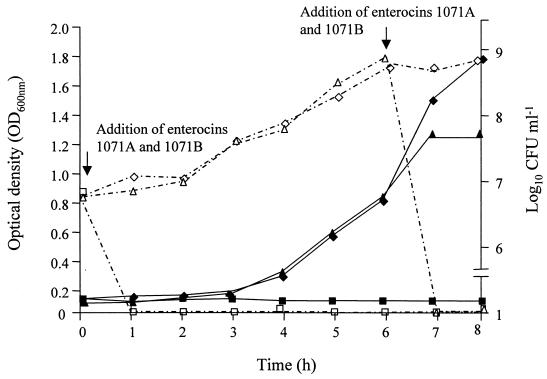
The addition of enterocins 1071A and 1071B to actively growing cells of E. faecalis LMG 13566 (6-h-old culture) or at the beginning of the lag phase completely inhibited cell growth (Fig. 1).
FIG. 1.
Effect of enterocins 1071A and 1071B on the growth of E. faecalis LMG 13566. Counts were made in the absence of enterocins 1071A and -B (◊) and in the presence of enterocins 1071A and -B added at the beginning of the lag phase (□) and after 6 h of growth (▵). Turbidity was determined for cells growing in the absence of enterocins 1071A and -B (⧫) and cells growing in the presence of enterocins 1071A and -B added at the beginning of the lag phase (■) and after 6 h of growth (▴).
No beta-hemolytic zones were detected when E. faecalis BFE 1071 was cultured on blood agar plates.
Isolation and purification of enterocins 1071A and 1071B.
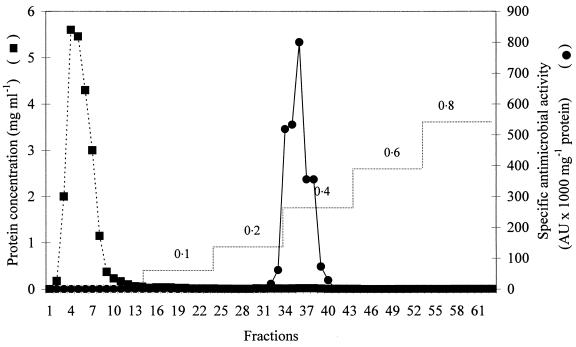
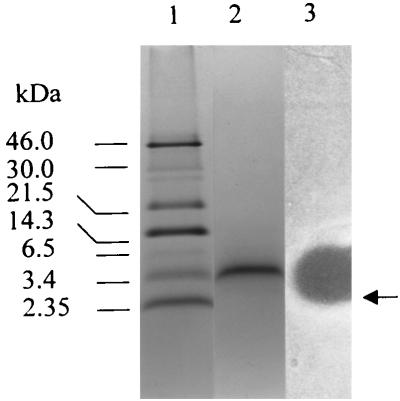
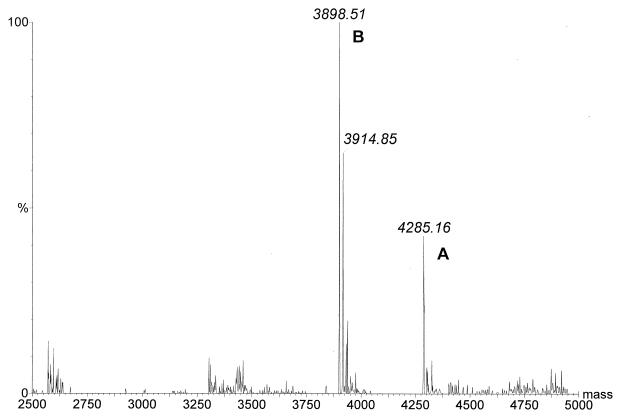
Precipitation of enterocins 1071A and 1071B by ammonium sulfate yielded a 97% recovery of antimicrobial activity and a 54-fold increase in specific antimicrobial activity (from 241.7 to 13,021 AU per mg of protein). Purification of the crude extract of enterocins 1071A and 1071B on an SP-Sepharose Fast Flow column (Fig. 2) yielded a 33,099-fold increase in specific antimicrobial activity compared with the activity in the culture supernatant (from 241.7 to 8 × 106 AU per mg of protein). Separation on tricine-SDS-PAGE yielded only one active peptide band (Fig. 3). Mass spectrometry analysis indicated that the single active peak collected from the SP-Sepharose column (Fig. 2) contained two peptides, enterocins 1071A and 1071B, with molecular masses of 4.285 and 3.899 kDa, respectively (Fig. 4).
