Abstract
The gene encoding the major intracellular tributyrin esterase of Lactococcus lactis was cloned using degenerate DNA probes based on 19 known N-terminal amino acid residues of the purified enzyme. The gene, named estA, was sequenced and found to encode a protein of 258 amino acid residues. The transcription start site was mapped 233 nucleotides upstream of the start codon, and a canonical promoter sequence was identified. The deduced amino acid sequence of the estA product contained the typical GXSXG motif found in most lipases and esterases. The protein was overproduced up to 170-fold in L. lactis by use of the nisin-controlled expression system recently developed for lactic acid bacteria. The estA gene was inactivated by chromosomal integration of a temperature-sensitive integration vector. This resulted in the complete loss of esterase activity, which could then be recovered after complementation of the constructed esterase-deficient strain with the wild-type estA gene. This confirms that EstA is the main enzyme responsible for esterase activity in L. lactis. Purified recombinant enzyme showed a preference for short-chain acyl esters, surprisingly also including phospholipids. Medium- and long-acyl-chain lipids were also hydrolyzed, albeit less efficiently. Intermediate characteristics between esterases and lipases make intracellular lactococcal EstA difficult to classify in either of these two groups of esterolytic enzymes. We suggest that, in vivo, EstA could be involved in (phospho)lipid metabolism or cellular detoxification or both, as its sequence showed significant similarity to S-formylglutathione hydrolase (FGH) of Paracoccus denitrificans and human EstD (or FGH), which are part of a universal formaldehyde detoxification pathway.
A large number of microbial lipolytic enzymes have been identified and characterized to date. These lipolytic enzymes, mainly esterases and lipases, belong to the general class of carboxylic ester hydrolases (EC 3.1.1) but differ in substrate specificity and type of enzyme kinetics (50). Applications of microbial lipolytic enzymes are widely found in the food, detergent, pharmaceutical, and chemical industries (19). The breakdown of milk fat by lipases and esterases is one of the main biochemical events that occur during cheese ripening, and it contributes to flavor development. The contribution of the native milk lipase to lipolysis in cheese is significant only in those varieties produced with raw milk, as this enzyme is inactivated during heat treatment. In soft blue-veined cheeses, the extracellular lipase from the mold Penicillium roquefortii contributes to the intense and characteristic flavor (26). In other varieties, like cheddar cheese, only a low level of milk fat hydrolysis occurs due to the weak lipolytic and esterolytic activities of the starter bacteria (7, 37). However, the small amounts of short-chain fatty acids are important because they have a low sensorial detection threshold (36) and they contribute to flavor balance (7).
Reports have been published on the purification and partial characterization of intracellular esterases of lactococci (5, 24, 48). The Lactococcus lactis ACA-DC 127 esterase, a monomeric 68-kDa enzyme, belongs to the class of serine esterases and hydrolyzes synthetic substrates (p-nitrophenyl [p-NP] esters shorter than or equal to C8) (48). The recently purified esterases of L. lactis NCDO 763 and E8 share several characteristics: they are intracellular, the estimated molecular mass of the monomer is 29 kDa, the optimal activity is at pH 7.0 to 8.0, and the reported N-terminal sequences are identical (5, 24). The enzyme from L. lactis NCDO 763 showed higher activities with p-NP-butanoate, like the esterase of ACA-DC 127, but it also hydrolyzed p-NP esters longer than C8 (5). On the other hand, the lactococcal enzyme described by Holland and Coolbear (24) had p-NP-butanoate, tributyrin, and milk fat hydrolase activity but was unable to hydrolyze triacylglycerols with long-chain fatty acids such as tripalmitin or triolein. To our knowledge, genetic characterization of the lipolytic enzymes produced by Lactococcus has not been reported up to now, in spite of the great developments in genetic engineering of the industrially important lactic acid bacteria in the past years.
This paper describes the cloning and characterization of the gene encoding the major tributyrin esterase of L. lactis subsp. lactis B1014, which is very similar to the E8 esterase (R. Holland and T. Coolbear, unpublished data). Further work was also carried out with Lactococcus lactis subsp. cremoris MG1363, a plasmid-free model strain of Lactococcus. Several L. lactis strains that overproduced either B1014 or MG1363 esterase were constructed by using the recently developed nisin-controlled expression system (12, 29, 30). The substrate selectivity of the esterases purified from the overproducing strains was defined by using p-NP-acyl ester substrates.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacteria and plasmids used or constructed for this study are listed in Table 1. Unless otherwise indicated, L. lactis strains were routinely grown without aeration at 30°C in M17 (Difco Laboratories, Detroit, Mich.) supplemented with 0.5% (wt/vol) glucose. Escherichia coli MC1061 (3) was used as a host for cloning experiments, and it was grown with aeration in L broth-based medium at 37°C (42). When antibiotics were added, the final concentrations used were as follows: ampicillin, 50 μg ml−1; chloramphenicol, 10 μg ml−1 for E. coli and 7.5 μg ml−1 for L. lactis; tetracycline, 10 μg ml−1 for E. coli and 5 or 2 μg ml−1 for L. lactis.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Source or reference |
|---|---|---|
| Strains | ||
| L. lactis subsp. lactis B1014 | Tributyrin esterase producer, wild type | New Zealand Dairy Research Institute culture collection |
| L. lactis subsp. cremoris | ||
| MG1363 | Plasmid-free, wild type | 17 |
| NZ9000 | MG1363; pepN::nisRK; host strain for esterase overexpression | 12 |
| NZ9800 | NZ9700 derivative containing Tn5276 (conjugative nisin-sucrose transposon); ΔnisA; non-nisin producer; host strain for esterase overexpression | 28 |
| NZ9340 | Tcr EstA− derivative of NZ9000; estA::pNZ9332 | This work |
| E. coli MC1061 | 3 | |
| Plasmids | ||
| pUC19 | Apr, lacZ | 54 |
| pUC18 | Apr, lacZ | 54 |
| pGEM-5Zf(+) | Apr | Promega Corp.b |
| pNZ8020 | Cmr, nisA transcriptional fusion vector | 12 |
| pNZ8030 | Cmr, nisA translational fusion vector | 12 |
| pGhost8 | Tcr, thermosensitive replicon | 35 |
| pNZ9301 | pUC18 derivative containing a 4.5-kb EcoRI-SstI fragment carrying the 5′ end of B1014 estA | This work |
| pNZ9302 | pUC19 derivative containing a 1.7-kb AseI-HindIII fragment carrying the 3′ end of B1014 estA | This work |
| pNZ9333 | pGEM-5Zf(+) derivative containing a 1.3-kb PCR product carrying MG1363 estA | This work |
| pNZ9308 | pNZ8020 derivative containing a 0.8-kb HincII-RsaI fragment carrying B1014 estA transcriptionally fused to the nisA promoter | This work |
| pNZ9330 | pNZ8020 derivative containing a 0.8-kb EcoRI-SspI fragment carrying MG1363 estA transcriptionally fused to the nisA promoter | This work |
| pNZ9310 | pNZ8030 derivative containing a 0.8-kb fragment carrying B1014 estA translationally fused to the nisA promoter | This work |
| pNZ9331 | pNZ8030 derivative containing a 0.8-kb fragment carrying MG1363 estA translationally fused to the nisA promoter | This work |
| pNZ9332 | pGhost8 derivative containing a 0.6-kb internal fragment of MG1363 estA | This work |
Apr, Cmr, Tcr, resistance to ampicillin, chloramphenicol, and tetracycline, respectively.
