Abstract
Adenosine-to-inosine (A-to-I) editing of a subset of RNAs in a eukaryotic cell is required in order to avoid triggering the innate immune system. Editing is carried out by ADAR1, which exists as short (p110) and long (p150) isoforms. ADAR1p150 is mostly cytoplasmic, possesses a Z-RNA binding domain (Zα), and is only expressed during the innate immune response. A structurally homologous domain to Zα, the Zβ domain, is separated by a long linker from Zα on the N-terminus of ADAR1 but its function remains unknown. Zβ does not bind to RNA in isolation, yet the binding kinetics of the segment encompassing Zα, Zβ and the 95-residue linker between the two domains (Zα-Zβ) are markedly different compared to Zα alone. Here we present the solution NMR backbone assignment of Zα-Zβ from H. Sapiens ADAR1. The predicted secondary structure of Zα-Zβ based on chemical shifts is in agreement with previously determined structures of Zα and Zβ in isolation, and indicates that the linker is intrinsically disordered. Comparison of the chemical shifts between the individual Zα and Zβ domains to the full Zα-Zβ construct suggests that Zβ may interact with the linker, the function of which is currently unknown.
Keywords: ADAR1, editing, Z-RNA, protein structure & dynamics, protein domains, backbone chemical shift assignment
Biological context
Distinguishing between self and non-self RNA is critical in controlling the innate immune response. In humans, self RNAs are edited by an adenosine deaminase that acts on RNA (ADAR1), which converts adenosines to inosines (Bass and Weintraub 1988; Wagner et al. 1989; Nishikura 2016). ADAR1 is constitutively expressed in most cells as a stable p110 isoform localized in the nucleus (O’Connell and Keller 1994; O’Connell et al. 1995; Patterson and Samual 1995). Upon invasion by a pathogen, the cell launches an interferon (IFN) response, resulting in the expression of a longer p150 isoform, which contributes to resisting the infection by editing self RNAs in the cytoplasm (O’Connell and Keller 1994; O’Connell et al. 1995; Patterson and Samual 1995; George and Samuel 1999) (Figure 1a). A-to-I editing is therefore augmented during the IFN response, primarily through the action of ADAR1p150 (Chung et al. 2018). In addition to becoming cytoplasmic, ADAR1p150 is also distinct from ADAR1p110 due to the presence of a N-terminal Zα domain, which is a member of a family of helix-turn-helix domains that recognize the unusual geometry of the Z-conformation in DNA or RNA, and binds to five base pairs in a symmetrical fashion (Herbert et al. 1998; Schwartz et al. 1999b; Brown et al. 2000; Placido et al. 2007) (Figure 1b).
Figure 1. The Zα and Zβ domains of ADAR1p150.
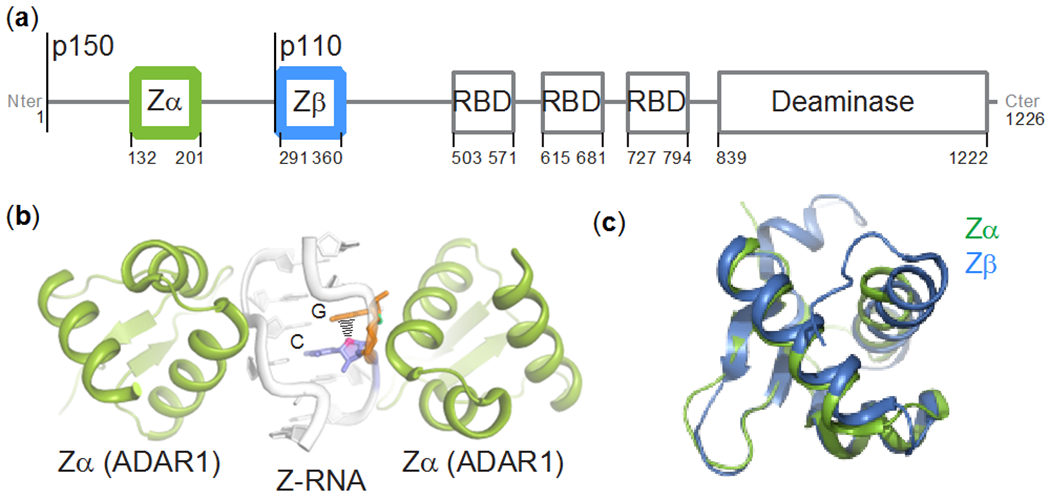
(a) Domain organization of ADAR1: Zα and Zβ are structurally homologous helix-turn-helix DNA-binding domains, RBD stands for double-stranded RNA binding domain. Both isoforms are indicated. (b) Crystal structure of (CpG)3 RNA bound to Zα from ADAR1 (PDB ID: 2GXB, (Placido et al. 2007)). (c) Structural alignment of the Zα (PDB ID: 2GXB) and Zβ (PDB ID: 1XMK, (Athanasiadis et al. 2018)) domains of ADAR1. The backbone RMSD between the two structures is 0.9 Å (excluding the termini).
