Abstract
We describe here the development of a single-reaction multiplex PCR assay for the enterotoxin genes from Staphylococcus aureus that utilizes a universal toxin gene primer in combination with toxin-specific primers to amplify characteristic toxin gene products. In combination with a new DNA purification method, the assay can detect enterotoxin genes A to E from a pure culture within 3 to 4 h. The test was used to characterize a diverse set of environmental S. aureus isolates, and a 99% correlation with toxin typing using standard immunological tests was found. The design of the assay allows it to be extended to include both newly characterized and as-yet-unknown toxin genes.
Staphylococcus aureus produces a variety of extracellular protein toxins and virulence factors which contribute to its pathogenic potential. Some S. aureus strains produce pyrogenic exotoxins, such as staphylococcal enterotoxins (SE) and toxic shock syndrome toxin 1 (TSST-1). The SE are exoproteins which, when ingested, induce gastroenteric syndrome in humans and can cause toxic shock (30). Staphylococcal food poisoning outbreaks are characterized by vomiting and diarrhea and occur quite frequently worldwide (4); hence, the detection of enterotoxins is epidemiologically essential.
Several types of staphylococcal enterotoxins have been distinguished serologically, biochemically, and by molecular genetic analysis (2, 5, 8, 11, 23), namely, SEA, SEB, SEC1, SEC2, SEC3, SED, and SEE. Overall, these well-recognized toxins fall into two groups: SEB, SEC1, SEC2, and SEC3, which have 66 to 98% amino acid sequence identity, and SEA, SED, and SEE, which have 53 to 81% identity. The tst gene coding for TSST-1 has little sequence homology with SE or pyrogenic exotoxins (30), although the toxins are structurally very similar.
There are other SE in addition to the five major serological groups. Culture supernatants containing these uncharacterized SE elicit an emetic response when administered to monkeys but do not react with antibodies produced against the characterized SE (25). It has been estimated that ca. 5% of staphylococcal food poisoning outbreaks are due to unidentified enterotoxins (25). In addition, the genes for four new enterotoxins, SEG (6, 32), SEH (36, 42), SEI (32), and SEJ (49), have been reported.
Various methods have been developed for detecting enterotoxin production (13, 14, 37, 48; T. C. Granade, K. M. Hurley, and P. A. Mied, Abstr. Annu. Meet. Am. Soc. Microbiol. 1986, abstr. P7, p. 276, 1986). Of these, it is reversed passive latex agglutination (RPLA) which is most commonly employed and is commercially available. In this test the enterotoxins are identified by antibodies specific for each of the enterotoxins. Cross-reactions between SEB and the SECs (27) and between SEA and SEE (28) have been reported; hence, kits are generally deficient in antibody specific for enterotoxin E, and SEE-producing strains would be classified as SEA producing.
RPLA also depends on sufficient amounts of toxin being produced in the absence of interfering bacterial products for successful detection. Toxin production requires long (e.g., 20 h) incubation periods and is also influenced by culture conditions, pH, water activity, and the substrate used. Insufficient production of toxins at levels below the threshold of these immunological assays leads to false-negative results.
More recently, oligonucleotide probes for specific detection of toxin genes have been developed by several workers (21, 45); however, hybridization techniques are laborious and time-consuming and, moreover, cross-reactions of the probe for SEA with the SEE enterotoxin gene have been reported (12). In addition, the hybridization assay requires enrichment steps for S. aureus, and thus the detection time could not be reduced significantly.
The PCR offers the possibility of detecting specific gene sequences by DNA amplification (38), and therefore bacterial enrichment is not required before a specific gene can be detected. PCR assays for the specific detection of enterotoxin genes, such as SEB and SEC (39), SEA and SEB (17), and SEA to SEE (22, 46; J. P. Rosec, J. P. Guiraud, C. Dalet, and N. Richard, Proc. 8th Int. Symp. Staphylococci Staphylococcal Infect., abstr. P94, p. 212, 1996) have been reported, but in all of these studies, a series of separate reactions is needed to identify a single gene or subset of these genes. In addition, Becker et al. (3) have reported the development of a multiplex PCR reaction for the detection of multiple staphylococcal enterotoxin genes which uses individual primer sets for each toxin gene. More recently, Monday and Bohach (31) have developed a multiplex PCR assay for all of the characterized enterotoxin genes (sea-sej) and tst, but again this requires unique primer sets for detection of individual genes.
In this study, we report the development of a rapid (3 to 4 h), single-reaction multiplex PCR assay which specifically detects genes for staphylococcal enterotoxins A to E in strains of toxigenic S. aureus from various environmental sources. This PCR reaction takes advantage of both conserved and unique regions of the toxin genes and uses one universal forward primer with reverse primers specific for each individual toxin gene. Template DNA was extracted from S. aureus strains using a new, simplified guanidinium thiocyanate method developed in this study. Gene identity is established by the characteristic size of the PCR products. This assay has the advantage that it potentially can be extended to detect uncharacterized toxin genes.
MATERIALS AND METHODS
Bacterial strains and culture media.
Staphylococcal strains (see Table 2) were obtained from the National Collection of Type Cultures (NCTC), London, England, except for FRI326 (provided by M. S. Bergdoll, Food Research Institute, University of Wisconsin–Madison). Other bacteria used were from the Nottingham Laboratory Culture Collection. The working cultures of all strains were prepared by inoculation from frozen glycerol stocks into brain heart infusion (BHI) broth and incubation at 37°C in a shaking incubator for 12 to 16 h.
TABLE 2.
