Abstract
Although natural selection appears to favor the elimination of gene redundancy in prokaryotes, multiple copies of each rRNA-encoding gene are common on bacterial chromosomes. Despite this conspicuous deviation from single-copy genes, no phenotype has been consistently associated with rRNA gene copy number. We found that the number of rRNA genes correlates with the rate at which phylogenetically diverse bacteria respond to resource availability. Soil bacteria that formed colonies rapidly upon exposure to a nutritionally complex medium contained an average of 5.5 copies of the small subunit rRNA gene, whereas bacteria that responded slowly contained an average of 1.4 copies. In soil microcosms pulsed with the herbicide 2,4-dichlorophenoxyacetic acid (2,4-D), indigenous populations of 2,4-D-degrading bacteria with multiple rRNA genes (x̄ = 5.4) became dominant, whereas populations with fewer rRNA genes (x̄ = 2.7) were favored in unamended controls. These findings demonstrate phenotypic effects associated with rRNA gene copy number that are indicative of ecological strategies influencing the structure of natural microbial communities.
Genes encoding the 5S, 16S, and 23S rRNAs are typically organized into an operon in members of the domain Bacteria. The copy number of rRNA operons per bacterial genome varies from 1 to as many as 15 (28). For example, the pathogenic bacteria Rickettsia prowazekii (2) and Mycoplasma pneumoniae (4) have one rRNA operon, while the enteric bacteria Escherichia coli (12) and Salmonella enterica serovar Typhimurium (1) each possess seven copies per genome. The greatest number of rRNA operons per genome known can be found among spore-forming bacteria isolated from soil; Bacillus subtilis (23) and Clostridium paradoxum (28) possess 10 and 15 copies, respectively. Several hypotheses have been proposed to explain the wide variation observed in rRNA operon copy number.
It is generally assumed that multiple copies of rRNA operons in prokaryotic organisms are required to achieve high growth rates. However, the short doubling time observed for certain bacteria with a single rRNA operon (37) and the marginal impact of rRNA operon inactivation on maximal growth rate (8, 27) suggest that the capacity for rapid growth is not the sole determinant of rRNA operon copy number. The number of transcripts that can be initiated at an rRNA operon promoter and the transcriptional rate of RNA polymerase set a maximum rate on the number of ribosomes that can be produced from a single rRNA operon. Calculations including promoter initiation efficiency and transcription rates indicate that one copy of the rRNA operon is insufficient to supply the number of ribosomes required to achieve maximal growth rates observed in E. coli (5).
Given the high demand for rRNA transcription and the central role of rRNAs in the regulation of ribosome synthesis, it is conceivable that the number of rRNA operons may dictate the rapidity with which microbes can synthesize ribosomes and respond to favorable changes in growth conditions (8, 30). Transcription of the rRNA operon is regulated to correspond with resource availability and can represent as much as 70% of total cellular transcription during rapid periods of growth (6). A proposed homeostatic model of ribosome biosynthesis provides a direct link between resource availability and the protein synthetic capacity of a bacterial cell (15). The concentrations of resources available for growth determine intracellular concentration of ATP and GTP, which in turn regulate the efficiency of transcription initiation at rRNA operons. The rRNA operon transcript is processed enzymatically to yield mature rRNAs that not only bind ribosomal proteins during assembly of the ribosome but also regulate translation of the ribosomal protein mRNAs (36). In E. coli, a positive relationship exists between the number of rRNA operons inactivated and the time required to increase growth in response to added resources (8). Condon et al. (8) suggested that E. coli maintains seven rRNA operons due to selective pressure on the ability to adapt quickly to environmental conditions. Therefore, the capacity to respond rapidly to fluctuating growth conditions may be more relevant than maximal growth rate for explaining the variation in rRNA operon multiplicity in different species of bacteria.
While multiple rRNA operons may provide an advantage under fluctuating conditions, constitutive expression from multiple rRNA operons would confer a metabolic expense on slower-growing cells due to the overproduction of ribosomes. Extra copies of plasmid-borne rRNA operons increase stable RNA concentrations while concomitantly decreasing growth rates in E. coli, indicating a potential cost associated with constitutive expression from multiple rRNA operons at low growth rates (30). The immediate degradation of ribosomes in starved cells of E. coli and Salmonella spp. also suggests that excess translation capacity is metabolically unfavorable in conditions of low nutrient availability (9, 20). For these reasons, we postulate that fewer rRNA operons represent a competitive advantage at low growth rates.