The Zβ domain of ADAR1, which is located C-terminal to Zα and separated by a flexible 95-residue linker, is structurally homologous to Zα (Athanasiadis et al. 2018) but lacks the critical Z-recognizing residues required for adoption of Z-DNA/RNA and B-Z/A-Z junctions (Athanasiadis 2012). In the following, we refer to the Zα-linker-Zβ segment as Zα-Zβ. While Zβ does not bind to DNA/RNA in isolation, Zα-Zβ is characterized by markedly different Z-DNA/RNA binding kinetics (Schwartz et al. 1999a). Zβ is resistant to proteolysis only in the context of Zα-Zβ and the flexible linker between the two folded domains becomes resistant to proteolysis when bound to Z-DNA (Schwartz et al. 1999a). These findings led to the proposal that Zα-Zβ may undergo a large structural rearrangement when bound to Z-DNA/Z-RNA or that Zα-Zβ functions as a bipartite binding domain with Zβ gaining the ability to bind nucleic acid only in the context of Zα-Zβ (Schwartz et al. 1999a). Based on its crystal structure as a dimer with a cadmium ion at the interface, it was also proposed that Zβ may act as a dimerization domain (Athanasiadis et al. 2018). The exact role of Zβ thus remains elusive, although it is clear that Zα, Zβ, and the linker region in between the two domains act in concert.
Here we report the NH/Cα/Cβ/CO solution NMR backbone assignments of Zα-Zβ as well as the NH/Cα/Cβ chemical shifts for the individual Zα and Zβ domains from ADAR1. While the assignments of the Zα-Zβ and Zβ domains from ADAR1 are novel, the Zα has been characterized by NMR previously (Schade et al. 1999a, b), however, the chemical shifts for ADAR1 Zα have not been deposited to the BMRB until now.
Methods and experiments
Protein expression and purification
The N-terminal Zα domain of Homo sapiens ADAR1 in the pet-28a(+) plasmid (N-terminal 6x His-tag and thrombin cleavage site between His tag and the Zα sequence) was a gift from Drs. Peter Dröge and Alekos Athanasiadis. Zα-Zβ and Zβ were ordered from GenScript (cloned into the same expression vector, pet-28a(+)) and prepared in the same way as Zα, which was expressed and purified similarly to (Placido et al. 2007; Kruse et al. 2020). Briefly, the plasmids were transformed and expressed in BL21(DE3) E. coli. The cell cultures were grown in M9 minimal media with 1 g/L 15N ammonium chloride and 1.5 g/L 13C glucose (Millipore-Sigma, Burlington, MA) induced with IPTG at a final concentration of 1 mM, and allowed to express Zα, Zβ, or Zα-Zβ for 4 hours at 37°C, then centrifuged to collect the cell pellets. Pellets were resuspended in lysis buffer (50 mM Tris-HCl (pH 8.0), 300 mM NaCl, 10 mM Imidazole, 5 mM β-Mercaptoethanol (BME)) and sonicated. Lysate was centrifuged and the supernatant was applied to a His-trap column, washed with 40 mL of lysis buffer, 80 mL of wash buffer (50 mM Tris-HCl (pH 8.0), 1 M NaCl, 10 mM Imidazole, 5 mM BME), and eluted in 20 mL of elution buffer (50 mM Tris-HCl (pH 8.0), 300 mM NaCl, 500 mM Imidazole, 1 mM BME). The eluents were concentrated to ~2 mL and applied to a Hiload 16/600 Superdex 75 Gel Filtration Column (GE Healthcare) and the peak corresponding to pure protein was collected and concentrated using an Amicon 3 kDa cutoff centrifugal filter (Millipore-Sigma, Burlington, MA). At this step, Zα-Zβ and to a lesser extent also Zβ showed concentration- and salt-dependent oligomerization. To prevent aggregation, more NaCl was added (to a final concentration of 100 mM for NMR measurements), with the concentration of NaCl being dependent upon the concentration of protein. The proteins were dialyzed and concentrated into the following buffers for NMR: 20 mM potassium phosphate (pH 6.4), 25 mM or 100 mM NaCl for Zα (2 mM protein), 20 mM potassium phosphate (pH 6.4), 25 mM NaCl or 100 mM NaCl for Zβ (2 mM protein), 20 mM potassium phosphate (pH 6.4), 100 mM NaCl for Zα-Zβ (680 μM protein). D2O was added to 5%. See Table 1 for specifics on which buffer was used for which NMR experiments.