Toxin analysis of control strains by PCR and SET-RPLA toxin assays
| Test straina | Toxin serotype | PCR toxin type |
|---|---|---|
| S. aureus NCTC 8532 | − | − |
| S. aureus NCTC 10652b | SEA, SED | SEA, SED |
| S. aureus NCTC 10654 | SEB | SEB |
| S. aureus NCTC 10655 | SEC | SEC |
| S. aureus NCTC 10656 | SED | SED |
| S. aureus NCTC 10657 | SEA, SEB | SEA, SEB |
| S. aureus F287 | SEA | SEA |
| S. aureus FRI326 | SEA | SEE |
| S. aureus NCTC 11963 (TSST-1)b | SEC | SEC |
| S. chromogenes NCTC 10530 | − | − |
| S. epidermidis NCTC 12100 | SEC | SEC |
| S. hyicus NCTC 10350 | − | − |
| S. intermedius NCTC 11048 | − | − |
| S. lentus NCTC 12102 | − | − |
| S. saprophyticus NCTC 7292 | − | − |
| S. sciuri NCTC 12103 | − | − |
| S. warneri NCTC 11044 | − | − |
S. aureus strains which produce enterotoxins A (F287), B (NCTC 10654), C (NCTC 10655), D (NCTC 10656), and E (FRI326) and nonenterotoxigenic strain (NCTC 8532) were used as reference standards. Strain F287 is from the Nottingham Laboratory Collection and is known to produce enterotoxin A only. Nonstaphylococcal strains used to demonstrate the specificity of the PCR assay were Bacillus cereus, Bacillus subtilis, Corynebacterium oris, Enterobacter spp., Escherichia coli, Klebsiella pneumoniae, Lactococcus lactis, Lactobacillus spp., Listeria monocytogenes, Micrococcus luteus, Proteus vulgaris, Pseudomonas aeruginosa, Salmonella spp., Shigella spp., and Yersinia enterocolitica.
The presence of toxins D and C in these strains was established by this study.
Isolation of genomic DNA.
Total genomic DNA was isolated using a modification of the method of Boom et al. (9). Briefly, 500 μl of overnight-grown staphylococcal culture was centrifuged in a microfuge (Biofuge 13; Heraeus Sepatech) at 12,000 × g for 2 min. The pellets were washed twice with 0.15 M NaCl–10 mM EDTA (pH 8.0) and resuspended in 500 μl of Tris-EDTA buffer. Lysostaphin (25 μg/ml, final concentration; Sigma) and 15 μl of 10% sodium dodecyl sulfate were added, and the cells were incubated at 37°C for 30 min. The lysed cells were added to 900 μl of lysis buffer (4 M GuSCN–0.1 M Tris-HCl (pH 6.4)–0.2 M EDTA (pH 8.0)–2.6% [wt/vol] Triton X-100) and 30 μl of a 20% diatom suspension (Celite; Janssen Chimica). The samples were mixed, incubated at room temperature for 10 min, mixed again, and then centrifuged at 13,000 × g for 20 s. The diatom-nucleic acid pellet was washed sequentially: twice with 1 ml of washing buffer (4 M GuSCN, 0.1 M Tris-HCl [pH 6.4]), twice with 1 ml of 70% ethanol (vol/vol), and once with 1 ml of acetone. Wash solutions were removed by centrifugation, and the pellets were dried by incubation at 56°C for 10 min. Purified DNA from this pellet was eluted by the addition of 100 μl of TE buffer, and diatoms were removed by centrifugation at 13,000 × g for 5 min. This DNA was used as template in PCR amplifications. The same procedure was used for extraction of genomic DNA from nonstaphylococcal strains except the lysostaphin treatment step was omitted. The DNA recovered was routinely sufficient for direct use in PCR amplifications without the need for further quantification.
Immunological detection of enterotoxins.
For enterotoxin detection, S. aureus and other staphylococcal strains were grown in BHI broth at 37°C for 16 to 18 h, the culture was filtered through 0.45-μm (pore-size) low-protein binding filter (Sterile Acrodisc; Gelman Sciences), and the filtrate was used for enterotoxin detection. The enterotoxins were detected by SET-RPLA (staphylococcal enterotoxin-RPLA detection kit for toxins A, B, C, and D; Oxoid, Ltd., Basingstoke, United Kingdom) used according to the manufacturer's instructions.
Oligonucleotide primers.
The oligonucleotide primers were designed by alignment of published DNA sequences of the S. aureus enterotoxin genes sea, seb, sec, sed, and see (2, 5, 8, 11, 23), tst (7), and type A streptococcal pyrogenic exotoxin (speA) (47). By comparison of both the DNA and the deduced amino acid sequences, one forward primer common for all enterotoxin genes and five reverse primers, each specific for one enterotoxin gene of S. aureus, were designed (synthesized by the Biopolymer Synthesis and Analysis Unit of Nottingham University and used without any further purification). The sequences and corresponding sequence locations of these oligonucleotide primers are shown in Table 1.
TABLE 1.
Details of primers and amplicons
| Primer name and size (nt)a | Description | Nucleotide sequence | Gene locationb | PCR product size (bp) |
|---|---|---|---|---|
| SA-U (20) | Universal forward primer | 5′-TGTATGTATGGAGGTGTAAC-3′ | – | |
| SA-A (18) | Reverse primer for sea | 5′-ATTAACCGAAGGTTCTGT-3′ | 639–657 | 270 |
| SA-B (18) | Reverse primer for seb | 5′-ATAGTGACGAGTTAGGTA-3′ | 564–582 | 165 |
| SA-C (20) | Reverse primer for sec | 5′-AAGTACATTTTGTAAGTTCC-3′ | 457–477 | 69 |
| ENT-C (25) | Reverse primer for sec | 5′-AATTGTGTTTCTTTTATTTTCATAA-3′ | 485–510 | 102 |
| SA-D (20) | Reverse primer for sed | 5′-TTCGGGAAAATCACCCTTAA-3′ | 676–696 | 306 |
| SA-E (16) | Reverse primer for see | 5′-GCCAAAGCTGTCTGAG-3′ | 584–600 | 213 |
nt, nucleotide.
–, The common “forward” primer corresponds to the region of each of the enterotoxin genes containing the conserved amino acid motif C-(x)-Y-G-G-(x)-T found at the following corresponding nucleotide locations in each gene sequence: sea, 388; seb, 418; sec, 409; sed, 391; and see, 388. The nucleotide locations given for each reverse primer correspond to the following published nucleotide sequences of the SE genes: sea (23), seb (8), sec (5), sed (11), and see (2).