Promega Corp. technical bulletin no. 150.
The plasmid vectors used in the cloning experiments with E. coli were pUC18 and pUC19 (54) and pGEM-5Zf(+) (Promega Corp.). In L. lactis, the plasmids pNZ8020 and pNZ8030 (12, 28, 30) and pGhost8 (35) were used. pGhost8 has a temperature-sensitive origin of replication; its replication is normally blocked at temperatures higher than 35°C.
DNA isolation and manipulation.
Isolation of plasmid DNA from E. coli and transformations of E. coli strains were carried out according to established procedures (42). Both plasmid and chromosomal DNAs of L. lactis were isolated as described previously (52). L. lactis cells were transformed by electroporation (53). DNA fragments were isolated from agarose gels by using the GlassMAX DNA isolation matrix system (BRL Life Technologies, Inc., Gaithersburg, Md.). Restriction enzymes, T4 DNA ligase, and other DNA-modifying enzymes were purchased from GIBCO/BRL Life Technologies, New England Biolabs Inc., or Promega Corp. and used as recommended by the manufacturer. Oligonucleotides were purchased from Pharmacia. Cloning procedures, agarose gel electrophoresis, radiolabeling of oligonucleotides, and Southern blot hybridizations were performed as described by Sambrook et al. (42). PCR was performed as described previously (27).
Cloning of the tributyrin esterase gene.
Chromosomal DNA from L. lactis B1014 was isolated and digested with several restriction enzymes. The B1014 tributyrin esterase gene was localized by using the degenerate oligonucleotides 5′-GCNGTNAT(ACT)AA(CT)AT(ACT)GA(AG)TA(CT)TA-3′ and 5′-TT(AG)TTNAC(CT)TT(ACT)C(GT)(AG)TTCATNCC-3′, which were based on parts of the first 19 N-terminal amino acids, i.e., AVINIEYY and GMNRKVNV, respectively, of the tributyrin esterase from L. lactis E8 (24). Southern hybridization revealed a 4.5-kb EcoRI-SstI fragment from chromosomal DNA that hybridized with both oligonucleotides, and fragments of approximately this size were cloned into the EcoRI and SstI restriction sites of pUC18. Colony blot hybridization localized a clone that contained the 4.5-kb EcoRI-SstI hybridizing fragment, which was named pNZ9301. Restriction analysis of the insert and Southern hybridization demonstrated that the tributyrin esterase gene was close to the EcoRI site of the fragment. Sequence analysis showed that the insert contained the 5′ end of the tributyrin esterase gene, but the whole sequence was not present in this fragment. The determined sequence of the cloned EcoRI-SstI fragment revealed an AseI restriction site close to the EcoRI site. A 1.7-kb AseI-HindIII fragment of chromosomal DNA from B1014 was subsequently isolated after hybridization with the LipE probe (5′-CCCAAACAGATATTCCGGTG-3′). This oligonucleotide was designed after the nucleotide sequence of the tributyrin esterase gene close to the EcoRI site that had been determined (nucleotides [nt] 392 to 411) (Fig. 1). This 1.7-kb AseI-HindIII fragment was cloned into the HincII-HindIII sites of pUC19, resulting in plasmid pNZ9302. Together, the two DNA chromosomal fragments present in pNZ9301 and pNZ9302 contained the complete open reading frame (ORF) encoding the B1014 tributyrin esterase gene (estA).
FIG. 1.
Nucleotide sequence of the L. lactis B1014 tributyrin esterase gene. The predicted amino acid sequence is given below the nucleotide sequence in the standard one-letter code. The stop codon is marked by an asterisk. The transcription initiation site mapped by primer extension is marked by a vertical arrow (nt 62). The putative Shine-Dalgarno site (SD), the conserved TG dinucleoside, and the −35 and −10 regions are underlined. The lipase/esterase consensus sequence at positions 119 to 123 is double underlined.
The estA gene from L. lactis MG1363 was amplified by PCR with oligonucleotides LipPCR1 (5′-GACTTCACAGCGCACACTTTG-3′) and Lip8 (5′-ATACCAGACCGAACACA-3′), designed on the basis of the nucleotide sequence of cloned chromosomal fragments from B1014, using the Advantage genomic polymerase mix (Clontech Laboratories, Inc., Palo Alto, Calif.). The amplified 1.6-kb PCR product, which contained the complete MG1363 esterase gene, was cloned in pGEM-5Zf(+) using the pGEM-T Vector System (Promega Corp.), resulting in pNZ9333.
DNA sequencing and nucleotide and deduced protein sequence analysis.
DNA sequence analysis was performed by the dideoxy chain termination procedure (43) with fluorescein-labeled primers and an AutoRead sequencing kit (Pharmacia). The nucleotide sequence was determined in both orientations with an ALF automatic DNA sequencer (Pharmacia Biotech). Computer analysis of DNA sequences and the deduced amino acid sequences was performed with the PC/GENE program, version 6.70 (IntelliGenetics). Database searching for related proteins was performed with BLAST (1), T/FASTA (39), and PROFILESEARCH (22). Multiple sequence alignment was performed with PILEUP (16).
RNA techniques and primer extension analysis.
RNA isolation and primer extension of the B1014 estA promoter were performed as described previously (28). The oligonucleotide with the sequence 5′-GTGGATTTTCTTCAATCCC-3′, which is complementary to positions 214 to 232 (Fig. 1), was used as a primer.
Construction of plasmids.
To overexpress the estA gene from the wild-type strains B1014 and MG1363 in NZ9800 and NZ9000, several plasmids containing these genes transcriptionally or translationally fused to the nisA promoter were constructed. An 845-bp HincII-RsaI fragment carrying the B1014 estA gene was cloned in pNZ8020 digested with SmaI, yielding pNZ9308. Similarly, pNZ9330 was obtained by cloning an 856-bp EcoRI-SspI fragment carrying the MG1363 estA gene in pNZ8020 digested with EcoRI and XbaI (Klenow fragment). The primers used for cloning the B1014 estA gene in pNZ8030 were NcoEst (5′-CGTCGCCATGGCAGTAATTAATATCGAATAC-3′), containing an NcoI site (underlined), and LipN1 (5′-CCATTGATAAACCAGCGA-3′). The PCR product obtained with these primers, which contained the 5′ end of B1014 estA, was digested with NcoI and EcoRI and cloned into pNZ8030. The missing part of the esterase gene was cloned as an EcoRI-RsaI (Klenow) fragment into the EcoRI-HindIII (Klenow) sites of the previous construction, yielding pNZ9310. A similar strategy was used to construct pNZ9331, which contains the MG1363 estA gene translationally fused to the nisA promoter. The primers used were NcoEst, as before, and EstB4 (5′-CTGGTCTAGACTTCAAGTTCAGGTTGGCG-3′). In this case, the PCR product contained the whole MG1363 estA gene and was digested with NcoI and SspI before being cloned in pNZ8030 digested with NcoI and HindIII (Klenow). The clones obtained were checked by restriction enzyme analysis and by PCR with several primers.
For disruption of the MG1363 estA gene, plasmid pNZ9332 was constructed by cloning a 581-bp internal fragment of this gene in pGhost8 digested with PstI and BamHI. The internal fragment was obtained by PCR with primers EstA1 (5′-CTCGGGATCCCTGAGTCAAGTAAGGTG-3′), containing a BamHI site (underlined), and EstA2 (5′-GTCCCTGCAGTTCTTCAACTCTGCTATTGC-3′), containing a PstI site (underlined).
