Abstract
Background
Racial bias is a key concern regarding the development, validation, and implementation of machine learning (ML) models in clinical settings. Despite the potential of bias to propagate health disparities, racial bias in clinical ML has yet to be thoroughly examined and best practices for bias mitigation remain unclear.
Objective
Our objective was to perform a scoping review to characterize the methods by which the racial bias of ML has been assessed and describe strategies that may be used to enhance algorithmic fairness in clinical ML.
Methods
A scoping review was conducted in accordance with the Preferred Reporting Items for Systematic Reviews and Meta-analyses (PRISMA) Extension for Scoping Reviews. A literature search using PubMed, Scopus, and Embase databases, as well as Google Scholar, identified 635 records, of which 12 studies were included.
Results
Applications of ML were varied and involved diagnosis, outcome prediction, and clinical score prediction performed on data sets including images, diagnostic studies, clinical text, and clinical variables. Of the 12 studies, 1 (8%) described a model in routine clinical use, 2 (17%) examined prospectively validated clinical models, and the remaining 9 (75%) described internally validated models. In addition, 8 (67%) studies concluded that racial bias was present, 2 (17%) concluded that it was not, and 2 (17%) assessed the implementation of bias mitigation strategies without comparison to a baseline model. Fairness metrics used to assess algorithmic racial bias were inconsistent. The most commonly observed metrics were equal opportunity difference (5/12, 42%), accuracy (4/12, 25%), and disparate impact (2/12, 17%). All 8 (67%) studies that implemented methods for mitigation of racial bias successfully increased fairness, as measured by the authors’ chosen metrics. Preprocessing methods of bias mitigation were most commonly used across all studies that implemented them.
Conclusions
The broad scope of medical ML applications and potential patient harms demand an increased emphasis on evaluation and mitigation of racial bias in clinical ML. However, the adoption of algorithmic fairness principles in medicine remains inconsistent and is limited by poor data availability and ML model reporting. We recommend that researchers and journal editors emphasize standardized reporting and data availability in medical ML studies to improve transparency and facilitate evaluation for racial bias.
Keywords: artificial intelligence, machine learning, race, bias, racial bias, scoping review, algorithm, algorithmic fairness, clinical machine learning, medical machine learning, fairness, assessment, model, diagnosis, outcome prediction, score prediction, prediction, mitigation
Introduction
Background
In recent years, artificial intelligence (AI) has drawn significant attention in medicine as machine learning (ML) techniques show an increasing promise of clinical impact. Driven by unprecedented data accessibility and computational capacity, ML has been reported to reach parity with human clinicians in a variety of tasks [1-3]. ML is poised to benefit patients and physicians by optimizing clinical workflows, enhancing diagnosis, and supporting personalized health care interventions [4-6]. Decision support tools based on ML have already been implemented across health systems [7,8], and the continued proliferation of clinical ML will impact patients in all fields of medicine.
However, despite its appeal, significant barriers remain to the full realization of clinically integrated ML. Key concerns include limited model transparency due to the “black box” of ML, inadequate reporting standards, and the need for prospective validation in clinical settings [1,9-12]. Racial bias in clinical ML is a crucial challenge arising from these limitations and must be addressed to ensure fairness in clinical implementation of ML. As ML is premised on prediction of novel outcomes based on previously seen examples, unintended discrimination is a natural consequence of algorithm development involving training data that reflect real-world inequities [13].
Equity in health care remains a continual pursuit [14,15]. Bias and disparities along dimensions of race, age, and gender have been shown to impact health care access and delivery, evident in varied settings, such as race correction in clinical algorithms or clinical trial enrollment and adverse event monitoring [16,17]. Considering the growing body of literature demonstrating profound adverse impacts of health care inequities on patient outcomes, mitigation of the numerous and insidious sources of potential bias in medicine requires remains a critical challenge to prevent harm to patients [14,17]. Thus, the potential for algorithms to perpetuate health disparities must be carefully weighed when incorporating ML models into clinical practice [18-20].
Algorithmic fairness is an area of ML research guiding model development with the aim of preventing discrimination involving protected groups, which are defined by attributes such as race, gender, religion, physiologic variability, preexisting conditions, physical ability, and sexual orientation [13,19]. However, application of algorithmic fairness principles in the medical ML literature remains nascent [20]. Greater awareness of the potential harms of bias in clinical ML as well as methods to evaluate and mitigate them is needed to support clinicians and researchers across the health care and data science disciplines, who must evaluate and implement clinical ML models with a critical eye toward algorithmic fairness. The objective of this study is to characterize the impact and mitigation of racial bias in clinical ML to date and describe best practices for research efforts extending algorithmic fairness to medicine.
Bias and Fairness in Machine Learning
In the setting of algorithmic fairness, bias is present when an algorithm systematically favors one outcome over another. Bias may be introduced into an ML algorithm throughout all steps of the development process, which involves data collection, data selection, model training, and model deployment [13]. Examples of these sources of bias are shown in Figure 1, and their definitions are given in Multimedia Appendix 1. Notably, historical bias may be present even if all steps of model development are optimally performed. This is of particular concern in the evaluation of racial bias in clinical ML, given the presence of existing and historical health care disparities [14].
Figure 1.
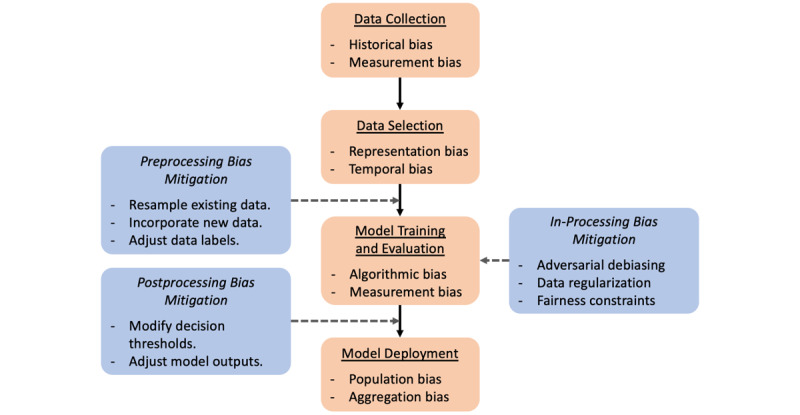
The clinical machine learning development workflow (orange boxes) offers several opportunities (blue boxes) to evaluate and mitigate potential biases introduced by the data set or model. Preprocessing methods seek to adjust the existing data set to preempt biases resulting from inadequate data representation or labeling. In-processing methods impose fairness constraints as additional metrics optimized by the model during training or present data in a structured manner to avoid biases in the sampling process. Postprocessing methods account for model biases by adjusting model outputs or changing the way they are used.
