Abstract
Sirtuin 1 (SIRT1) is a member of the sirtuin family that functions to deacetylate both histones and non-histone proteins. Previous studies have identified significant SIRT1 upregulation in eutopic endometrium from infertile women with endometriosis. However, SIRT1 function in the uterus has not been directly studied. Using immunochemistry analysis, we found SIRT1 to be most strongly expressed at GD4.5 and GD5.5 in decidualized cells and at GD7.5 in secondary decidual cells in mouse. To assess the role of SIRT1 in uterine function, we generated uterine Sirt1 conditional knockout mice (Pgrcre/+Sirt1f/f; Sirt1d/d). A 6-month fertility trial revealed that Sirt1d/d females were subfertile. Implantation site numbers were significantly decreased in Sirt1d/d mice compared with controls at GD5.5. Sirt1d/d implantation sites at GD4.5 could be divided into two groups, Group #1 with luminal closure and nonspecific COX2 expression compared with controls (14/20) and Group #2 with an open lumen and no COX2 (6/20). In Sirt1d/d Group #1, nuclear FOXO1 expression in luminal epithelial cells was significantly decreased. In Sirt1d/d Group #2, nuclear FOXO1 expression was almost completely absent, and there was strong PGR expression in epithelial cells. At GD5.5, stromal PGR and COX2 were significantly decreased in Sirt1d/d uterine in the areas surrounding the embryo compared with controls, indicating defective decidualization. An artificially induced decidualization test revealed that Sirt1d/d females showed defects in decidualization response. All together, these data suggest that SIRT1 is important for decidualization and contributes to preparing a receptive endometrium for successful implantation.
Keywords: implantation, subfertility, SIRT1, progesterone receptor
Introduction
Implantation is a critical event in the initiation of a successful pregnancy involving a series of complex mechanisms between the embryo and the maternal endometrium [1]. Decidualization, a critical components of this process, is the morphological and functional change of uterine stromal fibroblasts into large, epithelial-like, secretory decidual cells [2]. In humans, implantation is not necessary for the initiation of decidualization, but in mice, the blastocyst stimulates endometrial tissue to decidualize [3]. The murine implantation process involves the apposition of the blastocyst next to the endometrial luminal epithelium, the attachment of the trophectoderm layer of the blastocyst to the epithelium, and the invasion of the blastocyst through the epithelial layer [1, 4]. Upon embryo invasion, the uterine stroma surrounding the embryo undergoes remodeling through proliferation and differentiation as decidualization progresses [5–7]. The decidual process starts at the antimesometrial region of the endometrium, which becomes the primary decidual zone (PDZ) [8]. In mice, the PDZ starts to form on the afternoon of gestation day (GD) 4.5 (vaginal plug checked morning = GD 0.5) and becomes fully developed on the morning of GD 5.5 with the completion of cell proliferation [9, 10].
The ovarian steroid hormones estrogen (E2) and progesterone (P4) generate dynamic changes in the uterus to regulate endometrial receptivity, embryo implantation, and decidualization of stromal cells, all of which is essential for the establishment of a successful pregnancy [11]. P4 signaling through the progesterone receptor (PGR) is the major activation pathway for decidualization [12], and PGR signaling upregulation requires E2 action in the uterine stroma during the window of receptivity [13]. In mice, the uterus is in a prereceptive condition on GD 0.5–2.5 and becomes receptive to blastocyst implantation on GD 3.5. By the afternoon of GD 4.5, the endometrium is past the receptive window [14].
Decidualized stromal cells express a number of decidual marker genes in the implantation period in mice in coordination with simultaneous changes in the composition of the epithelium [15]. Cyclooxygenase-2 (COX2), an inducible proinflammatory gene, is an important marker for decidualization, and is highly expressed around the embryo invasion site. It was previously shown that COX2-deficient females have decidualization failure when fertilized embryos invade the endometrium [16]. E-cadherin, a critical cell adhesion molecule, constitutes epithelial cell–cell contact sites. In natural conditions, the expression of E-cadherin dissipates at the uterine luminal epithelial cells of the implantation sites in the mouse after embryo implantation due to entosis of epithelial cells by invading trophoblast [17]. Activation of signal transducer and activator of transcription 3 (STAT3) via phosphorylation is required for decidualization, directly interacting with PGR in the uterus at GD 5.5. In addition, mice lacking STAT3 in PGR positive cells fail to support embryo implantation and decidualization, indicating that crosstalk between endometrial STAT3 and PGR signaling is required for these critical events during early pregnancy [18]. Furthermore, previous studies have shown that tightly controlled spatiotemporal expression of PGR and forkhead box O1 (FOXO1) is critical for successful implantation and decidualization of stromal cells [19–21]. FOXO1 expression also increases with the reciprocal decrease of PGR in the nuclei of endometrial epithelia of both humans and mice during the time of embryo implantation, and FOXO1 plays an important role in uterine receptivity through regulation of epithelial integrity [21].
Sirtuin1 (SIRT1) is a globally expressed member of the sirtuin family of Class III histone deacetylases and has number of important roles in peripheral metabolic tissues, such as liver, muscle, and adipose tissue [22, 23]. In the ovary, SIRT1 is expressed in oocyte and granulosa cells in large follicles, and SIRT1 deletion can reduce the size of the ovaries during early stage follicular development [24]. The dysregulation of sirtuin family members has been known to result in abnormal reproductive phenotypes for over a decade, but the function of SIRT1 in uterine biology is still unclear [25]. In our previous analysis of SIRT1, we found it to be significantly upregulated in human and baboon endometrium affected by the presence of endometriosis [26]. This was consistent in both proliferative and secretory phase human biopsy samples, and the upregulation of SIRT1 was time-dependent after induction of endometriosis in baboons [26]. Therefore, we hypothesized that tight regulation of SIRT1 is important for normal uterine function.
In this study, we used uterine specific Sirt1 ablation in mice (Pgrcre/+Sirt1f/f, Sirt1d/d) to identify the role of SIRT1 in early pregnancy. Sirt1d/d females exhibited normal ovarian function and formed successfully developed blastocysts but delivered significantly decreased litter sizes, suggesting subfertility due to a uterine defect. In addition, they showed a decidual defect with altered COX2, PGR, and FOXO1 expression in the uterine stroma. Furthermore, Sirt1d/d uteri were unable to fully respond to artificial decidualization stimuli. Together, our data reveal SIRT1 to be a critical regulator of the endometrial microenvironment involved in PGR signaling for decidualization.
Materials and methods
Mouse procedures and tissue collection
All mice were maintained in the animal facility of the Institutional Animal Care and Use Committee of Michigan State University, and all animal experimental procedures were approved. Mice were bred on a mixed background of the mouse strains C57Bl/6 and SV129 under controlled humidity and temperature. Six-week-old female mice were used for artificially induced decidualization, and 8-week-old female mice were used for all other experiments. Sirt1f/f mice were used as controls, and Sirt1d/d (Sirt1 knockout, Pgrcre/+Sirt1f/f) mice were generated by mating Sirt1f/f mice with Pgr-cre mice resulting in the ablation of the floxed gene in uterine tissue [27, 28]. To collect uterine tissue from specific stages of pregnancy, female mice were mated with fertile wild-type males and designated as GD 0.5 when a vaginal plug was found in the morning. Uterine tissue was snap-frozen at the time of isolation and maintained at −80°C for Quantitative reverse transcription PCR (RT-qPCR) and Western blot analysis. For histological study, the uterine samples were fixed with 4% paraformaldehyde before paraffin embedding.
