Abstract
Cells sense a variety of extracellular growth factors and signaling molecules through numerous distinct receptor tyrosine kinases (RTKs) on the cell surface. In many cases, the same intracellular signaling molecules interact with more than one type of RTK. How signals from different RTKs retain the identity of the triggering receptor and how (or if) different receptors may synergize or compete remain largely unknown. Here we utilize an experimental strategy, combining microscale patterning and single-molecule imaging, to measure the competition between ephrin-A1:EphA2 and epidermal growth factor (EGF):EGF receptor (EGFR) ligand-receptor complexes for the shared downstream signaling molecules, Grb2 and SOS. The results reveal a distinct hierarchy, in which newly formed EGF:EGFR complexes outcompete ephrin-A1:EphA2 for Grb2 and SOS, revealing a type of negative crosstalk interaction fundamentally controlled by chemical mass action and protein copy number limitations.
Significance
Characterization of spatially resolved ephrin-A1:EphA2 and EGF:EGFR signaling clusters within the same cell reveal competition for Grb2 and SOS. Newly formed EGF:EGFR clusters outcompete already-active ephrin-A1:EphA2 clusters for Grb2 and SOS. Competition for Grb2 and SOS is driven by mass action and is dependent on receptor expression levels.
Introduction
There are 58 different receptor tyrosine kinases (RTKs) in the human genome (1), many of which can be expressed simultaneously on the same cell. These RTKs interact with a range of different growth factors and extracellular signaling molecules and trigger a variety of distinctive cellular responses. This broad collection of cell-surface RTKs feeds into a smaller number of conserved downstream signaling pathways, including Ras/mitogen-activated protein kinase (MAPK), phosphoinositide 3-kinase (PI-3K)/Akt, and ubiquitylation/deubiquitylation signaling (2, 3, 4). Kinases such as ERK, AKT, JNK, and LATS1, usually thought of as terminal nodes of their respective signaling cascades in the cytoplasm, then extend the RTK signals by interacting with various transcription factors (e.g., Fos, mammalian target of rapamycin [mTOR], Jun, p53, Yap), which drive cellular responses such as proliferation, differentiation, and apoptosis (2,5,6). Information traffic through RTK signaling pathways is a key determinant of cell fate, and dysregulation of these processes is known to cause cancer (7,8).
Here we introduce an experimental strategy to directly probe competition between different RTKs for the same downstream signaling molecules. We focus on epidermal growth factor (EGF):EGF receptor (EGFR) and ephrin-A1:EphA2, both of which interact with the adaptor protein Grb2 (binding to phosphotyrosine [pY] sites on the receptor) and the guanine nucleotide exchange factor SOS (binding to the Grb2 SH3 domain). SOS, in turn, activates Ras and initiates the MAPK pathway (9). Grb2 and SOS thus provide a key linkage between receptor triggering and MAPK activation. The EGF ligand for EGFR is a soluble molecule, whereas the ephrin-A1 ligand for EphA2 is a membrane surface protein, and ephrin-A1:EphA2 interactions naturally occur across the junction between two cells. The juxtacrine signaling geometry of ephrin-A1:EphA2 has been successfully reconstituted between ephrin-A1 functionalized supported lipid bilayers (SLBs) and live EphA2-expressing cells (10, 11, 12, 13, 14). The different signaling configurations between EphA2 and EGFR provide a convenient means to spatially segregate these RTKs from one another. This is achieved using a patterned supported membrane system (14, 15, 16, 17, 18, 19, 20, 21), in which circular corrals (3–5 μm diameter) of ephrin-A1 functionalized membrane are arrayed within a region of RGD-functionalized PEG (poly(ethylene glycol)), bound directly to the underlying solid silica substrate (13,22,23). Ephrin-A1 ligand diffuses freely within individual membrane corrals, thus allowing functional clustering and reorganization upon interaction with EphA2 on an interacting cell. The ephrin-A1, however, remains confined to the membrane corrals, leaving the RGD-functionalized regions of the surface free of ephrin-A1. Signaling clusters of ephrin-A1:EphA2 thus remain spatially concentrated, whereas EGF:EGFR are more randomly distributed throughout the cell surface. The RGD provides a substrate for formation of integrin focal adhesions and serves the purpose of maintaining a well-defined interface between the cell and substrate.
Single-molecule imaging and single-particle tracking photoactivated localization microscopy (sptPALM) are used to resolve Grb2 and SOS interactions with the two different activated receptors in real time. Constructs of full-length Grb2 and SOS, fused to the photoactivatable Eos molecule, were imaged in a streaming acquisition mode to reveal both the recruitment on-rate and dwell time. Although some long-lived species are present (24, 25, 26, 27), a majority of the Grb2 and SOS exhibit a dynamic equilibrium between receptor-bound and cytoplasmic states. A notable observation is that a stable amount of Grb2 and SOS is established at ephrin-A1:EphA2 signaling clusters in the absence of EGF stimulation of EGFR. Upon addition of EGF, a dramatic shift of both Grb2 and SOS away from the EphA2 signaling clusters to the newly active EGFR is evident in the MDA-MB-231 human breast epithelial cancer cell line. This appears to be largely driven by mass action and is reflective of the vast increase in the number of available pY sites for Grb2 binding after EGFR activation. In MCF-10A cells, which express far less EGFR than the MDA-MB-231 cells, EGFR activation by EGF does not produce a detectable impact on Grb2 localization to EphA2:ephrin-A1 signaling complexes. We perform detailed analyses of the kinetics of these processes and examine how differences in affinity between Grb2 and receptor could lead to a signaling hierarchy in which some receptors may be more competitive for downstream adaptors than others. Fundamentally, these experiments are enabled by the ability to spatially segregate receptors on the same cell surface, thus allowing direct observation of competitive interactions with the same pool of cytoplasmic signaling molecules.
Materials and methods
SLB
Phospholipid vesicles were prepared using previously described methods (28). The desired lipids were mixed in a chloroform solution and then the chloroform was evaporated using a rotary evaporator, resulting in the formation of a thin lipid film on the surface of the glass rotary. The lipids were thoroughly dried under a stream of N2 and hydrated with 2 mL of deionized water, resulting in a final lipid concentration of 0.5 mg/mL. The hydrated lipid solution was kept chilled on ice and sonicated using a probe-tip sonicator to break large vesicles into small unilamellar vesicles (SUVs). Debris was removed by centrifugation at 20,000 × g at 4°C for 3 h, and the supernatant containing the SUVs was transferred to a fresh glass tube. Lipid vesicles were stored at 4°C until further use. Two kinds of lipid mixtures were used in this study: one containing 96% 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC) plus 4% 1,2-dioleoylsn-glycero-3-[(N-(5-amino-1-carboxypentyl)iminodiacetic acid)] (Ni-NTA-DOGS) nickel salt lipids, and the other containing 99% DOPC plus 1% Marina Blue-1,2-dihexadecanoyl-sn-glycero-3-phosphoethanolamine (Marina Blue-DHPE).
To prepare SLBs on the substrate, glass coverslips were first cleaned by sonication in a 1:1 mixture of isopropanol and water for 30 min. After extensive rinsing with water, coverslips were further cleaned by incubating in 50% H2SO4 overnight, followed by storage in deionized water. Immediately prior to usage, glass coverslips were then treated with oxygen plasma for 2 min, rinsed with deionized water, and dried under an N2 stream. Lipid bilayers were self-assembled on the glass coverslips by incubating a 1:1 mixture of lipid vesicles and Tris-buffered saline (TBS; Sigma-Aldrich) for 5 min. The bilayer-containing coverslips were assembled into Attofluor Cell Chambers (Thermo Fisher) for further experiments.
Patterned substrate of SLB and immobilized polymer
The detailed methods used to prepare a hybrid pattern of fluid SLB and immobilized polymer have been published (13,23) in another journal. Briefly, clean coverslips were incubated with PLL-g-PEG-biotin (50%, Susos) for 2 h (Fig. S1 A). The coated coverslips were then UV etched with a designed photomask to generate patterned substrate with PLL-g-PEG-biotin and bare glass surfaces (Fig. S1 B). Lipid vesicles were then deposited onto the glass coverslips and bilayers were self-assembled on the bare glass surface only (Fig. S1 C). The patterned coverslips were each assembled into Attofluor Cell Chambers (Thermo Fisher) for further experiments.
Patterns were regular arrays of circles with diameters of D = 2–5 μm. The center-to-center distance between the neighboring circle is 3 × D. SLBs freely diffuse in the isolated circular patches, surrounded by immobilized PLL-g-PEG-biotin.