Depending on the context, bias in clinical ML may not be harmful and can even be used to overcome inequality [13]. In situations in which targeting a well-defined subpopulation above all others is desirable, an ML algorithm biased toward a particular group may be used to proactively mitigate existing disparities. However, bias may arise when ML models designed to serve the needs of a specific clinical population—such as a particular community or high-risk demographic—are inappropriately applied to other populations or when more general models are applied to specific populations. Additionally, ML algorithms tend to overfit to the data on which they are trained, which entails the learning of spurious relationships present in the training data set and may result in a lack of generalizability to other settings. As a result, a model that appears unbiased in one setting may display bias in another. Thus, bias in clinical ML must be considered in the light of the context and particular population of interest.
Bias in an ML model may lead to unfairness if not appropriately evaluated and accounted for. Fairness in ML is achieved when algorithmic decision-making does not favor an individual or group based on protected attributes. Research efforts have emphasized group fairness over individual fairness, given the need for algorithms that consider existing differences between populations—whether intrinsic or extrinsic—while preventing discrimination between groups [13,21]. Crucially, improving model fairness does not necessarily require compromising accuracy overall [22]. For instance, an unfair disease-screening tool might have poor sensitivity for disease detection in one low-risk population subgroup compared to another with higher risk; improving the fairness of this tool would entail adjusting the model to have more similar sensitivities between subgroups. In this study, we examine the racial bias of clinical ML in terms of model fairness with respect to race.
Assessing and Achieving Fairness in Machine Learning
Group fairness is quantified by evaluating the similarity of a given statistical metric between predictions made for different groups. Group fairness indicators encountered in this review are defined in Table 1. Critical examinations of different methods for evaluating fairness in ML, both in general application [13,23,24] and in the context of health care [21], have been previously described, though applications in clinical ML remain limited. It is important to note that fairness metrics may be at odds with one another, depending on the context and application [25]; thus, evaluation of an appropriate metric, given the clinical situation of interest, is paramount [26].
Table 1.
Group fairness metrics encountered in this review.
| Term | Description |
| AUROCa | Assesses overall classifier performance by measuring the TPRb and FPRc of a classifier at different thresholds. |
| Average odds | Compares the average of the TPR and FPR for the classification outcome between protected and unprotected groups. |
| Balanced accuracy | A measure of accuracy corrected for data imbalance, calculated as the average of sensitivity and specificity for a group. |
| Calibration | Assesses how well the risk score or probability predictions reflect actual outcomes. |
| Disparate impact | Measures deviation from statistical parity, calculated as the ratio of the rate of the positive outcome between protected and unprotected groups. Ideally, the disparate impact is 1. |
| Equal opportunity | For classification tasks in which one outcome is preferred over the other, equal opportunity is satisfied when the preferred outcome is predicted with equal accuracy between protected and unprotected groups. Ideally, the TPR or FNRd disparity between groups is 0. |
| Equalized odds | The TPR and FPR are equal between protected and unprotected groups. |
| Error rate | Compares the error rate of predictions, calculated as the number of incorrect predictions divided by the total number of predictions, between protected and unprotected groups. Ideally, the error rate disparity between groups is 0. |
| Statistical parity | Statistical parity (also known as demographic parity) is satisfied when the rate of positive outcomes is equal between protected and unprotected groups. |
aAUROC: area under the receiver operating characteristic curve.
bTPR: true-positive rate.
cFPR: false-positive rate.
dFNR: false-negative rate.
Approaches to bias mitigation fall into 3 major categories (Figure 1): preprocessing, in which inequities in data are removed prior to model training; in-processing, in which the model training process is conducted to actively prevent discrimination; and postprocessing, in which outputs of a trained model are adjusted to achieve fairness [13]. Preprocessing can be performed by resampling existing data, incorporating new data, or adjusting data labels. In-processing methods use adversarial techniques, impose constraints and regularization, or ensure fairness of underlying representations during training. Finally, postprocessing entails group-specific modification of decision thresholds or outcomes to ensure fairness in the application of model predictions. Different approaches may be optimal depending on the setting and stage of model development.
Methods
Study Design
We performed a scoping review of racial bias and algorithmic fairness in clinical ML models in accordance with the Preferred Reporting Items for Systematic Reviews and Meta-analyses (PRISMA) 2020 guidelines [27] and PRISMA Extension for Scoping Reviews [28]. The review protocol was not registered and is available upon request to the authors. The PubMed MEDLINE (National Library of Medicine), Scopus (Elsevier), and Embase (Elsevier) databases were queried by combining terminology pertaining to ML, race, and bias as keywords. Additional records were identified using Google Scholar search. The exact search strategy is detailed in Multimedia Appendix 1.
Study Selection
After duplicate record removal, studies were initially screened by title and abstract and then screened for final inclusion by full text review. All screening was performed independently by 2 reviewers. Studies were selected based on the following inclusion criteria: peer-reviewed original research, English language, full text available, development or evaluation of a clinically relevant ML model, and evaluation of bias of the model regarding racial or ethnic groups. Studies other than full-length papers were excluded. ML was defined as a computer algorithm that improves automatically via training on data [4]. Per PRISMA guidelines, any disagreements regarding study inclusion based on these criteria were reconciled by discussion.
Data Abstraction
Relevant data were abstracted from included papers by 1 reviewer. Data of interest included the clinical objective of ML models, identification of racial bias, impact of racial bias, metrics for bias assessment, mitigation of racial bias, methods for bias mitigation, data set size, data source, ML model architecture, and availability of computer code used for data preparation and ML model development. The methodological quality of included studies was not assessed, given the scoping nature of this review [28].
Results
Study Characteristics
The literature search was performed on September 8, 2021, and identified 635 records (Figure 2). Of these, 26 (4.1%) full-text papers were reviewed and 12 (46.2%) were included in the final analysis [29-40].
Figure 2.
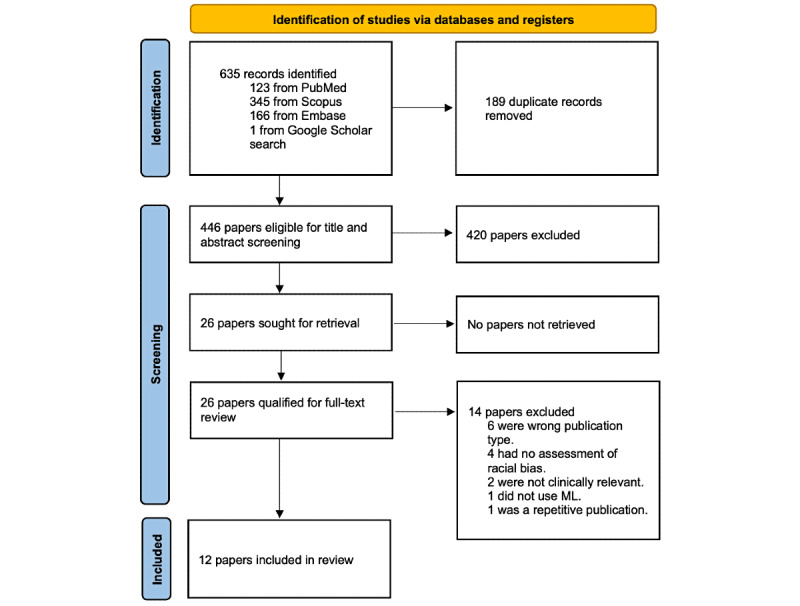
PRISMA flowchart of study inclusion. ML: machine learning; PRISMA: Preferred Reporting Items for Systematic Reviews and Meta-analyses.
