Abstract
Unwinding of the replication origin and loading of DNA helicases underlie the initiation of chromosomal replication. In Escherichia coli, the minimal origin oriC contains a duplex unwinding element (DUE) region and three (Left, Middle, and Right) regions that bind the initiator protein DnaA. The Left/Right regions bear a set of DnaA-binding sequences, constituting the Left/Right-DnaA subcomplexes, while the Middle region has a single DnaA-binding site, which stimulates formation of the Left/Right-DnaA subcomplexes. In addition, a DUE-flanking AT-cluster element (TATTAAAAAGAA) is located just outside of the minimal oriC region. The Left-DnaA subcomplex promotes unwinding of the flanking DUE exposing TT[A/G]T(T) sequences that then bind to the Left-DnaA subcomplex, stabilizing the unwound state required for DnaB helicase loading. However, the role of the Right-DnaA subcomplex is largely unclear. Here, we show that DUE unwinding by both the Left/Right-DnaA subcomplexes, but not the Left-DnaA subcomplex only, was stimulated by a DUE-terminal subregion flanking the AT-cluster. Consistently, we found the Right-DnaA subcomplex–bound single-stranded DUE and AT-cluster regions. In addition, the Left/Right-DnaA subcomplexes bound DnaB helicase independently. For only the Left-DnaA subcomplex, we show the AT-cluster was crucial for DnaB loading. The role of unwound DNA binding of the Right-DnaA subcomplex was further supported by in vivo data. Taken together, we propose a model in which the Right-DnaA subcomplex dynamically interacts with the unwound DUE, assisting in DUE unwinding and efficient loading of DnaB helicases, while in the absence of the Right-DnaA subcomplex, the AT-cluster assists in those processes, supporting robustness of replication initiation.
Abbreviations: BSA, bovine serum albumin; ChiDnaA, chimeric DnaA; DOR, DnaA oligomerization region; DUE, duplex unwinding element; EcoDnaA, Escherichia coli DNA; FL, full-length; ss, single-stranded; TmaDnaA, Thermotoga maritima DnaA
The initiation of bacterial DNA replication requires local duplex unwinding of the chromosomal replication origin oriC, which is regulated by highly ordered initiation complexes. In Escherichia coli, the initiation complex contains oriC, the ATP-bound form of the DnaA initiator protein (ATP–DnaA), and the DNA-bending protein IHF (Fig. 1, A and B), which promotes local unwinding of oriC (1, 2, 3, 4). Upon this oriC unwinding, two hexamers of DnaB helicases are bidirectionally loaded onto the resultant single-stranded (ss) region with the help of the DnaC helicase loader (Fig. 1B), leading to bidirectional chromosomal replication (5, 6, 7, 8). However, the fundamental mechanism underlying oriC-dependent bidirectional DnaB loading remains elusive.
Figure 1.
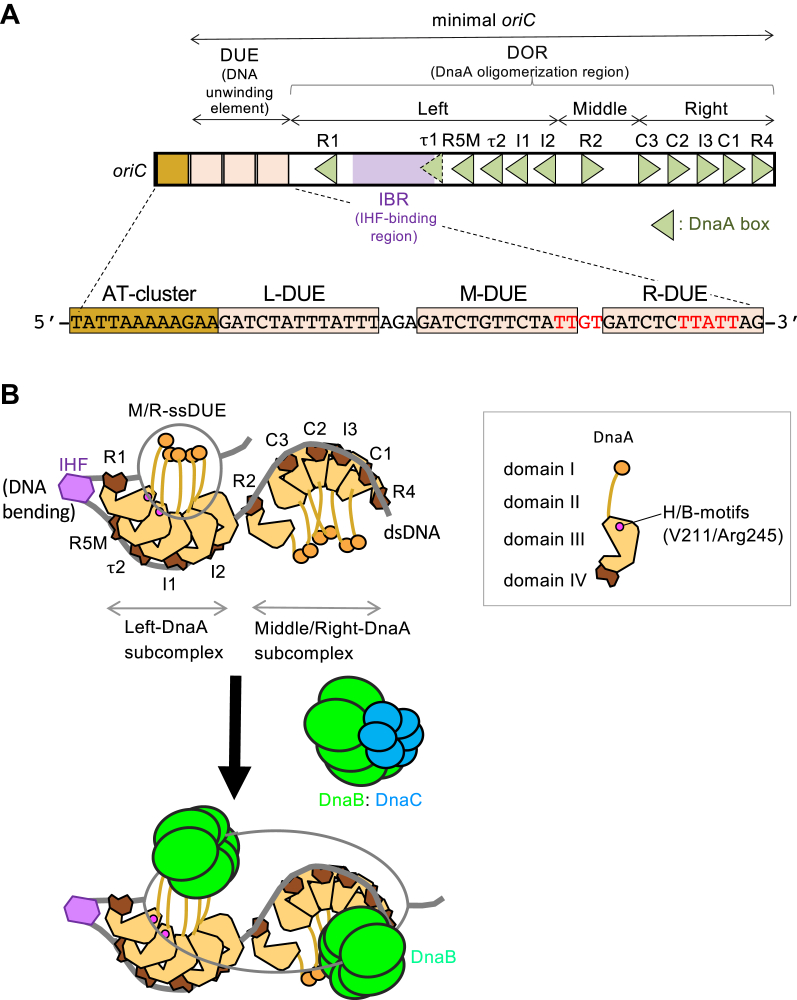
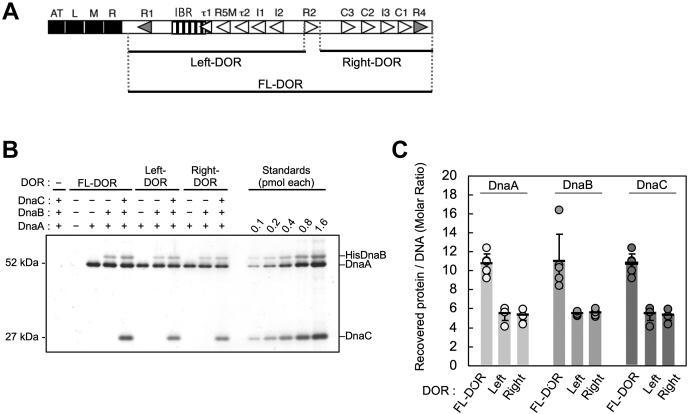
Schematic structures of oriC, DnaA, and the initiation complexes. A, the overall structure of oriC. The minimal oriC region and the AT-cluster region are indicated. The sequence of the AT-cluster−DUE (duplex-unwinding element) region is also shown below. The DUE region (DUE; pale orange bars) contains three 13-mer repeats: L-DUE, M-DUE, and R-DUE. DnaA-binding motifs in M/R-DUE, TT(A/G)T(T), are indicated by red characters. The AT-cluster region (AT cluster; brown bars) is flanked by DUE outside of the minimal oriC. The DnaA-oligomerization region (DOR) consists of three subregions called Left-, Middle-, and Right-DOR. B, model for replication initiation. DnaA is shown as light brown (for domain I–III) and darkbrown (for domain IV) polygons (right panel). ATP–DnaA forms head-to-tail oligomers on the Left- and Right-DORs (left panel). The Middle-DOR (R2 box)-bound DnaA interacts with DnaA bound to the Left/Right-DORs using domain I, but not domain III, stimulating DnaA assembly. IHF, shown as purple hexagons, bends DNA >160° and supports DUE unwinding by the DnaA complexes. M/R-DUE regions are efficiently unwound. Unwound DUE is recruited to the Left-DnaA subcomplex and mainly binds to R1/R5M-bound DnaA molecules. The sites of ssDUE-binding B/H-motifs V211 and R245 of R1/R5M-bound DnaA molecules are indicated (pink). Two DnaB homohexamer helicases (light green) are recruited and loaded onto the ssDUE regions with the help of the DnaC helicase loader (cyan). ss, single stranded.
The minimal oriC region consists of the duplex unwinding element (DUE) and the DnaA oligomerization region (DOR), which contains specific arrays of 9-mer DnaA-binding sites (DnaA boxes) with the consensus sequence TTA[T/A]NCACA (Fig. 1A) (3, 4). The DUE underlies the local unwinding and contains 13-mer AT-rich sequence repeats named L-, M-, and R-DUE (9). The M/R-DUE region includes TT[A/G]T(A) sequences with specific affinity for DnaA (10). In addition, a DUE-flanking AT-cluster (TATTAAAAAGAA) region resides just outside of the minimal oriC (Fig. 1A) (11). The DOR is divided into three subregions, the Left-, Middle-, and Right-DORs, where DnaA forms structurally distinct subcomplexes (Fig. 1A) (8, 12, 13, 14, 15, 16, 17). The Left-DOR contains high-affinity DnaA box R1, low-affinity boxes R5M, τ1−2, and I1-2, and an IHF-binding region (17, 18, 19, 20). The τ1 and IHF-binding regions partly overlap (17).
In the presence of IHF, ATP–DnaA molecules cooperatively bind to R1, R5M, τ2, and I1-2 boxes in the Left-DOR, generating the Left-DnaA subcomplex (Fig. 1B) (8, 17). Along with IHF causing sharp DNA bending, the Left-DnaA subcomplex plays a leading role in DUE unwinding and subsequent DnaB loading. The Middle-DOR contains moderate-affinity DnaA box R2. Binding of DnaA to this box stimulates DnaA assembly in the Left- and Right-DORs using interaction by DnaA N-terminal domain (Fig. 1B; also see below) (8, 12, 14, 16, 21). The Right-DOR contains five boxes (C3-R4 boxes) and cooperative binding of ATP–DnaA molecules to these generates the Right-DnaA subcomplex (Fig. 1B) (12, 18). This subcomplex is not essential for DUE unwinding and plays a supportive role in DnaB loading (8, 15, 17). The Left-DnaA subcomplex interacts with DnaB helicase, and the Right-DnaA subcomplex has been suggested to play a similar role (Fig. 1B) (8, 13, 16).
In the presence of ATP–DnaA, M- and R-DUE adjacent to the Left-DOR are predominant sites for in vitro DUE unwinding: unwinding of L-DUE is less efficient than unwinding of the other two (Fig. 1B) (9, 22, 23). Deletion of L-DUE or the whole DUE inhibits replication of oriC in vitro moderately or completely, respectively (23). A chromosomal oriC Δ(AT-cluster−L-DUE) mutant with an intact DOR, as well as deletion of Right-DOR, exhibits limited inhibition of replication initiation, whereas the synthetic mutant combining the two deletions exhibits severe inhibition of cell growth (24). These studies suggest that AT-cluster−L-DUE regions stimulate replication initiation in a manner concerted with Right-DOR, although the underlying mechanisms remain elusive.