FIG. 2.
Purification of enterocins 1071A and 1071B with ion-exchange chromatography (SP-Sepharose Fast Flow matrix). The values 0.1 to 0.8 refer to the NH4COOH gradient used, in moles per liter.
FIG. 3.
Separation of enterocins 1071A and 1071B by tricine-SDS-PAGE. Lane 1, Rainbow protein size markers; lane 2, enterocins 1071A and 1071B stained with Coomassie brilliant blue R250; lane 3, enterocins 1071A and 1071B overlaid with cells of E. faecalis LMG 13566 embedded in MRS agar (0.7% agar, wt/vol). The active peptide band is indicated by an arrow.
FIG. 4.
Molecular masses of enterocins 1071A (A) and 1071B (B), calculated from the electrospray ionization-mass spectroscopy multiple charged spectra.
Plasmid curing.
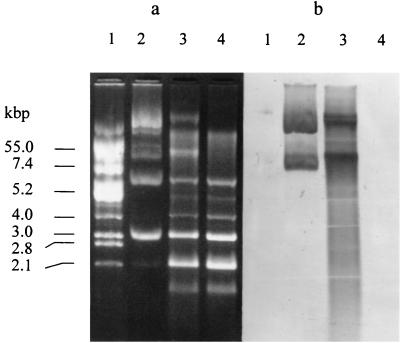
E. faecalis BFE 1071 contains at least five plasmids of about 2.1, 3.0, 6.0, 20.0, and 55.0 kbp (Fig. 5a). Curing with novobiocin yielded two mutants of E. faecalis BFE 1071, designated 1071/78 and 1071/79. Mutant 1071/78 produced enterocins and changed from vancomycin resistant to vancomycin sensitive. Mutant 1071/79 lost the ability to produce antimicrobial peptides, became sensitive to its own bacteriocins, and was also vancomycin sensitive. Southern hybridization results with a probe derived from the cloned EcoRV fragment of plasmid pEF1071 containing the genes encoding enterocins 1071A and 1071B revealed that mutant 1071/79 has lost a plasmid of about 50 kbp (Fig. 5b).
FIG. 5.
Plasmid profiles of E. faecalis BFE 1071 before and after plasmid curing. (a) Agarose gel electrophoresis of plasmids of E. coli V517 (lane 1), E. faecalis BFE 1071 (lane 2), mutant 1071/78 (lane 3), and mutant 1071/79 (lane 4). The numbers on the left indicate the sizes of the plasmids isolated from E. coli V517. (b) The same gel after Southern blotting onto a MagnaGraph nylon transfer membrane (MSI, Westboro, Mass.) and hybridized with a digoxigenin-labeled cloned EcoRV fragment of plasmid pEF1071 containing the genes encoding enterocins 1071A and 1071B.
Conjugative transfer experiments.
The E. faecalis OGX1 transconjugants contained a plasmid of about 50 kbp and produced enterocins 1071A and 1071B. Purification of the enterocins by ammonium sulfate precipitation and SP-Sepharose chromatography, followed by mass spectrometry, indicated the presence of two peptides with molecular masses of 4.285 and 3.899 kDa (data not shown). The spectrum of antimicrobial activity recorded for the peptides isolated from the conjugant (OXG1/pEF1071) was identical to that recorded for E. faecalis BFE 1071 (data not shown).
Isolation and manipulation of plasmid DNA.
Ligation of the EcoRV-digested plasmid pEF1071 to the EcoRV-restricted plasmid pTRKH2 and subsequent transformation into E. coli yielded recombinant plasmids with fragment sizes of 9 to 10 kbp.
DNA sequencing and analysis.