The observations above led us to question whether the number of rRNA operons in phylogenetically diverse bacteria reflected ecological strategies characterized by either rapid response to resource input (high copy number) or efficient allocation of resources under constant, slow-growth environments (low copy number). To determine whether the number of rRNA operons is of adaptive significance to bacteria rather than the result of genetic drift or coincidence, we explored the relationship between rRNA operon copy number, organismal phylogeny, and the capacity of bacteria to respond to added resources. Microbes isolated from soil were selected for this study because soils are inhabited by a rich diversity of microbes (33) and the soil matrix provides an array of microenvironments that can vary considerably with regard to resource availability. If rRNA operon copy number reflects the ecological strategy of bacteria in response to resource availability, bacterial populations with different numbers of rRNA operons are likely to coexist in soils and respond differently to perturbations.
MATERIALS AND METHODS
Colony response curves of soil bacteria.
Soil samples used for experimentation were obtained from the Long Term Ecological Research (LTER) site at Kellogg Biological Station, Hickory Corners, Mich., in May 1997. Soil cores (10-cm depth by 2.5-cm diameter) were removed from five locations within a conventional-till agricultural plot (plot T1; descriptions of plots may be accessed at http://lter.kbs.msu.edu). Sample cores were sieved (2-mm mesh), homogenized, and stored on wet ice for no more than 6 h before use. A 100-g portion of homogenized soil was suspended in 1.0 liter of 5 mM K2HPO4 buffer (pH 7.0) and shaken (22°C, 150 rpm, 15 min), and a 1-ml portion of the suspension was serially diluted. Aliquots from the dilution series were plated on 1.5% agar medium containing a 100-fold dilution of nutrient broth (Difco Inc.); bacterial colonies were enumerated at periodic time intervals and marked for subsequent isolation upon completion of the colony response curve. Hattori's laboratory has demonstrated that the pattern of colony formation by soil bacteria is reproducible and can be modeled by the superimposition of several (typically four) first-order reaction curves (16, 19). Bacteria from the time intervals designated I and IV (Fig. 1; groups described in reference 16) were isolated and characterized for rRNA operon copy number. Single colonies were picked and streaked for isolation a minimum of six times on dilute nutrient agar plates (Difco); culture purity was also confirmed via light microscopy and PCR amplification of the 16S rRNA gene (rDNA; see below). Isolates from rice paddy soils were obtained from Tsutomu Hattori (Institute of Genetic Ecology, Tohoku University, Sendai, Japan) (16, 26).
FIG. 1.
Correlation between time of colony appearance and rRNA operon copy number. (A) Colony appearance curve for isolates from conventional-tilled agricultural soil in Michigan (▵) and from rice paddy soils in Japan (adapted from reference 16) (▴). Each point represents the arithmetic average of colonies observed on a minimum of three agar plates at that time interval. Bacteria from the time intervals designated I and IV (groups described in reference 16) were isolated and characterized for rRNA operon copy number. (B) Mean number of rRNA operons for bacterial isolates from group I (early colony formers) and group IV (late colony formers) are presented as rice paddy isolates (filled bars; n = 6 [early] or 7 [late]), conventional-tilled soil isolates (open bars; n = 5 [early] or 6 [late]). Error bars are 1 standard deviation above the sample mean. Statistical analyses of early- and late-appearing sample populations were performed using Student's t test, assuming unequal sample variances (α = 0.05, df = 10).
rRNA operon copy number determination for soil isolates.
Genomic DNA was obtained from each soil isolate, independently digested with at least three different restriction enzymes (AccI, BstEII, PinA1, PvuII, PstI, or SacI; Gibco/BRL Co.), and separated on a 1.0% agarose gel using standard methods (3, 25). rRNA operon copy numbers were determined by Southern hybridization analysis of gel-separated restriction digests using a digoxigenin-dUTP-labeled DNA probe complementary to a conserved region (positions 8 to 536) of the E. coli 16S rDNA. Alternative arrangements of rRNA genes into operons are known, but individual rRNA genes are usually present in stoichiometric quantities. Therefore, the number of 16S rDNA copies (number of bands with equal intensity hybridizing to the 16S rDNA probe) was considered to be a reasonable estimator of the number of rRNA operon equivalents per genome. In cases where enzymatic digestion failed to resolve hybridized fragments or bands of equal intensity could not be discriminated, results were discarded and additional analyses were performed with different restriction endonucleases. Genomic DNA isolated from E. coli and digested with PvuII was included on each Southern hybridization gel as a positive control.
