Abstract
We previously demonstrated that increased monoclonal antibody productivity in dihydrofolate reductase (DHFR)-amplified CHO cells correlates with phosphorylated transcription factor-cytomegalovirus (CMV) promoter interactions. In this article, we extend the characterization to include CMV promoter methylation and its influence on NFκB and CREB1 transcription factor binding to the CMV promoter in two families of DHFR-amplified CHO cell lines. CMV promoter methylation was determined using bisulfite sequencing. To overcome Sanger-sequencing limitations due to high CG bias and multiple transgenes copies, pyrosequencing was used to determine the frequency of methylated cytosines in regions proximal to and containing the NFκB and CREB1 transcription-factor consensus binding sites. Chromatin immunoprecipitation was performed to interrogate transcription factor–DNA interactions. Antibodies to CREB1 and NFκB were used to immunoprecipitate formaldehyde-crosslinked protein-DNA fractions, followed by reverse transcription quantitative real-time polymerase chain reaction to quantitate the number of copies of CMV-promoter DNA bound to the various transcription factors. The relative unmethylated fraction at the CREB1 and NFκB consensus binding sites determined by pyrosequencing was correlated with transcription factor binding as determined by chromatin immunoprecipitation. Azacytidine treatment reduced methylation in all treated samples, though not at all methylation sites, while increasing transcription. Distinct promoter methylation patterns arise upon clonal selection in different families of cell lines. In both cell line families, increased methylation was observed upon amplification. In one family, the NFκB binding-site methylation was accompanied by increased CREB1 interaction with the promoter. In the other cell line family, lower methylation frequency at the NFκB consensus binding site was accompanied by more NFκB recruitment to the promoter region.
Keywords: Transcriptional activation, azacytidine, DNA methylation, CMV promoter
Statement of Significance: In this work, we have characterized the methylation pattern in various positions along the cytomegalovirus (CMV) promoter driving the expression of the mAb heavy chain and light chain in different clones. The interplay between the methylated regions of transcription-factor consensus binding sites and the nuclear proteins influences the transcript levels, leading to higher productivity phenotypes. Our workflow demonstrates a way to identify influencers of high transcriptional rates and identifies opportunities for transcription factor screening and rational synthetic promoter design, toward enhancing specific productivity in commercially applicable manufacturing processes.
INTRODUCTION
Every year, the global monoclonal antibody (mAb) market exceeds all previous estimates, surpassing US$143.5 billion in 2020 with an expected compound annual growth rate of 14.4% from 2020 to 2027 [1]. The sustained, rapid growth of this sector is driven by the relatively high success rate of recombinant protein drugs in clinical trials. Recombinant therapeutic proteins are increasingly produced in CHO cell culture, partly due to the large safety dataset and FDA approvals using these cells, and perhaps largely due to the astounding 10 g/L or greater titers and yields possible [2].
Still, cell line development is inherently a demanding task that necessitates screening of several hundred to thousands of clones in the quest for a suitable higher producer, only to be repeated yet again for each new campaign. While there is still potential for improvement by further leveraging media and process engineering, as we enter the age of continuous manufacturing, success now depends on cell-line-development teams identifying cells with greater specific productivity to offset cost of goods in larger perfusion cultures [3]. However, molecular determinants of high specific productivity are often clouded by biological noise in CHO cell lines, where the genetic instability is a significant concern [4]. Most recombinant protein expression is driven by employing vectors with a strong viral promoter. Among these, the human cytomegalovirus (CMV) has been the choice of a promoter in most classical vector designs that are still utilized today. However, in an evolutionary response to viral infections, highly active viral promoters are often silenced by DNA methylation in mammalian cells [5, 6], which can interfere with transcription factor binding and inhibit transcription [7]. Even after integration at a high productivity locus, transgene silencing may still be observed [8]. The degree of expression from a stably integrated insert depends on the number of copies incorporated, the accessibility of the region to transcriptional machinery, the higher order chromatin assembly, etc., and several promoter and chromatin engineering approaches have been employed to improve productivity. Uniform chromatin opening elements and matrix attachment regions have been successfully used to influence the state of the chromatin and thereby increase transcription [9]. In addition, CRISPR strategies have been employed to influence the methylation of both endogenous promoters and the CMV promoters driving transgene expression in CHO cells. Notably, downregulation of the CMV promoter-controlled transgene was more difficult than downregulation of the endogenous promoters [10].