DNA sequence analysis of the insert of one of the recombinant plasmids, named pEco2, revealed two open reading frames (ORFs) encoding bacteriocinlike prepeptides, designated enterocins 1071A and 1071B, with a common promoter region and their own ribosome binding sites (Fig. 6). The prepeptides have the consensus G-G-Xaa-processing site (Fig. 6). The first ORF (ent1071A) encodes a prepeptide of 57 amino acids, while the second ORF (ent1071B) encodes a prepeptide of 62 amino acids. The estimated molecular masses of the deduced mature peptides (39 and 34 amino acids) are 4.259 and 3.899 kDa, respectively.
FIG. 6.
Nucleotide sequence of the regions encoding enterocin 1071A and enterocin 1071B of E. faecalis BFE 1071 and the deduced amino acid sequences. The putative promoter region and transcription initiation sites are indicated in bold, and the ribosome binding sites (RBS) are in italics and underlined. The arrows indicate the processing sites of the peptides.
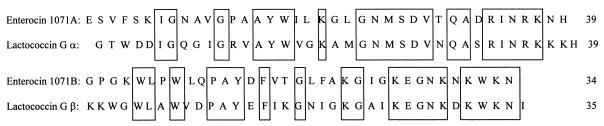
The BLASTX protein database homology search on the deduced mature peptides, enterocin 1071A and enterocin 1071B, showed 64% homology with the lactococcin G α and 61% homology with the lactococcin G β peptide (Fig. 7). No DNA homology was found with any DNA sequences reported for bacteriocins using the BLASTN DNA database.
FIG. 7.
Alignment of the peptides of enterocin 1071A and lactococcin G α and of enterocin 1071B and lactococcin G β. Identical amino acids are boxed.
DISCUSSION
In this paper, we have described the purification and genetic characterization of two new plasmid-encoded bacteriocins produced by E. faecalis BFE 1071, which was isolated from minipigs in Göttingen. This is the first report of two new antimicrobial peptides representative of a fourth type of E. faecalis bacteriocin.
Based on the spectrum of antimicrobial activity recorded for enterocins 1071A and 1071B (Table 1), they are in all aspects different from other bacteriocins thus far described for E. faecalis and its subspecies. E. faecalis BFE 1071 did not show any hemolytic activity, which is a characteristic shared by producers of hemolysin/bacteriocins described for E. faecalis subsp. zymogenes DS16 (type 1 enterocins) (6, 11, 14, 15). The antimicrobial activity spectrum is also narrower than recorded for the cyclic peptide antibiotic AS-48 produced by E. faecalis subsp. liquefaciens S-48 (20, 21) and bacteriocin 21 produced by E. faecalis 39-5SA (type 2 enterocins) (9, 35), but broader than described for bacteriocin 31 produced by E. faecalis YI17 (type 3 enterocins) (34). The small size, antilisterial activity, and heat stability suggested that enterocins 1071A and 1071B belong to the class II group of bacteriocins, according to the classification of Klaenhammer (18).
Bacteriocins from enterococci may be either plasmid encoded (8, 11, 20, 34, 35) or located on the genome (2, 5). Loss of antimicrobial activity and immunity against its own bacteriocin after plasmid curing, and expression and secretion of two antimicrobial peptides of exact molecular masses (4.854 and 3.899 kDa, respectively) by the transconjugant strain OGX1, indicated that the genes coding for the production of and immunity against enterocins 1071A and 1071B are located on a plasmid designated pEF1071.
Sequencing of an approximately 9-kbp EcoRV fragment of plasmid pEF1071 indicated that the genes encoding enterocins 1071A (ent1071A) and 1071B (ent1071B) are arranged in one operon (Fig. 6). The prepeptides had putative signal sequences of 18 and 27 amino acids, respectively, at the N terminus and the G-G-Xaa-processing site (Fig. 6). This has also been described for other bacteriocin prepeptides (18).