Surface functionalization
After lipid bilayer deposition, substrates were incubated with a 0.05% bovine serum albumin (BSA; Sigma-Aldrich) solution for 2 h to block non-specific binding sites. The hybrid substrates were then incubated with a 1 μg/mL solution of DyLight-405 NeutrAvidin or NeutrAvidin (Thermo Fisher) for 30 min. The cell chamber was then thoroughly rinsed, and substrates were incubated for 60 min with a solution of 5 nM ephrin-A1-Alexa 680 and 1 μg/mL of RGD-PEG-PEG-biotin (Peptides International) (Fig. S1 D and E). Ephrin-A1 was expressed with a 10-histidine tag and purified from insect cell culture as published earlier (12), and labeled with Alexa 680 fluorophore (Thermo Fisher) following the vendor’s protocol. Ephrin-A1 density on the supported membrane was calibrated with quantitative fluorescence microscopy as published previously (29).
In all cell experiments, the protein incubation solutions were exchanged by rinsing the substrate with imaging buffer (25 mM Tris, 140 mM NaCl, 3 mM KCl, 2 mM CaCl2, 1 mM MgCl2, and 5.5 mM D-glucose) and warmed up to 37°C. Cells were then added into the chamber and allowed to engage and interact with the surface at the conditions indicated.
Cell culture, transfection, and preparation for live cell experiments
MDA-MB-231, MCF-10A, HEK239 cells were grown in DMEM (high glucose) (Thermo Fisher) supplemented with 10% fetal bovine serum (FBS) (Thermo Fisher) and 1% penicillin/streptomycin (Thermo Fisher), at 37°C and in an atmosphere of 5% CO2. MCF-10A cells were cultured in DMEM/F12 (Thermo Fisher) supplemented with 5% horse serum (Thermo Fisher), 20 ng/mL EGF (Sigma), 0.5 mg/mL hydrocortisone (Sigma), 100 ng/mL cholera toxin (Sigma), 19 μg/mL insulin (Sigma), and 1% penicillin/streptomycin.
MDA-MB-231 cells were first seeded on 35-mm culture dishes. One day before experiments, cells were transfected with 0.2–1 μg of the plasmid of interest with Lipofectamine 2000 (Thermo Fisher). Grb2-tdEos, SHC1 SH2-tdEos, and Myr (myristolation tag)-tdEos, were generously provided by Dr. Bruce J. Mayer’s and Ji Yu’s labs (University of Connecticut Health Center, USA). SOS-mEos3.2 and CAAX-mEos were constructed in the lab. Plasmid transfection was followed by the vendor’s protocol. Cell imaging was performed within 24 h after transfection.
For live cell experiments, cells were washed with PBS and detached by incubating with enzyme free, Hank’s-based cell dissociation buffer (Thermo Fisher) for 5–10 min at 37°C. Detached cells were gently pipetted to produce single cells. The cell suspension was centrifuged, and the cells were resuspended in the imaging buffer. The cells were then added into pre-warmed cell chambers and allowed to interact with the substrates.
Cell imaging
An Eclipse Ti inverted microscopy (Nikon) with a CSU-X1 confocal spinning disk unit (Yokogawa) and Evolve electron multiplying charge-coupled device (EMCCD) camera (photometrics) were used for most imaging experiments. For epifluorescence, reflection interference contrast microscopy (RICM), and total internal reflection fluorescence (TIRF) imaging, an Eclipse Ti inverted microscope (Nikon) with the same camera was used. Images were collected in Metamorph (Molecular Devices) and analyzed with ImageJ (National Institutes of Health) and Matlab. TIRF microscopy was performed with a 100× TIRF objective with an extra 1.5× magnification with a numerical aperture of 1.49 (Nikon) and an iChrome MLE-L multilaser engine as a laser source (Topica Photonics). Throughout live cell imaging, the cell chamber was kept at 37°C and CO2 environment. The fluorescence recovery after photobleaching (FRAP) experiment was performed in the W1 spinning disk confocal microscope, with Nikon Eclipse Ti-E inverted microscope body. Localized laser exposure and imaging acquisition were controlled with iLAS and Metamorph software.
Immunostaining and western blotting
Following 1–2 h of incubation on the substrates with imaging buffer, samples were rinsed with Dulbecco’s PBS. The cells were fixed with 4% paraformaldehyde for 15 min at 37°C, and the cell membrane was permeabilized with 0.1% Triton-X for 3 min at room temperature. After blocking with 1 % BSA overnight at 4°C, cells were immunostained with primary antibodies and fluorescent secondary antibodies. The primary antibodies used were rabbit anti-EphA2 (CST, 7997S), rabbit anti-pY588-EphA2 (CST, 12677S), and Alexa 488-labeled phalloidin (Thermo Fisher A12379). Secondary antibodies were goat anti-rabbit or goat anti-mouse antibodies conjugated with Alexa fluorophores (Alexa 488, Alexa 568, Alexa 594, Alexa 647; Thermo Fisher).
Western blot analysis of EphA2 and EGFR phosphorylation was done after 1-h incubation of cells with substrates. Cells were lysed by lysis buffer followed by standard western blotting procedures. Densitometry analysis of chemiluminescence for quantification of phosphorylation levels was performed using ImageJ.
Single-molecule imaging
To improve the accuracy of measuring Grb2 single-molecule dynamics on the cell membrane, we clarified the non-membrane-bound Grb2-tdEos molecules from membrane-bound Grb2 with the right TIR angle. Thus, tdEos was tagged with or without a myristoylation membrane targeting tag. Membrane-bound myr-tdEos showed a clear bright dot, but cytoplasmic tdEos looked blurred under TIR illumination. Here, we defined membrane-bound Eos tagged molecules as a clear diffraction limit dot having more than two consecutive frames. Thus, non-specific blur dots including one-frame dots were filtered out in trajectory analysis.
To minimize the inevitable photobleaching effect and maximize signal to noise, the laser power and acquisition settings including video rate had incorporated each other an optimal manner. Obviously, the photobleaching time of fluorescence protein is significantly and inversely proportional to the illumination power. Thus, the same optical conditions were carefully set when compared with the dwell time between each dataset for statistical analysis. Under the same TIRF conditions, photobleaching times of individual myr-tdEos molecules were measured and compared with dwell times of Grb2-tdEos. Photobleaching time of myr-tdEos was significantly longer than Grb2-tdEos dwell time (Fig. S16 B). These experimental conditions verify that the measured dwell time (λoff−1) can reflect the protein-protein interaction time on the cell membrane.
To access the kinetic equilibrium of Grb2-tdEos and SOS-mEos3.2 in ephrin-A1:EphA2 clusters in SLB, the initial aggregated and non-specific auto fluorescence signals were photobleached until single-molecule fluorescence dots appeared and disappeared at constant rate in prior to the addition of EGF (Video S9) (Fig. S13 A–C).
Single-particle tracking
Using a standard cross-correlation single-particle tracking method (30, 31, 32), the position of the centroid of the bright dot was defined and xy coordinates were connected through frame to frame using the nearest-neighbor method. The searching area for the position of the particle in the subsequent image frames was estimated by calculating the diffusion surface area using reported diffusion coefficients of SH2 domains proteins on phosphorylated EGFR (33). To avoid crowdedness and trajectory intermixing, we kept the molecular density on the image to less than 0.4 molecules per μm2 via a pulse of UV 404 nm beam.
In vivo sptPALM analysis
A 2D projection of xy coordinates of the individual molecules was used to generate a single-particle tracking photoactivated localization microscopy (sptPALM) image. We defined two sptPALM binding maps here; sptPALM-total binding map and sptPALM-new binding map. A concept similar to the normal sptPALM, which was the event counting basis (34), the sptPALM-total binding map was created by replacing the xy coordinates of every single-molecule event with a Gaussian point spread function (PSF) of 50 nm FWHM (full width half maximum) (Octan plugin in ImageJ, by Dr. Ji Yu). The original pixel size was 10 times sub-pixelated. FWHM and pixel size can be adjustable. By integrating these events for a certain amount of image sequences in a single-molecule movie, sptPALM-total binding map sequences were created to capture the spatiotemporal dynamics changes of Grb2 and SOS on the membrane. In this paper, the term sptPALM is equivalent to sptPALM-total binding map.
The concept of sptPALM-new binding map bases the molecule identification, so thus trajectory number basis. The first xy coordinates of individual trajectories were replaced by a Gaussian PSF. sptPALM-new binding sequences were created by integrating these events for a certain amount of image sequences from single-molecule movies. Thus, this sequence map directly reflects the new binding rate γon(t) for acquisition time.
Trajectory classification inside and outside SLB
The trajectories of molecules were classified into subgroups of SLB and background regions. To do this, a binary image of SLB locations was created and used as a mask. Using polygonal functions implemented in Matlab, all trajectories were tested for whether they resided inside, outside, or crossing SLBs.