It is therefore important to gain an understanding of the local epigenetic changes surrounding the integrated transgene as well as to identify mechanisms that lead to improved specific productivity. To ensure valid comparisons between higher and lower productivity cell lines and obtain industrially relevant observations, suspension-adapted industrial clones and their dihydrofolate reductase (DHFR)-amplified progeny were selected for this study. The vector design incorporated euchromatic elements and enhancer elements. Further, the cell lines have known sites of integration and well-characterized copy numbers [11, 12]. Following our previous characterization of the effects of transcription factor–transgene interactions on productivity [13], in this study, we investigated the role of DNA CpG methylation in various positions along the CMV promoter in high-producing CHO cells. Our results indicate that despite vector elements and repeated rounds of MTX screening, higher producers show a counterintuitive increased methylation of the CMV promoter region, particularly near transcription factor binding sites. Treatment with 5′-azacytidine (azacytidine) leads to hypomethylation in the sites upstream of transcription factor consensus sequences. Azacytidine-induced improved transcriptional availability is correlated with increased RNA copies, increased yield and improved specific productivity. Elucidating the mechanism for these improvements will pave the way for more cell selection strategies leveraging demethylation of promoter regions similar to that described by Weinguny et al. [14]. In addition, this mechanistic understanding may influence design of synthetic promoters [15] and further the scope of next-generation cell line development [16].
METHODS AND MATERIALS
Cell lines
Chinese hamster ovary cell lines that produce a recombinant monoclonal humanized IgG with different specific productivities were a generous gift from an industrial collaborator. These cell lines were developed by co-transfecting two plasmids, one containing IgG heavy chain (HC) and DHFR genes and the other containing IgG light chain (LC) and neomycin phosphotransferase (Neo) genes. Transfected cell lines were initially selected in medium containing 400 μg/mL neomycin (G418). After selection, the neomycin was removed, and all subsequent cultures were performed in the absence of neomycin. Subsequently, gene amplification was performed by stepwise selection with increasing MTX concentrations. For these studies, two lower productivity parental cell lines, A0 and CO, and their DHFR-amplified higher producing progeny, A1 and C1, respectively, were chosen for investigation. These cell lines have been previously described [11, 12]. Cells were cultured in a nonproprietary, serum-free medium [17] containing hydrolysate and 5 mg/L recombinant human insulin.
Cell culture
Cells from liquid nitrogen storage were added to a 15 mL centrifuge tube, with 4 mL of medium. The tubes were then centrifuged for 5 min; the supernatant was discarded, and the pellet was then resuspended in 5 mL fresh medium. The resuspended cells were then added to 5 mL of fresh medium prepared in a T-25 flask, and the cultures were incubated at 37 °C, 6% CO2, with shaking at 130 rpm. The T-25 cell culture flasks were arranged in a standing position. Cells were passaged twice weekly prior to the experiment.
Batch culture experiments with and without azacytidine addition
Prior to batch studies, cells were counted using a Countess cell counter (Thermo Fisher Scientific). Cells were then resuspended in fresh flasks and medium at an initial cell density of 0.1 × 106 cells/mL, in a total volume of 11 mL. Cell were grown in the presence and absence of azacytidine, in triplicate biological replicates. A total of 10 mg of azacytidine was dissolved initially in 100 μL DMSO, to which 900 μL of MilliQ water was subsequently added. The solution was filter sterilized with a 0.2 μm filter. Azacytidine dissolved in 0.1X DMSO was added to the relevant flasks at the beginning of the batch culture for a final concentration of 100 μM. An equivalent volume of 0.1X DMSO in water was added to the flasks without azacytidine, to control for the potential effects of the small amount of DMSO present.
A total of 0.2 mL of culture was taken each day for 6 days for cell counts, and on Days 3, 5 and 6, 1.5 mL of cell culture was removed from each flask, centrifuged, and both the supernatant and pellet retained for ELISA and DNA extraction, respectively.
ELISA
ELISA was performed for supernatants taken on Days 3, 5 and 6 using STEMCELL™ Technologies Human IgG ELISA Antibody Pair Kit as per the protocol.
qP calculation
Specific productivity was calculated as the slope of cell titer over integrated viable cell density (IVCD). IVCD was calculated using the trapezoid rule (the summation of all areas under the curve between each viable cell density (VCD) data point, as approximated by a trapezoid):
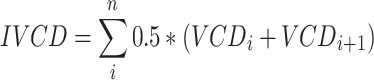 |
DNA extraction
DNA extractions were performed using the DNeasy mini kit (Qiagen) as per the manufacturer’s protocol. Samples were treated with RNAse A (Ambion) and Proteinase K (Sigma) to remove residual RNA and protein followed by ethanol precipitation. DNA concentrations were determined using a Nanodrop spectrophotometer. DNA quality was verified by gel electrophoresis using a 1% agarose gel in 1 x TAE buffer.
Bisulfite conversion and pyrosequencing
Bisulfite conversion was carried out on 500 ng of genomic DNA using the EZ DNA methylation kit (Zymo Research Corporation, CA) according to the manufacturer’s instructions [18–20]. Pyrosequencing assays were designed in-house using PyroMark Assay Design Software 2.0 (Qiagen, Crawley, UK). Primers used were as follows: forward, 5′ TTTTATTGAAGTTAATGGGTGGAGTATTT 3′; reverse, 5’ ATTTTAATACCAAAACAAACTCCC 3′; sequencing, 5′ GTATTAGTTATAGTTATTATTATGG 3′.