Phylogenetic analyses.
The 16S rDNA was amplified from early- and late-appearing soil isolates using primers 8f (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492r (5′-GGTTACCTTGTTACGACTT-3′) with reaction conditions described previously (22, 35). Partial sequences were obtained from soil isolates with an automated DNA sequencer (ABI 373A) using fluorescent dideoxy dye terminator chemistry and overlapping sequencing primers 8f and 519r (5′-GTATTACCGCGGCTGCTGG-3′). Sequences were initially aligned using the ARB software package (http://www.biol.chemie.tu-muenchen.de) automated aligner and then verified manually against known secondary structures (31). Soil isolate partial sequences (between positions 28 and 519 of the E. coli 16S rDNA consensus) were added to a Ribosomal Database Project (24) subtree, using parsimony with the ARB software package (31). Branch lengths were right aligned for presentation purposes in ARB and therefore do not necessarily represent the actual evolutionary distance, but the branching topology is preserved.
Soil microcosm amendment experimentation.
Soil microcosms were established from homogenized soil collected from the top 10 cm of a fallow agricultural plot (LTER, Kellogg Biological Station) which had no previous documented exposure to the herbicide 2,4-dichlorophenoxyacetic acid (2,4-D). Ten to fifteen soil samples from a 6-m2 area were pooled and sieved (2-mm mesh). For each microcosm, 243 g of soil (8% moisture content) was transferred to a polyethylene bag, while 27-g samples of soil was dried (100°C, overnight) to serve as a carrier for liquid amendments. Each 27-g portion of carrier soil was mixed with 2,4-D dissolved in 0.1 M Na2HPO4 buffer (pH 7.0) or buffer alone such that each microcosm received identical concentrations of sodium phosphate but either 0, 10, or 100 μg of 2,4-D per g of soil (final concentration). Final moisture content of soil in each microcosm was adjusted to 25% (wt/wt) with sterile, distilled water. Microcosms were incubated for 1 week, during which samples were periodically removed for isolation of 2,4-D-degrading bacteria. Colonies able to degrade 2,4-D were identified by autoradiography based on the ability to incorporate 14C from 14C-ring-labeled 2,4-D into biomass (10). rRNA operon copy numbers were determined as described above with Southern hybridization analysis of genomic DNA digested with EcoRI or PvuII.
Amplified rDNA restriction analysis of 2,4-D-degrading isolates.
Bacterial species able to degrade 2,4-D were identified based on restriction fragment length polymorphism (RFLP) patterns resulting from gel electrophoresis of enzymatically digested, PCR-amplified 16S rDNA. 16S rDNA was amplified using primers 8f and 1492r and reaction conditions described previously (22, 35). Amplified DNA from isolates was independently digested with MspI, CfoI, AluI, or HaeIII. Digested DNA was electrophoresed through 2.75% Metaphor agarose gels (FMC Bioproducts, Inc.), and the RFLP patterns of all isolates were compared. Isolates whose 16S rDNA restriction pattern differed with at least one enzyme were defined as different species.
Nucleotide sequence accession numbers.
The nucleotide sequences for rice paddy isolates have been previously deposited in the GenBank database under accession no. D84561, D84564, D84568, D84570, D84577, D84597, D84604, D84635, D84639, D84640, D84641, D84644, and D84645. Nucleotide sequences for isolates obtained from the Kellogg Biological Station LTER site are deposited in GenBank under accession no. AF183149 to AF183159.
RESULTS AND DISCUSSION
In environments with periodic resource fluctuations, lag time (L, the time before initiation of cell division) and maximal growth rate (μmax) are important components of fitness (21, 34). Populations that can rapidly achieve high maximal growth rates (short L, high μmax) are able to utilize available resources before competing populations. In contrast, lag time does not impose a fitness advantage in environments with a constant supply of resources (18, 32). Multiple rRNA operons allow transcriptional initiation from multiple loci, permitting a rapid increase in the intracellular concentration of rRNA, thereby effectively decreasing lag time. A potential tradeoff for a rapid up-shift capacity is the metabolic expense of rRNA overproduction at low growth rates, apparently due to inadequate regulation of rRNA operons (30). In agreement with these observations, bacteria isolated from low-nutrient aquatic environments share the characteristics of slow growth and few (typically one to two) rRNA operons (7, 13).