Table 1:
NMR experiments and sample information.
| Construct identity | Measured NMR experiments | Field strength | Sample concentration | Sample temperature | Buffer conditions | Molecular weight |
|---|---|---|---|---|---|---|
| Zα |
15N-HSQC HNCACB |
900 MHz 600 MHz |
2 mM | 25°C | 20 mM potassium phosphate (pH 6.4), 25 mM NaCl | 9.2 kDa |
| Zα | 15N-HSQC | 900 MHz | 2 mM | 35°C | 20 mM potassium phosphate (pH 6.4), 100 mM NaCl | 9.2 kDa |
| Zβ |
15N-HSQC HNCACB |
900 MHz 900 MHz |
2 mM | 25°C | 20 mM potassium phosphate (pH 6.4), 25 mM NaCl | 10.9 kDa |
| Zβ | 15N-HSQC | 900 MHz | 2 mM | 35°C | 20 mM potassium phosphate (pH 6.4), 100 mM NaCl | 10.9 kDa |
| Zα-Zβ |
15N-HSQC TROSY-HNCACB TROSY-HN(CO)CACB TROSY-HN(CA)CO TROSY-HNCO HNN |
900 MHz 600 MHz 600 MHz 600 MHz 600 MHz 900 MHz |
680 μM | 35°C | 20 mM potassium phosphate (pH 6.4), 100 mM NaCl | 27.2 kDa |
Perdeuterated Zα-Zβ was prepared in the same way as non-perdeuterated Zα-Zβ except that the M9 minimal media culture contained 99.8 % D2O instead of water and had uniformly deuterated 13C glucose (Cambridge Isotope Laboratories, Tewksbury, MA). Additionally, the E. coli were D2O adapted before expression following the protocol from (Cai et al. 2016).
NMR spectroscopy
The TROSY backbone experiments for the assignment of Zα-Zβ and the HNCACB for the isolated Zα domain were carried out on a Bruker 600 MHz spectrometer equipped with a 5/3 mm triple resonance 1H/13C/15N/19F cryoprobe (CP2.1 TCI). All other experiments were done on a Varian 900 MHz spectrometer equipped with a 5 mm triple resonance 1H/13C/15N cold probe with a Z-axis gradient.
15N-HSQC spectra of the individual Zα and Zβ domains were recorded both at 25°C and with 25 mM NaCl, and at 35°C with 100 mM NaCl. The 15N-HSQC of Zα-Zβ was recorded at 35°C with 100 mM NaCl. All 15N-HSQC spectra were collected with 1024 (1H) x 120 (15N) complex points, a 1.6 s recycle delay, and 16 scans. The spectral widths were 16 and 35 ppm for the 1H and 15N dimensions, respectively.
Assignment of the individual Zα and Zβ domains was achieved through HNCACB experiments measured at 25°C and 25 mM NaCl with 1024 (1H) x 96 (13C) x 80 (15N) complex points (2306 of the points were collected following a 30% non-uniform sampling scheme from the Wagner website: http://gwagner.med.harvard.edu/intranet/hmsIST/gensched_new.html, (Hyberts et al. 2012)), a 1 s recycle delay, and 8 scans. The spectral widths were 15.6, 70, and 35 ppm for the 1H, 13C, and 15N dimensions, respectively.
