Abstract
Apolipoprotein E (APOE) is a key regulator of lipoprotein metabolism, and consequently, affects the plasma and tissue lipid contents. The aim of the present study was to investigate the parallel effects of APOE genetic variants and promoter methylation levels of six CpGs on the risk of diabetic dyslipidemia. A total of 204 Palestinian type 2 diabetes (T2D) patients (mean age ± SD: 62.7±10.2) were enrolled in the present study (n=96 with dyslipidemia and n=108 without dyslipidemia). Next generation sequencing was performed to analyze five single nucleotide polymorphisms: Two variants rs7412 and rs429358 that determine APOE ε alleles, and three variants in the promoter region (rs769446, rs449647, and rs405509). For all subjects, the most common genotype was ε3/ε3 (79.4%). No statistical differences were observed in the APOE ε polymorphisms and the three promoter variants among T2D patients with and without dyslipidemia (P>0.05). A comparison of lipid parameters between ε3/ε3 subjects and ε4 carriers in both groups revealed no significant differences in the mean values of LDL-C, HDL-C, TG, and TC levels (P>0.05). Six CpG sites in the APOE promoter on chromosome 19:44905755-44906078 were identified, and differential DNA methylation in these CpGs were observed between the study groups. Logistic regression analysis revealed a significant association of DNA methylation level at the six CpGs with an increased risk of diabetic dyslipidemia (odds ratio, 1.038; 95% confidence interval, 1.012-1.064; P=0.004). In conclusion, the present study revealed that DNA methylation levels in six CpGs in the APOE promoter region was associated with the risk of diabetic dyslipidemia independently of the APOE ε4 variant which could be a potential therapeutic target to reverse the methylation of the APOE promoter.
Keywords: apolipoprotein E polymorphisms, apolipoprotein E promoter, methylation, CpG, type 2 diabetes, dyslipidemia
Introduction
Apolipoprotein E (ApoE) is a constituent of chylomicrons, and plasma very low density lipoproteins (VLDL), and high density lipoproteins (HDL). Human ApoE-encoded by the APOE gene-is a polymorphic protein with three common isoforms (ApoE2, ApoE3, and ApoE4) defined by two single nucleotide polymorphisms (rs429358 and rs7412) in the coding region of exon 4 that overlaps with a well-defined CpG island (CGI) (1). These isoforms have differing affinities for binding to LDL receptors: ApoE3 and ApoE4 bind with similar affinity, while ApoE2 has only 2% of this binding affinity. ApoE3 is the most commonly expressed isoform in healthy people (2). ApoE protein plays a key role in lipid metabolism, including the redistribution of lipoproteins and maintenance of cholesterol homeostasis by stabilization of lipoprotein particles in the circulation and enhancing uptake by the liver and other tissues. Different genetic studies have revealed the association between APOE and its ε2/ε3/ε4 alleles with several pathological conditions and disorders, including Alzheimer's disease (AD) (2), cardiovascular disease (3), familial dysbetalipoproteinemia (4), metabolic syndrome and diabetes (5). The ε4 allele has been associated with higher total and LDL cholesterol levels than in patients with the ε3 allele, and thus reported as the strongest genetic factor for AD and coronary artery disease in several studies and across several ethnic groups (1).
Furthermore, it has been reported that ε4 carriers are less responsive to lipid lowering therapy (i.e Statins) but more sensitive to a low fat diet and physical activity (6). A population-based study reported that ε4/ε4 carriers had a 2.28-fold increased risk of discontinuing a statin therapy compared to ε3/ε3 carriers (7), which has been attributed to therapeutic inefficacy and severe side effect (8). Moreover, ε2 carriers had a better response in TG, TC, and LDL-C reduction percentage in response to statins treatment, compared with ε4 carriers (9). Thus, knowing the APOE genotype may be useful in guiding the choice of treatment, in assessing the potential side effects, and in suggesting the complementary approaches that can be used to control metabolic variations in ε4 carriers. In addition, three common single nucleotide polymorphisms (SNPs) rs449647 (A491-T), rs769446 (C427-T), and rs405509 (G219-T) located in the promoter region of the APOE gene have been reported to influence APOE transcriptional activity, most likely through differential binding of transcription factors and thus differential APOE gene expression (10).
Conversely, it is well known that epigenetic mechanisms such as DNA methylation regulate the transcription of several genes and has been linked to the development of various diseases. Several studies investigated the regulatory mechanisms underlying APOE gene regulation (11). Promoters and regulatory element activities (i.e enhancers) are affected by cytosine methylation at CpG sites in the genome (12). Gene expression inhibition is correlated with hypermethylated promoters and non-promoter sites that are located within enhancer regions leading to loss of enhancer activity and consequently transcriptional inactivation of specific genes (13). Thus, the pleiotropic roles of APOE may lie in its unique epigenetic properties (1). To the best of our knowledge, no previous studies have investigated the association of APOE gene variants and methylation with the risk of dyslipidemia in type 2 diabetes (T2D) patients. Therefore, the present study aimed to determine the allelic and genotypic frequencies of the APOE gene polymorphisms and the three promoter variants to investigate whether APOE genotypes influenced lipid profiles in Palestinian T2D patients with and without dyslipidemia. Moreover, whether DNA methylation of the APOE promoter region differed in diabetic dyslipidemia patients compared with those without dyslipidemia was also assessed. This genetic study may provide additional information regarding the Palestinian population beyond traditional risk factors.
Materials and methods
Study participants
A total of 204 unrelated hospitalized T2DM patients were randomly recruited from Ramallah hospital between January and April 2019. The median age of the subjects studied was 62 years (age range 40-97 years), 125 were males and 79 were females. Among these subjects, 96 were diagnosed with diabetic dyslipidemia and 108 T2DM subjects without dyslipidemia, and none of these patients had been previously treated with lipid lowering drugs. Diagnosis of T2D was based on World Health Organization criteria: Fasting blood glucose ≥126 mg/dl and/or currently on use of antidiabetic medication (14). Patients who were diagnosed with type 1 diabetes were excluded from the study. Dyslipidemia was defined as a level of total cholesterol (TC) >200 mg/dl, triglycerides (TG) >50 mg/dl, LDL-C >130 mg/dl, HDL-C <40 mg/dl, or under medication of lipid lowering drugs (15). The study was conducted according to the guidelines expressed in the Declaration of Helsinki (16), and written informed consent was obtained from all enrolled participants. The study procedure was approved by the local ethical committee at Al-Quds University (East Jerusalem, Palestine; approval no. 71/REC).