Characteristics of the included studies are summarized in Table 2. Data sets and models used are summarized in Multimedia Appendix 1. Of the 12 studies, 3 (25%) were published in 2019, 5 (42%) in 2020, and 4 (33%) in 2021. In addition, 9 (75%) studies originated from the United States, 1 (8%) from Canada, 1 (8%) from Sweden, and 1 (8%) from both the United Kingdom and Nigeria. Applications of ML were varied and involved diagnosis, outcome prediction, and clinical score prediction performed on data sets including images, diagnostic studies, clinical text, and clinical variables. Furthermore, 1 (8%) study described a model in routine clinical use [36], 2 (17%) examined prospectively validated clinical models [35,39], and the remaining 9 (75%) described internally validated models.
Table 2.
Study characteristics.
| Author (year) | Clinical objective | How was fairness evaluated? | Was racial bias identified? | How was the AIa model biased? | Was racial bias mitigated? | Protected class |
| Abubakar et al (2020) [29] | Identification of images of burns vs healthy skin | Accuracy | Yes | Poor accuracy of models trained on a Caucasian data set and validated on an African data set and vice versa | Yes | Dark-skinned patients, light-skinned patients |
| Allen et al (2020) [30] | Intensive care unit (ICU) mortality prediction | Equal opportunity difference (FNRb disparity) | N/Ac | N/A | Yes | Non-White patients |
| Briggs and Hollmén (2020) [31] | Prediction of future health care expenditures of individual patients | Balanced accuracy, statistical parity, disparate impact, average odds, equal opportunity | N/A | N/A | Yes | Black patients |
| Burlina et al (2021) [32] | Diagnosis of diabetic retinopathy from fundus photography | Accuracy | Yes | Lower diagnostic accuracy in darker-skinned individuals compared to lighter-skinned individuals | Yes | Dark-skinned patients |
| Chen et al (2019) [33] | ICU mortality prediction, psychiatric readmission prediction | Error rate (0-1 loss) | Yes | Differences in error rates in ICU mortality between racial groups | No | Non-White patients |
| Gianattasio et al (2020) [34] | Dementia status classification | Sensitivity, specificity, accuracy | Yes | Existing algorithms varying in sensitivity and specificity between race/ethnicity groups | Yes | Hispanic, non-Hispanic Black patients |
| Noseworthy et al (2020) [35] | Prediction of left ventricular ejection fraction ≤35% from the electrocardiogram (ECG) | AUROCd | No | N/A | No | Non-White patients |
| Obermeyer et al (2019) [36] | Prediction of future health care expenditures of individual patients | Calibration | Yes | Black patients with a higher burden than White patients at the same algorithmic risk score | Yes | Black patients |
| Park et al (2021) [37] | Prediction of postpartum depression and postpartum mental health service utilization | Disparate impact, equal opportunity difference (TPRe disparity) | Yes | Black women with a worse health status than White women at the same predicted risk level | Yes | Black patients |
| Seyyed-Kalantari et al (2021) [38] | Diagnostic label prediction from chest X-rays | Equal opportunity difference (TPR disparity) | Yes | Greater TPR disparity in Hispanic patients | No | Non-White patients |
| Thompson et al (2021) [39] | Identification of opioid misuse from clinical notes | Equal opportunity difference (FNR disparity) | Yes | Greater FNR in the Black subgroup than in the White subgroup | Yes | Black patients |
| Wissel et al (2019) [40] | Assignment of surgical candidacy score for patients with epilepsy using clinical notes | Regression analysis of the impact of the race variable on the candidacy score | No | N/A | No | Non-White patients |
aAI: artificial intelligence.
bFNR: false-negative rate.
cN/A: not applicable.
dAUROC: area under the receiver operating characteristic curve.
eTPR: true-positive rate.
Of the 12 studies, 5 (42%) published code used for analysis, 3 (25%) made model development code available [34,36,39], 2 (17%) published bias analysis code [33,36], 1 (8%) published code relevant to debiasing [30], and 1 (8%) published data selection code [33]. In addition, 1 (8%) study used publicly available code for analysis [31], and code was specified as available upon request in 1 (8%) study [35]. Bias of an ML model was evaluated using an external database in 8 (67%) studies [30-34,37,38], single-institutional data in 3 (25%) studies [35,36,40], and data from 2 institutions in 2 (17%) studies [29,39]. No institutional data sets were published. Convolutional neural networks (CNNs) were the predominant ML modeling technique used (5/12, 42%), followed by logistic regression (3/12, 25%), least absolute shrinkage and selection operator (LASSO; 2/12, 17%), and extreme gradient boosting (XGBoost; 2/12, 17%). In addition, 3 (25%) studies evaluated models adapted from existing neural network architectures: ResNet50 in 2 (17%) studies [29,32] and DenseNet in the other [38].
Of the 12 studies, 9 (75%) evaluated a model developed internally by the same researchers [29-33,35,37,39,40], 2 (17%) evaluated a model developed externally by separate researchers [36,38], and 1 (8%) evaluated both internally and externally developed models [34]. In addition, 8 (67%) studies concluded that racial bias was present [29,32-34,36-39], 2 (17%) concluded that bias was not present [35,40], and 2 (17%) assessed the implementation of bias mitigation strategies without comparison to a baseline model [30,31]. A variety of methods were used to assess the presence of algorithmic racial bias: 3 (25%) studies used multiple metrics to assess fairness [31,34,37], while the remaining 9 (75%) used a single metric. The most commonly used fairness metrics were equal opportunity difference [41], defined either as the difference in the true-positive rate (TPR) or the false-negative rate (FNR) between subgroups (5/12, 42%) [30,31,38,39]; accuracy (4/12, 25%) [29,31,32,34]; and disparate impact (2/12, 17%) [31,37].
The approaches and efficacy of bias mitigation methods used in the studies evaluated are summarized in Table 3. All 8 (67%) studies that implemented methods for mitigation of racial bias successfully increased fairness, as measured by the authors’ chosen metrics [29-32,34,36,37,39]. Preprocessing bias mitigation was the most commonly used strategy (7/13, 54%). In addition, 1 (8%) study removed race information from the training data, though superior improvements in disparate impact and equal opportunity difference were achieved by reweighing [37]. Furthermore, 2 (17%) studies performed in-processing bias mitigation using the prejudice remover regularizer [42] or adversarial debiasing during model training [31,37]. However, in both studies, in-processing was ineffective in reducing bias and was outperformed by other bias mitigation methods. Finally, 1 (8%) study evaluated multiple types of ML models for bias during the development process, concluding that a LASSO model was preferable to conditional random forest, gradient boosting, and ensemble models for racially unbiased dementia ascertainment [34].
Table 3.