DnaA consists of four functional domains (Fig. 1B) (4, 25). Domain I supports weak domain I–domain I interaction and serves as a hub for interaction with various proteins such as DnaB helicase and DiaA, which stimulates ATP–DnaA assembly at oriC (26, 27, 28, 29, 30). Two or three domain I molecules of the oriC–DnaA subcomplex bind a single DnaB hexamer, forming a stable higher-order complex (7). Domain II is a flexible linker (28, 31). Domain III contains AAA+ (ATPase associated with various cellular activities) motifs essential for ATP/ADP binding, ATP hydrolysis, and DnaA–DnaA interactions in addition to specific sites for ssDUE binding and a second, weak interaction with DnaB helicase (1, 4, 8, 10, 19, 25, 32, 33, 34, 35). Domain IV bears a helix-turn-helix motif with specific affinity for the DnaA box (36).
As in typical AAA+ proteins, a head-to-tail interaction underlies formation of ATP–DnaA pentamers on the DOR, where the AAA+ arginine-finger motif Arg285 recognizes ATP bound to the adjacent DnaA protomer, promoting cooperative ATP–DnaA binding (Fig. 1B) (19, 32). DnaA ssDUE-binding H/B-motifs (Val211 and Arg245) in domain III sustain stable unwinding by directly binding to the T-rich (upper) strand sequences TT[A/G]T(A) within the unwound M/R-DUE (Fig. 1B) (8, 10). Val211 residue is included in the initiator-specific motif of the AAA+ protein family (10). For DUE unwinding, ssDUE is recruited to the Left-DnaA subcomplex via DNA bending by IHF and directly interacts with H/B-motifs of DnaA assembled on Left-DOR, resulting in stable DUE unwinding competent for DnaB helicase loading; in particular, DnaA protomers bound to R1 and R5M boxes play a crucial role in the interaction with M/R-ssDUE (Fig. 1B) (8, 10, 17). Collectively, these mechanisms are termed ssDUE recruitment (4, 17, 37).
Two DnaB helicases are thought to be loaded onto the upper and lower strands of the region including the AT-cluster and DUE, with the aid of interactions with DnaC and DnaA (Fig. 1B) (25, 38, 39). DnaC binding modulates the closed ring structure of DnaB hexamer into an open spiral form for entry of ssDNA (40, 41, 42, 43). Upon ssDUE loading of DnaB, DnaC is released from DnaB in a manner stimulated by interactions with ssDNA and DnaG primase (44, 45). Also, the Left- and Right-DnaA subcomplexes, which are oriented opposite to each other, could regulate bidirectional loading of DnaB helicases onto the ssDUE (Fig. 1B) (7, 8, 35). Similarly, recent works suggest that the origin complex structure is bidirectionally organized in both archaea and eukaryotes (1, 46). In Saccharomyces cerevisiae, two origin recognition complexes containing AAA+ proteins bind to the replication origin region in opposite orientations; this, in turn, results in efficient loading of two replicative helicases, leading to head-to-head interactions in vitro (46). Consistent with this, origin recognition complex dimerization occurs in the origin region during the late M-G1 phase (47). The fundamental mechanism of bidirectional origin complexes might be widely conserved among species.
In this study, we analyzed various mutants of oriC and DnaA in reconstituted systems to reveal the regulatory mechanisms underlying DUE unwinding and DnaB loading. The Right-DnaA subcomplex assisted in the unwinding of oriC, dependent upon an interaction with L-DUE, which is important for efficient loading of DnaB helicases. The AT-cluster region adjacent to the DUE promoted loading of DnaB helicase in the absence of the Right-DnaA subcomplex. Consistently, the ssDNA-binding activity of the Right-DnaA subcomplex sustained timely initiation of growing cells. These results indicate that DUE unwinding and efficient loading of DnaB helicases are sustained by concerted actions of the Left- and Right-DnaA subcomplexes. In addition, loading of DnaB helicases are sustained by multiple mechanisms that ensure robust replication initiation, although the complete mechanisms are required for precise timing of initiation during the cell cycle.
Results
The L-DUE regions stimulate DUE unwinding dependent upon Right-DOR
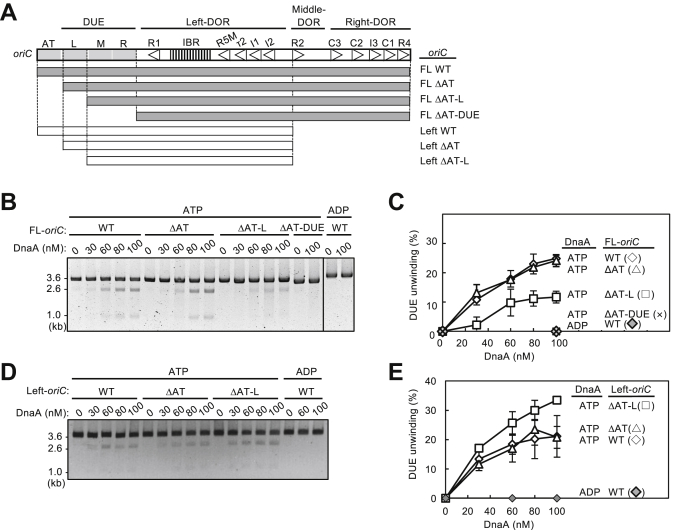
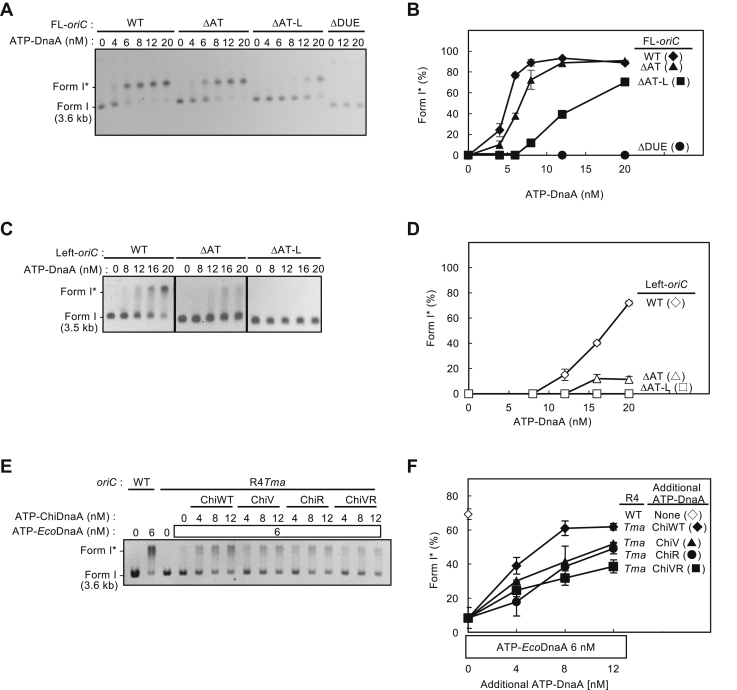
To investigate the regulatory roles of the oriC Right-DOR and AT-cluster region in DUE unwinding, we combined a series of plasmids carrying full-length (FL) or Middle/Right-DOR–deleted (Left) oriC with deletions in the AT-cluster–DUE subregions and assessed unwinding activities by P1 nuclease assay (Fig. 2A). In FL-oriC, deletion of the whole AT-cluster–DUE region (FL ΔAT-DUE) totally abolished the unwinding activity, whereas deletion of the AT-cluster region (FL ΔAT) preserved the activity (Fig. 2, B and C). In addition, deletion of AT-cluster–L-DUE (FL ΔAT-L) moderately inhibited unwinding. These results are consistent with previously reported data and with the idea that L-DUE is an auxiliary unwinding region (9, 22, 23).
Figure 2.
Deletion analysis for oriC substructures in DUE unwinding activity.A, structures of oriC variants. The DNA regions included are shown by bars under the oriC structure. Specific motifs are indicated according to Figure 1. AT; AT-cluster. B–E, DUE unwinding assays using oriC variants. The indicated amounts of ATP/ADP–DnaA were incubated for 3 min at 38 °C with IHF (32 nM) and plasmid (3.4 nM) bearing FL-oriC variants (B and C) or Left-oriC variants (D and E), followed by serial incubations with P1 nuclease and AlwNI restriction enzyme. DNA fragments were analyzed by 1% agarose gel electrophoresis. Two independent experiments were carried out, and a representative gel image in black/white-inverted mode is shown (B and D). Percentages of P1 nuclease-digested oriC DNA molecules per the input DNA are shown as “DUE unwinding (%)”. Mean values with SDs (n = 2) are also shown (C and E). DUE, duplex unwinding element; FL, full length.
Notably in Left-oriC, deletion of the AT-cluster–L-DUE (Left ΔAT-L) preserved full unwinding activity (Fig. 2, D and E). These results suggest that the stimulatory effect of the L-DUE depends on the Right-DnaA subcomplex, whereas the Left-DnaA subcomplex interacts with M/R-DUE elements to facilitate DUE unwinding. This is consistent with the presence of DnaA-binding TT[A/G]T(T) motifs within M/R-DUE and with the ssDUE recruitment mechanism (10).