Assignment of Zα-Zβ was achieved by measurement of 3D TROSY-HNCACB, 3D TROSY-HN(CO)CACB, 3D TROSY-HN(CA)CO, 3D TROSY-HNCO, and 3D HNN experiments on a perdeuterated sample. The four TROSY experiments were measured with 1024 (1H) x 96 (13C) x 80 (15N) complex points (1274 of the points were collected following a 16% sampling scheme from the Wagner site), a 1.9 s recycle delay (except for the TROSY-HNCO, which had a delay of 1 s), and 16 scans. The spectral widths were 18 (1H), 80 (13C), and 35 (15N) ppm for the TROSY-HNCACB and TROSY-HN(CO)CACB experiments, and 18 (1H), 14 (13C), and 35 (15N) ppm for the TROSY-HN(CA)CO and TROSY-HNCO experiments. The HNN experiment was measured with 1024 (1H) x 96 (15N) x 80 (15N) complex points (1927 of the points were collected following a 25% NUS sampling scheme from the Wagner site), a 1 s recycle delay, and 16 scans. The spectral widths were 15.6 & 70 & 35 ppm for the 1H, 13C, and 15N dimensions. The HNN experiment, which correlates 15N and 1HN of residue i with the 15N of residues of i+1 and i-1, was helpful for assigning overlapped regions of which there were many in the linker region of Zα-Zβ. Residues 205-212 have chemical shifts identical to those of residues 253-260, as they are part of a repeat sequence within the linker region (with the sequence NQHSGVVRP).
The 3D NUS-spectra were constructed using the hmsIST software (Hyberts et al. 2012), and the linearly acquired 2D spectra were subject to NUS zero-filling as an alternative to linear prediction. A solvent subtraction function was applied in the direct dimension. Further data processing and visualization were performed using NMRPipe/NMRDraw (Delaglio et al. 1995) and NMRFAM Sparky (Lee et al. 2015). Resonance assignment was performed using the CCPNmr analysis software version 2.4.2 (Vranken et al. 2005).
Assignment and data deposition
Initial 3D experiments on 15N, 13C isotopically enriched Zα-Zβ at 25°C only resolved peaks from the linker region and the flexible termini. Thus, we turned to 2H, 15N, 13C isotope labeling and measured TROSY versions of the standard suite of backbone experiments (Cavanagh 2007) at 35°C, which had enough signal to be able to assign 98% of the entire Zα-Zβ construct (Table 2). The assignments of the Zα and Zβ domains in isolation were helpful in narrowing down the search area of the peaks within the full construct. While the backbone assignments of the Zα and Zβ domains were done at 25°C, we were able to assign the 15N-1H HSQC spectra measured at 35°C through nearest neighbor assignment. The 15N-1H HSQC of the three constructs and the peak assignments are shown (Figure 2). We have deposited the chemical shifts for Zα, Zβ and Zα-Zβ under BMRB accession numbers: 50714, 50713, and 50715 respectively.
Table 2:
Backbone assignment statistics of Zα, Zβ, and Zα-Zβ.
| Construct identity with numbering of relevant residues | Total number of relevant residues* | Total number of relevant non-proline residues | % backbone resonances assigned (number of backbone atoms assigned)$ |
|---|---|---|---|
| Zα (139-202) | 64 | 61 | 97.6% (61 15N, 63 Cα, 57 Cβ) |
| Zβ (290-365) | 76 | 75 | 98.7% (74 15N, 75 Cα, 72 Cβ) |
| Zα-Zβ (139-365) | 227 | 212 | 98.2% (208 15N, 221 Cα, 205 Cβ, 220 CO) |
All constructs have 20 extra non-relevant residues from cloning and the His-tag.
Backbone assignment % as extracted from CCPNmr (Vranken et al. 2005).
Figure 2. Assigned 1H-15N HSQC spectra of Zα, Zβ, and Zα-Zβ.