For the fertility trial, female fertility was assessed by mating Sirt1d/d (n = 5) and Sirt1f/f females (n = 5) with fertile C57Bl/6 males continuously for 6 months and counting the numbers of litters and pups produced by each female mouse. To evaluate the ovarian functions in Sirt1d/d mice, virgin female mice were mated with wild-type male mice. Serum samples were collected at GD3.5 and the serum levels of estrogen and progesterone were examined by University of Virginia Center for Research in Reproduction Ligand Assay and Analysis Core. Embryos were collected at GD3.5 by uterine flushing with M2 medium (Sigma-Aldrich), rinsed into microdrops of M2 medium, and counted. Collected embryos were examined to evaluate preimplantation embryonic development. Ovaries were fixed with 4% paraformaldehyde for histological study.
RT-qPCR
Total RNA was isolated from frozen uterine tissues with QIAGEN RNeasy Mini kit (QIAGEN, 74106). Complementary DNA (cDNA) was synthesized with M-MLV Reverse Transcriptase (Invitrogen, 28,025-013) by using 1 μg of total RNA primed with random hexamer primer according to the manufacturer’s instructions. Quantitative real-time-PCR was performed in a Step One Plus Real-Time PCR System (Applied Biosystem), and quantification was performed with SYBR green (PowerUp SYBR Green Master Mix, Applied Biosystems, A25742) with a primer set specific for Sirt1 (Forward: 5′- ACAGAACGTCACACGCCAG −3′, Reverse: 5′- TTGAGGGTCTGGGAGGT CTG −3′). Expression was normalized with the housekeeping gene Rpl7 (Forward: 5′- TCAATGGAGTAAGCCCAAAG-3′, Reverse: 5′-CAAGAGACCGAGCAATCAAG-3′).
Western blot analysis
Protein isolation from uterine tissue and Western blot analysis was performed as previously described [29]. For total protein extraction, 600 μl of lysis buffer (50 mM Tris–HCl (pH 8.0), 1 mM EDTA, 150 mM NaCl, 0.5% NP40, 10% Glycerol, and 2 mM MgCl2) with a protease inhibitor cocktail (Roche, Indianapolis, IN) and a phosphatase inhibitor cocktail (Sigma Aldrich, St. Louis, MO) were added to the tube with the uterine tissue. Uterine tissue in lysis buffer was homogenized to get a high protein yield. After centrifuging the tubes for 15 minutes with maximal speed, the supernatant was collected in a fresh tube. The Bradford protein assay was used to measure the concentration of total protein in the samples. About 5 μg of proteins were separated on sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) gels and transferred to Polyvinylidene difluoride (PVDF) membrane (Millipore, Bedford, MA). The membranes were blocked by incubation for 1 hour in 0.5% (wt/vol) casein (Sigma Aldrich, St. Louis, MO) in PBS with 0.1% Tween 20 and incubated overnight at 4°C with primary antibodies against Actin (SC-1616, 1:1000 concentration, Santa Cruz Biotechnology, Dallas, TX), SIRT1 (9475S, Cell Signaling, Danvers, MA, 1:5000 concentration). After washing with Phosphate-Buffered Saline with 0.1% Tween (PBST), the membranes were incubated with horseradish peroxidase-linked secondary antibody.
Histology and immunostaining
For histological analysis, endometrial sections were stained with Hematoxylin (Hematoxylin Stain, Mayer’s solution, VOLU-SOL) and Eosin (Eosin-Y, Fisherbrand). Immunohistochemistry was performed as previously described [30] with specific commercially available primary antibodies for COX2 (1:500, Cayman, 160,112), FOXO1 (1:500, Cell Signaling, 2880), PGR (1:1000, Cell Signaling, 8757), E-cadherin (1:2000, BD Bioscience, 610 181), and Ki67 (1:1000, BD Pharmingen, 550 609). Goat Anti-Rabbit IgG Biotinylated (BA-1000) or Goat Anti-Mouse IgG Biotinylated (BA-9200, Vector Laboratories) was used after administration of primary antibodies overnight. Horseradish peroxidase-conjugated streptavidin substrate (Streptavidin Horseradish Peroxidase [HRP] Conjugate, Invitrogen, 434 323) positive immunoreactivity was detected with the Vectastain Elite DAB kit (Vector Laboratories, Burlingame, CA). For immunohistochemistry staining comparison, a semiquantitative grade (H-score) was measured by adding the percentage of intensively stained nuclei (3×), the percentage of moderately stained nuclei (2×), and the percentage of weakly stained nuclei (1×) in 100 cells of 5 different images; the score range is 0–300 [31]. FOXO1 is localized in the nucleus of the luminal and glandular epithelium of the endometrium at GD4.5 to regulate endometrial receptivity [21]. Subcellular localization of FOXO1 proteins at implantation sites was semi-quantitatively examined by counting stained nuclei and cytoplasm in 100 uterine luminal epithelial cells at GD4.5 (n = 5).
Artificially induced decidualization
For artificial decidualization, Sirt1f/f (control) and Sirt1d/d female mice (n = 3/genotype) were ovariectomized under anesthesia at 6 weeks of age. After resting at least 2 weeks to remove endogenous ovarian hormones completely, the mice were administered daily subcutaneous injections (s.c.) of E2 (100 ng) for 3 days. The mice were administered daily 1 mg of P4 s.c. and 6.7 ng of E2 s.c. for 3 days after undergoing 2 days of rest. A single horn of each mouse was mechanically stimulated the full length of the antimesometrial lumen 6 h after the third P4 and E2 injection, while the other horn was left unstimulated as a control. For stimulation, a 25-gauge sterile needle was inserted into the uterine lumen through the uterine wall near the tubal utero junction, and the end of the needle was scratched against the antimesometrial uterine wall. Mice were treated with meloxicam for analgesia before the skin incision was made. Mice were anaesthetized with isoflurane gas by inhalation. To induce a strong decidual reaction, daily treatment with s.c. injections of 1 mg P4 and 6.7 ng E2 continued for 5 days after the mechanical trauma. The wet weights of the stimulated and control uterine horns of each mouse were measured. Uterine tissues were then isolated from both horns and fixed in 4% paraformaldehyde (vol/vol) before paraffin embedding.
Statistical analysis
Statistical significance analyses were performed with one-way Analysis of variance (ANOVA) analysis, Tukey’s post hoc multiple range test, and Student’s t-tests using GraphPad software (San Diego, CA). p < 0.05 was considered statistically significant.
Results
Increased SIRT1 during early pregnancy in the mouse uterus
To examine the function of SIRT1 in mouse uterine tissue, we profiled the SIRT1 protein level during early pregnancy. SIRT1 was weakly expressed in uterine stromal cells and luminal and glandular epithelial cells at GD 0.5–3.5 (Figure 1a-c). At GD 4.5, SIRT1 expression increased in nuclei of decidual stromal cells, mainly in the PDZ (Figure 1d and e). At GD 5.5, SIRT1 expression is strongest, with high expression levels in both the PDZ and secondary decidual zone (SDZ). (Figure 1f and g). At GD 7.5, expression of SIRT1 was similar to GD 5.5, but it was weakly localized to nuclei of decidual cells and weaker in the PDZ than the SDZ (Figure 1h-j). SIRT1 is also highly expressed in the embryos (Figure 1e, g, and j).