Dwell time λoff−1(t) measurement
The dwell time λoff−1 of individual Grb2-tdEos and SOS-mEos3.2 molecules was calculated by multiplying the length of trajectory by the time interval of the single-molecule movie. Time-dependent mean λoff−1(t) was obtained by integrating individual λoff−1 for a specific number of image sequences before and after EGF. The distribution of λoff−1 at each time bin was fitted with a second-order exponential decay function, providing two characteristic time constants (Fig. S16 B). The fast time constant was almost the same for every dataset of molecules due to the dominant short photobleaching time of the fluorescence protein. However, the slower time constant varied significantly from data to data and before and after EGF stimulation. Thus, here we used the slower time constant for dataset comparison. The one-frame trajectories were excluded due to the predominant fast photobleaching population and possible non-specific membrane-binding or membrane-closing events. However, note that even including one-frame populations, the slower time constant value was not changed.
New binding rate γon(t) measurement
The new binding rate γon (t) (#/s/μm2) of specific molecules was calculated by counting the total number of trajectories in the ephrin-A1:EphA2 or EGF:EGFR clusters for the specific number of image sequences. The density of the new binding frequency was obtained by dividing the observed areas. Although we tried to control the similar density of single-molecule events by a UV 405 nm beam, it is difficult to synchronize the number of photoactivated molecules from cell to cell. Therefore, the comparisons between different samples and molecules are not meaningful here. However, a comparison of time-varying γon−1(t) in the same sample, such as γon-Grb2−1(t) and γon-sos−1(t) before and after EGF stimulation is reasonable.
Results
Spatially resolved ephrin-A1:EphA2 and EGF:EGFR signaling clusters on the same cell surface
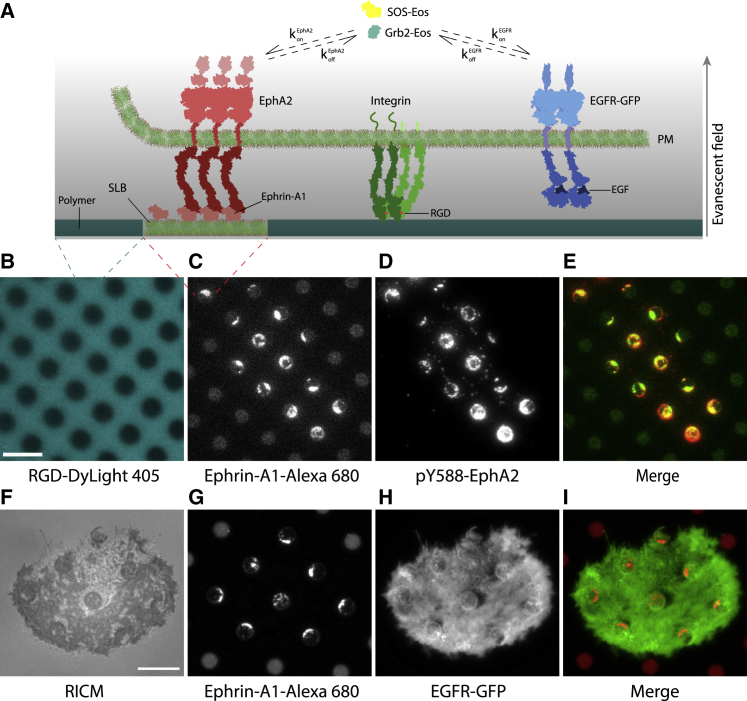
In order to differentiate signaling activity from EphA2 and EGFR RTKs on the same cell, we developed a micropatterned-SLB system to isolate the membrane-bound ephrin-A1 signaling clusters with EphA2 from soluble EGF signaling clusters with EGFR (Fig. 1 A). The micropatterned SLB was conjugated to ephrin-A1-Alexa 680 via Ni-NTA-His interaction, and the ephrin-A1 ligands freely diffused on the SLB surface as indicated by the rapid fluorescence recovery after photobleaching (Figs. S1 F and S3). To guide integrin signal-induced cell spreading on the substrate, the background region was functionalized with Arg-Gly-Asp (RGD) peptides through labeled with DyLight 405 (Fig. 1 B, blue). The detailed protocols of SLB and patterning are in the section “Materials and methods” (Fig. S1) and previous reports (13,22). We first chose to examine human breast cancer cells, MDA-MB-231, because this cell line overexpresses both EGFR and EphA2 receptors (35,36).
Figure 1.
Spatial segregation of EphA2 from EGFR signaling complexes. (A) Schematic representation of the micropatterned SLB substrate platform used here. Fluorescently labeled ephrin-A1 functionalized on SLB corrals binds to EphA2 receptors on the live cell. The ephrin-A1:EphA2 complexes form clusters, which recruit cytoplasmic proteins Grb2 and SOS, among others. Soluble EGF binds to and activates EGFR, resulting in the recruitment of Grb2 and SOS to EGF:EGFR complexes. Receptor clustering and activation and membrane recruitment of Grb2 and SOS can be monitored in real time by TIRF imaging of individual, live cells interacting with the micropatterned substrate. Polymer-coated regions of the substrates, which surround the membrane corrals, are functionalized with RGD peptide to allow integrin-mediated cell interaction and spreading on the micropatterned substrate. (B–E) Epifluorescence images of polymer-functionalized RGD-DyLight 405 (B) and SLB-functionalized ephrin-A1-Alexa 680 (C), TIRF image (immunofluorescence) of a cell stained with anti-pY588 EphA2 antibody (D) and merge of ephrin-A1-Alexa 680 and anti-pY588 EphA2 (E). (F–I) RICM image of an MDA-MB-231 cell interacting with a micropatterned hybrid substrate showing adhesion (F), epifluorescence image of SLB-functionalized ephrin-A1-Alexa 680 (G), TIRF image of EGFR-GFP in the absence of EGF (H), and merge of ephrin-A1-Alexa 680 and EGFR-GFP (I). Bars are 5 μm in (B) and 10 μm in (F). To see this figure in color, go online.
To verify ephrin-A1:EphA2 signal cluster formation between the cell and patterned SLB, we performed multi-channel time-lapse imaging including TIRF and RICM. RICM indicates both cell adhesion to the substrate (reflected by out-of-phase contrast dark signal in outside SLB) (Fig. 1 F) and ephrin-A1 binding and clustering with cell-surface EphA2, revealed by adhesions within the SLB corrals (Figs. 1 F and S3 A) (37). No adhesions were observed within the membrane corral regions in control SLBs without ephrin-A1 (Fig. S6). Next, in the blue channel, we confirmed that the RGD-DyLight 405 signal was evenly distributed in the background with a sharp edge at the SLB boundaries (Fig. 1 B, blue). In the red channel, we observed ephrin-A1-Alexa 680 signals only in SLBs, which were clustered in only those corrals that came in contact with cells; SLB corrals outside cellular contacts showed homogeneously distributed ephrin-A1 (Fig. 1 C and G). Using an EphA2-selective anti-phosphotyrosine antibody (pY588) labeled with Alexa Fluor 488 (Fig. 1 D), we confirmed ephrin-A1 and pY-EphA2 signals were colocalized (Fig. 1 E).
We next examined the physical interference of the ephrin-A1:EphA2 clusters on EGFR membrane dynamics by transfecting the cells with GFP-labeled EGFR. In the absence of EGF, the EGFR-GFP signal was homogeneously distributed throughout the cell membrane, including over the SLB corrals (Fig. 1 H and I). Additionally, we measured similar diffusion rates of EGFR inside (D = 0.13 μm2s−1) and outside SLB corrals (D = 0.10 μm2s−1) (Fig. S2) (Video S1), suggesting the patterned ephrin-A1:EphA2 signaling clusters imposed minimal structural interference with EGFR mobility within the membrane.