Bias mitigation methods among reviewed studies.
| Description of strategies used | Effectiveness | |
| Preprocessing | ||
|
|
Reweighing training data |
|
|
|
Combining data sets to increase heterogeneity |
|
|
|
Generating synthetic minority class data |
|
|
|
Adjusting label selection |
|
|
|
Removing race information from training data |
|
| In-processing | ||
|
|
Use of a regularizer during training |
|
|
|
Adversarial debiasing |
|
| Postprocessing | ||
|
|
Calibration |
|
|
|
Reject option-based classification |
|
|
|
Varying cut-point selection | |
aFNR: false-negative rate.
bTPR: true-positive rate.
Discussion
Principal Findings
Given the pressing issue of equity in health care and the rapid development of medical ML applications, racial bias must be thoroughly evaluated in clinical ML models in order to protect patient safety and prevent the algorithmic encoding of inequality. Algorithmic fairness is a relatively novel field within the discipline of ML, and its application to medical ML remains nascent. In our evaluation of the literature describing mitigation of racial bias in clinical ML, we identified a variety of bias mitigation methods, which when applied successfully increase fairness and demonstrate the feasibility and importance of racial bias evaluation in the medical ML development process. Based on our findings, there is a need for heightened awareness of algorithmic fairness concepts, increased data availability, and improved reporting transparency in medical ML development to ensure fairness in clinical ML.
Impact of Racial Bias in Clinical Machine Learning
The broad scope of medical ML applications and potential patient harms following deployment across health care systems demand an increased emphasis on evaluation and mitigation of racial bias in clinical ML. Screening and outcome prediction tasks are commonly examined among reviewed studies. Racial bias in such tasks is particularly concerning as decisions made from flawed models trained on data, which reflect historical inequities in disease diagnosis and care delivery, may perpetuate inequalities by shaping clinical decision-making [14,19]. Evaluation and mitigation of potential biases must occur throughout the model development life cycle to protect patients from algorithmic unfairness.
Reviewed studies frequently identified racial bias in clinical ML models. Notably, 1 algorithm in clinical use for prediction of future health care expenditures was found to discriminate against Black patients when compared to White patients, potentially contributing to disparities in health care delivery [36]. Other ML models that possibly demonstrate racial bias remain in preclinical states of development. Several studies have explicitly studied racial bias against Black patients compared to White patients. For example, 2 studies demonstrated that ML algorithms predicted similar risk scores in Black and White patients, though the Black patients were less healthy [36,37], and another demonstrated that an opioid misuse classifier had a higher FNR for Black patients [39]. Disparities in mortality prediction and X-ray diagnosis were identified in other races and ethnic groups [33,34,38], as well as disparities in burn identification and diabetic retinopathy identification in dark-skinned versus lighter-skinned patients [29,32]. Although conclusions cannot be drawn regarding the prevalence of racial bias among published clinical ML studies, the broad scope of clinical ML models susceptible to racial bias in this review exposes the potential of racial bias encoded in ML models to negatively impact patients across all aspects of health care.
Assessment of Racial Bias
Clinical ML models must be carefully evaluated for potential biases imposed upon patients. Different fairness metrics may highlight different aspects of fairness relevant to a particular clinical setting; therefore, evaluation of all appropriate fairness metrics is needed when evaluating for potential bias. For example, calibration is particularly important to models performing risk prediction, while equal opportunity and disparate impact are relevant to screening and diagnostic settings. Inconsistent choice of fairness metrics among studies included in this review shows the need for a more standardized assessment process of racial bias in clinical ML. Some studies assessed fairness using metrics such as accuracy, area under the receiver operating characteristic curve (AUROC), and correlation of outcome with race, which may not sufficiently evaluate fairness [21]. Moreover, there are inherent trade-offs to the use of different fairness metrics [25], and static fairness criteria may even lead to delayed harms in the long term [43].
Obermeyer et al [36] present an example of using model calibration in conjunction with varied outcome labels to successfully de-bias an algorithm used to manage population health, and case studies have examined trade-offs of bias evaluation metrics in other settings, such as criminal justice [44], which may also serve as useful frameworks for clinical ML researchers. Use of “causal models,” which allow for closely tailored examination of discriminatory relationships in data, is another opportunity for investigation and mitigation of biased model behavior [45]. An increased focus from medical journals on bias evaluation checklists applicable to clinical ML models, such as the Prediction Model Risk of Bias Assessment Tool (PROBAST), is desirable to further emphasize vigilance regarding biased ML models [46]. Ultimately, more thorough analysis of fairness criteria in clinical ML will allow researchers to better contextualize and act on potential biases.
Clinical ML researchers should also be aware of potential barriers to ML fairness when adapting pretrained models and data representations. For instance, deep neural networks performing image processing tasks are frequently pretrained on large data sets and then fine-tuned to adapt to other tasks. Methods for removal of spurious variations from such models have been described, such as joint learning and unlearning algorithms, which account for contributions of undesirable variations during model development [47]. Language models trained in an unsupervised manner on vast amounts of text may learn biases present in training data [48]. Similarly, biases have been described in word embeddings [49], which are vectorized word representations used as inputs to ML models. Identification of bias in embeddings raises concerns about performance disparities in clinical applications of natural language processing if the bias is not screened for and appropriately addressed [50]. The lack of interpretability often inherent to ML models heightens the need for thorough evaluation of their potential biases.
Creating Fair Models
Preprocessing and postprocessing methods of bias mitigation were successfully implemented among the publications reviewed for this study. Postprocessing methods appear to be easier to implement and may allow tailoring of imperfect models to new settings [51]. However, using preprocessing and in-processing to create unbiased data sets and algorithms at the outset of model development is desirable to facilitate the creation of fair, generalizable models. Continued evaluation of these techniques in clinical contexts is needed to inform best practices.
As data quality is generally the limiting factor to development of robust ML models, improvements to data generally translates directly into model performance improvements. Supplementation of data sets using generative models to synthesize patient data may be a viable approach to address data limitations. A study by Burlina et al [32] illustrated this fact by using a generative adversarial network to synthesize fundoscopy images while reducing class imbalance. However, though data limitations may contribute to disparities in model performance across racial groups, algorithmic unfairness may arise from other underlying biases in data as well [38]. Publications included in this review demonstrated improved fairness in ML models using multisource data sets, which may mitigate biases in the data collection process of single-source data sets [29,38]. Moreover, care must also be taken to ensure that multi-institutional data sets are appropriately prepared and used due to evidence that site-specific signatures contribute to bias in ML models [52]. Finally, protected attributes should not simply be ignored during model development, an approach called “fairness through unawareness,” as models may be able to infer protected group membership from other data features. Additionally, omission of protected attributes may cause bias if a legitimate relationship exists between the attribute and outcome of interest [19].
Several online resources aggregate examples and code implementations of published fairness evaluation and bias mitigation methods. Some examples of these resources include Aequitas, Artificial Intelligence Fairness 360 (IBM, Armonk, NY, United States), and Fairlearn (Microsoft Corporation, Redmond, WA, United States) [53,54]. Additionally, TensorFlow, a popular deep learning framework, includes a tool for evaluation of fairness indicators. Work by Briggs et al [31] highlights the feasibility and positive impact of standardized methodologies for addressing bias using a variety of performance indicators and mitigation techniques. Greater adoption of these and other strategies in fairness evaluation and bias mitigation will help set standard benchmarks for fairness in clinical ML.