Bisulfite-converted DNA was amplified using the PyroMark PCR kit (Qiagen, Crawley, UK). Each 25 μL reaction mix consisted of 12.5 μL master mix, 2.5 μL coral load, 5.5 μL nuclease-free water, 1.25 μL each of 10 μM forward and reverse primers and 2 μL each of bisulfite-converted DNA. PCR was carried out under the following conditions: initial hot start, 95 °C for 15 min, followed by 45 cycles of 30s at 94 °C, 30 s at 56 °C, and 30s at 72 °C, with final elongation for 10 min at 72 °C. The PCR products were verified via gel electrophoresis (1% w/v agarose gel) prior to pyrosequencing analysis to check the size of DNA fragments and as a quality control to check samples for contamination. DNA methylation levels in samples were examined using the PyroMark Q24 Pyrosequencing platform as per the manufacturer’s recommendations (Qiagen, Crawley, UK). Enzymes, substrates and nucleotides from the PyroMark Gold Q96 kit (Qiagen UK) were used. Levels of methylation at each CpG site were analyzed using the PyroMark Q24 software [21, 22] as carried out previously [23, 24]. The degree of methylation at each CpG site is expressed as the percentage of methylated cytosine over the sum of methylated and unmethylated cytosine. Each round of pyrosequencing was performed in triplicate, on triplicate biological samples from Day 5.
Chromatin immunoprecipitation
Chromatin immunoprecipitation was performed using the ChIP-IT kit (53008 Active Motif, Carlsbad CA) according to the manufacturer’s instructions. Briefly, 4 × 107 cells from each cell line were harvested at Day 3 and incubated with 30 mL fresh medium containing 1.5 mL 36% formaldehyde (47630 Sigma-Aldrich, St Louis, MO) for 10 min to crosslink the DNA-associated proteins to the chromatin. The reaction was stopped by washing the cells with phosphate buffered saline (PBS) and incubating with Glycine Stop-fix solution for 10 min. A final PBS wash step was used to clean the cell pellet. A sonicator (450D Branson, Danbury CT) fitted with a microtip was employed to disrupt the cells and shear the DNA to 500–1500-base pair fragments. The sonicator settings were set in accordance with the tip manufacturer’s instructions and kept at 40% amplitude. The shearing was verified by separating the sheared DNA on a 1.8% agarose gel. In subsequent steps, the Protein-DNA complex was immunoprecipitated using antibodies to CREB1, NFκB, Sp1 or RNA polymerase II. After treatment with Proteinase K and RNase to remove cellular proteins and RNA, DNA fragments were purified by using silica spin columns provided with the kit. The final elution volume in each fraction was 100 μL. This volume was concentrated to 20 μL by using a SpeedVac DNA concentrator (BC-SDNA11 Savant, GMI Inc. Ramsey, Minnesota) to obtain an adequate concentration of DNA template for reverse transcription quantitative real-time polymerase chain reaction (RT-qPCR). ChIP was performed on three separate dates with duplicate PCR analysis for each sample.
RT-qPCR
RT-PCR was performed using the Roche LightCycler® 480 Real-Time PCR System and the LightCycler 480 Mastermix (04707494001 Roche, Indianapolis, IN). For quantification of CMV, the probe/primers combinations were as follows: forward primer: gcagagctcgtttagtgaacc; reverse primer: gaggtcaaaacagcgtggat; Universal ProbeLibrary probe: #80 (cat.no. 04689038001, Roche, Indianapolis, IN). Reaction conditions were set up according to the manufacturer’s instructions. Crossing points (Ct) were generated from the LightCycler Software. Relative quantification of the CMV promoter and GAPDH bound to the transcription factors was performed using the 2^delta delta Ct method [25]. All samples were normalized to the respective input DNA for the ChIP reaction (e.g. A0 cell line, CMV copies in input DNA) and then to sample 3 of the A0 CREB1 immunoprecipitated for CMV.
Statistical analysis
All statistical analyses were performed using GraphPad Prism version 9.1.2 for Windows, GraphPad Software, San Diego, CA, USA, www.graphpad.com.