Demographic, anthropometric, and biochemical measurements
Demographic information including sex and age was collected from the patient's medical record using a well-structured questionnaire. Body mass index (BMI) was calculated as weight divided by height squared (kg/m2). Blood pressure was measured with a mercury sphygmomanometer at the time of blood sampling. Blood samples (5 ml) were drawn after a 12-h minimum fast. Plasma glucose, Glycated Hb (HbA1C), cholesterol, HDL cholesterol, and triglyceride were determined using standard methods of commercial kits. LDL cholesterol was calculated based on the Friedewald formula (17).
DNA extraction and genotyping
Genomic DNA was extracted from whole blood (300 µl) using a genomic QIAamp DNA purification kit according to the manufacturer's instructions (Qiagen GmbH). The DNA concentration was determined using a NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific, Inc.) and was stored at -20˚C until required for further analysis. Amplicon-based next generation sequencing (NGS) was used for genotyping as previously described (18). Briefly, three primers (two forward and one reverse) were used in multiplex PCR to target the two SNPs of APOE rs429358 [C/T] and rs7412 [C/T]. Based on these 2 SNPs, the APOE alleles (ε2, ε3, and ε4) were determined.
Moreover, four primers (two forward and two reverse) were used to target the three SNPs in APOE promoter region rs769446 [T/C], rs449647[A/T], and rs405509 [G/T]. All primers were modified with over hanged Illumina adaptor sequences at the 5' ends that were added to the forward (5'-CGTCGGCAGCGTCAGATGTGTATAAGAGACA-3') and reverse primers (5'-GTCTCGTGGGCTCGGAGATGTGTATAAGAGA-3'). The primer sequences and the final length of target sequences are shown in Table I. The reaction was carried out using 3 µl of the extracted DNA in a final volume of 25 µl, which contained 12.5 µl PCRBIO HS Taq Mix Red (PCR Biosystems, Ltd.), 8.5 µl double distilled water (dH2O) and 0.2 µM of each primer. The amplification conditions were as follows: Initial denaturation at 95˚C for 5 min followed by 32 cycles of 95˚C for 30 sec, 65˚C for 30 sec, and 72˚C for 40 sec, with a final extension step of 72˚C for 6 min. The PCR products of all samples were visualized on 1.5% agarose gel, cleaned using the Agencourt AMPure XP system (X1, cat. no. A63881; Beckman Coulter Genomics), and eluted in 25 µl elution buffer. DNA library preparation and barcoding were performed using a Nextera XT Index Kit (Illumina, Inc.) as described previously (18). Samples were deep sequenced on a NextSeq 500/550 machine using the 150-cycle Mid Output Kit (Illumina, Inc.). The obtained DNA sequences were analyzed using the Galaxy program (https://usegalaxy.org/). A total of 10 virtual probe sequences (four for the two APOE gene variants and six for the three APOE promoter variants) were used to identify the targeted polymorphisms (Table I). The genotypes were determined, based on the calculated ratio between the read counts for wild type and mutant alleles, for all SNPs (APOE ε and promoter polymorphisms) in each sample. All genotyping personnel were blinded to the clinical data.
Table I.
Sequences of the primers and virtual probes used in PCR and sequence analysis.
| SNP | Primer name | Primer sequence, 5'-3'a | PCR target/bp |
|---|---|---|---|
| rs429358T/C and rs7412C/T | APOEF1 | TCCAAGGAGCTGCAGGCGGCGCA | APOE gene/120 and 218 bp |
| APOEF2 | AGAGCACCGAGGAGCTG | ||
| APOER | GCCCCGGCCTGGTACACTGCCA | ||
| rs449647 T/A, rs769446 C/T and rs405509 A/C | ProAPOEF1 | CACGCCTGGCTAACTTTTGT | APOE Promoter/248 and 225 bp |
| ProAPOEF2 | AAGGACAGGGTCAGGAAAGG | ||
| ProAPOER1 | TCCTGGATCCCAGAAAGAAA | ||
| ProAPOER2 | AGGTGGGGCATAGAGGTCTT | ||
| Methylation primers | APOEPIF | GAGGGGTTATTTTTAGGAGTAT | APOE promoter/324 bp |
| APOEPIR | TCCCAATCCTAAAATTCAAATT | ||
| Gene | Probe Name | Probes sequences | Targeted SNP |
| APOE ε probes | rs429358T | GACGTGTGCGGC | T |
| rs429358C | GACGTGCGCGGC | C | |
| rs7412C | GCAGAAGCGCCTGG | C | |
| rs7412T | GCAGAAGTGCCTGG | T | |
| APOE promoter probes | rs449647A | TCTCAAACTCCTG | A |
| rs449647T | TCTCAATCTCCTG | T | |
| rs769446T | GTGAGCTACCGC | T | |
| rs769446C | GTGAGCCACCGC | C | |
| rs405509A | GTCTGTATTACTG | T | |
| rs405509C | GTCTGGATTACTG | G |
aThe targeted SNPs are in bold.
DNA methylation analysis
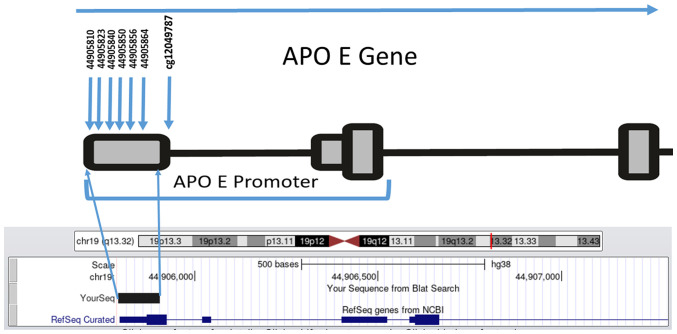
Based on the availability and quantity of DNA samples, a sub group of 119 samples (58 with dyslipidemia and 61 without dyslipidemia) were randomly selected to examine whether APOE promoter methylation was associated with a risk of dyslipidemia. The median age of the subjects studied was 62 years (age range 40-88 years), 70 were males and 49 were females. A minimum concentration of genomic DNA >50 ng was used in the methylation analysis and bisulfite converted using (CiTi Converter DNA Methylation kit, Gdansk) according to the manufacturer's protocol, by which the unmethylated cytosine residues were converted into thymine residues, while the methylated cytosines remained unchanged. A total of six CpG dinucleotides in the APOE promoter, (CpG1-CpG6; Fig. 1) located on chromosome 19 (44905755-44906078) were studied. This region was selected based on a previous study which showed that CpG (cg12049787) within the APOE promoter was associated with AD and correlated with APOE expression (cg26190885, cg08955609) (19). Thus, a fragment of 150 bp that contained six CpGs upstream of the CpG (cg12049787), was amplified using two newly designed primers (Fig. 1).