Response time of soil isolates and rRNA operon copy number.
To test whether rRNA operon copy number is correlated with the response time (a function of μmax and L) of bacterial populations in soil to resource availability, heterotrophic bacteria appearing early and late on agar media were isolated from soils from an agricultural research site in Michigan and a rice paddy near Sendai, Japan (16) (Fig. 1A). Early-appearing isolates possessed, on average, a significantly greater number of rRNA operons (x̄ = 5.5 copies) than late-appearing isolates (x̄ = 1.4 copies) (Fig. 1B). Of the early-appearing isolates, 6 of 11 contained five or more copies of the rRNA operon per genome, while 12 of 13 late-appearing species contained two or fewer copies. The time required for colony formation was a phenotype retained by isolates upon subsequent transfer on solid media (personal observation and reference 26). The biased distribution of diverse soil bacteria with high rRNA operon copy numbers appearing early on two different complex culture media suggests that the response of bacteria to favorable growth conditions reflects ecological strategies and not solely the ability to utilize a particular limiting resource.
The ability of bacteria with high rRNA operon copy number to rapidly respond to nutrient enrichment likely influences their population dynamics in soil (discussed below) and their recovery from soil by common enrichment techniques. Our capacity to culture only a small proportion of the diversity in soil (estimated between 0.1 and 0.5% [33]) may result from the inability of bacteria with low rRNA operon copy number to form visible colonies in a short period of time.
Relationship of rRNA operon copy number to phylogeny and genome size.
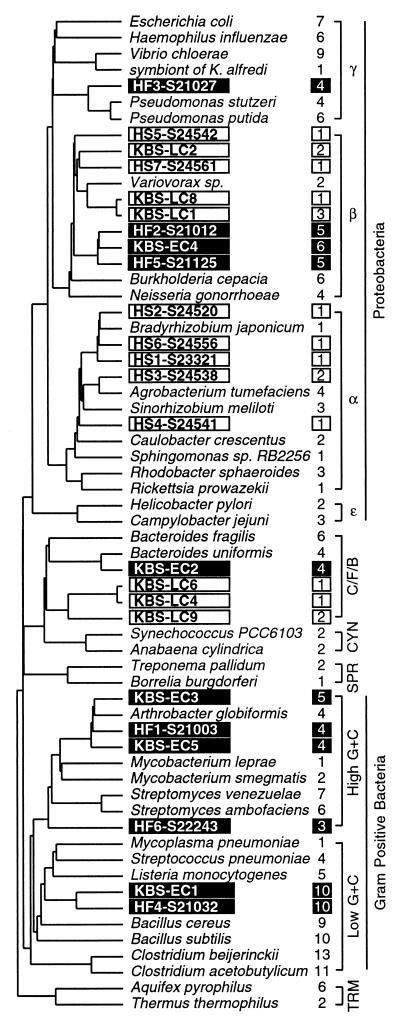
The phylogeny of isolates was reconstructed to preclude the possibility that the number of rRNA operons per isolate reflected evolutionary history alone. A statistical correlation between rRNA operon copy number and response time assumes that species were removed independently from the same distribution (14). Since bacteria were isolated based on the phenotypic parameter of response time, it was possible that the correlation with rRNA operon copy number resulted from two monophyletic groups of bacteria with either high or low numbers of rRNA operons per genome. The distribution of rRNA operon copy number within a collection of phylogenetically diverse bacteria indicated no obvious evolutionary constraint on the number of rRNA operons per genome (Fig. 2). The occurrence of bacteria with the same number of rRNA operons in disparate phylogenetic lineages appears to have arisen from convergent evolution, driven by adaptation to similar selective pressures influencing the fitness of bacteria in different environments.
FIG. 2.