Figure 1.
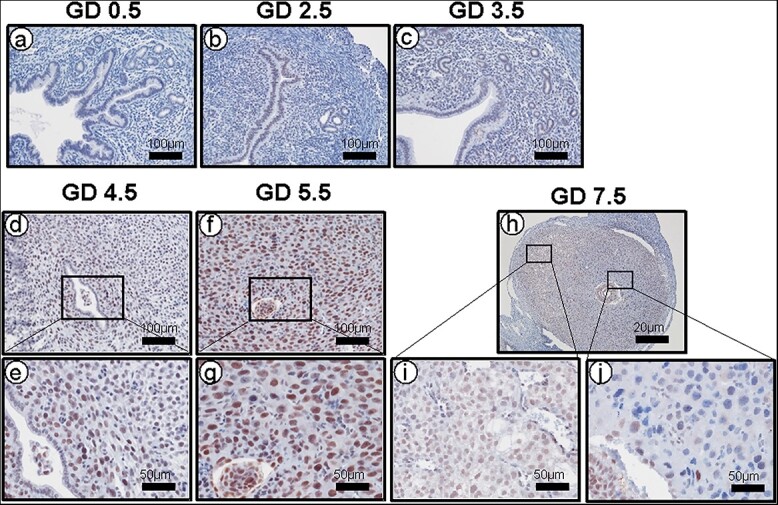
Increased SIRT1 during early pregnancy in the mouse uterus. Representative images of SIRT1 IHC in uterine tissue of natural pregnant mice (n = 3). At GD 0.5–3.5, SIRT1 shows low expression (a-c), but at GD 4.5 (d-e) and 5.5 (f-g) SIRT1 expression was strong in decidualized cells. At GD7.5, SIRT1 was also expressed in secondary decidual cells (h-j).
Fertility defect of mice with ablation of Sirt1 in the PGR-expressing cells
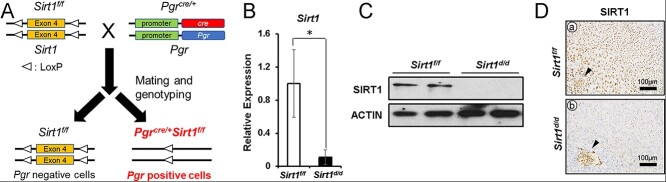
Global deletion of Sirt1 in mice results in perinatal lethality in inbred backgrounds [21, 32]. To assess the function of Sirt1 in the female reproductive tract but avoid this lethal phenotype, we generated uterine-specific Sirt1 knockout mice by crossing Pgr-cre (Pgrcre/+) and Sirt1 floxed female mice (Sirt1f/f) (Figure 2A) [27, 28]. We confirmed the ablation of uterine SIRT1 at GD 5.5 when SIRT1 is normally strongest, with RT-qPCR showing that Sirt1 mRNA was significantly decreased in the uterus of Sirt1d/d mice compared with control mice (Figure 2B). Western blot analysis demonstrated loss of SIRT1 proteins in the uteri of Sirt1d/d mice (Figure 2C). To confirm the loss of SIRT1 in all uterine compartments, we performed immunohistochemistry analysis in the implantation site region of GD 5.5. In Sirt1d/d mice, there was weak SIRT1 expression except for in the embryo (Figure 2D). These results demonstrate the successful ablation of Sirt1 in the murine uterus.
Figure 2.
Generation of Sirt1d/d mice. Sirt1d/d female mice do not express SIRT1 in any uterine cell layer. (A) The schematic shows the strategy for generating Pgrcre/+Sirt1f/f (Sirt1d/d) mice. (B) Relative expression of uterine Sirt1 at GD 5.5, mRNA normalized to Rpl7 (n = 5, *p < 0.05). The results represent the mean ± SEM. *p < 0.05. (C) Uterine SIRT1 protein expression at GD 5.5 normalized to ACTIN. (D) Representative images of SIRT1 IHC in uterine tissue at GD 5.5 (n = 3).
To investigate the impact of ablation of Sirt1 on female fertility, female control (Sirt1f/f) and Sirt1d/d mice were mated with wild-type C57Bl/6 male mice for 6 months. Sirt1d/d mice had only 2.40 ± 0.60 litters/mouse compared with 5.40 ± 0.30 litters/mouse from control mice (Table 1). Control mice had an average 6.63 ± 0.13 pups/litter, whereas female Sirt1d/d mice had an average 4.20 ± 0.81 pups/litter. Sirt1d/d mice produced significantly fewer litters per mouse (*p < 0.05) and pups per litter (***p < 0.001), revealing that Sirt1d/d female mice are subfertile.
Table 1.
Subfertility of Sirt1d/d female mice
| Number of mice tested | Number of litters | Number of pups | Average pups/litter | Average number of litters/mouse | |
|---|---|---|---|---|---|
| Sirt1f/f | 5 | 27 | 179 | 6.63 ± 0.13 | 5.40 ± 0.30 |
| Sirt1d/d | 5 | 10 | 42 | 4.20 ± 0.81 | 2.40 ± 0.60 |
To test for an ovarian cause of subfertility, female Sirt1d/d mice were examined for proper ovarian histology, ovulation, and steroid hormone production at GD 3.5. Blastocysts generated by natural mating were flushed from the uterus at GD3.5. We isolated 7.00 ± 0.71 and 6.20 ± 0.49 embryos from uteri of Sirt1f/f and Sirt1d/d mice, respectively (n = 5; Supplementary Figure 1A). The serum levels of E2 and P4 were 16.22 ± 3.05 pg/ml and 10.53 ± 1.85 ng/ml, respectively, in control mice, meanwhile 17.58 ± 4.08 pg/ml and 8.44 ± 1.75 ng/ml, respectively, in Sirt1d/d mice, not significantly differing between the mice at GD 3.5 (n = 5/genotype) (Supplementary Figure 1 B and C). In addition, histological analysis of the intact ovaries from Sirt1f/f and Sirt1d/d mice revealed no morphological defect (Supplementary Figure 1D). These results demonstrate that ovarian morphology and function were not affected in the Sirt1d/d females suggesting that the subfertility is primarily due to a uterine defect.
Implantation defect in Sirt1d/d mice
To determine the cause of subfertility in Sirt1d/d mice, 8-week-old female Sirt1f/f and Sirt1d/d mice were mated with fertile control male mice. The uteri were examined at GD 5.5 to examine the ability of embryos to implant. Sirt1f/f female mice showed average 7.57 ± 0.43 implantation sites per mouse (n = 7). However, Sirt1d/d uteri contained significantly fewer implantation sites (5.00 ± 1.04) than control mice (n = 8) (Figure 3A and B).
Figure 3.
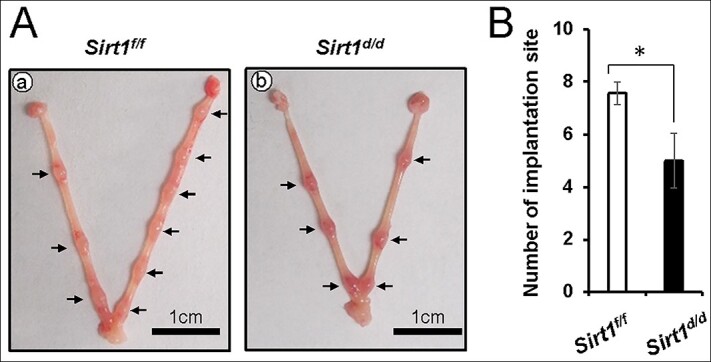
Implantation defect in GD 5.5 Sirt1d/d mice. (A) Representative images of GD 5.5 uterus gross morphology from Sirt1f/f (a) and Sirt1d/d (b) mice. (B) The average implantation site number in Sirt1f/f and Sirt1d/d uteri. The results represent the mean ± SEM. *p < 0.05.