Induction of estA expression by nisin.
For overexpression of B1014 and MG1363 estA genes, overnight cultures of L. lactis NZ9000 and NZ9800 harboring pNZ9308, pNZ9310, pNZ9330, or pNZ9331 were transferred (1%) into fresh medium with chloramphenicol and grown until a cell density equivalent to an A600 of 0.5 was reached. The cells were induced with 3 ng of nisin A ml−1 and grown for another 120 min (30). Uninduced cultures were used as controls. Cells were then harvested and resuspended in 100 mM potassium phosphate buffer, pH 7.0. Cell extracts were prepared with a bead beater and used to assay esterase activity.
Construction of an estA mutant of L. lactis and complementation analysis.
The MG1363 estA gene was inactivated by single crossover integration of pNZ9332 into the NZ9000 chromosome. Strain NZ9000 was chosen to allow subsequent complementation of the obtained esterase-deficient derivative. Plasmid pNZ9332 was electroporated into NZ9000 competent cells, and a single colony was used to generate integrants following the procedure described by Maguin et al. (35). Integrants were selected at 37°C (nonpermissive temperature for plasmid replication) in the presence of 2 μg of tetracycline per ml. Integration of pNZ9332 in NZ9000, resulting in strain NZ9340, was confirmed by Southern hybridization and PCR with the oligonucleotides TetR (5′-CCCAGTTTGTAATTCCAGGAGTAG-3′, complementary to the 5′ end of the tetracycline resistance gene present in pGhost8), EstB1 (5′-CAGCCCCGGGAACAAGAAGGAGAAAAAGAA-3′, complementary to the chromosomal sequence upstream the MG1363 estA gene), and EstB4 (complementary to the chromosomal sequence downstream of the MG1363 estA gene; positions 1364 to 1383 in Fig. 1). Strain NZ9340 was grown in subsequent experiments at 37°C. The translational fusion plasmid pNZ9331 constructed previously was used to deliver the wild-type MG1363 estA gene into the esterase-deficient mutant for complementation analysis. This plasmid was transformed into NZ9340, and one transformant was used to assay esterase activity after induction with nisin A as described above.
Enzyme assays.
For p-NP esterase, reaction mixtures in a total volume of 2.0 ml contained 50 mM sodium phosphate buffer (pH 7.5), purified tributyrin esterase (0.2 μg/ml) or a suitable amount of cell extract, and substrates at appropriate concentrations. Stock solutions (50 mM) of p-NP esters of ethanoate (C2), butanoate (C4), hexanoate (C6), octanoate (C8), decanoate (C10), dodecanoate (C12), tetradecanoate (C14), and hexadecanoate (C16) were dissolved in dimethyl sulfoxide prior to addition to the reaction mixture. Initial rates of p-nitrophenol release at 30°C from the ester substrates were quantitated by measuring absorbance at 410 nm (Σ410 = 12.2 at pH 7.5). For tributyrin esterase, the reaction mixtures (2.5 ml) contained 0 to 20 mM tributyrin, 2.4% gum arabic, 0.1 M HEPES buffer (pH 8.0), 0.1 M NaCl, 0.004 M CaCl2, and purified tributyrin esterase (4 μg/ml). Incubations were carried out at 30°C for 20 min. Samples (0.5 ml) were taken and added to a stop mixture comprising 0.5 ml of water, 1.0 ml of ethanol, and 0.1 ml of 2.5 M H2SO4. Tridecanoic acid (C13) was added as an internal standard. Butanoic acid released in the enzyme reaction was extracted and quantified by gas-liquid chromatography. For phospholipase, activity on monomeric dihexanoyl-dithio-phosphatidylcholine(diC6dithioPC) was determined in 200 mM Tris (pH 8.0)–100 mM NaCl–100 mM CaCl2, with 5,5′-dithiobis-(2-nitrobenzoic acid) as a chromogenic reagent as described previously (51). Initial substrate concentrations ranged between 0 and 2.4 mM. Activity on micellar didodecanoyl-dithio-phosphatidylcholine (diC12dithioPC) was determined in the same manner except that the substrate was mixed with equimolar amounts of sodium taurodeoxycholate.
Tributyrin esterase purification.
Tributyrin esterase was purified from a 4-liter culture of L. lactis NZ9800 harboring pNZ9308. Growth was monitored, and nisin was added (3 ng ml−1, final concentration) when the culture reached an A600 of 0.7. The culture was harvested at the beginning of the stationary phase. Enzyme purification was carried out by the method described for the purification of tributyrin esterase from L. lactis subsp. cremoris E8 (24), except that the three chromatography steps used were (in order of application) anion exchange (Q-Sepharose; 11- by 5-cm column; Pharmacia LKB Biotechnology, Uppsala, Sweden), hydrophobic interaction (Alkyl-Superose HR 10/10 column; Pharmacia), and gel filtration (Sephadex G150 Superfine, 90- by 1.6-cm column; Pharmacia).
PAGE.
Polyacrylamide gel electrophoresis (PAGE) (with sodium dodecyl sulfate and nondenaturing) was performed as described for tributyrin esterase preparations from L. lactis subsp. cremoris E8 (24).
Protein determinations.
Protein quantities were estimated by the method of Bradford (2) with a Bio-Rad kit and bovine serum albumin as the protein standard.
Amino acid sequencing.
The N-terminal sequence of B1014 EstA was determined at the Protein Sequencing facility in Delft (The Netherlands).
Nucleotide sequence accession numbers.
The nucleotide sequences reported in this paper have been submitted to GenBank and assigned accession numbers AF157484 and AF157601 for Lactococcus lactis subsp. lactis B1014 and L. lactis subsp. cremoris MG1363, respectively.
RESULTS
Cloning of the tributyrin esterase gene, estA.
Two oligonucleotides based on the reported N-terminal sequence of the tributyrin esterase from L. lactis E8 (24) were used to screen for the tributyrin esterase gene from L. lactis B1014. Two chromosomal DNA fragments, together encoding the putative tributyrin esterase, were cloned (see Materials and Methods), and the corresponding gene was named estA. The L. lactis MG1363 estA gene was cloned by using two primers which were based on the nucleotide sequence surrounding the B1014 tributyrin esterase gene.
Nucleotide sequence of L. lactis estA.
The complete nucleotide and the deduced amino acid sequences of the estA gene of L. lactis B1014 obtained by PCR were identical to that of the two initially cloned fragments and are shown in Fig. 1. One complete 774-bp ORF (ATG initiation codon at nt 295 and TAA termination codon at nt 1072) was identified in the sequence. This ORF could encode a polypeptide of 258 amino acids with a deduced molecular weight of 29,640. A good potential Shine-Dalgarno sequence, 5′-AAGGAG (ΔG0 [25°C] = −12.8 kcal [1 kcal = 4.184 kJ/mol]) (47), which is complementary to the 3′ end of the L. lactis 16S rRNA (4), was found 13 bases upstream from the initiation codon ATG. The transcription initiation site was determined by primer extension analysis of the upstream region of the B1014 estA gene and was localized 233 nt upstream from the ATG start codon at a guanine residue (nt 62) (data not shown). This transcription start site is preceded by the canonical −35 and −10 promoter sequences at positions 25 to 30 and 50 to 55, respectively, that are spaced by 20 nt. The putative −10 region is immediately preceded by the sequence TGN, which is often found in lactococcal promoters (14). The sequence downstream of the stop codon lacks any inverted repeats. No additional ORF was found between the promoter and the transcription start site of B1014 estA. Analysis of the ORF indicated that the protein lacks a classical secretion signal sequence at the N terminus of the esterase. The amino acid sequence GLSMG, starting at residue 649, fits the GXSXG motif found in most bacterial and eukaryotic serine hydrolases, like lipases and esterases (8). The sequencing of the PCR-amplified product from MG1363 chromosomal DNA cloned in pNZ9333 revealed an ORF of the same size as and with 84% similarity to B1014 estA, preceded by an identical putative Shine-Dalgarno sequence. The deduced product of MG1363 estA also contained the typical GXSXG motif.