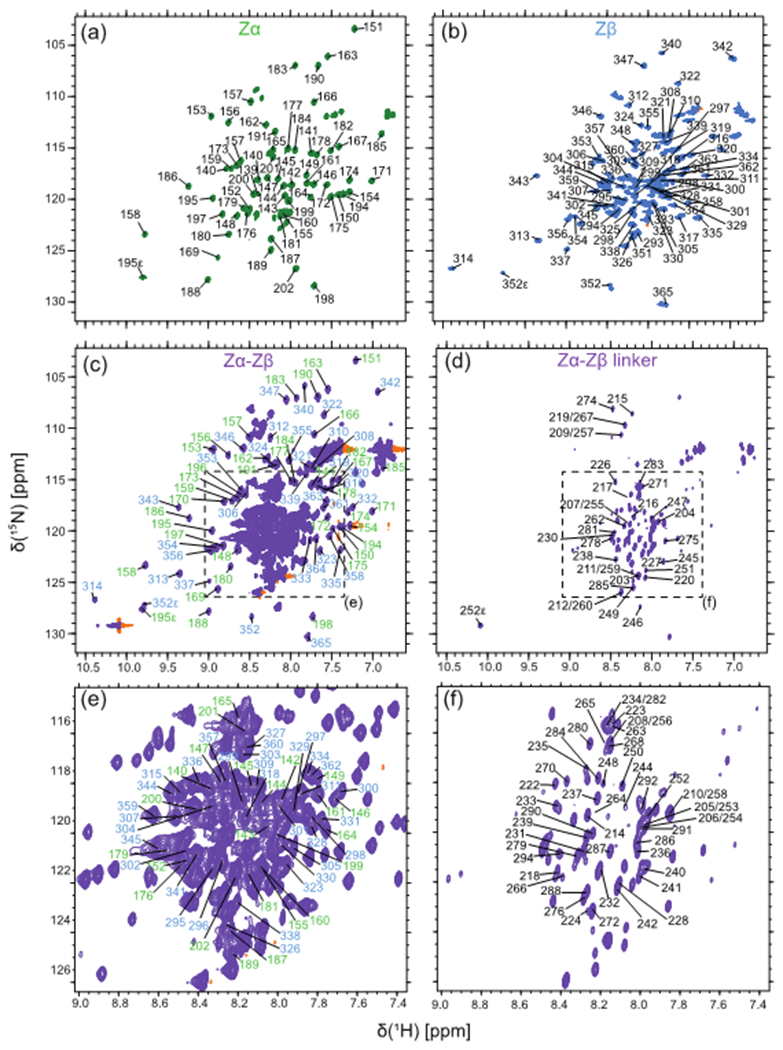
Shown are the 1H-15N HSQC spectra of Zα (green, a), Zβ (blue, b), and Zα-Zβ (purple, c and d) measured at 900 MHz, 35°C, and in 20 mM potassium phosphate (pH 6.4), 100 mM NaCl. The spectrum of Zα-Zβ in (c) has a low contour level cutoff, where only the Zα and Zβ domain assignments are shown in green and blue, respectively, and a high contour level cutoff in (d) to highlight the linker residues. (e) and (f) are blown-up regions from (c) and (d), respectively, to more clearly show the assignments within the crowded region of the spectra.
Chemical shift analysis
Several conclusions could immediately be drawn from the spectra and assignments of Zα-Zβ. First, the linker between the Zα and Zβ domains is intrinsically disordered, as determined by its low chemical shift dispersion and favorable relaxation properties (Figure 2). The Secondary Structure Propensity Score (SSP) (Marsh et al. 2006) of the linker region mostly fluctuates between 0 and 0.2 (a score of 1 indicates a fully formed α-helix, while −1 indicates a β-sheet), confirming that the linker is indeed intrinsically disordered with some potential α-helical propensity (Figure 3). In addition, the SSP score shows that the secondary structure of the Zα and Zβ domains is well-folded and in agreement with the structures of the isolated domains (Zα PDB IDs: 2GXB (Placido et al. 2007), 1QGP (Schade et al. 1999a); Zβ PDB ID: 1XMK (Athanasiadis et al. 2018)) (Figure 3). This suggests that the Zα and Zβ domains within the context of the larger Zα-Zβ construct adopt a similar structure as they do in isolation.
Figure 3. Secondary Structure Propensity Score of Zα-Zβ of ADAR1.