To understand the molecular basis of the implantation defect in Sirt1d/d mice, we examined the uterine histology and COX2 expression [33] at GD 4.5. The endometrium in control mice normally undergoes luminal epithelial closure at the time of embryo implantation [34], and COX2 is strongly and specifically expressed in stromal cells that are beginning to decidualize near the embryo invasion region [35]. However, our histological and immunochemistry analysis revealed two types of abnormal implantation sites in Sirt1d/d mice (Figure 4). Group #1 (14/20, 70%) showed a partially closed luminal epithelial layer, decreased COX2 expression in stromal cells near implantation sites, and abnormal COX2 expression in luminal epithelial cells (Figure 4h). Group #2 (6/20, 30%) had a more severe phenotype compared with Group #1, exhibiting a fully opened luminal epithelial layer with floating embryos (Figure 4f). In addition, COX2 expression was nearly absent (Figure 4i). To determine if the two different phenotypes in Sirt1d/d mice originated due to regionally differing Sirt1 knockout efficiency, we examined the expression of SIRT1 at implantation sites of two different groups. However, SIRT1 was absent around implantation sites in both groups of Sirt1d/d mice (Figure 4k and l).
Figure 4.
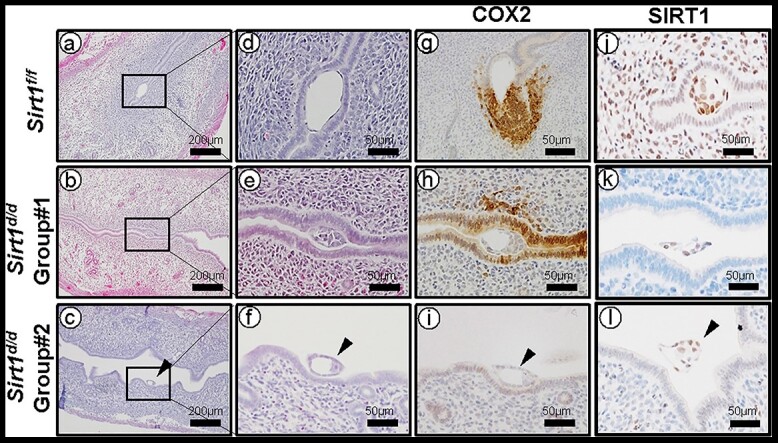
Altered stromal cell decidualization in GD 4.5 Sirt1d/d uteri. Representative images of H&E (a-f), COX2 IHC (g-i), and SIRT1 IHC (j-l) at GD 4.5 Sirt1f/f (a, d, g, and j), Sirt1d/d Group #1 (b, e, h, and k), Sirt1d/d Group #2 (c, f, i, and l) implantation sites.
Altered FOXO1 and PGR expression in GD 4.5 Sirt1d/d uteri
Proper interplay between PGR and FOXO1 around the implantation window is another key to successful embryo implantation [21]. At GD 4.5, FOXO1 was normally localized in nuclei of luminal epithelial cells in control mice, and PGR was negative (Figure 5Aa, d, g, and j). However, Sirt1d/d females showed two types of altered expression patterns for PGR and FOXO1 proteins. In GD 4.5 Sirt1d/d Group #1, FOXO1 was still expressed in luminal epithelial cells (Figure 5Ae), but nuclear FOXO1 was significantly decreased, whereas cytoplasmic FOXO1 expression was upregulated (Figure 5B). These data show there is diminished translocalization of FOXO1 in the absence of SIRT1. Sirt1d/d Group #2 showed a more severe phenotype, where the luminal epithelial layer was negative for nuclear FOXO1 (Figure 5Ac and f), and PGR was strongly expressed (Figure 5Ai, l and 5C). To determine if FOXO1 regulates SIRT1 expression, we performed SIRT1 immunohistochemistry in Pgrcre/+Foxo1f/f mice [21]. Although FOXO1 was not detected in Pgrcre/+Foxo1f/f mice, SIRT1 proteins were strongly detected at stromal and luminal epithelial cells of Pgrcre/+Foxo1f/f mice (Supplementary Figure 2). These results suggest that SIRT1 loss disrupts the normal balance of FOXO1 and PGR at GD 4.5.
Figure 5.
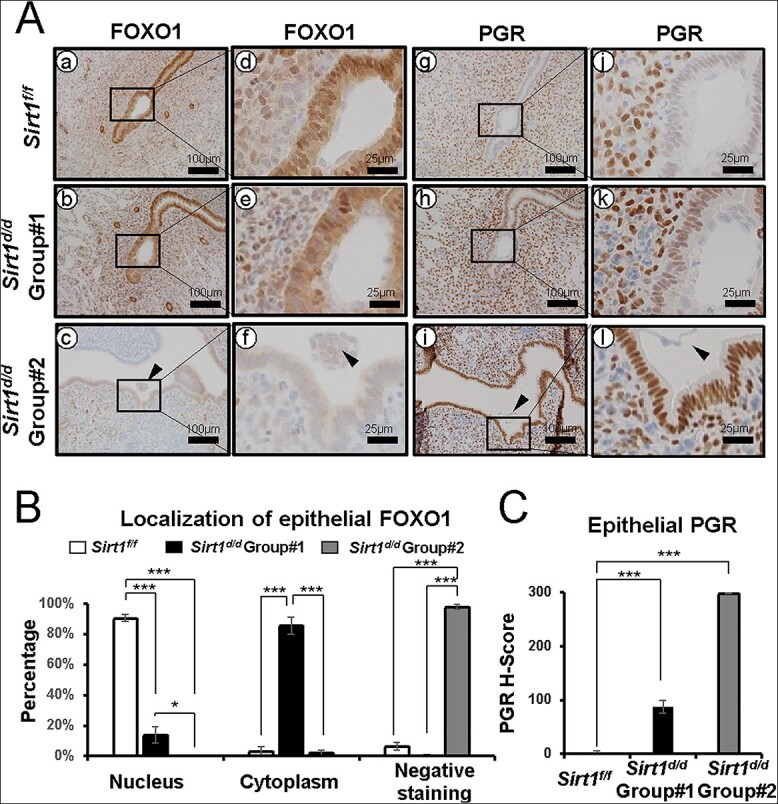
Altered FOXO1 and PGR expression in GD 4.5 Sirt1d/d uteri. (A) Representative images of FOXO1 (a-f) and PGR (g-l) IHC at GD 4.5 Sirt1f/f (a, d, g, and j), Sirt1d/d Group #1 (b, e, h, and k), Sirt1d/d Group #2 (c, f, I, and l) implantation sites. (B) Quantification of the localization of FOXA2 at Sirt1f/f, Sirt1d/d Group #1, and Sirt1d/d Group #2 implantation sites. (C) Semi-quantitative H-score for epithelial PGR at Sirt1f/f, Sirt1d/d Group #1, and Sirt1d/d Group #2 implantation sites. The results represent the mean ± SEM. *p < 0.05; ***p < 0.001.