Grb2 and SOS recruitment dynamics to ephrin-A1:EphA2 signal clusters
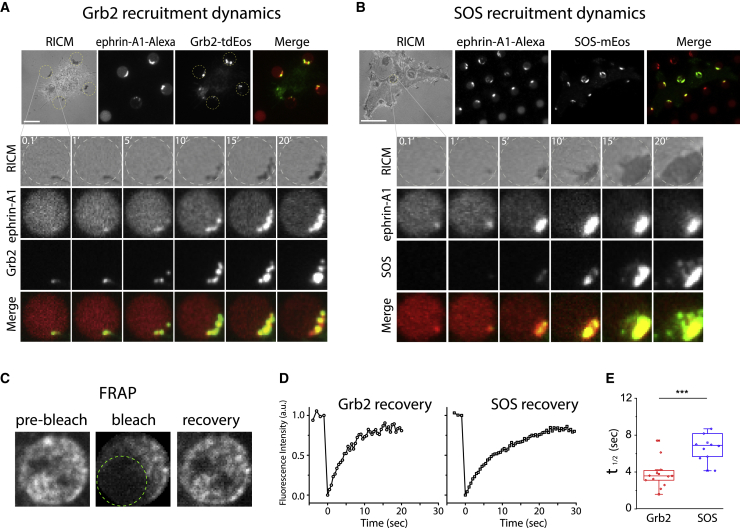
We next characterized the recruitment dynamics of Grb2 and SOS, which are upstream activators of the Ras-Raf-MEK/ERK pathway, to ephrin-A1:EphA2 clusters in the SLB. For these experiments, MDA-MB-231 cells expressing Grb2-tdEos or SOS-mEos3.2 fusion proteins were spread on ephrin-A1-Alexa 680 functionalized SLB-hybrid substrates. In parallel, we used multi-channel time-lapse TIRF and RICM imaging to capture cell spreading and identify SLB corrals (Fig. S3) (Videos S2 and S3). At the same time, ephrin-A1 clustering and Grb2-tdEos or SOS-mEos3.2 SLB recruitment were monitored, respectively (Fig. 2 A and Video S2 for Grb2-tdEos, Fig. 2 B; Video S3 for SOS-mEos3.2). The multi-channel image sequences extracted from the movies visualized ephrin-A1 engagement with cell-surface EphA2 receptors and recruitment of Grb2 and SOS (Fig. 2 A and B). From the time course intensity analysis, we found a gradual accumulation of ephrin-A1-Alexa 680 at the cell-contacted point with SLB (Fig. S4 A and B), resulting in a symmetrical decrease in the remaining SLB region and the formation of ephrin-A1:EphA2 complexes (Figs. 2 A and B, and S3). The ephrin-A1:EphA2 cluster dynamics were closely mirrored by Grb2-tdEos accumulation dynamics (Fig. S4 A), suggesting rapid activation of the receptor and recruitment of the adaptor protein Grb2. However, there was a measurable time delay between ephrin-A1 clustering and SOS-mEos3.2 recruitment dynamics (Fig. S4 B and C).
Figure 2.
Recruitment of Grb2 and SOS to ephrin-A1:EphA2 clusters. (A and B) Multi-channel time-lapse images of MDA-MB-231 cells expressing Grb2-tdEos (A) or SOS-mEos3.2 (B) interacting with SLB-functionalized ephrin-A1. RICM image sequences show cell-SLB contacts (dark signal), while ephrin-A1 channel shows accumulation of the respective proteins at the cell-SLB contact. Merged images show colocalization of ephrin-A1 with Grb2-tdEos or SOS-mEos3.2. Full time-lapse are shown in Videos S2 and S3. (C) Point-source (green dotted circle) FRAP of Grb2-tdEos in individual SLBs. (D) Representative fluorescence recovery curves after photobleaching of Grb2-tdEos (left) and SOS-mEos3.2 (right). (E) Box chart showing fluorescence recovery half-live time of Grb2-tdEos and SOS-mEos3.2. The mean values are 3.8 s (standard deviation [SD]: 1.5) and 7.2 s (SD: 2.2) for Grb2 (n = 16 cells) and SOS (n = 14 cells), respectively. Paired sample t-test confirmed statistically significant differences between the population means. Bars are 5 μm in (A) and 10 μm in (B). To see this figure in color, go online.
We further characterized the population exchange rate of Grb2 and SOS in the steady state of ephrin-A1:EphA2 clusters. Using a point-source FRAP experiment (Fig. 2 C), we found that the Grb2-tdEos and SOS-mEos3.2 signals both exhibited dynamic turnover with residency half-lives of a few seconds (Fig. 2 D). These fast timescales suggest that the primary exchange is with cytosolic molecules since the EphA2 clusters are quite densely packed with minimal lateral diffusion of the receptor itself (we measured the diffusion rate of ephrin-A1:EphA2 cluster-bound Grb2 to be ∼0.003 μm2s−1). The Grb2 recovery time was roughly two times faster than SOS (Figs. 2 E, S4 D and E). We noted that these are composite dwell time distributions from many molecules, likely exhibiting different binding configurations. Thus, the observed recovery time differences likely represented population differences with respect to binding configuration more so than they corresponded with individual molecular kinetics. Nonetheless, the Grb2 and SOS recruitment to ephrin-A1:EphA2 signaling clusters was indicative of likely signaling activity in this pathway. We observed that the ephrin-A1:EphA2:Grb2 signaling clusters persisted for more than 3 h (Fig. S5). This long timescale was possibly indicative of a thwarted endocytosis process, as has previously been reported when ephrin-A1:EphA2 cluster size was physically limited (12). However, all experiments were performed over shorter timescales. As a control, we did not find localized Grb2-tdEos or SOS-mEos3.2 signals in SLB samples that were not functionalized with ephrin-A1 ligands (Fig. S6).
EGFR activation depletes Grb2 and SOS from pre-established ephrin-A1:EphA2 clusters
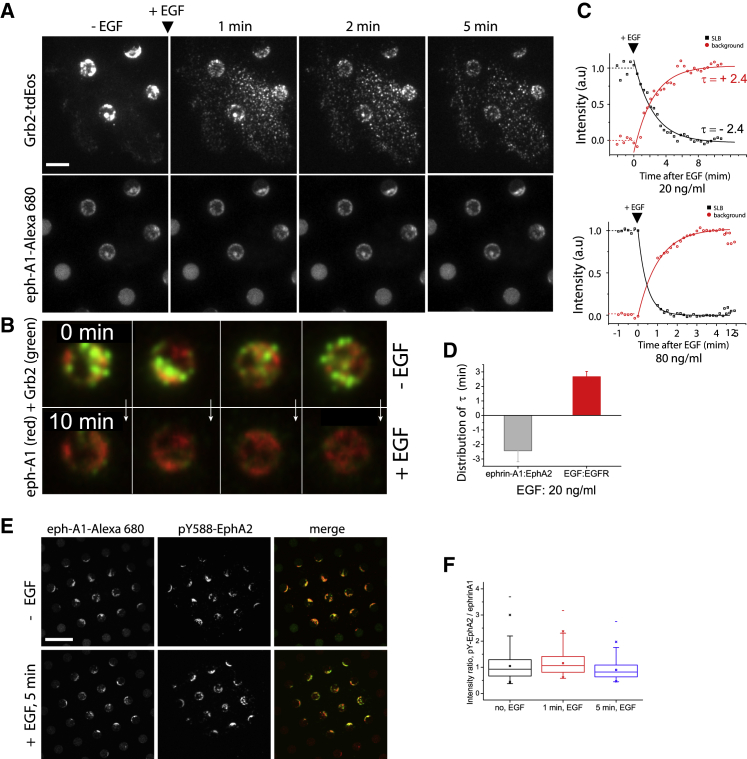
We next examined how Grb2 and SOS recruited to pre-established ephrin-A1:EphA2 clusters was affected by EGFR activation with EGF. We started experiments with cells that were in the fully spread state (∼1 h adhesion), but not in the motile phase. We recorded time-lapse images of Grb2-tdEos and SOS-mEos3.2 before and after addition of EGF at a low imaging rate of 0.1–0.5 Hz to avoid photobleaching effects of the native EOS fluorescence signal. During recording, EGF (10–80 ng/mL) was added abruptly, and the recording continued for several minutes more (Videos S4 and S5 for Grb2-tdEos, Videos S6 and S7 for SOS-mEos3.2). Here, we defined time zero, t = 0, as the moment of EGF addition. Before adding EGF (t < 0), the Grb2-tdEos or SOS-mEos3.2 signal in the ephrin-A1:EphA2 clusters in SLB was constant (Figs. 3 C and S7 C, t < 0). Upon adding EGF, both Grb2-tdEos and SOS-mEos3.2 signals localized with ephrin-A1:EphA2 signaling clusters decreased (Figs. 3 A and B, S7 A and B) (Videos S4, S5, S6, and S7). At the same time, numerous small puncta of Grb2-tdEos or SOS-mEos3.2 signals appeared throughout the cell membrane, corresponding to recruitment at developing EGF:EGFR signaling clusters (Figs. 3 A and S7 A, upper panels), consistent with previous reports (Oh et al., 2014). We rarely observed any of these newly formed puncta in the SLB regions (Fig. S8), indicating again no direct interactions between EGF:EGFR and ephrin-A1:EphA2. We also observed the shape and intensity of the ephrin-A1-Alexa 680 clusters in SLB remained unchanged (Figs. 3 A and S7 B, lower panels), indicating that the additional EGF treatment did not physically affect ephrin-A1:EphA2 binding and clustering. Taken together, these results suggest that the newly formed EGF:EGFR signaling complexes outcompete ephrin-A1:EphA2 for the limited supply of Grb2 and SOS, possibly attenuating or replacing ephrin-A1:EphA2-induced signaling through Ras to the MAPK pathway. As an additional control experiment, we confirmed by direct imaging localization of Grb2 to the cell membrane and corresponding depletion from the cytosol upon stimulation with EGF (Fig. S9). Finally, we also confirmed that, even when EGF stimulation occurred simultaneously with ephrin-A1, EphA2 complexation and clustering, and the ensuing ephrin-A1:EphA2 signaling clusters, were unchanged (Fig. S10).