Figure 1.
Genomic map of the six CpG sites studied in the APOE promoter, cg12049787 CpG was studied previously (19).
The sequences of the primers, modified with over hanged Illumina adaptors at the 5' ends, that were used for amplification of our target fragment are shown in Table I. PCR was performed in a final volume of 25 µl consisting of 12.5 µl (X2) hot start master mix (PCRBIO HS Taq MixRed, PCR Biosystems, Ltd.), 10 µM of each primer and 5 µl bisulfite converted DNA. DNA library preparation and barcoding were performed as described above. The methylation ratios were determined using a Python script (methratio.py) and MethylDackel free program on the UseGalaxy.eu website.
Statistical analysis
All data were analyzed using SPSS version 20.0 (IBM Corp). Demographic characteristics are reported as the mean ± SD or n (%). Comparisons between the two groups were performed using an unpaired Student's t-test or a χ2 test, as appropriate and confirmed by a nonparametric analysis using Mann-Whitney test. Allele and genotype differences between groups and deviations from Hardy-Weinberg (HW) equilibrium were assessed using a χ2 test. Stepwise logistic regression analysis was applied to assess whether DNA methylation levels at the six selected CpG dinucleotides were associated with dyslipidemia.
Results
Study participants
The study included 204 T2DM individuals; the anthropometric, clinical, and biochemical measurements for T2D patients with and without dyslipidemia are presented in Table II. Mean BMI, TC, TG, and LDL-C were significantly higher in participants with dyslipidemia than in those without dyslipidemia (P<0.05) and confirmed by a nonparametric analysis using Mann-Whitney test (Table SI). No significant differences were found in age, sex, systolic and diastolic blood pressure, fasting blood glucose level, and HbA1C (P>0.05).
Table II.
Demographics and biochemical characteristics of study subjects.
| Variable | With dyslipidemia, n=96e | Without dyslipidemia, n=108e | P-value |
|---|---|---|---|
| Age | 62.3 (9.86) | 63.1 (10.53) | 0.62c |
| Sex female/male, n | 34/62 | 45/63 | 0.36d |
| Body mass index, kg/m2 | 29.9 | 25 | 2.43x10-18b,c |
| Systolic blood pressure, mmHg | 135.02 (18.75) | 139.85 (20.49) | 0.08a,d |
| Diastolic blood pressure, mmHg | 77.97 (13.11) | 81.06 (11.15) | 0.07a,c |
| HbA1C, % | 8.02 (1.3) | 8.13 (1.5) | 0.59d |
| Fasting blood glucose, g/dl | 241.62 (108.66) | 227.22 (92.82) | 0.31d |
| Total triglyceride, mg/dl | 252.6 (148.79) | 149.68 (60.32) | 4.18x10-9b,d |
| Total cholesterol, mg/dl | 237.38 (86.47) | 161.78 (50.7) | 5.34x10-12b,d |
| High-density lipoprotein cholesterol, mg/dl | 36.07 (13.17) | 49.58 (17.99) | 3.94x10-9b,d |
| Low-density lipoprotein cholesterol, mg/dl | 161.51 (54.32) | 96.18 (41.64) | 1.06x10-17b,d |
aP<0.05,
bP<0.0001.
cUnpaired Student's t-test.
dχ2-test.
eData are presented as the mean ± SD.
APOE genotypes and allele frequencies
The genotype and allelic distributions of the APOE gene are described in Table III. The distribution of APOE genotypes was in HW equilibrium in both groups (P>0.05). No significant differences were observed in the APOE genotypes and alleles frequencies between the two groups (P>0.05). In all subjects, the frequency of each genotype in descending order was (ε3/ε3, 79.4%; ε2/ε3, 9.3%; ε3/ε4, 7.4%; ε2/ε2, 2.5%; ε2/ε4, 1.5%). No ε4/ε4 genotype was detected. The ε3 allele was the most common (81.3%) followed by the ε2 allele (11.2%) and then the ε4 allele (7.4%).
Table III.
APOE genotypes and allele frequencies and their distribution in type 2 diabetes mellitus patients with and without dyslipidemia.
| Genotype | All subjects, n (%) | With dyslipidemia, n (%) | Without dyslipidemia, n (%) | P-valuea |
|---|---|---|---|---|
| ε2/ε2 | 5 (2.5) | 4 (4.2) | 1 (0.9) | 0.19 |
| ε2/ε3 | 19 (9.3) | 8 (8.3) | 11 (10.2) | 0.81 |
| ε2/ε4 | 3 (1.5) | 2 (2.1) | 1 (0.9) | 0.60 |
| ε3/ε3 | 162 (79.4) | 75 (78.1) | 87 (80.6) | 0.73 |
| ε3/ε4 | 15 (7.4) | 7 (7.3) | 8 (7.4) | 1 |
| ε2 | 27 (11.2) | 14 (12.4) | 13 (10.2) | 0.68 |
| ε3 | 196 (81.3) | 90 (79.6) | 106 (82.8) | 0.62 |
| ε4 | 18 (7.4) | 9(8) | 9(7) | 0.81 |
aFisher's exact test.
Association between APOE genotypes with the risk of dyslipidemia
Logistic regression adjusted for age, sex, and BMI showed no association between APOE genotypes and the risk of dyslipidemia (Table IV). A comparison of lipid parameters between ε3/ε3 subjects and ε4 allele carriers in both groups revealed no significant differences in mean values for the LDL-C, HDL-C, TG, and TC levels (Table V).
Table IV.
Association of APOE genotypes with the risk of dyslipidemia in type 2 diabetes mellitus patients.
| Genotype | All subjects, n (%) | With dyslipidemia, n (%) | Without dyslipidemia, n (%) | Adjusted OR 95% CIa | P-value | OR 95% CI | P-value |
|---|---|---|---|---|---|---|---|
| ε2/ε2 | 5 (2.5) | 4 (4.2) | 1 (0.9) | 0.28 (0.01-3.66) | 0.36 | 4.6 (0.51-42) | 0.17 |
| ε2/ε3 | 19 (9.3) | 8 (8.3) | 11 (10.2) | 1.25 (0.39-4.17) | 0.71 | 0.8 (0.32-2.2) | 0.72 |
| ε2/ε4 | 3 (1.5) | 2 (2.1) | 1 (0.9) | 1.34 (0.05-20.20) | 0.83 | 2.3 (0.21-26) | 0.5 |
| ε3/ε3 | 162 (79.4) | 75 (78.1) | 87 (80.6) | 1.00 | NA | 1.00 | NA |
| ε3/ε4 | 15 (7.4) | 7 (7.3) | 8 (7.4) | 0.67 (0.17-2.67) | 0.57 | 1.02 (0.35-2.9) | 0.98 |
aAsjusted for age, sex and body mass index. OR, odds ratio; CI, confidence interval.