RESULTS
Specific productivity increases with azacytidine treatment
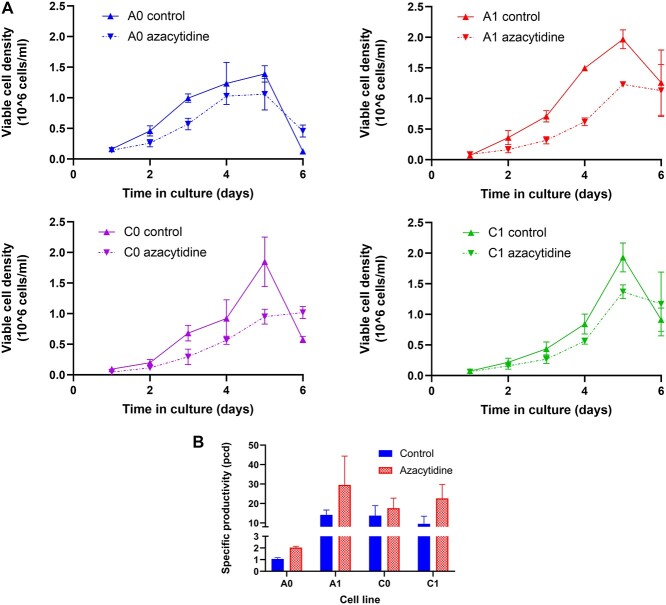
Cell growth was characterized in batch culture for the cell clones under study, two parental cell lines (A0 and C0) and their DHFR-amplified progeny (A1 and C1, respectively, Fig. 1a). In every case, there was a slight reduction in growth rate and final cell density after azacytidine treatment, with the A1 clone showing the greatest reduction in growth rate and peak VCD. Specific productivity was also increased in all cell lines (Fig. 1b, P = 0.0071, two-way analysis of variance (ANOVA)), but by varying amounts. An increase in specific productivity after azacytidine treatment has been previously reported [5], but there has been no direct link to the methylation state around the transgene. Without direct exploration of methylation changes in the areas proximal to the transgene, it cannot be determined how azacytidine and demethylation alter productivity.
Figure 1.
Growth and specific productivity of control and azacytidine treated cultures. aA: VCD in control and azacytidine-treated cell cultures over time. Cells were treated with 100 μM azacytidine beginning at Day 0. B: Specific productivity of control cell cultures and azacytidine-treated cell cultures determined by ELISA assay.
Transcriptional increase upon azacytidine treatment
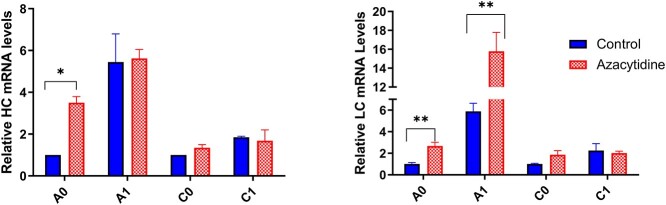
To determine whether the increased specific productivity was due to increased transcription of the monoclonal antibody, qRT-PCR for the mAb HC and LC was performed on RNA samples from cultures with and without azacytidine treatment (Fig. 2). An approximate 3-fold increase in HC mRNA and 2-fold increase in LC mRNA were seen in the A0 cell line, and an approximately 2.5-fold increase in LC mRNA was seen in the A1 cell line upon azacytidine treatment. A small increase in HC and LC mRNA was seen in the C0 cell line, and no discernable increase in mRNA levels was observed in the C1 cell line, overall showing some, but not complete agreement with the changes in specific productivity (Fig. 1b, Supplementary Fig. S1).
Figure 2.
qPCR assay for HC and LC mRNA levels of the expressed monoclonal antibody in the control cell cultures and azacytidine treated cell cultures. mRNA levels are normalized to the mRNA levels in the untreated parental control (A0 or C0). *P < 0.05, **P < 0.01, unpaired t-test.
Characterization of CpG methylation in various positions along the CMV promoter
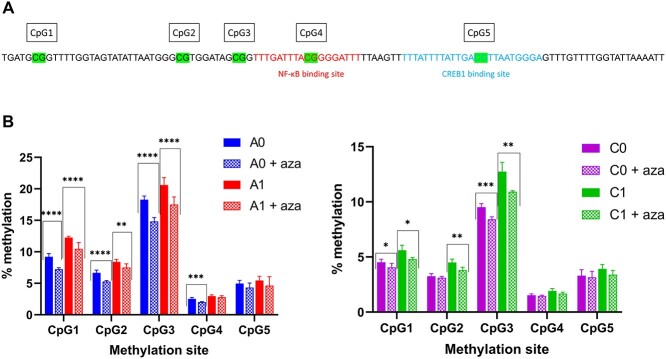
To elucidate the mechanisms behind the cell-line-specific responses to azacytidine, bisulfite treatment of genomic DNA and subsequent pyrosequencing was performed to elucidate methylation differences in various positions along the CMV promoter. As industrial CHO cells, particularly those isolated by DHFR/methotrexate selection and amplification, may contain hundreds of copies of the CMV promoter [12], it is critical to determine the fraction of those CMV promoters that are methylated, which requires pyrosequencing of bisulfite-treated DNA, rather than traditional Sanger sequencing. The percent methylation was determined at five sites in the CMV promoter upstream of and spanning the putative NFκB and CREB1 binding sites (Fig. 3a). The overall level of methylation was relatively low (Figs 3b and c) with a maximum of ~20% methylation observed at CpG3 in the A1 cell line. Notably, the level of methylation was significantly greater in the A family cell lines than in the C family cell lines (P ≤ 0.0001 for all sites, t-test), particularly at CpG1, CpG2 and CpG3, which are upstream of the putative transcription factor binding sites. Higher levels of methylation were also seen in the amplified progeny compared with the parental cell lines. Azacytidine treatment reduced the methylation fraction in all samples, particularly at the CpG1 and CpG3 sites. The effect was more substantial in the A family cell lines than in the C family cell lines. Interestingly, CpG4 and CpG5, which occur inside the putative transcription factor binding sites for NFκB and CREB1, respectively, show the lowest levels of methylation in both families (less than 3% methylation of CpG4 in the A cell lines and less than 2% methylation of CpG4 in the C cell lines; ~5% methylation of CpG5 in the A cell lines and 3–4% methylation of CpG5 in the C cell lines), suggesting that transcription factor binding to the DNA may interfere with DNA methylation.