Right-DnaA subcomplex binds ssDUE
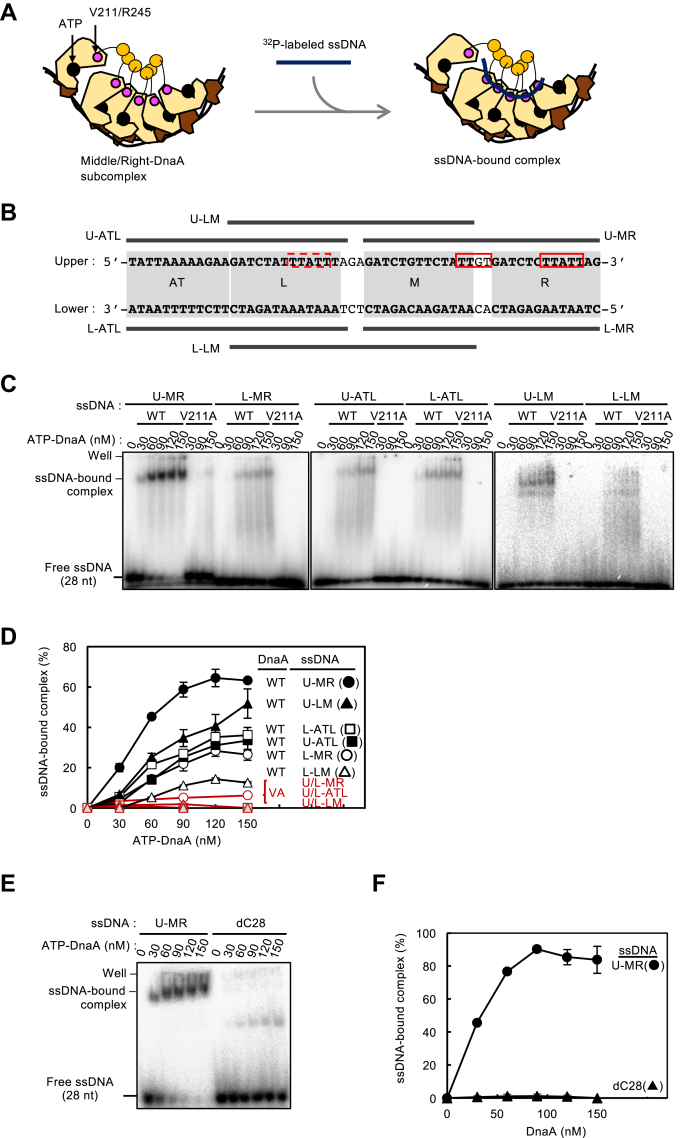
ATP–DnaA complexes constructed on the FL- or Left-oriC sustain ssDUE-binding activity in a manner dependent on the DnaA AAA+ domain H/B-motifs (Val211 and Arg245). Here, we addressed the specificity of ssDNA binding to the Right-DnaA subcomplex. ATP–DnaA WT and ATP–DnaA V211A constructed similar complexes with a Middle/Right-DOR DNA fragment containing DnaA boxes R2–R4 (Fig. S1, A and B). The resultant Middle/Right-DnaA subcomplexes were further incubated with a series of 32P-labeled ssDUE fragments, and formation of the ternary complexes composed of ATP–DnaA, Middle/Right-DOR, and ssDUE was assessed by EMSA (Fig. 3A). The U-MR ssDUE carried the upper strand of the 28-mer M/R-DUE sequence, which includes the specific DnaA-binding sequences TTGT and TTATT (Fig. 3B). Our previous study demonstrated that ATP–DnaA complexes constructed on WT DOR binds to this U-MR region most efficiently in DUE subregions (10). Further analyses suggested that R1/R5M-bound DnaA protomers within the Left-DnaA subcomplex bind to the U-MR sequence, stabilizing ssDUE–DnaA–DOR ternary complexes (17).
Figure 3.
The Right-DnaA complex binds ssDUE. A, schematic of EMSA for assessing ssDUE recruitment using Middle/Right-DOR. Modeled structure of DnaA is shown according to Figure 1. Positions of ATP and Val211/Arg245 residues are also shown. B, sequences of both upper and lower strands of ssDUE used in this study and their positions in the DUE. The AT-cluster region and L, M, and R 13-mers are indicated by bold capital letters and gray boxes, respectively. DnaA-binding sequence [TT[A/G]T(T)] in the M-R regions are indicated by red boxes. Similar sequence in the L region is also indicated by a broken-line red box. The oligonucleotides including ssDUE regions (U-MR, L-MR, U-LM, L-LM, U-ATL, and L-ATL) are indicated by black bars. C and D, EMSA with Middle/Right-DOR DNA in the presence of 32P-labeled ssDNA fragments and ATP–DnaA. Indicated amounts of ATP–DnaA WT or V211A were incubated for 5 min at 4 °C with the Middle/Right-DOR DNA (35 nM), followed by further incubation for 10 min at 30 °C with or without each ssDNA (16 nM). A representative gel image is shown (C). The amounts of ssDUE bound to DOR–DnaA complexes were quantified as “ssDNA-bound complex (%)” (D). Mean values with SDs (n = 2) are shown. VA, V211A. E and F, the binding activity to Middle/Right-DOR–DnaA complexes are also analyzed using U-MR and nonspecific ssDNA (dC28), 28-mer oligo-dC. A representative gel image is shown (E) and the mean amounts of U-MR or dC28 bound to the Middle/Right DOR–DnaA complexes were shown as with SDs (n = 2) (F). DUE, duplex unwinding element; DOR, DnaA oligomerization region; ss, single stranded.
Similar to ATP–DnaA–WT-DOR complexes, ATP–DnaA–Middle/Right-DOR subcomplexes efficiently bound to the U-MR ssDUE in a manner dependent upon DnaA V211 (Fig. 3, C and D). By contrast, when we used ssDNA containing the upper or lower strands of AT-cluster–L-DUE (U-ATL or L-ATL, respectively) and the upper strand of L/M-DUE (U-LM), we observed moderate binding that depended on V211 (Fig. 3, C and D). The relatively higher affinity for U-LM might be due to the presence of TTATT sequences in the L-DUE region (Fig. 3B). The A stretch in U-ATL could slightly inhibit the TTATT sequences in DnaA binding by altering the ssDNA secondary structure. In contrast to U-LM, the lower strand of L/M-DUE (L-LM) exhibited only faint and unstable binding (Fig. 3, C and D). These experiments included higher concentrations of ssDUE than our previous experiments (10), which enabled detection of these moderate binding activities. In addition, nonspecific ssDNA, 28-mer oligo-dC (dC28), did not substantially bind to the Middle/Right-DOR–DnaA complexes: dC28-specific faint binding signals could be caused by nonspecific binding of dsDNA-free DnaA molecules (Figs. 3, E and F and S1, C and D).
Role for DnaA box R4–bound DnaA in ssDUE binding
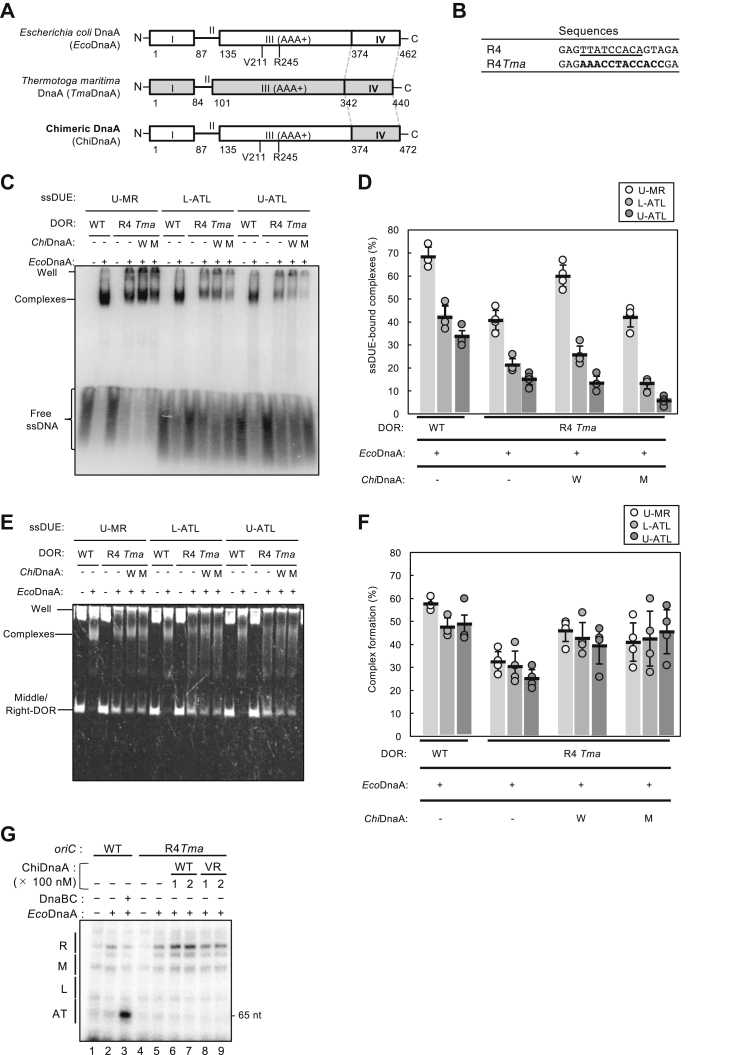
To further address the mechanism of ssDUE binding to the Right-DnaA subcomplex, we used chimeric DnaA (ChiDnaA) in which E. coli DnaA (EcoDnaA) domains I–III were integrated with Thermotoga maritima DnaA homolog (TmaDnaA) domain IV (Fig. 4A). Domain III of EcoDnaA bears ssDUE binding H/B-motifs, Val211 and Arg245. As described previously (15, 16, 17, 48), TmaDnaA has an alternative 12-mer DnaA-binding sequence, AAACCTACCACC, termed the TmaDnaA box (Fig. 4B). Thus, ChiDnaA specifically binds to the TmaDnaA box but not to the EcoDnaA box.
Figure 4.
Role for DnaA box R4 in ssDUE bindingby Middle/Right-DOR–DnaA complex. A, basic structure of ChiDnaA. Schematic structures of Escherichia coli DnaA (EcoDnaA), Thermotoga maritima DnaA (TmaDnaA), and ChiDnaA are shown. Domains of EcoDnaA and TmaDnaA are indicated by open and gray boxes, respectively. B, substitution of DnaA box R4. Sequences of original oriC R4 box (underline) and substituted TmaDnaA box (bold letters) are shown. In R4Tma: oriC, the TmaDnaA box replaces the R4 box. C–F, EMSA using the Middle/Right-DOR with or without substitution of the TmaDnaA box for the R4 box. Middle/Right-DOR WT or R4Tma (35 nM) was incubated for 5 min on ice in the presence (120 nM) (+) or absence (−) of ATP-EcoDnaA, ATP-ChiDnaA WT (W), or ATP–ChiDnaA V211A/R245A (M), followed by further incubation for 10 min at 30 °C with 32P-labeled ssDNA U-MR, U-ATL, or L-ATL (16 nM) in buffer including 100 mM potassium glutamate (see Experimental procedures). Resultant complexes were analyzed by radioactive imaging or GelStar staining, respectively (C and E). The amounts of ssDNA bound to the DnaA–DOR complexes were quantified as “ssDNA-bound complex (%)”(D). Band intensities of Middle/Right-DOR–DnaA complexes were quantified as “Complex formation (%)” (F). Three to four independent experiments were carried out, and each data and mean values with SDs (n = 3–4) are shown in the graphs. When EcoDnaA and the DORs bearing the R4Tma substitution were coincubated, abnormal complexes such as aggregates may have formed, including λ DNA, resulting in material remaining in the gel wells. G, KMnO4 modification experiments. Plasmid bearing oriC (5 nM) with the WT R4 box (WT) or the substituted TmaDnaA box sequence (R4Tma) were incubated for 10 min at 37 °C with IHF (100 nM) in the presence (+) or absence (−) of EcoDnaA (100 nM), ChiDnaA WT or V211A/R245A (VR) (100 or 200 nM), DnaB K236A (300 nM), and DnaC (300 nM), followed by further incubation with 10 mM KMnO4. Modified DNAs were analyzed by primer extension and 7.5% denaturing mini-gel PAGE. Sequencing reactions were performed using the same primer, and determined positions of sequence motifs (L, M, R, and AT) are shown using the oriC WT KMnO4 modification data (Fig. S3). Similar results were shown in repeated experiments. DUE, duplex unwinding element; ChiDnaA, chimeric DnaA; EcoDnaA, Escherichia coli DNA; TmaDnaA, Thermotoga maritima DnaA; ss, single stranded.