Table V.
Comparison of the lipid profiles between ε3/ε3 genotype subjects and ε4 allele carriersd.
| With dyslipidemia | Without dyslipidemia | |||||
|---|---|---|---|---|---|---|
| Variable | ε3/ε3 | ε4 | P-value | ε3/ε3 | ε4 | P-value |
| Age, yearse | 62.4 (9.8) | 61.1(11) | 0.61a | 64.3 (10.3) | 57.6 (10.4) | 0.009a,b |
| Sex, female/ male, n | 28/47 | 12/5 | 0.544b | 37/50 | 8/12 | 0.521c |
| Body mass index, kg/m2e | 29.7(4) | 30.8 (2.7) | 0.289a | 24.9 (3.6) | 23.7 (3.3) | 0.184b |
| Total cholesterol, mg/dle | 238.37(91) | 232.44 (77.95) | 0.84a | 160.78 (49.19) | 162.78 (66.16) | 0.93b |
| Total triglyceride, mg/dle | 251.53 (161.56) | 251.56 (116.82) | 1a | 150.15 (62.02) | 131.67 (37.19) | 0.21b |
| High-density lipoprotein, mg/dle | 36.57 (13.93) | 36.44 (8.38 ) | 0.97a | 49.01 (18.42) | 48.33 (14.44) | 0.90b |
| Low-density lipoprotein, mg/dle | 164.23 (56.55) | 136 (46.81) | 0.12a | 90.89 (35.17) | 114.44 (68.68) | 0.34b |
aP<0.01.
bUnpaired Student's t-test.
cχ2 test.
dε4 carrier=ε2/ε4+ε3/ε4.
eData are presented as the mean ± SD.
Allelic and genotypic frequency of APOE promoter variants and the risk of dyslipidemia
The genotypes and alleles distributions of the three promoter variants (rs769446, rs449647, and rs405509) are shown in Table VI. No deviations from the expected HW frequencies were found in both groups (P>0.05). The genotype distribution and allele frequency of the three promoter variants were not statistically different between the two studied groups (P>0.05; Table VI). Moreover, no statistical differences were observed in the genotype frequencies of the three APOE promoter variants stratified according to the ε2, ε3/ε3, and ε4 status (data not shown).
Table VI.
Genotypes and alleles frequencies of APOE promoter variants among type 2 diabetes mellitus patients with and without dyslipidemia.
| SNP rs | Genotype/ allele | Total, n (%) | With dyslipidemia, n (%) | Without dyslipidemia, n (%) | Odds ratioa | 95% Confidence intervala | P-value |
|---|---|---|---|---|---|---|---|
| rs769446 | TT | 181 (88.7) | 87(91) | 94(87) | 1.00 | 0.14 | |
| TC | 23 (11.3) | 9(9) | 14(13) | 0.44 | (0.14-1.34) | ||
| T | 385(94) | 183(95) | 202(94) | 1.00 | 0.14 | ||
| C | 23(6) | 9(5) | 14(6) | 0.44 | (0.14-1.34) | ||
| rs449647 | AA | 84 (41.2) | 42(44) | 42(39) | 1.00 | 0.07 | |
| AT | 92 (45.1) | 41(43) | 51(47) | 0.42 | (0.19-0.92) | ||
| TT | 28 (13.7) | 13(14) | 15(14) | 0.81 | (0.29-2.30) | ||
| A | 260 (63.7) | 125(65) | 135(62) | 0.28 | |||
| T | 148 (36.3) | 67(35) | 81(38) | 0.76 | (0.46-1.25) | ||
| rs405509 | GG | 57 (27.9) | 26(27) | 31(29) | 1.00 | 0.58 | |
| GT | 99 (48.5) | 48(50) | 51(47) | 0.96 | (0.42-2.19) | ||
| TT | 48 (23.5) | 22(23) | 26(24) | 1.51 | (0.56-4.09) | ||
| G | 213 (52.2) | 100(52) | 113(52) | 1.00 | 0.44 | ||
| T | 195 (47.8) | 92(48) | 103(48) | 1.22 | (0.74-2.00) |
aAdjusted for age, sex and BMI.
DNA methylation level of the APOE promoter region
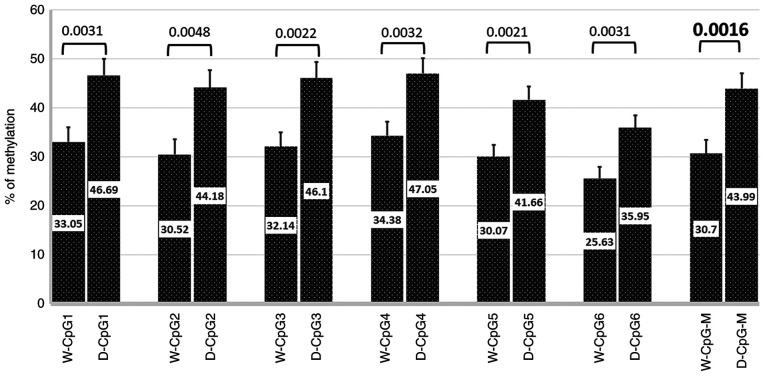
A sub group of 119 patients was included to study the methylation pattern among T2D patients with and without dyslipidemia. The clinical characteristics of this subgroup (n=119; 58 with dyslipidemia and 61 without dyslipidemia) are described in Table SII. The mean BMI was higher in patients with dyslipidemia (30.2±3.8 kg/m2) compared with those without dyslipidemia (24.8±3.6 kg/m2) (P<0.05). The lipid profiles including TC, TG, and LDL were also significantly higher in patients with dyslipidemia compared with those without dyslipidemia (P<0.05; Table SII). Percent methylation for each CpG (1-6) across the APOE promoter was compared between diabetic patients with and without dyslipidemia. The percent methylation level at any CpG site was significantly higher in patients with dyslipidemia compared with those without dyslipidemia: CpG1, P=0.0031; CpG2, P=0.0048; CpG3, P=0.0022; CpG4, P=0.0032; CpG 5, P=0.0021, CpG6, P=0.0031, mean CpG (1-6) methylation, P=0.0016 (Fig. 2).
Figure 2.
Comparison of the methylation pattern in the APOE promoter CpG sites between diabetic patients with (n=58) and without dyslipidemia (n=61). Numbers on bars indicate the mean methylation level at each CpG site. Error bars represent the standard deviation of methylation level at each CpG site. APOE, apolipoprotein E; D, patients with dyslipidemia; W, patients without dyslipidemia; M, mean methylation across the six CpGs.