Phylogenetic distribution of bacteria characterized for rRNA operon copy number. Filled boxes indicate soil isolates that appeared early, while open boxes indicate isolates that appeared late. Isolates from conventional-tilled soils in Michigan (designated by prefix “KBS”) and rice paddy soils in Japan (designated by prefix “HF” or “HS”) are included. Values to the right of species' names indicate the number of rRNA operon equivalents per chromosome. Major phylogenetic divisions are indicated on the far right with abbreviations as follows: C/F/B, Cytophaga/Flexibacter/Bacteroides; CYN, cyanobacteria; SPR, spirochetes; TRM, thermophiles. Strain designations and literature references for 16S rRNA sequences and rRNA operon copy numbers used for this analysis are available online at the Ribosomal RNA Operon Copy Number Database (http://rdp.cme.msu.edu/rrn).
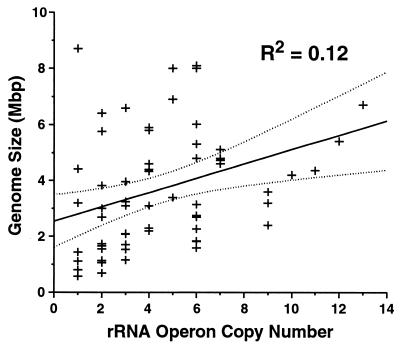
A simple relationship is also not evident between the number of rRNA operons and genome size as might be expected if recombination between rRNA operons leads to increases in genome size (1, 17) or if multiple rRNA operons are required to provide translation capacity for an increased number of protein-encoding genes in larger genomes. A linear regression provides only weak evidence for a positive correlation between genome size and rRNA operon copy number (Fig. 3). While the ability to explain changes in genome size based on increased rRNA operon copy number is low (r2 = 0.12), the hypothesis that no relationship exists between these variables cannot be rejected (P < 0.01). It is possible that the strength of this relationship is biased toward easily cultivable bacteria or those that are easy to manipulate genetically. One group in particular that may weaken the relationship between genome size and rRNA operon copy number are the limited number of bacteria (n = 4) with more than 10 rRNA operons (Fig. 3).
FIG. 3.
Relationship between genome size and rRNA operon copy number. Phylogenetic groups represented: Proteobacteria subgroups α (n = 8), β (n = 3), γ (n = 9), and ɛ (n = 4), Cytophaga/Flexibacter/Bacteroides (n = 7), cyanobacteria (n = 4), spirochetes (n = 4), high-G+C gram-positive bacteria (n = 7), low-G+C gram-positive bacteria (n = 16), and thermophiles (n = 3). A linear regression for all data points was calculated using the least-squares method (P < 0.01 that no relationship exists), upper and lower 95% confidence bounds are indicated by dotted lines. The data used are available online at the Ribosomal RNA Operon Copy Number Database (http://rdp.cme.msu.edu/rrn).
Effects of selection in soil microcosms.
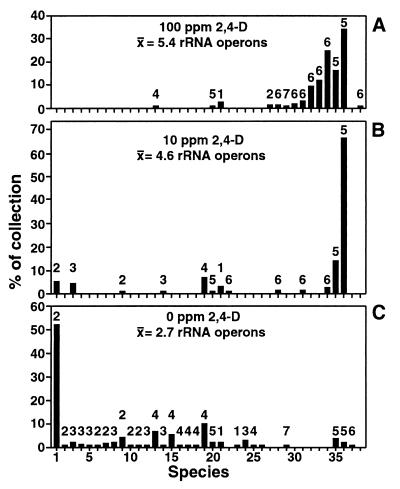
The potential adaptive significance of rRNA gene copy number was tested directly in soil microcosms by examining the dynamics of indigenous bacterial populations competing for the herbicide 2,4-D. The relative abundance of bacterial populations able to use 2,4-D as a sole carbon and energy source was measured before and after selection in nine soil microcosms (11). In unamended microcosms, there were 103 CFU of 2,4-D-degrading bacteria per g soil. Not surprisingly, the total number of 2,4-D-degrading bacteria increased to 105 or 106 over 7 days following a one-time pulse with either 10 or 100 ppm (final concentration), respectively, of 2,4-D (11). Although there was a dramatic change in the number of 2,4-D-degrading bacteria, the total number of readily cultured bacteria remained at approximately 107 CFU/g of soil in each of the nine microcosms. A total of 837 isolates representing 38 2,4-D-degrading species were isolated from the microcosms. The effect of selection for rRNA operon copy number among 2,4-D-degrading populations is clearly apparent between unamended and amended microcosms (Fig. 4). The most abundant species in unamended microcosms were minor components of microcosms amended with 10 or 100 ppm of 2,4-D, while several species at low abundance in the controls developed into numerically dominant populations in the pulsed microcosms.