Amino acid sequence comparisons of the deduced mature peptides indicated that they are unique among the enterococcal bacteriocins. In spite of the high protein sequence homology with lactococcin G α and β peptides (64 and 61%, respectively), enterocins 1071A and 1071B are not active against L. lactis subsp. lactis IL1403 (the indicator organism for lactococcin G) (Table 1). Although enterocins 1071A and 1071B have antilisterial activity, they do not contain the highly conserved YGNGVxC motif found in the N-terminal part of most of the pediocin-like bacteriocins (18). Enterocin B (5) and enterocin I (8) are also exceptions. Enterocins 1071A and 1071B also lack cysteine residues, which are usually present in the pediocin-like bacteriocins, including enterocins, and they have a medium spectrum of activity (Table 1) according to Jack et al. (16).
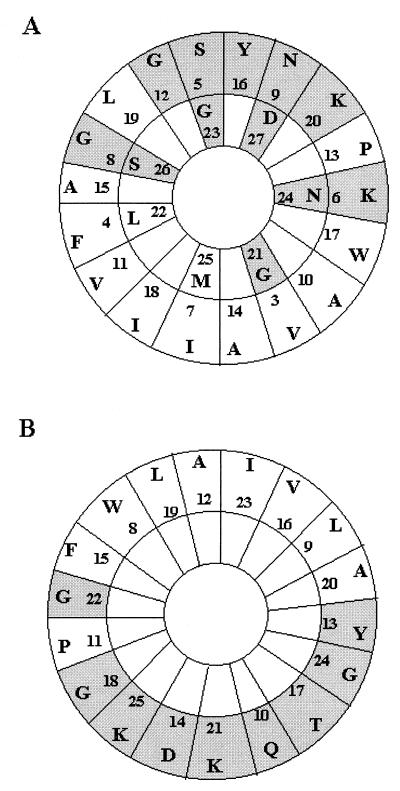
Based on the deduced amino acid sequences of the mature enterocin 1071A and enterocin 1071B peptides, it is very likely that they act as pore-forming toxins that create cell membrane channels through a “barrel-stave” mechanism and thus produce an ionic imbalance in the cell (26). The decrease in CFU, concomitant with a decrease in turbidity of E. faecalis LMG 13566 cells treated with enterocin 1071A and enterocin 1071B (Fig. 1), supports the latter hypothesis. A region in the deduced amino acid sequence of enterocin 1071A, starting with amino acid residue 4 and ending with residue 27, may form an amphiphilic α-helix, as shown when displayed on an Edmundson α-helical wheel (Fig. 8). The polar amino acids are found almost completely on one side of the α-helix, whereas the nonpolar residues are found on the opposite side of the helix, except for a proline (residue 13) between the polar amino acids and two glycines (residue 8 and 21) between the nonpolar residues (Fig. 8). However, glycine may be considered relatively neutral with respect to its hydrophobic-hydrophilic character, and the replacement of an amino acid by one of opposite hydrophobicity may not represent an intolerable disruption of the amphiphilic character of a peptide (26). The amphiphilic distribution of the amino acid is similar for the enterocin 1071B peptide displayed on the Edmundson wheel, starting with amino acid residue 8 and ending with residue 25. The exception is a proline (residue 11) between the polar residues (Fig. 8). The 25-amino-acid-long amphiphilic region of enterocin 1071A should be long enough to span a membrane, as a minimum of about 20 residues is needed to form a membrane-spanning α-helix (26). The 18-amino-acid-long amphiphilic region of enterocin 1071B may be less than required to span the cell membrane. However, in front of the amphiphilic region, the N-terminal part of the peptide is hydophobic, which is presumably also part of the transmembrane region. The C-terminal part of both peptides is hydrophilic, and therefore one might expect that these regions will be located outside the membrane. The C-terminal part of enterocin 1071A ends with a histidine, and it is interesting that lactococcin G α, lactococcin A, and lactococcin S, bacteriocins produced by L. lactis LMG 2081, Lactococcus lactis subsp. cremoris, and Lactobacillus sake, respectively, also end with histidine (13, 24, 25).
FIG. 8.