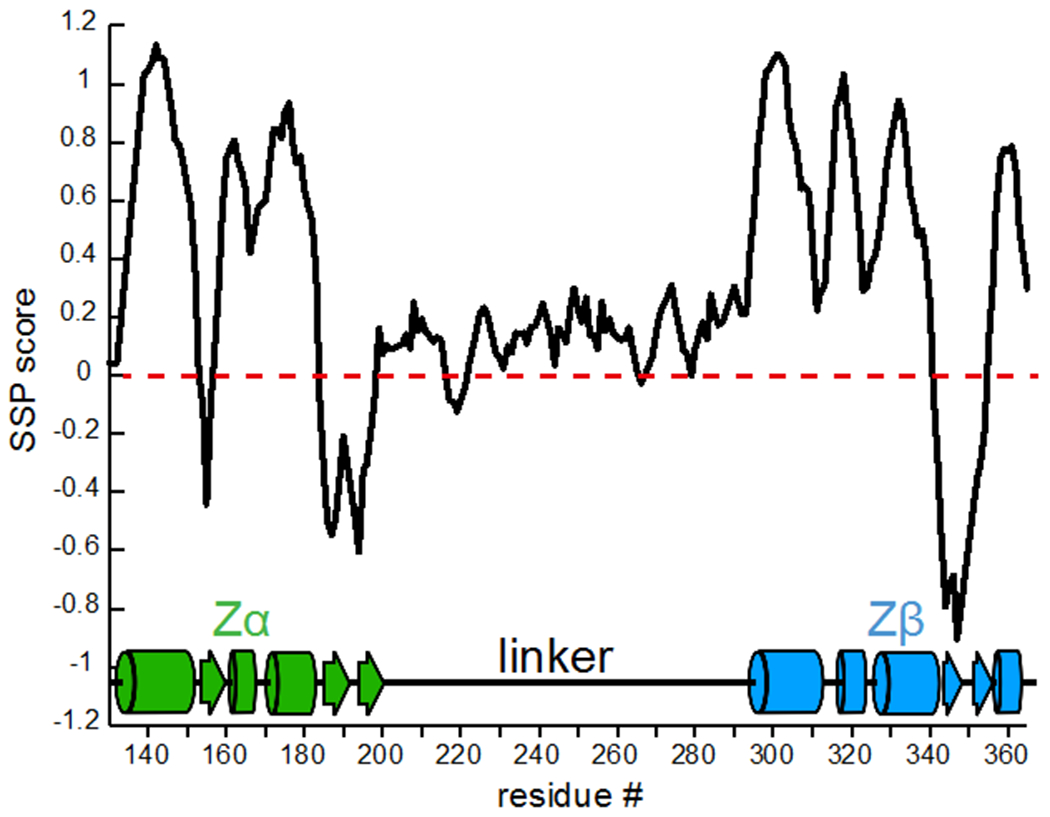
The Secondary Structure Propensity score (SSP) calculated from the assigned HN, N, Cα, Cβ, and CO chemical shifts (Marsh et al. 2006) for Zα-Zβ is shown. The residue # is on the x-axis while SSP score is on the y-axis.
An overlay of the Zα, Zβ, and Zα-Zβ 15N-1H HSQC shows that for the most part, the peaks from the Zα and Zβ domains in isolation match well to those within the context of Zα-Zβ (Figure 4a). However, there are deviations between the constructs, especially in the Zβ domain. In order to investigate this in greater depth, we calculated Chemical Shift Perturbations (CSPs) (Montaville et al. 2008; Williamson 2013) between the isolated domains and the full construct (Figure 4b) according to the following equation:
Figure 4. Chemical shift perturbations between Zα and Zβ in isolation versus Zα-Zβ.
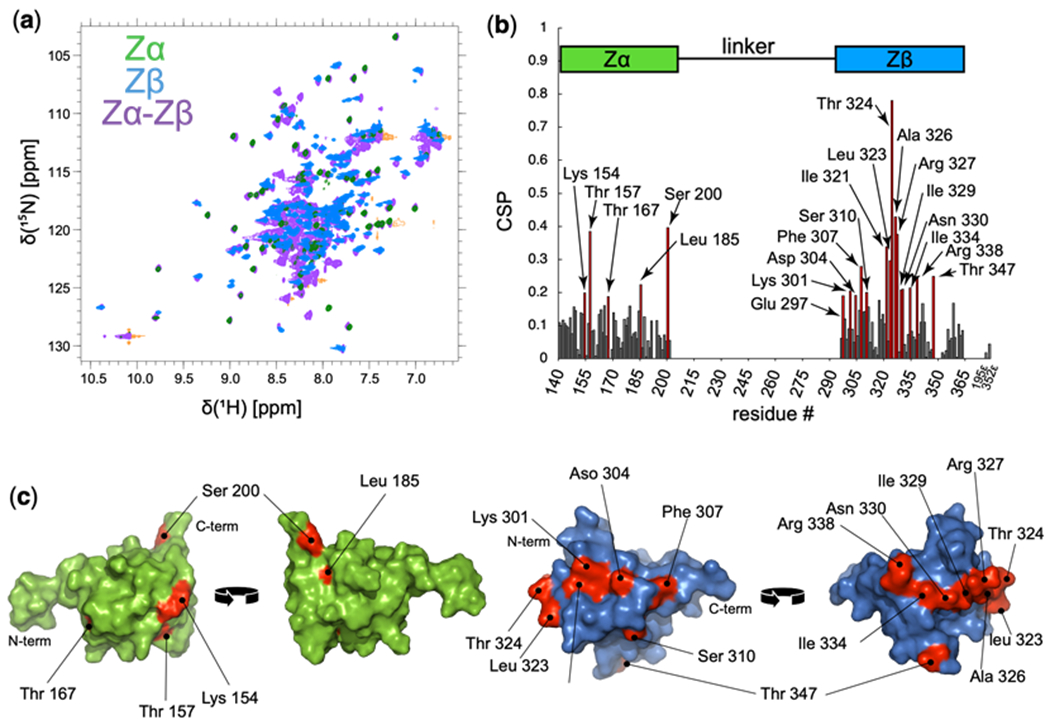
(a) Overlay of the 1H-15N HSQC spectra of Zα, Zβ and Zα-Zβ in green, blue, and purple, respectively. (b) Chemical shift perturbations (CSPs) (Montaville et al. 2008; Williamson 2013) between the isolated Zα and Zβ domains versus the domains within the context of Zα-Zβ. Residues which showed CSP values above 0.19 were considered to be significant. (c) Residues which showed significant CSPs from (b) are plotted (residues colored red) onto the structures of Zα (green, PDB ID: 2GXB, (Placido et al. 2007)) and Zβ (blue, PDB ID: 1XMK, (Athanasiadis et al. 2018)).