Figure 3.
Competition between ligand-activated EphA2 and EGFR receptors for Grb2 recruitment on the same cell surface. (A) Time-lapse Grb2-tdEos images (in the green channel) before and after EGF (80 ng/mL) (full version, Video S4) show a signal decrease in ephrin-A1 SLB regions but simultaneous increase in regions outside of SLBs (top row). Corresponding ephrin-A1-Alexa 680 images show similar intensity and cluster shape before and after EGF (bottom row). Scale bar, 5 μm. (B) SLB collection from the time-lapse movie highlighted Grb2-tdEos signal (green) depletion in ephrin-A1:EphA2 clusters (red) after EGF stimulation (see Video S5). (C) Representative normalized intensity profiles of Grb2-tdEos inside (black, ephrin-A1:EphA2 clusters) and outside of SLB corrals (red, EGF:EGFR clusters) before (t < 0) and after EGF (t > 0) with 20 ng/mL (upper) and 80 ng/mL (lower). Exponential decay function fitting to the curves provided characteristic time constants of ꞇEphA2 = - 0.5 (± 0.01) min (80 ng/mL EGF) and ꞇEphA2 = −2.4 (± 0.12) min (20 ng/mL EGF) inside SLBs and ꞇEGFR = +1.0 (± 0.01) min (80 ng/mL EGF) and +2.4 (± 0.25) min (20 ng/mL EGF) outside SLB respectively. A negative sign means a decrease in intensity. (D) The distribution of the time constants from three replicates and seven cells with 20 ng/mL of EGF. Mean values are ꞇEphA2 = −2.5 (± 0.7) and ꞇEGFR = +2.6 (± 0.3) min. (E) Simultaneous fluorescence images of ephrin-A1-Alexa 608 (first column) and pY588-GFP antibody (second), and their merge (third) before (top row) and 5 min after EGF (80 ng/mL) (bottom row) show signal colocalization. (F) Quantification of the ratio between pY-EphA2 signals and ephrin-A1-Alexa 680 signals as a function of EGF stimulation time shows the minimal effect of EGF in EphA2 activation. Mean values were 1.0 (SD: 0.5), 1.1 (SD: 0.4), and 0.9 (SD: 0.3) for no EGF, 1 min EGF, and 5 min EGF, respectively. The total number of SLBs analyzed in 30 cells was n = 731, n = 714, and n = 628 for no EGF, 1 min EGF, and 5 min EGF, respectively. Scale bar, 10 μm. To see this figure in color, go online.
Next, to quantify and compare the dynamics, we normalized time-lapse intensity of Grb2-tdEos inside and outside the SLB region, corresponding to the ephrin-A1:EphA2 and EGF:EGFR clusters respectively. We found the anti-symmetric dynamics of Grb2-tdEos in the ephrin-A1:EphA2 and EGF:EGFR clusters with time constant of ꞇ = −2.4 and ꞇ = 2.4 min respectively (Fig. 3 C, upper panel). Using 20 ng/mL of EGF (replicated three times) we found a mean value of the time constant of ꞇEphA2 = −2.5 (±0.7) and ꞇEGFR = +2.6 (±0.3) min respectively (Fig. 3 D, n = 7 cells). The curves reached a plateau at ∼10 min (Fig. 3 C, upper panel). The switching dynamics of Grb2 between EphA2 and EGFR depended on the EGF dose, with higher doses switching faster (Fig. 3 C, lower panel). We found the same anti-symmetric dynamics between EphA2 and EGFR were observed for SOS-mEos3.2 (Fig. S7 C). We noted that, although both Grb2 and SOS levels at ephrin-A1:EphA2 clusters went down upon EGF activation of EGFR, they did not go to zero and appeared to establish a new balance.
Finally, to test our hypothesis that the Grb2/SOS shift from EphA2 to EGFR was largely driven by mass action and chemical equilibrium, we confirmed that the signaling state of EphA2 was unchanged by EGFR activation. For these experiments, we performed measurement of EphA2 phosphorylation (using a phosphotyrosine antibody specific to EphA2 pY588) calibrated by simultaneous measurements of ephrin-A1 content in the clusters. In addition, western blot data showed no change in phosphorylation of EphA2 with EGF stimulation (Fig. S11 A) but significant increase in EGFR phosphorylation (Fig. S11 B). The results revealed no changes in EphA2 phosphorylation levels before and after EGF stimulation (Fig. 3 E and F, n = 30 cells), suggesting that binding sites for Grb2 were still present. This result is consistent with previous studies of cell lysis assays data (35). The observed depletion is likely the result of simple competition with newly formed pY binding sites for Grb2 on EGFR, and SOS followed Grb2.
SHC1 is not linked to Grb2 depletion in ephrin-A1:EphA2 clusters by EGF stimulation
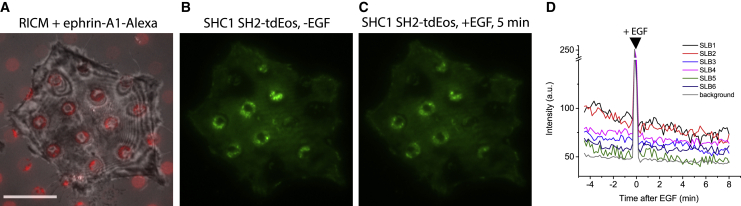
Previous cell lysate data showed that Grb2 bound to EphA2 through SHC1, which bound to phosphorylated EphA2 via either PTB (phosphotyrosine binding domain) or SH2 domain (38). However, not many studies have been performed in pY-EphA2 binding assay with SH2 domain containing proteins in vivo. Therefore, we sought to test if EGF stimulation could deplete SHC1 in the ephrin-A1:EphA2 clusters. We generated a tdEos fusion with the SH2 domain of SHC1 and transfected this into MBA-MB-231 cells (Fig. 4 A–C). Parallel ensemble and single-molecule imaging experiments were performed. We found a significant SHC1 SH2-tdEos signal in ephrin-A1:EphA2 clusters in the absence of EGF (Figs. 4 B and S12 A and B), which, upon addition of EGF (80 ng/mL), remained largely unchanged (Fig. 4 C and D) (Video S8). In addition, the binding rate, dwell time, and diffusion rate of individual SHC1 SH2-tdEos were also not significantly changed (Fig. S12 C–E). Thus, this indicated that Grb2 depletion from the ephrin-A1:EphA2 clusters with EGF stimulation was due to direct binding between Grb2 and pY in EphA2.
Figure 4.
SHC1 SH2 domain kinetics in ephrin-A1:EphA2 clusters was not affected by EGF stimulation. (A) RICM image of MDA-MB-231 cells expressing SHC1 SH2-tdEos spread on ephrin-A1-Alexa 680 SLB array (red) substrate. (B and C) Fluorescence images of SHC1 SH2-tdEos show no significant change in intensity before (B) and 5 min after (C) EGF (80 ng/mL). (D) Intensity profiles of SHC1 SH2-tdEos in SLBs and background before (t < 0) and after EGF (t > 0). The intensity jump at 0 min was due to brief exposure to external room light, which we used as an indicator to differentiate the signals before and after EGF. Scale bar, 10 μm. To see this figure in color, go online.