Edmundson α-helical wheel representation of the amphiphilic regions in enterocin 1071A (A) and enterocin 1071B (B). For enterocin 1071A, the amphiphilic region starts with residue 4 and ends with residue 27; for enterocin 1071B, the amphiphilic region starts with residue 8 and ends with residue 25. The letters with shaded and white backgrounds indicate polar and nonpolar residues, respectively.
ACKNOWLEDGMENT
We thank W. H. van Zyl for critical reading of the manuscript.
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aymerich T, Holo H, Håvarstein L S, Hugas M, Garriga M, Nes I F. Biochemical and genetic characterization of enterocin A from Enterococcus faecium, a new antilisterial bacteriocin in the pediocin family of bacteriocins. Appl Environ Microbiol. 1996;62:1676–1682. doi: 10.1128/aem.62.5.1676-1682.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhunia A K, Johnson M C, Ray B. Purification, characterization and antimicrobial spectrum of a bacteriocin produced by Pediococcus acidilactici. J Appl Bacteriol. 1988;62:261–268. doi: 10.1111/j.1365-2672.1988.tb01893.x. [DOI] [PubMed] [Google Scholar]
- 4.Burger J H, Dicks L M T. Technique for isolating plasmids from exopolysaccharide producing Lactobacillus spp. Biotechnol Tech. 1994;8:769–772. [Google Scholar]
- 5.Casaus P, Nilsen T, Cintas L M, Nes I F, Hernandez P E, Holo H. Enterocin B, a new bacteriocin from Enterococcus faecium T136 which can act synergistically with enterocin A. Microbiology. 1997;143:2287–2294. doi: 10.1099/00221287-143-7-2287. [DOI] [PubMed] [Google Scholar]
- 6.Clewell D B, Tomich P K, Gawron-Burke M C, Franke A E, Yagi Y, An F Y. Mapping of Streptococcus faecalis plasmids pAD1 and pAD2 and studies relating to transposition of Tn917. J Bacteriol. 1982;152:1220–1230. doi: 10.1128/jb.152.3.1220-1230.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dunny G M, Clewell D B. Transmissible toxin (hemolysin) plasmid in Streptococcus faecalis and its mobilization of a noninfectious drug resistance plasmid. J Bacteriol. 1975;124:784–790. doi: 10.1128/jb.124.2.784-790.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Floriano B, Ruiz-Barba J L, Jimenez-Diaz R. Purification and genetic characterization of enterocin I from Enterococcus faecium 6T1a, a novel antilisterial plasmid-encoded bacteriocin which does not belong to the pediocin family of bacteriocins. Appl Environ Microbiol. 1998;64:4883–4890. doi: 10.1128/aem.64.12.4883-4890.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fujimoto S, Tomita H, Wakamatsu E, Tanimoto K, Ike Y. Physical mapping of the conjugative bacteriocin plasmid pPD1 of Enterococcus faecalis and identification of the determinant related to the pheromone response. J Bacteriol. 1995;177:5574–5581. doi: 10.1128/jb.177.19.5574-5581.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gálvez A, Maqueda M, Martínez-Bueno M, Valdivia E. Bactericidal and bacteriolytic action of peptide antibiotic AS-48 against Gram-positive and Gram-negative bacteria and other organisms. Res Microbiol. 1989;140:57–68. doi: 10.1016/0923-2508(89)90060-0. [DOI] [PubMed] [Google Scholar]
- 11.Gilmore M S, Segarra R A, Booth M C, Bogie C P, Hall L R, Clewell D B. Genetic structure of the Enterococcus faecalis plasmid pAD1-encoded cytolytic toxin system and its relationship to lantibiotic determinants. J Bacteriol. 1994;176:7335–7344. doi: 10.1128/jb.176.23.7335-7344.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holck A, Axelsson L, Birkeland S-E, Aukrust T, Blom H. Purification and amino acid sequence of sakacin A, a bacteriocin from Lactobacillus sake Lb706. J Gen Microbiol. 1992;138:2715–2720. doi: 10.1099/00221287-138-12-2715. [DOI] [PubMed] [Google Scholar]