The Role of Transparency and Data Availability
ML is often characterized as a black box due to its limited interpretability, which is particularly problematic when attempting to address and prevent racial biases in clinical ML [55]. Although research in recent years has yielded significant progress in explainable ML methods [56], publication of model development code and data sets remains the most straightforward approach to transparency. Regrettably, medical ML research falls far short of these standards [57,58]. Code and data availability was inconsistent among the publications included in this review, and the majority of studies evaluated racial bias using publicly available data sets, including the Medical Information Mart for Intensive Care (MIMIC) [30,33,38], Kaggle EyePACS [32], and Dissecting Bias [31]. Considering the vast number of private, institutional data sets used to develop clinical ML models, there is a crucial need for future publications to maximize transparency, ensuring the ability to evaluate for fairness in clinical ML.
Increased publication of institutional data sets would facilitate the interdisciplinary collaboration needed to translate concepts of fairness in ML into the realm of medicine. Improved availability of data sets would also enable researchers to more easily validate existing models and perform fairness evaluations on different patient populations, translating benefits of ML across populations. Additionally, collaboration between institutions to maintain diverse, broadly representative data sets would facilitate the development of generalizable models free of the biases inherent to single-institutional data. However, ethical and patient confidentiality considerations may limit publication of clinical data. In contrast, publication of code and trained models, which are infrequently made available in the clinical ML literature [1,59], would similarly allow researchers to assess clinical ML on diverse populations without limitations imposed by patient privacy standards or institutional data-sharing regulations. Another possible paradigm to mitigate bias by training on diversely representative data sets while maintaining data privacy is federated learning, which involves piecewise training of an ML model on separate data sets and removes the need for data sharing during model development [60].
Moreover, increased emphasis on fairness in clinical ML through adoption of model development and reporting guidelines is needed [59,61]. Reporting guidelines for medical ML studies are inconsistently adopted, due in part to a lack of editorial policies among medical journals [1]. Moreover, reporting of demographic information needed to assess biases due to data sets is lacking [62,63]. The proposed Minimum Information for Medical AI Reporting guideline addresses these concerns by recommending that clinical ML studies report information necessary for understanding potential biases, including relevant demographic information of patient data used for model development [64]. In conjunction with upcoming reporting guidelines tailored to clinical ML [61], efforts to improve reporting quality will contribute to a standardized framework for fairness evaluation and bias mitigation in clinical ML.
Limitations
As with any literature review, there are limitations to this study. Given the heterogeneity of terminology used to describe ML and racial bias, our search may have overlooked relevant publications. Additionally, we were limited by publication bias as we excluded publications other than full-length manuscripts, and researchers may be less likely to publish results confirming the absence of racial bias in a clinical ML model. Finally, the novelty of ML fairness in medicine and the resulting paucity of literature on this topic, as well as the breadth of relevant subjects encompassed, prevented us from obtaining the quantity and quality of data required to perform a systematic review or meta-analysis. In particular, the lack of standardized methods to evaluate and mitigate bias precludes any definitive conclusions regarding their suitability in clinical ML applications. However, the scoping review provides a methodological framework for critical evaluation of a previously uncharacterized area of research and draws attention to the lack of standardization regarding racial bias mitigation in clinical ML development. We emphasize the need for further work to build on this important aspect of the medical ML literature.
Conclusion
Algorithmic fairness in clinical ML is a primary concern in its ethical adoption. As medical ML applications continue to approach widespread adoption across a multitude of clinical settings, potential racial biases in ML models must be proactively evaluated and mitigated in order to prevent patient harm and propagation of inequities in health care. The adoption of algorithmic fairness principles in medicine remains nascent, and further research is needed to standardize best practices for fairness evaluation and bias mitigation. We recommend that researchers and journal editors emphasize standardized reporting and data availability in ML studies to improve transparency and facilitate future research. Continued interrogation of biases in clinical ML models is needed to ensure fairness and maximize the benefits of ML in medicine.
Abbreviations
- AI
artificial intelligence
- AUROC
area under the receiver operating characteristic curve
- FNR
false-negative rate
- FPR
false-positive rate
- LASSO
least absolute shrinkage and selection operator
- ML
machine learning
- PRISMA
Preferred Reporting Items for Systematic Reviews and Meta-analyses
- TPR
true-positive rate
Supplementary data file containing bias definitions, search strategy, and a table with study data set characteristics.
Footnotes
Authors' Contributions: No part of this work has been previously published.
Conflicts of Interest: None declared.
References
- 1.Nagendran M, Chen Y, Lovejoy CA, Gordon AC, Komorowski M, Harvey H, Topol EJ, Ioannidis JPA, Collins GS, Maruthappu M. Artificial intelligence versus clinicians: systematic review of design, reporting standards, and claims of deep learning studies. BMJ. 2020 Mar 25;368:m689. doi: 10.1136/bmj.m689. http://www.bmj.com/lookup/pmidlookup?view=long&pmid=32213531 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McKinney SM, Sieniek M, Godbole V, Godwin J, Antropova N, Ashrafian H, Back T, Chesus M, Corrado GC, Darzi A, Etemadi M, Garcia-Vicente F, Gilbert FJ, Halling-Brown M, Hassabis D, Jansen S, Karthikesalingam A, Kelly CJ, King D, Ledsam JR, Melnick D, Mostofi H, Peng L, Reicher JJ, Romera-Paredes B, Sidebottom R, Suleyman M, Tse D, Young KC, De Fauw J, Shetty S. International evaluation of an AI system for breast cancer screening. Nature. 2020 Jan 01;577(7788):89–94. doi: 10.1038/s41586-019-1799-6. [DOI] [PubMed] [Google Scholar]