Figure 3.
Pyrosequencing to determine methylation frequency of the CpG sites in the CpG island containing the NFκB and CREB1 transcription factor consensus binding sites in the CMV promoter. a: Locations of the CpG sites evaluated and the relative locations of the transcription factor binding sites. b: Methylation fraction in A0 and A1 cell lines in control cell and azacytidine treated cultures. c: Methylation fraction in C0 and C1 cell lines in control cell and azacytidine treated cultures. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, unpaired t-test.
Differential association of transcriptional factors in the A and C cell lines
We had previously observed differential transcription factor binding of CREB1 in the A0 and A1 cell lines [13] which we ascribed, in part, to differential activation of CREB1 due to altered phosphorylation. As we observed differential methylation of the CMV promoter upstream of the NFκB binding site (CpG3) between the A family cell lines and the C family cell lines, we hypothesized that this might impact transcription factor binding between the different cell families and between parental cells and DHFR-amplified progeny. Chromatin immunoprecipitation (ChIP) was performed to determine the relative in vivo binding of transcription factors to the CMV promoter (Fig. 4).
Figure 4.
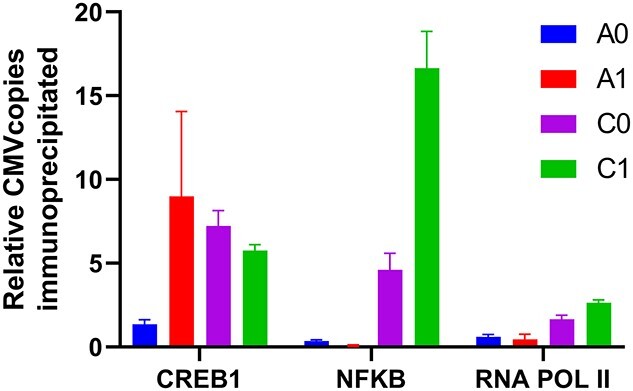
Relative number of copies of CMV associated with transcription factors determined by chromatin immunoprecipitation. All values are normalized by the number of copies of CMV bound to CREB1 in the A0 cell line.
As seen in Figure 4, in ChIP performed using an antibody directed against the NFκB transcription factor, the C family cell lines showed increased NFκB association with the CMV promoter compared with the A family cell lines. In addition, the DHFR-amplified C1 cell line, showed approximately 3.6-fold more CMV binding than the parental C0 cell line (P < 0.001, Tukey’s multiple comparison test). No increased binding to NFκB was observed in the A1 cell line compared with the A0 cell line. In contrast to the NFκB results, ChIP performed using an antibody against CREB1 showed no significant difference in CMV binding between the A1, C0 or C1 cell lines (P = 0.37, one way ANOVA), but did show an approximately 6.6-fold increase in CMV binding in the A1 cell line compared with the A0 cell line (P < 0.001, Tukey’s multiple comparison test) as previously reported [13]. RNA polymerase II (RNA Pol II), the key enzyme responsible for transcription, showed more than a 2-fold increase in association with the CMV promoter in the C-family of cell lines compared with the A family cell lines, consistent with the overall higher specific productivity in the C0 cell line compared with the A0 cell line. The increased binding to RNA Pol II may be due to improved recruitment of the polymerase due to binding of both CREB1 and NFκB transcription factors by the C cell lines.
We hypothesize that interplay between transgene methylation, posttranslational changes in transcription factors and their binding to promoter regions plays a concerted role toward enhancing transcription. Our results suggest that the greater level of methylation in the A family cell lines at CpG3 (A0 has 1.9-fold higher methylation fraction than C0, and A1 has 1.6-fold higher methylation fraction than C1, Fig. 3b and c) interferes with binding of the NFκB transcription factor to the CMV promoter in the A family cell lines (Fig. 4). A similar phenomenon has been previously observed, with methylation of the CRE consensus sequence (CpG5 in Fig. 3a) interfering with transcription factor binding and transcriptional activity in PC12 and HeLa cells [26]. Interestingly, even though the C1 cell line has ~1.3-fold higher methylation fraction than the C0 parental cell line at CpG3, it still shows 3.6-fold greater CMV binding than the parental cell line, suggesting that methylation is not the only factor determining transcription factor binding. Notably, while the A1 cell line shows much greater CREB1 binding to CMV than the A0 parental cell line, a similar increase is not seen in the C1/C0 cell lines, suggesting that the C family cell lines do not increase their productivity by increased engagement with the CREB1 transcription factor.