FIG. 4.
Distribution of rRNA operon copy number among 2,4-D-degrading bacteria isolated from amended and unamended soil microcosms. Species are identified based on similarity of 16S rRNA restriction patterns. The height of each bar reflects the abundance of that species relative to all isolates from that microcosm of 247 (A), 263 (B), or 327 (C) isolates. Numbers above bars indicate the rRNA operon copy number for each species; the mean number of rRNA operons per isolate is indicated for each treatment. Different isolates of the same species exhibited similar rRNA operon copy numbers (data not shown). Each treatment represents data from three replicate microcosms.
The majority of 2,4-D-degrading bacteria contained between one and four rRNA operons in unamended microcosms (Fig. 4C). In contrast, among the species detected in microcosms amended with 10 or 100 ppm of 2,4-D, 7 of 13 and 11 of 14 species, respectively, possessed between five and seven rRNA operon copies (Fig. 4A and B). Despite a reduction in species diversity, the effect of selection for 2,4-D-degrading species with higher rRNA operon copy number in amended microcosms is significant with (P < 0.001) or without (P < 0.01) consideration of species abundance. (A one-tailed Student's t test assuming unequal sample variance was used to test the hypothesis that selection for higher rRNA operon copy number was greater in the microcosms receiving 10- and 100-ppm 2,4-D amendment than in the unamended [0 ppm] microcosms. A nonparametric [Wilcoxon ranked-sum] test yielded similar results [P < 0.02] when population abundance was excluded from the analysis to reduce the contribution of highly abundant species.)
The positive correlation between rRNA operon copy number and 2,4-D concentration supports an association between rRNA operon copy number and competitive fitness. In fact, the ability of 2,4-D-degrading populations with high rRNA gene copy number to respond rapidly to new resource conditions was a general characteristic of these populations, as demonstrated by comparison of the growth rates in liquid media of a number of 2,4-D-degrading populations on a variety of substrates (succinate or acetate as the limiting carbon and energy source, and in complex medium) other than 2,4-D (data not shown).
Conclusions.
Experiments described above elucidate the potential role of rRNA operon multiplicity by providing a direct correlation between rRNA operon copy number and the time required for soil bacteria to form colonies (a function of μmax and L) in response to resource availability. The potential adaptive significance of rRNA operon multiplicity was demonstrated in soil microcosms by the reproductive success of diverse 2,4-D-degrading bacteria containing a significantly greater number of rRNA operons per genome during competition for a pulse of 2,4-D. We propose that the number of rRNA operons in a bacterial genome represent one trait among a group of interdependent traits that comprise a strategy for responding to the availability of resources.
As genomic information rapidly accumulates for the Bacteria from whole-genome sequencing projects, our model of bacterial competitiveness becomes increasingly complex as individual genes are considered in the context of the entire genome and, ultimately, the organism. Certainly no single gene product can determine bacterial competitiveness in all environments. However, gene products involved in the regulation of central metabolism and cellular growth may establish a basic foundation for the competitive success of a bacterial species. Genes directly involved in the response of bacteria to specific selective pressures from the environment will undoubtedly further shape the competitive fitness or life history strategy of a species. The correlation between the copy number of rRNA genes and the response rate of diverse bacteria to a variety of growth substrates indicates an evolutionary linkage between the number of rRNA genes and the basic competitive ability of bacterial species. While other genes may enhance this basic ability, the multiplicity of rRNA genes in the Bacteria provides a genetic indicator of the general ecological strategy of a bacterial species for exploitation of nutrients.
ACKNOWLEDGMENTS
We thank R. E. Lenski for providing perspective on the interplay between microbial ecology and evolution, T. Hattori for providing rice paddy soil isolates, and B. Stevenson, D. Buckley, and J. Breznak for thoughtful discussion of the manuscript.
The U.S. Department of Energy and National Science Foundation (IBN-9875254), including graduate research fellowships from the NSF Center for Microbial Ecology (BIR91-20006) at Michigan State University (J.A.K. and J.M.D.), supported this research.