Comparison of amino acid sequences.
Database searches revealed several (putative) proteins with 30 to 35% sequence identities to EstA from L. lactis B1014. These homologous proteins could be divided into three groups, as summarized in Table 2. The highest similarity (group A) was with XynC, an acetyl esterase of similar size (266 residues) from the thermophile Caldocellum saccharolyticum (34), and also with the N-terminal domain of XynZ, a much larger protein of 837 residues from the thermophile Clostridium thermocellum (20, 21). Group B contains many related proteins from various Mycobacterium species (of which only a few are listed in Table 2) and from Corynebacterium glutamicum, all of which are listed as extracellular α-antigens with a possible function in fibronectin binding (10, 25, 38, 40, 46). Group C contains the human esterase EstD (31, 55), which is identical to S-formylglutathione hydrolase (FGH) (15); this group also includes the FGHs of Paracoccus denitrificans, Anabaena azollae, and Saccharomyces cerevisiae (9, 23, 44) and several highly related putative proteins that have recently been discovered through genome sequencing of Arabidopsis thaliana, Synechocystis, Haemophilus influenzae, and E. coli. All of the related group C enzymes are presumably intracellular, since they do not contain a signal sequence, and they are all similar in size (266 to 299 residues) to L. lactis B1014 EstA.
TABLE 2.
Encoded proteins with highest sequence similarity to EstA of L. lactis B1014
| Group | Organism | EMBL identification | Protein | No. of residues | Signal peptide present | Function |
|---|---|---|---|---|---|---|
| A | Lactococcus lactis B1014 | AF157484 | EstA | 258 | − | Esterase/lipase ? |
| Caldocellum saccharolyticum | CSXYNAB | XynC | 266 | − | Acetyl esterase | |
| Clostridium thermocellum | CTXYLZ | XynZ | 837 | + | ? (N domain), xylanase (C domain) | |
| B | Mycobacterium scrofulaceum | MSAAA | A-α | 330 | + | α-Antigen, fibronectin binding |
| Mycobacterium avium | MAALANT | A85B | 330 | + | α-Antigen, fibronectin binding | |
| Mycobacterium leprae | ML85CPRA | A85C | 333 | + | α-Antigen, fibronectin binding | |
| Corynebacterium glutamicum | CGCOP1G | PS1 | 657 | + | Antigen? (N domain) | |
| C | Homo sapiens | HSETRD | EstD | 282 | − | FGH, esterase D |
| Saccharomyces cerevisiae | SCXCDNA | YJLO68c | 299 | − | FGH | |
| Arabidopsis thaliana | AC002510 | T32G6.5 | 284 | − | Esterase ? | |
| Anabaena azollae | AF035558 | FGH | 278 | − | FGH | |
| Paracoccus denitrificans | PD34346 | FGH | 279 | − | FGH | |
| Haemophilus influenzae | HI0184 | 0184 | 275 | − | Esterase? | |
| Synechocystis sp. | SSD904 | ? | 276 | − | Esterase | |
| Escherichia coli | ECU00007 | YeiG | 278 | − | Esterase? | |
| Escherichia coli | ECU73857 | YaiM | 277 | − | Esterase ? |
A multiple sequence alignment of the most homologous regions of these (putative) proteins is shown in Fig. 2. In the central section, Ser121 of L. lactis B1014 is in the consensus sequence GXSXG around the catalytic serine typical of all lipases and esterases. Near the C terminus, a conserved His231 is found that is predicted to be the catalytic histidine (8). The catalytic Asp/Glu is more difficult to identify, since the surrounding sequence in lipases and esterases is not highly conserved (8). However, the alignment suggests that Asp202 may be the catalytic residue, since this residue is conserved in groups A and C but not in group B of binding proteins. The sequence around residues His44 and Gly45 of B1014 esterase closely resembles the oxyanion hole region of lipases (11). The deduced amino acid sequence of the product of the MG1363 estA gene is very similar (less than 7% difference) to EstA from B1014 (data not shown).
FIG. 2.
Multiple-sequence alignment of (putative) proteins with sequence homology to EstA of L. lactis B1014 (LL EstA), Caldocellum saccharolyticum acetyl esterase XynC (CS XynC), Clostridium thermocellum XynZ (CT XynZ), Mycobacterium scrofulaceum α-antigen (MS A-α), Mycobacterium avium antigen 85B (MA A85B), Mycobacterium leprae antigen 85C (ML A85C), Corynebacterium glutamicum PS1 protein (CG PS1), human esterase D (HS EstD), Anabaena azollae FGH (AA FGH), an Arabidopsis thaliana T32G6.5 hypothetical gene product (AT T32G6.5), Saccharomyces cerevisiae FGH (SC YJL068c), P. denitrificans FGH (PD FGH), an H. influenzae 0184 hypothetical gene product (HI 0184), a Synechocystis sp. hypothetical gene product (SS ?), and E. coli YeiG and YaiM hypothetical gene products (EC YeiG and EC YaiM). Only relevant regions of highest homology are shown. The numbering at the top is that of EstA, while numbers before and after each sequence indicate the numbers of additional N-terminal and C-terminal residues, respectively. The consensus sequence shows fully conserved residues in uppercase and highly conserved residues in lowercase letters. The putative catalytic Ser, Asp, and His residues are indicated in boldface and by asterisks.
Overproduction of esterase in L. lactis.
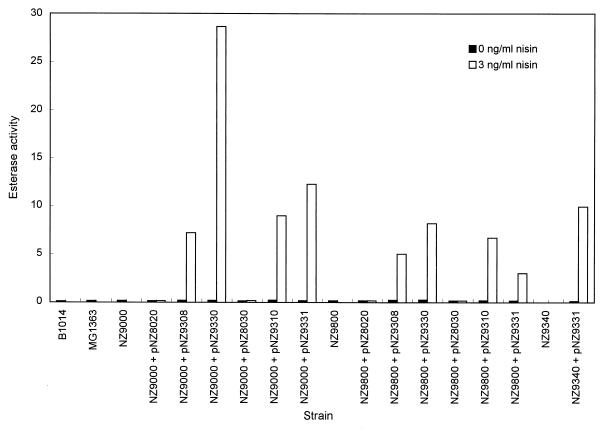
The estA coding sequences of B1014 and MG1363 were cloned in two expression vectors under the control of the nisA promoter. This expression system allows the transcription of the cloned genes to be activated after induction with subinhibitory amounts of nisin A (29). Plasmids pNZ9308 and pNZ9330 are derivatives of pNZ8020 and carry the estA gene from B1014 and MG1363, respectively, translationally fused to the nisA promoter. These constructs were introduced into L. lactis NZ9000 and NZ9800. Cell extracts of the transformants were used to determine the esterase activity with p-NP-butanoate as the substrate after induction with 3 ng of nisin A ml−1 and in the absence of nisin A. Cultures of the wild-type strains were used as controls; the specific esterase activities were 0.06 μmol mg−1 min−1 for B1014 esterase and 0.10 μmol mg−1 min−1 for MG1363 esterase (Fig. 3). The esterase activity in strain NZ9000 harboring pNZ9308, which contains the estA gene from B1014, was about 50 times higher after nisin induction than in the uninduced control, which had a level of activity comparable to the wild-type strain. The overproduction of EstA in NZ9000 with pNZ9330, which contains the estA gene from MG1363, was even higher, accounting for about a 170-fold increase. When these plasmids were introduced in strain NZ9800, the esterase activity also increased after induction with nisin, but only 28-fold for pNZ9308 and 50-fold for pNZ9330. Overproduction of EstA was found as well in NZ9000 and NZ9800 carrying these estA genes translationally fused to the start codon of nisA. Analysis of the intracellular protein of all these constructs on sodium dodecyl sulfate-PAGE gels revealed overproduction of the 29-kDa tributyrin esterase protein after induction with nisin A (data not shown). Esterase activity was also assayed with p-NP-butanoate and tributyrin as substrates in whole cells, cell homogenates, cell-free supernatants, and cell-free pellet fractions prepared from both NZ9800 containing pNZ9308 (estA from B1014) and B1014. Esterase activity in the cell homogenate was 80- to 100-fold higher in strain NZ9800 containing pNZ9308 than in wild-type B1014 when expressed as specific activity based upon the amount of protein (data not shown).