DISCUSSION
When subjected to amplification and adaption stresses, cells ensure their survival by various genomic, proteomic and phenotypic changes. The final high-producing clones therefore may have increases in protein production due to a combination of mechanisms. In the studies of the A and C cell clones, we have observed the following:
(A) Amplified cell lines have increased transcription-factor association with the CMV promoter upstream of the transgene.
(B) Amplified cell lines have changes in the methylation patterns of the CMV promoter region, particularly upstream of the transcription factor binding sites.
(C) Addition of the hypomethylation agent azacytidine can alter the methylation state and lead to increased transgene production from the cells.
Historically, the CMV promoter has been employed quite successfully to drive high levels of recombinant protein production, despite the observation that methylation influences the production from this promoter [5]. Indeed, there have been many approaches to understanding the epigenetic mechanisms that lead to loss of promoter activity [27], and to identify the reasons for reductions in titers in manufacturing processes [28]. A variety of promoter engineering strategies have been employed to increase production stability, including engineering in a CpG island from an endogenous housekeeping gene upstream of the CMV promoter [29] and alternatively, generating a CpG-free version of a promoter comprised of the mouse CMV enhancer, the human elongation factor 1 alpha core promoter and a synthetic intron at the 5′ untranslated region [30]. Interestingly, the addition of the CpG island upstream of the CMV promoter led to improved stability, although it decreased initial productivity. Removing the CpG sites from the synthetic promoter resulted in a larger fraction of highly productive clones upon initial isolation, but no significant improvement in long-term expression stability, highlighting the complicated role of DNA methylation in stability of transgene expression.
The epigenetic approaches described above have been directed toward understanding or reducing production instability. While cell line instability has long plagued the biopharmaceutical industry [4], promoter methylation may influence aspects of productivity beyond stability. In this study, we focused on variations in CMV promoter methylation that appear to alter rates of transcription between clones in a non-time-dependent manner. We demonstrated differences in methylation patterns in clones derived from different populations (A and C cell lines) expressing the same mAb.
A noteworthy observation is that the degree of methylation is not the only determinant of transcriptional activity from the promoter. The A family cell lines show higher methylation levels at every CpG site examined in this study, particularly the sites upstream of the transcription factor consensus binding sites, but the A1 cell line shows comparable or greater protein production than the C0 and C1 cell lines and notably higher mRNA levels. Brown and coworkers investigated the relationships between the NFκB and CREB binding sites in the CMV promoter in transient gene expression in CHO cells [31]. The two sites appear to act synergistically, with transcription factor decoys to either site reducing expression 40 to 50% and decoys to both sites reducing expression nearly 80%. This synergy suggests that the methylation in the A family cell lines that may interfere with NFκB binding is compensated for by increased CREB binding in the amplified cells lines. Interestingly, we had observed in our early studies of these cell lines that the C family of cell lines showed greater productivity per gene copy than the A family of cell lines [11]. This observation was true for both the parental and the amplified progeny cell lines. It is possible that the accessibility of the C family cell lines to both the NFκB and CREB transcription factors (Fig. 4) leads to this increased transcription, which would also be consistent with the observed increased interaction with RNA polymerase.
In both of the studied cell families (A and C), we have amplification at the same locus [12], but analysis of the methylation patterns reveals site-specific differences. Incorporating the observations from the parent and progeny methylation patterns in the design of site-directed integration workflows will be important, as we are cognizant that different methylation is possible even when the insert is amplified in the same locus. Finally, we observe that when treated with hypomethylation agent, there is reduction in site-specific methylation and increased productivity seen in all clones. This indicates that while specific transcription factors may improve the transcript levels (CREB1 in A cell lines), there are other global improvements that can be leveraged to obtain further increases in specific productivity. Leveraging DNA methyltransferases [32] and even synthetic promoters [33] may further enhance transcription in higher producing cells.
While these results are specific for the cell lines studied and cannot necessarily be generalized to all CHO cell lines using strong viral promoters, these observations suggest that the nuclear proteome, promoter engineering and DNA methylation mechanisms might be used in a combinatorial fashion to help derive the next generation of high specific productivity clones for implementation in intensified and continuous processes, potentially reducing the high cost of recombinant protein therapeutics. It is our belief that the route to increased specific productivity lies in the understanding of the interplay between the nuclear proteome, and the local epigenetic changes surrounding the transgene.
AUTHOR CONTRIBUTIONS
H.D. performed the qPCR and ChIP experiments, analyzed the data and drafted the manuscript. S.A. performed the pyrosequencing and reviewed the manuscript. S.N. performed qPCR on the mRNA light chain and initiated methylation studies. M.H. performed cell growth and DNA purification. D.L.M. designed the pyrosequencing primers, analyzed the pyrosequencing results and edited the manuscript. S.S. designed the experimental plan, assisted with DNA purification, analyzed the data and edited the manuscript.