Single-molecule kinetic study of Grb2 and SOS at EphA2 and EGFR signaling clusters
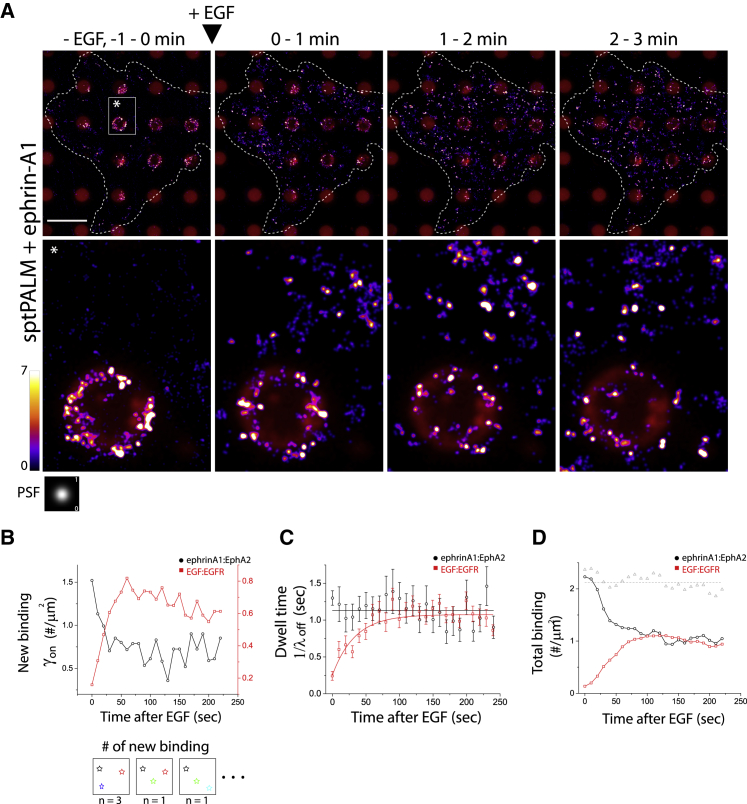
Under steady, low-level activation with 405 nm illumination, newly arriving Grb2 or SOS molecules can be individually detected and counted at EphA2 and EGFR signaling clusters (Video S9) (Fig. S13). The localization images of Grb2-tdEos using trajectories (Fig. S14) were reconstructed into sptPALM images (Fig. 5 A; Fig. S15 A for SOS-mEos3.2). Upon addition of EGF, we observed a depletion in binding activity at EphA2 clusters and a corresponding rise in membrane binding in the areas of EGFR signaling cluster formation (Figs. 5 A and B, and S15 A and B for SOS-mEos3.2; Video S10 for Grb2-tdEos, Video S11 for SOS-mEos3.2). Analysis of the single-molecule binding dwell time, defined as the characteristic time constant of distribution of Grb2-tdEos or SOS-mEos3.2 trajectory lengths (Fig. S16 B), revealed no changes at EphA2 clusters, with a mean dwell time of ∼1 s for Grb2 (Fig. 5 C, black) and 0.6 s for SOS (S15 C, black). In contrast, initial dwell times at EGFR began around a few hundred milliseconds and steadily increased to 1 s over the course of about 1 min (Fig. 5 C, red for Grb2 and S15 C for SOS). Monovalent Grb2:pY and SOS:Grb2 mean binding dwell times were in the sub ∼100 ms timescale under our illumination conditions, so the elongated dwell times we observed on both EphA2 and EGFR indicate some form of entrapment, rebinding, or multivalency. In the case of EGFR, it takes ∼1 min for this state to form with a high dose of EGF (80 ng/mL). Examination of the total number of binding events on EphA2 and EGFR by sptPALM image analysis (Figs. 5 A and S15 A) and numerical calculations (Figs. 5 D and S15 D; Eq. 3 in supporting material) confirmed the same reciprocal relocation of Grb2 and SOS observed in bulk measurements.
Figure 5.
Single-molecule kinetics measurements recapitulate Grb2 switching from ephrin-A1:EphA2 clusters to newly formed EGF:EGFR clusters. (A) The Grb2-tdEos sptPALM (fire color) image sequences superimposed with the ephrin-A1-Alexa 680 SLBs (red) capture spatiotemporal changes in total Grb2-tdEos binding after EGF (80 ng/mL), which decreases in SLBs (equivalent to ephrin-A1:EphA2 complexes) but simultaneously increases in the background (equivalent to EGF:EGFR complexes). The 2D xy coordinates of all trajectories (except one frame length) were replaced by Gaussian point spread function (PSF) with maxima of 1 and 50 nm FWHM, and integrated every minute (600 image frames). A portion of the single-molecule movie used here is shown in Video S10. (B) Time-dependent new binding rate, γon(t), of Grb2-tdEos per area in SLB (black) and background (red) after EGF. Here, only newly emerged molecules (trajectory-based) through frame to frame were counted (cartoon illustration), integrated for 100 images sequence each (10 s bin), and divided by observed SLB and background areas respectively. (C) Corresponding time-dependent mean dwell time (1/λoff(t), λoff = off rate) (see Fig. S16) of Grb2-tdEos in ephrin-A1:EphA2 (black) and EGF:EGFR clusters (red). The 1/λoff-EphA2(t) fluctuated slightly against 1120 ms, but 1/λoff-EGFR(t) gradually increased to saturate 1000 ms. (D) Computed total binding of Grb2-tdEos using measured γon(t) and λoff-EGFR(t) with Eq. 3 (supporting material) in eprhinA1:EphA2 (black) and EGF:EGFR clusters (red). The curves recapitulate the asymmetric kinetics of Grb2 in ephrin-A1:EphA2 and EGF:EGFR clusters upon EGF. However, the sum of membrane signals (gray dotted lines) remained constant, indicating the Grb2 mass flow from the EphA2 signaling complex to the EGFR signaling complex upon EGF. Scale bar, 10 μm. To see this figure in color, go online.
EphA2 and EGFR competition for Grb2 and SOS depends on receptor copy number
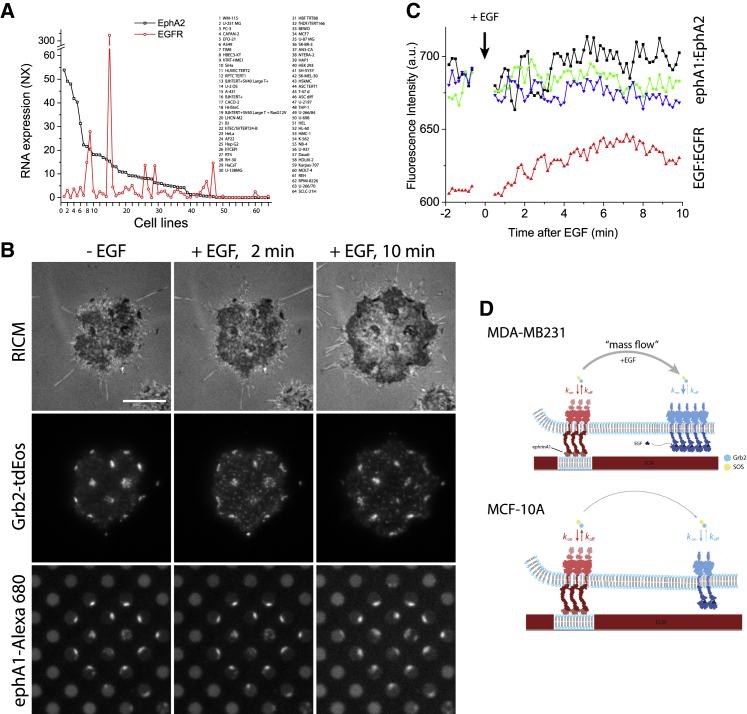
Based on the proteomic database (39), the expression levels of EphA2 and EGFR vary considerably among different cancer and immortalized cell lines (Fig. 6 A). If the competitive crosstalk between EphA2 and EGFR is a simple result of kinetic mass action, in which the large number of newly formed Grb2 binding sites on EGFR drives a re-equilibration with EphA2, then we expect this effect to be dependent on EGFR expression level. To test this hypothesis, we first examined the human breast cell line MCF-10A, which expresses EphA2 at levels similar to that of MDA-MB-231 cells, but expresses significantly less EGFR (40, 41, 42). In ensemble fluorescence measurements, we observed similar Grb2-tdEos (Fig. 6 B) and SOS-mEos3.2 signals (Fig. S17) in the ephrin-A1:EphA2 clusters as observed with the MDA-MB-231 cells. However, after introducing EGF (20 ng/mL), we did not find a significant reduction in the Grb2-tdEos signal at the EphA2 clusters, but we did detect a gradual, albeit relatively low, increase across the cell membrane from the smaller levels of EGFR (Fig. 6 B and C) (Video S12). The same result was obtained with a high dose of EGF (80 ng/mL) in a single-molecule experiment (Video S13). Moreover, the dwell time of Grb2-tdEos in ephrin-A1:EphA2 clusters was similar to that found in MDA-MB-231 cells (Fig. S18). Consistently, we did not find EGF-induced depletion of Grb2 from ephrin-A1:EphA2 clusters in the cells that expressed a higher amount of EphA2 than EGFR (Fig. S19; Video S14).
Figure 6.