FIG. 3.
Expression levels of estA in different L. lactis strains and constructs. B1014 and MG1363 are wild-type tributyrin esterase producer strains, NZ9000 and NZ9800 are nisin-sensitive strains for induction of gene expression, and NZ9340 is an esterase-deficient mutant. Plasmids pNZ9308 (B1014 estA) and pNZ9330 (MG1363 estA) are derivatives of pNZ8020 (transcriptional fusion vector), while pNZ9310 (B1014 estA) and pNZ9331 (MG1363 estA) are derivatives of pNZ8030 (translational fusion vector). The esterase activity was determined in cell extracts after induction with 3 ng of nisin A ml−1 and in the absence of nisin A, and it is expressed as micromoles of p-nitrophenol liberated from p-NP-butanoate per minute per milligram of protein.
Disruption of estA.
The chromosomal estA gene of NZ9000 was inactivated by gene disruption with a temperature-sensitive integration vector. This vector can be introduced and maintained extrachromosomally at the permissive temperature (28°C), but it is lost when the temperature of the culture is shifted above 35°C, allowing the selection of integrants (35). To direct the integration of the vector into the chromosomal estA gene, a pGhost8 derivative was constructed by cloning a 581-bp internal fragment from the L. lactis MG1363 estA gene in this vector, resulting in pNZ9332. This construction was introduced into NZ9000 by electroporation, and one of the tetracycline-resistant (Tcr) transformants, selected at 28°C, was grown at 37°C in medium containing 2 μg of tetracycline per ml to select for the integration of the plasmid. Digested chromosomal DNA from several isolates that were able to grow at the nonpermissive temperature in the presence of tetracycline was analyzed by Southern hybridization with the same 581-bp internal fragment as a probe. The hybridization profile showed that the chromosomal copy of the estA gene had been disrupted by integration of multiple tandem copies of pNZ9332 (data not shown). The integration was also confirmed by PCR; a 1.2-kb PCR fragment was obtained when oligonucleotides TetR and EstB4 (complementary to pGhost8 and the sequence downstream of the MG1363 estA gene, respectively) were used, while no PCR product was obtained with oligonucleotides EstB1 and EstB4, which flank the integration site. One of the integrants, named NZ9340, was chosen for further characterization. The cell extract of this integrant was tested for esterase activity with p-NP-butanoate as the substrate and showed complete loss of this activity (Fig. 3). The esterase activities of cell extracts obtained from control cultures of wild-type MG1363 and NZ9000 grown at either 37 or 30°C did not differ, indicating that the absence of enzyme activity in NZ9340 was not due to the growth temperature. Also, this strain did not show any detectable level of activity when tributyrin was used as the substrate (data not shown). To further confirm this result, we complemented the esterase deficiency in this integrant. This was done by transforming integrant NZ9340 (which had no measurable esterase activity) with the translational fusion plasmid pNZ9331, which contains the MG1363 estA gene. As shown in Fig. 3, NZ9340 containing pNZ9331 recovered the esterase activity measured with p-NP-butanoate as the substrate. The activity was 0.07 μmol mg−1 min−1 without nisin induction and 9.9 μmol mg−1 min−1 after induction with nisin A.
Purification and substrate specificity of EstA from B1014.
The overproducing strain NZ9800 containing pNZ9308, which carries the estA gene from B1014 translationally fused to the nisA promoter, was used as the source of tributyrin esterase. Enzyme purification was monitored by the assay of p-NP-butanoate and tributyrin esterase activities at each step. From an initial 500 mg of protein in a cell homogenate prepared from a 4-liter culture of strain NZ9800 containing pNZ9308, the final yield was 8 mg of essentially pure tributyrin esterase protein. The first 20 amino acid residues of the purified B1014 EstA were identical to the N-terminal sequence of the enzyme purified from L. lactis E8 (24). B1014 tributyrin esterase purified from the overproducing strain behaved in the same manner during purification and PAGE under either denaturing or nondenaturing conditions as the enzyme isolated from strain E8.
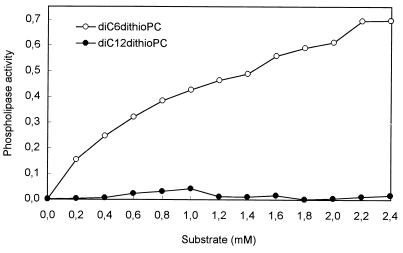
The substrate specificity of purified EstA was determined by use of p-NP esters of straight-chain fatty acids ranging in chain length from C2 (ethanoate) to C16 (hexadecanoate). Highest activity was observed with p-NP-hexanoate (C6). Km and Vmax values were calculated from Lineweaver-Burk plots using a least-squares best fit of the Michaelis-Menten equation (Table 3). The calculated Vmax values were substantially higher than the observed maximum rates of reaction, particularly with p-NP esters of fatty acids of longer chain length. EstA was not active on p-NP ester substrates with an acyl chain length greater than C12 (dodecanoate). The triacylglycerol tributyrin was also a hydrolytic substrate for EstA from B1014. The maximum specific activity measured with tributyrin as the substrate was 8 μmol of butyrate released per min per mg of enzyme protein. Moreover, activity was assayed with the phospholipid analogs diC6dithioPC and diC12dithioPC (Fig. 4). The enzyme showed relatively good phospholipase activity on the monomeric diC6dithioPC, which increased gradually with the substrate concentration, but no activity was detected on micellar diC12dithioPC at any of the concentrations assayed (up to 2.4 mM).
TABLE 3.
Kinetic parameters of purified EstA on p-NP esters of fatty acidsa
| p-NP ester substrate | Km (mM) | Vmax (μmol min−1 mg of protein−1) |
|---|---|---|
| p-NP-ethanoate | 0.87 | 170 |
| p-NP-butanoate | 3.39 | 531 |
| p-NP-hexanoate | 0.09 | 332 |
| p-NP-octanoate | 0.06 | 297 |
| p-NP-decanoate | 0.01 | 75 |
| p-NP-dodecanoate | 0.01 | 26 |
| p-NP-tetradecanoate | — | — |
| p-NP-hexadecanoate | — | — |
Km and Vmax were calculated from Lineweaver-Burk plots of enzyme activity on p-NP ester substrates by using a least-squares best fit of the Michaelis-Menten equation. —, no enzyme activity.