- 13.Holo H, Nilssen O, Nes I F. Lactococcin A, a new bacteriocin from Lactococcus lactis subsp. cremoris: isolation and characterization of the protein and its gene. J Bacteriol. 1991;173:3879–3887. doi: 10.1128/jb.173.12.3879-3887.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ike Y, Clewell D B, Segarra R A, Gilmore M S. Genetic analysis of the pAD1 hemolysin/bacterocin determinant in Enterococcus faecalis: Tn917 insertional mutagenesis and cloning. J Bacteriol. 1990;172:155–163. doi: 10.1128/jb.172.1.155-163.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ike Y, Clewell D B. Evidence that the hemolysin/bacteriocin phenotype of Enterococcus faecalis subsp. zymogenes can be determined by plasmids in different incompatibility groups as well as by the chromosome. J Bacteriol. 1992;174:8172–8177. doi: 10.1128/jb.174.24.8172-8177.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jack R W, Tagg J R, Ray B. Bacteriocins of gram-positive bacteria. Microbiol Rev. 1995;59:171–200. doi: 10.1128/mr.59.2.171-200.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Joosten H M L J, Nuñez M, Devreese B, van Beeumen J, Marugg J D. Purification and characterization of enterocin 4, a bacteriocin produced by Enterococcus faecalis INIA 4. Appl Environ Microbiol. 1996;62:4220–4223. doi: 10.1128/aem.62.11.4220-4223.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klaenhammer T R. Genetics of bacteriocins produced by lactic acid bacteria. FEMS Microbiol Rev. 1993;12:39–86. doi: 10.1111/j.1574-6976.1993.tb00012.x. [DOI] [PubMed] [Google Scholar]
- 19.Maisnier-Patin S, Forni E, Richard J. Purification, partial characterization and mode of action of enterococcin EFS2, an antilisterial bacteriocin produced by a strain of Enterococcus faecalis isolated from cheese. Int J Food Microbiol. 1996;30:255–270. doi: 10.1016/0168-1605(96)00950-6. [DOI] [PubMed] [Google Scholar]
- 20.Martínez-Bueno M, Gálvez A, Valdivia E, Maqueda M. A transferable plasmid associated with AS-48 production in Enterococcus faecalis. J Bacteriol. 1990;172:2817–2818. doi: 10.1128/jb.172.5.2817-2818.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martínez-Bueno M, Maqueda M, Gálvez A, Samyn B, van Beeumen J, Coyette J, Valdivia E. Determination of the gene sequence and the molecular structure of the enterococcal peptide antibiotic AS-48. J Bacteriol. 1994;176:6334–6339. doi: 10.1128/jb.176.20.6334-6339.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marugg J D, Gonzalez C F, Kunka B S, Ledeboer A M, Pucci M J, Toonen M Y, Walker S A, Zoetmulder L C M, Vandenbergh P A. Cloning, expression, and nucleotide sequence of genes involved in production of pediocin PA-1, a bacteriocin from Pediococcus acidilactici PAC1.0. Appl Environ Microbiol. 1992;58:2360–2367. doi: 10.1128/aem.58.8.2360-2367.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McCormick J K, Poon A, Sailer M, Gao Y, Roy K L, McMullen L M, Vederas J C, Stiles M E, Van Belkum M J. Genetic characterization and heterologous expression of brochocin-C, an antibotulinal, two-peptide bacteriocin produced by Brochothrix campestris ATCC 43754. Appl Environ Microbiol. 1998;64:4757–4766. doi: 10.1128/aem.64.12.4757-4766.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mortvedt C I, Nissen-Meyer J, Nes I F. Purification and amino acid sequence of lactocin S, a bacteriocin produced by Lactobacillus sake L45. Appl Environ Microbiol. 1991;57:1829–1834. doi: 10.1128/aem.57.6.1829-1834.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nissen-Meyer J, Holo H, Håvarstein L S, Slette K, Nes I F. A novel lactococcal bacteriocin whose activity depends on the complementary action of two peptides. J Bacteriol. 1992;174:5685–5692. doi: 10.1128/jb.174.17.5686-5692.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ojcius D M, Young D-E. Cytolytic pore-forming proteins and peptides: is there a common structural motif? Trends Biochem Sci. 1991;16:225–229. doi: 10.1016/0968-0004(91)90090-i. [DOI] [PubMed] [Google Scholar]