To assess the requirement of R4-bound DnaA in ssDUE binding, we replaced DnaA box R4 with the TmaDnaA box sequence (R4Tma) in the Middle/Right-DOR DNAs (Fig. 4, C–G). The binding activities of ssDNA U-MR, U-ATL, or L-ATL were analyzed by EMSA using these DOR DNAs, EcoDnaA WT, and ChiDnaA WT or V211A/R245A double mutant. In the presence of EcoDnaA WT and ChiDnaA WT, these ssDUEs formed complexes with the Middle/Right-DOR–R4Tma DNAs at levels comparable to EcoDnaA WT with the WT forms of the DOR DNAs (Fig. 4, C and D). In the absence of ChiDnaA, the ssDUE-binding levels decreased moderately. Residual EcoDnaA binding to the Middle/Right-DOR–R4Tma DNAs might support this interaction, moderately stabilizing the tertiary complexes (Fig. 4, E and F) (15). In the presence of ssDUEs, formation of Middle/Right-DOR–DnaA complexes were similar except for the case using Middle/Right-DOR–R4Tma in the absence of ChiDnaA (Fig. 4, E and F). Binding of the ssDUE was similarly moderately reduced using the ChiDnaA mutant, suggesting that DnaA bound at R4 plays a supporting role in ssDUE binding in these DnaA subcomplexes (Fig. 4, C and D). Basically, similar results were shown using Right-DOR (Fig. S2): changes in the band shapes of free ssDNA could be caused by difference of potassium salts included in reaction buffer (see Experimental procedures).
In addition, we performed KMnO4 modification experiments to further analyze the role of DnaA box R4-bound DnaA in DUE unwinding. Pyrimidine residues (mainly thymine) within stably unwound DNA regions or ssDNA can be specifically oxidized by KMnO4, which can be detected by primer extension experiments. Here, the lower strand of AT-cluster–DUE regions was analyzed (Figs. 4G and S3). In WT oriC, unwinding of R-DUE was detected in the presence of IHF and EcoDnaA (Fig. 4G, lanes one and 2). The ATPase-defective DnaB helicase mutant K236A can engage in loading onto unwound oriC but is inactive with respect to migration on ssDNA (39). Addition of DnaB K236A and DnaC induced unwinding of the AT-cluster region (Fig. 4G, lane 3), consistent with previous reports (39, 49) (see also below). In the oriC R4Tma construct, EcoDnaA moderately induced R-DUE unwinding, possibly with the support of the Left-DnaA subcomplex. Further addition of ChiDnaA WT stimulated unwinding (Fig. 4G, lanes 4–7), whereas addition of the ChiDnaA VR mutant did not (Fig. 4G, lanes eight and 9), suggesting that ssDUE binding of R4-bound DnaA stabilizes the unwound state of DUE. In P1 nuclease assays, reaction times with the nuclease are very short (e.g., 200 s), which might make it difficult to distinguish changes in stability (see Discussion). Taken together, these findings suggest that Right-DOR plays an assistive role in stable DUE unwinding.
Role for the Right-DOR in binding DnaB helicase
Previously, we demonstrated that DnaA complexes constructed on the DOR stably bind DnaB helicases, although DnaA monomers cannot form a stable complex with DnaB helicase (8, 30). DnaA complexes have multiple binding sites for the homohexamer DnaB helicase, and binding is dramatically stimulated in a cooperative manner (28, 30). We revealed that a patch including Glu21 and Phe46 of DnaA domain I constitutes the primary DnaB-binding site (28, 30). DnaB Leu160, located on the lateral surface of this helicase, was suggested to interact with DnaA Phe46 (7). In addition, our previous pull-down experiments suggest that the DnaA complexes constructed on the FL-DOR bind two DnaB helicases, whereas those on the Left-DOR bind only a single DnaB helicase (8). Hence, we directly investigated the role of the Right-DnaA subcomplex in the binding activity of DnaB helicase.
We performed pull-down experiments using FL-, Left-, and Right-DOR (Fig. 5A). Consistent with the previous results (7, 8, 30), FL- and Left-DOR bound about ten and five monomeric DnaB molecules per DOR, respectively, in the presence of DnaC (Fig. 5, B and C). Considering experimental loss of proteins during washing steps, these numbers are consistent with the idea that one or two DnaB hexamers bound to the Left- or FL-DOR, respectively. In the case of Right-DOR, recovery of DnaB was comparable to that of Left-DOR (Fig. 5, B and C), supporting the idea that the Right-DnaA subcomplex binds a single DnaB helicase or DnaB–DnaC complex independently of Left-DOR. DnaC moderately promoted the recovery of DnaB, consistent with previous results for FL-DOR (7, 30). In these experiments, the specificity of DnaA binding to DnaB was supported by the observation that DnaA B/H-motifs were dispensable for DnaB binding to the Right-DOR (Fig. S4), whereas DnaA residue Phe46 was essential (Fig. S5).
Figure 5.
Right-DnaA subcomplex binds DnaBC. A, biotinylated DOR fragments are shown by bars below the oriC structure. For symbols in the oriC structure, see Figure 1A. B, ATP–DnaA (10 pmol: 0.4 μM), His-DnaB (10 pmol as monomer), and DnaC (10 pmol) were incubated in the presence or absence (−) of biotinylated FL-, Left- or Right-DOR fragment (250 fmol, 10 nM) (Fig. 2A), followed by pull-down assay. Proteins bound to DNA were analyzed by SDS–11% PAGE and silver staining. +, presence; −. absence. C, amounts of recovered proteins (DnaA, DnaB, and DnaC) were determined using standard curves, and values from the negative control excluding the DOR were subtracted. About 150 fmol DNA was recovered. The means and SDs of the numbers of DnaA, DnaB, and DnaC molecules recovered per DNA are indicated by light-to-dark graybars, respectively. Five independent experiments were carried out, and the mean values with SDs (n = 5) are shown in the graphs with each data as corresponding color dots. DOR, DnaA oligomerization region; FL, full length.
DnaB helicase loading is redundantly supported by the Right-DOR and AT-cluster–L-DUE regions
Next, to clarify the oriC elements involved in DnaB loading, we assessed DnaB loading activities by form I∗ assay using a series of oriC deletion mutants. In this in vitro reconstituted assay, DnaB loading and helicase activity on the unwound oriC of supercoiled circular DNA (form I) causes positive supercoiling, which is resolved by DNA gyrase and produces plasmid DNA with a higher supercoiling state, called form I∗ (35, 43, 50). Form I∗ migrates in agarose gel electrophoresis at a different rate from form I, which is faster or slower (35, 43, 50). This could be affected by product quality of agarose, conditions of electrophoresis, or formation of specific sequence-specific structures such as hairpins and cruciforms which can be stimulated by higher supercoiling states of DNA. Whether it is faster or slower, the meaning is the same in indication of DnaB loading.
In FL-oriC derivatives, deletion of the AT-cluster–L-DUE regions, but not the AT-cluster region alone, moderately inhibited the formation of form I∗ (Fig. 6, A and B), consistent with the results obtained from the DUE-unwinding assay (Fig. 2, B and C). Even at the saturation level of DnaA (20 nM) for FL-oriC WT, formation form I∗ by oriC ΔAT–L was only moderate.
Figure 6.
DnaB loading activity of Right-DnaA subcomplex. Form I∗ assay of FL-oriC (A and B), Left-oriC (C and D), and FL-oriC R4Tma variants (E and F). For DNA regions, see Figure 2. Representative gel images are shown in black/white-inverted mode, and DNA migration positions of form I and form I∗ are indicated. The amount of form I∗ relative to the input DNA, quantified as “Form I∗ (%)”, is shown on the right panel. Indicated amounts of ATP–DnaA (ATP–EcoDnaA) or mixtures with ATP–ChiDnaA were incubated for 15 min at 30 °C in the presence of DnaB, DnaC, IHF, gyrase, and SSB with plasmid bearing FL-oriC WT or AT-cluster–DUE mutants (A), Left-oriC WT or AT-cluster–DUE mutants (C), or FL-oriC WT or mutant with the R4Tma substitution (E). Two independent experiments were carried out, and a representative gel image in a black/white-inverted mode is shown. (B, D, and E) Percentages of form I∗ molecules per total DNA are shown as “Form I∗ (%)”, quantified from Figure 6, A, C, and E, respectively. Mean values with SDs (n = 2) are also shown. ChiDnaA, chimeric DnaA; DUE, duplex unwinding element; EcoDnaA, Escherichia coli DNA; FL, full length.
Next, we used a similar approach to analyze deletion mutants using Left-oriC plasmids. As previously suggested (8), Left-oriC basically sustains form I∗ formation activity but requires a higher concentration of DnaA for full activity than FL-oriC. Notably, form I∗ formation was severely inhibited in Left-oriC ΔAT and completely inhibited in Left-oriC ΔAT–L (Fig. 6, C and D), even though these oriC plasmids fully retained DUE-unwinding activity (Fig. 2, D and E). Taken together, these results can be explained by the functionally redundant role of the Right-DOR and AT-cluster–L-DUE regions in facilitating DnaB loading. In the absence of the Right-DnaA subcomplex, the AT-cluster region is inferred to support DnaB loading by stimulating DNA unwinding by heat and superhelicity, which is assisted by the L-DUE region.
To directly investigate the role of the R4-bound DnaA protomer in DnaB loading, we performed the form I∗ assay using plasmids bearing oriC R4Tma derivatives (Fig. 6, E and F). In oriC R4Tma, EcoDnaA alone could not produce a substantial amount of form I∗, whereas addition of ChiDnaA induced efficient formation of form I∗ at a level comparable to that induced by oriC-WT with EcoDnaA. By contrast, introduction of B/H-motif mutants (V, R, or VR) in ChiDnaA moderately decreased the formation of form I∗ relative to that achieved by ChiDnaA WT. These results are consistent with the results described above (Figs. 6, A–D and 4D) and suggest that the Right-DnaA subcomplex interacts with ssDUE to stimulate stable unwinding and efficient DnaB loading.