The relationship between mean methylation levels and the risk of dyslipidemia was analyzed using a logistic regression model adjusted for potential confounding factors. the analysis showed a significant association of methylation level at the CpGs (1-6) with increased risk of dyslipidemia odds ratio (OR)=1.038; 95% confidence interval (CI)=1.012-1.064); P=0.004 (adjusted for age, sex, BMI, HDL, LDL levels, and ε4 allele, Table VII). Moreover, LDL levels were revealed to be an independent risk factor for dyslipidemia development (OR=1.025; 95% CI=1.013-1.038); P<0.001 (adjusted for age, sex, BMI, HDL, and ε4 allele). BMI was positively associated with the risk of dyslipidemia (OR=1.458; 95% CI=1.212-1.754); P<0.001 (adjusted for age, sex, LDL, HDL, and ε4 allele).
Table VII.
Logistic regression analysis of the association between each variable and risk of dyslipidemia.
| Variable | Odds ration | 95% confidence interval | Adjusted P-value |
|---|---|---|---|
| Mean CpG(1-6) | 1.038 | 1.012-1.064 | 0.004b |
| Age | 1.049 | 0.987-1.115 | 0.126 |
| Sex | 1.466 | 0.463-4.641 | 0.515 |
| Body mass index | 1.458 | 1.212-1.754 | 0.00a |
| High-density lipoprotein | 1 | 0.954-1.047 | 0.988 |
| Low-density lipoprotein | 1.025 | 1.013-1.038 | 0.00a |
| ε4 carrier | 1.63 | 0.376-7.067 | 0.514 |
aP<0.01,
bP<0.0001.
Stratification of LDL and BMI mean values by sex and age showed no significant differences in each group with and without dyslipidemia (Table SIII).
Discussion
It is now well established that the APOE gene plays an important role in lipid metabolism and its genetic variations are associated with cognitive function, diabetes and cardiovascular disease (20,21). To the best of our knowledge, the effect of APOE polymorphisms and methylation profile on diabetic dyslipidemia has never been studied in the Palestinian population. First, we investigated the relationship between the APOE polymorphisms and the risk of dyslipidemia in T2D patients. The current study showed that there was no significant difference in the distribution of ε2, ε3, and ε4 alleles among T2D patients with and without dyslipidemia. In all T2D subjects, the ε3 allele was the most common (81.3%), followed by the ε2 allele (11.2%) and then the ε4 allele (7.4%), which is different from that reported in Turkish (22), Egyptian (23) and Saudi diabetic patients (24). The differences in APOE allele frequency in our Palestinian population could indicate differences in disease risk. A Palestinian study conducted on healthy men (n=140) reported that the overall prevalence of dyslipidemia was 66.4% and hypo HDL was the most prevalent type of dyslipidemia (59.3%), which was followed by hypertriglyceridemia (20%) (25). In contrast, dyslipidemia prevalence was 78.7% in men and 80.4% in women in Turkish adults (26), 33% in Saudi adults (27), and 36.8% in Egyptian adults (28), which may reflect different genetic predispositions, socioeconomic status, and lifestyles of the studied subjects. Another Palestinian study revealed that hypo HDL was highly prevalent among obese and normal-weight adults indicating that factors other than BMI and central obesity could influence hypo HDL in Palestinians; the study revealed a significant association of hypo HDL with marital status, tobacco smoking, and occupational exposure to pesticides (29).
Although the three ApoE isoforms differ in their binding affinity to LDL receptors resulting in differences in clearance and uptake of lipoproteins and lipid levels (30), our results showed no differences in serum lipid levels (TC, TG, LDL-C, and HDL-C) between the ε4 and ε3/ε3 carriers in both groups with and without dyslipidemia. In agreement with these results, Al-Shammari et al (31) reported no association between APOE allelic patterns and the blood lipid levels in healthy Kuwaiti Arab subjects and patients with combined hyperlipidemia. A recent study conducted in 2021 on Indonesian T2D patients reported no association between TC, TG, and LDL-C levels with APOE polymorphisms, but showed that ε2 and ε4 carriers had lower levels of HDL-C (32). Other studies revealed that APOE polymorphisms were associated with serum lipid levels in healthy controls but not in T2D patients (33,34). A meta-analysis of 16 studies revealed an association of APOE polymorphisms with the levels of TC, TG, HDL-C, and LDL-C (35). However, the inconsistent results may be attributed to the differences in the included populations, the sample sizes, genotyping methods, and other risk factors. In the present study, we used amplicon based NGS assay, which provides DNA sequences with minimum genotyping error compared with other traditional genotyping methods. DNA was sequenced with a quality score of >20, representing an error rate of 1 in 100 with a corresponding call accuracy of 99%. Moreover, no significant associations were found between the promoter variants rs769446, rs449647, and rs405509 with the risk of dyslipidemia or changes in the lipid profile. Notably, among the ε3/ε3 subjects who had rs405509 GG genotype, there was a trend towards higher LDL and triglyceride levels than in TT carries, but this did not reach statistical significance (data not shown). These findings were previously described in the general population (36). Hence, future studies with larger sample sizes including non-diabetic subjects are warranted.
No previous studies have investigated APOE methylation in relation to diabetic dyslipidemia in Palestinians, to the best of our knowledge. Therefore, to clarify the contribution of APOE promoter methylation to the risk of diabetic dyslipidemia, we used 119 available DNA samples and evaluated the levels of APOE promoter methylation in the T2D patients with and without dyslipidemia. A total of six CpG sites were identified in the APOE promoter within the area 44905755-44906078 on chromosome 19 which were differentially methylated between the studied groups. We observed that the APOE promoter was significantly hypermethylated in T2D patients with dyslipidemia compared with those without dyslipidemia for the six CpG sites. These results indicated that DNA methylation changes in the APOE promoter may be involved in the progression of diabetic dyslipidemia. These results could be attributed to the fact that the APOE promoter region has important epigenetic regulatory functions in which hypermethylation of this region could modify gene expression and consequently influence blood lipid levels and participate in the pathogenesis of dyslipidemia. However, the function of this candidate element should be tested in vivo to validate its transcriptional regulatory activity and to investigate its effect on the ApoE level. A recent study revealed a negative correlation between the levels of total APOE RNA and DNA methylation at the APOE CGI in the frontal lobe tissues (37). Another APOE genotyping and sequencing study conducted on 10,369 individuals revealed that dementia risk increased with decreasing ApoE levels caused by rare genetic variations other than the common ε2/ε3/ε4 polymorphisms (38).