REFERENCES
- 1.Anderson P, Roth J. Spontaneous tandem genetic duplications in Salmonella typhimurium arise by unequal recombination between rRNA (rrn) cistrons. Proc Natl Acad Sci USA. 1981;78:3113–3117. doi: 10.1073/pnas.78.5.3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson S G E, Zomorodipour A, Winkler H H, Kurland C G. Unusual organization of the rRNA genes in Rickettsia prowazekii. J Bacteriol. 1995;177:4171–4175. doi: 10.1128/jb.177.14.4171-4175.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Smith J A, Seidman J G, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1987. [Google Scholar]
- 4.Bercovier H, Kafri O, Sela S. Mycobacteria possess a surprisingly small number of ribosomal RNA genes in relation to the size of their genome. Biochem Biophys Res Commun. 1986;136:1136–1141. doi: 10.1016/0006-291x(86)90452-3. [DOI] [PubMed] [Google Scholar]
- 5.Bremer H. Parameters affecting the synthesis of ribosomes and RNA polymerase in bacteria. J Theor Biol. 1975;53:115–124. doi: 10.1016/0022-5193(75)90106-x. [DOI] [PubMed] [Google Scholar]
- 6.Bremer H, Dennis P P. Modulation of chemical composition and other parameters of the cell by growth rate. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. 2nd ed. Washington, D.C.: American Society for Microbiology; 1996. pp. 1553–1569. [Google Scholar]
- 7.Button D K, Robertson B R, Lepp P W, Schmidt T M. A small, dilute-cytoplasm, high-affinity, novel bacterium isolated by extinction culture and having kinetic constants compatible with growth at ambient concentrations of dissolved nutrients in seawater. Appl Environ Microbiol. 1998;64:4467–4476. doi: 10.1128/aem.64.11.4467-4476.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Condon C, Liveris D, Squires C, Schwartz I, Squires C L. rRNA operon multiplicity in Escherichia coli and the physiological implications of rrn inactivation. J Bacteriol. 1995;177:4152–4156. doi: 10.1128/jb.177.14.4152-4156.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davis B D, Luger S M, Tai P C. Role of ribosome degradation in the death of starved Escherichia coli cells. J Bacteriol. 1986;166:439–445. doi: 10.1128/jb.166.2.439-445.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dunbar J, Wong D C L, Yarus M J, Forney L J. Autoradiographic method for isolation of diverse microbial species with unique catabolic traits. Appl Environ Microbiol. 1996;62:4180–4185. doi: 10.1128/aem.62.11.4180-4185.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dunbar J D. Ph.D. thesis. East Lansing: Michigan State University; 1996. [Google Scholar]
- 12.Ellwood M, Nomura M. Deletion of a ribosomal ribonucleic acid operon in Escherichia coli. J Bacteriol. 1980;143:1077–1080. doi: 10.1128/jb.143.2.1077-1080.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fegatella F, Lim J, Kjelleberg S, Cavicchioli R. Implications of rRNA operon copy number and ribosome content in the marine oligotrophic ultramicrobacterium Sphingomonas sp. strain RB2256. Appl Environ Microbiol. 1998;64:4433–4438. doi: 10.1128/aem.64.11.4433-4438.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Felsenstein J. Phylogenies and the comparative method. Am Nat. 1985;125:1–15. [Google Scholar]
- 15.Gaal T, Bartlett M S, Ross W, Trunbough C L, Jr, Gourse R L. Transcription regulation by initiating NTP concentration: rRNA synthesis in bacteria. Science. 1997;278:2092–2097. doi: 10.1126/science.278.5346.2092. [DOI] [PubMed] [Google Scholar]
- 16.Gorlach K, Shingaki R, Morisaki H, Hattori T. Construction of eco-collection of paddy field soil bacteria for population analysis. J Gen Appl Microbiol. 1994;40:509–517. [Google Scholar]
- 17.Hancock J M. Simple sequences and the expanding genome. Bioessays. 1996;18:421–425. doi: 10.1002/bies.950180512. [DOI] [PubMed] [Google Scholar]
- 18.Hansen S R, Hubble S P. Single-nutrient microbial competition: qualitative agreement between experimental and theoretically forecast outcomes. Science. 1980;207:1491–1493. doi: 10.1126/science.6767274. [DOI] [PubMed] [Google Scholar]