For the Zα domain, five residues showed CSPs above the noise level (we took the noise level to be at ~0.19) which included Lys154, Thr157, Thr167, Leu185, and Ser200 (Figure 4b). The Zβ domain showed significantly more CSPs above the noise, with 15 residues including Glu297, Lys301, Asp304, Phe307, Ser310, Ile321, Leu323, Thr324, Ala326, Arg 327, Ile329, Asp330, Ile334, Arg338, and Thr347 (Figure 4b). Plotting these residues on the structures of Zα and Zβ revealed that while their location on Zα appears to be random (and therefore difficult to determine whether they are of functional importance), on Zβ, they collectively form a belt that stretches almost 360° around the protein (Figure 4c). Such an interface suggests that Zβ may interact with the linker region of Zα-Zβ in some way, a hypothesis which we are planning to test in the near future.
Acknowledgements
The authors thank David Jones (University of Colorado, Aurora) for help with NMR spectroscopy, and Jeffrey Kieft for support. This project was supported by NIH Grant R01GM130694-01A1, NSF Grant 1917254 for Infrastructure Innovation for Biological Research, and a start-up package by the University of Colorado to B.V., and University of Colorado Cancer Center Support Grant P30 CA046934 and NIH Biomedical Research Support Shared Grant S10 OD025020-01.
Footnotes
Accession numbers
The chemical shift assignments for Zα (BMRB 50714), Zβ (BMRB 50713) and Zα-Zβ (BMRB 50715) have been deposited in the Biological Magnetic Resonance Data Bank.
Conflict of interest
The authors declare they have no conflict of interest.
References
- Athanasiadis A (2012) Zalpha-domains: At the intersection between RNA editing and innate immunity. Semin Cell Dev Biol 23:275–280 [DOI] [PubMed] [Google Scholar]
- Athanasiadis A, Placido D, Maas S, et al. (2018) The Crystal Structure of the Z β Domain of the RNA-editing Enzyme ADAR1 Reveals Distinct Conserved Surfaces Among Z -domains [DOI] [PubMed] [Google Scholar]
- Bass BL, Weintraub H (1988) An unwinding activity that covalently modifies its double-stranded RNA substrate. Cell. 10.1016/0092-8674(88)90253-X [DOI] [PubMed] [Google Scholar]
- Brown BA, Lowenhaupt K, Wilbert CM, et al. (2000) The Zα domain of the editing enzyme dsRNA adenosine deaminase binds left-handed Z-RNA as well as Z-DNA. Proc Natl Acad Sci U S A. 10.1073/pnas.240464097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai M, Huang Y, Yang R, et al. (2016) A simple and robust protocol for high-yield expression of perdeuterated proteins in Escherichia coli grown in shaker flasks. J Biomol NMR. 10.1007/s10858-016-0052-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh J (2007) Protein NMR spectroscopy : principles and practice. Academic Press [Google Scholar]
- Chung H, Calis JJA, Wu X, et al. (2018) Human ADAR1 Prevents Endogenous RNA from Triggering Translational Shutdown. Cell. 10.1016/j.cell.2017.12.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, et al. (1995) NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6:277–293. 10.1007/BF00197809 [DOI] [PubMed] [Google Scholar]
- George CX, Samuel CE (1999) Human RNA-specific adenosine deaminase ADAR1 transcripts possess alternative exon 1 structures that initiate from different promoters, one constitutively active and the other interferon inducible. Proc Natl Acad Sci U S A. 10.1073/pnas.96.8.4621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert A, Schade M, Lowenhaupt K, et al. (1998) The Zα domain from human ADAR1 binds to the Z-DNA conformer of many different sequences. Nucleic Acids Res. 10.1093/nar/26.15.3486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyberts SG, Milbradt AG, Wagner AB, et al. (2012) Application of iterative soft thresholding for fast reconstruction of NMR data non-uniformly sampled with multidimensional Poisson Gap scheduling. J Biomol NMR. 10.1007/s10858-012-9611-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse H, Mrazikova K, D’Ascenzo L, et al. (2020) Short but Weak: The Z-DNA Lone-Pair⋯π Conundrum Challenges Standard Carbon Van der Waals Radii. Angew Chemie - Int Ed. 10.1002/anie.202004201 [DOI] [PubMed] [Google Scholar]