Right-DnaA subcomplexes stimulate DnaB loading and stable unwinding of the AT-cluster region
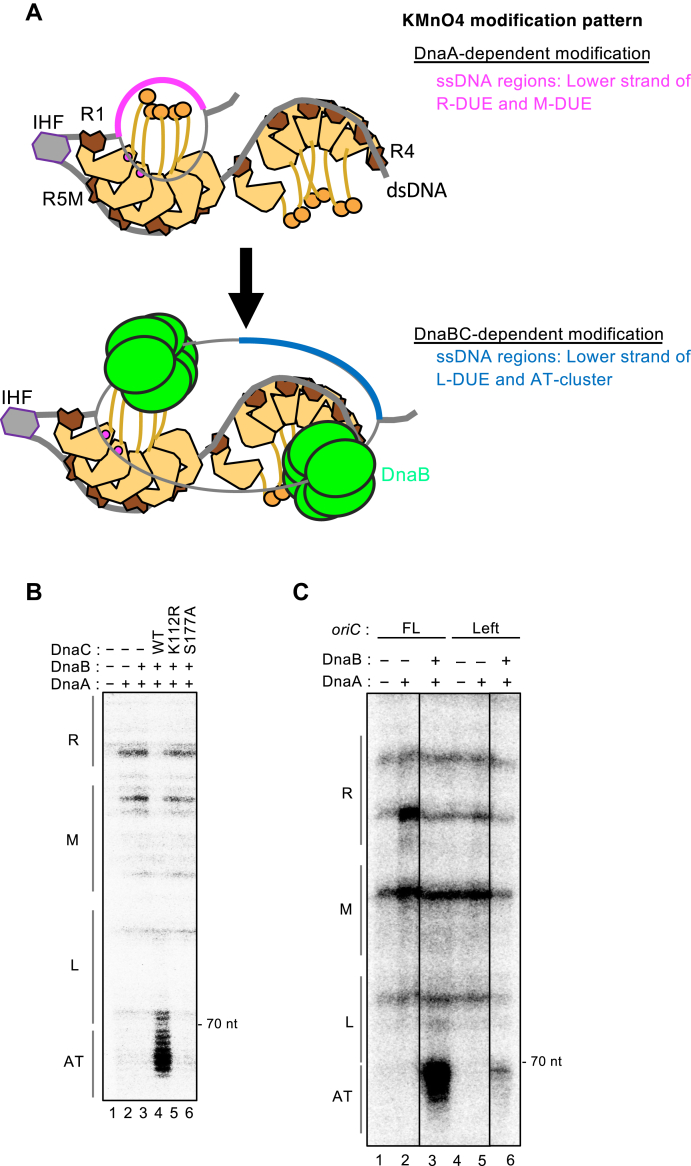
In previous studies, unwinding of the AT-cluster region was detected by KMnO4 modification and primer extension experiments using DnaB K236A, DnaA, and DnaC (39, 49). This reaction was similarly assessed using the Left-oriC variant. DnaB loaded on the lower strand is thought to become a physical obstacle inhibiting modification of the lower-strand M/R-DUE (Fig. 7A) (39). Another upper strand–loaded DnaB may become a physical obstacle to maintain expansion of unwound regions into the AT-cluster region (39). Consistent with this, both site-specific protection and enhancement of modification in the lower strand were detected in the presence of DnaA, DnaB K236A, and DnaC WT (Fig. 7B, lane 4), but not in the absence of either DnaB or DnaC (Fig. 7, lanes two and 3). Next, to assess the requirement of DnaC-dependent changes of DnaB complexes, we used two DnaC AAA+ domain mutants: K112R in the Walker A motif and S177A in the ssDNA-interaction site (which corresponds to the DnaA H-motif). Purified DnaC K112R and S177A mutant proteins sustained DnaB interaction (Fig. S6) but were defective in DNA-dependent ATPase (Figs. 7B and S6), consistent with a previous study using DnaC S177D mutant (42). In contrast to DnaC WT, unwinding of the AT-cluster region was totally impaired in the presence of the DnaC K112R or S177A mutant (Fig. 7B, lanes five and 6), supporting the idea that the unwound state of the AT-cluster region is stabilized by DnaB loaded onto the ssDUE regions. Similar results were reported previously for DnaC K112R (49).
Figure 7.
Unwinding of the AT-cluster region is stimulated by loaded DnaB and the Right-DnaA subcomplex. A, schematic of KMnO4 modification dependent on DUE unwinding by the oriC–DnaA complex (upper panel) and subsequent DnaB loading (lower panel). Loaded DnaB molecules likely protect the M/R-DUE region and enhance modification of the AT-cluster–L-DUE region in the lower strand. B, KMnO4 modification experiments using DnaC mutants. Plasmid pBSoriC-bearing oriC (5 nM) was incubated for 10 min at 37 °C with IHF (100 nM) in the presence of EcoDnaA (400 nM), DnaB WT (600 nM), and DnaC WT, K112R, or S177A (600 nM), followed by further incubation with 10 mM KMnO4. The lower strand was analyzed. Similar results were shown in repeated experiments. C, KMnO4 modification experiments using oriC and/or DnaB mutants. Plasmids pBSoriC and pBSleftoriC bearing FL- or Left-oriC (5 nM) were incubated for 15 min at 30 °C with IHF (100 nM) in the presence of EcoDnaA (400 nM), DnaB K236A (600 nM), and DnaC (600 nM), followed by KMnO4 modification. The lower strand was analyzed. Similar results were shown in repeated experiments. DUE, duplex unwinding element; EcoDnaA, Escherichia coli DNA; FL, full length.
Next, we addressed the role of the Right-DnaA subcomplex in DnaB loading, which induces unwinding of the AT-cluster region (Fig. 7C). Relative to FL-oriC, Left-oriC had much lower unwinding activity in the AT-cluster region (Fig. 7C, lanes three and 6). Also, the protection/modifications patterns of the data support a possibility that a pair of DnaB molecules are loaded only by the Left-DnaA subcomplex even with reduced efficiency (Fig. 7C, lanes five and 6) (see Discussion). These results provide further evidence of the significant role of the Right-DnaA subcomplex in efficient DnaB loading.
Role of DnaA bound at DnaA box R4 in facilitating ssDUE binding in cells
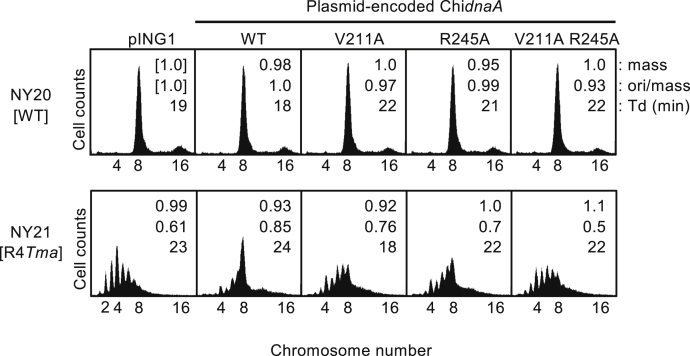
To address the significance of DnaA bound to DnaA box R4 in the process of replication initiation, we introduced plasmid bearing the ChiDnaA WT, V211A, R245A, or V211A/R245A double mutant gene into cells carrying the chromosomal oriC-R4Tma mutation (NY21) (15). The resultant cells, harboring the plasmid encoding ChiDnaA WT or mutants, grew at 30 °C, 37 °C, or 42 °C on LB plates at rates comparable to those of oriC-WT cells (NY20) harboring the vector plasmid (Fig. S7), consistent with previous studies using cells with the chromosomal Right-DOR deletions or R4 box substitutions (15, 24, 51).
Next, we performed flow cytometry analysis using oriC-WT or R4Tma cells. In these experiments, the cells were exponentially grown at 37 °C in LB medium, followed by run-out replication of the chromosome in the presence of rifampicin and cefalexin. The flow cytometry peaks corresponded to the number of oriC copies in a single cell, an indicator of replication initiation regulation. In oriC-WT cells (NY20) harboring the pING1 vector, the majority of cells contained eight oriCs, and the remainder had sixteen (Fig. 8). Expression of ChiDnaA WT or mutants had little effect on the initiation frequency in NY20 cells. In oriC-R4Tma cells (NY21) harboring pING1, the majority of cells contained two to eight oriCs, and asynchronous initiations were observed (Fig. 8), consistent with previous studies (15, 24, 51), indicating that timely initiation was disturbed in NY21 cells. In NY21 cells, expression of ChiDnaA WT from the plasmid significantly restored synchronous replication initiation, and the majority of peaks shifted to eight oriCs (Fig. 8), indicating that ChiDnaA specifically bound R4Tma, consistent with our previous findings (15). By contrast, expression of ChiDnaA V211A/R245A double mutants did not restore initiation. In addition, expression of ChiDnaA V211A or R245A only moderately stimulated initiation (Fig. 8). Taken together, these results suggest that ssDNA-binding by R4-bound DnaA is important for efficient initiation, even in vivo.
Figure 8.
Role of B/H-motifs of DnaA bound at DnaA box R4 in timely replication initiation. Cells harboring the oriC-WT (NY20) or R4Tma mutant (NY21) along with pECTM derivatives encoding ChiDnaA WT or mutants (V211A, R245A, V211A/R245A), or its parental vector (pING1), were grown at 37 °C in LB medium, followed by further incubation for run-out replication. DNA contents were quantified by flow cytometry and are indicated with the equivalent chromosome numbers on the x axes. Cell sizes (mass) at the time of drug addition were quantified with a Coulter counter, and the mass and ori/mass ratio relative to those of NY20 cells with pING1 and the doubling time (Td) of cells are indicated at the top right of each panel. ChiDnaA, chimeric DnaA.
Discussion
To uncover the molecular regulation of the efficient loading of DnaB helicases onto E. coli oriC, we investigated the roles of the oriC Right-DOR and AT-cluster–DUE regions. Our in vitro experiments demonstrated that deletions of either the Right-DOR or AT-cluster–L-DUE regions had minimal or moderate effects on DUE unwinding. By contrast, oriC with double deletion of Right-DOR and AT-cluster regions was largely defective in DnaB loading. Based on the observation that the Right-DOR was active in DnaA-dependent binding of ssDUE and DnaB, we infer that the oriC Right-DOR and AT-cluster regions play independent essential roles in the process of DnaB loading onto ssDUE, that is, the sequences are functionally redundant but in mechanistically different ways. Our findings suggest that binding of ssDUE by the Right-DnaA subcomplex is crucial for sustaining precise initiation timing in vivo, supporting that these mechanisms are important for regulation of replication initiation during the cell cycle.