FIG. 4.
Phospholipase activity of purified EstA from L. lactis B1014 on the chromogenic substrates diC6dithioPC (short-chain phospholipid substrate) and diC12dithioPC (long-chain phospholipid substrate). The enzyme activity is expressed as micromoles of thiol groups released from the substrate per minute per milligram of protein.
DISCUSSION
The estA genes of L. lactis B1014 and MG1363 were cloned via reverse genetics with oligonucleotides designed on the basis of the published amino-terminal sequence of the purified tributyrin esterase from L. lactis E8 (24). The similarity of the estA sequences (84%) from B1014 and MG1363 is slightly higher than that of other genomic sequences from L. lactis subsp. lactis and L. lactis subsp. cremoris, the divergence of which is estimated to be between 20 and 30% (18). It is noteworthy that the N-terminal sequences of the E8, B1014, and MG1363 esterases are very similar to the one reported recently for L. lactis NCDO 763 (5). The predicted size of the estA gene product (29 kDa) is in accordance with that published for the monomers of the purified enzymes from L. lactis E8 and NCDO 763, which are organized as tetramers and trimers, respectively (5, 24). The identity between the deduced N-terminal sequence of EstA from B1014 and MG1363 and that of the enzymes purified from E8 and NCDO 763 (5, 24) shows that the N-terminal part of EstA is not subjected to any modification, except for the removal of the N-terminal methionine. The absence of a classical N-terminal secretion signal sequence in EstA is in agreement with the published intracellular location of the lactococcal esterases (5, 24). The promoter −35 and −10 regions, according to the identified transcription initiation site, conform to the promoter consensus sequence for lactococci and other gram-positive bacteria, including a TG dinucleoside located 1 bp upstream of the −10 sequence (14). Although rho-independent terminators, such as inverted repeat sequences potentially capable of forming stem-loop structures, are common in L. lactis, no obvious transcription termination structure was found downstream of the TAA stop codon. The absence of a putative stem-loop structure in this part of the sequence could result in a higher turnover of mRNA, which could explain the lack of success in detecting clear estA transcripts (data not shown). It is tempting to speculate that the noncoding region of the estA mRNA (233 bp upstream of the estA start codon) could play a role in the control of gene expression. Nucleotide sequences located between the promoter and the structural gene have been shown to be involved in a variety of regulatory mechanisms for many metabolic genes in gram-positive bacteria (6, 33, 41).
Lactococcal EstA has similarity with various proteins that have been grouped on the basis of their function and/or origin, i.e., xylan/cellulose-degrading enzymes including an acetyl esterase (20, 34), extracellular fibronectin-binding proteins (38, 40), and a group including various intracellular esterases (15, 23, 44) (Table 2). Alignment of these proteins, which showed highest homology to L. lactis B1014 EstA, revealed that the longest conserved sequence contained the typical GXSXG box that is found in serine hydrolases such as lipases, esterases, and some proteinases (8). This region has been suggested as the presumptive active site of human EstD, based on homology studies with several esterases (55). Some serine group reagents, such as phenylmethylsulfonyl fluoride or diisopropyl-fluorophosphate and Pefabloc, have been shown to inhibit the enzymatic activity of purified esterases of L. lactis ACA-DC 127 and NCDO 763, while inhibitors of metalloenzymes and cysteine enzymes did not have any effect (5, 48). These results, taken together, suggest that Ser121 of L. lactis B1014 EstA is the nucleophilic serine of the active site, which is usually well conserved in many hydrolytic enzymes and forms an acyl intermediate with the substrate. The EstA residues Asp202 and His231 present in conserved sequences located in the carboxyl terminus presumably also participate in catalysis as members of the triad; however, they have not been identified as such for any of the proteins listed in Table 2. The importance of a histidine residue for the esterase activity of L. lactis NCDO 763 has been pointed out by Chich et al. (5), who reported a strong inhibition of this activity by 3,4-dichloroisocoumarin, which binds to the His residue of the catalytic triad of serine enzymes. The residues Ser121, Asp202, and His231 of EstA conform to the typical catalytic triad of lipases and esterases that generally consists of Ser-Asp/Glu-His (8). No disulfide bridge is present in the lactococcal EstA sequence, which contains only a single cysteine residue at position 198, in contrast to several lipases and esterases that contain two or three conserved disulfide bridges important for substrate binding and/or recognition (8). EstA Cys198 forms no part of the active site, but it could be important for the enzymatic activity of EstA. This is supported by the observed inhibition of enzymatic activity in lactococcal esterases by Hg2+, which reacts with thiol groups (5, 48). A sulfhydryl group is also essential for the enzymatic activity of human EstD (32).
The estA genes from both lactococcal strains B1014 and MG1363 were overexpressed efficiently in L. lactis under the control of the nisA promoter with the nisin-controlled expression system (12, 13, 30). Hydrolysis of p-NP-butanoate by these esterase-overproducing strains increased from 28- to 170-fold, respectively, after induction with nisin compared with the uninduced cultures. Higher hydrolysis of tributyrin was also detected in NZ9800 carrying pNZ9308, which contains the estA gene from B1014 transcriptionally fused to the nisA promoter. These results indicate that tributyrin and p-NP-butanoate hydrolysis are catalyzed by the same enzyme. The disruption of the estA gene in MG1363 with an integration vector containing a thermosensitive replicon resulted in the complete loss of esterase activity. This activity was successfully restored when the esterase-deficient mutant (NZ9340) was complemented with a plasmid containing the estA gene from the wild-type strain. Consequently, our results demonstrate that EstA is the major esterase in L. lactis.
The substrate specificity of B1014 esterase was studied by using purified enzyme from the overproducing strain NZ9800 carrying pNZ9308. EstA from B1014 showed the highest activity against low-molecular-weight substrates (p-NP-butanoate and tributyrin). This preference for esters of short-chain fatty acids is characteristic of esterases, and it is shared by other lactococcal esterases (5, 24, 48). Lactococcal EstA hydrolyzed phospholipids with medium-chain fatty acid residues (diC6dithioPC), and the activity showed a typical hyperbolic dependence on substrate concentration similar to that of pancreatic phospholipase A2 on monomeric substrates (51). However, no increase in enzyme activity was observed at high diC6dithioPC concentrations, which will favor micelle formation, or against micellar diC12dithioPC. This is in contrast to the behavior of pancreatic phospholipase A2, whose catalytic efficiency rises abruptly when similar substrates are in the aggregated form (51). This indicates that lactococcal EstA lacks, at least with the phospholipid substrates assayed, the property of interfacial activation that is also characteristic of lipases.