- 19.Hashimoto T, Hattori T. Grouping of soil bacteria by analysis of colony formation on agar plates. Biol Fertil Soils. 1989;7:198–201. [Google Scholar]
- 20.Hsu D, Shih L M, Zee Y C. Degradation of rRNA in Salmonella strains: a novel mechanism to regulate the concentrations of rRNA and ribosomes. J Bacteriol. 1994;176:4761–4765. doi: 10.1128/jb.176.15.4761-4765.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koch A L. The adaptive responses of Escherichia coli to a feast and famine existence. Adv Microb Physiol. 1971;6:147–217. doi: 10.1016/s0065-2911(08)60069-7. [DOI] [PubMed] [Google Scholar]
- 22.Laguerre G, Allard M, Revoy F, Amarger N. Rapid identification of rhizobia by restriction fragment length polymorphism analysis of PCR-amplified 16S rRNA genes. Appl Environ Microbiol. 1994;60:56–63. doi: 10.1128/aem.60.1.56-63.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Loughney K, Lund E, Dahlberg J E. Deletion of an rRNA gene set in Bacillus subtilis. J Bacteriol. 1983;154:529–532. doi: 10.1128/jb.154.1.529-532.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maidak B L, Olsen G J, Larsen N, Overbeek R, McCaughey M J, Woese C R. The RDP (Ribosomal Database Project) Nucleic Acids Res. 1997;25:109–110. doi: 10.1093/nar/25.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 26.Mitsui H, Gorlach K, Lee H-J, Hattori R, Hattori T. Incubation time and media requirements of culturable bacteria from different phylogenetic groups. J Microbiol Methods. 1997;30:103–110. [Google Scholar]
- 27.Pisabarro A, Correia A, Martin J F. Characterization of the rrnB operon of the plant pathogen Rhodococcus fascians and targeted integrations of exogenous genes at rrn loci. Appl Environ Microbiol. 1998;64:1276–1282. doi: 10.1128/aem.64.4.1276-1282.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rainey F A, Ward-Rainey N L, Janssen P H, Hippe H. Clostridium paradoxum DSM 7308(T) contains multiple 16S rRNA genes with heterogeneous intervening sequences. Microbiology. 1996;142:2087–2095. doi: 10.1099/13500872-142-8-2087. [DOI] [PubMed] [Google Scholar]
- 29.Schmidt T M. Multiplicity of ribosomal RNA operons in prokaryotic genomes. In: de Bruijn F J, Lupski J R, Weinstock G M, editors. Bacterial genomes: physical structure and analysis. New York, N.Y: Chapman and Hall Co.; 1997. pp. 221–229. [Google Scholar]
- 30.Stevenson B S, Schmidt T M. Growth rate-dependent expression of RNA from plasmid-borne rRNA operons in Escherichia coli. J Bacteriol. 1997;180:1970–1972. doi: 10.1128/jb.180.7.1970-1972.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Strunk O, Gross O, Reichel B, May M, Hermann S, Struckmann N, Nonhoff B, Lenke M, Vilbig A, Ludwig T, Bode A, Schleifer K H, Ludwig W. ARB: a software environment for sequence data. Munich, Germany: Department of Microbiology, Technical University of Munich; 1998. [Google Scholar]
- 32.Tilman D. Tests of resource competition theory using four species of Lake Michigan algae. Ecology. 1981;62:802–815. [Google Scholar]
- 33.Torsvik V, Goksoyr J, Daae F L. High diversity in DNA of soil bacteria. Appl Environ Microbiol. 1990;56:782–787. doi: 10.1128/aem.56.3.782-787.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vasi F, Travisano M, Lenski R E. Long-term experimental evolution in Escherichia coli. II. Changes in life-history traits during adaptation to a seasonal environment. Am Nat. 1994;144:432–456. [Google Scholar]
- 35.Weisburg W G, Barns S M, Pelletier D A, Lane D J. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zengel J M, Lindahl L. Diverse mechanisms for regulating ribosomal protein synthesis in Escherichia coli. Prog Nucleic Acid Res Mol Biol. 1994;47:331–370. doi: 10.1016/s0079-6603(08)60256-1. [DOI] [PubMed] [Google Scholar]
- 37.Zillig W. The order Thermococcales. In: Balows A, Truper H G, Dworkin M, Harder W, Schleifer K-H, editors. The prokaryotes. New York, N.Y: Springer-Verlag; 1992. pp. 702–706. [Google Scholar]