Altered E-cadherin, COX2, FOXO1, and PGR expression in GD 5.5 Sirt1d/d uteri
To further characterize the role of SIRT1 in implantation and decidualization, we analyzed uterine histology and screened important molecular markers at GD 5.5 (Figure 6A). In normal control mice, the luminal epithelium at the embryo invasion site undergoes degradation by this time [17], and therefore we observed no E-cadherin-positive epithelium at implantation sites (Figure 6A a and d). FOXO1 was also not expressed (Figure 6Ag), but PGR and COX2 were abundant in nuclei of decidual stromal cells in the PZD (Figure 6Ba and d) [16, 21, 36]. In contrast, the Sirt1d/d uterus showed structural and molecular defects. Like at GD 4.5, Sirt1d/d implantation sites exhibited two distinct phenotypes. Group #1 showed normal implantation structure, and E-cadherin (Figure 6Ab and e) and FOXO1 (Figure 6Ah) were negative. However, the expression of COX2 and PGR was significantly downregulated in the PZD (Figure 6Bb and e; C). Group #2 had a more critical defect. Immunohistochemistry results revealed intact epithelial cells positive for E-cadherin and FOXO1 (Figure 6Afand i). This phenotype matches that of GD 4.5 control mice [17]. Similar to Group #1, PGR and COX2 were significantly decreased in the PDZ (Figure 6Bc and f; C). However, strong E-cadherin staining was constantly detected at inter implantation sites of control and both groups of Sirt1d/d mice (Figure 6Aj-l). All these data underscore the importance of SIRT1 in the mechanisms of epithelial degradation and stromal decidualization during implantation.
Figure 6.
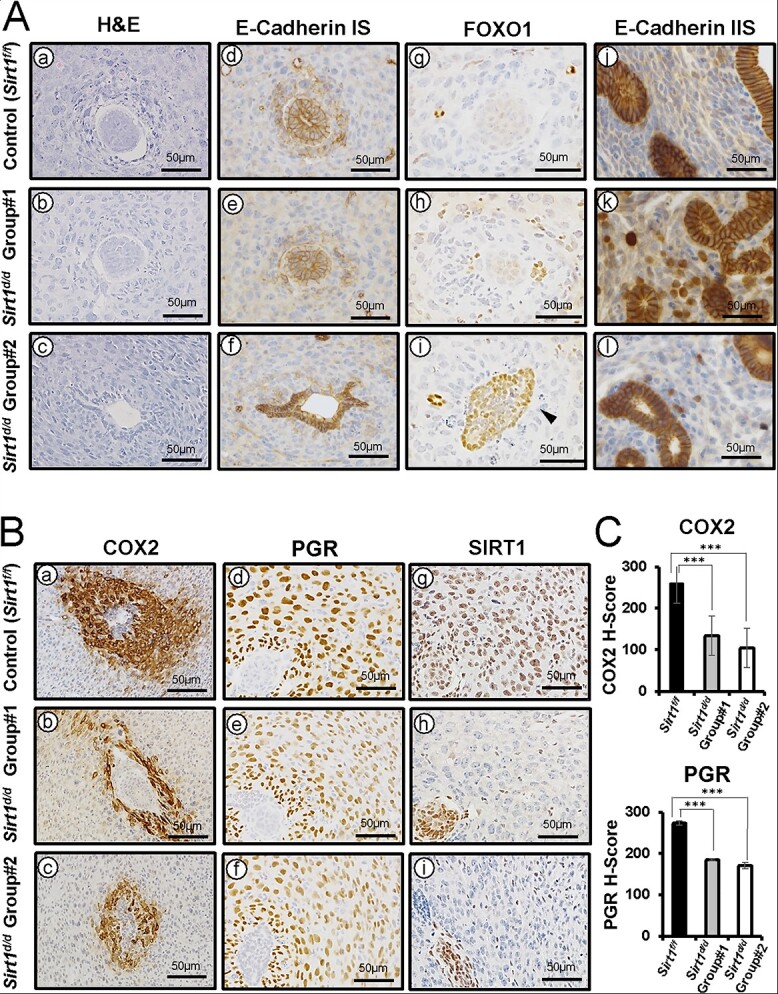
Altered E-Cadherin, FOXO1, COX2, and PGR expression in GD 5.5 Sirt1d/d uteri (A) Representative images of H&E (a-c), E-cadherin (d-f and j-l), and FOXO1 (g-i) IHC at implantation sites (IS) and inter implantation sites of Sirt1f/f (a, d, g, and j), Sirt1d/d Group #1 (b, e, h, and k), and Sirt1d/d Group #2 (c, f, i, and l). (B) Representative images of COX2 (a-c), PGR (d-f), and SIRT1 (g-i) IHC at GD 5.5 Sirt1f/f (a, d, and g), Sirt1d/d Group #1 (b, e, and h), Sirt1d/d Group #2 (c, f, and i) ISs. (C) Semi-quantitative H-score for COX2 and PGR at Sirt1f/f, Sirt1d/d Group #1, and Sirt1d/d Group #2 ISs. The results represent the mean ± SEM. ***p < 0.001.
Defect of decidualization response in Sirt1d/d mice
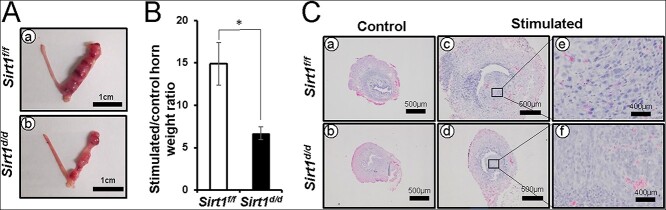
To determine the effect of SIRT1 ablation on decidualization, we used a mouse model of artificially induced decidualization. In the stimulated uterine horn of control mice, the decidual response was fully induced after hormone administration and mechanical trauma (Figure 7Aa). However, the stimulated uterine horn of Sirt1d/d mice showed a partial defect of decidual response (Figure 7Ab), and the stimulated/control horn weight ratio was significantly decreased (Figure 7B). In addition, histological analysis of control and Sirt1d/d uteri showed decidual cell morphology, but the size of stimulated horn was decreased due to a defective decidual response in Sirt1d/d uteri (Figure 7C). These data reveal that SIRT1 is required for a complete decidualization response.
Figure 7.
Defect of decidualization response in Sirt1d/d mice. (A) Uterine morphology of artificially induced decidualized uteri of Sirt1f/f and Sirt1d/d mice. The right side horn received mechanical stimulation. (B) A significant decrease in stimulated/unstimulated (control) horn weight ratio in Sirt1d/d mice as compared with Sirt1f/f mice. (C) H&E staining of control and stimulated horns in Sirt1f/f (a and c) and Sirt1d/d (b and d) mice at day 5 of decidualization. The results represent the mean ± SEM. *p < 0.05.
Discussion
We utilized the Pgr-cre driver to determine the effect of Sirt1 deletion in the mouse uterus on fertility. A 6-month fertility trial showed that female Sirt1d/d mice were subfertile. Because Pgr is also expressed in ovarian granulosa cells, we needed to rule out a potential ovarian defect caused by Sirt1 deletion. The average number of blastocysts, and serum P4 and E2 levels in Sirt1d/d mice at GD3.5 were not different than control mice. PGR is not expressed in the oocyte, and Sirt1d/d mice showed normal ovarian function. Together, Sirt1d/d mice exhibit normal ovarian function, and the subfertility of Sirt1d/d females is due to a uterine defect.