It can be concluded that EstA does not seem to be an essential enzyme for L. lactis, as no difference in growth was observed between the wild-type and mutant strains in the medium and conditions used in this study, i.e., M17 medium supplemented with glucose and 30 or 37°C. It is difficult to ascertain the physiological role of this lactococcal esterase, but some options may be considered. Esterases from Lactococcus may be involved in the metabolism or turnover of bacterial lipids, taking into account that the enzyme has the ability to hydrolyze different soluble lipid substrates, including triacylglycerols and phospholipids. Lactic acid bacteria are also able to produce free fatty acids from mono- and diacylglycerides (45). This hypothesis is supported by an early study on lactococcal lipolytic activity that revealed rather intense activity in Lactococcus against its own neutral lipids (49). Interestingly, the clear homology found between the lactococcal esterase and FGH from humans, also known as esterase D (15), and Anabaena azollae, Saccharomyces cerevisiae, and P. denitrificans FGHs (9, 23, 44) suggests that this enzyme could play a role in detoxification. FGH is part of a glutathione-dependent formaldehyde detoxification pathway (23). The wide distribution of FGH (yeasts, bacteria, and mammals) has led these authors to suggest that this is a universal detoxification pathway, and it is possible that this pathway also plays a role in Lactococcus.
The availability of lactococcal esterase in high quantities will facilitate further kinetic and structural characterization of the enzyme and a closer definition of its natural lipid substrates. In addition, esterase-overproducing and esterase-deficient strains will allow us to determine the impact of this lactococcal enzyme on flavor formation in dairy products. The assessment of this function has been hampered up to now by the lack of isogenic strains with various esterase activities.
ACKNOWLEDGMENTS
We thank Frans Boer for expert technical assistance with protein purification. We are grateful to Michiel Kleerebezem and Gerrit Smit for critically reading the manuscript. We thank Niek Dekker and Ruud Dijkman (CBLE, University of Utrecht, Utrecht, The Netherlands) for the generous gift of diC6dithioPC and diC12dithioPC.
Leonides Fernández was the recipient of a grant from the Ministerio de Educación y Cultura of Spain and acknowledges the financial support of the Comisión Interministerial de Ciencia y Tecnología (CICYT project ALI95-0820) of Spain.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Bradford M M. A rapid and sensitive method for quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 3.Casadaban M J, Cohen S N. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J Mol Biol. 1980;138:179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- 4.Chiaruttini C, Millet M. Gene organization, primary structure and RNA processing analysis of a ribosomal RNA operon in Lactococcus lactis. J Mol Biol. 1993;230:57–76. doi: 10.1006/jmbi.1993.1126. [DOI] [PubMed] [Google Scholar]
- 5.Chich J F, Marchesseau K, Gripon J C. Intracellular esterase from Lactococcus lactis subsp. lactis NCDO 763: purification and characterization. Int Dairy J. 1997;7:169–174. [Google Scholar]
- 6.Chopin A. Organization and regulation of genes for amino acid biosynthesis in lactic acid bacteria. FEMS Microbiol Rev. 1993;12:21–38. doi: 10.1111/j.1574-6976.1993.tb00011.x. [DOI] [PubMed] [Google Scholar]
- 7.Crow V L, Holland R, Pritchard G G, Coolbear T. The diversity of potential cheese ripening characteristics of lactic acid starter bacteria. 2. The levels and subcellular distributions of peptidase and esterase activities. Int Dairy J. 1994;4:723–742. [Google Scholar]
- 8.Cygler M, Schrag J D, Sussman J L, Harel M, Silman I, Gentry M K, Doctor B P. Relationship between sequence conservation and three-dimensional structure in a large family of esterases, lipases, and related proteins. Protein Sci. 1993;2:366–382. doi: 10.1002/pro.5560020309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Degrassi G, Uotila L, Klima R, Venturi V. Purification and properties of an esterase from the yeast Saccharomyces cerevisiae and identification of the encoding gene. Appl Environ Microbiol. 1999;65:3470–3472. doi: 10.1128/aem.65.8.3470-3472.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Mendonca Lima L, Content J, van Heuverswyn H, Degrave W. Nucleotide sequence of the gene coding for the 85-B antigen of Mycobacterium leprae. Nucleic Acids Res. 1991;19:5789. doi: 10.1093/nar/19.20.5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Derewenda Z S. Structure and function of lipases. Adv Protein Chem. 1994;45:1–52. doi: 10.1016/s0065-3233(08)60637-3. [DOI] [PubMed] [Google Scholar]
- 12.de Ruyter P G G A, Kuipers O P, de Vos W M. Controlled gene expression systems for Lactococcus lactis with the food-grade inducer nisin. Appl Environ Microbiol. 1996;62:3662–3667. doi: 10.1128/aem.62.10.3662-3667.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Ruyter P G G A, Kuipers O P, Beerthuyzen M M, van Alen-Boerrigter I, de Vos W M. Functional analysis of promoters in the nisin gene cluster of Lactococcus lactis. J Bacteriol. 1996;178:3434–3439. doi: 10.1128/jb.178.12.3434-3439.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Vos W M, Simons G. Gene cloning and expression systems in lactococci. In: Gasson M J, de Vos W M, editors. Genetics and biotechnology of lactic acid bacteria. Glasgow, United Kingdom: Blackie Academic and Professional; 1994. pp. 52–105. [Google Scholar]
- 15.Eiberg H, Mohr J. Identity of the polymorphisms for esterase D and S-formylglutathione hydrolase in red blood cells. Hum Genet. 1986;74:174–175. doi: 10.1007/BF00282085. [DOI] [PubMed] [Google Scholar]
- 16.Feng D F, Doolittle R F. Progressive sequence alignment as a prerequisite to correct phylogenetic trees. J Mol Evol. 1987;25:351–360. doi: 10.1007/BF02603120. [DOI] [PubMed] [Google Scholar]
- 17.Gasson M J. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J Bacteriol. 1983;154:1–9. doi: 10.1128/jb.154.1.1-9.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goddon J J, Delorme C, Ehrlich S D, Renault P. Divergence of genomic sequences between Lactococcus lactis subsp. lactis and Lactococcus lactis subsp. cremoris. Appl Environ Microbiol. 1992;58:4045–4047. doi: 10.1128/aem.58.12.4045-4047.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Godfrey A. Lipases for industrial use. Lipid Technol. 1995;7:58–61. [Google Scholar]
- 20.Grépinet O, Chebrou M C, Béguin P. Purification of Clostridium thermocellum xylanase Z expressed in Escherichia coli and identification of the corresponding product in the culture medium of C. thermocellum. J Bacteriol. 1988;170:4576–4581. doi: 10.1128/jb.170.10.4576-4581.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grépinet O, Chebrou M C, Béguin P. Nucleotide sequence and deletion analysis of the xylanase gene (xynZ) of Clostridium thermocellum. J Bacteriol. 1988;170:4582–4588. doi: 10.1128/jb.170.10.4582-4588.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gribskov M, McLachlan A D, Eisenberg D. Profile analysis: detection of distantly related proteins. Proc Natl Acad Sci USA. 1987;84:4355–4358. doi: 10.1073/pnas.84.13.4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harms N, Ras J, Reijnders W N M, van Spanning R J M, Stouthamer A H. S-Formylglutathione hydrolase of Paracoccus denitrificans is homologous to human esterase D: a universal pathway for formaldehyde detoxification? J Bacteriol. 1996;178:6296–6299. doi: 10.1128/jb.178.21.6296-6299.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holland R, Coolbear T. Purification of tributyrin esterase from Lactococcus lactis subsp. cremoris. J Dairy Res. 1996;63:131–140. doi: 10.1017/s0022029900031605. [DOI] [PubMed] [Google Scholar]