- 3.Shen J, Zhang CJP, Jiang B, Chen J, Song J, Liu Z, He Z, Wong SY, Fang P, Ming W. Artificial intelligence versus clinicians in disease diagnosis: systematic review. JMIR Med Inform. 2019 Aug 16;7(3):e10010. doi: 10.2196/10010. https://medinform.jmir.org/2019/3/e10010/ v7i3e10010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Topol EJ. High-performance medicine: the convergence of human and artificial intelligence. Nat Med. 2019 Jan 7;25(1):44–56. doi: 10.1038/s41591-018-0300-7.10.1038/s41591-018-0300-7 [DOI] [PubMed] [Google Scholar]
- 5.Abul-Husn NS, Kenny EE. Personalized medicine and the power of electronic health records. Cell. 2019 Mar 21;177(1):58–69. doi: 10.1016/j.cell.2019.02.039. https://linkinghub.elsevier.com/retrieve/pii/S0092-8674(19)30222-3 .S0092-8674(19)30222-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Obermeyer Z, Emanuel EJ. Predicting the future: big data, machine learning, and clinical medicine. N Engl J Med. 2016 Sep 29;375(13):1216–9. doi: 10.1056/NEJMp1606181. http://europepmc.org/abstract/MED/27682033 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Domingo J, Galal G, Huang J, Soni P, Mukhin V, Altman C, Bayer T, Byrd T, Caron S, Creamer P, Gilstrap J, Gwardys H, Hogue C, Kadiyam K, Massa M, Salamone P, Slavicek R, Suna M, Ware B, Xinos S, Yuen L, Moran T, Barnard C, Adams JG, Etemadi M. Preventing delayed and missed care by applying artificial intelligence to trigger radiology imaging follow-up. NEJM Catalyst. 2022 Mar 16;3(4) doi: 10.1056/cat.21.0469. [DOI] [Google Scholar]
- 8.Avati A, Jung K, Harman S, Downing L, Ng A, Shah NH. Improving palliative care with deep learning. BMC Med Inform Decis Mak. 2018 Dec 12;18(Suppl 4):122. doi: 10.1186/s12911-018-0677-8. https://bmcmedinformdecismak.biomedcentral.com/articles/10.1186/s12911-018-0677-8 .10.1186/s12911-018-0677-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.He J, Baxter SL, Xu J, Xu J, Zhou X, Zhang K. The practical implementation of artificial intelligence technologies in medicine. Nat Med. 2019 Jan;25(1):30–36. doi: 10.1038/s41591-018-0307-0. http://europepmc.org/abstract/MED/30617336 .10.1038/s41591-018-0307-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilkinson J, Arnold KF, Murray EJ, van Smeden M, Carr K, Sippy R, de Kamps M, Beam A, Konigorski S, Lippert C, Gilthorpe MS, Tennant PWG. Time to reality check the promises of machine learning-powered precision medicine. Lancet Digital Health. 2020 Dec;2(12):e677–e680. doi: 10.1016/s2589-7500(20)30200-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kelly CJ, Karthikesalingam A, Suleyman M, Corrado G, King D. Key challenges for delivering clinical impact with artificial intelligence. BMC Med. 2019 Oct 29;17(1):195. doi: 10.1186/s12916-019-1426-2. https://bmcmedicine.biomedcentral.com/articles/10.1186/s12916-019-1426-2 .10.1186/s12916-019-1426-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Challen R, Denny J, Pitt M, Gompels L, Edwards T, Tsaneva-Atanasova K. Artificial intelligence, bias and clinical safety. BMJ Qual Saf. 2019 Mar 12;28(3):231–237. doi: 10.1136/bmjqs-2018-008370. http://qualitysafety.bmj.com/lookup/pmidlookup?view=long&pmid=30636200 .bmjqs-2018-008370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mehrabi N, Morstatter F, Saxena N, Lerman K, Galstyan A. A survey on bias and fairness in machine learning. ACM Comput Surv. 2021 Jul;54(6):1–35. doi: 10.1145/3457607. [DOI] [Google Scholar]
- 14.Bailey ZD, Feldman JM, Bassett MT. How structural racism works: racist policies as a root cause of U.S. racial health inequities. N Engl J Med. 2021 Feb 25;384(8):768–773. doi: 10.1056/nejmms2025396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rodriguez JA, Clark CR, Bates DW. Digital health equity as a necessity in the 21st Century Cures Act era. JAMA. 2020 Jun 16;323(23):2381–2382. doi: 10.1001/jama.2020.7858.2766776 [DOI] [PubMed] [Google Scholar]
- 16.Unger JM, Vaidya R, Albain KS, LeBlanc M, Minasian LM, Gotay CC, Henry NL, Fisch MJ, Lee SM, Blanke CD, Hershman DL. Sex differences in risk of severe adverse events in patients receiving immunotherapy, targeted therapy, or chemotherapy in cancer clinical trials. JCO. 2022 May 01;40(13):1474–1486. doi: 10.1200/jco.21.02377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vyas DA, Eisenstein LG, Jones DS. Hidden in plain sight: reconsidering the use of race correction in clinical algorithms. N Engl J Med. 2020 Aug 27;383(9):874–882. doi: 10.1056/NEJMms2004740. http://paperpile.com/b/Mk3QOF/QN3U . [DOI] [PubMed] [Google Scholar]
- 18.Paulus JK, Kent DM. Predictably unequal: understanding and addressing concerns that algorithmic clinical prediction may increase health disparities. NPJ Digit Med. 2020 Jul 30;3(1):99. doi: 10.1038/s41746-020-0304-9. doi: 10.1038/s41746-020-0304-9.304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rajkomar A, Hardt M, Howell MD, Corrado G, Chin MH. Ensuring fairness in machine learning to advance health equity. Ann Intern Med. 2018 Dec 04;169(12):866. doi: 10.7326/m18-1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen IY, Pierson E, Rose S, Joshi S, Ferryman K, Ghassemi M. Ethical machine learning in healthcare. Annu Rev Biomed Data Sci. 2021 Jul 20;4(1):123–144. doi: 10.1146/annurev-biodatasci-092820-114757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fletcher RR, Nakeshimana A, Olubeko O. Addressing fairness, bias, and appropriate use of artificial intelligence and machine learning in global health. Front Artif Intell. 2020;3:561802. doi: 10.3389/frai.2020.561802. doi: 10.3389/frai.2020.561802.561802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wick M, Panda S, Tristan J. Unlocking fairness: a trade-off revisited. Advances in Neural Information Processing Systems 32: Annual Conference on Neural Information Processing Systems; December 2019; Vancouver, BC, Canada. 2019. https://jtristan.github.io/papers/neurips19.pdf . [Google Scholar]
- 23.Verma S, Rubin J. Fairness definitions explained. Proceedings of the International Workshop on Software Fairness; 2018; Gothenburg, Sweden. 2018. [DOI] [Google Scholar]
- 24.Friedler S, Scheidegger C, Venkatasubramanian S, Choudhary S, Hamilton E, Roth D. A comparative study of fairness-enhancing interventions in machine learning. Proceedings of the Conference on Fairness, Accountability, Transparency; 2019; Atlanta, GA. 2019. [DOI] [Google Scholar]