In addition, this redundant mechanism is considered to be a “fail-safe” system in DnaB loading, supporting robustness of initiation. In cases when ATP–DnaA fails to form functional subcomplexes in the Right-DOR, the AT-cluster region facilitates efficient DnaB loading. Consistent with this, previous reports have suggested the significance of the oriC AT-cluster region for replication activity of oriC plasmid (11), and of the chromosomal oriC (24), for example, chromosomal oriC mutant cells carrying a double deletion of the AT-cluster–L-DUE, and Right-DOR (DnaA box C3-R4) exhibited severe inhibition of cell growth (24), similar to the failure of the Left-oriCΔ(AT–L-DUE) construct used in this study to initiate in vitro.
Taking the present and previously reported results together, we suggest a mechanistic model for oriC complex dynamics and DnaB loading (Fig. 9). First, M/R-DUE is unwound in the Left-DnaA subcomplex, resulting in an open complex. In this complex, the TT[A/G]T(T) sites within the M/R-ssDUE bind DnaA, as demonstrated for the ssDUE recruitment mechanism. The AT-cluster–L-DUE region is only unstably unwound, and unwinding of L-DUE is stimulated by interaction with the Right-DnaA subcomplex, resulting in an expanded open complex. This additional interaction also stabilizes the unwound state of M/R-DUE. The AT-cluster region also assists in the unwinding of L-DUE, which becomes specifically important in the absence of the Right-DnaA subcomplex. Based on the potential binding of the Right-DnaA to the upper strand of AT-cluster–L-DUE, the two DnaA subcomplexes might swivel using the space of the Middle-DOR region, temporally taking on a closed bilobed configuration (Fig. 9, left panels). This hypothesis must be further tested, but it is consistent with our previous study suggesting that the relative orientation of the two DnaA subcomplexes is important for efficient DnaB loading (16) (also see below). Electron microscopic analysis revealed morphological variation of oriC–DnaA complexes, ranging from globular shapes to longer kidney-like shapes, which could reflect flexible conformational changes of the complexes (52).
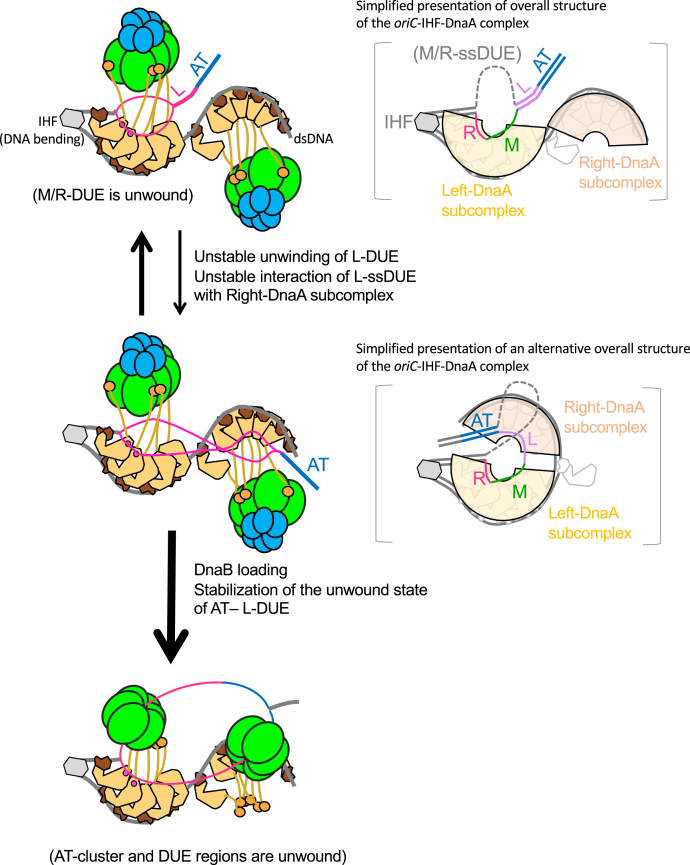
Figure 9.
Model for the concerted action of the DUE and Right-DnaA subcomplex to promote efficient DnaB loading. First, oriC DUE unwinding is promoted by interaction between M/R-DUE and the Left-DnaA subcomplex (Open complex, top left panel). DnaBC complexes are recruited by specific binding of DnaA domain I and DnaB. These complexes are stable and change dynamically to the next complexes by unstable unwinding of L-DUE and unstable interaction of the upper strand of L-DUE and Right-DnaA subcomplex (Expanded open complex, middle left panel). In the right panels with brackets, possible overall structures of the oriC-DnaA-IHF complexes are shown in an alternative simplified manner. DnaBC complexes and DnaA domains I-II are omitted for simplicity. Overall Left/Right-DnaA subcomplexes are surrounded by arches. The two DnaA subcomplexes could swivel using the linker DNA region, bringing the ssDUE and Right-DnaA subcomplex into proximity to facilitate their interaction (right panels). Binding of the Right-DnaA subcomplex to ssDUE could change dynamically, allowing temporal binding to the upper or lower strand of L-DUE region. Unstable unwinding of the AT-cluster region takes over for L-DUE when the Right-DnaA subcomplex is nonfunctional. Two DnaB molecules are loaded onto the lower-strand M/R-DUE region and the upper-strand AT-cluster–L-DUE region via a specific interaction with DnaA domain III (DnaB-loaded complex, bottom panel). DUE, duplex unwinding element; ss, single stranded.
During the initiation process, two DnaB–DnaC complexes bind respectively to the Right- and Left-DnaA subcomplexes via direct interaction between DnaB and DnaA domain I, resulting in various configurations depending on the unwound state of the DUE (Fig. 9). The precise timing of DnaB–DnaC binding to the oriC–DnaA complex during the cell cycle remains elusive. When DnaB helicases are loaded onto the unwound DUE region, the AT-cluster region is stably unwound, which might be caused indirectly by physical obstruction by the loaded DnaB helicase. Stabilization of the unwound state of L-DUE by the Right-DnaA subcomplex should support the most efficient loading of DnaB helicases. In the absence of the Right-DnaA subcomplex, the AT-cluster assists in unwinding of the DUE to promote DnaB loading. As such, DnaB helicases loaded onto each DUE strand migrate bidirectionally to initiate bidirectional replication.
Based on our observation that an oriC mutant bearing a 10 bp deletion, but not a five or 18 bp deletion, between R2 and C3 boxes in the Middle-DOR region retained DnaB-loading activity, we suggested that the relative orientation of the two DnaA subcomplexes is crucial for efficient DnaB loading (16). As shown by our EMSA data from this study, the sequence specificity of the Right-DnaA in binding ssDUE regions is not very strict. Consistently, our model suggests that the Right-DnaA subcomplex binds the ssDUE region dependent upon its spatial positioning, but not only on the DUE sequence (Fig. 9). Binding of the Right-DnaA subcomplex to ssDUE could change dynamically, allowing temporal binding to the upper strand of L/M-DUE and the upper or lower strand of AT-cluster–L-DUE, supporting the unwound state.
The mechanisms that regulate the direction and selection of the DUE strand during DnaB loading remain to be elucidated. Based on the observations that the two DnaA subcomplexes form with opposing orientations and that interaction of DnaB with DnaA domain IIII His136 is required for DnaB loading onto ssDUE, a specific DnaB–DnaA domain III interaction should be important for such regulation. In addition, the Left-DnaA subcomplex preferentially binds the upper strand of M/R-DUE and could direct DnaB loading to the lower-strand DUE (7, 8, 53). Based on these, we hypothesize that DnaB helicase bound to the Left-DnaA subcomplex is loaded onto the lower strand of R/M-DUE and migrates outward from oriC (Fig. 9). Even if the Right-DnaA subcomplex is absent, the Left-DnaA subcomplex would bind the second DnaB molecule, loading it onto the upper-strand DUE, although the total efficiency of DnaB loading is reduced, as shown here and in previous reports. The coordinated actions of the two DnaA subcomplexes would support the efficient directional loading of DnaB helicases, and this should be further examined in the future.
The bilobed structures of oriC are also conserved in Helicobacter pylori and Bacillus subtilis, which are evolutionarily distant from E. coli (2). The oriC regions in these bacteria are separated by insertion of the dnaA gene, and each oriC fragment containing clusters of DnaA boxes is required for initiation. It is possible that the two DnaA subcomplexes constructed in each oriC fragment operate in concert to promote bidirectional loading of the replicative helicases, where a common mechanistic principle is conserved.
Experimental procedures
Proteins, DNAs, and E. coli strains
The proteins used in this study were prepared as described previously (10, 15, 17).
Construction of M13oriCMS9, pBSoriC, and pBSleftoriC (formerly pBS-ΔR4-R2) was described previously (8, 15, 16). Oligonucleotides, plasmids, and DOR fragments amplified by PCR are listed in Tables S1–S3, respectively. The DUE-truncated pBSoriC and pBSleftoriC constructs, except for pBSoriCΔDUE, were constructed by inverse PCR using pBSoriC or pBSleftoriC as a template, primer pairs M28 and delAT or delATL (10), and self-ligation to yield pBSoriCΔAT, pBSoriCΔATL, pBSleftoriCΔAT, and pBSleftoriCΔATL. pBSoriCΔDUE was constructed by inverse PCR using pBSoriC as a template and primers ori-1 and M28R1r (8). DNA was digested with HincII and self-ligated, yielding pBSoriCΔDUE. To construct mutant oriC plasmids containing the TmaDnaA binding sequence at DnaA box R4, DNA fragments were amplified by PCR using M13oriCMS9 or pBSoriC as a template DNA and primers R4tmafb and R4tma12rb, followed by self-ligation (15), yielding M13oriCMS9 R4Tma or pBSoriCR4Tma. Plasmids for overproduction of TmaDnaA mutant proteins, pECTM-V211A, pECTM-R245A, and pECTM-V211A/R245A, were described previously (17). An overproduction plasmid for DnaC was constructed using pBAD/HisB and a dnaC-bearing fragment that was amplified by PCR using primers yy03 and yy04, yielding pBAD/His-dnaC. Mutations were introduced by PCR using pBAD/His-dnaC and primers yy15 and yy16 (for K112R) or yyc9 and yyc10 (for S177A) (Table S1). Overproduced His-DnaC proteins were purified using an Ni-NTA affinity column, resulting in the purity of >90% as determined by SDS-PAGE.
E. coli strains are listed in Table S4. Strains bearing mutant oriC were constructed using the λRed site-directed recombination system as previously described (15, 17).