DNA amplification.
PCR reactions were performed in a reaction buffer (50 mM KCl, 10 mM Tris-HCl [pH 9.0], 4 mM MgCl2, 0.01% gelatin) in a total volume of 50 μl containing 1 μl (∼1 ng) of template DNA, 20 to 30 pmol each of primers SA-U, SA-A, SA-B, SA-C/ENT-C, SA-D, and SA-E, and 0.2 mM of each deoxynucleotide triphosphate (dATP, dTTP, dGTP, and dCTP). This mixture was heated to 94°C for 5 min before the addition of 1.0 U of a thermostable Taq DNA polymerase (Advanced Biotechnologies). Twenty-five amplification cycles (94°C for 30 s, 50°C for 30 s, and 72°C for 30 s) were performed using a Techne thermal cycler (PHC-3) with a final extension cycle of 2 min at 72°C. The PCR products were stored at −20°C until analyzed.
Detection of amplified DNA.
A 10-μl aliquot of the amplified PCR product was analyzed on 2% TAE agarose NA (Pharmacia) gel containing ethidium bromide (0.5 μg/ml). The electrophoresis was carried out in Anachem Origo Horizontal gel tanks at 80 V for 1 h or until the desired resolution was obtained. Alternatively, 5 μl of PCR product was electrophoresed on a 6% polyacrylamide gel (1). Gels were viewed by UV transillumination (UVP) and photographed by using a 35-mm camera. AluI-digested pBR322 DNA or 100-bp ladder (Pharmacia Biotech) were used as molecular size markers in all gels.
Purification of PCR products and restriction enzyme analysis.
Individual PCR products were purified from gels using QIAquick PCR purification kit according to the manufacturer's instructions or by the freeze extraction method of Qian and Wilkinson (35). Restriction enzymes were purchased from Promega and used according to the manufacturer's recommendations.
Southern hybridization.
DNA samples were electrophoresed on a 1.5% agarose NA (Pharmacia) gel with an appropriate molecular size marker. Southern blots (41) were performed by using alkaline vacuum transfer and Hybond N+ membrane (Amersham). Hybridization and subsequent detection were performed by using a nonradioactive digoxigenin DNA labeling and detection kit (Boehringer Mannheim) according to the manufacturer's instructions. The PCR products to be labeled were band extracted from SeaPlaque (FMC) low-melting-point agarose gel by using the freeze extraction method of Qian and Wilkinson (35).
RESULTS
Selection and specificity of primers targeting toxin genes.
By alignment of the known toxin gene sequences it was possible to identify a highly conserved region of enterotoxin genes which was used to design a universal “forward” primer (SA-U; Table 1) which could be used in combination with toxin gene-specific reverse primers. In spite of the high degree of relatedness among enterotoxin genes (∼50% for seb or sec to sea, 84% for sea to see, and ∼54% for sed to sea or see) it was possible to identify DNA sequences unique to each of the specific enterotoxin genes, and these were used to design several oligonucleotide primers which could be used for the amplification and differentiation of toxin genes A to E. These primers were termed SA-A, SA-B, SA-C–ENT-C, SA-D, and SA-E, respectively. Initially, the primer SA-C was used to detect the presence of the sec gene, yielding a PCR product of 69 bp, but this could only be reliably resolved for size estimation with acrylamide gels. Therefore, for convenience, a second primer (ENT-C) was designed, giving a PCR product of 102 bp, although both primers were able to amplify sec gene sequences equally well.
Specificity of oligonucleotide primers.
Individually each of the reverse primers was tested in combination with the universal forward primer by using a panel of known enterotoxigenic S. aureus strains (see Materials and Methods and Table 2) to ensure that a PCR product of the expected size was produced and that no additional or nonspecific products were generated. In each case a PCR product of the expected size was generated (Fig. 1, lanes 1 to 6; Table 1); however, when using primer SA-D, an extra product of approximately 790 bp was produced in addition to the expected SED amplicon of 306 bp (Fig. 1, lane 5). To investigate the origin of this additional band, both PCR products were purified and used as a template for PCR amplification with the SA-D and SA-U primers. Only one band of 306 bp was obtained when the 306-bp band was used as a template but, again, products of 306 and 790 bp were generated when the 790-bp band was used as a template (data not shown). This indicated that the 306-bp fragment is an internal part of the 790-bp product and suggested that the 790-bp product is probably formed by mismatching of either of these two primers. However, the amplification of this additional product did not appear to interfere with the amplification of the desired sed gene sequences.
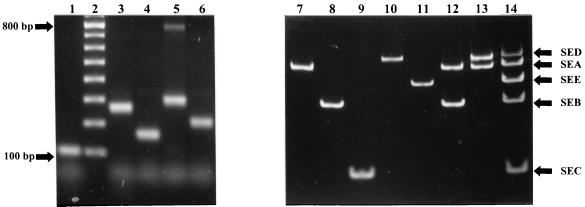
FIG. 1.
Gel analysis of PCR-amplified toxin gene sequences. The individual toxin gene products were characterized by comparison with standard molecular size markers. Lane 1, SEC; lane 2, 100-bp ladder; lane 3, SEA; lane 4, SEB; lane 5, SED; lane 6, SEE. These were combined to create a standard set of toxin gene products (lane 14). Genomic DNA was prepared from toxigenic strains of S. aureus and analyzed using the multiplex PCR assay, including primer SEC for gene sec (lanes 7 to 14). The S. aureus strains used were as follows: lane 7, F287 (SEA); lane 8, NCTC 10654 (SEB); lane 9, NCTC 10655 (SEC); lane 10, NCTC 10656 (SED); lane 11, FRI326 (SEE); lane 12, NCTC 10657 (SEA and SEB); and lane 13, NCTC 10652 (SEA and SED). The additional 790-bp product produced by the SA-D–SA-U primer pair is visible in lane 5.