- 25.Joliff G, Mathieu L, Hahn V, Bayan N, Duchiron F, Renaud M, Shechter E, Leblon G. Cloning and nucleotide sequence of the csp1 gene encoding PS1, one of the two major secreted proteins of Corynebacterium glutamicum: the deduced N-terminal region of PS1 is similar to the Mycobacterium antigen 85 complex. Mol Microbiol. 1992;6:2349–2362. doi: 10.1111/j.1365-2958.1992.tb01410.x. [DOI] [PubMed] [Google Scholar]
- 26.Kinsella J E, Hwang D H. Enzymes of Penicillium roquefortii involved in the biosynthesis of cheese flavor. Crit Rev Food Sci Nutr. 1976;8:191–228. doi: 10.1080/10408397609527222. [DOI] [PubMed] [Google Scholar]
- 27.Kuipers O P, Boot H J, de Vos W M. Improved site-directed mutagenesis method using PCR. Nucleic Acids Res. 1991;19:4558. doi: 10.1093/nar/19.16.4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuipers O P, Beerthuyzen M M, Siezen R J, de Vos W M. Characterization of the nisin gene cluster nisABTCIPR of Lactococcus lactis: requirement of expression of the nisA and nisI genes for development of immunity. Eur J Biochem. 1993;216:281–291. doi: 10.1111/j.1432-1033.1993.tb18143.x. [DOI] [PubMed] [Google Scholar]
- 29.Kuipers O P, de Ruyter P G G A, Kleerebezem M, de Vos W M. Controlled overproduction of proteins by lactic acid bacteria. Trends Biotechnol. 1997;15:135–140. doi: 10.1016/S0167-7799(97)01029-9. [DOI] [PubMed] [Google Scholar]
- 30.Kuipers O P, de Ruyter P G G A, Kleerebezem M, de Vos W M. Quorum sensing-controlled gene expression in lactic acid bacteria. J Biotechnol. 1998;64:15–21. [Google Scholar]
- 31.Lee E Y H, Lee W H. Molecular cloning of the human esterase D gene, a genetic marker of retinoblastoma. Proc Natl Acad Sci USA. 1986;83:6337–6341. doi: 10.1073/pnas.83.17.6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee W-H, Bookstein R, Wheatley W, Benedict W F, Lee E Y H P. Purification, biochemical characterization, and biological function of human esterase D. Proc Natl Acad Sci USA. 1986;83:6790–6794. doi: 10.1073/pnas.83.18.6790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luesink E J, Kuipers O P, de Vos W M. Regulation of the carbohydrate metabolism in Lactococcus lactis and other lactic acid bacteria. Lait. 1998;78:69–76. [Google Scholar]
- 34.Lüthi E, Love D R, McAnulty J, Wallace C, Caughey P A, Saul D, Bergquist P L. Cloning, sequence analysis, and expression of genes encoding xylan-degrading enzymes from the thermophile “Caldocellum saccharolyticum.”. Appl Environ Microbiol. 1990;56:1017–1024. doi: 10.1128/aem.56.4.1017-1024.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maguin E, Prévost H, Ehrlich S D, Gruss A. Efficient insertional mutagenesis in lactococci and other gram-positive bacteria. J Bacteriol. 1996;178:931–935. doi: 10.1128/jb.178.3.931-935.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McNeill G P, Connolly J F. A method for the quantification of individual free fatty acids in cheese: application to ripening of cheddar-type cheeses. Irish J Food Sci Technol. 1989;13:119–128. [Google Scholar]
- 37.Meyers S A, Cuppett S L, Hutkins R W. Lipase production by lactic acid bacteria and activity on butter oil. Food Microbiol. 1996;13:383–389. [Google Scholar]
- 38.Ohara N, Matsuo K, Yamaguchi R, Yamazaki A, Tasaka H, Yamada T. Cloning and sequencing of the gene for the α antigen from Mycobacterium avium and mapping of B-cell epitopes. Infect Immun. 1993;61:1173–1179. doi: 10.1128/iai.61.4.1173-1179.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pearson W R, Lipman D J. Improved tools for biological sequence analysis. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rinke de Wit T F, Bekelie S, Osland A, Wieles B, Janson A A M, Thole J E R. The Mycobacterium leprae antigen 85 complex gene family: identification of the genes for the 85A, 85C, and related MPT51 proteins. Infect Immun. 1993;61:3642–3647. doi: 10.1128/iai.61.9.3642-3647.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rutberg B. Antitermination of transcription of catabolic operons. Mol Microbiol. 1997;23:413–421. doi: 10.1046/j.1365-2958.1997.d01-1867.x. [DOI] [PubMed] [Google Scholar]
- 42.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 43.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shaw W H, Arioli T, Plazinski J. Cloning and sequencing of a S-formylglutathione hydrolase (FGH) gene from the cyanobacterium Anabaena azollae (accession no AF035558) (PGR98-024) Plant Physiol. 1998;116:868. [Google Scholar]
- 45.Stadhouders J, Veringa H A. Fat hydrolysis by lactic acid bacteria in cheese. Neth Milk Dairy J. 1973;27:77–91. [Google Scholar]
- 46.Takano M, Ohara N, Mizuno A, Yamada T. Cloning, sequencing and expression in Escherichia coli of the gene for α antigen from Mycobacterium scrofulaceum. Scand J Immunol. 1994;40:165–170. doi: 10.1111/j.1365-3083.1994.tb03446.x. [DOI] [PubMed] [Google Scholar]
- 47.Tinoco I, Jr, Borer P N, Dengler B, Levine M D, Uhlenbeck O C, Crothers D M, Gralla J. Improved estimation of secondary structure in ribonucleic acids. Nat New Biol. 1973;246:40–41. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]
- 48.Tsakalidou E, Kalantzopoulos G. Purification and partial characterization of an esterase from Lactococcus lactis ssp. lactis strain ACA-DC 127. Lait. 1992;72:533–543. doi: 10.1111/j.1365-2672.1992.tb01828.x. [DOI] [PubMed] [Google Scholar]
- 49.Umemoto Y, Sato Y. Lipolysis by intracellular lipase of Streptococcus lactis against its neutral lipids obtained by growth at low temperature. Agric Biol Chem. 1978;42:221–225. [Google Scholar]
- 50.Verger R. ‘Interfacial activation’ of lipases: facts and artifacts. Trends Biotechnol. 1997;15:32–38. [Google Scholar]
- 51.Volwerk J J, Dedieu A G R, Verheij H M, Dijkman R, de Haas G H. Hydrolysis of monomeric substrates by porcine pancreatic (pro)-phospholipase A2. The use of a spectrophotometric assay. Recl Trav Chim Pays-Bas Belg. 1979;98:214–220. [Google Scholar]
- 52.Vos P, van Asseldonk M, van Jeveren F, Siezen R J, Simons G, de Vos W M. A maturation protein is essential for the production of active forms of Lactococcus lactis SK11 serine proteinase located in or secreted from the cell envelope. J Bacteriol. 1989;171:2795–2802. doi: 10.1128/jb.171.5.2795-2802.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wells J M, Wilson P W, Le Page R W F. Improved cloning vectors and transformation procedure for Lactococcus lactis. J Appl Bacteriol. 1993;74:629–636. doi: 10.1111/j.1365-2672.1993.tb05195.x. [DOI] [PubMed] [Google Scholar]
- 54.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 55.Young L J S, Lee E Y H P, To H, Bookstein R, Shew J Y, Donoso L A, Sery T, Giblin M, Shields J A, Lee W H. Human esterase D gene: complete cDNA sequence, genomic structure, and application in the genetic diagnosis of human retinoblastoma. Hum Genet. 1988;79:137–141. doi: 10.1007/BF00280552. [DOI] [PubMed] [Google Scholar]