P1 nuclease assay for DUE unwinding
The assay was performed essentially as described previously (15, 16, 17). Briefly, ATP– or ADP–DnaA was incubated for 3 min at 38 °C in 20 μl buffer containing a derivative of 1.32 nM M13oriCMS9 or 3.4 nM pBSoriC, 32 nM IHF, and 100 mM potassium chloride, followed by further incubation with 4 units of P1 nuclease (Wako) for 200 s. The resultant DNA samples were analyzed by 1% agarose gel electrophoresis and GelStar (Lonza) staining.
Electrophoretic mobility shift assay
EMSA for analyzing DnaA oligomer formation was performed as described previously (13, 15, 17). Briefly, DNA fragments (35 nM) were incubated for 10 min at 30 °C in 10 μl buffer containing ATP– or ADP–EcoDnaA, 0.25 mg/ml bovine serum albumin (BSA), and 200 ng λ phage DNA as a competitor, followed by 2% agarose gel electrophoresis at 4 °C, GelStar (Lonza) staining, and densitometric scanning.
EMSA-based ssDUE recruitment assay was performed as described previously (8, 10, 15, 17). Briefly, WT or mutant EcoDnaA and/or ChiDnaA, preincubated with 3 μM ATP or ADP, were incubated with 35 nM DOR derivatives for 5 min on ice, followed by incubation with 16 nM 32P-labeled ssDUE derivatives (MR28, renamed as U-MR; MR28rev, renamed as L-MR; U-ATL; L-ATL; U-LM; L-LM) at 30 °C for 10 min in 5 μl buffer containing 80 mM potassium chloride or 100 mM potassium glutamate, 0.25 mg/ml BSA, 2 mM ATP, and 25 ng λ phage DNA, followed by 4% PAGE at 4 °C.
KMnO4 modification-primer extension assay for ssDUE unwinding
This experiment was performed essentially as described previously (39). The indicated amounts of ATP–DnaA, DnaB K236A ATPase-defective mutant, and DnaC were incubated for 10 min at 37 °C in 20 μl buffer containing 5 nM pBSoriC and its derivatives, 20 mM Tris–HCl (pH 7.5), 10 mM magnesium acetate, 125 mM potassium glutamate, 3 mM ATP, 0.1 mg/ml BSA, and 100 nM IHF. To this mixture was added 1 μl of 0.2 M KMnO4, followed by further incubation for 3 min. DNA modification points were analyzed by primer extension reaction using 5′-32P-labeled primer KWSmaIoriCFwd, 7.5% mini-gel or sequencing-gel electrophoresis, and a BAS-2500 image analyzer (Fuji). The position of each DUE element (L, M, and R) and the AT-cluster region was determined using Sanger sequence data shown by the identical primer and a DNA size marker made from the lower strand of the ssDUE treated with KMnO4.
Form I∗ assay for DnaB loading
This assay was performed as previously described (8, 34, 46). The indicated amounts of ATP–EcoDnaA and ATP–ChiDnaA were incubated for 15 min at 30 °C in Form I∗ buffer (12.5 μl) containing 3 mM ATP, 1.6 nM pBSoriC or pBSoriCR4Tma, 42 nM IHF, 100 nM His-DnaB, 100 nM His-DnaC, 76 nM GyrA, 100 nM His-GyrB, and 760 nM SSB. The reaction was stopped by addition of 0.5% SDS, and DNA was purified by phenol–chloroform extraction. Samples were analyzed by 0.65% agarose gel electrophoresis with 0.5xTris-borate-EDTA buffer for 15 h at 23V, followed by ethidium bromide staining. When M13oriCMS9 was used in this assay, the reaction was performed in buffer that was identical except for the concentration of potassium glutamate (150 mM).
DOR pull-down assay
This assay was performed as previously described with minor modifications (7, 8). DOR fragments were amplified by PCR, using pBSoriC (for Left-DOR, Middle/Right-DOR, and Right-DOR) and pBSoriCR4Tma (for Middle/Right-DOR R4Tma) as templates and primers listed in Table S3. Biotinylated DOR DNA fragments (250 fmol) were incubated for 10 min at 4 °C in pull-down buffer (20 mM Hepes–KOH at pH 7.6, 1 mM EDTA, 4 mM DTT, 5 mM magnesium acetate, 40 mM ammonium sulfate, 20 mM NaCl, 10% [v/v] glycerol, 0.1% TritonX-100, 1 mM ATP, and 0.1 mg/ml BSA), containing DnaA (10 pmol), His-DnaB (10 pmol), and DnaC (10 pmol). Biotinylated DNA-bound materials were recovered using streptavidin-coated beads (Invitrogen), washed in pull-down buffer (25 μl) excluding BSA, dissolved in SDS sample buffer, and analyzed by SDS–11% PAGE and silver staining. In parallel, quantitative control DNA was used for ethanol precipitation and electrophoresis. Sixty percent of the input DNA (about 150 fmol) was recovered in the bead-bound fraction in the experiments.
Flow cytometry analysis
The analysis was performed essentially as described previously (17, 35). Briefly, cells were grown exponentially until the absorbance of the culture (A660) reached 0.1. Portions of the cultures were immediately chilled in 70% ethanol and used for analysis of cell mass (volume) on a Multisizer 3 Coulter counter (Beckman Coulter). Incubation of remaining portions of the cultures was continued for 4 h in the presence of 300 μg/ml rifampicin and 10 μg/ml cephalexin for run-out replication of the chromosomal DNA, followed by DNA staining with SYTOX Green (Life Technologies) and analysis of the cellular DNA content on a FACS Calibur flow cytometer (BD Biosciences).
Data availability
All data is available in the article.
Supporting information
This article contains supporting information (8, 15, 16, 17).
Conflict of interest
The authors declare that they have no conflicts of interest related to the content of this article.
Acknowledgments
We thank Yusuke Yamano for construction of DnaC overproducing plasmids and purification of DnaC proteins.
Author contributions
Y. S., M. N., and R. Y. investigation; Y. S., M. N., K. K., S. O., and T. K. writing–original draft; R. Y. validation; R. Y., K. K., S. O., and T. K. writing–review and editing; T. K. supervision.
Funding and additional information
This study was supported by a Grant-in-aid for Scientific Research, JSPS KAKENHI Grant numbers: JP20H03212, JP17H03656, JP20K15717, and JP19K23729, in addition to JSPS pre-doctoral fellowships (to Y. S. and R. Y.): JP16J02075 and JP22J11077.
Edited by Patrick Sung
Footnotes
Present address for Yukari Sakiyama: Daiichi Sankyo Co Tokyo, Japan.
Supporting information
References
- 1.Costa A., Hood I.V., Berger J.M. Mechanisms for initiating cellular DNA replication. Annu. Rev. Biochem. 2013;82:25–54. doi: 10.1146/annurev-biochem-052610-094414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wolański M., Donczew R., Zawilak-Pawlik A., Zakrzewska-Czerwińska J. oriC-encoded instructions for the initiation of bacterial chromosome replication. Front. Microbiol. 2014;5:735. doi: 10.3389/fmicb.2014.00735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leonard A.C., Rao P., Kadam R.P., Grimwade J.E. Changing perspectives on the role of DnaA-ATP in orisome function and timing regulation. Front. Microbiol. 2019;10:2009. doi: 10.3389/fmicb.2019.02009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Katayama T., Kasho K., Kawakami H. The DnaA cycle in Escherichia coli: activation, function and inactivation of the initiator protein. Front. Microbiol. 2017;8:1–15. doi: 10.3389/fmicb.2017.02496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O’Donnell M., Langston L., Stillman B. Principles and concepts of DNA replication in bacteria, archaea, and eukarya. Cold Spring Harb. Perspect. Biol. 2013;5:a010108. doi: 10.1101/cshperspect.a010108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chodavarapu S., Kaguni J.M. Replication initiation in bacteria. Enzymes. 2016;39:1–30. doi: 10.1016/bs.enz.2016.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hayashi C., Miyazaki E., Ozaki S., Abe Y., Katayama T. DnaB helicase is recruited to the replication initiation complex via binding of DnaA domain I to the lateral surface of the DnaB N-terminal domain. J. Biol. Chem. 2020;295:1131–1143. doi: 10.1074/jbc.RA120.014235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ozaki S., Katayama T. Highly organized DnaA-oriC complexes recruit the single-stranded DNA for replication initiation. Nucl. Acids Res. 2012;40:1648–1665. doi: 10.1093/nar/gkr832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bramhill D., Kornberg A. Duplex opening by dnaA protein at novel sequences in initiation of replication at the origin of the E. coli chromosome. Cell. 1988;52:743–755. doi: 10.1016/0092-8674(88)90412-6. [DOI] [PubMed] [Google Scholar]
- 10.Ozaki S., Kawakami H., Nakamura K., Fujikawa N., Kagawa W., Park S.-Y., et al. A common mechanism for the ATP-DnaA-dependent formation of open complexes at the replication origin. J. Biol. Chem. 2008;283:8351–8362. doi: 10.1074/jbc.M708684200. [DOI] [PubMed] [Google Scholar]
- 11.Asai T., Takanami M., Imai M. The AT richness and gid transcription determine the left border of the replication origin of the E. coli chromosome. EMBO J. 1990;9:4065–4072. doi: 10.1002/j.1460-2075.1990.tb07628.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rozgaja T.A., Grimwade J.E., Iqbal M., Czerwonka C., Vora M., Leonard A.C. Two oppositely oriented arrays of low-affinity recognition sites in oriC guide progressive binding of DnaA during Escherichia coli pre-RC assembly. Mol. Microbiol. 2011;82:475–488. doi: 10.1111/j.1365-2958.2011.07827.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ozaki S., Noguchi Y., Hayashi Y., Miyazaki E., Katayama T. Differentiation of the DnaA-oriC subcomplex for DNA unwinding in a replication initiation complex. J. Biol. Chem. 2012;287:37458–37471. doi: 10.1074/jbc.M112.372052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaur G., Vora M.P., Czerwonka C.A., Rozgaja T.A., Grimwade J.E., Leonard A.C. Building the bacterial orisome: high-affinity DnaA recognition plays a role in setting the conformation of oriC DNA. Mol. Microbiol. 2014;91:1148–1163. doi: 10.1111/mmi.12525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Noguchi Y., Sakiyama Y., Kawakami H., Katayama T. The Arg fingers of key DnaA protomers are oriented inward within the replication origin oriC and stimulate DnaA subcomplexes in the initiation complex. J. Biol. Chem. 2015;290:20295–20312. doi: 10.1074/jbc.M115.662601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shimizu M., Noguchi Y., Sakiyama Y., Kawakami H., Katayama T., Takada S. Near-atomic structural model for bacterial DNA replication initiation complex and its functional insights. Proc. Natl. Acad. Sci. U. S. A. 2016;113:E8021–E8030. doi: 10.1073/pnas.1609649113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sakiyama Y., Kasho K., Noguchi Y., Kawakami H., Katayama T. Regulatory dynamics in the ternary DnaA complex for initiation of chromosomal replication in Escherichia coli. Nucl. Acids Res. 2017;45:12354–12373. doi: 10.1093/nar/gkx914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McGarry K.C., Ryan V.T., Grimwade J.E., Leonard A.C. Two discriminatory binding sites in the Escherichia coli replication origin are required for DNA strand opening by initiator DnaA-ATP. Proc. Natl. Acad. Sci. U. S. A. 2004;101:2811–2816. doi: 10.1073/pnas.0400340101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kawakami H., Keyamura K., Katayama T. Formation of an ATP-DnaA-specific initiation complex requires DnaA Arginine 285, a conserved motif in the AAA+ protein family. J. Biol. Chem. 2005;280:27420–27430. doi: 10.1074/jbc.M502764200. [DOI] [PubMed] [Google Scholar]