To simply confirm that the amplified products arose from the expected toxin gene sequences, restriction digests of the PCR products were performed, and fragments of the expected sizes were obtained for each PCR product (Fig. 2). Although this is not absolute proof that the gene sequences amplified are from the target gene, consideration of both the size of the amplicon and the presence of the expected restriction sites provides a rapid and economic way of confirming product identity without recourse to DNA sequence analysis.
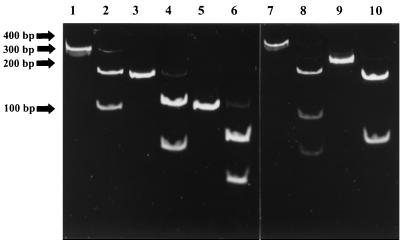
FIG. 2.
Restriction analysis of toxin gene amplicons. Analysis of the gene sequences identified restriction enzymes that would have either one or two sites in each of the toxin PCR products. The toxin gene fragments and their restriction fragments were analyzed on a 6% acrylamide gel. Lane 1, SEA (270 bp); lane 2, SEA and RsaI (172 and 98 bp); lane 3, SEB (165 bp); lane 4, SEB and RsaI (107 and 58 bp); lane 5, SEC (102 bp); lane 6, SEC and RsaI (65 and 37 bp); lane 7, SED (306 bp); lane 8, SED and RsaI (166, 83, and 54 bp); lane 9, SEE (213 bp); and lane 10, SEE and MboI (150 and 63 bp). Relative positions of the molecular size markers are given to the left of the gels.
Development of a single PCR multiplex toxin assay.
Once the specificity of the primer pairs had been determined, PCR conditions, buffers, and primer concentrations were optimized to establish conditions under which the primers could be combined into a single PCR reaction without affecting the ability of the primer pairs to generate a gene-specific amplicon (see Materials and Methods for PCR conditions). The results (Fig. 1, lanes 7 to 13) show that using these conditions the amplification of the staphylococcal enterotoxin genes sea, seb, sec, sed, and see was specific with a unique band of the predicted size present in strains producing a single toxin. Strain NCTC 10657 harboring both toxin genes A and B produced the corresponding 270- and 306-bp products, indicating that more than one toxin gene can be detected specifically in one reaction without any cross-reaction between primers. Correlation of PCR assay results with toxin production (Table 2) indicated that (with the exception of FRI326, which produces toxin E and cross-reacts with antibody to toxin A) there was 100% correlation between the PCR typing of reference strains encoding the corresponding toxins (genotype) and toxin production as defined by the SET-RPLA (phenotype). This was true even for strain NCTC 10652, from which a 306-bp PCR product was produced, indicating the presence of toxin gene D (Fig. 1, lane 10). Previously, this strain had not been indicated as a toxin D-producing strain in the NCTC catalogue; however, production of this toxin by this strain was confirmed by SET-RPLA, demonstrating the phenotypic expression of this gene. Similarly, strain NCTC 11963, designated as TSST-1 producing by the NCTC catalogue, was found to harbor toxin gene C (data not shown), and again toxin production could be identified by SET-RPLA.
In control experiments DNA from other toxigenic staphylococcal strains (Table 2) was tested for the presence of toxin gene sequences, but only Staphylococcus epidermidis NCTC 12100, which had been previously identified as enterotoxin C positive, gave a positive enterotoxin C PCR assay result and a corresponding positive result using SET-RPLA. Because of the close relationship of streptococcal pyrogenic exotoxin A to both SEB and SEC1, strains of Streptococcus pneumoniae (NCTC 7465) and Streptococcus pyogenes were also tested, but no amplicons were generated (data not shown). Nonstaphylococcal strains (see Table 2) were also screened but again did not produce any detectable enterotoxins by SET-RPLA, and none generated any amplicons using the PCR assay (data not shown).
Detection of toxin genes from unknown S. aureus strains of various animal origins.
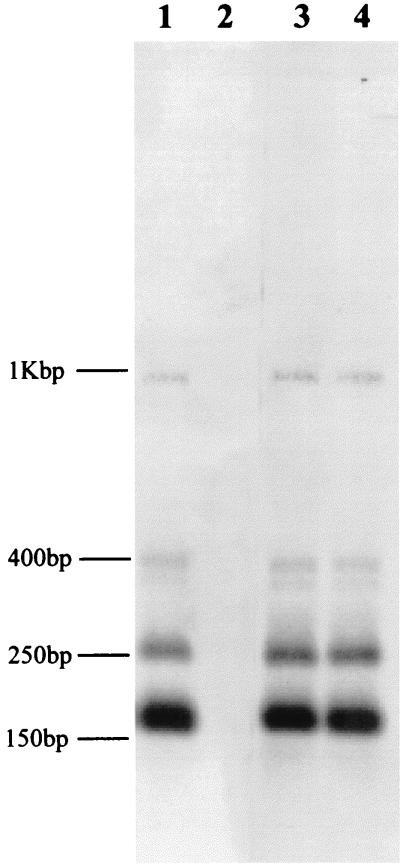
The multiplex PCR reaction was then used to determine the toxin type of 157 randomly selected S. aureus strains of ovine (67 strains), bovine (11 strains), poultry (40 strains), and human (39 strains) origins which had not been previously characterized for toxin gene production. The toxin type of any given S. aureus strain was determined by comparison of the pattern of PCR band(s) against the standard set of toxin-specific PCR products. In a parallel, “blind” study the strains were also tested by SET-RPLA so that the effectiveness of the PCR assay could be compared with normal phenotypic testing. The results (Table 3) show that, except for two ovine strains, the PCR assay results agreed with the serotyping in all cases. The two strains with the apparent false-positive tests were further analyzed by DNA hybridization to determine whether they actually contained a sec gene. The 102-bp sec-specific PCR product from the control strain NCTC 10655 was labeled and hybridized to RsaI-digested chromosomal DNA from the two ovine strains and control strains NCTC 10655 (toxin type C) and NCTC 8532 (nonenterotoxigenic). Both of the ovine strains and NCTC 10655 gave identical hybridization patterns of the expected fragment sizes (Fig. 3), confirming that they harbored toxin gene C, but this gene was not expressed at levels detectable by SET-RPLA.