- Lee W, Tonelli M, Markley JL (2015) NMRFAM-SPARKY: enhanced software for biomolecular NMR spectroscopy. Bioinformatics 31:1325–7. 10.1093/bioinformatics/btu830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh JA, Singh VK, Jia Z, Forman-Kay JD (2006) Sensitivity of secondary structure propensities to sequence differences between α- and γ-synuclein: Implications for fibrillation. Protein Sci. 10.1110/ps.062465306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montaville P, Coudevylle N, Radhakrishnan A, et al. (2008) The PIP2 binding mode of the C2 domains of rabphilin-3A. Protein Sci. 10.1110/ps.073326608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikura K (2016) A-to-I editing of coding and non-coding RNAs by ADARs. Nat Rev Mol Cell Biol 17:83–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell MA, Keller W (1994) Purification and properties of double-stranded RNA-specific adenosine deaminase from calf thymus. Proc Natl Acad Sci U S A. 10.1073/pnas.91.22.10596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell MA, Krause S, Higuchi M, et al. (1995) Cloning of cDNAs encoding mammalian double-stranded RNA-specific adenosine deaminase. Mol Cell Biol. 10.1128/mcb.15.3.1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson JB, Samual SE (1995) Expression and regulation by interferon of a double-stranded-RNA-specific adenosine deaminase from human cells: evidence for two forms of the deaminase. Mol Cell Biol 15:5376–5388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Placido D, Brown BA, Lowenhaupt K, et al. (2007) A Left-Handed RNA Double Helix Bound by the Zα Domain of the RNA-Editing Enzyme ADAR1. Structure. 10.1016/j.str.2007.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schade M, Turner CJ, Kühne R, et al. (1999a) The solution structure of the Za domain of the human RNA editing enzyme ADAR1 reveals a prepositioned binding surface for Z-DNA. Proc Natl Acad Sci U S A. 10.1073/pnas.96.22.12465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schade M, Turner CJ, Lowenhaupt K, et al. (1999b) Structure-function analysis of the Z-DNA-binding domain Zα of dsRNA adenosine deaminase type I reveals similarity to the (α + β) family of helix-turn-helix proteins. EMBO J. 10.1093/emboj/18.2.470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz T, Lowenhaupt K, Kim YG, et al. (1999a) Proteolytic dissection of Zab, the Z-DNA-binding domain of human ADAR1. J Biol Chem. 10.1074/jbc.274.5.2899 [DOI] [PubMed] [Google Scholar]
- Schwartz T, Rould MA, Lowenhaupt K, et al. (1999b) Crystal structure of the Zalpha domain of the human editing enzyme ADAR1 bound to left-handed Z-DNA. Science (80- ) 11:1841–1845 [DOI] [PubMed] [Google Scholar]
- Vranken WF, Boucher W, Stevens TJ, et al. (2005) The CCPN data model for NMR spectroscopy: Development of a software pipeline. Proteins Struct Funct Genet 59:687–696. 10.1002/prot.20449 [DOI] [PubMed] [Google Scholar]
- Wagner RW, Smith JE, Cooperman BS, Nishikura K (1989) A double-stranded RNA unwinding activity introduces structural alterations by means of adenosine to inosine conversions in mammalian cells and Xenopus eggs. Proc Natl Acad Sci U S A. 10.1073/pnas.86.8.2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson MP (2013) Using chemical shift perturbation to characterise ligand binding. Prog. Nucl. Magn. Reson. Spectrosc [DOI] [PubMed] [Google Scholar]