In addition, a review study reported that the common and rare APOE variants alongside environmental factors and epigenetics are associated with variations in lipids and lipoprotein levels affecting the clinical presentation of dyslipidemia (39). Together, these results indicated that molecular screening of the APOE gene and study of epigenetic variations are crucial to understanding the implications of the APOE variants in each of these diseases, which would be possible using high throughput sequencing, i.e NGS. In the present study, logistic regression analysis revealed that higher DNA methylation levels at the six CpGs in the promoter region of APOE increases the odds of dyslipidemia independently of the APOE ε4 carrier. Karlsson et al (40) also reported that APOE allelic variation and increased methylation levels in the promoter region may act independently to increase the risk of dementia. Another case-control study showed that a higher DNA methylation level at two CpGs in the APOE promoter was independently associated with atherosclerotic cerebral infarction (41). In contrast to our results, Mur et al (42) showed that DNA methylation levels at five CpGs in the APOE promoter region were higher in ε4 carriers compared to ε3 carriers.
It is well known that the endogenous pattern of DNA methylation is influenced by environmental factors such as environmental pollutants, and the social environment including early life stressors (maternal stress) that have a marked effect on DNA methylation during fetal development and throughout life. These epigenetic changes may affect neuronal structure and function and thus an organism's health behaviors (43,44) which are however beyond the scope of this study. In the current study we confirmed the known effect of age on methylation levels, a group of non-diabetic healthy individuals (n=58) of younger age (mean age ± SD, 45.6±8.5) tended to have lower methylation levels at the six analyzed CpG sites (mean ± SD, 11.595±7.944) (data not shown).
However, additional detailed study is required to investigate the association between DNA methylation and age between the two sex subgroups. Furthermore, LDL levels and BMI were revealed as independent risk factors for dyslipidemia, which confirmed that the progression of diabetic dyslipidemia is a consequence of genetic and other risk factors that affect lipid levels such as age, smoking, and a high fat intake (45).
In addition to the small sample size, some limitations should be addressed in our study. First, the selected six CpGs in the APOE promoter region may not represent the methylation pattern of the whole APOE gene, other CpGs may also be related to the risk of dyslipidemia. Second, confounding factors such as smoking, energy intake, folic acid supplementation, dyslipidemia/antidiabetic medications, and family history might affect DNA methylation patterns. A recent study reported that statin use was an independent factor of higher ABCG1 methylation (at the cg06500161 site) and this was correlated with the differential expression of genes involved in both lipid metabolism and glycemic pathways (46). Unfortunately, no information on the treatment or family history of diabetes and dyslipidemia was available for all study participants; therefore we could not include these confounding factors in our statistical analysis.
A study conducted on stunted growth revealed a significant association between low dietary intakes (lower intakes of protein and carbohydrate) and increased global DNA methylation (47). Generally, the DNA methylation process is catalyzed by DNA methyltransferases enzymes (DNMTs) in which the methyl group is transferred from S-adenosylmethionine and incorporated into carbon 5 of the cytosine residue followed by a release of s-adenosylhomocysteine (48). Therefore, any genetic deletion or silencing of these enzymes will induce passive demethylation of the CpG sites in gene promoters and subsequently aberrant gene expression (48). Furthermore, some dietary intakes that act as methyl donors may influence DNA methylation either by inhibiting DNMTs enzymes or by changing the availability of substrates required for these enzymatic reactions (49).
In conclusion, this study demonstrates that APOE ε genotypes and the promoter variants (rs769446, rs449647, and rs405509) are not associated with an increased risk of diabetic dyslipidemia. Our study revealed that APOE DNA methylation levels at the six CpGs in the promoter region are associated with diabetic dyslipidemia. Differential DNA methylation at these loci might serve as a biomarker for diabetic dyslipidemia and represent an interesting possibility for future research to identify a new therapeutic target to reverse the methylation of APOE promoter and may improve the clinical management of diabetic dyslipidemia.
Supplementary Material
Acknowledgements
Not applicable.
Funding Statement
Funding: This work was funded by the Deanship of Scientific Research (Al-Quds University-Palestine, Abu Dis, Palestine).
Availability of data and materials
The original data generated using next generation sequencing dataset has been deposited in the National Center for Biotechnology Information Sequence Read Archive (BioProject ID: PRJNA830334, http://www.ncbi.nlm.nih.gov/bioproject/830334; and BioProject ID: PRJNA830330, http://www.ncbi.nlm.nih.gov/bioproject/830330). The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
SE and AN conceived the study. SC and AN performed the analysis. KE, ME, MG and AS performed the experiments and curated the data. SE wrote the manuscript. SC and AN reviewed and edited the manuscript. All authors read and approved the final manuscript. SE and AN confirm the authenticity of all the raw data.