TABLE 3.
Toxin analysis of uncharacterized strains by PCR and SET-RPLA assay
| Origin of S. aureus strainsa | No. tested (% positive) | No. of toxin serotypes | PCR toxin types |
|---|---|---|---|
| Human | 39 (31) | 2 SEA, 1 SEB, 2 SEC, 3 SEA-SED, 3 SEC-SED, 1 SEB-SED | 2 SEA, 1 SEB, 2 SEC, 3 SEA-SED, 3 SEC-SED, 1 SEB-SED |
| Bovine | 11 (9) | 1 SEC | 1 SEC |
| Ovine | 67 (75) | 2 SEA, 39 SEC, 6 SEA-SEC | 2 SEA, 41 SEC, 6 SEA-SEC |
| Poultry | 40 (50) | 20 SED | 20 SED |
Human isolates came from 21 cases of non-line scepticemia, 4 cases of line-associated scepticemia, and 14 healthy adults. Bovine strains were recovered from mastitic cattle, ovine strains were from mastitic sheep, and poultry strains were from various sources within a poultry processing plant (both environmental and carcasses).
FIG. 3.
Southern hybridization detection of sec gene. Staphylococcal chromosomal DNA was restricted with RsaI and separated on a 2% (wt/vol) agarose gel. The DNA was hybridized at high stringency with digoxigenin-labeled, PCR-amplified toxin C gene. Lane 1, NCTC 10655 (SEC); lane 2, NCTC 8532 (nonenterotoxigenic); lanes 3 and 4, ovine strains. The two strongly hybridizing bands of 160 and 260 bp are those predicted from an analysis of the published gene sequence of sec (8). The fainter-hybridizing bands correspond in size to predicted partial restriction digest fragments. The relative positions of the molecular size markers are indicated on the left.
DISCUSSION
The determination of staphylococcal enterotoxin type has a long history of successful use in epidemiological studies in both clinical and environmental microbiology studies. As our knowledge of the molecular genetic structure of these organisms increases, it becomes increasingly more difficult to test for all of the known phenotypes, with genotype analysis often providing the only way the diversity of different subspecies types can be identified. Oligonucleotide primers for specific detection of enterotoxin genes sea, seb, sec, sed, and see have previously been reported (22, 43, 44); these were used in individual PCR assays, thus requiring several PCRs for each sample to screen for the presence of all of the enterotoxin genes. Monday and Bohach (31) have recently described a multiplex PCR assay for the detection of all of the staphylococcal enterotoxin genes, but again this assay uses separate primer pairs for each toxin gene to be detected.
Here we used a series of novel primers, comprising one universal “forward” and five “reverse” primers together in a single reaction, and the results we present show that it is possible, using a single PCR assay, to rapidly screen staphylococcal isolates for the presence of the different enterotoxin genes which are routinely detected by serological test. The assay was found to be specific and detected only staphylococcal enterotoxin genes, including those found in species other than S. aureus, with no cross-reaction with other toxin-producing genera or nontoxigenic strains. Since the identity of the gene is given by the characteristic size of the PCR product(s) generated, the assay can detect and characterize the presence of multiple toxin genes present in one strain. This fact led us to the finding that two of the reference strains chosen, NCTC 10652 (toxin A positive) and NCTC 11963 (TSST-1 positive), contained additional previously unreported toxin genes, namely, sed in NCTC 10652 and sec in NCTC 11963, and the use of SET-RPLA assays confirmed that the genes were expressed in these two strains. Another advantage of this PCR assay is that it allows good differentiation between toxin genes sea and see. In the SET-RPLA test, enterotoxin E is detected by cross-reaction with the enterotoxin A-specific antibody, leading to ambiguous results. Recently, Lakner et al. (26) have reported the development of a specific antibody-based test for the presence of SEE; however, this now means that five individual tests must be performed on each isolate to be toxin typed and, as the number of toxin types characterized increases, an increasing number of tests will be required for a full analysis of toxigenotype in the future.
The limitation of all genotypic tests is that the presence of the gene does not always necessarily mean that the toxin will be produced. Like previous workers (e.g., reference 33), we identified the presence of an sec gene in two strains which did not produce detectable levels of SEC toxin when we used the SET-RPLA assay. This may be due to low-level production of enterotoxin below the threshold of detection for the immunoassay. Since the production of enterotoxins by staphylococcal strains can be affected by the growth conditions used (inoculum level, temperature, pH, and water activity [16]), it is possible that for these particular isolates the standard culture conditions specified for the SET-RPLA assay are suboptimal for gene expression. Alternatively, the sec gene may not be expressed due to mutations either in the coding region or in a regulatory region (either the promoter or an accessory regulator, e.g., agr). However, one major application of the SET-RPLA assay is the toxin typing of strains for epidemiological purposes when it is not usually essential to know whether or not a gene is expressed. Alternatively, the SET-RPLA test is used for the detection of preformed toxin in processed foods when the toxin-producing cells have been killed. The advantage of a PCR test in this latter case is that even nonviable cells serve as suitable targets for PCR amplification, and the test may be applied to rapidly screen for the presence of toxin-producing cells in a food sample, thus avoiding the problems of false-positive results which have been reported elsewhere. For example, Park et al. (34) found that nonstaphylococcal isolates, such as species of Enterobacter, Proteus, Pseudomonas, and Serratia, may give false-positive reactions during the detection of staphylococcal enterotoxins in foods using an immunological assay if the test organisms were not preheated.
The intrinsic robustness of staphylococcal cells and the presence of thermostable nucleases, originating either from the food matrix or from the cells themselves, means that reproducibly extracting good template DNA from environmental isolates is often difficult. Thus, many of the methods described for recovering DNA from environmental samples of gram-positive bacteria use phenols and other organic solvents, and therefore the protocols are often both lengthy and difficult for routine use in food analysis laboratories. The method we describe here, based on a method developed by Boom et al. (9) for the extraction of DNA from gram-negative bacteria, circumvents that problem and provides a rapid method which can be used to recover DNA from cells either from purified cultures or from food extractions.