Competition between ephrin-A1:EphA2 and EGF:EGFR for Grb2 recruitment depends on the level of receptor expression. (A) Relative RNA expression levels of EphA2 (black) and EGFR (red) in a range of cancer and immortalized cell lines (n = 64 cell lines from 12 organs) obtained from the Human Protein Atlas (39). Normalized NX (consensus normalized expression) values were used, and NX value of 1.0 is defined as a threshold for expression of the corresponding proteins based on database information. Most of the cancer cells in this database show higher RNA expression of EphA2 than EGFR, excluding cell lines such as hTERT-HME1 (mammary gland), A431 (skin), hTCEpi (cornea), HaCaT (skin), and U-2197 (subcutis). (B) MCF-10A cells (top row) expressing Grb2-tdEos (second) spread on ephrin-A1-Alexa 680-coated SLB (bottom) hybrid substrate, before (first column), 2 min (second) and 10 min (third) after EGF (20 ng/mL). The Grb2-tdEos fluorescence signals and ephrin-A1-Alexa 680 signals in SLBs did not change significantly. However, Grb2-tdEos signals increased gradually in the background. Video S12 was used for this analysis. (C) Intensity profiles corresponding in SLBs (equivalent to ephrin-A1:EphA2 clusters) and background (equivalent to EGF:EGFR clusters). (D) A proposed model for the Grb2/SOS mass flow from a lower activated RTK to a higher activated RTK depending on the receptor expression level of different cell lines. Here, the cognate ligand fully activates corresponding RTK. To see this figure in color, go online.
Discussion
Here, we report and quantify the real-time intracellular competition between ephrin-A1:EphA2 and EGF:EGFR signaling complexes for their shared downstream signaling molecules Grb2 and SOS. A key element in this study is a direct comparison of time-dependent Grb2 or SOS kinetics in spatially segregated ephrin-A1:EphA2 and EGF:EGFR clusters on the membrane surface of a single living cell. Overall, the results illustrate a dynamic redistribution of Grb2 and SOS among activated receptors, generally following the law of kinetic mass action (Fig. 6 D). Competition for the limited cytoplasmic supply of Grb2 and SOS—which can be expressed endogenously at relatively low levels (43)—establishes a hierarchy in which receptor signaling complexes with higher affinity/avidity for Grb2, larger numbers of pY binding sites, or simply higher expression levels may be able to significantly reduce the ability of other receptors to continue to signal through the Ras/MAPK pathway. We observe these effects to be relatively rapid, revealing that the apparently stable association of signaling molecules with the ephrin-A1:EphA2 complexes (and likely other systems as well) remains highly responsive to other activities elsewhere in the cell. This occurs when a significant fraction of the associated signaling molecules exhibit fast on-off kinetics and are in a dynamic equilibrium state, as we observed for Grb2 and SOS at the ephrin-A1:EphA2 complexes.
In the specific case of EGFR, recent reports of a protein condensation phase transition involving multivalent engagement of Grb2 and SOS (44) suggest that high degrees of multivalent interactions within the EGFR condensates may play a significant role in establishing stronger engagement of Grb2 and SOS, and the ability to outcompete EphA2 for these molecules. EphA2 signaling clusters, by contrast, are mediated through direct interactions among ephrin-A1:EphA2 complexes (45) and are not known to involve extensive multivalency of Grb2 and SOS. However, one can certainly imagine how this balance could shift in different cell types and under different conditions.
Previously, extensive cell lysate phosphorylation assays have shown that ligand-activated EphA2 can attenuate EGFR-promoted proliferation signals involving Ras, Raf, MEK1/2, and Erk1/2 activation in some types of cancer (36,46,47). Additionally, blocking EphA2 has been shown to overcome acquired resistance to EGFR kinase inhibitors in lung cancer (48). Kinetic mass action represents a chemical driving force exerting equal and opposite effects on both types of receptor cluster. Thus, activation of EphA2 will impose the same competitive force on EGFR that we observed here in the EGFR-driven depletion of Grb2/SOS at EphA2 signaling clusters. Of course, crosstalk between EphA2 and EGFR also occurs in more complex manners. For example, ligand-activated EGFR increases EphA2 expression (35,47), and EphA2 overexpression in murine adenocarcinoma upregulates EGFR (49). Our data add to the story by revealing another way that these changes in expression level can rebalance competitive (or cooperative) signaling among different RTKs.
Author contributions
D.O., M.P.S., and J.T.G. designed the research. D.O. and Z.C. prepared samples and performed experiments. K.H.B. conducted western blotting experiments. D.O., Z.C., and J.T.G. analyzed the results. D.O., F.B., and H.T.O. wrote the computer codes for trajectory analysis. D.O. and J.T.G. wrote the manuscript.
Declaration of interests
The authors declare no competing interests.
Acknowledgments
We thank Adrienne Greene (J.T.G.’s laboratory, University of California, Berkeley) for her contribution of purified ephrin-A1 proteins, as well as helpful discussions. We thank Jean K. Jung for helping in SLB substrate hybridization. We thank members in J.T.G.’s laboratory and M.P.S.’s laboratory for stimulating discussions and sharing of reagents. This work was supported by the Novo Nordisk Foundation Challenge Program under the Center for Geometrically Engineered Cellular Systems. Collaborative work at the Mechanobiology Institute, National University of Singapore, was supported by CRP001-084. Z.C. is funded by Shanghai Municipal Science and Technology Major Project ZJLab (2019SHZDZX02).
Editor: Sarah L. Veatch.
Footnotes
Dongmyung Oh and Zhongwen Chen contributed equally to this work.
Supporting material can be found online at https://doi.org/10.1016/j.bpj.2022.04.015.
Contributor Information
Michael P. Sheetz, Email: misheetz@utmb.edu.
Jay T. Groves, Email: jtgroves@lbl.gov.
Supporting material
References
- 1.Robinson D.R., Wu Y.M., Lin S.F. The protein tyrosine kinase family of the human genome. Oncogene. 2000;19:5548–5557. doi: 10.1038/sj.onc.1203957. [DOI] [PubMed] [Google Scholar]
- 2.Lemmon M.A., Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2010;141:1117–1134. doi: 10.1016/j.cell.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu P., Cheng H., et al. Zhao J.J. Targeting the phosphoinositide 3-kinase pathway in cancer. Nat. Rev. Drug Discov. 2009;8:627–644. doi: 10.1038/nrd2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haglund K., Dikic I. The role of ubiquitylation in receptor endocytosis and endosomal sorting. J. Cell Sci. 2012;125:265–275. doi: 10.1242/jcs.091280. [DOI] [PubMed] [Google Scholar]
- 5.Pawson T. Protein modules and signalling networks. Nature. 1995;373:573–580. doi: 10.1038/373573a0. [DOI] [PubMed] [Google Scholar]
- 6.Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2000;103:211–225. doi: 10.1016/s0092-8674(00)00114-8. [DOI] [PubMed] [Google Scholar]
- 7.Tandon M., Vemula S.V., Mittal S.K. Emerging strategies for EphA2 receptor targeting for cancer therapeutics. Expert Opin. Ther. Targets. 2011;15:31–51. doi: 10.1517/14728222.2011.538682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Normanno N., De Luca A., et al. Salomon D.S. Epidermal growth factor receptor (EGFR) signaling in cancer. Gene. 2006;366:2–16. doi: 10.1016/j.gene.2005.10.018. [DOI] [PubMed] [Google Scholar]
- 9.Orton R., Sturm O., et al. Kolch W. Computational modelling of the receptor-tyrosine-kinase-activated MAPK pathway. Biochem. J. 2005;392:249–261. doi: 10.1042/bj20050908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Salaita K., Nair P.M., et al. Groves J.T. Restriction of receptor movement alters cellular response: physical force sensing by EphA2. Science. 2010;327:1380–1385. doi: 10.1126/science.1181729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu Q., Lin W.C., et al. Groves J. EphA2 receptor activation by monomeric Ephrin-A1 on supported membranes. Biophys. J. 2011;101:2731–2739. doi: 10.1016/j.bpj.2011.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Greene A., Lord S., et al. Groves J. Spatial organization of EphA2 at the cell-cell interface modulates trans-endocytosis of ephrinA1. Biophys. J. 2014;106:2196–2205. doi: 10.1016/j.bpj.2014.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen Z., Oh D., et al. Groves J.T. Spatially modulated ephrinA1:EphA2 signaling increases local contractility and global focal adhesion dynamics to promote cell motility. Proc. Natl. Acad. Sci. U S A. 2018;115:E5696–E5705. doi: 10.1073/pnas.1719961115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Biswas K.H., Groves J.T. Hybrid live cell-supported membrane interfaces for signaling studies. Annu. Rev. Biophys. 2019;48:537–562. doi: 10.1146/annurev-biophys-070317-033330. [DOI] [PubMed] [Google Scholar]