Previous research by our group identified that SIRT1 was overexpressed in eutopic endometrium from women with endometriosis. In women, this SIRT1 overexpression was not dependent on the menstrual cycle stage; rather, overexpression was consistent at all stages [26]. Therefore, SIRT1 is a potential target of endometriosis treatment. On the other hand, one study showed that SIRT1 was reduced in damaged human endometrial stromal cells, which may be the cause of repeated implantation failures [37]. Therefore, it appears that a properly regulated amount of SIRT1 is important for normal uterine processes such as decidualization.
The invasion of the blastocyst to the luminal epithelium triggers the process of decidualization that includes the differentiation of the fibroblastic stromal cells into morphologically different decidual cells [38]. These differentiated cells have a special biosynthetic and secretory properties [39]. In the normal implantation period, the stromal decidual cell marker COX2 [16] induces prostaglandin, an important molecule for angiogenesis [40]. However, we found that COX2 was not sufficiently induced in Sirt1d/d implantation sites. In Sirt1d/d Group #1, expression of COX2 was not limited to the stromal cells but also extended to epithelial cells and was not specific to the invasion site. On the other hand, COX2 was almost negative in Sirt1d/d Group #2. Therefore, endogenous decidualization was disrupted at the molecular level by loss of SIRT1.
It is well-known that ovarian hormones are critical for decidualization, especially P4 signaling through PGR, which is necessary to maintain the decidual tissue [41]. There are two different ways to activate the PGR: (1) ligand binding dependent [42] and (2) ligand binding independent [43]. The amount of ovarian progesterone was similar in the GD 3.5 Sirt1f/f and Sirt1d/d, so it can be assumed that the decrease in PGR was not due to a decrease in the ligand. Therefore, our results suggest that changes in PGR-related decidual signaling in stromal cells lead to abnormal implantation phenotypes in Sirt1d/d mice [36, 44, 45]. FOXO1 is one of the proteins that interact most tightly with PGR [46], and FOXO1 and PGR were established as important to inducing in vitro decidualization [19, 20, 47, 48]. Our results showed that SIRT1 contributes to the activation of the FOXO1-PGR axis, and ablation of SIRT1 decreases decidual cell proliferation. Together, these data suggest that SIRT1 contributes to the regulation of the FOXO1-PGR axis at implantation.
At the beginning of the implantation period, GD 4.5, the withdrawal of PGR in epithelial cells and the rapid accumulation of FOXO1 in the endometrial epithelium is necessary to start the epithelial degradation necessary for embryo implantation [21, 48]. However, in Sirt1d/d mice, PGR was still localized to the nucleus in the epithelial region at this stage, and FOXO1 failed to translocate to the nuclei [46]. Also, Sirt1d/d Group #2 implantation regions showed an intact FOXO1 positive epithelium compared with no luminal epithelial cells in control and Sirt1d/d Group #1 implantation regions. We thus speculate that the subfertility of Sirt1d/d females is due to biological adaptation of individual PGR signaling mechanisms at specific endometrial regions.
Our fertility experiment revealed that Sirt1d/d mice exhibit subfertility instead of infertility, and delayed embryo implantation could have a ripple effect on reducing litter sizes. Ovariectomy before embryo implantation results in blastocyst dormancy and delayed implantation in the mouse [49]. These conditions are maintained by continued P4 treatment but can be terminated with an injection of E2 leading to blastocyst activation and subsequent implantation. The window for successful implantation defines as a limited time span when the activated stage of the blastocyst is superimposed on the receptive state of the uterus [49]. Although the serum P4 level is normal in Sirt1d/d mice, Group #2 of Sirt1d/d mice showed delayed progression of implantation. The expression of SIRT1 is weak in uterine epithelium and stroma until GD 4.5 (Figure 1), and the effect of Sirt1 knock-out on artificial decidualization response is only a partially defect. These results suggest that Sirt1 ablation in uterine cells using Pgr-cre model could result in a compensation or redundancy effect of other SIRT1 family proteins.
Sirt1d/d mice had only 2.40 ± 0.60 litters/mouse compared with 5.40 ± 0.30 litters/mouse from control mice. This result suggests that the mating latency from cohabitation to mating for Sirt1d/d mice is remarkably longer than control mice. We also found the varying phenotypes in Sirt1d/d mice at GD 4.5 and GD 5.5 that appear to represent failures of embryo attachment and decidualization process at two different stages. Although this study could not determine the source of these variations in Sirt1d/d mice, it could possibly have resulted from functional redundancy or compensation of other family members, variables related to SIRT1 deficiency, or slightly different timing from mating event to sample collection between mice. There are also possible pregnancy losses that led to reduced number of litters per mouse. Therefore, further study would be needed to investigate pathophysiological effect of SIRT1 loss during pregnancy.
Decidualization is stromal cell proliferation and differentiation into specialized type of cells (decidual cells) with polyploidy [7]. The decidual cell polyploidy is characterized by the formation of large mono or bi-nucleated cells, a characteristic of nuclear endoreduplication, consisting of DNA with four, eight, and even higher multiples of the haploid complement [39, 50, 51]. Although control mice revealed abundant decidual cell morphology in the stimulated horn, decidual cells were decreased in the stimulated horn of Sirt1d/d mice. Although we did not provide experimental results on the molecular effects of SIRT1 deficiency on our artificial decidual experiment, the phenotypes of Sirt1d/d mice at GD4.5 and GD5.5 suggest a critical function of SIRT1 in decidualization. Therefore, further study would be needed to determine the molecular mechanisms of SIRT1 in decidualization using artificial decidual experiments as well as in vitro decidualization of primary stromal cells.
In conclusion, SIRT1 has a role in implantation and decidualization. Ablation of Sirt1 disrupts decidualization via dysregulating PGR/FOXO1 proteins. Failure of decidualization is one cause leading to infertility. Further research determining the mechanism of SIRT1’s involvement in uterine dysfunction associated with infertility and endometriosis will be critical to understanding both of these common uterine diseases to design future therapies.
Supplementary Material
Acknowledgement
The authors thank Dr. Francesco J. DeMayo at the National Institute of Environmental Health Sciences for the unstained slides of Pgrcre/+Foxo1f/f mice. The University of Virginia Center for Research in Reproduction Ligand Assay and Analysis Core is supported by the Eunice Kennedy Shriver NICHD/NIH Grant R24HD102061.
Contributor Information
Yeon Jeong Hwang, Department of Obstetrics, Gynecology and Reproductive Biology, Michigan State University, Grand Rapids, MI, USA; Division of Developmental Biology and Physiology, Department of Biotechnology, Institute of Basic Sciences, Sungshin Women’s University, Seoul, South Korea.
Gi-Jun Sung, Department of Obstetrics, Gynecology and Reproductive Biology, Michigan State University, Grand Rapids, MI, USA.
Ryan Marquardt, Department of Obstetrics, Gynecology and Reproductive Biology, Michigan State University, Grand Rapids, MI, USA; Cell and Molecular Biology Program, College of Natural Science, Michigan State University, East Lansing, MI, USA.
Steven L Young, Department of Obstetrics and Gynecology, Division of Reproductive Endocrinology and Infertility, University of North Carolina, Chapel Hill, NC, USA.
Bruce A Lessey, Department of Obstetrics and Gynecology, Division of Reproductive Endocrinology and Infertilithy, Atrium Health, Wake Forest Baptist, Winston-Salem, NC, USA.