The design of this multiplex PCR assay also makes it suitable to be extended to include newly characterized enterotoxin genes. Analysis of the gene sequences of seg (32) and seh (EMBL accession no. U11702) shows that both proteins contain the conserved peptide motif and that the DNA sequences have 17/20 and 16/20 matches with the universal forward primer, respectively, with the last two 3′ nucleotides being exact matches, suggesting that effective annealing of the forward primer to the template for PCR amplification would be achieved. It is interesting to note that while the DNA sequence of sei (32) does contain some (50%) homology with the universal primer, the conserved peptide motif is not present in the derived protein sequence. A second conserved peptide motif [(Y/F)-K-(x)-(K/E)-V-D-(x)-(F/Y)] is found upstream (5′) of the universal primer region, but a third conserved peptide motif found near the end of the gene (3′ of the universal primer region; Y-(x)-D-N-K) is also missing. Since the sei gene is known to produce a functional toxin (32), it would appear that either SEI is structurally distinct from the other toxins or there is an anomaly in the sequence leading to a mistranslation.
The presence of these other conserved peptide motifs does raise the possibility that a toxin gene-positive reaction could be introduced into this PCR assay. Currently, the assay only detects genes for which the complete gene sequence is known. By including a universal reverse primer in the multiplex PCR reaction, for example, one homologous to the conserved sequences found at the 3′ end of the gene (e.g., the region encoding the Y-(x)-D-N-K peptide motif), two products would be generated for each known toxin genes (one toxin-specific product and one toxin-gene positive product). The presence of uncharacterized toxin genes would be detected by the presence of a single toxin gene-positive PCR product but the absence of a toxin-specific PCR product. Including a toxin-positive element into the multiplex PCR assay would have the further advantage of reducing the chances of false-negative results caused by mutations occurring in the toxin-specific primer regions.
To check the reproducibility of the PCR toxin assay, we screened a range of different staphylococcal isolates which had been isolated from a variety of sources and, again, good correlation was found between the PCR assay and the SET-RPLA results. In agreement with the work of Hirooka et al. (19), we found that enterotoxin C was most commonly associated with ovine and bovine enterotoxigenic S. aureus strains. Although only one of the bovine isolates was found to be enterotoxin positive, it is perhaps significant that enterotoxin C was detected, since our bovine strains were recovered from cases of bovine mastitis, and the involvement of enterotoxin C-positive S. aureus strains with cases of subclinical and clinical bovine mastitis has been well documented by other workers (15, 24, 29). Of the poultry strains studied, 50% of the isolates carried enterotoxin D, and this was the only enterotoxin detected. Harvey et al. (18) have previously reported a predominance of enterotoxin D-positive strains in their study on chicken isolates, and Shiozawa et al. (40) found that 37.5% of enterotoxigenic strains from poultry in Japan (16 samples) produced enterotoxin D alone. Hence, the production of enterotoxin D may be characteristic of enterotoxigenic strains of poultry origin.
Although 31% of the human isolates were enterotoxin positive, the majority of these (10 of 12) were from non-line-associated septicemia cases, and the remaining two were from healthy volunteers. This result supports the concepts of S. aureus as an opportunistic pathogen and the importance of the enterotoxins in the establishment of infection. For instance, Childs et al. (10) have previously reported an association between the presence of enterotoxins B, C, and D, either alone or in combination with TSST-1, with S. aureus infections of burn wounds in children and, similarly, a higher incidence of enterotoxin production was found among isolates from patients with septicemia (63%) than those from nasal isolates of healthy subjects (11%; 20). The fact that none of the isolates tested here from line-associated cases were enterotoxin positive suggests that, when given access to the body by a physical route, expression of the enterotoxins is less significant in allowing the bacterium to cause infection.
In combination with the simplified DNA extraction procedure described here, the application of this single multiplex PCR assay should enable more samples to be rapidly characterized for enterotoxin production for epidemiological studies. Having tested the assay on a wide range of isolates from different environmental sources, we can also recommend its use as a screening test for the presence of enterotoxin genes and as a confirmatory test for enterotoxins actually elicited, as determined by immunological assays.
ACKNOWLEDGMENTS
N.K.S. was supported by a Commonwealth Scholarship.