- 15.Groves J.T., Ulman N., Boxer S.G. Micropatterning fluid lipid bilayers on solid supports. Science. 1997;275:651–653. doi: 10.1126/science.275.5300.651. [DOI] [PubMed] [Google Scholar]
- 16.Mossman K.D., Campi G., et al. Dustin M.L. Altered TCR signaling from geometrically repatterned immunological synapses. Science. 2005;310:1191–1193. doi: 10.1126/science.1119238. [DOI] [PubMed] [Google Scholar]
- 17.DeMond A.L., Mossman K.D., et al. Groves J.T. T cell receptor microcluster transport through molecular mazes reveals mechanism of translocation. Biophys. J. 2008;94:3286–3292. doi: 10.1529/biophysj.107.119099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Manz B.N., Jackson B.L., et al. Groves J. T-cell triggering thresholds are modulated by the number of antigen within individual T-cell receptor clusters. Proc. Natl. Acad. Sci. U S A. 2011;108:9089–9094. doi: 10.1073/pnas.1018771108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Biswas K.H., Hartman K.L., et al. Groves J.T. E-cadherin junction formation involves an active kinetic nucleation process. Proc. Natl. Acad. Sci. U S A. 2015;112:10932–10937. doi: 10.1073/pnas.1513775112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Biswas K.H., Hartman K., et al. Groves J.T. Sustained alpha-catenin activation at E-cadherin junctions in the absence of mechanical force. Biophys. J. 2016;111:1044–1052. doi: 10.1016/j.bpj.2016.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wong J.J., Chen Z., et al. Jardetzky T.S. EphrinB2 clustering by Nipah virus G is required to activate and trap F intermediates at supported lipid bilayer-cell interfaces. Sci. Adv. 2021;7:eabe1235. doi: 10.1126/sciadv.abe1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Biswas K.H., Cho N.J., Groves J.T. Fabrication of multicomponent, spatially segregated DNA and protein-functionalized supported membrane microarray. Langmuir. 2018;34:9781–9788. doi: 10.1021/acs.langmuir.8b01364. [DOI] [PubMed] [Google Scholar]
- 23.Chen Z., Oh D., et al. Groves J.T. Probing the effect of clustering on EphA2 receptor signaling efficiency by subcellular control of ligand-receptor mobility. Elife. 2021;10:e67379. doi: 10.7554/elife.67379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iversen L. Molecular kinetics. Ras activation by SOS: allosteric regulation by altered fluctuation dynamics. Science. 2014;345:50–54. doi: 10.1126/science.1250373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang W.Y.C., Yan Q., et al. Groves J.T. Phosphotyrosine-mediated LAT assembly on membranes drives kinetic bifurcation in recruitment dynamics of the Ras activator SOS. Proc. Natl. Acad. Sci. U S A. 2016;113:8218–8223. doi: 10.1073/pnas.1602602113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Christensen S.M., Tu H.L., et al. Groves J.T. One-way membrane trafficking of SOS in receptor-triggered Ras activation. Nat. Struct. Mol. Biol. 2016;23:838–846. doi: 10.1038/nsmb.3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee Y.K., Low-Nam S.T., et al. Groves J.T. Mechanism of SOS PR-domain autoinhibition revealed by single-molecule assays on native protein from lysate. Nat. Commun. 2017;8:15061. doi: 10.1038/ncomms15061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin W.C., Yu C.H., et al. Groves J.T. Supported membrane formation, characterization, functionalization, and patterning for application in biological science and technology. Curr. Protoc. Chem. Biol. 2010;2:235–269. doi: 10.1002/9780470559277.ch100131. [DOI] [PubMed] [Google Scholar]
- 29.Galush W.J., Nye J.A., Groves J.T. Quantitative fluorescence microscopy using supported lipid bilayer standards. Biophys. J. 2008;95:2512–2519. doi: 10.1529/biophysj.108.131540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gelles J., Schnapp B.J., Sheetz M.P. Tracking kinesin-driven movements with nanometre-scale precision. Nature. 1988;331:450–453. doi: 10.1038/331450a0. [DOI] [PubMed] [Google Scholar]
- 31.Oh D., Yu Y., Lee H., et al. Ritchie K. Dynamics of the serine chemoreceptor in the Escherichia coli inner membrane: a high-speed single-molecule tracking study. Biophys. J. 2014;106:145–153. doi: 10.1016/j.bpj.2013.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ritchie K., Kusumi A. [27] Single-particle tracking image microscopy. Methods Enzymol. 2003;360:618–634. doi: 10.1016/s0076-6879(03)60131-x. [DOI] [PubMed] [Google Scholar]
- 33.Jadwin J.A., Oh D., et al. Mayer B.J. Time-resolved multimodal analysis of Src Homology 2 (SH2) domain binding in signaling by receptor tyrosine kinases. Elife. 2016;5:e11835. doi: 10.7554/elife.11835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Manley S., Gillette J.M., et al. Lippincott-Schwartz J. High-density mapping of single-molecule trajectories with photoactivated localization microscopy. Nat. Methods. 2008;5:155–157. doi: 10.1038/nmeth.1176. [DOI] [PubMed] [Google Scholar]
- 35.Larsen A.B., Pedersen M.W., et al. Poulsen H.S. Activation of the EGFR gene target EphA2 inhibits epidermal growth factor-induced cancer cell motility. Mol. Cancer Res. 2007;5:283–293. doi: 10.1158/1541-7786.mcr-06-0321. [DOI] [PubMed] [Google Scholar]
- 36.Macrae M., Neve R.M., et al. McCormick F. A conditional feedback loop regulates Ras activity through EphA2. Cancer Cell. 2005;8:111–118. doi: 10.1016/j.ccr.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 37.Sengupta K., Limozin L. Adhesion of soft membranes controlled by tension and interfacial polymers. Phys. Rev. Lett. 2010;104:088101. doi: 10.1103/physrevlett.104.088101. [DOI] [PubMed] [Google Scholar]
- 38.Pratt R.L., Kinch M.S. Activation of the EphA2 tyrosine kinase stimulates the MAP/ERK kinase signaling cascade. Oncogene. 2002;21:7690–7699. doi: 10.1038/sj.onc.1205758. [DOI] [PubMed] [Google Scholar]
- 39.Ponten F., Schwenk J.M., et al. Edqvist P.H.D. The Human Protein Atlas as a proteomic resource for biomarker discovery. J. Intern. Med. 2011;270:428–446. doi: 10.1111/j.1365-2796.2011.02427.x. [DOI] [PubMed] [Google Scholar]
- 40.Wymant J.M., Hiscox S., et al. Jones A.T. The role of BCA2 in the endocytic trafficking of EGFR and significance as a prognostic biomarker in cancer. J. Cancer. 2016;7:2388–2407. doi: 10.7150/jca.15055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zelinski D.P., Zantek N.D., et al. Kinch M.S. EphA2 overexpression causes tumorigenesis of mammary epithelial cells. Cancer Res. 2001;61:2301–2306. [PubMed] [Google Scholar]
- 42.Noblitt L.W., Bangari D.S., et al. Mittal S.K. Immunocompetent mouse model of breast cancer for preclinical testing of EphA2-targeted therapy. Cancer Gene Ther. 2005;12:46–53. doi: 10.1038/sj.cgt.7700763. [DOI] [PubMed] [Google Scholar]
- 43.Shi T., Niepel M., et al. Wiley H.S. Conservation of protein abundance patterns reveals the regulatory architecture of the EGFR-MAPK pathway. Sci. Signal. 2016;9:rs6. doi: 10.1126/scisignal.aaf0891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin C N.L., Stinger B., et al. Groves J.T. A two-component protein condensate of EGFR and Grb2 regulates Ras activation at the membrane. bioRxiv. 2021 doi: 10.1073/pnas.2122531119. Preprint at. PPR:PPR431355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Himanen J.P., Yermekbayeva L., et al. Dhe-Paganon S. Architecture of Eph receptor clusters. Proc. Natl. Acad. Sci. U S A. 2010;107:10860–10865. doi: 10.1073/pnas.1004148107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miao H., Li D.Q., et al. Wang B. EphA2 mediates ligand-dependent inhibition and ligand-independent promotion of cell migration and invasion via a reciprocal regulatory loop with Akt. Cancer Cell. 2009;16:9–20. doi: 10.1016/j.ccr.2009.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Larsen A.B., Stockhausen M.T., Poulsen H.S. Cell adhesion and EGFR activation regulate EphA2 expression in cancer. Cell Signal. 2010;22:636–644. doi: 10.1016/j.cellsig.2009.11.018. [DOI] [PubMed] [Google Scholar]
- 48.Amato K.R., Wang S., et al. Chen J. EPHA2 blockade overcomes acquired resistance to EGFR kinase inhibitors in lung cancer. Cancer Res. 2016;76:305–318. doi: 10.1158/0008-5472.can-15-0717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.De Robertis M., Loiacono L., et al. Fazio V.M. Dysregulation of EGFR pathway in EphA2 cell subpopulation significantly associates with poor prognosis in colorectal cancer. Clin. Cancer Res. 2017;23:159–170. doi: 10.1158/1078-0432.ccr-16-0709. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.