Tae Hoon Kim, Department of Obstetrics, Gynecology and Reproductive Biology, Michigan State University, Grand Rapids, MI, USA.
Yong-Pil Cheon, Division of Developmental Biology and Physiology, Department of Biotechnology, Institute of Basic Sciences, Sungshin Women’s University, Seoul, South Korea.
Jae-Wook Jeong, Department of Obstetrics, Gynecology and Reproductive Biology, Michigan State University, Grand Rapids, MI, USA.
Conflict of Interest
S.L.Y., B.A.L., and J-W.J. have shared intellectual property interests in technology related to SIRT1 and its role in progesterone resistance and as a biomarker for endometrial receptivity.
Funding
Research reported in this publication was supported by the Eunice Kennedy Shriver National Institute of Child Health & Human Development of the National Institutes of Health under Award Number P01HD106485, R01HD102170, R01HD101243, F31HD101207, and T32HD087166 as well as MSU AgBio Research and Michigan State University.
Data availability
The data underlying this article are available in the article and in its online supplementary material.
References
- 1. Carson DD, Bagchi I, Dey SK, Enders AC, Fazleabas AT, Lessey BA, Yoshinaga K. Embryo implantation. Dev Biol 2000; 223:217–237. [DOI] [PubMed] [Google Scholar]
- 2. Ramathal CY, Bagchi IC, Taylor RN, Bagchi MK. Endometrial decidualization: of mice and men. Semin Reprod Med 2010; 28:17–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dey SK, Lim H, Das SK, Reese J, Paria BC, Daikoku T, Wang H. Molecular cues to implantation. Endocr Rev 2004; 25:341–373. [DOI] [PubMed] [Google Scholar]
- 4. PrabhuDas M, Bonney E, Caron K, Dey S, Erlebacher A, Fazleabas A, Fisher S, Golos T, Matzuk M, McCune JM, Mor G, Schulz L et al. Immune mechanisms at the maternal-fetal interface: perspectives and challenges. Nat Immunol 2015; 16:328–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lee KY, DeMayo FJ. Animal models of implantation. Reproduction 2004; 128:679–695. [DOI] [PubMed] [Google Scholar]
- 6. Lee KY, Jeong JW, Tsai SY, Lydon JP, DeMayo FJ. Mouse models of implantation. Trends Endocrinol Metab 2007; 18:234–239. [DOI] [PubMed] [Google Scholar]
- 7. Tan J, Raja S, Davis MK, Tawfik O, Dey SK, Das SK. Evidence for coordinated interaction of cyclin D3 with p21 and cdk6 in directing the development of uterine stromal cell decidualization and polyploidy during implantation. Mech Dev 2002; 111:99–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Krehbiel RH. Cytological studies of the decidual reaction in the rat during early pregnancy and in the production of deciduomata. Physiol Zool 1937; 10:212–234. [Google Scholar]
- 9. Yuan J, Aikawa S, Deng W, Bartos A, Walz G, Grahammer F, Huber TB, Sun X, Dey SK. Primary decidual zone formation requires scribble for pregnancy success in mice. Nat Commun 2019; 10:5425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhou C, Bao H-L, Kong S-B, Lu J-H, Wang H-B. Developmental aspect of decidual patterning. Reprod Dev Med 2017; 1:100–114. [Google Scholar]
- 11. Kelleher AM, Peng W, Pru JK, Pru CA, DeMayo FJ, Spencer TE. Forkhead box a2 (FOXA2) is essential for uterine function and fertility. Proc Natl Acad Sci U S A 2017; 114:E1018–E1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jeong JW, Lee HS, Lee KY, White LD, Broaddus RR, Zhang YW, Vande Woude GF, Giudice LC, Young SL, Lessey BA, Tsai SY, Lydon JP et al. Mig-6 modulates uterine steroid hormone responsiveness and exhibits altered expression in endometrial disease. Proc Natl Acad Sci U S A 2009; 106:8677–8682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Marquardt RM, Kim TH, Shin JH, Jeong JW. Progesterone and estrogen signaling in the endometrium: what goes wrong in endometriosis? Int J Mol Sci 2019; 20:3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Burnum KE, Cornett DS, Puolitaival SM, Milne SB, Myers DS, Tranguch S, Brown HA, Dey SK, Caprioli RM. Spatial and temporal alterations of phospholipids determined by mass spectrometry during mouse embryo implantation. J Lipid Res 2009; 50:2290–2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cha J, Sun X, Dey SK. Mechanisms of implantation: strategies for successful pregnancy. Nat Med 2012; 18:1754–1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lim H, Gupta RA, Ma WG, Paria BC, Moller DE, Morrow JD, DuBois RN, Trzaskos JM, Dey SK. Cyclo-oxygenase-2-derived prostacyclin mediates embryo implantation in the mouse via PPARdelta. Genes Dev 1999; 13:1561–1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li Y, Sun X, Dey SK. Entosis allows timely elimination of the luminal epithelial barrier for embryo implantation. Cell Rep 2015; 11:358–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lee JH, Kim TH, Oh SJ, Yoo JY, Akira S, Ku BJ, Lydon JP, Jeong JW. Signal transducer and activator of transcription-3 (Stat3) plays a critical role in implantation via progesterone receptor in uterus. FASEB J 2013; 27:2553–2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Vasquez YM, Mazur EC, Li X, Kommagani R, Jiang L, Chen R, Lanz RB, Kovanci E, Gibbons WE, DeMayo FJ. FOXO1 is required for binding of PR on IRF4, novel transcriptional regulator of endometrial stromal decidualization. Mol Endocrinol 2015; 29:421–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Takano M, Lu Z, Goto T, Fusi L, Higham J, Francis J, Withey A, Hardt J, Cloke B, Stavropoulou AV, Ishihara O, Lam EW et al. Transcriptional cross talk between the forkhead transcription factor forkhead box O1A and the progesterone receptor coordinates cell cycle regulation and differentiation in human endometrial stromal cells. Mol Endocrinol 2007; 21:2334–2349. [DOI] [PubMed] [Google Scholar]
- 21. Vasquez YM, Wang X, Wetendorf M, Franco HL, Mo Q, Wang T, Lanz RB, Young SL, Lessey BA, Spencer TE, Lydon JP, DeMayo FJ. FOXO1 regulates uterine epithelial integrity and progesterone receptor expression critical for embryo implantation. PLoS Genet 2018; 14:e1007787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Frye RA. Characterization of five human cDNAs with homology to the yeast SIR2 gene: Sir2-like proteins (sirtuins) metabolize NAD and may have protein ADP-ribosyltransferase activity. Biochem Biophys Res Commun 1999; 260:273–279. [DOI] [PubMed] [Google Scholar]
- 23. Pfluger PT, Herranz D, Velasco-Miguel S, Serrano M, Tschop MH. Sirt1 protects against high-fat diet-induced metabolic damage. Proc Natl Acad Sci U S A 2008; 105:9793–9798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tao X, Zhang X, Ge SQ, Zhang EH, Zhang B. Expression of SIRT1 in the ovaries of rats with polycystic ovary syndrome before and after therapeutic intervention with exenatide. Int J Clin Exp Pathol 2015; 8:8276–8283. [PMC free article] [PubMed] [Google Scholar]