- 25.Kleinberg J, Mullainathan S, Raghavan M. Inherent trade-offs in the fair determination of risk scores. 8th Innovations in Theoretical Computer Science Conference (ITCS 2017); January 2017; Berkeley, CA. 2017. [DOI] [Google Scholar]
- 26.McCradden MD, Joshi S, Mazwi M, Anderson JA. Ethical limitations of algorithmic fairness solutions in health care machine learning. Lancet Digital Health. 2020 May;2(5):e221–e223. doi: 10.1016/s2589-7500(20)30065-0. [DOI] [PubMed] [Google Scholar]
- 27.Page MJ, McKenzie JE, Bossuyt PM, Boutron I, Hoffmann TC, Mulrow CD, Shamseer L, Tetzlaff JM, Akl EA, Brennan SE, Chou R, Glanville J, Grimshaw JM, Hróbjartsson A, Lalu MM, Li T, Loder EW, Mayo-Wilson E, McDonald S, McGuinness LA, Stewart LA, Thomas J, Tricco AC, Welch VA, Whiting P, Moher D. The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. BMJ. 2021 Mar 29;372:n71. doi: 10.1136/bmj.n71. http://www.bmj.com/lookup/pmidlookup?view=long&pmid=33782057 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tricco AC, Lillie E, Zarin W, O'Brien KK, Colquhoun H, Levac D, Moher D, Peters MD, Horsley T, Weeks L, Hempel S, Akl EA, Chang C, McGowan J, Stewart L, Hartling L, Aldcroft A, Wilson MG, Garritty C, Lewin S, Godfrey CM, Macdonald MT, Langlois EV, Soares-Weiser K, Moriarty J, Clifford T, Tunçalp Ö, Straus SE. PRISMA Extension for Scoping Reviews (PRISMA-ScR): checklist and explanation. Ann Intern Med. 2018 Sep 04;169(7):467. doi: 10.7326/M18-0850. [DOI] [PubMed] [Google Scholar]
- 29.Abubakar A, Ugail H, Bukar AM. Assessment of human skin burns: a deep transfer learning approach. J Med Biol Eng. 2020 Apr 24;40(3):321–333. doi: 10.1007/s40846-020-00520-z. [DOI] [Google Scholar]
- 30.Allen A, Mataraso S, Siefkas A, Burdick H, Braden G, Dellinger RP, McCoy A, Pellegrini E, Hoffman J, Green-Saxena A, Barnes G, Calvert J, Das R. A racially unbiased, machine learning approach to prediction of mortality: algorithm development study. JMIR Public Health Surveill. 2020 Oct 22;6(4):e22400. doi: 10.2196/22400. https://publichealth.jmir.org/2020/4/e22400/ v6i4e22400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Briggs E, Hollmén J. Mitigating discrimination in clinical machine learning decision support using algorithmic processing techniques. In: Appice A, Tsoumakas G, Manolopoulos Y, Matwin S, editors. International Conference on Discovery Science. Vol 12323. Cham: Springer International; 2020. pp. 19–33. [Google Scholar]
- 32.Burlina P, Joshi N, Paul W, Pacheco KD, Bressler NM. Addressing artificial intelligence bias in retinal diagnostics. Transl Vis Sci Technol. 2021 Feb 05;10(2):13. doi: 10.1167/tvst.10.2.13. https://tvst.arvojournals.org/article.aspx?doi=10.1167/tvst.10.2.13 .2772266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen I, Szolovits P, Ghassemi M. Can AI help reduce disparities in general medical and mental health care? AMA J Ethics. 2019 Feb 01;21(2):E167–179. doi: 10.1001/amajethics.2019.167. https://journalofethics.ama-assn.org/article/can-ai-help-reduce-disparities-general-medical-and-mental-health-care/2019-02 .amajethics.2019.167 [DOI] [PubMed] [Google Scholar]
- 34.Gianattasio K, Ciarleglio A, Power M. Development of algorithmic dementia ascertainment for racial/ethnic disparities research in the US Health and Retirement Study. Epidemiology (Cambridge, Mass) 2020;31(1):126–133. doi: 10.1097/ede.0000000000001101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Noseworthy PA, Attia ZI, Brewer LC, Hayes SN, Yao X, Kapa S, Friedman PA, Lopez-Jimenez F. Assessing and mitigating bias in medical artificial intelligence: the effects of race and ethnicity on a deep learning model for ECG analysis. Circ Arrhythm Electrophysiol. 2020 Mar;13(3):e007988. doi: 10.1161/CIRCEP.119.007988. http://europepmc.org/abstract/MED/32064914 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Obermeyer Z, Powers B, Vogeli C, Mullainathan S. Dissecting racial bias in an algorithm used to manage the health of populations. Science. 2019 Oct 25;366(6464):447–453. doi: 10.1126/science.aax2342.366/6464/447 [DOI] [PubMed] [Google Scholar]
- 37.Park Y, Hu J, Singh M, Sylla I, Dankwa-Mullan I, Koski E, Das AK. Comparison of methods to reduce bias from clinical prediction models of postpartum depression. JAMA Netw Open. 2021 Apr 01;4(4):e213909. doi: 10.1001/jamanetworkopen.2021.3909. https://jamanetwork.com/journals/jamanetworkopen/fullarticle/10.1001/jamanetworkopen.2021.3909 .2778568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seyyed-Kalantari L, Liu G, McDermott M, Chen I, Ghassemi M. CheXclusion: fairness gaps in deep chest X-ray classifiers. Pacific Symp Biocomput. 2021;26:232–243. doi: 10.1142/9789811232701_0022. [DOI] [PubMed] [Google Scholar]
- 39.Thompson H, Sharma B, Bhalla S, Boley R, McCluskey C, Dligach D, Churpek MM, Karnik NS, Afshar M. Bias and fairness assessment of a natural language processing opioid misuse classifier: detection and mitigation of electronic health record data disadvantages across racial subgroups. J Am Med Inform Assoc. 2021 Oct 12;28(11):2393–2403. doi: 10.1093/jamia/ocab148. http://europepmc.org/abstract/MED/34383925 .6349190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wissel BD, Greiner HM, Glauser TA, Mangano FT, Santel D, Pestian JP, Szczesniak RD, Dexheimer JW. Investigation of bias in an epilepsy machine learning algorithm trained on physician notes. Epilepsia. 2019 Sep 23;60(9):e93–e98. doi: 10.1111/epi.16320. http://europepmc.org/abstract/MED/31441044 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hardt M, Price E, Srebro N. Equality of opportunity in supervised learning. Advances in Neural Information Processing Systems 29: Annual Conference on Neural Information Processing Systems; December 2016; Barcelona, Spain. 2016. [Google Scholar]
- 42.Kamishima T, Akaho S, Asoh H, Sakuma J. Fairness-aware classifier with prejudice remover regularizer. Joint European Conference on Machine Learning and Knowledge Discovery in Databases; September 2012; Berlin, Heidelberg. 2012. [DOI] [Google Scholar]
- 43.Liu L, Dean S, Rolf E, Simchowitz M, Hardt M. Delayed impact of fair machine learning. Proceedings of the Twenty-Eighth International Joint Conference on Artificial Intelligence; August 10-16, 2019; Macao. 2019. [DOI] [Google Scholar]
- 44.Rodolfa K, Salomon E, Haynes L, Mendieta I, Larson J, Ghani R. Case study: predictive fairness to reduce misdemeanor recidivism through social service interventions. Proceedings of the 2020 Conference on Fairness, Accountability, and Transparency; 2020; Barcelona, Spain. 2020. [DOI] [Google Scholar]