- 20.Kasho K., Oshima T., Chumsakul O., Nakamura K., Fukamachi K., Katayama T. Whole-genome analysis reveals that the nucleoid protein IHF predominantly binds to the replication origin oriC specifically at the time of initiation. Front. Microbiol. 2021;12:697712. doi: 10.3389/fmicb.2021.697712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller D.T., Grimwade J.E., Betteridge T., Rozgaja T., Torgue J.J., Leonard A.C. Bacterial origin recognition complexes direct assembly of higher-order DnaA oligomeric structures. Proc. Natl. Acad. Sci. U. S. A. 2009;106:18479–18484. doi: 10.1073/pnas.0909472106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hwang D.S., Kornberg A. Opening of the replication origin of Escherichia coli by DnaA protein with protein HU or IHF. J. Biol. Chem. 1992;267:23083–23086. [PubMed] [Google Scholar]
- 23.Hwang D.S., Kornberg A. Opposed actions of regulatory proteins, DnaA and IciA, in opening the replication origin of Escherichia coli. J. Biol. Chem. 1992;267:23087–23091. [PubMed] [Google Scholar]
- 24.Stepankiw N., Kaidow A., Boye E., Bates D. The right half of the Escherichia coli replication origin is not essential for viability , but facilitates multi-forked replication. Mol. Microbiol. 2009;74:467–479. doi: 10.1111/j.1365-2958.2009.06877.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaguni J.M. Replication initiation at the Escherichia coli chromosomal origin. Curr. Opin. Chem. Biol. 2011;15:606–613. doi: 10.1016/j.cbpa.2011.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sutton M.D., Carr K.M., Vicente M., Kaguni J.M. Escherichia coli DnaA protein. The N-terminal domain and loading of DnaB helicase at the E. coli chromosomal origin. J. Biol. Chem. 1998;273:34255–34262. doi: 10.1074/jbc.273.51.34255. [DOI] [PubMed] [Google Scholar]
- 27.Felczak M.M., Simmons L.A., Kaguni J.M. An essential tryptophan of Escherichia coli DnaA protein functions in oligomerization at the E. coli replication origin. J. Biol. Chem. 2005;280:24627–24633. doi: 10.1074/jbc.M503684200. [DOI] [PubMed] [Google Scholar]
- 28.Abe Y., Jo T., Matsuda Y., Matsunaga C., Katayama T., Ueda T. Structure and function of DnaA N-terminal domains: specific sites and mechanisms in inter-DnaA interaction and in DnaB helicase loading on oriC. J. Biol. Chem. 2007;282:17816–17827. doi: 10.1074/jbc.M701841200. [DOI] [PubMed] [Google Scholar]
- 29.Keyamura K., Fujikawa N., Ishida T., Ozaki S., Su’etsugu M., Fujimitsu K., et al. The interaction of DiaA and DnaA regulates the replication cycle in E. coli by directly promoting ATP–DnaA-specific initiation complexes. Genes Dev. 2007;21:2083–2099. doi: 10.1101/gad.1561207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keyamura K., Abe Y., Higashi M., Ueda T., Katayama T. DiaA dynamics are coupled with changes in initial origin complexes leading to helicase loading. J. Biol. Chem. 2009;284:25038–25050. doi: 10.1074/jbc.M109.002717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nozaki S., Ogawa T. Determination of the minimum domain II size of Escherichia coli DnaA protein essential for cell viability. Microbiology. 2008;154:3379–3384. doi: 10.1099/mic.0.2008/019745-0. [DOI] [PubMed] [Google Scholar]
- 32.Erzberger J.P., Mott M.L., Berger J.M. Structural basis for ATP-dependent DnaA assembly and replication-origin remodeling. Nat. Struct. Mol. Biol. 2006;13:676–683. doi: 10.1038/nsmb1115. [DOI] [PubMed] [Google Scholar]
- 33.Felczak M.M., Kaguni J.M. The box VII motif of Escherichia coli DnaA protein is required for DnaA oligomerization at the E. coli replication origin. J. Biol. Chem. 2004;279:51156–51162. doi: 10.1074/jbc.M409695200. [DOI] [PubMed] [Google Scholar]
- 34.Duderstadt K.E., Chuang K., Berger J.M. DNA stretching by bacterial initiators promotes replication origin opening. Nature. 2011;478:209–213. doi: 10.1038/nature10455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sakiyama Y., Nishimura M., Hayashi C., Akama Y., Ozaki S., Katayama T. The DnaA AAA+ Domain His136 residue directs DnaB replicative helicase to the unwound region of the replication origin, oriC. Front. Microbiol. 2018;9:1–17. doi: 10.3389/fmicb.2018.02017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fujikawa N., Kurumizaka H., Nureki O., Terada T., Shirouzu M., Katayama T., et al. Structural basis of replication origin recognition by the DnaA protein. Nucl. Acids Res. 2003;31:2077–2086. doi: 10.1093/nar/gkg309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grimwade J.E., Leonard A.C. Blocking, bending, and binding: regulation of initiation of chromosome replication during the Escherichia coli cell cycle by transcriptional modulators that interact with origin DNA. Front. Microbiol. 2021;12:732270. doi: 10.3389/fmicb.2021.732270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Soultanas P. Loading mechanisms of ring helicases at replication origins. Mol. Microbiol. 2012;84:6–16. doi: 10.1111/j.1365-2958.2012.08012.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fang L., Davey M.J., O’Donnell M. Replisome assembly at oriC, the replication origin of E. coli, reveals an explanation for initiation sites outside an origin. Mol. Cell. 1999;4:541–553. doi: 10.1016/s1097-2765(00)80205-1. [DOI] [PubMed] [Google Scholar]
- 40.Arias-Palomo E., O’Shea V.L., Hood I.V., Berger J.M. The bacterial DnaC helicase loader is a DnaB ring breaker. Cell. 2013;153:438–448. doi: 10.1016/j.cell.2013.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chodavarapu S., Jones A.D., Feig M., Kaguni J.M. DnaC traps DnaB as an open ring and remodels the domain that binds primase. Nucl. Acids Res. 2016;44:210–220. doi: 10.1093/nar/gkv961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Arias-Palomo E., Puri N., O’Shea Murray V.L., Yan Q., Berger J.M. Physical basis for the loading of a bacterial replicative helicase onto DNA. Mol. Cell. 2019;74:173–184.e4. doi: 10.1016/j.molcel.2019.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nagata K., Okada A., Ohtsuka J., Ohkuri T., Akama Y., Sakiyama Y., et al. Crystal structure of the complex of the interaction domains of Escherichia coli DnaB helicase and DnaC helicase loader: structural basis implying a distortion-accumulation mechanism for the DnaB ring opening caused by DnaC binding. J. Biochem. 2020;167:1–14. doi: 10.1093/jb/mvz087. [DOI] [PubMed] [Google Scholar]
- 44.Makowska-Grzyska M., Kaguni J.M. Primase directs the release of DnaC from DnaB. Mol. Cell. 2010;37:90–101. doi: 10.1016/j.molcel.2009.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Felczak M.M., Chodavarapu S., Kaguni J.M. DnaC, the indispensable companion of DnaB helicase, controls the accessibility of DnaB helicase by primase. J. Biol. Chem. 2017;292:20871–20882. doi: 10.1074/jbc.M117.807644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Coster G., Diffley J.F.X. Bidirectional eukaryotic DNA replication is established by quasi-symmetrical helicase loading. Science. 2017;357:314–318. doi: 10.1126/science.aan0063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Amin A., Wu R., Cheung M.H., Scott J.F., Wang Z., Zhou Z., et al. An essential and cell-cycle-dependent ORC dimerization cycle regulates eukaryotic chromosomal DNA replication. Cell Rep. 2020;30:3323–3338.e6. doi: 10.1016/j.celrep.2020.02.046. [DOI] [PubMed] [Google Scholar]
- 48.Sugiyama R., Kasho K., Miyoshi K., Ozaki S., Kagawa W., Kurumizaka H., et al. A novel mode of DnaA-DnaA interaction promotes ADP dissociation for reactivation of replication initiation activity. Nucl. Acids Res. 2019;47:11209–11224. doi: 10.1093/nar/gkz795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Davey M.J., Fang L., McInerney P., Georgescu R.E., O’Donnell M. The DnaC helicase loader is a dual ATP/ADP switch protein. EMBO J. 2002;21:3148–3159. doi: 10.1093/emboj/cdf308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Baker T.A., Kornberg A. Transcriptional activation of initiation of replication from the E. coli chromosomal origin: an RNA-DNA hybrid near oriC. Cell. 1988;55:113–123. doi: 10.1016/0092-8674(88)90014-1. [DOI] [PubMed] [Google Scholar]
- 51.Weigel C., Messer W., Preiss S., Welzeck M., Morigen, Boye E. The sequence requirements for a functional Escherichia coli replication origin are different for the chromosome and a minichromosome. Mol. Microbiol. 2001;40:498–507. doi: 10.1046/j.1365-2958.2001.02409.x. [DOI] [PubMed] [Google Scholar]
- 52.Crooke E., Thresher R., Hwang D.S., Griffith J., Kornberg A. Replicatively active complexes of DnaA protein and the Escherichia coli chromosomal origin observed in the electron microscope. J. Mol. Biol. 1993;233:16–24. doi: 10.1006/jmbi.1993.1481. [DOI] [PubMed] [Google Scholar]
- 53.Weigel C., Seitz H. Strand-specific loading of DnaB helicase by DnaA to a substrate mimicking unwound oriC. Mol. Microbiol. 2002;46:1149–1156. doi: 10.1046/j.1365-2958.2002.03232.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data is available in the article.