Ethics approval and consent to participate
The study was conducted according to the guidelines expressed in the Declaration of Helsinki, and written informed consent was obtained from all enrolled participants. The study procedure was approved by the local ethical committee at Al-Quds University (East Jerusalem, Palestine; approval no. 71/REC).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Yu CE, Foraker J. Epigenetic considerations of the APOE gene. [corrected] Biomol Concepts. 2015;6:77–84. doi: 10.1515/bmc-2014-0039. [DOI] [PubMed] [Google Scholar]
- 2.Belloy ME, Napolioni V, Greicius MD. A Quarter century of APOE and Alzheimer's disease: Progress to date and the path forward. Neuron. 2019;101:820–838. doi: 10.1016/j.neuron.2019.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ashiq S, Ashiq K. The association of apolipoprotein-E (APOE) gene polymorphisms with coronary artery disease: A systematic review and meta-analysis. Egypt J Med Hum Genet. 2021;22(16) [Google Scholar]
- 4.Smelt AH, de Beer F. Apolipoprotein E and familial dysbetalipoproteinemia: Clinical, biochemical, and genetic aspects. Semin Vasc Med. 2004;4:249–257. doi: 10.1055/s-2004-861492. [DOI] [PubMed] [Google Scholar]
- 5.Sun Y, Wei R, Yan D, Xu F, Zhang X, Zhang B, Yimiti D, Li H, Sun H, Hu C, et al. Association between APOE polymorphism and metabolic syndrome in Uyghur ethnic men. BMJ Open. 2016;6(e010049) doi: 10.1136/bmjopen-2015-010049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Villeneuve S, Brisson D, Marchant NL, Gaudet D. The potential applications of apolipoprotein E in personalized medicine. Front Aging Neurosci. 2014;6(154) doi: 10.3389/fnagi.2014.00154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maitland-van der Zee AH, Stricker BH, Klungel OH, Mantel-Teeuwisse AK, Kastelein JJ, Hofman A, Leufkens HG, van Duijn CM, de Boer A. Adherence to and dosing of beta-hydroxy-beta-methylglutaryl coenzyme A reductase inhibitors in the general population differs according to apolipoprotein E-genotypes. Pharmacogenetics. 2003;13:219–223. doi: 10.1097/00008571-200304000-00006. [DOI] [PubMed] [Google Scholar]
- 8.Cai C, Wen Z, Li L. The relationship between ApoE gene polymorphism and the efficacy of statins controlling hyperlipidemia. Am J Transl Res. 2021;13:6772–6777. [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang L, He S, Li Z, Gan X, Li S, Cheng X, Yang N, Zheng F. Apolipoprotein E polymorphisms contribute to statin response in Chinese ASCVD patients with dyslipidemia. Lipids Health Dis. 2019;18(129) doi: 10.1186/s12944-019-1069-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bizzarro A, Seripa D, Acciarri A, Matera MG, Pilotto A, Tiziano FD, Brahe C, Masullo C. The complex interaction between APOE promoter and AD: An Italian case-control study. Eur J Hum Genet. 2009;17:938–945. doi: 10.1038/ejhg.2008.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bekris LM, Lutz F, Yu CE. Functional analysis of APOE locus genetic variation implicates regional enhancers in the regulation of both TOMM40 and APOE. J Hum Genet. 2012;57:18–25. doi: 10.1038/jhg.2011.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ciceri F, Rotllant D, Maes T. Understanding epigenetic alterations in Alzheimer's and Parkinson's disease: Towards targeted biomarkers and therapies. Curr Pharm Des. 2017;23:839–857. doi: 10.2174/1381612823666170124121140. [DOI] [PubMed] [Google Scholar]
- 13.Bae MG, Kim JY, Choi JK. Frequent hypermethylation of orphan CpG islands with enhancer activity in cancer. BMC Med Genomics. 2016;9 (Suppl 1)(S38) doi: 10.1186/s12920-016-0198-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kumar R, Nandhini LP, Kamalanathan S, Sahoo J, Vivekanadan M. Evidence for current diagnostic criteria of diabetes mellitus. World J Diabetes. 2016;7:396–405. doi: 10.4239/wjd.v7.i17.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jellinger PS, Dickey RA, Ganda OP, Mehta AE, Nguyen TT, Rodbard HW, Seibel JA, Shepherd MD, Smith DA. AACE medical guidelines for clinical practice for the diagnosis and treatment of dyslipidemia and prevention of atherogenesis. Endocr Pract. 2000;6:162–213. AACE Lipid Guidelines Committee. The American Association of Clinical Endocrinologists. [PubMed] [Google Scholar]
- 16.Goodyear MD, Krleza-Jeric K, Lemmens T. The declaration of Helsinki. BMJ. 2007;335:624–625. doi: 10.1136/bmj.39339.610000.BE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krishnaveni P, Gowda VM. Assessing the validity of friedewald's formula and anandraja's formula for serum LDL-cholesterol calculation. J Clin Diagn Res. 2015;9:BC01–BC04. doi: 10.7860/JCDR/2015/16850.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ereqat S, Cauchi S, Eweidat K, Elqadi M, Nasereddin A. Estrogen receptor 1 gene polymorphisms (PvuII and XbaI) are associated with type 2 diabetes in Palestinian women. PeerJ. 2019;7(e7164) doi: 10.7717/peerj.7164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shao Y, Shaw M, Todd K, Khrestian M, D'Aleo G, Barnard PJ, Zahratka J, Pillai J, Yu CE, Keene CD, et al. DNA methylation of TOMM40-APOE-APOC2 in Alzheimer's disease. J Hum Genet. 2018;63:459–471. doi: 10.1038/s10038-017-0393-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shatwan IM, Winther KH, Ellahi B, Elwood P, Ben-Shlomo Y, Givens I, Rayman MP, Lovegrove JA, Vimaleswaran KS. Association of apolipoprotein E gene polymorphisms with blood lipids and their interaction with dietary factors. Lipids Health Dis. 2018;17(98) doi: 10.1186/s12944-018-0744-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Satizabal CL, Samieri C, Davis-Plourde KL, Voetsch B, Aparicio HJ, Pase MP, Romero JR, Helmer C, Vasan RS, Kase CS, et al. APOE and the association of fatty acids with the risk of stroke, coronary heart disease, and mortality. Stroke. 2018;49:2822–2829. doi: 10.1161/STROKEAHA.118.022132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Duman BS, Oztürk M, Yilmazer S, Hatemi H. Apolipoprotein E polymorphism in Turkish subjects with Type 2 diabetes mellitus: Allele frequency and relation to serum lipid concentrations. Diabetes Nutr Metab. 2004;17:267–274. [PubMed] [Google Scholar]
- 23.Galal AA, Abd Elmajeed AA, Elbaz RA, Wafa AM, Elshazli RM. Association of apolipoprotein E gene polymorphism with the risk of T2DM and obesity among Egyptian subjects. Gene. 2021;769(145223) doi: 10.1016/j.gene.2020.145223. [DOI] [PubMed] [Google Scholar]
- 24.Alharbi KK, Khan IA, Syed R. Association of apolipoprotein E polymorphism with type 2 diabetes mellitus in a Saudi population. DNA Cell Biol. 2014;33:637–641. doi: 10.1089/dna.2014.2461. [DOI] [PubMed] [Google Scholar]