DATA AVAILABILITY
The data underlying this article will be shared on reasonable request to the corresponding author.
ACKNOWLEDGMENT
We would like to thank Ranya Pranomphon for assistance with ELISA and Professor Nathaniel Cady for useful suggestions about the data analysis.
FUNDING
This work was supported in part by a grant from the US National Science Foundation [CBET-0967821].
CONFLICT OF INTEREST STATEMENT
The authors declare no conflict of interest.
ETHICS AND CONSENT
No patients were involved in these studies.
ANIMAL RESEARCH
All cell lines used in these studies were established, immortalized cells and no animal experiments were performed.
Supplementary Material
Contributor Information
Hussain Dahodwala, National Institute for Innovation in Manufacturing Biopharmaceuticals, Newark, DE 19713, USA.
Sophia D Amenyah, School of Biomedical Sciences, Ulster University, Coleraine, Co. Londonderry, BT52 1SA, Northern Ireland, UK.
Sarah Nicoletti, College of Nanoscale Science and Engineering, SUNY Polytechnic Institute, Albany, NY 12203, USA.
Matthew N Henry, Australian Institute for Bioengineering and Nanotechnology (AIBN), The University of Queensland, St. Lucia, QLD 4072 , Australia.
Diane J Lees-Murdock, School of Biomedical Sciences, Ulster University, Coleraine, Co. Londonderry, BT52 1SA, Northern Ireland, UK.
Susan T Sharfstein, College of Nanoscale Science and Engineering, SUNY Polytechnic Institute, Albany, NY 12203, USA.
References
- 1. Lu R-M, Hwang Y-C, Liu I-J et al. Development of therapeutic antibodies for the treatment of diseases. J Biomed Sci 2020; 27: 1.Lu R-M, Hwang Y-C, Liu I-Jet al. Development of therapeutic antibodies for the treatment of diseases. J Biomed Sci 2020; 27: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Stolfa, G, Smonskey, MT, Boniface, R et al. CHO-omics review: the impact of current and emerging technologies on Chinese hamster ovary based bioproduction. Biotechnol J 2018; 13: 1700227. [DOI] [PubMed] [Google Scholar]
- 3. Pollock, J, Ho, SV, Farid, SS. Fed-batch and perfusion culture processes: economic, environmental, and operational feasibility under uncertainty. Biotechnol Bioeng 2013; 110: 206–19. [DOI] [PubMed] [Google Scholar]
- 4. Dahodwala, H, Lee, KH. The fickle CHO: a review of the causes, implications, and potential alleviation of the CHO cell line instability problem. Curr Opin Biotechnol 2019; 60: 128–37. [DOI] [PubMed] [Google Scholar]
- 5. Wippermann, A, Noll, T. DNA methylation in CHO cells. J Biotechnol 2017; 258: 206–10. [DOI] [PubMed] [Google Scholar]
- 6. Fernandez, AF, Esteller, M. Viral epigenomes in human tumorigenesis. Oncogene 2010; 29: 1405–20. [DOI] [PubMed] [Google Scholar]
- 7. Krebs, AR. Studying transcription factor function in the genome at molecular resolution. Trends Genet 2021; 37: 798–806. 10.1016/j.tig.2021.03.008. [DOI] [PubMed] [Google Scholar]
- 8. Hilliard, W, Lee, KH. Systematic identification of safe harbor regions in the CHO genome through a comprehensive epigenome analysis. Biotechnol Bioeng 2021; 118: 659–75. [DOI] [PubMed] [Google Scholar]
- 9. Betts, Z, Dickson, AJ. Ubiquitous chromatin opening elements (UCOEs) effect on transgene position and expression stability in CHO cells following methotrexate (MTX) amplification. Biotechnol J 2016; 11: 554–64. [DOI] [PubMed] [Google Scholar]
- 10. Marx, N, Dhiman, H, Schmieder, V et al. Enhanced targeted DNA methylation of the CMV and endogenous promoters with dCas9-DNMT3A3L entails distinct subsequent histone modification changes in CHO cells. Metab Eng 2021; 66: 268–82. [DOI] [PubMed] [Google Scholar]
- 11. Jiang, Z, Huang, Y, Sharfstein, ST. Regulation of recombinant monoclonal antibody production in Chinese hamster ovary cells: a comparative study of gene copy number, mRNA level, and protein expression. Biotechnol Prog 2006; 22: 313–8. [DOI] [PubMed] [Google Scholar]
- 12. Jiang, Z, Sharfstein, ST. Characterization of gene localization and accessibility in DHFR-amplified CHO cells. Biotechnol Prog 2009; 25: 296–300. [DOI] [PubMed] [Google Scholar]
- 13. Dahodwala, H, Kaushik, P, Tejwani, V et al. Increased mAb production in amplified CHO cell lines is associated with increased interaction of CREB1 with transgene promoter. Curr Res Biotechnol 2019; 1: 49–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Weinguny, M, Klanert, G, Eisenhut, P et al. Directed evolution approach to enhance efficiency and speed of outgrowth during single cell subcloning of Chinese hamster ovary cells. Comput Struct Biotechnol J 2020; 18: 1320–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Johari, YB, Mercer, AC, Liu, Y et al. Design of synthetic promoters for controlled expression of therapeutic genes in retinal pigment epithelial cells. Biotechnol Bioeng 2021; 118: 2001–15. [DOI] [PubMed] [Google Scholar]