We are indebted to Will Waites and the late G. S. A. B. Stewart for intellectual contributions to the development of the work and to David Fowler for technical support.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 1. New York, N.Y: John Wiley & Sons, Inc.; 1994. [Google Scholar]
- 2.Bayles K W, Iandolo J J. Genetic and molecular analyses of the gene encoding staphylococcal enterotoxin D. J Bacteriol. 1989;171:4799–4806. doi: 10.1128/jb.171.9.4799-4806.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Becker K, Roth R, Peters G. Rapid and specific detection of toxigenic Staphylococcus aureus: use of two multiplex PCR enzyme immunoassays for amplification and hybridization of staphylococcal enterotoxin genes, exfoliative toxin genes, and toxic shock syndrome toxin 1 gene. J Clin Microbiol. 1998;36:2548–2553. doi: 10.1128/jcm.36.9.2548-2553.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bergdoll M S. Staphylococcus aureus. In: Doyle M P, editor. Food-borne bacterial pathogens. New York, N.Y: Marcel Dekker, Inc.; 1989. pp. 463–523. [Google Scholar]
- 5.Betley M, Mekalanos J J. Nucleotide sequence of type A staphylococcal enterotoxin gene. J Bacteriol. 1988;170:34–41. doi: 10.1128/jb.170.1.34-41.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Betley M J, Borst D W, Regassa L B. Staphylococcal enterotoxins, toxic shock syndrome toxin and streptococcal pyrogenic exotoxins: a comparative study of their molecular biology. Chem Immunol. 1992;55:1–35. [PubMed] [Google Scholar]
- 7.Bloomster-Hautamaa D A, Kreiswirth B N, Kornblum J S, Novick R P, Schlievert P M. The nucleotide and partial amino acid sequence of toxic shock syndrome toxin-1. J Biol Chem. 1986;261:15783–15786. [PubMed] [Google Scholar]
- 8.Bohach G A, Schlievert P M. Nucleotide sequence of the staphylococcal enterotoxin C1 gene and relatedness to other pyrogenic toxins. Mol Gen Genet. 1987;209:15–20. doi: 10.1007/BF00329830. [DOI] [PubMed] [Google Scholar]
- 9.Boom R, Sol C J A, Salimans M M M, Jansen C L, Wertheim van Dillen P M E, vander Noorda J. Rapid and simple method for purification of nucleic acids. J Clin Microbiol. 1990;28:495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Childs C, Edwards-Jones V, Heathcote D M, Dawson M, Davenport P J. Patterns of Staphylococcus aureus colonization, toxin production, immunity and illness in burned children. Burns. 1994;20:514–521. doi: 10.1016/0305-4179(94)90011-6. [DOI] [PubMed] [Google Scholar]
- 11.Couch J L, Soltis M T, Betley M J. Cloning and nucleotide sequence of the type E staphylococcal enterotoxin gene. J Bacteriol. 1988;170:2954–2960. doi: 10.1128/jb.170.7.2954-2960.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ewald S, Heavelman C J, Notermans S. The use of DNA probes for confirming enterotoxin produced by Staphylococcus aureus and micrococci. Int J Food Microbiol. 1990;11:251–258. doi: 10.1016/0168-1605(90)90018-z. [DOI] [PubMed] [Google Scholar]
- 13.Fey H, Pfister H, Ruegg O. Comparative evaluation of different ELISA systems for the detection of staphylococcal enterotoxins A, B, C, and D. J Clin Microbiol. 1984;19:34–38. doi: 10.1128/jcm.19.1.34-38.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fujikawa H, Igarashi H. Rapid latex agglutination test for detection of staphylococcal enterotoxins A to E that uses high-density latex particles. Appl Environ Microbiol. 1988;54:2345–2348. doi: 10.1128/aem.54.10.2345-2348.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garcia M L, Moreno B, Bergdoll M S. Characterization of staphylococci isolated from mastitic cows in Spain. Appl Environ Microbiol. 1980;39:543–548. doi: 10.1128/aem.39.3.548-553.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Genigeorgis C A. Present state of knowledge on staphylococcal intoxication. Int J Food Microbiol. 1989;9:327–360. doi: 10.1016/0168-1605(89)90100-1. [DOI] [PubMed] [Google Scholar]
- 17.Grabovetsky V V, Ignatov K B, Chistyakova L G, Dyadchenko M A, Vinogradov S V, Kiselev V I. Revealing of genes, that control the enterotoxigenic properties of Staphylococcus aureus strains by means of polymerase chain reaction. Konnektnb Abtopob. 1995;5:75–79. [Google Scholar]
- 18.Harvey J, Patterson J T, Gibbs P A. Enterotoxigenicity of Staphylococcus aureus strains isolated from poultry: raw poultry carcasses as a potential food-poisoning hazard. J Appl Bacteriol. 1982;52:251–258. doi: 10.1111/j.1365-2672.1982.tb04847.x. [DOI] [PubMed] [Google Scholar]
- 19.Hirooka E Y, Muller E E, Freitas J C, Vicente E, Yoshimoto Y, Bergdoll M S. Enterotoxigenicity of Staphylococcus intermedius of canine origin. Int J Food Microbiol. 1988;7:185–191. doi: 10.1016/0168-1605(88)90036-0. [DOI] [PubMed] [Google Scholar]
- 20.Humphreys H, Keane C T, Hone R, Pomeroy H, Russell R J, Arbuthnott J P, Coleman D C. Enterotoxin production by Staphylococcus aureus isolates from cases of septicaemia and from healthy carriers. J Med Microbiol. 1989;28:163–172. doi: 10.1099/00222615-28-3-163. [DOI] [PubMed] [Google Scholar]
- 21.Jaulhac B, Bes M, Bornstein N, Piemont Y, Brun Y, Fleurette J. Synthetic DNA probes for detection of genes for enterotoxins A, B, C, D, E and for TSST-1 in staphylococcal strains. J Appl Bacteriol. 1992;72:386–392. doi: 10.1111/j.1365-2672.1992.tb01851.x. [DOI] [PubMed] [Google Scholar]
- 22.Johnson W M, Tyler S D, Ewan E P, Ashton F E, Pollard D R, Rozee K R. Detection of genes for enterotoxins, exfoliative toxins, and toxic shock syndrome toxin 1 in Staphylococcus aureus by the polymerase chain reaction. J Clin Microbiol. 1991;29:426–430. doi: 10.1128/jcm.29.3.426-430.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones C L, Khan S A. Nucleotide sequence of the enterotoxin B gene from Staphylococcus aureus. J Bacteriol. 1986;166:29–33. doi: 10.1128/jb.166.1.29-33.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kenny K, Reiser R F, Bastida-Corcuera F D, Norcross N L. Production of enterotoxins and toxic shock syndrome toxin by bovine mammary isolates of Staphylococcus aureus. J Clin Microbiol. 1993;31:706–707. doi: 10.1128/jcm.31.3.706-707.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kokan N P, Bergdoll M S. Detection of low-enterotoxin producing Staphylococcus aureus strains. Appl Environ Microbiol. 1987;53:2675–2676. doi: 10.1128/aem.53.11.2675-2676.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lakner M, Schneider E, Usleber E, Dietrich R, Becker H, Martlbauer E. Development and application of immunochromatographic tests for the detection of staphylococcal enterotoxin E. Food Agric Immunol. 1998;10:249–257. [Google Scholar]