- 25.Ali I, Kharma A, Samara M, Odeh S, Jaradat N, Zaid AN, Ahmad MA. Prevalence of dyslipidemia in undiagnosed palestinian men: A cross-sectional study. J Lipids. 2019;2019(3473042) doi: 10.1155/2019/3473042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bayram F, Kocer D, Gundogan K, Kaya A, Demir O, Coskun R, Sabuncu T, Karaman A, Cesur M, Rizzo M, et al. Prevalence of dyslipidemia and associated risk factors in Turkish adults. J Clin Lipidol. 2014;8:206–216. doi: 10.1016/j.jacl.2013.12.011. [DOI] [PubMed] [Google Scholar]
- 27.Alzahrani GS, Aljehani SM, Al-Johani JJ. Risk factors of dyslipidemia among Saudi population, 2017. Egypt J Hosp Med. 2018;71:2262–2265. [Google Scholar]
- 28.Reda A, Ragy H, Saeed K, Alhussaini MA. A semi-systematic review on hypertension and dyslipidemia care in Egypt-highlighting evidence gaps and recommendations for better patient outcomes. J Egypt Public Health Assoc. 2021;96(32) doi: 10.1186/s42506-021-00096-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Damiri B, Aghbar A, Alkhdour S, Arafat Y. Characterization and prevalence of metabolic syndrome among overweight and obese young Palestinian students at An-Najah National University. Diabetes Metab Syndr. 2018;12:343–348. doi: 10.1016/j.dsx.2017.12.021. [DOI] [PubMed] [Google Scholar]
- 30.Mahley RW. Central nervous system lipoproteins: ApoE and regulation of cholesterol metabolism. Arterioscler Thromb Vasc Biol. 2016;36:1305–1315. doi: 10.1161/ATVBAHA.116.307023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Al-Shammari S, Fatania H, Al-Radwan R, Akanji AO. Apolipoprotein E polymorphism and lipoprotein levels in a Gulf Arab population in Kuwait: A pilot study. Ann Saudi Med. 2004;24:361–364. doi: 10.5144/0256-4947.2004.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maratni NP, Saraswati MR, Dewi NN, Yasa I, Eka Widyadharma IP, Putra IB, Suastika K. Association of apolipoprotein E gene polymorphism with lipid profile and ischemic stroke risk in type 2 diabetes mellitus patients. J Nutr Metab. 2021;2021(5527736) doi: 10.1155/2021/5527736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Srirojnopkun C, Kietrungwilaikul K, Boonsong K, Thongpoonkaew J, Jeenduang N. Association of APOE and CETP TaqIB polymorphisms with type 2 diabetes mellitus. Arch Med Res. 2018;49:479–485. doi: 10.1016/j.arcmed.2019.02.005. [DOI] [PubMed] [Google Scholar]
- 34.Seo JY, Youn BJ, Cheong HS, Shin HD. Association of APOE genotype with lipid profiles and type 2 diabetes mellitus in a Korean population. Genes Genomics. 2021;43:725–735. doi: 10.1007/s13258-021-01095-y. [DOI] [PubMed] [Google Scholar]
- 35.Zhou T, Li H, Zhong H, Zhong Z, Lin S. Association of apoE gene polymorphisms with lipid metabolism in renal diseases. Afr Health Sci. 2020;20:1368–1381. doi: 10.4314/ahs.v20i3.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Radwan ZH, Wang X, Waqar F, Pirim D, Niemsiri V, Hokanson JE, Hamman RF, Bunker CH, Barmada MM, Demirci FY, Kamboh MI. Comprehensive evaluation of the association of APOE genetic variation with plasma lipoprotein traits in U.S. whites and African blacks. PLoS One. 2014;9(e114618) doi: 10.1371/journal.pone.0114618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee EG, Tulloch J, Chen S, Leong L, Saxton AD, Kraemer B, Darvas M, Keene CD, Shutes-David A, Todd K, et al. Redefining transcriptional regulation of the APOE gene and its association with Alzheimer's disease. PLoS One. 2020;15(e0227667) doi: 10.1371/journal.pone.0227667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rasmussen KL, Tybjaerg-Hansen A, Nordestgaard BG, Frikke-Schmidt R. APOE and dementia-resequencing and genotyping in 105,597 individuals. Alzheimers Dement. 2020;16:1624–1637. doi: 10.1002/alz.12165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Khalil YA, Rabès JP, Boileau C, Varret M. APOE gene variants in primary dyslipidemia. Atherosclerosis. 2021;328:11–22. doi: 10.1016/j.atherosclerosis.2021.05.007. [DOI] [PubMed] [Google Scholar]
- 40.Karlsson IK, Ploner A, Wang Y, Gatz M, Pedersen NL, Hägg S. Apolipoprotein E DNA methylation and late-life disease. Int J Epidemiol. 2018;47:899–907. doi: 10.1093/ije/dyy025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang H, Zhao X, Wang C, Du R, Wang X, Fu J, Sun Q. A preliminary study of the association between apolipoprotein E promoter methylation and atherosclerotic cerebral infarction. J Stroke Cerebrovasc Dis. 2019;28:1056–1061. doi: 10.1016/j.jstrokecerebrovasdis.2018.12.027. [DOI] [PubMed] [Google Scholar]
- 42.Mur J, McCartney DL, Walker RM, Campbell A, Bermingham ML, Morris SW, Porteous DJ, McIntosh AM, Deary IJ, Evans KL, Marioni RE. DNA methylation in APOE: The relationship with Alzheimer's and with cardiovascular health. Alzheimers Dement (NY) 2020;6(e12026) doi: 10.1002/trc2.12026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mitchell C, Schneper LM, Notterman DA. DNA methylation, early life environment, and health outcomes. Pediatr Res. 2016;79:212–219. doi: 10.1038/pr.2015.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cacabelos R, Torrellas C. Epigenetics of aging and Alzheimer's disease: Implications for pharmacogenomics and drug response. Int J Mol Sci. 2015;16:30483–30543. doi: 10.3390/ijms161226236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chaudhary R, Likidlilid A, Peerapatdit T, Tresukosol D, Srisuma S, Ratanamaneechat S, Sriratanasathavorn C. Apolipoprotein E gene polymorphism: Effects on plasma lipids and risk of type 2 diabetes and coronary artery disease. Cardiovasc Diabetol. 2012;11(36) doi: 10.1186/1475-2840-11-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu Y, Shen Y, Guo T, Parnell LD, Westerman KE, Smith CE, Ordovas JM, Lai CQ. Statin use associates with risk of type 2 diabetes via epigenetic patterns at ABCG1. Front Genet. 2020;11(622) doi: 10.3389/fgene.2020.00622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Iqbal MS, Rahman S, Haque MA, Bhuyan MJ, Faruque ASG, Ahmed T. Lower intakes of protein, carbohydrate, and energy are associated with increased global DNA methylation in 2- to 3-year-old urban slum children in Bangladesh. Matern Child Nutr. 2019;15(e12815) doi: 10.1111/mcn.12815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hervouet E, Peixoto P, Delage-Mourroux R, Boyer-Guittaut M, Cartron PF. Specific or not specific recruitment of DNMTs for DNA methylation, an epigenetic dilemma. Clin Epigenetics. 2018;10(17) doi: 10.1186/s13148-018-0450-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mahmoud AM, Ali MM. Methyl donor micronutrients that modify DNA methylation and cancer outcome. Nutrients. 2019;11(608) doi: 10.3390/nu11030608. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original data generated using next generation sequencing dataset has been deposited in the National Center for Biotechnology Information Sequence Read Archive (BioProject ID: PRJNA830334, http://www.ncbi.nlm.nih.gov/bioproject/830334; and BioProject ID: PRJNA830330, http://www.ncbi.nlm.nih.gov/bioproject/830330). The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.