- 25. Tatone C, Di Emidio G, Barbonetti A, Carta G, Luciano AM, Falone S, Amicarelli F. Sirtuins in gamete biology and reproductive physiology: emerging roles and therapeutic potential in female and male infertility. Hum Reprod Update 2018; 24:267–289. [DOI] [PubMed] [Google Scholar]
- 26. Yoo JY, Kim TH, Fazleabas AT, Palomino WA, Ahn SH, Tayade C, Schammel DP, Young SL, Jeong JW, Lessey BA. KRAS activation and over-expression of SIRT1/BCL6 contributes to the pathogenesis of endometriosis and progesterone resistance. Sci Rep 2017; 7:6765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Soyal SM, Mukherjee A, Lee KY, Li J, Li H, DeMayo FJ, Lydon JP. Cre-mediated recombination in cell lineages that express the progesterone receptor. Genesis 2005; 41:58–66. [DOI] [PubMed] [Google Scholar]
- 28. Cheng HL, Mostoslavsky R, Saito S, Manis JP, Gu Y, Patel P, Bronson R, Appella E, Alt FW, Chua KF. Developmental defects and p53 hyperacetylation in Sir2 homolog (SIRT1)-deficient mice. Proc Natl Acad Sci U S A 2003; 100:10794–10799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kim BG, Yoo JY, Kim TH, Shin JH, Langenheim JF, Ferguson SD, Fazleabas AT, Young SL, Lessey BA, Jeong JW. Aberrant activation of signal transducer and activator of transcription-3 (STAT3) signaling in endometriosis. Hum Reprod 2015; 30:1069–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kim TH, Yoo JY, Wang Z, Lydon JP, Khatri S, Hawkins SM, Leach RE, Fazleabas AT, Young SL, Lessey BA, Ku BJ, Jeong JW. ARID1A is essential for endometrial function during early pregnancy. PLoS Genet 2015; 11:e1005537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ishibashi H, Suzuki T, Suzuki S, Moriya T, Kaneko C, Takizawa T, Sunamori M, Handa M, Kondo T, Sasano H. Sex steroid hormone receptors in human thymoma. J Clin Endocrinol Metab 2003; 88:2309–2317. [DOI] [PubMed] [Google Scholar]
- 32. McBurney MW, Yang X, Jardine K, Hixon M, Boekelheide K, Webb JR, Lansdorp PM, Lemieux M. The mammalian SIR2alpha protein has a role in embryogenesis and gametogenesis. Mol Cell Biol 2003; 23:38–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lim H, Paria BC, Das SK, Dinchuk JE, Langenbach R, Trzaskos JM, Dey SK. Multiple female reproductive failures in cyclooxygenase 2-deficient mice. Cell 1997; 91:197–208. [DOI] [PubMed] [Google Scholar]
- 34. Matsumoto L, Hirota Y, Saito-Fujita T, Takeda N, Tanaka T, Hiraoka T, Akaeda S, Fujita H, Shimizu-Hirota R, Igaue S, Matsuo M, Haraguchi H et al. HIF2alpha in the uterine stroma permits embryo invasion and luminal epithelium detachment. J Clin Invest 2018; 128:3186–3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chakrabarty A, Tranguch S, Daikoku T, Jensen K, Furneaux H, Dey SK. MicroRNA regulation of cyclooxygenase-2 during embryo implantation. Proc Natl Acad Sci U S A 2007; 104:15144–15149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jeong JW, Lee KY, Kwak I, White LD, Hilsenbeck SG, Lydon JP, DeMayo FJ. Identification of murine uterine genes regulated in a ligand-dependent manner by the progesterone receptor. Endocrinology 2005; 146:3490–3505. [DOI] [PubMed] [Google Scholar]
- 37. Li J, Qi J, Yao G, Zhu Q, Li X, Xu R, Zhu Z, Zhao H, Wang Y, Ding Y, Sun Y. Deficiency of sirtuin 1 impedes endometrial decidualization in recurrent implantation failure patients. Front Cell Dev Biol 2021; 9:598364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lopata A. Blastocyst-endometrial interaction: an appraisal of some old and new ideas. Mol Hum Reprod 1996; 2:519–525. [DOI] [PubMed] [Google Scholar]
- 39. Ansell JD, Barlow PW, McLaren A. Binucleate and polyploid cells in the decidua of the mouse. J Embryol Exp Morphol 1974; 31:223–227. [PubMed] [Google Scholar]
- 40. Matsumoto H. Molecular and cellular events during blastocyst implantation in the receptive uterus: clues from mouse models. J Reprod Dev 2017; 63:445–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lee K, Jeong J, Tsai MJ, Tsai S, Lydon JP, DeMayo FJ. Molecular mechanisms involved in progesterone receptor regulation of uterine function. J Steroid Biochem Mol Biol 2006; 102:41–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Xu R, Feiner H, Li P, Yee H, Inghirami G, Delgado Y, Perle MA. Differential amplification and overexpression of HER-2/neu, p53, MIB1, and estrogen receptor/progesterone receptor among medullary carcinoma, atypical medullary carcinoma, and high-grade invasive ductal carcinoma of breast. Arch Pathol Lab Med 2003; 127:1458–1464. [DOI] [PubMed] [Google Scholar]
- 43. Li X, O'Malley BW. Unfolding the action of progesterone receptors. J Biol Chem 2003; 278:39261–39264. [DOI] [PubMed] [Google Scholar]
- 44. Boonyaratanakornkit V, Bi Y, Rudd M, Edwards DP. The role and mechanism of progesterone receptor activation of extra-nuclear signaling pathways in regulating gene transcription and cell cycle progression. Steroids 2008; 73:922–928. [DOI] [PubMed] [Google Scholar]
- 45. Cheon YP, Li Q, Xu X, DeMayo FJ, Bagchi IC, Bagchi MK. A genomic approach to identify novel progesterone receptor regulated pathways in the uterus during implantation. Mol Endocrinol 2002; 16:2853–2871. [DOI] [PubMed] [Google Scholar]
- 46. Adiguzel D, Celik-Ozenci C. FoxO1 is a cell-specific core transcription factor for endometrial remodeling and homeostasis during menstrual cycle and early pregnancy. Hum Reprod Update 2021; 27:570–583. [DOI] [PubMed] [Google Scholar]
- 47. Kajihara T, Brosens JJ, Ishihara O. The role of FOXO1 in the decidual transformation of the endometrium and early pregnancy. Med Mol Morphol 2013; 46:61–68. [DOI] [PubMed] [Google Scholar]
- 48. Labied S, Kajihara T, Madureira PA, Fusi L, Jones MC, Higham JM, Varshochi R, Francis JM, Zoumpoulidou G, Essafi A, Fernandez de Mattos S, Lam EW et al. Progestins regulate the expression and activity of the forkhead transcription factor FOXO1 in differentiating human endometrium. Mol Endocrinol 2006; 20:35–44. [DOI] [PubMed] [Google Scholar]
- 49. Paria BC, Huet-Hudson YM, Dey SK. Blastocyst's state of activity determines the "window" of implantation in the receptive mouse uterus. Proc Natl Acad Sci U S A 1993; 90:10159–10162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Moulton BC. Effect of progesterone on DNA, RNA and protein synthesis of deciduoma cell fractions separated by velocity sedimentation. Biol Reprod 1979; 21:667–672. [DOI] [PubMed] [Google Scholar]
- 51. Sachs L, Shelesnyak MC. The development and suppression of polyploidy in the developing and suppressed deciduoma in the rat. J Endocrinol 1955; 12:146–151. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are available in the article and in its online supplementary material.