- 16. Hamdi, A, Szeliova, D, Ruckerbauer, DE et al. Key challenges in designing CHO chassis platforms. Processes 2020; 8: 26. [Google Scholar]
- 17. Dahodwala, H, Nowey, M, Mitina, T et al. Effects of clonal variation on growth, metabolism, and productivity in response to trophic factor stimulation: a study of Chinese hamster ovary cells producing a recombinant monoclonal antibody. Cytotechnology 2012; 64: 27–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Delaney, C, Garg, SK, Yung, R. Analysis of DNA methylation by pyrosequencing. Methods Mol Biol 2015; 1343: 249–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ehrich, M, Zoll, S, Sur, S et al. A new method for accurate assessment of DNA quality after bisulfite treatment. Nucleic Acids Res 2007; 35: e29–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Costello, JF, Plass, C. Methylation matters. J Med Genet 2001; 38: 285–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Caffrey, A, McNulty, H, Rollins, M, Prasad, G, Gaur, P, Talcott, J.B, Witton, C, Cassidy, T, Marshall, B, Dornan, J et al. Effects of maternal folic acid supplementation during the second and third trimesters of pregnancy on neurocognitive development in the child: An 11-year follow-up from a randomised controlled trial. BMC Med. 2021; 19: 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Royo, JL, Hidalgo, M, Ruiz, A. Pyrosequencing protocol using a universal biotinylated primer for mutation detection and SNP genotyping. Nat Protoc 2007; 2: 1734–9. [DOI] [PubMed] [Google Scholar]
- 23. Amenyah, SD, McMahon, A, Ward, M et al. Riboflavin supplementation alters global and gene-specific DNA methylation in adults with the MTHFR 677 TT genotype. Biochimie 2020; 173: 17–26. [DOI] [PubMed] [Google Scholar]
- 24.Amenyah, SD, Ward, M, McMahon, A et al. DNA methylation of hypertension-related genes and effect of riboflavin supplementation in adults stratified by genotype for the MTHFR C677T polymorphism. Int J Cardiol 2022; 322: 233–9. [DOI] [PubMed] [Google Scholar]
- 25. Rao, X, Huang, X, Zhou, Z et al. An improvement of the 2ˆ(−delta delta CT) method for quantitative real-time polymerase chain reaction data analysis. Biostat Bioinforma Biomath 2013; 3: 71–85. [PMC free article] [PubMed] [Google Scholar]
- 26. Iguchi-Ariga, SM, Schaffner, W. CpG methylation of the cAMP-responsive enhancer/promoter sequence TGACGTCA abolishes specific factor binding as well as transcriptional activation. Genes Dev 1989; 3: 612–9. [DOI] [PubMed] [Google Scholar]
- 27. Kim, M, O’Callaghan, PM, Droms, KA et al. A mechanistic understanding of production instability in CHO cell lines expressing recombinant monoclonal antibodies. Biotechnol Bioeng 2011; 108: 2434–46. [DOI] [PubMed] [Google Scholar]
- 28. Osterlehner, A, Simmeth, S, Gopfert, U et al. Promoter methylation and transgene copy numbers predict unstable protein production in recombinant Chinese hamster ovary cell lines. Biotechnol Bioeng 2011; 108: 2670–81. [DOI] [PubMed] [Google Scholar]
- 29. Zuniga, RA, Gutierrez-Gonzalez, M, Collazo, N et al. Development of a new promoter to avoid the silencing of genes in the production of recombinant antibodies in chinese hamster ovary cells. J Biol Eng 2019; 13: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ho, SCL, Koh, EYC, Soo, BPC et al. Evaluating the use of a CpG free promoter for long-term recombinant protein expression stability in Chinese hamster ovary cells. BMC Biotechnol 2016; 16: 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Brown, AJ, Sweeney, B, Mainwaring, DO et al. NF-κB, CRE and YY1 elements are key functional regulators of CMV promoter-driven transient gene expression in CHO cells. Biotechnol J 2015; 10: 1019–28. [DOI] [PubMed] [Google Scholar]
- 32. Weinguny, M, Eisenhut, P, Klanert, G et al. Random epigenetic modulation of CHO cells by repeated knockdown of DNA methyltransferases increases population diversity and enables sorting of cells with higher production capacities. Biotechnol Bioeng 2020; 117: 3435–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gutiérrez-González, M, Latorre, Y, Zúñiga, R et al. Transcription factor engineering in CHO cells for recombinant protein production. Crit Rev Biotechnol 2019; 39: 665–79. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article will be shared on reasonable request to the corresponding author.