- 27.O'Sullivan D J, Klaenhammer T R. High- and low-copy-number Lactococcus shuttle cloning vectors with features for clone screening. Gene. 1993;137:227–231. doi: 10.1016/0378-1119(93)90011-q. [DOI] [PubMed] [Google Scholar]
- 28.Pot B, Vandamme P, Kersters K. Analysis of electrophoretic whole-organism protein fingerprints. In: Goodfellow M, O'Donnell A G, editors. Chemical methods in prokaryotic systematics. Chichester, U.K: John Wiley and Sons Ltd.; 1994. pp. 493–521. [Google Scholar]
- 29.Reichelt T, Kennes J, Krämer J. Co-transfer of two plasmids determining bacteriocin production and sucrose utilization in Streptococcus faecium. FEMS Microbiol Lett. 1984;23:147–150. [Google Scholar]
- 30.Ruiz-Barba J L, Piard J C, Jimènez-Díaz R. Plasmid profiles and curing of plasmids in Lactobacillus plantarum strains isolated from green olive fermentations. J Appl Bacteriol. 1991;71:417–421. doi: 10.1111/j.1365-2672.1991.tb03810.x. [DOI] [PubMed] [Google Scholar]
- 31.Sahl H-G, Bierbaum G. Lantibiotics: biosynthesis and biological activities of uniquely modified peptides from Gram-positive bacteria. Annu Rev Microbiol. 1998;52:41–79. doi: 10.1146/annurev.micro.52.1.41. [DOI] [PubMed] [Google Scholar]
- 32.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 33.Schägger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 34.Tomita H, Fujimoto S, Tanimoto K, Ike Y. Cloning and genetic organization of the bacteriocin 31 determinant encoded on the Enterococcus faecalis pheromone-responsive conjugative plasmid pYI17. J Bacteriol. 1996;178:3585–3593. doi: 10.1128/jb.178.12.3585-3593.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tomita H, Fujimoto S, Tanimoto K, Ike Y. Cloning and genetic sequence analysis of the bacteriocin 21 determinant encoded on the Enterococcus faecalis pheromone-responsive conjugative plasmid pPD1. J Bacteriol. 1997;179:7843–7855. doi: 10.1128/jb.179.24.7843-7855.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Van Belkum M J, Hayema B J, Jeeninga R E, Kok J, Venema G. Organization and nucleotide sequences of two lactococcal bacteriocin operons. Appl Environ Microbiol. 1991;57:492–498. doi: 10.1128/aem.57.2.492-498.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Van Reenen C A, Dicks L M T. Evaluation of numerical analysis of random amplified polymorphic DNA (RAPD)-PCR as a method to differentiate Lactobacillus plantarum and Lactobacillus pentosus. Curr Microbiol. 1996;32:183–187. doi: 10.1007/s002849900033. [DOI] [PubMed] [Google Scholar]
- 38.Van Reenen C A, Dicks L M T, Chikindas M, Verellen T L J, Vandamme E J. Isolation, purification and partial characterization of plantaricin 423, a bacteriocin produced by Lactobacillus plantarum. J Appl Microbiol. 1998;84:1131–1137. doi: 10.1046/j.1365-2672.1998.00451.x. [DOI] [PubMed] [Google Scholar]
- 39.Venema K, Abee T, Haandrikman A J, Leenhouts K J, Kok J, Konings W N, Venema G. Mode of action of lactococcin B, a thiol-activated bacteriocin from Lactococcus lactis. Appl Environ Microbiol. 1993;59:1041–1048. doi: 10.1128/aem.59.4.1041-1048.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Villani F, Salzano G, Sorrentino E, Pepe O, Marino P, Coppola S. Enterocin 226NWC, a bacteriocin produced by Enterococcus faecalis 226, active against Listeria monocytogenes. J Appl Bacteriol. 1993;74:380–387. doi: 10.1111/j.1365-2672.1993.tb05142.x. [DOI] [PubMed] [Google Scholar]