- 45.Kusner MJ, Loftus JR. The long road to fairer algorithms. Nature. 2020 Feb 04;578(7793):34–36. doi: 10.1038/d41586-020-00274-3.10.1038/d41586-020-00274-3 [DOI] [PubMed] [Google Scholar]
- 46.Wolff RF, Moons KG, Riley RD, Whiting PF, Westwood M, Collins GS, Reitsma JB, Kleijnen J, Mallett S. PROBAST: a tool to assess the risk of bias and applicability of prediction model studies. Ann Intern Med. 2019 Jan 01;170(1):51. doi: 10.7326/m18-1376. [DOI] [PubMed] [Google Scholar]
- 47.Alvi M, Zisserman A, Nellåker C. Turning a blind eye: explicit removal of biases and variation from deep neural network embeddings. 15th European Conference on Computer Vision; September 2018; Munich, Germany. 2018. [DOI] [Google Scholar]
- 48.Vig J, Gehrmann S, Belinkov Y. Investigating gender bias in language models using causal mediation analysis. Advances in Neural Information Processing Systems 33: Annual Conference on Neural Information Processing Systems; December 2020; Virtual Conference. 2020. https://proceedings.neurips.cc/paper/2020/file/92650b2e92217715fe312e6fa7b90d82-Paper.pdf . [Google Scholar]
- 49.Straw I, Callison-Burch C. Artificial intelligence in mental health and the biases of language based models. PLoS One. 2020 Dec 17;15(12):e0240376. doi: 10.1371/journal.pone.0240376. https://dx.plos.org/10.1371/journal.pone.0240376 .PONE-D-20-06032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bolukbasi T, Chang K, Zou J, Saligrama V, Kalai A. Man is to computer programmer as woman is to homemaker? Debiasing word embeddings. Advances in Neural Information Processing Systems 29: Annual Conference on Neural Information Processing Systems; December 2016; Barcelona, Spain. 2016. https://papers.nips.cc/paper/2016/file/a486cd07e4ac3d270571622f4f316ec5-Paper.pdf . [Google Scholar]
- 51.Huang Y, Li W, Macheret F, Gabriel R, Ohno-Machado L. A tutorial on calibration measurements and calibration models for clinical prediction models. J Am Med Inform Assoc. 2020 Apr 01;27(4):621–633. doi: 10.1093/jamia/ocz228. http://europepmc.org/abstract/MED/32106284 .5762806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Howard FM, Dolezal J, Kochanny S, Schulte J, Chen H, Heij L, Huo D, Nanda R, Olopade OI, Kather JN, Cipriani N, Grossman RL, Pearson AT. The impact of site-specific digital histology signatures on deep learning model accuracy and bias. Nat Commun. 2021 Jul 20;12(1):4423. doi: 10.1038/s41467-021-24698-1. doi: 10.1038/s41467-021-24698-1.10.1038/s41467-021-24698-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bellamy RKE, Dey K, Hind M, Hoffman Sc, Houde S, Kannan K, Lohia P, Martino J, Mehta S, Mojsilovic A, Nagar S, Ramamurthy Kn, Richards J, Saha D, Sattigeri P, Singh M, Varshney Kr, Zhang Y. AI Fairness 360: an extensible toolkit for detecting and mitigating algorithmic bias. IBM J Res Dev. 2019 Jul 1;63(4/5):4:1–4:15. doi: 10.1147/jrd.2019.2942287. [DOI] [Google Scholar]
- 54.Wexler J, Pushkarna M, Bolukbasi T, Wattenberg M, Viegas F, Wilson J. The what-if tool: interactive probing of machine learning models. IEEE Trans Visual Comput Graphics. 2020;26(1):56–65. doi: 10.1109/tvcg.2019.2934619. [DOI] [PubMed] [Google Scholar]
- 55.Rudin C. Stop explaining black box machine learning models for high stakes decisions and use interpretable models instead. Nat Mach Intell. 2019 May 13;1(5):206–215. doi: 10.1038/s42256-019-0048-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tjoa E, Guan C. A survey on explainable artificial intelligence (XAI): toward medical XAI. IEEE Trans Neural Netw Learning Syst. 2021 Nov;32(11):4793–4813. doi: 10.1109/tnnls.2020.3027314. [DOI] [PubMed] [Google Scholar]
- 57.McDermott MBA, Wang S, Marinsek N, Ranganath R, Foschini L, Ghassemi M. Reproducibility in machine learning for health research: still a ways to go. Sci Transl Med. 2021 Mar 24;13(586):eabb1655. doi: 10.1126/scitranslmed.abb1655.13/586/eabb1655 [DOI] [PubMed] [Google Scholar]
- 58.Roberts M, Driggs D, Thorpe M, Gilbey J, Yeung M, Ursprung S, Aviles-Rivero AI, Etmann C, McCague C, Beer L, Weir-McCall JR, Teng Z, Gkrania-Klotsas E, Rudd JHF, Sala E, Schönlieb C. Common pitfalls and recommendations for using machine learning to detect and prognosticate for COVID-19 using chest radiographs and CT scans. Nat Mach Intell. 2021 Mar 15;3(3):199–217. doi: 10.1038/s42256-021-00307-0. [DOI] [Google Scholar]
- 59.Huang J, Shlobin N, DeCuypere M, Lam S. Deep learning for outcome prediction in neurosurgery: a systematic review of design, reporting, and reproducibility. Neurosurgery. 2022;90(1):16–38. doi: 10.1227/neu.0000000000001736. [DOI] [PubMed] [Google Scholar]
- 60.Sheller MJ, Edwards B, Reina GA, Martin J, Pati S, Kotrotsou A, Milchenko M, Xu W, Marcus D, Colen RR, Bakas S. Federated learning in medicine: facilitating multi-institutional collaborations without sharing patient data. Sci Rep. 2020 Jul 28;10(1):12598. doi: 10.1038/s41598-020-69250-1. doi: 10.1038/s41598-020-69250-1.10.1038/s41598-020-69250-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wawira Gichoya J, McCoy LG, Celi LA, Ghassemi M. Equity in essence: a call for operationalising fairness in machine learning for healthcare. BMJ Health Care Inform. 2021 Apr 28;28(1):e100289. doi: 10.1136/bmjhci-2020-100289. https://informatics.bmj.com/lookup/pmidlookup?view=long&pmid=33910923 .bmjhci-2020-100289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bozkurt S, Cahan E, Seneviratne M, Sun R, Lossio-Ventura JA, Ioannidis JPA, Hernandez-Boussard T. Reporting of demographic data and representativeness in machine learning models using electronic health records. J Am Med Inform Assoc. 2020 Dec 09;27(12):1878–1884. doi: 10.1093/jamia/ocaa164. http://europepmc.org/abstract/MED/32935131 .5906103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Guo LN, Lee MS, Kassamali B, Mita C, Nambudiri VE. Bias in, bias out: underreporting and underrepresentation of diverse skin types in machine learning research for skin cancer detection-a scoping reviewA scoping review. J Am Acad Dermatol. 2021 Jul 10; doi: 10.1016/j.jaad.2021.06.884.S0190-9622(21)02086-7 [DOI] [PubMed] [Google Scholar]
- 64.Hernandez-Boussard T, Bozkurt S, Ioannidis J, Shah N. MINIMAR (MINimum Information for Medical AI Reporting): developing reporting standards for artificial intelligence in health care. J Am Med Inform Assoc. 2020 Dec 09;27(12):2011–2015. doi: 10.1093/jamia/ocaa088. http://europepmc.org/abstract/MED/32594179 .5864179 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data file containing bias definitions, search strategy, and a table with study data set characteristics.