- 27.Lee A C-M, Robbins R N, Bergdoll M S. Isolation of specific and common antibodies to staphylococcal enterotoxins A and E by affinity chromatography. Infect Immun. 1978;21:387–391. doi: 10.1128/iai.21.2.387-391.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee A C-M, Robbins R N, Bergdoll M S. Isolation of specific and common antibodies to staphylococcal enterotoxins B, C1, and C2. Infect Immun. 1980;27:431–434. doi: 10.1128/iai.27.2.431-434.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lopes C A, Moreno G, Curi P R, Gottschalk A F, Modolo J R, Correa A, Pavan C. Characteristics of Staphylococcus aureus from subclinical bovine mastitis in Brazil. Br Vet J. 1990;146:443–448. doi: 10.1016/0007-1935(90)90033-y. [DOI] [PubMed] [Google Scholar]
- 30.Marrack P, Kappler J. The staphylococcal enterotoxins and their relatives. Science. 1990;248:705–711. doi: 10.1126/science.2185544. [DOI] [PubMed] [Google Scholar]
- 31.Monday S R, Bohach G A. Use of multiplex PCR to detect classical and newly described pyrogenic toxin genes in staphylococcal isolates. J Clin Microbiol. 1999;37:3411–3414. doi: 10.1128/jcm.37.10.3411-3414.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Munson S H, Tremaine M T, Betley M J, Welch R A. Identification and characterization of staphylococcal enterotoxin types G and I from Staphylococcus aureus. Infect Immun. 1998;66:3337–3348. doi: 10.1128/iai.66.7.3337-3348.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neill R J, Fanning G R, Delahoz F, Wolff R, Gemski P. Oligonucleotide probes for detection and differentiation of Staphylococcus aureus strains containing genes for enterotoxins A, B, and C and toxic shock syndrome toxin 1. J Clin Microbiol. 1990;28:1514–1518. doi: 10.1128/jcm.28.7.1514-1518.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park C E, Akhtar M, Rayman M K. Non-specific reactions of a commercial enzyme-linked immunosorbent assay kit (TECRA) for detection of staphylococcal enterotoxins in food. Appl Environ Microbiol. 1992;58:2509–2512. doi: 10.1128/aem.58.8.2509-2512.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qian L, Wilkinson M. DNA fragment purification: removal of agarose ten minutes after electrophoresis. BioTechniques. 1991;10:736–737. [PubMed] [Google Scholar]
- 36.Ren K, Bannan J D, Pancholi V, Cheung A L, Robbins J C, Fischetti V A, Zabriskie J B. Characterization and biological properties of a new staphylococcal exotoxin. J Exp Med. 1994;180:1675–1683. doi: 10.1084/jem.180.5.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robbins R N, Gould S, Bergdoll M S. Detecting the enterotoxigenicity of Staphylococcus aureus strains. Appl Microbiol. 1974;28:946–950. doi: 10.1128/am.28.6.946-950.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saiki R K, Gelfand D H, Stoeffel S, Scharf S J, Higuchi R, Horn G T, Mullis K B, Erlich H A. Primer-directed enzymic amplification of DNA with a thermostable DNA polymerase. Science. 1988;239:487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- 39.Schmitz F J, Steiert M, Hofmann B, Verhoef J, Hadding U, Heinz H P, Kohrer K. Development of a multiplex-PCR for direct detection of the genes for enterotoxin B and C, and toxic shock syndrome toxin-1 in Staphylococcus aureus isolates. J Med Microbiol. 1998;47:335–340. doi: 10.1099/00222615-47-4-335. [DOI] [PubMed] [Google Scholar]
- 40.Shiozawa K E, Kato E, Shimizu A. Enterotoxigenicity of Staphylococcus aureus strains isolated from chickens. J Food Prot. 1980;43:683–685. doi: 10.4315/0362-028X-43.9.683. [DOI] [PubMed] [Google Scholar]
- 41.Southern E M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 42.Su Y C, Wong A C L. Identification and purification of a new staphylococcal enterotoxin, H. Appl Environ Microbiol. 1995;61:1438–1443. doi: 10.1128/aem.61.4.1438-1443.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsen H-Y, Chen T-R. Use of the polymerase chain reaction for specific detection of type A, D and E enterotoxigenic Staphylococcus aureus in foods. Appl Microbiol Biotechnol. 1992;37:685–690. doi: 10.1007/BF00240750. [DOI] [PubMed] [Google Scholar]
- 44.Tsen H-Y, Chen T-R, Yu G-K. Detection of B, C types of enterotoxigenic Staphylococcus aureus using polymerase chain reaction. J Chinese Agric Chem Soc. 1994;32:322–331. [Google Scholar]
- 45.Tsen H-Y, Yang R-Y, Huang F-Y. Novel oligonucleotide probes for identification of enterotoxigenic Staphylococcus aureus. J Ferment Bioeng. 1993;76:7–13. [Google Scholar]
- 46.Tsen H-Y, Yu G-K, Lin I-T. Plasmid profiles and pulsed-field gel electrophoresis for type A enterotoxigenic Staphylococcus aureus isolated from foods. J Food Prot. 1995;58:147–153. doi: 10.4315/0362-028X-58.2.147. [DOI] [PubMed] [Google Scholar]
- 47.Weeks C R, Ferretti J J. Nucleotide sequence of the type A streptococcal enterotoxin (erythrogenic toxin) gene from Streptococcus pyogenes. Infect Immun. 1986;52:144–150. doi: 10.1128/iai.52.1.144-150.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yamada S, Igarashi H, Terayama T. Improved reversed passive hemagglutination for simple and rapid detection of staphylococcal enterotoxins A to E in food. Microbiol Immunol. 1977;21:675–682. doi: 10.1111/j.1348-0421.1977.tb00336.x. [DOI] [PubMed] [Google Scholar]
- 49.Zhang S, Iandolo J J, Stewart G C. The enterotoxin D plasmid of Staphylococcus aureus encodes a second enterotoxin determinant (sej) FEMS Microbiol Lett. 1998;168:227–233. doi: 10.1111/j.1574-6968.1998.tb13278.x. [DOI] [PubMed] [Google Scholar]